Abstract
Leaf and tuber extracts of Curcuma wenyujin contain a mixture of curcuminoids. However, the curcuminoid constituents and their molecular mechanisms are poorly understood, and the relevant curcumin synthases remain unclear. In this study, we comprehensively compared the metabolite profiles of the leaf and tuber tissues of C. wenyujin. A total of 11 curcuminoid metabolites were identified and exhibited differentially changed contents in the leaf and tuber tissues. An integrated analysis of metabolomic and transcriptomic data revealed the proposed biosynthesis pathway of curcuminoid. Two candidate type Ⅲ polyketide synthases (PKSs) were identified in the metabolically engineering yeasts, indicating that CwPKS1 and CwPKS2 maintained substrate and product specificities. Especially, CwPKS1 is the first type Ⅲ PKS identified to synthesize hydrogenated derivatives of curcuminoid, dihydrocurcumin and tetrehydrocurcumin. Interestingly, the substitution of the glycine at position 219 with aspartic acid (G219D mutant) resulted in the complete inactivation of CwPKS1. Our results provide the first comparative metabolome analysis of C. wenyujin and functionally identified type Ⅲ PKSs, giving valuable information for curcuminoids biosynthesis.
Keywords: Metabolome, Curcuminoid, Transcriptome, Hydrogenated curcumin, Polyketide synthase
1. Introduction
Curcuminoids are a class of polyphenol coloring compounds that exist in the plant Curcuma species, such as Curcuma longa, C. zedoaria, C.Wenyujin, etc. They are a mixture of curcumin and its derivatives, including demethoxycurcumin (DMC), bisdemethoxycurcumin (Bis-DMC), and others [1]. These compounds have been verified to exert many important pharmacological and biological effects, such as anti-inflammatory, antioxidant, cardio-protective, antimicrobial, hepato-protective, nephro-protective, anti-neoplastic, hypoglycemic, immunomodulatory and anti-rheumatic effects, as well as delay the development of type 2 diabetes [2,3]. In particular, curcumin can induce multiple cytotoxic effects in tumor cells including cell cycle arrest, apoptosis, autophagy, signaling disruption, and radiation effect enhancement [2]. Additionally, curcumin has the advantages of a low molecular weight, low toxicity and clean anti-tumor effects, which makes it widely considered an ideal anticancer chemical. Moreover, curcumin is becoming a popular dietary supplement in the Eastern and Western countries [4].
Type Ⅲ polyketide synthase (PKS) family generally utilizes CoA thioesters as substrates and accomplishes an entire series of decarboxylative condensations and cyclization reactions, giving rise to certain important secondary metabolites such as flavonoids, benzalacetones, stilbenes, xanthones, chromones, acridones, cannabinoids, aliphatic waxes, anthrones, and pyrrones [5,6]. It has been unequivocally established that type Ⅲ PKSs in C. longa are responsible for the formation of diarylheptanoids scaffold [6]. C. longa diketide-CoA synthase (ClDCS) and curcumin synthase (ClCURS), consecutively condense two molecular of feruloyl-CoA or p-coumaroyl-CoA and one molecular of malonyl-CoA to form the three major curcuminoids. Recently, a novel type Ⅲ PKS from Oryza sativa (OsCUS) can catalyze the one-pot formation of Bis-DMC from two p-coumaroyl-CoA and one malonyl-CoA [7]. A new curcumin synthase from Zingiber officinale (ZoCURS) and a polyketide synthase from Aquilaria sinensis (AsPECPS), co-incubation with DCS, are capable of direct synthesizing tetrahydrobisdemethoxycurcumin (THBDC) by condensation two molecular of dihydro-coumaroyl-CoA and malonyl-CoA [8,9]. In addition, analysis of co-regulated metabolite modules in C. longa revealed that some type Ⅲ PKSs with different substrate selectivities may contribute to hydroxylated diarylheptanoids biosynthesis [10,11]. Type Ⅲ PKS family has a versatility and exceptionally broad substrate specificity, which makes it an ideal candidate for the engineering of biocatalysts. Hence, screening and characterization of new functional type Ⅲ PKSs would be advantageous to deeply understand the biosynthesis of curcuminoids in plants.
Curcuma wenyujin Y.H. Chen et C. Ling, belonging to the Zingiberaceae family, has been widely used as a spice, food preservative, dietary supplement, natural dye, and traditional medicine for over a thousand years [12]. In China, C. wenyujin is widely distributed in the tropical and subtropical regions and has been officially approved as a multifunctional medicinal herb. It has roles in blood activation and stasis elimination, Qi replenishment, meridian dredging, pain relief, etc. [13]. C. wenyujin is rich in volatile oils, diterpenoids, curcuminoids and other compounds. Among them, volatile oils have been extensively studied for their use, phytochemistry, and pharmacology [12]. We found few reports on curcuminoids in C. wenyujin, whereas the curcuminoids in C. longa have been well-studied [14]. Shen et al reported the contents and composition of only three major curcuminoids from C. wenyujin [15]. Zhang et al. quantitated eight compounds from Curcuma rhizomes using quantitative thin layer chromatography [16]. Owing to the increasing demand in the pharmaceutical and food industries, there is a pressing need to understand the productivity of curcuminoids in C. wenyujin. Metabolome analysis is widely used to identify and quantitate all metabolites in a particular tissue or organism. For example, a GC–MS approach was used to investigate the chemical profile of volatile oils of C. wenyujin collected from different growing areas and provide a valuable reference for the quality control [17]. Liquid chromatography–MS (LC–MS) was applied to analyze metabolites in turmeric [10]. Another factor to be taken into account is the presence of a wide variance in secondary metabolites that may be caused by genotype, differential geography, cultivation practices, climate, season, stage of maturity, storage, extraction, and analysis methods [18]. Until now, there was a lack of a deep investigation of C. wenyujin to gain insight into its curcuminoid constituents and their biosynthetic pathway.
In our previous study, the transcriptome sequencing of the leaf and tuber tissues of C. wenyujin has been well understood [19]. Hence, in this study, we applied a widely targeted liquid chromatography–electrospray ionization–tandem mass spectrometry (LC–ESI–MS/MS)-based metabolome approach to assess the metabolic changes between the leaf and tuber tissues of C. wenyujin. Moreover, we performed an integrative analysis of the metabolome and transcriptome to gain insight into the curcuminoid metabolic pathway in C. wenyujin. Furthermore, two candidate type Ⅲ PKS genes were screened and characterized in engineered yeasts. This paper will provide the foundation for us to understand the molecular and metabolic bases of curcuminoid biosynthesis in this important medicinal plant.
2. Materials and methods
2.1. Plant materials
C. wenyujin was cultivated in a field located at both sides of the Feiyun River in Ruian City, Wenzhou, China (14 m altitude, 27° 47′ N, 120° 37′ E). The original plants and their macroscopic characteristics were authenticated by Professor Zengxi Guo from the Institute of Food and Drug Control in Zhejiang, China. Fresh leaf and tuber samples of C. wenyujin were collected from adult plants as previously described [19], and stored at −80 °C until further analysis. Each sample had three biological replicates.
2.2. Sample preparation and extraction
The leaf and tuber samples of C. wenyujin were freeze-dried by a vacuum freeze-dryer (Scientz-100F) and then crushed by a mixer mill (MM 400; Retsch, Haan, Germany) with zirconia beads for 1.5 min at 30 Hz. Then, 100 mg of powder was extracted with 1.2 mL 70% methanol solution (v/v) at 4 °C overnight. After centrifugation to remove the undissolved residues, the extracts were absorbed by solid phase extraction with a CNWBOND Carbon-GCB SPE Cartridge (ANPEL, Shanghai, China), filtered using a SCAA-104 filter with a 0.22 μm pore size (ANPEL, Shanghai, China), and followed by LC–ESI−MS/MS analysis.
2.3. LC–ESI–MS/MS analysis
Metabolite profiling analysis was performed by Metware Biotechnology Co., Ltd. (Wuhan, China)
In brief, the extracts were analyzed using a LC–ESI–MS/MS system (UPLC, Shimadzu Nexera X2 system; MS, 4500 Q TRAP; Applied Biosystems, Foster City, CA, USA) coupled with an SB–C18 column (Agilent, 2.1 mm × 100 mm, 1.8 μm). The mobile phase consisted of solvent A, pure water with 0.1% formic acid, and solvent B, acetonitrile with 0.1% formic acid. The gradient program was set as follows: initial conditions of 95% A: 5% B; a linear ramp to 5% A: 95% B over 9 min; maintaining at 5% A: 95% B for 1 min; returning to initial conditions over 1.1 min and keeping for 2.9 min. The flow rate was 0.35 mL/min; The column temperature was set to 40 °C; The injection volume was 4 μL. The effluent was connected to an ESI–triple quadrupole linear ion trap (Q TRAP)–MS.
