Abstract
Total milk solid (TMS) content directly reflects the quality of milk. Rumen bacteria ferment dietary components, the process of which generates the precursors for the synthesis of milk solid, therefore, the variation in rumen bacterial community could be associated with milk solid in dairy cows. In this study, 45 healthy mid-lactation Holstein dairy cows with the similar body weight, lactation stage, and milk yield were initially used for the selection of 10 cows with high TMS (HS) and 10 cows with low TMS (LS). All those animals were under the same feeding management, and the individual milk yield was recorded for 14 consecutive days before milk and rumen fluid were sampled. Rumen fluid was used to determine bacterial community by 16S rRNA gene sequencing technique. The HS cows had significantly greater feed intake and milk TMS, fat, protein content than LS cows (P < 0.05). Among the volatile fatty acids (VFA), propionic acid and valeric acid concentrations were significantly greater in HS cows than those in LS cows (P < 0.05). There was no significant difference in the concentrations of acetate, butyrate, isobutyrate, valerate, and the total VFA (P > 0.05), nor was the acetate-to-propionate ratio, pH value, ammonia nitrogen and microbial crude protein concentrations (P > 0.05). Significant differences in the relative abundances of some bacterial genera were found between HS and LS cows. Spearman’s rank correlation analysis revealed that TMS content was correlated positively with the abundances of Ruminococcaceae UCG-014, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, Butyrivibrio 2, Prevotellaceae UCG-003, Candidatus Saccharimonas, Ruminococcus 2, Lachnospiraceae XPB1014 group, probable genus 10, Eubacterium ventriosum group, but negatively correlated with Pyramidobacte. In addition, Ruminococcaceae UCG-014, Ruminococcus 2, Ruminococcaceae UCG001, probable genus 10 and Eubacterium ventriosum group might boost the total VFA production in the rumen. In conclusion, the dry matter intake of dairy cows and some special bacteria in rumen were significantly associated with TMS content, which suggests the potential function of rumen bacteria contributing to TMS content in dairy cows.
Keywords: Dairy cow, Total milk solid content, Rumen bacteria, Milk fat
1. Introduction
Total milk solid (TMS) is the sum of milk fat, milk protein, lactose, and minerals and a direct index to evaluate milk quality. It is also the pricing index in milk trade in some countries. The precursors for TMS include volatile fatty acids (VFA), long-chain fatty acids, amino acids (AA), peptides, glucose, glycerin etc., which are primarily produced in the rumen by bacterial degradation of dietary components (Hurtaud et al. 1998; Bauman and Griinari, 2003; Chen et al. 2011). Therefore, there is a relationship between the precursor production in the rumen and the uptake by the mammary gland. The nutritional value of milk would be unbalanced if regulating one of the ingredients in milk, such as milk protein and milk fat (Jenkins and McGuire, 2006; Nichols et al., 2018). Thus, increasing TMS content is the effective measure to improve milk quality and production efficiency.
A number of factors have impact on TMS content, such as diet, environment, and animal’s genetics (Dechow et al., 2007; Kmicikewycz and Heinrichs, 2015). An appropriate proportion of high-quality roughage in total mixed ration (TMR) is the key to ensure TMS content and milk fat (Kmicikewycz and Heinrichs, 2015). Seasons have an influence on TMS, and TMS content is lower in summer but higher in winter (Bernabucci et al., 2015). Dechow et al. (2007) reported that TMS content of cross dairy cows (Holstein × Brown Swiss) was significantly higher than that in Holstein cows. Within a breed, there were considerable individual variations in the feed efficiency, milk yield, and milk protein yield when cows were fed the same diet (Shabat et al., 2016; Xue et al., 2019). Understanding the mechanisms that determine these individual variations is important to increase TMS and milk quality. It has been found that these individual variations are related to the key ruminal bacterial species and rumen fermentation profiles, which influence the precursors for the synthesis of milk protein, fat, and lactose (Shabat et al., 2016; Schären et al., 2018; Xue et al., 2019). For example, the abundance of Megasphaera elsdenii was significantly higher in cows with high feed efficiency than cows with low feed efficiency, and the abundance of Streptococcus, unclassified Enterobacteriaceae, Ruminobacter, Treponema, and unclassified Bacteroidaceae were all higher in cows with a lower milk yield (Shabat et al., 2016; Zhao et al., 2019), indicating that the ruminal bacterial profile plays a key role in the conversion of feed to milk.
