Abstract
N-terminal acetylation is widespread in the eukaryotic proteome but in bacteria is restricted to a small number of proteins mainly involved in translation. It was long known that elongation factor Tu (EF-Tu) is N-terminally acetylated, whereas the enzyme responsible for this process was unclear. Here, we report that RimI acetyltransferase, known to modify ribosomal protein S18, is likewise responsible for N-acetylation of the EF-Tu. With the help of inducible tufA expression plasmid, we demonstrated that the acetylation does not alter the stability of EF-Tu. Binding of aminoacyl tRNA to the recombinant EF-Tu in vitro was found to be unaffected by the acetylation. At the same time, with the help of fast kinetics methods, we demonstrate that an acetylated variant of EF-Tu more efficiently accelerates A-site occupation by aminoacyl-tRNA, thus increasing the efficiency of in vitro translation. Finally, we show that a strain devoid of RimI has a reduced growth rate, expanded to an evolutionary timescale, and might potentially promote conservation of the acetylation mechanism of S18 and EF-Tu. This study increased our understanding of the modification of bacterial translation apparatus.
Keywords: ribosome, post-translational modification, bacteria, EF-Tu
Abbreviations: BSA, bovine serum albumin; EF-Tu, elongation factor Tu; IF, initiation factor; MS, mass spectrometry
Post-translational modifications are paramount for the regulation of protein activity, stability, and localization. Acetylation is among the most abundant protein modifications in eukaryotes (1), while bacteria have generally few acetylated proteins (2). A number of Escherichia coli proteins are N-acetylated (3), such as ribosomal proteins S5 (4), S18 (5), and L12 (6) as well as the translation elongation factor Tu (EF-Tu) (7).
Protein acetylation is commonly carried out by the GCN5-related (GNAT) family of enzymes (2). A family of acetyltransferases RimJ (8), RimI (8), and RimL (9) were found to be responsible for the N-terminal acetylation of ribosomal proteins. Acetylation of S5 was found to be involved in ribosome assembly (10, 11), in a way that its inactivation leads to bacterial growth cold sensitivity. RimL is known to catalyze ribosomal protein L12 acetylation (9). Recently, RimL ascribed an additional function of microcin C acetylation resulting in a ground state E. coli resistance level toward this antibiotic (12). While the phenotype of rimL inactivation is moderate (9, 13), rimL expression was shown to be increased at the stationary phase of bacterial culture (13). RimI-dependent acetylation of the ribosomal protein S18 (8, 14) was characterized structurally and biochemically (15).
EF-Tu, the most abundant bacterial protein (16), is N-terminally acetylated. Its abundance and conservation in bacteria allowed it to be recognized by the innate immunity system of plants as one of pathogen-associated molecular pattern (17), and its acetylated N-terminal fragment is used as a recognition element (18). While there are indications that acetylation of EF-Tu may happen at internal lysine residues and this modification is under a growth stage control in Bacillus subtilis (19), neither regulation nor function of N-terminal acetylation of EF-Tu was studied. The enzyme responsible for acetylation of EF-Tu has eluded identification for almost 4 decades. In this work, we have demonstrated that S18-specific acetyltransferase RimI is in addition responsible for the N-terminal acetylation of EF-Tu, thus being a dual-specificity enzyme.
Results and discussion
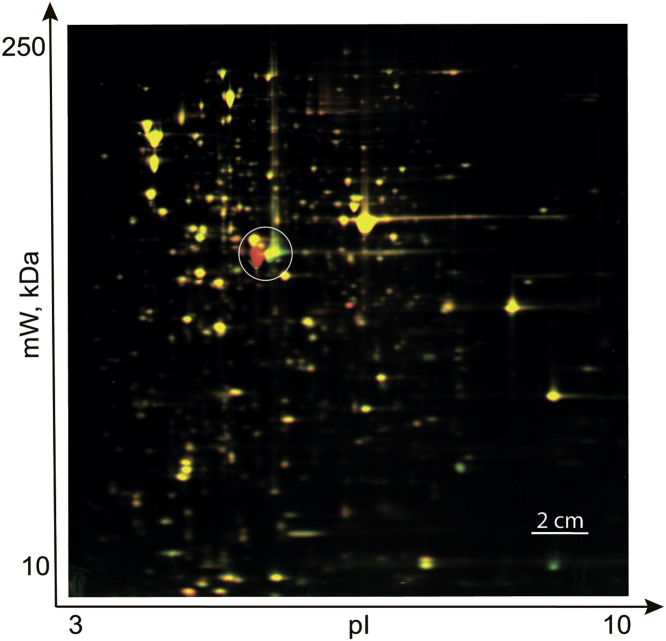
The majority of eukaryotic protein acetyltransferases is capable to modify multiple targets, while similar enzymes of bacteria are usually considered to be single protein specific. Since amino group acetylation removes a single positive charge, it might be detected by 2D protein gel electrophoresis. We compared total proteins extracted from ΔrimI strain from Keio knockout collection (20), labeled by Cy2 green fluorescent dye with that from the isogenic parental strain (21) labeled with the Cy5 red fluorescent dye with the help of 2D protein electrophoresis (Fig. 1). We expected that proteins that differed by the acetylation would be represented by a couple of green and red spots of the same molecular mass but different isoelectric points. We readily observed such an effect when comparing proteins from the WT and ΔrimI strains (Fig. 1). The protein spots with an isoelectric point dependent on the RimI were excised from the gel and identified by MALDI mass spectrometry (MS) following tryptic digestion. Both spots appeared to correspond to the EF-Tu protein. While intensity of several protein spots on the gel (Fig. 1) appears to depend on rimI gene functionality, they do not demonstrate the pattern that would indicate they represent the substrates of acetylation.
Figure 1.
Identification of novel substrates for known ribosomal protein N-acetyltransferases. 2D protein gel electrophoresis of total proteome from the WT strain labeled by Cy5 and Cy2-labeled proteome from ΔrimI strain. Horizontal axis corresponds to isoelectrofocusing, whereas vertical axis to separation by molecular mass (scale shown). Yellow spots correspond to the proteins with unchanged expression level. Red spots reflect proteins underrepresented in ΔrimI strain. Green spots correspond to proteins overrepresented in ΔrimI strain. Circled are the spots corresponding to EF-Tu—the new substrate of RimI. EF-Tu, elongation factor Tu.