LIT and triple quadrupole (QQQ) scans were acquired on Q TRAP, equipped with an ESI turbo ion spray interface, operated in electrospray ionization–positive/negative mode and controlled by Analyst 1.6.3 software (AB Sciex). The ESI source operation parameters were set as previously described [20]. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with the collision gas (nitrogen) set to medium. DP and CE for the individual MRM transitions were performed with further DP and CE optimization. A specific set of MRM transitions was monitored for each period according to the metabolites eluted within that particular period [20].
2.4. Qualitative and quantitative analysis of metabolites
Based on the self-built database MWDB (MetWare, Wuhan, China), qualitative and quantitative analysis of the metabolites in the leaf and tuber tissues of C. wenyujin was performed according to previously described methods [20]. In brief, metabolites were identified by comparing the m/z values, the retention time (RT) and the fragmentation patterns with the standards. Unsupervised principal component analysis (PCA) of the significantly changed metabolites (SCMs) was performed by R package to study metabolite variety-specific accumulation (www.r-project.org/). Hierarchical cluster analysis (HCA) of metabolites was presented as heatmap with dendrograms, while Pearson correlation coefficients (PCCs) between samples were calculated by the cor function in R and were presented as only heatmap. Both HCA and PCC were carried out by R package Complex Heatmap. For HCA, normalized signal intensities of metabolites (unit variance scaling) are visualized as a color spectrum [20].
Significantly changed metabolites were filtered according to VIP ≥1 and absolute
log2 (fold change) ≥1.
2.5. Identification of the genes involved in curcuminoid biosynthesis in C.wenyujin
The transcriptome dataset of C.wenyujin titled “CRA003702” (https://ngdc.cncb.ac.cn/gsa/) was obtained from our previous study [19]. Transcriptome annotation was carried out against the databases of NR, SWISS-PROT, PFAM, and KEGG databases to mine their possible functions as previously described [19]. FPKM was used to quantify the gene/transcript expression levels with the R statistical package software EdgeR [21]. Differently expressed genes (DEGs) were screened by the absolute value of |log2Ratio| ≥ 1 with a statistically significant value of P < 0.05. Multiple sequence alignments were performed using ClustalW software with default parameters, and then the phylogenetic tree was generated by the neighbor-joining method with 1000 bootstrap replicates using MEGA 7 [22]. Correlations between metabolites and transcripts were generated by R Package psych, heatmap, and reshape (https://cran.r-project.org) [23].
2.6. Functional identification of type Ⅲ PKSs in engineered yeasts
To evaluate the enzymatic activities of the candidate genes, their ORFs were first amplified by PCR using the specific primers designed on the basis of the transcript sequences (Supplemental Table 1) and Hi Fi Taq DNA Polymerase (Thermo Fisher, San Jose, USA). These genes were individually cloned into the vector pEASY-Blunt Zero (Transgen, Beijing, China). And then, candidate Type Ⅲ PKSs were amplified and ligated into the yeast expression vector pYES2, generating the pYES2:CwPKS. Because the substrates p-coumaroyl-diketide-CoA and feruloyl-diketide-CoA were not commercially available, the functional assays of candidate CwPKSs were carried out in engineered yeast coexpressing At4CL (Arabidopsis thaliana p-coumaroyl: CoA ligase; ID: AY376729) and ClDCS (ID: AB495006.1). The sequences of At4CL and ClDCS were obtained from the NCBI database, codon-optimized for yeast, ordered as synthetic DNA and ligated into a pESC-LEU vector, generating the pESC-LEU: At4CL-ClDCS. The two plasmids with control group (group 1: pESC-LEU and pYES2 empty vectors; group 2:pYES2 empty vector and pESC-LEU: At4CL-ClDCS plasmid) were transformed into Saccharomyces cerevisiae BY4741 cells using lithium acetate method [24].
Transformants were cultured in synthetic dropout medium SD-Ura-Leu (SD media without uracil and leucine, with 2% glucose as the carbon source). A single clone was selected and seeded into a flask containing 5 mL SD -Ura-Leu liquid medium and grown at 30 °C and 220 rpm for 24 h. Then, the resulting exponential culture was diluted to an initial OD600 of 0.2 in 5 mL of fresh SD -Ura-Leu medium and cultivated at 30 °C and 220 rpm for 12 h until an OD600 of approximately 5.0. Next, aliquots were diluted to an initial OD600 of 0.05 in 20 mL SD -Ura-Leu medium and cultivated at 30 °C and 220 rpm. After 2 days, each strain was collected and re-inoculated in 20 mL of SD -Ura-Leu medium using 2% galactose as the carbon source instead of the glucose. At 10 h of growth, 200 μM ferulic acid or p-coumaric acid was added to the media. After 24 h of inoculation, the changes in color of the culture media were observed and the yeasts were collected for products extraction and analysis.
To investigate the catalytic abilities of CwPKSs toward substrate analogues, 200 μM dihydro-ferulic acid, dihydro-coumaric acid, and 4-methoxy-feruloyl acid were added separately. Further, the combinations of dihydro-ferulic acid and p-coumaric acid, dihydro-coumaric acid and ferulic acid, dihydro-ferulic acid and ferulic acid were also investigated.
2.7. LC-MS analysis of curcuminoids in engineered yeasts
The yeasts were homogenized in 1800 bar using a high-pressured homogenizer and then were mixed with an equal volume of ethyl acetate for curcuminoids extraction. The mixtures were centrifuged at 3000 × g for 10 min. Collecting the organic phase, then the aqueous phase was re-extracted using ethyl acetate three times. The extracts were concentrated by solvent evaporation in a fume hood, and then suspended in1000 μL of acetonitrile, followed by LC–MS analysis. LC–MS was performed using an Agilent 1260 Infinity Ⅱ system with a C18 column (150 × 4.6 mm). Mobile phase A was composed of 0.1% (v/v) of trifluoroacetic acid in water, and mobile phase B was composed of acetonitrile. For curcuminoid quantification, a gradient of 40–43% mobile phase B for 15 min, 43–70% for 3 min, 70–40% for 4 min and 40% mobile phase B for an additional 5 min was used. Curcumin, DMC, Bis-DMC, dihydrocurcumin and tetrehydrocurcumin were commercially available and used as authentic standards. Curcumin, DMC and Bis-DMC were detected at an absorbance wavelength of 425 nm and quantified with m/z [M − H]- 367, 337, 307, respectively. Owing to lacking authentic standards, THBDC, dihydro-DMC and dihydro Bis-DMC were detected at absorbance wavelengths of 280, 360, 360 nm, and quantified with m/z [M − H]- 311, 369, 339, respectively [25].
2.8. In silico modeling of CwPKS1 and simulation analysis
Homology modeling of CwPKS1 was carried out using SWISS-MODEL, a fully automated protein structure homology modeling server (https://swissmodel.expasy.org/) [26]. The crystal structure of ClCURS1 protein (PDB ID: 3ov2) with a sequence identity of 94% was chosen as the best template. The best structural model was sorted by the discrete optimized protein energy (DOPE) score. The docking of double-substrate (β-keto acid and ferulic acid) into CwPKS1 was performed by Autodock program (https://autodock.scripps.edu/). The enzyme-substrate complex structures were employed for 50 ns (nm) molecular dynamics (MD) simulations using Amber 16 program under the ff99SB force field (https://ambermd.org) [27]. The root-mean-square deviations (RMSD) of the atomic positions and atom distance measurement were computed using the CPPTRAJ program [28].