This study aimed to explore the relationships between the rumen bacterial profile and TMS content. We selected 2 groups of cows with high or low TMS content to measure their rumen bacterial community by using 16S rRNA gene sequencing and the real-time qPCR techniques, and analyzed the relationships between the microbial profile and TMS content.
2. Materials and methods
The use of the animals and the experimental procedures were approved by the Animal Care Committee of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China) in accordance with the guidelines for animal experimental welfare and ethical inspection in China (No: IAS 2019 - 28).
2.1. Animals, grouping, and management
This trial was carried out in Mengde Dairy Station, Tianjin, China. There were over 1,500 Holstein lactation dairy cows in the farm, and their individual milk yield were recorded daily. All dairy cows were fed in the same cowshed with the same TMR and free access to water. On the basis of the productive records, 45 healthy lactating cows were selected with the similar body weight, parity (parity = 1.30 ± 0.74), day in milk (153 ± 16 d) and daily milk yield (34 ± 6 kg). Milk samples were taken from those cows (as detailed below) to measure milk protein, milk fat, lactose, and TMS content using MilkoScan FT120 (Foss, Hillerød, Denmark). The TMS content varied from 10.4% to 14.1% in those animals. Ten cows with the highest or lowest TMS content respectively were further selected by the mean and standard deviation (SD) based on the method of previously research (Nkrumah et al., 2006; Ramos and Kerley, 2013). The SD values above and below the mean were used to group cows into high TMS (HS, TMS > mean + 0.7 × SD) phenotype and low TMS (LS, TMS < mean – 0.7 × SD) phenotype. The aim of this study was to explore the relationships between the ruminal bacteria and TMS in cows at a similar level of milk production. A great variation in milk yield could have a confound effect on milk solid. To minimize other potential effect besides to TMS between 2 groups, some cows in each group with extremely high or low milk yield were removed (Appendix Table 1). In addition, the cows that were failed to obtain rumen fluid samples were also removed. Finally, there were 6 cows (n = 6) in both groups with a similar milk yield.
All the selected 45 cows were maintained at the same farming condition and fed the same TMR containing 36% forage and 64% concentration (dry matter basis) for 14 d continuous stable feeding. The TMR consisted of corn silage, alfalfa hay, alfalfa silage, corn, soybean meal, orange peel granules, expanded soybean, distillers dried grains with soluble and wheat bran. Cows were kept in individually tethered stalls in a barn and fed 3 times a day at 07:00, 13:00, and 18:00. Fresh drinking water was available all times. The feed offered and the refusal of cows were recorded daily to calculate the daily feed intake of individual animals. Cows were milked 3 times a day at 06:30, 12:30, and 17:30 using an automatic milking system, and the daily milk yield were recorded by the automatic milking system.
2.2. Sampling
Milk and rumen fluid were sampled at the end of the 14-d continuous stable feeding. After each milking, milk samples (50 mL each) were collected and stored at 4 °C. The samples collected in the morning, afternoon, and evening were then mixed at a ratio of 4:3:3, and subsampled as a daily sample referenced a previously article by Wu et al. (2018). The aliquots of the samples were used to analyze milk components (see below).
Rumen fluid samples (about 200 mL each) from each cow were collected after morning milking before feeding. Sample was collected using an oral stomach tube (Shen et al. 2012). The pH value was immediately measured using a pH meter (PHS-100 portable acidity meter, Tianqi Mdt InfoTech Ltd., Shanghai, China). Then, samples were filtered through a 4-layer cheesecloth, and aliquoted into 6 sterilized tube (5 mL), then stored at -80 °C until the further analysis.
2.3. Measurement of rumen fermentation parameters
The rumen liquid sample was thawed on ice, then centrifuged at 12,000 × g at 4 °C for 10 min. The supernatant was harvested for the determination of VFA, ammonia nitrogen (NH3-N), and microbial crude protein (MCP) concentrations. A colorimetric method was used to detect NH3-N and MCP as described by Wang et al. (2019). The VFA concentration was measured using a liquid chromatographic method (Agilent 7890A-7000B, Agilent, Beijing, China).