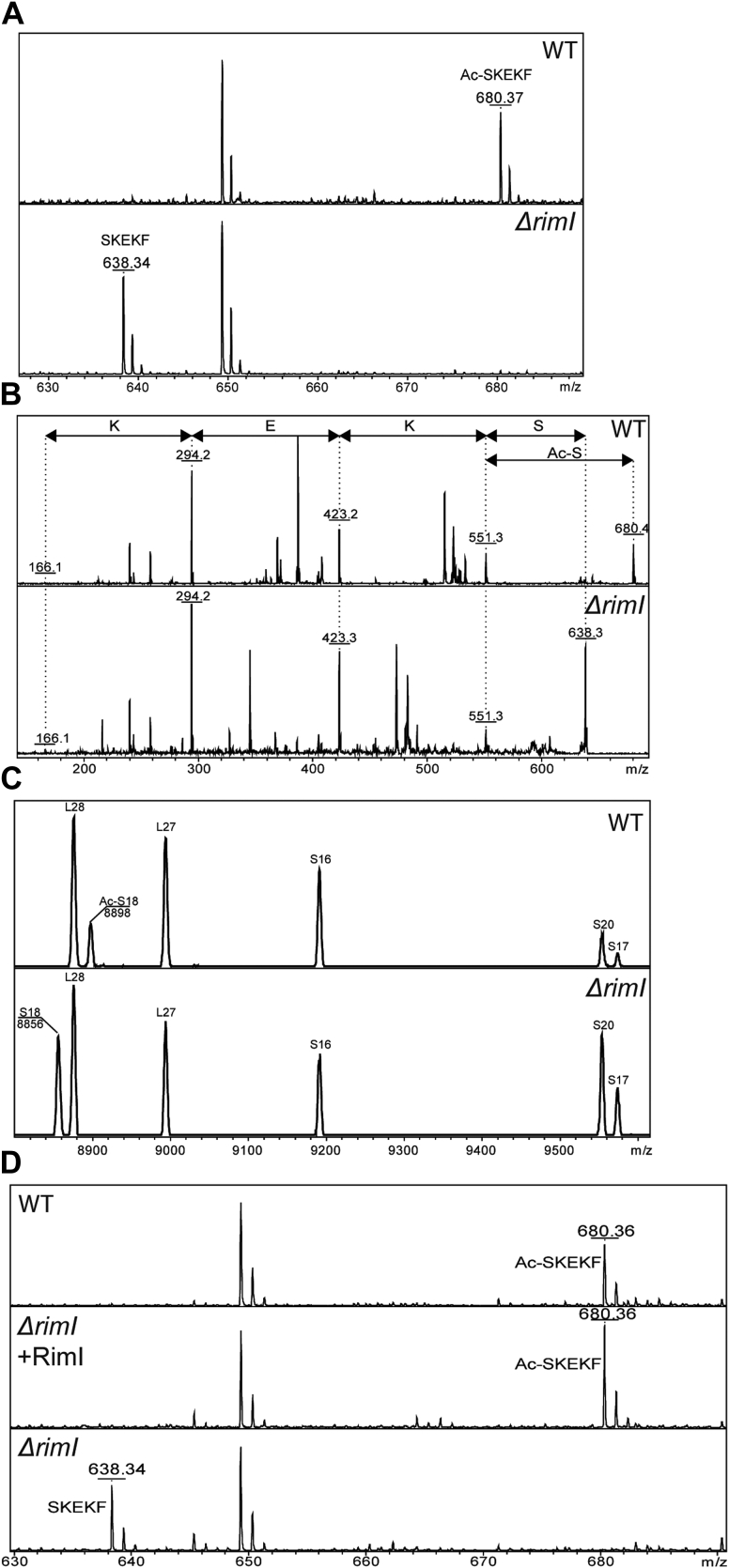
To identify EF-Tu amino acid that is acetylated by RimI, we transformed ΔrimI strain with the plasmid carrying tufA gene with the C-terminal hexahistidine tag and used the resulting culture for EF-Tu preparation by metallochelate chromatography. WT strain transformed with the same plasmid was used for purification of the control EF-Tu sample. Tryptic hydrolysate of recombinant EF-Tu was subjected to MALDI MS analysis, which failed to reveal the modification site presumably because N-terminal end of EF-Tu is rich with basic amino acids, and digestion with trypsin results in a very short tryptic peptide. To overcome this difficulty, we applied chymotrypsin hydrolysis followed by MALDI MS analysis (Fig. 2A) and readily identified that EF-Tu from the WT, but not from ΔrimI strain, contains acetyl group attached to the N-terminal SKEKF peptide. Further fragmentation of this peptide in mass spectrometer (Fig. 2B) indicated that the N-terminal serine group is acetylated. This result corroborated earlier findings that EF-Tu is N-terminally acetylated (7). Thus, RimI is a dual-specificity protein acetyltransferase responsible for the N-terminal modification of two proteins, S18 and EF-Tu.
Figure 2.
Mass spectrometry analysis of protein modification in the WT and ΔrimI strain (panels are labeled accordingly).A, mass spectrometry analysis of EF-Tu modification. EF-Tu hydrolysate by chymotrypsin was analyzed. Panels corresponding to the WT and ΔrimI strain are labeled. Shown are peaks corresponding to the N-terminal SKEKF peptide. B, fragmentation analysis of the N-terminal peptide of EF-Tu. Panels corresponding to the WT and ΔrimI strain are labeled. Mass differences between the fragments are labeled by amino acids removed. C, mass spectrometry analysis of S18 protein. Total ribosomal proteins were used for the analysis. The panels correspond to the WT strain (upper panel) and the ΔrimI strain (lower panel), labeled accordingly. Peaks corresponding to ribosomal proteins are marked. D, genetic complementation of the ΔrimI strain by the plasmid coding for RimI. Shown are peaks corresponding to the N-terminal SKEKF peptide. EF-Tu, elongation factor Tu.
To verify that rimI gene is indeed inactivated in the ΔrimI strain from Keio collection and since S18 protein is too small to be resolved by the standard 2D gel, we compared the mass of S18 protein, the known target of RimI (8), from knockout and the WT strains (Fig. 2C). As expected, the mass of S18 from the knockout strain was found to be lower by precisely the mass of the acetyl group. To check that it is inactivation of rimI gene responsible for the difference in the EF-Tu isoelectric point, but not some hypothetic secondary mutation, we transformed ΔrimI strain with the plasmid coding for RimI protein. Analysis of EF-Tu by MALDI MS confirmed that reintroduction of rimI gene restored acetylation of N-terminal serine (Fig. 2D). Thus, we demonstrated that RimI is necessary for EF-Tu N terminus acetylation. It is most likely that RimI directly acetylates EF-Tu, although we should formally acknowledge that a possibility might exist that an influence of RimI on EF-Tu acetylation could be indirect.