3. Results
3.1. Widely targeted metabolomics and differential metabolites
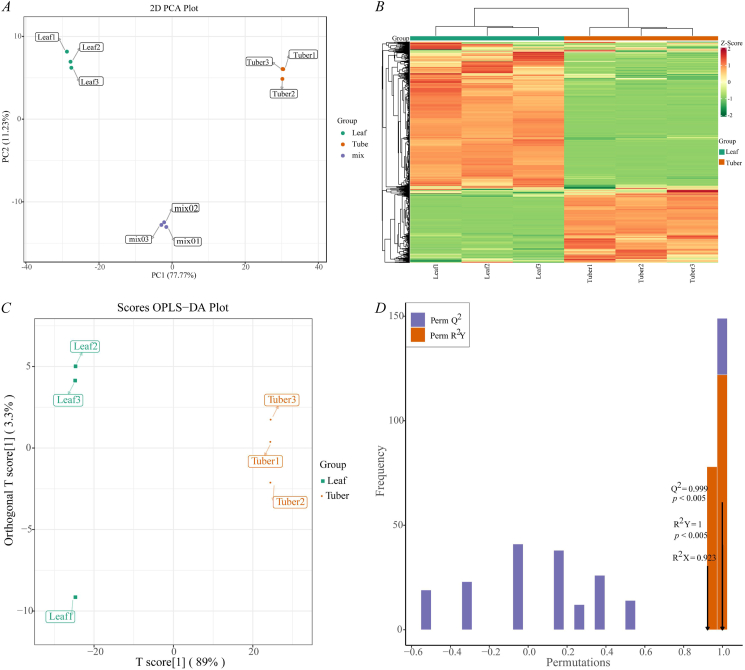
To explore the curcuminoids in the leaf and tuber tissues of C. wenyujin, the secondary metabolites of samples were analyzed by widely targeted metabolomics. A total of 823 compounds were detected and fell into 12 Class Ⅰ (Supplemental file 1). PCA was conducted with 823 metabolites, and 77.77% of the variation was explained by PC1 and 11.23% of the variation was explained by PC2. PCA is clearly separated into two groups with a significance of 0.01 (Fig. 1A). Cluster analysis of the stratified heatmap showed that the three biological replicates of both the leaf and tuber samples were grouped together, which indicated high reliability of the resulting metabolome data. It was observed that a clear distinction in metabolite concentration between the leaf and tuber samples, suggesting a distinct difference in their metabolite characteristics (Fig. 1B). The PLS-DA score map exhibited a distinct separation between the groups and obvious clustering within each group, suggesting a significant difference between the two groups [29] (Fig. 1C). The quality parameters of the model with two principal components were as follows: Q2 = 0.999 (p < 0.005), R2Y = 1 (p < 0.005), and R2x = 0.923, suggesting that the current model could interpret and predict the data (Fig. 1D). Using VIP ≥1.0 and | Log2FC | ≥ 1 as thresholds for significant differences, 177 up-changed and 387 down-changed metabolites in the tubers comparing with leaves were identified, while 259 metabolites had no significantly different concentrations (Supplemental file 2). There were 83 and 174 uniquely accumulated metabolites in the tubers and leaves, respectively. By KEGG enrichment analysis, the 158 significantly changed metabolites (SCMs) were gathered into 74 groups. The results showed that the largest numbers of SCMs were associated with metabolic pathways and biosynthesis of secondary metabolites (Supplemental file 3). Therefore, the differences in metabolic profiles obtained from the leaves and tubers of C. wenyujin could reflect their biological differences.
Fig. 1.
Analysis of metabolites in the leaf and tuber tissues of Curcuma wenyujin. (A) PCA score plot; (B) clustering heatmap of all metabolites; each column represents a sample and each row represents a metabolite. Red and green colors indicate higher or lower metabolite contents, respectively. (C) OPLS- DA score plot; (D) OPLS-DA validation plot.
3.2. Curcuminoid profiles in the leaf and tuber tissues of C. wenyujin
Due to the pharmaceutically important properties of curcuminoids, metabolic profiles in the leaf and tuber samples of C. wenyujin were analyzed. By widely targeted metabolomics analysis, 11 curcuminoids were detected, including Bis-DMC, dihydrodidemethoxy curcumin, DMC, dihydrodemethoxy curcumin, curcumin, dihydrocurcumin, tetrahydrocurcumin, hexahydrocurcumin, octahydrocurcumin, 5′-hydroxyhexahydrocurcumin and dimethylcurcumin (Supplemental file 4). Among them, Bis-DMC, DMC and dimethylcurcumin were not detected in the leaves, while the others were detected in both the leaf and tuber samples of C. wenyujin (Table 1). By screening with VIP ≥1.0 and | log2FC | ≥ 1, five curcuminoids showed the marked increase in the tubers compared with that in leaves; conversely, tetrahydrocurcumin showed a significantly reduced accumulation level in the tubers (Table 1). The content of curcumin in the tubers was 792.8 times higher than that in the leaves (log2FC = 9.51, p < 0.001). These results are in accordance with those reported previously [15]. Notably, several hydrogenated curcuminoids were detected and reported for the first time in C. wenyujin. Moreover, this study is the first to report that 5′-hydroxyhexahydrocurcumin accumulated in the leaf and tuber samples. Similarly, Xie et al. found 12 diarylheptanoids in turmeric rhizomes, including four same compounds (curcumin, DMC, Bis-DMC and dihydrocurcumin) found in C. wenyujin [10].
Table 1.
The curcuminoids identified in the leaf and tuber tissues of Curcuma wenyujin.
| Compounds (Index) | Molecular structure | content |
VIP | Log2FC (tuber/leaf) | Trend | P-value | |
|---|---|---|---|---|---|---|---|
| Leaf | Tuber | ||||||
| Bisdemethoxycurcumin (MWSmce231) | 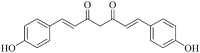 |
9 | 241773 | 1.06 | 14.71 | up | <0.001 |
| Demethoxycurcumin (Hmpp004047) | 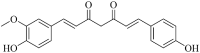 |
9 | 8013300 | 1.06 | 19.76 | up | <0.001 |
| Dihydrocurcumin (Hmjp004041) | 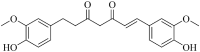 |
9059667 | 21962333 | 1.06 | 1.28 | up | <0.001 |
| Curcumin (pme2296) | 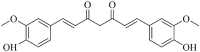 |
29922 | 21837333 | 1.06 | 9.51 | up | <0.001 |
| Tetrahydrocurcumin (MWSmce311) | 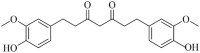 |
6630033 | 2010100 | 1.06 | −1.72 | down | <0.001 |
| Dimethylcurcumin (MWSmce086) | 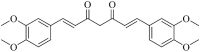 |
9.0 | 159237 | 1.06 | 14.11 | up | <0.01 |
| Dihydrobisdemethoxy curcumin (Hmjp003854) | 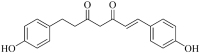 |
361530 | 470427 | 0.99 | 0.38 | insig | <0.05 |
| Dihydrodemethoxy Curcumin (Hmjp003948) | 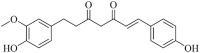 |
4291200 | 6956400 | 1.05 | 0.70 | insig | <0.001 |
| Hexahydrocurcumin (MWSmce374) | 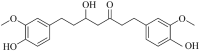 |
6778733 | 4215900 | 1.05 | −0.69 | insig | <0.001 |
| Octahydrocurcumin (MWSmce496) | 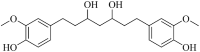 |
8691733 | 9238400 | 0.96 | 0.09 | insig | <0.05 |
| 5′-Hydroxyhexahydro curcumin (Hmpp002596) | 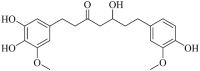 |
26873667 | 37578333 | 1.05 | 0.48 | insig | <0.001 |
3.3. Metabolites and transcripts involved in curcuminoid precursors pathway
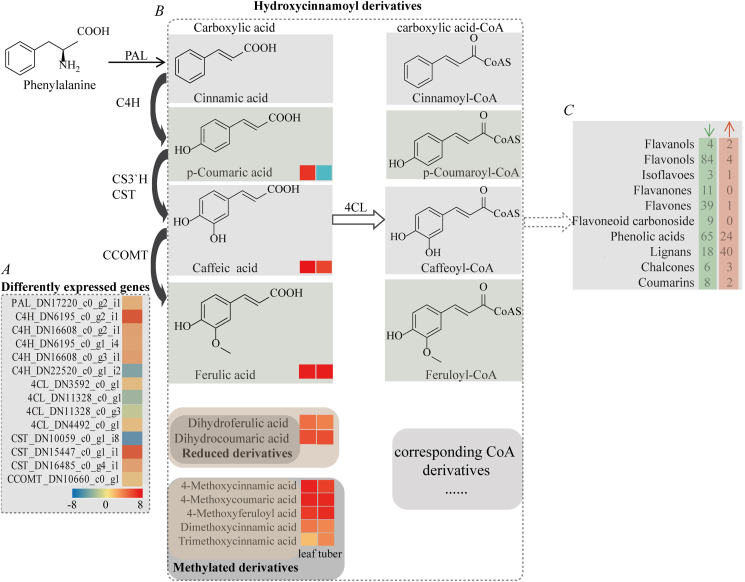
It is known that curcuminoid biosynthesis utilizes carboxylic acid-CoA precursors from general phenylpropanoid biosynthesis. By LC–ESI–MS/MS-based metabolomics analysis, diverse carboxylic acids were detected in the leaf or tuber tissues of C. wenyujin. For instance, the abundances of important intermediates, p-coumaric acid, 2-hydroxycinnamic acid, hydroxycinnamic acids, were too low to be detected by LC–ESI–MS/MS in the tubers but existed in leaves. The amount of caffeic acid was significantly decreased in the tubers vs. the leaves. Some methylated derivatives and reduced derivatives were also detected (Supplemental file 1, 2). These carboxylic acids are activated as their CoA esters, subsequently contributing to an enormous array of secondary metabolites, such as phenolic acids, coumarins, quinate derivatives, flavonoids, cinnamoyl cholines, and lignin [30]. Interestingly, a large proportion of branch metabolites from the phenylpropanoid pathway showed markedly decreasing levels in the tubers vs. the leaves, especially flavonoids and phenolic acids (Fig. 2). A total of 150 flavonoids showed significantly decreased levels, whereas only 8 flavonoids increased in the tubers vs. the leaves (Supplemental file 5). These phenylpropanoid pathway-associated metabolites in the tubers exhibited lower accumulation levels, in agreement with the hypothesis from ginger and turmeric rhizomes that the primary biochemical function of this pathway was to convert into curcuminoids and gingerols instead of flavonoids at appreciable levels [31].