2.4. Acquisition of DNA and construction of library
The genomic DNA in rumen microbiota in each rumen fluid sample was extracted with a cetyltrimethylammonium bromide (CTAB) regent. The purity of extracted DNA was checked using 1% agarose gel, and the DNA concentration was determined using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA).
The purified DNA was used as the template, and 341F (5'-CCTACGGGNGGCWGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') were the primers. The V3 to V4 of bacterial 16S rRNA gene was amplified using an ABI GeneAmp 9700 PCR thermocycler (ABI, CA, USA) (Parada et al., 2016). The PCR protocol was 94 °C for 2 min, followed by 30 cycles at 98 °C for 10 s and 62 °C for 30 s. The PCR reaction solution consisted of 10 × KOD Buffer 5 μL, 2 mmol/L dNTP 5 μL, 25 mmol/L MgSO4 3 μL, 10 μmol/L primer 1.5 μL respectively, KOD Polymerase (TOYOBO, Japan) 1 μL, and the template DNA 100 ng. Each PCR reaction was performed in triplicate. Once the PCR reaction was completed, the amplified products were extracted using 2% agarose gel, further purified through the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). Finally, the purified products were quantified using a Quantus Fluorometer (Promega, USA), and pooled in equimolar, then paired-end sequenced (2 × 300 bp) at Illumina Hiseq 2500 PE250 platform (Illumina, San Diego, USA) under the standard protocols by GENE DENOVO. (Guangzhou, China). The raw reads of 16S rRNA gene sequencing were deposited into the NCBI Sequence Read Archive database (Accession Number: PRJNA722820).
All raw reads analysis and the quality control were referenced to the methods as reported by Huang et al. (2020). UPARSE (7.1 version, http://drive5.com/uparse/) was used to cluster the operational taxonomic unit (OTU) with 97% similarity cutoff (Liu et al., 2017), then identified, and removed the chimeric sequences. The longest read was deemed as a representative sequence of the taxonomy for each OTU. Further, those representative sequences were identified against to the Silva SSU128 database using RDP Classifier.
2.5. Real-time qPCR
The copy numbers of these bacterial species that were related with fiber, protein, or starch degradation as described by Satoshi and Yasuo, (2001) and Stevenson and Weimer, (2007) were determined by real-time qPCR in a BioRad CFX96 (Hercules, CA, USA). The same DNA samples in 16S rRNA gene sequencing were used for real-time qPCR. Primers were selected from the published papers (Table 1). The real-time qPCR was performed with SYBR Premix Ex Taq II assay kit (TaKaRa Bio Inc., Shiga, Japan) and calculated with the standard curve method. The standard curve for each bacterial species was generated using the method as described by Stubner, 2002 with plasmid DNA that contained the cloned marker loci. All standard curves met the requirements for further analysis (R2 > 0.98, 90% > E > 120%).
Table 1.
Primers used for real-time qPCR.
| Bacteria | Sequence (5’ to 3’) | Annealing temperature, °C | Size, bp | References |
|---|---|---|---|---|
| Butyrivibrio | F: GTGCCAGCMGCCGCGG | 65 | 371 | Stiverson et al. (2011) |
| R: TGCGGCACYGACTCCCTATG | ||||
| Fibrobacter succinogenes | F: GGTATGGGATGAGCTTGC | 53 | 446 | Satoshi and Yasuo (2001) |
| R: GCCTGCCCCTGAACTATC | ||||
| Ruminococcus albus | F: GTTTTAGGATTGTAAACCTCTGTCTT | 58 | 170 | Li et al. (2008) |
| R: CCTAATATCTACGCATTTCACCGC | ||||
| Ruminococcus genus | F: GAGTGAAGTAGAGGTAAGCGGAATTC | 60 | 75 | Petri et al. (2013) |
| R: GCCGTACTCCCCAGGTGG | ||||
| Prevotella bryantii | F: ACTGCAGCGCGAACTGTCAGA | 57 | 421 | Bekele et al. (2010) |
| R: ACCTTACGGTGGCAGTGTCTC | ||||
| Prevotella genus | F: GGTTCTGAGAGGAAGGTCCCC | 61 | 121 | Stevenson and Weimer (2007) |
| R: TCCTGCACGCTACTTGGCTG |
2.6. Statistical analysis
The comparison of the differences in the milk component content, rumen fermentation parameters, and bacterial abundances between HS and LS groups were performed using the t-test procedure of SAS (Version 9.4, USA). Spearman correlation analysis was used to analyze the relationship of the bacterial abundance with the milk component content, ruminal NH3-N, MCP, and VFA concentrations. The results were presented as the mean and standard error of means (SEM). Significant and extremely significant differences were declared at P < 0.05 and P < 0.01, respectively.