The structure of Salmonella typhimurium RimI protein in a complex with bisubstrate inhibitor CoA-S-acetyl-ARYFRR containing a peptide part mimicking the N-terminal peptide of S18 ribosomal protein was determined by Blanchard et al. (15) previously. Interaction of RimI with S18 N-terminal peptide included several sequence-specific contacts, such as electrostatic interaction of Arg2 with Glu103 residue of RimI. The amino acid of the peptide inhibitor equivalent to Tyr4 of S18 interacts with Pro131 of RimI, whereas Phe4 formed Van der Waals contacts with Trp27, Thr31, Phe67, and Asn35 of RimI. Curiously, a comparison of the N-terminal peptides of S18 and EF-Tu revealed little resemblance. Of note is the fact that both proteins possess a small terminal amino acid, Ala1 in S18 and Ser1 in EF-Tu, whereas the second residue is positively charged, Arg2 in S18 and Lys2 in EF-Tu.
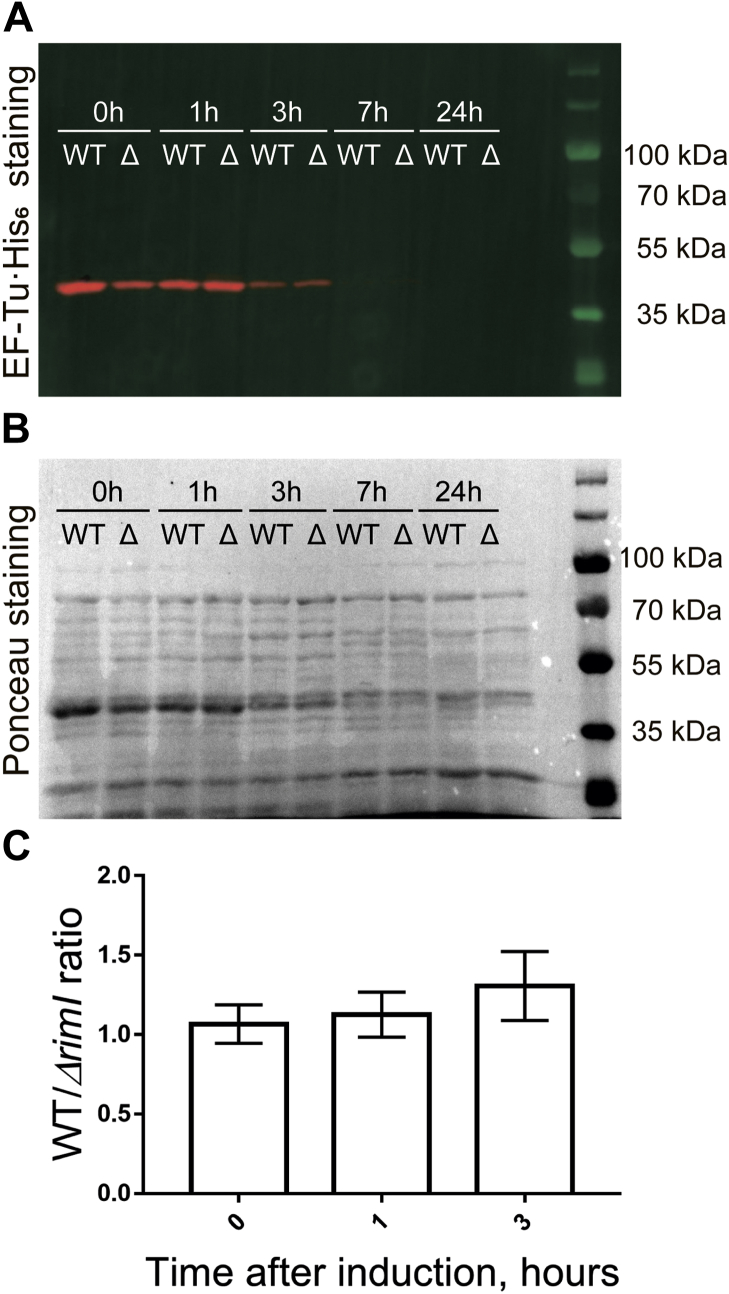
Since protein N-terminal acetylation is considered to predetermine protein stability at least in eukaryotes (22), we decided to check whether acetylation of EF-Tu by RimI influences the stability of the factor. To this end, we used ΔrimI strain transformed by the plasmid coding for EF-Tu·His6 under the control of anhydrotetracycline-inducible Tet promoter. Expression of EF-Tu·His6 was switched on for 30 min at the early exponential growth phase (absorbance of 0.1 at 600 nm) followed by washing the cells with medium lacking inducer. The amount of EF-Tu·His6 in cells after cessation of expression was monitored by immunoblotting with anti-His6 antibodies (Fig. 3A) and revealed that acetylation does not alter EF-Tu stability.
Figure 3.
Phenotype of the ΔrimI strain. The amount of EF-Tu·His6 in the cells after cessation of its expression monitored by immunoblotting with anti-His6 antibodies. The time after expression switch off is indicated above the lanes. Results of representative immunoblotting are shown at (A) and (B). A, immunoblotting with anti-His6 antibodies (red color) and visualization of prestained molecular weight markers (green color). B, Ponceau staining of the membrane. C, analysis of relative EF-Tu stability (EF-Tu·His6 signal ratio in WT and ΔrimI cells normalized to the total protein level). Shown are results of three independent experiments. EF-Tu, elongation factor Tu.