Fig. 2.
Phenylpropanoid pathway and its branch metabolites. (A) The heatmap of differentially expressed transcripts in the phenylpropanoid pathway was generated using log2FC(tuber/leaf) values. (B) Carboxylic acids were detected in the leaf and tuber tissues in Curcuma wenyujin. Up-changed (red) and down-changed (blue) metabolites are shown with the rectangle. (C) Branch metabolites from the phenylpropanoid pathway. The numbers present up-changed (upward arrow in red) and down-changed metabolites (downward arrow in green) in the tubers vs. the leaves.
Furthermore, the transcripts mapped in the phenylpropanoid pathway (ko00940) in C. wenyujin were screened and found eight phenylalanine ammonia-lyases (PALs), nine cinnamic acid 4-hydroxylases (C4Hs), nine 4-coumarate: CoA ligases (4CLs), five coumaroyl shikimate transferases (CSTs), three cinnamate 3-monooxygenases (C3′Hs), four caffeic acid 3-O-methyltransferases (C3OMTs), two caffeoyl-CoA O-methyltransferases (CCOMTs) (Fig. 2; Supplemental file 6). PAL, C4H and 4CL catalyze the first three steps of the general phenylpropanoid pathway and direct the carbon flow to the various branches for general phenylpropanoid metabolism [32]. These transcripts were abundant and may play a role in depleting substrate pools to an undetectable level in the tubers. Differentially expression levels analysis indicated that one PAL transcript (DN17220_c0_g2_i1), five C4H and four 4CL transcripts were up-regulated in the tubers vs. the leaves. One CCOMT (DN10660_c0_g1_i1) was notably expressed at a higher level (FC(tuber/leaf) = 2.29) in the tubers than in the leaves, in consistence with a role for CCOMT in the formation of xylem development [33]. The high expression level of CCOMT indicated that caffeic acid and caffeoyl-CoA could be easily converted into the curcuminoid precursors feruloyl acid and feruloyl-CoA, which in turn converted to curcuminoid [34].
3.4. Putative type Ⅲ PKSs involved in curcuminoid biosynthesis
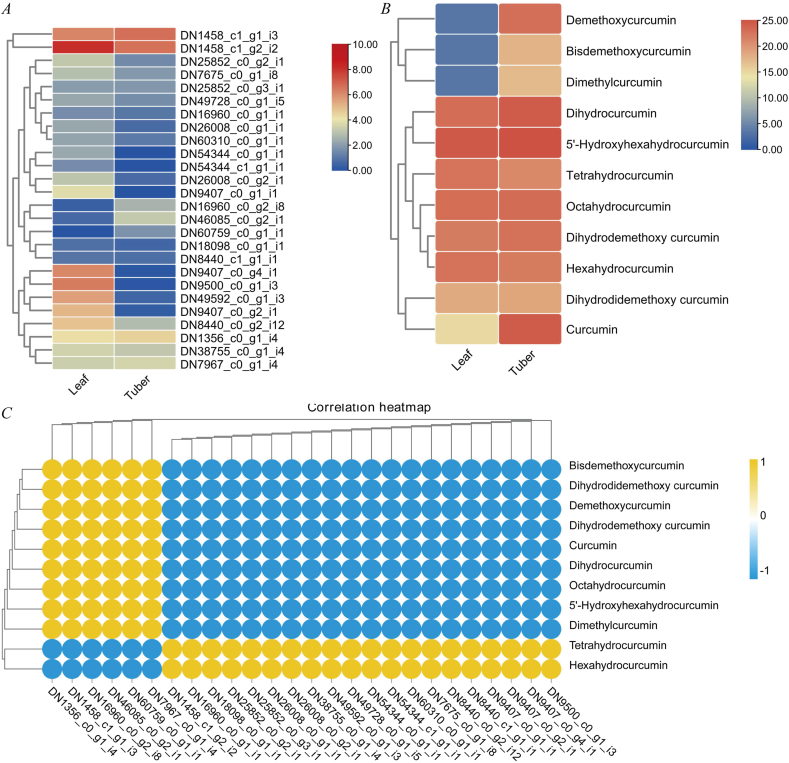
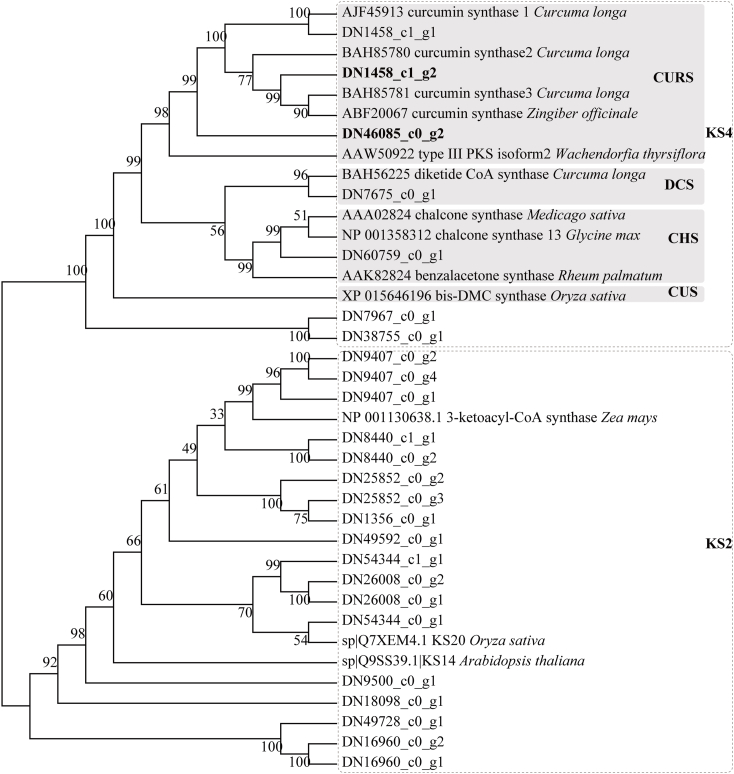
To further investigate the roles of type Ⅲ PKSs in C. wenyujin, we identified a total of 26 putative genes with seed file (PF02797) about the Type Ⅲ PKSs domain by HMMER 3.0 software [35]. The heatmap analysis revealed that DN16960_c0_g2_i8 and DN46085_c0_g2_i1 were specifically expressed in the tuber (Fig. 3A). DN46085_c0_g2_i1 had a significantly up-regulated abundance in the tubers with a log2 FC (tuber/leaf) value of 4.31. Next, the correlation between curcuminoid metabolites and -expression levels of candidate Type III PKSs indicated that curcumin had a close positive correlation with six transcripts, including DN1356_c0_g1_i4, DN1458_c1_g1_i3, DN16960_c0_g2_i8, DN46085_c0_g2_i1, DN67059_c0_g1_i1 and DN7967_c0_g1_i4 (Fig. 3B and C). Finally, the phylogenetic relationship tree was generated using type Ⅲ PKSs from C. wenyujin and previously reported PKSs in the literature. Our estimated phylogenetic tree analysis revealed that these type Ⅲ PKSs were grouped into two major sub-families, KS2 and KS4, based on their function (Fig. 4). Nineteen transcripts from C. wenyujin were grouped into KS2, which usually play roles in long-chain fatty acid elongation or condensation in plants [36]. Seven transcripts from C. wenyujin were grouped into KS4, a large fraction of which were chalcone synthases, stilbene synthases, type Ⅲ PKSs, and naringenin-chalcone synthases. DN7675_c0_g1 was clustered together with C. longa DCS, which formed β-diketide-CoA as the curcuminoid precursor [37]. DN1458_c1_g1, fully identical to C. longa CURS1, exhibited high expression levels in both tissues. Purified ClCURS1 enzyme in vitro synthesized curcumin, Bis-DMC and DMC by utilizing both feruloyl-CoA and p-coumaroyl-CoA as substrates [37]. DN1458_c1_g2, high sequence similarities with C. longa CURS3 (ClCURS3) and ZoCURS, had a significantly up-regulated abundance in the leaves with a log2FC(tuber/leaf) value of -1.12. DN46085_c0_g2 shared moderate similarities with ClCURS3 and maintained the conserved Cys-His-Asn catalytic triad. Collectively, DN46085_c0_g2 and DN1458_c1_g2 might serve biological functions in curcumin biosynthesis in C. wenyujin.