3. Results
3.1. Feed intake and milk composition
As shown in Table 2, the dry matter intake (DMI) differed between HS and LS cows (P < 0.05), and the DMI in LS cows was 11.0% lower than that in HS. Moreover, crude protein (CP) intake, neutral detergent fiber (NDF) intake and acid detergent fiber (ADF) intake were all significantly higher in HS group than that in LS group (P = 0.012, P = 0.011 and P = 0.010, respectively). The milk yield was similar between 2 groups (P = 0.932). The TMS (both the content [%] and yield (kg); P < 0.001, P = 0.001), milk fat (%, P < 0.001), milk protein (%, P = 0.002), and the fat-to-protein ratio (P = 0.001) were all significantly greater in HS cows than those in LS group. Meanwhile, there was no significant difference in lactose (%) and non-fat milk solid (%) between 2 groups (P > 0.05).
Table 2.
Milk composition of lactating cows selected for high or low milk solid.
| Item | HS | LS | SEM | P-value |
|---|---|---|---|---|
| DMI, kg/d | 23.89 | 21.30 | 0.558 | 0.011 |
| CP intake, kg/d | 2.49 | 2.25 | 0.051 | 0.012 |
| NDF intake, kg/d | 13.78 | 12.28 | 0.322 | 0.011 |
| ADF intake, kg/d | 3.87 | 3.41 | 0.100 | 0.010 |
| Milk yield, kg/d | 33.66 | 33.55 | 0.639 | 0.932 |
| TMS yield, kg/d | 4.38 | 3.81 | 0.118 | 0.007 |
| Moisture, % | 86.99 | 88.62 | 0.271 | 0.001 |
| Milk fat, % | 4.41 | 2.77 | 0.269 | 0.000 |
| Milk protein, % | 3.34 | 2.99 | 0.066 | 0.002 |
| Milk fat-to-protein ratio | 1.32 | 0.93 | 0.072 | 0.001 |
| Lactose, % | 4.84 | 4.98 | 0.044 | 0.112 |
| TMS, % | 13.01 | 11.38 | 0.272 | 0.000 |
| Non-fat milk solid, % | 8.60 | 8.61 | 0.077 | 0.976 |
| Percentage to TMS, %rowhead | ||||
| Milk fat | 33.89 | 24.23 | 1.639 | 0.000 |
| Milk protein | 25.66 | 26.32 | 0.367 | 0.398 |
| Lactose | 37.21 | 43.86 | 1.064 | 0.000 |
| Non-fat milk solid | 56.11 | 75.77 | 1.639 | 0.000 |
HS = high total milk solid group; LS = low total milk solid group; SEM = standard error of the mean; DMI = dry matter intake; CP = crude protein; NDF = neutral detergent fiber; ADF = acid detergent fiber; TMS = total milk solid.
When expressing milk fat, protein, and lactose as the percentages to TMS, HS cows had significantly greater milk fat (P < 0.001), but lower lactose (P < 0.001), and similar percentage of milk protein (P > 0.05) compared with those in LS cows (Table 2).
3.2. Rumen fermentation parameters
NH3-N, MCP, pH and VFA concentration in rumen fluid were shown in Table 3. There was no significant difference in pH, NH3-N, and MCP concentrations between 2 groups (P > 0.05). Among the VFA concentrations, propionic acid and valeric acid concentrations were significantly greater in HS cows than those in LS group (P < 0.05). Otherwise, there was no significant difference in the concentrations of acetate, butyrate, isobutyrate, valerate, and the total VFA (P > 0.05), nor was the acetate-to-propionate ratio (P > 0.05). There was no significant difference in the molar percentage of any of these VFA (P > 0.05).