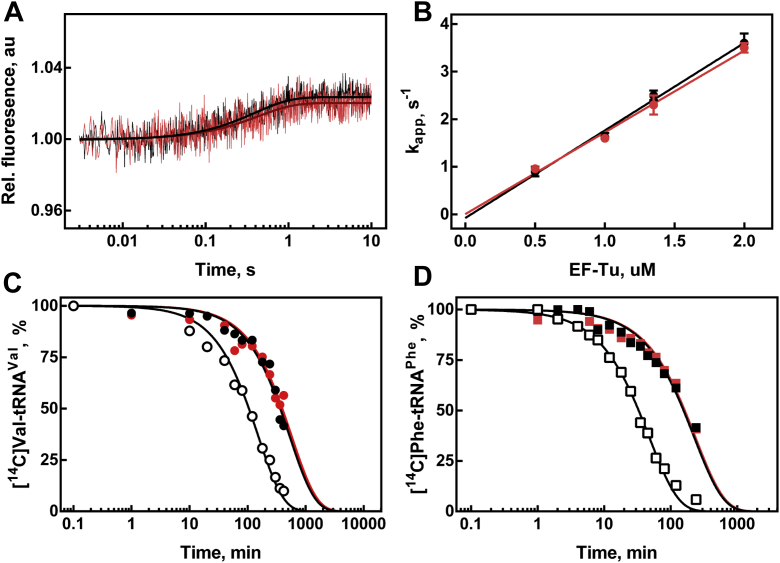
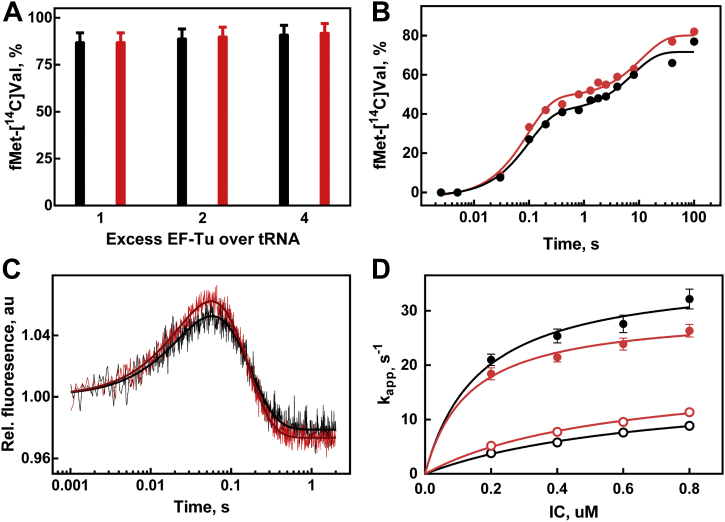
Finally, to assess the performance of unacetylated EF-Tu in partial reactions of protein biosynthesis, we used C-terminally His6-tagged factors purified from the WT (EF-Tu) and ΔrimI strains (EF-Tu-Ac). First, we questioned whether an interaction of aminoacylated tRNA with EF-Tu depends on the acetylation of the latter. Kinetics of the ternary complex formation by EF-Tu·GTP or EF-Tu-Ac·GTP with Phe-tRNAPhe has been studied by a stopped-flow experiment monitoring proflavine fluorescence of Phe-tRNAPhe(Prf16/17). The resultant fluorescence changes were fitted to single exponential curves (Fig. 4A). Linear fitting of the concentration dependence of apparent association rate constants (Fig. 4B) yielded indistinguishable within the error rate constants (kon = 1.8 ± 0.1 μM−1 s−1 for EF-Tu, kon = 1.7 ± 0.1 μM−1 s−1 for EF-Tu-Ac, koff were close to zero in both cases). Since within the structure of ternary EF-Tu·GTP–aminoacyl-tRNA complex (23), the aminoacylated 3′-terminal nucleotide of tRNA is located close to the N terminus of EF-Tu, we hypothesized that nonacetylated terminal amino group might facilitate aminoacyl-tRNA hydrolysis. To this end, we tested the stability of Val-tRNAVal and Phe-tRNAPhe in the complex with EF-Tu·GTP or EF-Tu-Ac·GTP with the help of filter-binding assay (Fig. 4, C and D). The hydrolysis time dependencies were fitted into single exponential curves. As expected, binding to the EF-Tu·GTP protected aminoacyl-tRNAs from spontaneous hydrolysis, but no significant difference in the spontaneous Val-tRNAVal or Phe-tRNAPhe hydrolysis in a complex with EF-Tu·GTP or EF-Tu-Ac·GTP has been detected.
Figure 4.
EF-Tu acetylation does not appear to impact the interaction with aminoacyl-tRNA.A, time courses of ternary complex formation of 0.25 μM Phe-tRNAPhe(Prf16/17) and 1.35 μM EF-Tu·GTP (black trace, black fitting curve) or EF-Tu-Ac·GTP (red trace, dark red fitting curve). B, concentration dependence of ternary complex formation with EF-Tu·GTP (black circles) and EF-Tu-Ac·GTP (red circles). C, portion of unhydrolyzed Val-tRNAVal at specific time points in the presence of EF-Tu·GTP (black circles, kapp = 0.114 ± 0.006 h−1), EF-Tu-Ac·GTP (red circles, kapp = 0.105 ± 0.008 h−1), and in the absence of EF-Tu (open circles, kapp = 0.40 ± 0.02 h−1). D, portion of unhydrolyzed Phe-tRNAPhe at specific time points in the presence of EF-Tu·GTP (black squares, kapp = 0.27 ± 0.02 h−1), EF-Tu-Ac·GTP (red squares, kapp = 0.26 ± 0.02 h−1), and in the absence of EF-Tu (open squares, kapp = 1.27 ± 0.04 h−1). EF-Tu, elongation factor Tu.
Next, we addressed the possible role of EF-Tu N-terminal acetylation in EF-Tu-dependent steps of translation on the ribosome. Initiation complexes programmed with mRNA coding for MVF (Met-Val-Phe) or MF (Met-Phe) peptides and containing fMet-tRNAfMet in the P-site were preformed with the complete set of translation initiation factors (IFs). Analysis of the dipeptide formation yield has been done at proportions of Val-tRNAVal to EF-Tu ranging from 1:1 to 1:4 (Fig. 5A). In all cases, dipeptide formation was almost quantitative. We did not observe any difference in dipeptide yield for the ternary complexes of either WT EF-Tu or EF-Tu-Ac. While the end points of dipeptide formation demonstrated no dependence on the acetylation of EF-Tu, we set up to determine whether rate constants of dipeptide formation, which reflects binding and accommodation rates of aminoacyl-tRNA brought to the ribosome by EF-Tu·GTP, might depend on EF-Tu modification. Millisecond resolution of automatic quench-flow mixing of initiation ribosomal complexes with Val-tRNAVal·EF-Tu·GTP or Val-tRNAVal·EF-Tu-Ac·GTP (Fig. 5B) allowed us to calculate the rate constants of the apparently biphasic dipeptide formation process. However, the rate constants kapp1 = 10 ± 2 s−1, kapp2 = 0.13 ± 0.03 s−1 for Val-tRNAVal·EF-Tu·GTP and kapp1 = 10 ± 1 s−1, kapp2 = 0.09 ± 0.02 s−1 for Val-tRNAVal·EF-Tu-Ac·GTP were found to be the same within the error limits. In addition, we used a stop flow fluorescence detection assay to monitor movements of the Phe-tRNAPhe(Prf16/17) brought to the ribosome with the vacant A-site in a form of either Phe-tRNAPhe(Prf16/17)·EF-Tu·GTP or Phe-tRNAPhe(Prf16/17)·EF-Tu-Ac·GTP (Fig. 5C). An increase in a characteristic biphasic fluorescence change represents the following steps: initial binding of the ternary complex to the ribosome and subsequent codon reading and recognition, which induces GTPase activation and results in GTP hydrolysis by EF-Tu. The subsequent decay of the high fluorescence intermediate is related to the release of tRNA from the GDP-bound form of EF-Tu and accommodation of the tRNA in the A site of the ribosome (24). Two exponential fitting of the fluorescence curves allowed us to deduce the rate constants of ternary complex binding and A-site accommodation for a number of concentrations of the initiation complexes (Fig. 5D). Interestingly, we have observed that acetylated form of EF-Tu performs slightly better ensuring faster delivery of aminoacyl-tRNA to the translating ribosome (k1 = 37 ± 3 s−1 for EF-Tu and k1 = 30 ± 2 s−1 for EF-Tu-Ac). At the same time, the rates of tRNA accommodation were the same for both variants of EF-Tu (k2 = 17 ± 1 s−1 for EF-Tu and k2 = 19 ± 2 s−1 for EF-Tu-Ac).