Fig. 3.
(A) Heatmap of candidate type Ⅲ PKSs. (B) Heatmap of curcuminoids; (C) The correlation analysis of curcuminoids and putative type Ⅲ PKSs.
Fig. 4.
Phylogenetic tree of the putative Type Ⅲ PKSs from Curcuma wenyujin with known plant PKSs. Alignment of sequences was generated with ClustalW, and the phylogenetic tree was constructed with MEGA7 software using a neighbor-joining algorithm. The reliability of the tree was measured by bootstrap analysis with 1000 replicates. CHS, chalcone synthase; DCS, diketide-CoA synthase; CURS, curcumin synthase; CUS, curcuminoid synthase; KS, ketoacyl-CoA synthase.
3.5. Functional characterization of candidate type Ⅲ PKSs
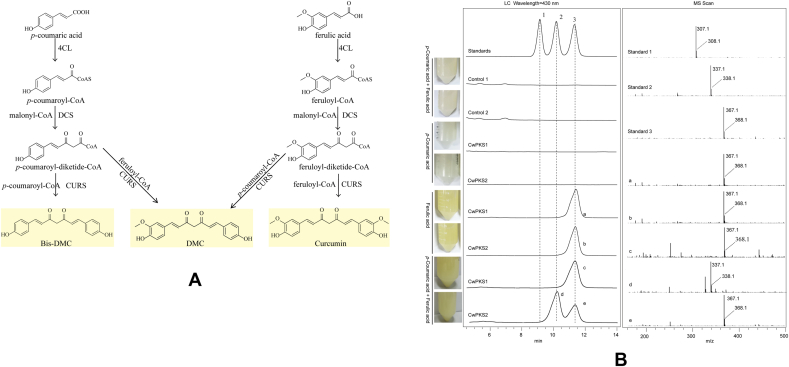
To investigate the biochemical activities of these candidate type Ⅲ PKSs, we first introduced the plasmid pESC-LEU:At4CL-ClDCS into S. cerevisiae BY4741 to provide the precursor diketide-CoA (Fig. 5A). At4CL could transform several carboxylic acids into the corresponding CoA esters [37]. ClDCS catalyzed the formation of feruloyldiketideCoA or p-coumaroyldiketide-CoA from feruloyl-CoA or p-coumaroyl CoA and malonyl-CoA in vitro [38]. DN1458_c1_g2 (namely CwPKS1) and DN46085_c0_g2 (namely CwPKS2) were amplified and then introduced into the engineering yeasts, respectively. By galactose-induction, curcuminoids were accumulated in a liquid medium and portrayed a bright yellow color when ferulic acid was added separately or with p-coumaric acid. The culture medium did not turn yellow when p-coumaric acid was added separately. LC–MS analysis verified that the two engineered yeasts did not yield any curcuminoid towards p-coumaric acid as the sole precursor whereas they produced the curcumin with ferulic acid. When p-coumaric acid and ferulic acid were fed simultaneously, CwPKS2 gave the DMC and curcumin, while CwPKS1 only gave curcumin (Fig. 5B). These results indicated that the precursor of Bis-DMC and DMC, p-coumaroyl-diketide-CoA, cannot be synthesized by ClDCS in the engineering yeasts due to its lower efficiency toward p-coumaroyl-CoA as the starter substrate [38]. In addition, the intracellular pH of a yeast cell is typically mildly acidic whereas the pH optima of ClDCS was 6.5–7.5, so ClDCS exhibit no activity toward p-coumaric acid in the engineering yeasts [38].
Fig. 5.
(A), Curcuminoid biosynthetic pathway. (B), LC–MS analysis of the fermentation products in the engineering yeasts by feeding the p-coumaric acid and ferulic acid precursor. The yellow color of the tubes indicates the presence of curcuminoid. Control 1, the yeast harboring pESC-LEU and pYES2 empty vectors; Control 2, the yeast harboring pYES2 empty vector and pESC-LEU: At4CL-ClDCS plasmid; CwPKS1, the yeast harboring pESC-LEU: At4CL-ClDCS and pYES2:CwPKS1; CwPKS2; the yeast harboring pESC-LEU:At4CL-ClDCS and pYES2:CwPKS2; 1, Bis-DMC; 2, DMC; 3. curcumin.a–e: the MS spectra.
3.6. Substrate promiscuity assays of candidate type Ⅲ PKSs
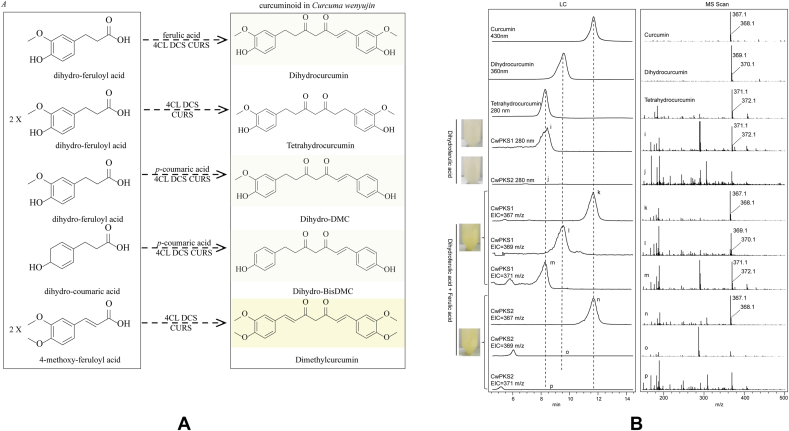
Compound analogues may differ by mass shifts that represent common biosynthetic modifications such as reduction (+2), dehydrogenation (−2), oxidation (+16), hydration (+18), methylation (+14), and methoxylation (+30). Compound analogues can be affiliated with their compound classes, highly interconnected, and presented structural and biosynthetic correlation [10]. Hence, we deduced possible substrates of these curcuminoids (Fig. 6A) and performed promiscuous catalytic activities of these type Ⅲ PKSs in the engineered yeasts. When ferulic acid and dihydro-ferulic acid were fed simultaneously, the cultures became yellowish and LC–MS analysis unveiled that CwPKS1 enabled the production of curcumin, dihydrocurcumin and tetrahydrocurcumin whereas only curcumin was detectable in the yeast overexpressed CwPKS2. The addition of dihydro-ferulic acid individually resulted in unchanged color of the cultures because the deduced product tetrahydrocurcumin is an off-white color. LC–MS results verified that CwPKS1 gave tetrahydrocurcumin using dihydro-ferulic acid as substrate but CwPKS2 could not (Fig. 6B).
Fig. 6.
(A), Proposed substrates related to curcuminoid biosynthesis in Curcuma wenyujin. (B), LC–MS analysis of curcuminoids produced in the engineering yeasts by feeding promiscuous substrates. 3, curcumin; i-j, the UV absorption spectrum (280 nm) and MS spectra for m/z 371.1; k, curcumin; l-p, Extracted ion chromatogram (EIC) and MS spectra.
To investigate the abilities to produce dihydro-DMC and dihydro-BisDMC of CwPKSs, the combinations of p-coumaric acid and dihydro-ferulic acid or dihydro-coumaric acid were used as precursors (Fig. 6a). However, no absorption peak at 360 nm and no corresponding proton peaks of m/z [M − H]- (339 and 309) were observed, indicating that there were no dihydro-DMC and dihydro-BisDMC (Fig. S1). Surprisingly, an absorption peak at 280 nm and a proton peak of m/z [M − H]- 311 were present. Presumably, THBDC was generated in the fermentation products of the two engineered yeasts (Fig. S2). We inferred CwPKSs together with At4CL and ClDCS could condense two molecular dihydro-coumaric acid and malonyl-CoA. This agreed with previous results reported in other type Ⅲ PKSs, ZoCURS and AsPECPS [8,9].
4-methoxy-feruloyl acid is presumably the precursor of dimethylcurcumin according to its chemical structure. What is more, 4-methoxy-feruloyl acid showed a significant down-accumulation level in the tuber but dimethylcurcumin was significantly up-accumulated in the tuber, compared with that in the leaf (Supplemental file 1). However, no obvious yellow colors were observed in the engineered yeasts when 4-methoxy-feruloyl acid was fed. No absorption peak at 430 nm was observed and no proton peak was quantified by m/z [M − H]- 395, meaning that dimethylcurcumin was not yielded by the engineered yeasts (Fig. S3).