Table 3.
Rumen fermentation parameters in lactating cows selected for high or low milk solid.
| Item | HS | LS | SEM | P-value |
|---|---|---|---|---|
| pH | 6.84 | 6.79 | 0.094 | 0.772 |
| NH3-N, mg/dL | 9.74 | 10.11 | 0.784 | 0.829 |
| MCP, mg/mL | 0.31 | 0.28 | 0.015 | 0.532 |
| VFA concentration, mmol/Lrowhead | ||||
| Acetic acid | 10.91 | 8.70 | 0.637 | 0.081 |
| Propionic acid | 4.40 | 3.10 | 0.335 | 0.044 |
| Isobutyric acid | 0.72 | 0.50 | 0.057 | 0.054 |
| Butyric acid | 2.75 | 1.99 | 0.266 | 0.164 |
| Isovaleric acid | 0.41 | 0.28 | 0.071 | 0.386 |
| Valerate acid | 0.38 | 0.29 | 0.022 | 0.045 |
| Acetate-to-propionate ratio | 2.55 | 2.79 | 0.102 | 0.261 |
| Total VFA, mmol/L | 19.58 | 14.87 | 1.294 | 0.065 |
| Molar proportion, %rowhead | ||||
| Acetic acid | 56.36 | 58.31 | 0.966 | 0.335 |
| Propionic acid | 22.25 | 21.20 | 0.580 | 0.389 |
| Isobutyric acid | 3.63 | 3.45 | 0.165 | 0.623 |
| Butyric acid | 13.78 | 13.21 | 0.686 | 0.698 |
| Isovaleric acid | 2.02 | 1.81 | 0.282 | 0.726 |
| Valerate acid | 2.00 | 2.02 | 0.097 | 0.790 |
HS = high total milk solid group; LS = low total milk solid group; SEM = standard error of the mean; NH3-N = ammonia nitrogen; MCP = microbial crude protein; VFA = volatile fatty acid.
3.3. Composition of rumen bacteria
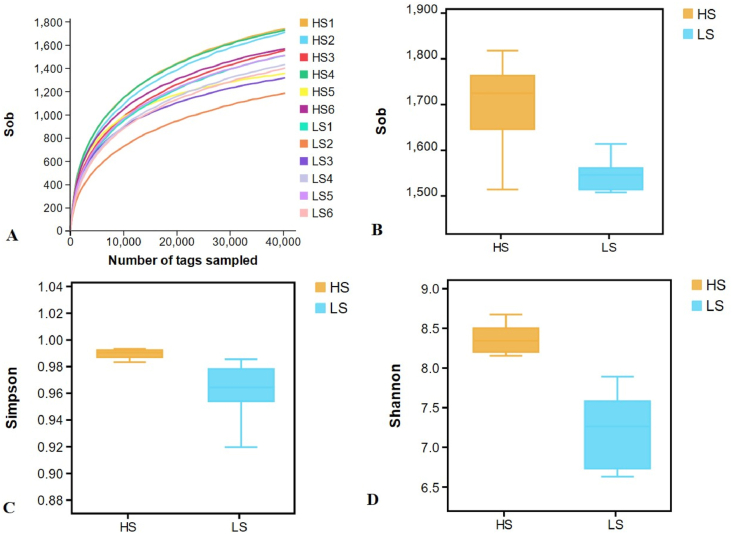
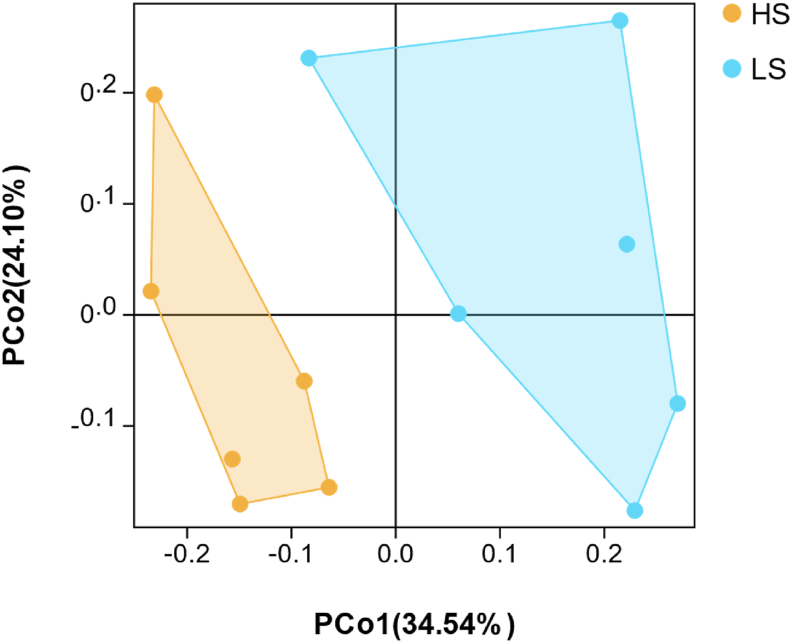
Through the 16S rRNA gene sequencing analysis, we acquired 1,221,314 high-quality sequences, with the average of 101,776 ± 4,697 sequences per sample which belonged to 1,608 ± 127 OTU (Appendix Fig. 1). A total of 1,302 OTU were commonly shared by HS and LS cows (Appendix Fig. 2). Rarefaction curve showed that samples in this experiment covered the majority of rumen bacteria (Fig. 1A). The richness index values of bacteria (Sobs, Simpson and Shannon) were all significantly greater in HS cows than those in LS cows (P < 0.05; Fig. 1B, C, and D). The similarity between and within groups were exhibited in a PCoA plot with unweighted UniFrac distance metrics (Fig. 2). The plot showed a clear differentiation in bacteria between 2 groups, and PC1 explained 34.5% of the variation and PC2 explained 24.1% of the variation.
Fig. 1.
Alpha diversity analysis of rumen bacteria in lactating cows of HS and LS. (A) Number of tags sampled; (B) Sob index; (C) Simpson index; (D)Shannon indexes. HS = high total milk solid group; LS = low total milk solid group.
Fig. 2.
Principal coordinate analysis (PCoA). The unweighted UniFrac distance metrics analysis between the rumen bacteria in lactating cows of HS or LS. HS = high total milk solid group; LS = low total milk solid group.
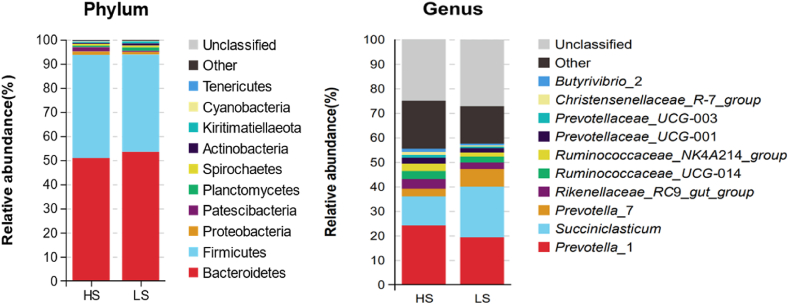
A total of 26 phyla were identified in the rumen fluid by the taxonomic analysis, and only 0.15% bacterial phylum in HS cows and 0.09% in LS cows were unclassified. As shown in Fig. 3, Bacteroidetes and Firmicutes were 2 dominant phyla both in HS and LS cows, and the relative abundances of these 2 phyla accounted for approximately 93.0% (Bacteroidetes 53.0% and Firmicutes 40.0%). At genus level, 227 genera were identified, Prevotella 1 and Succiniclasticum were the dominant genera (Appendix Table 2). The relative abundance of Prevotella 1 (belonging to Bacteroidetes) was 25.1% and 16.8% in HS and LS cows respectively; the relative abundance of Succiniclasticum (belonging to Firmicutes) was 12.4% and 25.5% in HS and LS cows respectively. The average relative abundances of Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-014 and Rikenellaceae RC9 gut group were 2.20%, 2.41%, and 3.20% respectively.