Figure 5.
Influence of EF-Tu acetylation on the interaction of ternary aminoacyl-tRNA·EF-Tu·GTP complex with the ribosomal A-site.A, the extent of dipeptide formation upon mixing of Val-tRNAVal·EF-Tu·GTP (black bars) or Val-tRNAVal·EF-Tu-Ac·GTP (red bars) with the ribosomal initiation complex. Excess of EF-Tu over Val-tRNAVal used is indicated below the bars. B, time dependence of dipeptide formation upon mixing of Val-tRNAVal·EF-Tu·GTP (black circles) or Val-tRNAVal·EF-Tu-Ac·GTP (red circles) with the ribosomal initiation complex. C, relative fluorescence change upon mixing of 0.1 μM Phe-tRNAPhe(Prf16/17)·EF-Tu·GTP (black trace, black fitting curve) or 0.1 μM Phe-tRNAPhe(Prf16/17)·EF-Tu-Ac·GTP (red trace, dark red fitting curve) with the 0.6 μM ribosomal initiation complex. D, dependencies of the kapp1 and kapp2 rate constants for the process of EF-Tu–dependent Phe-tRNAPhe(Prf16/17) binding and accommodation into the ribosomal A-site on the concentration of the initiation complexes. Black symbols correspond to the WT EF-Tu, whereas red symbols correspond to EF-Tu-Ac. Filled symbols correspond to kapp1, and open symbols correspond to kapp2 rate constants. EF-Tu, elongation factor Tu.
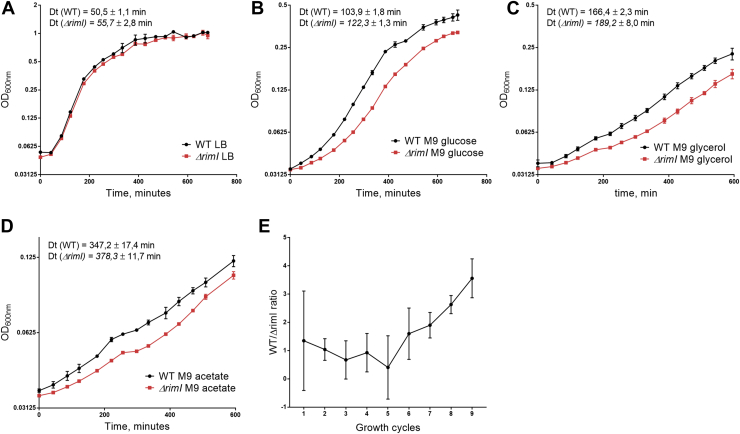
Finally, to assess whether acetylation of EF-Tu that mildly increases the rate of aminoacyl-tRNA delivery to the ribosome confers advantage to bacterial protein biosynthesis, we monitored the growth curves of the WT and ΔrimI strains (Fig. 6, A–D). Application of this assay revealed a moderate growth disadvantage of the ΔrimI strain. While the difference is small in rich medium (LB) (Fig. 6A), in the minimal medium, growth defects become more pronounced (Fig. 6, B–D). In a large timescale, growth advantage granted by the system of S18 and EF-Tu acetylation would lead to the fixation of this trait in bacterial population. To prove this assumption, we performed a growth competition assay (Fig. 6E). Equal numbers of the kanamycin-sensitive parental strain cells and kanamycin-resistant ΔrimI strain cells were mixed and incubated for 24 h at 37 °C in M9 minimal medium supplemented with glucose (0.4%). At the end of the growth cycle, 1/1000th of the medium containing the mixture of the cells was used for inoculation of the fresh portion M9 medium at 1000 times of dilution. After each growth cycle, the cells were serially diluted and plated on either LB agar or LB agar plates supplemented with kanamycin. The increase of WT to ΔrimI cells ratio in the mixture was slow, but detectable, and suggests that knockout of rimI compromises cell fitness. Beneficial influence of RimI activity on cell growth may explain nearly universal presence of rimI gene in bacterial genomes (25). Thus, we suggest that EF-Tu acetylation accelerates tRNA arrival to the A-site of the ribosome that finally results in improvement of bacterial growth rate, which is especially evident for the growth in poor media when aminoacyl-tRNA availability might be limiting.
Figure 6.
Growth curves (log2scale) of the WT (black) and ΔrimI strains (red) in different growth conditions.A, LB medium. B, M9 medium supplemented with 0.4% glucose. C, M9 medium supplemented with 0.4% glycerol. D, M9 medium supplemented with 0.4% acetate. Each point represents three independent replicates. Doubling times (Dt) are indicated. E, growth competition between the WT strain and ΔrimI strain. Shown is the ratio of WT to ΔrimI cells in the mixture. Each point corresponds to a 24 h growth cycle and represents three independent replicates.
However, it should be noted that it is difficult to estimate the direct impact of EF-Tu acetylation on cell fitness, since RimI is responsible for modification of S18 as well. It is most likely that the absence of acetylation on both S18 and EF-Tu may contribute to the observed growth defects in ΔrimI strain.
Experimental procedures
Strains, medium, and plasmids
E. coli strain BW25113 (21) used as WT and the isogenic strain JW4335 (20), lacking the rimI gene, were kindly provided by Hironori Niki, National Institute of Genetics, Japan. E. coli strains were grown at 37 °C in LB medium and supplied with 50 μg/ml kanamycin if required. For the measurement of growth curves, LB or M9 minimal medium was inoculated by a single colony of either the WT or ΔrimI strains and incubated for 24 h at 37 °C. The culture was diluted to absorbance of 0.01 at 600 nm (final culture volume of 400 μl) with fresh LB or M9 minimal medium and grown at 37 °C for 12 h in 24-well microtiter plates with constant shaking at 200 rpm. The absorbance at 600 nm of culture was measured every 40 min. M9 minimal medium was supplemented with either glucose (0.4%), glycerol (0.4%), or sodium acetate (0.4%).
A growth competition experiment was performed using M9 medium supplemented with glucose (0.4%) at 37 °C. Equal amounts of kanamycin-sensitive BW25141 parental strain and kanamycin-resistant ΔrimI deletion strain cells were used for simultaneous inoculation of M9 medium. At each 24 h growth cycle, the medium was diluted 1000-fold. At the same time, serial dilutions of the bacterial culture were made until a dilution of 10−7 was reached. Equal 5 μl aliquots from the diluted cultures were plated to LB agar and LB agar supplemented with kanamycin (50 μg/ml). Plates were incubated at 37 °C overnight, and the colonies were counted and compared.