Taken together, our results reveal that the CwPKS2 accepts p-coumaric acid, dihydro-coumaric acid, and ferulic acid as substrates except dihydroferulic acid, while CwPKS1 utilizes dihydro-coumaric acid, ferulic acid and dihydro-ferulic acid except p-coumaric acid. Particularly, the enzymatic production of dihydrocurcumin and tetrahydrocurcumin from dihydro-ferulic acid by wild plant PKSs has not been previously reported.
3.7. Mutants of CwPKS1 analysis
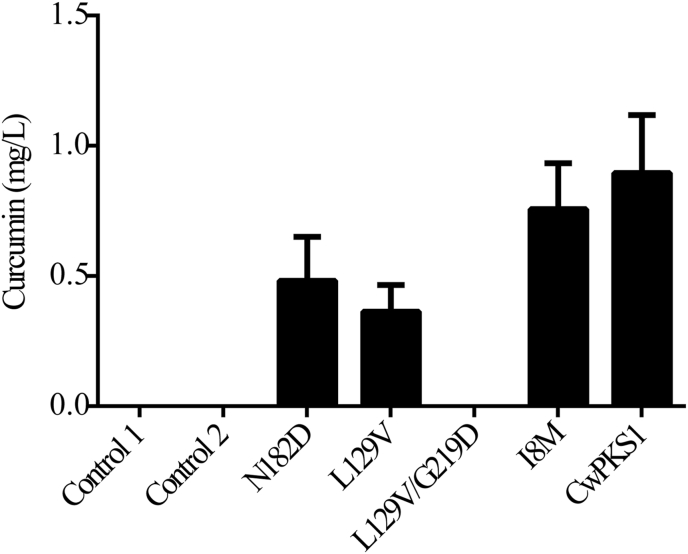
Interestingly, four mutants of CwPKS1 (N182D, L129V, I8M, L129V/G219D) were simultaneously obtained by a high-fidelity PCR using the primers designed based on DN1458_c1_g2. They were introduced into the yeast carried pESC-LEU: At4CL-ClDCS, respectively. After galactose-induction and ferulic acid-feeding, the engineering yeast respectively expressed N182D, L129V and I8M could convert ferulic acid into curcumin. The yield of curcumin catalyzed by CwPKS1 was 0.90 ± 0.22 mg/L while other mutants exhibited low contents. But the culture medium of the yeast carrying L129V/G219D showed no color change. LC–MS analysis verified no curcumin was yielded. These results indicated that the L129V/G219D completely lost its activity. L129V decreased curcumin production by the proximal half compared with the wild type (Fig. 7). Nevertheless, L129 and G219 were not crucial for catalysis or substrate/product binding sites by conserved domain analysis. To better understand the mechanism of inactivation of L129V/G219D mutant, we next constructed the model of enzyme-substrates complexes.
Fig. 7.
The curcumin titer of the engineering yeasts carrying CwPKS1 mutants. The data are averages of three biological replicates with error bars representing standard deviations.
3.8. Proposed inactivation mechanism of the G219D mutant
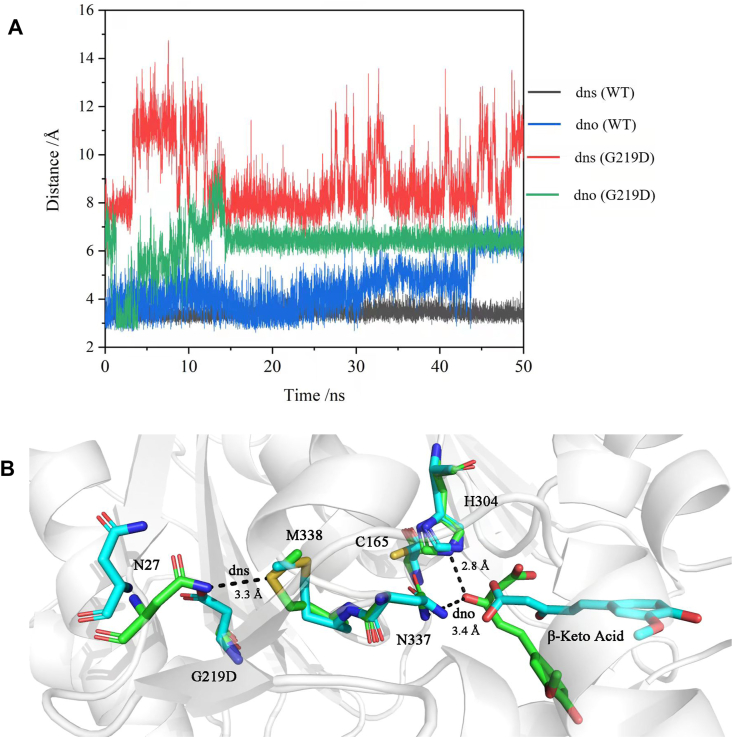
The overall structure of CwPKS1 exhibited the typical type III PKS folds. The CoA-binding tunnel and the conserved catalytic triad of Cys165-His304-Asn337 of CwPKS1 were also similar to those of other plant type III PKSs in locations and orientations [39,40]. L129 was far away from the enzyme active center, so we focused on G219D for further analysis (Fig. S4). The results of 50 ns MD simulations of CwPKS1 WT and substrates indicated that the N atom of Asn337 side chain was hydrogen-bonded to the carbonyl O atom of the substrate, β-keto acid (Fig. 8A, dno(WT), leading to immobilization of the β-keto acid. The amide N atom of Asn27 formed an atypical hydrogen bond with the S atom of Met338 with a distance of 3.41 ± 0.22 Å (Fig. 8A, dns(WT). Whereas, the distance between the N atom of Asn27 and the S atom of Met338 was shown to be always above 7 Å in G219D mutant (Fig. 8A, dns(G219D). Because aspartic acid had a larger side chain (-CH2COOH) than glycine (-H), we inferred that the steric hindrance of the group (-CH2COOH) disrupted the atypical hydrogen bond between Asn27 and Met338, thus leading to spatial flexibility increasing of Met338 and juxtaposition next to Asn337 located in the loop (Fig. 8B). Subsequently, the hydrogen bond between Asn337 and β-keto acid was destroyed, thus β-keto acid could not be anchored into the long-chain binding tunnel, thereby causing a loss in activity(Fig. 8A, dos(G219D).
Fig. 8.
(A), Key atom distances analysis of CwPKS1 WT and G219D mutant by MD simulations; (B), Comparison of the key residues of WT (green) and G219D (cyan blue) interacting with β-keto acid in the best model.
4. Discussion
Curcuminoids have been used in traditional medicine and as food additives in many Asian countries especially China and India for thousands of years and are beneficial to human health. Curcuminoids are present in large amounts in C. longa but also occur in other Curcuma species. However, the contents and composition of curcuminoids from various Curcuma species are different. Considering the important applications of C. wenyujin as a traditional Chinese medicine, we present the first metabolite examination of the tuber and leaf tissues of C. wenyujin using UPLC-ESI-MS/MS which has the advantages of high sensitivity and throughput, fast separation, and wide coverage. Through clustering analysis, PCA, and OPLS-DA, the metabolites were clearly separated from the tuber and leaf tissues in C. wenyujin. Therefore, metabolite analysis based on the UPLC–ESI–MS/MS platform is an effective method to identify differentially changed metabolites. In this study, 11 curcuminoids were detected in the tuber and leaf tissues of C. wenyujin via widely targeted metabolomics analysis. In addition to the three well-known curcuminoids, six hydrogenated curcuminoids were also found in C. wenyujin. These reduced derivatives of curcuminoids demonstrate higher solubility at physiological pH and a longer half-life in plasma than curcumin [41]. In particular, tetrahydrocurcumin has a greater potential for anti-inflammatory, anti-diabetic, anti-oxidant and anti-hyperlipidemic activities than curcumin [42]. A new Curcuma cultivar, C. longa cv. Okinawa Ougon (Ougon) is considered a promising functional food additive candidate owing to its higher concentrations of reduced curcuminoid derivatives [43]. Moreover, dimethylcurcumin is present only in the tubers of C. wenyujin; this compound is capable of delaying the onset of cancer and inhibiting cancer cell proliferation with notable efficacy [44,45]. Besides, we identified a diarylheptanoid with an ortho-diol moiety on an aromatic ring, 5′-hydroxyhexahydrocurcumin. Such hydroxylated curcuminoids were also found in turmeric, such as 3-hydroxy-bisdemethoxycurcumin and 3-hydroxy-demethoxycurcumin. However, their functions have not yet been elucidated and their biosynthetic pathways are proposed to be different [10].
Although the range of plant secondary metabolites is huge, many metabolites can be affiliated with their compound classes and exhibit high interconnection. Their biosynthetic principles are highly conserved and associated with a set of specific enzyme families. Hence, metabolome integrated with transcriptome analysis has become a powerful tool to mine natural product biosynthesis pathways and their producers [46,47]. It is well known that curcuminoids are synthesized from phenylalanine and malonyl-CoA in plants [48]. In this study, transcriptome analysis of the leaf and tuber tissues of C. wenyujin identified candidate genes involved in the curcuminoid biosynthesis process and showed differentially expressive levels. The enzymes involved in the phenylpropanoid pathway were screened by KEGG annotation and functional annotation (Supplemental Table 2). The core set of candidate genes for curcuminoid biosynthesis was type III PKSs including DCS and CURS. To date, plant type III PKS is becoming an important tool for bio synthesizing certain important secondary metabolites [6,49]. So screening and identification of the PKSs in C. wenyujin have important functional implications. Further, these functional enzymes facilitate synthetic biology of important metabolites [50]. Next, the correlations between candidate type III PKSs and 11 curcuminoids were calculated by the Pearson correlation coefficients. When the correlation coefficient is greater than ±0.8 to ±1, two metabolites exist a strong linear relationship [50]. Therefore, two type Ⅲ PKSs were screened out and exhibited differentially expressive levels, which resulted in different contents and constituents of curcuminoids in the tuber and leaf tissues of C. wenyujin. A type Ⅲ PKS shared high sequence similarities with C.longa CURS3, while a new type III PKS (CwPKS2, DN46085_c0_g2) shared moderate similarities with CwPKS1 and maintained the conserved Cys-His-Asn catalytic triad. These genes would be good candidates for the study of curcuminoid synthesis in C.wenyujin.
In our study, we gave insights into the functions of the key curcumin synthases in curcuminoid biosynthesis in C. wenyujin. CwPKS2 can utilize ferulic acid and p-coumaric acid, and give rise to the synthesis of curcumin and DMC in the engineering yeast. Meanwhile, CwPKS2 (DN46085_c0_g2) had a significantly up-regulated abundance in the tubers. So CwPKS2 is mainly responsible for curcumin and DMC synthesis in the tuber of C. wenyujin. CwPKS1 showed substrate promiscuity and could utilize the ferulic acid and dihydro-ferulic acid, in charge of the synthesis of curcumin, dihydrocurcumin and tetrahydrocurcumin in the engineering yeast. Additionally, these hydrogenated derivatives of curcuminoids are considered to be formed from the selective reduction of an unsaturated double bond of the major curcuminoids by a reductase. It has also been proven that curcumin can be reduced to its multi-hydrogenated forms by curcumin reductase from commensal Escherichia coli (EcCurA) and Vibrio vulnificus [51,52]. Further, 11 transcripts were screened out from the transcriptome of C. wenyujin and were found to share 32.6–45.8% amino acid sequence similarities with EcCurA, which belongs to the zinc-independent medium-chain dehydrogenases/reductases superfamily. Of them, six were annotated as NADPH-dependent double-bond reductases or 2-alkenal reductases that can catalyze the reduction of the α, β-unsaturated double bond [53] (Supplemental file 7). Therefore, we infer that an alternative biosynthetic pathway is that consecutive reduction of the double bonds of curcumin results in the formation of dihydrocurcumin and tetrahydrocurcumin by curcumin reductase. Furthermore, the reduction of the carbonyl group of tetrahydrocurcumin could give hexahydrocurcumin (MWSmce374) and octahydrocurcumin (MWSmce496). However, further verification by in vitro or in vivo experiments is required. Despite CwPKSs showing substrate promiscuity, CwPKS1 could not utilize the 4-methoxy-ferulic acid and p-coumaric acid while CwPKS2 could not utilize 4-methoxy-ferulic acid and dihydroferulic acid. There may be unidentified PKSs to play roles in these uncommon curcuminoids biosynthesis.
Overall, CwPKS1 is the first polyketide synthase identified to synthesize dihydrocurcumin and tetrehydrocurcumin by condensation of ferulic acid and/or dihydro-ferulic acid. These findings are significantly different from what was previously reported.
Data availability statement
The raw sequence data are publicly accessible at https://bigd.big.ac.cn/gs under accession number CRA003702. All supporting data are included in Supplemental files.
CRediT authorship contribution statement
Rong Chen: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition. Tianyuan Hu: Methodology, Investigation, Writing – original draft. Ming Wang: Methodology, Formal analysis. Yuhan Hu: Methodology, Formal analysis. Shu Chen: Data curation. Qiuhui Wei: Formal analysis. Xiaopu Yin: Supervision, Project administration. Tian Xie: Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82173919, 82104320), and the Natural Science Foundation of Zhejiang Province (LY21C050004, LQ22H280013).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.04.006.
Contributor Information
Rong Chen, Email: rongchen1984@hznu.edu.cn.
Tianyuan Hu, Email: hutianyuan007@126.com.
Ming Wang, Email: 2021112012295@stu.hznu.edu.cn.
Yuhan Hu, Email: hyh423@hotmail.com.
Shu Chen, Email: 2021112012298@stu.hznu.edu.cn.
Qiuhui Wei, Email: weiqiuhui@hznu.edu.cn.
Xiaopu Yin, Email: yinxp@hznu.edu.cn.
Tian Xie, Email: tianxie@hznu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yuandani Jantan I. Rohani as, sumantri IB. Immunomodulatory effects and mechanisms of curcuma species and their bioactive compounds: a review. Front Pharmacol. 2021;643119:12. doi: 10.3389/fphar.2021.643119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatamipour M., Ramezani M., Tabassi S.A.S., Johnston T.P., Sahebkar A. Demethoxycurcumin: a naturally occurring curcumin analogue for treating non-cancerous diseases. Cell Physiol. 2019;234(11):19320–19330. doi: 10.1002/jcp.27029. [DOI] [PubMed] [Google Scholar]
- 3.Pivari F., Mingione A., Brasacchio C., Soldati L. Curcumin and type 2 diabetes Mellitus: prevention and treatment. Nutrients. 2019;1837(8):11. doi: 10.3390/11081837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotha R.R., Luthria D.L. Curcumin: biological, pharmaceutical, nutraceutical and analytical aspects. Molecules. 2019;24(16):2930. doi: 10.3390/molecules24162930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim Y.P., Go M.K., Yew W.S. Exploiting the biosynthetic potential of type III polyketide synthases. Molecules. 2016;806(6):21. doi: 10.3390/molecules21060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisht R., Bhattacharyya A., Shrivastava A., Saxena P. An overview of the medicinally important plant type III PKS derived polyketides. Front Plant Sci. 2021;12:746908. doi: 10.3389/fpls.2021.746908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita H., Wanibuchi K., Nii H., et al. Structural basis for the one-pot formation of the diarylheptanoid scaffold by curcuminoid synthase from Oryza sativa. Proc Natl Acad Sci U S A. 2010;107(46):19778–19783. doi: 10.1073/pnas.1011499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Gao B., Wang X., Zhang Z., Liu X., Wang J., et al. Identification of a new curcumin synthase from ginger and construction of a curcuminoid-producing unnatural fusion protein diketide-coa synthase:curcumin synthase. RSC Adv. 2016;6(15) doi: 10.1039/C5RA23401H. [DOI] [Google Scholar]
- 9.Wang X.H., Gao B.W., Nakashima Y., Mori T., Zhang Z.X., Kodama T., et al. Identificatinof a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood. Nat Commun. 2022;13(1):348. doi: 10.1038/s41467-022-27971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z., Ma X., Gang D.R. Modules of co-regulated metabolites in turmeric (Curcuma longa) rhizome suggest the existence of biosynthetic modules in plant specialized metabolism. J Exp Bot. 2009;60(1):87–97. doi: 10.1093/jxb/ern263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kita T., Imai S., Sawada H., Seto H. Isolation of dihydrocurcuminoids from cell clumps and their distribution in various parts of turmeric (Curcuma longa) Biosci Biotechnol Biochem. 2009;73(5):1113–1117. doi: 10.1271/bbb.80871. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Wu Y., Li Y., Guo F. Review of the traditional uses, phytochemistry, and pharmacology of Curcuma wenyujin Y. H. Chen et C. Ling. J Ethnopharmacol. 2021;269 doi: 10.1016/j.jep.2020.113689. [DOI] [PubMed] [Google Scholar]