To track EF-Tu stability over time, DNA sequence coding for EF-Tu·His6 under control of anhydrotetracycline-inducible Tet promoter was cloned (26) into the pASK-IBA4 plasmid with the following oligonucleotides: Forward: 5′-AAAAATCTAGATAACGAGGGCAAAAAATGTCTAAAGAAAAATTTGAACGTACAAAACC-3′, Reverse: 5′-CTTCACAGGTCAAGCTTAGTTAGATATCTTAATGATGATGATGATGGTGGCCCAGAACTTTAGCAACAA-3′. For the recombinant EF-Tu purification, DNA sequence coding for EF-Tu·His6 under control of IPTG-inducible lac promoter was cloned into the plasmid pET-33b(+) with the following oligonucleotides: Forward: 5′-CACACAGAATTCATTAAAGAGGAGAAATTAACTATGTCTAAAGAAAAATTTGAACGTACAAAACC-3′, Reverse: 5′-CCTTAGCGGCCGCTTAATGGTGATGGTGATGGTGCCCGCCCAGAACTTTAGCAAC-3′.
2D protein gel electrophoresis
2D difference gel electrophoresis analysis was performed as previously described (27, 28). Briefly, E. coli cells were lysed with 2D sample buffer (1 M Tris–HCl [pH 8.5], 7 M urea, 2 M thiourea, 4% [w/v] CHAPS), 50 μg of total protein was labeled with Cy2 or Cy5 (Lumiprobe) and applied for isoelectric focusing on a self-prepared nonlinear pH gradient (pH 3.5–9.5) gel. The second dimension was performed with gradient (8–16%) gels.
Purification of EF-Tu and other components
For the recombinant EF-Tu purification, 10 μl overnight cultures of the WT BW25113 or JW4335 ΔrimI strains transformed with the EF-Tu·His6 IPTG-inducible pCA24N plasmid were used for inoculation of the 500 ml LB with 0.34 g/l chloramphenicol. At an absorbance of 0.7 at 600 nm, expression of EF-Tu gene was induced by 1 mM IPTG (final concentrations are given throughout the article). After 3 h of induction, cells were harvested by centrifugation, resuspended in the buffer 25 mM Tris–HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 5% glycerol, 50 μM GDP, 5 mM β-mercaptoethanol, 1× protease inhibitor cocktail (Roche), DNAse I 0.1 μ/μl, and lysed by a French press. EF-Tu was prepared from cleared lysates by Akta start (GE) FPLC with Protino nickel–iminodiacetic acid (Macherey-Nagel) and HiTrapQ (GE) columns at 4 °С. Buffer 25 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 2.5% glycerol, 50 μM GDP, and 5 mM β-mercaptoethanol with segmented gradient of imidazole concentration 0 to 300 mM was used for nickel–iminodiacetic acid chromatography. Buffer 25 mM Tris–HCl (pH 7.5), 70 mM NH4Cl, 7 mM MgCl2, 2.5% glycerol, and 2.5 mM DTT with linear gradient of KCl concentration 5 to 1000 mM were used for ion-exchange chromatography. Fractions containing EF-Tu, according to the SDS gel electrophoresis, were pulled together. Pure EF-Tu was concentrated and transferred to storage buffer by Amicon Centrifugal Filter (Millipore), storage buffer conditions 25 mM Tris–HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 10% glycerol, and 2.5 mM DTT.
Ribosomes, IFs (IF1, IF2, and IF3), fMet-tRNAfMet, [14C]Phe-tRNAPhe, [14C]Val-tRNAVal, and tRNAPhe(Prf16/17) were prepared according to previously published techniques (29, 30, 31, 32, 33, 34, 35). mRNA MF containing sequence 5′- …ACU·AUG·UUU …-3′ coding for Met-Phe-… and mRNA MVF containing sequence 5′-…ACU·AUG· GUU·UUU…-3′ and coding for Met-Val-Phe-… were obtained by T7 transcription in vitro and used throughout the in vitro experiments if not indicated otherwise. A sequence of the template for T7 transcription of mRNA MVF: 5′-CGAATTTAATACGACTCACTATAGGGAATTCAAAAATTTAAAAGTTAACAGGTATACATACTATGGTTTTTATTACTACGATCTTCTTCACTTAATGCGTCTGCAGGCATGCAAC-3′. A sequence of the template for T7 transcription of mRNA MF 5′-CGAATTTAATACGACTCACTATAGGGAATTCAAAAATTTAAAAGTTAACAGGTATACATACTATGTTTACGATTACTACGATCTTCTTCACTTAATGCGTCTGCAGGCATGCAAGC-3′. The sequence of T7 promoter is shown in italics, sequences coding for methionine-valine-phenylalanine or methionine-phenylalanine are shown in bold. All in vitro reactions were performed in TAKM7 buffer 50 mM Tris–HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2 at 37 °С.
For a typical 1 ml reaction of initiation complex formation, 1 μM of the ribosomes were incubated with 3 μM mRNA, 2 μM fMet-tRNAfMet, IF1, IF2, and IF3, 1.5 μM in TAKM7 with 1 mM GTP and 2 mM DTT for 1 h at 37 °C. Ternary complexes of aminoacyl-tRNA·EF-Tu·GTP were prepared by preincubation of the WT EF-Tu or EF-Tu-Ac with 1 mM GTP, 3 mM phosphoenolpyruvate, 2 mM DTT, 1% pyruvate kinase in TAKM7 for 15 min at 37 °С, followed by addition of aminoacyl-tRNA and 1 min incubation. fMet-tRNAfMet, Phe-tRNAPhe, and Val-tRNAVal were beforehand purified by HPLC and stored at −80 °C, whereas Phe-tRNAPhe(Prf16/17) was prepared immediately before ternary complex formation. For this, 8 μM tRNAPhe(Prf16/17) was incubated with 0.2 mM Phe, 3 mM ATP, 6 μM β-mercaptoethanol, 40 nM phenylalanine-tRNA synthetase, and 40 nM tRNA nucleotidyltransferase in buffer TAKM7 for 30 min at 37 °C. In the rapid kinetics experiments, 60 to 70 μl of the initiation complex and 60 to 70 μl of the ternary complex were used per reaction. For spontaneous aminoacyl-tRNA deacylation experiments, 20 μl of the ternary complex were used per reaction. In the experiments on dipeptide formation, 20 μl of the initiation complex and 20 μl of the ternary complex were used per reaction.