- 13.Xia Q., Wang X., Xu D.J., Chen X.H., Chen F.H. Inhibition of platelet aggregation by curdione from curcuma wenyujin essential oil. Thromb Res. 2012;130:409–414. doi: 10.1016/j.thromres.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Kocaadam B., Şanlier N. Curcumin. An active component of turmeric (Curcuma longa)and its effects on health. Crit Rev Food Sci Nutr. 2017;57(13):2889–2895. doi: 10.1080/10408398.2015.1077195. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y., Han C., Chen X., Hou X., Long Z. Simultaneous determination of three Curcuminoids in Curcuma wenyujin Y.H.chen et C.Ling. by liquid chromatography-tandemmass spectrometry combined with pressurized liquid extraction. Pharm BiomedAnal. 2013;81–82:146–150. doi: 10.1016/j.jpba.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J.S., Guan J., Yang F.Q., Liu H.G., Cheng X.J., Li S.P. Qualitative and quantitative analysis of four species of Curcuma rhizomes using twice development thin layer chromatography. Pharm Biomed Anal. 2008;48(3):1024–1028. doi: 10.1016/j.jpba.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Wu P., Dong X.M., Song G.Q., Wei M.M., Fang C., Zheng F.B., et al. Bioactivity-guided discovery of quality control markers in rhizomes of Curcuma wenyujin based on spectrum-effect relationship against human lung cancer cells. Phytomedicine. 2021;86:153559. doi: 10.1016/j.phymed.2021.153559. [DOI] [PubMed] [Google Scholar]
- 18.Dosoky N.S., Setzer W.N. Chemical composition and biological activities of essentia oils of curcuma species. Nutrients. 2018;10(9):1196. doi: 10.3390/nu10091196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R., Wei Q., Liu Y., Wei X., Chen X., Yin X., Xie T. Transcriptome sequencing and functional characterization of new sesquiterpene synthases from Curcuma wenyujin. Arch Biochem Biophys. 2021;709:108986. doi: 10.1016/j.abb.2021.108986. [DOI] [PubMed] [Google Scholar]
- 20.Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., Yu S., Xiong L., Luo J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. 2013 Nov;6(6):1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 21.Robinson M.D., McCarthy D.J., Smyth G.K., edgeR A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revelle W.R. Northwestern University; Evanston, IL: 2018. psych: procedures for personality and psychological research.https://CRAN.R-project.org/package=psych [Google Scholar]
- 24.Shao Z., Zhao H. Zhao. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37(2):1–10. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan S., Rupasinghe T.W., Tull D.L., Boughton B., Oliver C., McSweeny C., Gras S.L., Augustin M.A. Degradation of curcuminoids by in vitro pure culture fermentation. J Agric Food Chem. 2014;62(45):11005–11015. doi: 10.1021/jf5031168. [DOI] [PubMed] [Google Scholar]
- 26.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013;3:198–210. doi: 10.1002/wcms.1121. [DOI] [Google Scholar]
- 28.Roe D.R., Cheatham I.T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theor Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 29.Thévenot E.A., Roux A., Xu Y., Ezan E., Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res. 2015;14(8):3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- 30.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3(1):2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 31.Koo H.J., McDowell E.T., Ma X., Greer K.A., Kapteyn J., Xie Z., et al. Ginger and turmeric expressed sequence tags identify signature genes for rhizome identity and development and the biosynthesis of curcuminoids, gingerols and terpenoids. BMC Plant Biol. 2013;13:27. doi: 10.1186/1471-2229-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng J., Wang B., He G., Wang Y., Tang X., Wang S., et al. Metabolomics integrated with transcriptomics reveals redirection of the phenylpropanoids metabolic flux in ginkgo biloba. Agric Food Chem. 2019;67(11):3284–3291. doi: 10.1021/acs.jafc.8b06355. [DOI] [PubMed] [Google Scholar]
- 33.Do C.T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., et al. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226(5):1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez-Ahumada Mdel C., Timmermann B.N., Gang D.R. Biosynthesis of curcuminoidsand gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): identification of curcuminoid synthase and hydroxycinnamoyl-Co A thioesterases. Phytochemistry. 2006;67(18):2017–2029. doi: 10.1016/j.phytochem.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 35.Eddy S.R. Accelerated profile HMM searches. PLoS Comput Biol. 2011;(10):7. doi: 10.1371/journal.pcbi.1002195. e1002195.10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Kelly E.E., Masluk R.P., Nelson C.L., Cantu D.C., Reilly P.J. Structural classification and properties of ketoacyl synthases. Protein Sci. 2011;20(10):1659–1667. doi: 10.1002/pro.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa M.A., Bedgar D.L., Moinuddin S.G., Kim K.W., Cardenas C.L., Cochrane F.C., et al. Characterization in vitro and in vivo of the putative multigene 4-coumarate: CoA ligasenetwork in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formatin. Phytochemistry. 2005;66(17):2072–2091. doi: 10.1016/j.phytochem.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Katsuyama Y., Kita T., Funa N., Horinouchi S. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. Biol Chem. 2009;284(17):11160–11170. doi: 10.1074/jbc.M900070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsui T., Kodama T., Mori T., Tadakoshi T., Noguchi H., Abe I., Morita H. 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases. J Biol Chem. 2017;292(22):9117–9135. doi: 10.1074/jbc.M117.778977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drufva E.E., Hix E.G., Bailey C.B. Site directed mutagenesis as a precision tool to enable synthetic biology with engineered modular polyketide synthases. Synth Syst Biotechnol. 2020;5(2):62–80. doi: 10.1016/j.synbio.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.36Kao Y.W., Hsu S.K., Chen J.Y., Lin I.L., Chen K.J., Lee P.Y., et al. Curcumin metabolite tetrahydrocurcumin in the treatment of eye diseases. Int J Mol Sci. 2020;22(1):212. doi: 10.3390/ijms22010212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai C.S., Ho C.T., Pan M.H. The cancer chemopreventive and therapeutic potential of tetrahydrocurcumin. Biomolecules. 2020;831(6):10. doi: 10.3390/biom10060831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka K., Arita M., Li D., Ono N., Tezuka Y., Kanaya S. Metabolomic characterization of a low phytic acid and high anti-oxidative cultivar of turmeric. Nat Prod Commun. 2015;10(2):329–334. doi: 10.1177/193457. 8X1501000231. [DOI] [PubMed] [Google Scholar]
- 44.Hu H., Zhou H., Xu D. A review of the effects and molecular mechanisms of dimethylcurcumin (ASC-J9) on androgen receptor-related diseases. Chem Biol Drug Des. 2021;97(4):821–835. doi: 10.1111/cbdd.13811. [DOI] [PubMed] [Google Scholar]
- 45.Steward W.P., Gescher A.J. Curcumin in cancer management: recent results of analoguedesign and clinical studies and desirable future research. Mol Nutr Food Res. 2008;52(9):1005–1009. doi: 10.1002/mnfr.200700148. [DOI] [PubMed] [Google Scholar]
- 46.Jiang T., Guo K., Liu L., Tian W., Xie X., Wen S., Wen C. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci Rep. 2020 Oct 1;10(1) doi: 10.1038/s41598-020-73274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber T., Kim H.U. The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1(2):69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsuyama Y., Kita T., Horinouchi S. Identification and characterization of multiple curcumin synthases from the herb Curcuma longa. FEBS Lett. 2009;583(17):2799–2803. doi: 10.1016/j.febslet.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 49.Park D., Swayambhu G., Lyga T., Pfeifer B.A. Complex natural product production methods and options. Synth Syst Biotechnol. 2021;6(1):1–11. doi: 10.1016/j.synbio.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Chen W., Chen H., Zhong Q., Yun Y., Chen W. Metabolomics analysis of the deterioration mechanism and storage time limit of tender coconut water during storage. Foods. 2020;9(1):46. doi: 10.3390/foods9010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park S.B., Bae D.W., Clavio N.A.B., Zhao L., Jeong C.S., Choi B.M., et al. Structural and biochemical characterization of the curcumin-reducing activity of CurA from vibri ovulnificus. Agric Food Chem. 2018;66(40):10608–10616. doi: 10.1021/acs.jafc.8b03647. [DOI] [PubMed] [Google Scholar]
- 52.Hassaninasab A., Hashimoto Y., Tomita-Yokotani K., Kobayashi M. Discovery of the curcumin metabolic pathway involving a unique enzyme in an intestinal microorganism. Proc Natl Acad Sci U S A. 2011;108(16):6615–6620. doi: 10.1073/pnas.1016217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y.F., Zheng H.B., Liu X.Y., Cheng A.X., Lou H.X. Molecular diversity of alkenal double bond reductases in the liverwort Marchantia paleacea. Molecules. 2018;23(7):1630. doi: 10.3390/molecules23071630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data are publicly accessible at https://bigd.big.ac.cn/gs under accession number CRA003702. All supporting data are included in Supplemental files.