MS
MS analysis was carried out as follows: protein zones of interest were cut out of gels, rinsed twice with ultrapure water, rinsed twice with washing buffer (50 mM NH4HCO3 and 40% acetonitrile) for 20 min, dehydrated in acetonitrile for 5 min, and dried in the open air. Dried pieces of the gel were incubated with 5 ng/μl trypsin (Trypsin Gold; Promega) solution in 100 mM NH4HCO3 for 4 h at 37 °С. After incubation, an equal volume of 0.5% trifluoroacetic acid was added; the samples were loaded on a support, dried in the open air, and analyzed by MALDI MS using an Ultraflex II mass spectrometer (Bruker). For MS analysis of EF-Tu modification status, protein extracts from the WT and ΔrimI strains were subjected to 2D difference gel electrophoresis. The protein spots of interest were identified with trypsin digestion and subsequent MALDI-TOF analysis. For MS analysis of S18 modification status, total ribosomal proteins from the WT and ΔrimI strains were subjected to MALDI-TOF analysis directly without fragmentation. To identify an exact modification site of EF-Tu, protein spots were treated by 50 ng/μl сhymotrypsin (Sigma) solution supplemented with 10 mM of CaCl2. Reaction conditions and buffer composition were the same as in trypsin digestion protocol. The mass spectra were analyzed with the FlexAnalysis program.
Monitoring of EF-Tu stability
WT and ΔrimI cells were transformed with a plasmid coding for EF-Tu·His6 under the control of anhydrotetracycline-inducible Tet promoter. The expression of EF-Tu·His6 was switched on (100 μg/l) for 30 min at the early exponential growth phase (absorbance of 0.1 at 600 nm) followed by washing off the cells with the medium without inducer. Samples were taken from the culture after indicated time points and lysed by 2D sample buffer. Protein extracts were separated by SDS-PAGE, and semidry transfer was performed for 30 min at 20 V using Amersham Hybond P Western blotting membranes, polyvinylidene difluoride. The membrane was blocked with bovine serum albumin (BSA) and incubated with primary monoclonal anti-His6 antibodies (sc-8036; Santa Cruz; 1:5000 dilution, 16 h, 4 °C, 2% BSA, and Tris-buffered saline with Tween-20). Secondary horseradish peroxidase–conjugated antimouse antibodies were used (1:5000 dilution, room temperature, 2 h, 2% BSA, and Tris-buffered saline with Tween-20), and the bands were developed using the ECL Western Blotting Detection Kit (Amersham) in the chemiluminescence mode in ChemiDoc MP (Bio-Rad). Gel loading control was done by Ponceau S staining prior to membrane blocking with BSA.
Rapid kinetics experiments
Rapid kinetics were measured using an SX-20 stopped-flow apparatus (Applied Photophysics). Proflavin fluorescence was excited at 460 nm and measured after passing a cutoff filter KV495 nm (Schott). Samples were rapidly mixed in equal volumes. Time courses depicted in the figures were obtained by averaging 5 to 7 individual transients. Calculations were performed using Prism 6.02 software (GraphPad Software, Inc). Standard deviations were calculated using the same software.
To study the kinetics of ternary complex formation, Phe-tRNAPhe(Prf16/17) was rapidly mixed with increasing amounts of EF-Tu·GTP or EF-Tu-Ac·GTP prepared as described previously. Data were evaluated by fitting to a single-exponential function with a characteristic time constant (kapp), amplitude (A), and final signal amplitude (F∞) according to equation F = F∞ + A ∗ exp(−kapp ∗ t), where F is the fluorescence at time t. A linear function was used to analyze the concentration dependence of the ternary complex formation apparent rate constants from the concentration of EF-Tu·GTP.
To monitor the time course of A-site aminoacyl-tRNA accommodation, we rapidly mixed Phe-tRNAPhe(Prf16/17)·EF-Tu·GTP or Phe-tRNAPhe(Prf16/17)·EF-Tu-Ac·GTP with the increasing amounts of the initiation complexes programmed with mRNA MF at 20 °C. Data were evaluated by fitting to a two-exponential function with two characteristic time constants (kapp1 and kapp2), amplitudes (A1 and A2), according to equation F = F∞ + A1 ∗ exp(−kapp1 ∗ t) + A2 ∗ exp(−kapp2 ∗ t). The hyperbolic function was used to analyze the concentration dependence of the A-site reactions.
Spontaneous aminoacyl-tRNA deacylation
Ternary complexes of aminoacyl-tRNA·EF-Tu·GTP with either [14C]Val-tRNAVal or [14C]Phe-tRNAPhe and either EF-Tu or EF-Tu-Ac were prepared as described previously with two-fold excess of EF-Tu over aminoacyl-tRNA or without the factor in no EF-Tu control. After incubation at 37 °C, reactions were terminated by trichloroacetic acid, filtered through the nitrocellulose membrane, and counted at the scintillation counter.
Dipeptide formation
To assess the endpoint and kinetics of dipeptide formation, 0.5 μM initiation complexes programmed with MVF mRNA were mixed with 0.25 μM [14C]Val-tRNAVal·EF-Tu·GTP and [14C]Val-tRNAVal·EF-Tu-Ac·GTP at the final proportions EF-Tu:aa-tRNA 1:1, 2:1, and 4:1 for the endpoint monitoring and 1.5:1 for the monitoring of kinetics for dipeptide formation. For the endpoint determination, the reactions were incubated for 5 min after mixing. Then samples were quenched with 1/10 volume of 5 M KOH and hydrolyzed for 30 min at 37 °C. Samples were neutralized with 1/10 volume of glacial acetic acid and analyzed by reversed-phase HPLC. The percentage of the synthesized dipeptide was determined by the incorporation of the radioactive label. To measure the kinetics of dipeptide formation, the reactants were mixed in a quench-flow instrument KinTek RQF-3 (KinTek Corporation). The reaction was stopped at certain time points (0.0025 s–5 min) by the addition of 0.8 M KOH, and subsequent procedures were performed as described previously. The reaction rate constants were calculated using two exponential equations.
Data availability
All data are available in the main text.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Moscow State University development program and the National Research Centre “Kurchatov Institute”—Petersburg Nuclear Physics Institute institutional research project 121060200127.
Author contributions
A. A. B. and P. V. S. conceptualization; P. I. P., O. S., S. E., M. F. S., M. V. S., I. A. O., and A. P. methodology; P. I. P., O. S., S. E., V. T., I. A. O., and A. P. investigation; P. V. S. writing–original draft; S. E., A. P., A. L. K., and P. V. S. writing–review & editing; A. P., O. A. D., A. L. K., and P. V. S. supervision; O. A. D. funding acquisition.
Funding and additional information
This work was supported by the Russian Science Foundation grant 21-64-00006 (to O. A. D.).
Edited by Karin Musier-Forsyth
Contributor Information
Andrey L. Konevega, Email: konevega_al@pnpi.nrcki.ru.
Petr V. Sergiev, Email: petya@genebee.msu.ru.
References
- 1.Verdin E., Ott M. 50 Years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 2.Favrot L., Blanchard J.S., Vergnolle O. Bacterial GCN5-related N -acetyltransferases: From resistance to regulation. Biochemistry. 2016;55:989–1002. doi: 10.1021/acs.biochem.5b01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nesterchuk M.V., Sergiev P.V., Dontsova O.A. Posttranslational modifications of ribosomal proteins in Escherichia coli. Acta Naturae. 2011;3:22–33. [PMC free article] [PubMed] [Google Scholar]
- 4.Wittmann-Liebold B., Greuer B. The primary structure of protein S5 from the small subunit of the Escherichia coli ribosome. FEBS Lett. 1978;95:91–98. doi: 10.1016/0014-5793(78)80059-3. [DOI] [PubMed] [Google Scholar]
- 5.Yaguchi M. Primary structure of protein S18 from the small Escherichia coli ribosomal subunit. FEBS Lett. 1975;59:217–220. doi: 10.1016/0014-5793(75)80378-4. [DOI] [PubMed] [Google Scholar]
- 6.Terhorst C., Möller W., Laursen R., Wittmann-Liebold B. The primary structure of an acidic protein from 50-S ribosomes of Escherichia coli which is involved in GTP hydrolysis dependent on elongation factors G and T. Eur. J. Biochem. 1973;34:138–152. doi: 10.1111/j.1432-1033.1973.tb02740.x. [DOI] [PubMed] [Google Scholar]
- 7.Laursen R.A., L'Italien J.J., Nagarkatti S., Miller D.L. The amino acid sequence of elongation factor Tu of Escherichia coli. The complete sequence. J. Biol. Chem. 1981;256:8102–8109. [PubMed] [Google Scholar]
- 8.Yoshikawa A., Isono S., Sheback A., Isono K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 1987;209:481–488. doi: 10.1007/BF00331153. [DOI] [PubMed] [Google Scholar]
- 9.Isono S., Isono K. Ribosomal protein modification in Escherichia coli. III. Studies of mutants lacking an acetylase activity specific for protein L12. Mol. Gen. Genet. 1981;183:473–477. doi: 10.1007/BF00268767. [DOI] [PubMed] [Google Scholar]
- 10.Roy-Chaudhuri B., Kirthi N., Kelley T., Culver G.M. Suppression of a cold-sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis. Mol. Microbiol. 2008;68:1547–1559. doi: 10.1111/j.1365-2958.2008.06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cumberlidge A.G., Isono K. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J. Mol. Biol. 1979;131:169–189. doi: 10.1016/0022-2836(79)90072-x. [DOI] [PubMed] [Google Scholar]
- 12.Kazakov T., Kuznedelov K., Semenova E., Mukhamedyarov D., Datsenko K.A., Metlitskaya A., Vondenhoff G.H., Tikhonov A., Agarwal V., Nair S., Van Aerschot A., Severinov K. The RimL transacetylase provides resistance to translation inhibitor microcin C. J. Bacteriol. 2014;196:3377–3385. doi: 10.1128/JB.01584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordiyenko Y., Deroo S., Zhou M., Videler H., Robinson C.V. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 2008;380:404–414. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- 14.Isono K., Isono S. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol. Gen. Genet. 1980;177:645–651. doi: 10.1007/BF00272675. [DOI] [PubMed] [Google Scholar]
- 15.Vetting M.W., Bareich D.C., Yu M., Blanchard J.S. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008;17:1781–1790. doi: 10.1110/ps.035899.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furano A.V. Content of elongation factor Tu in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S., Kondo N., Yoshida M., Nishiyama M., Kosono S. Dynamic changes in lysine acetylation and succinylation of the elongation factor Tu in Bacillus subtilis. Microbiology (Reading) 2019;165:65–77. doi: 10.1099/mic.0.000737. [DOI] [PubMed] [Google Scholar]
- 20.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen K.T., Mun S.-H., Lee C.-S., Hwang C.-S. Control of protein degradation by N-terminal acetylation and the N-end rule pathway. Exp. Mol. Med. 2018;50:91. doi: 10.1038/s12276-018-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissen P., Kjeldgaard M., Thirup S.R., Polekhina G., Reshetnikova L., Clark B.F.C., Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 24.Pape T. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen D.G., Meyer J.G., Baumgartner J.T., D'Souza A.K., Nelson W.C., Payne S.H., Kuhn M.L., Schilling B., Wolfe A.J. Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio. 2018;9:e01905–e01918. doi: 10.1128/mBio.01905-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bond S.R., Naus C.C. RF-Cloning.org: An online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 2012;40:W209–W213. doi: 10.1093/nar/gks396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoch P.G., Burenina O.Y., Weber M.H.W., Elkina D.A., Nesterchuk M.V., Sergiev P.V., Hartmann R.K., Kubareva E.A. Phenotypic characterization and complementation analysis of Bacillus subtilis 6S RNA single and double deletion mutants. Biochimie. 2015;117:87–99. doi: 10.1016/j.biochi.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Pletnev P., Guseva E., Zanina A., Evfratov S., Dzama M., Treshin V., Pogorel’skaya A., Osterman I., Golovina A., Rubtsova M., Serebryakova M., Pobeguts O.V., Govorun V.M., Bogdanov A.A., Dontsova O.A., et al. Comprehensive functional analysis of Escherichia coli ribosomal RNA methyltransferases. Front. Genet. 2020;11:97. doi: 10.3389/fgene.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieden H.-J., Gromadski K., Rodnin D., Rodnina M.V. Mechanism of elongation factor (EF)-Ts-catalyzed nucleotide exchange in EF-Tu. J. Biol. Chem. 2002;277:6032–6036. doi: 10.1074/jbc.M110888200. [DOI] [PubMed] [Google Scholar]
- 30.Cunha C.E., Belardinelli R., Peske F., Holtkamp W., Wintermeyer W., Rodnina M.V. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation (Austin) 2013;1 doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milon P., Konevega A.L., Peske F., Fabbretti A., Gualerzi C.O., Rodnina M.V. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 32.Vinogradova D.S., Zegarra V., Maksimova E., Nakamoto J.A., Kasatsky P., Paleskava A., Konevega A.L., Milón P. How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson J.R., Uhlenbeck O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtkamp W., Cunha C.E., Peske F., Konevega A.L., Wintermeyer W., Rodnina M.V. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wintermeyer W., Schleich H.G., Zachau H.G. Incorporation of amines or hydrazines into tRNA replacing wybutine or dihydrouracil. Methods Enzymol. 1979;59:110–121. doi: 10.1016/0076-6879(79)59073-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available in the main text.