Fig. 3.
Relative abundances of rumen bacteria at phylum and genus levels in lactating cows of HS and LS. HS = high total milk solid group; LS = low total milk solid group.
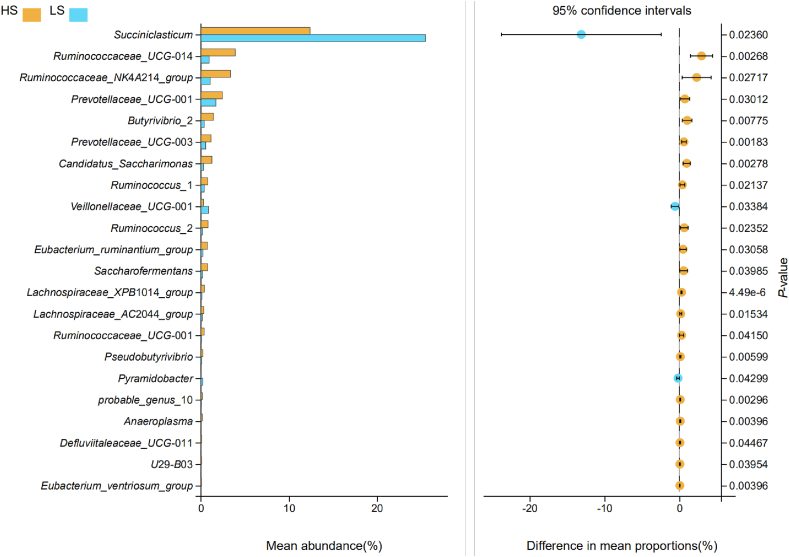
3.4. Comparison of rumen bacteria between HS and LS cows
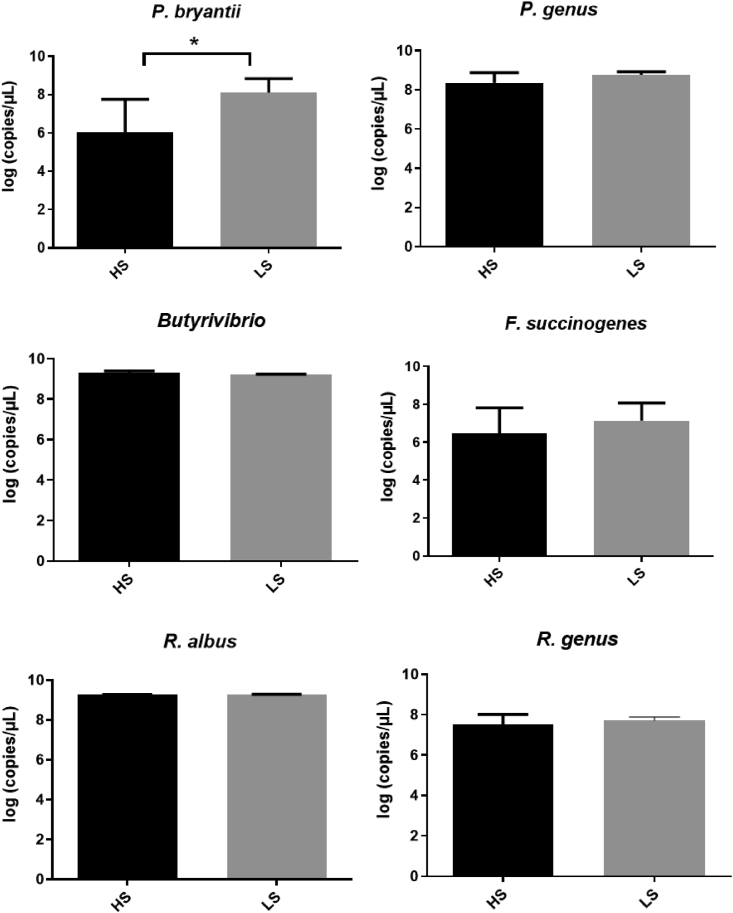
At genus level, there were a total of 22 bacterial genera, the abundance of which differed significantly between HS and LS cows, and all these genera were assigned to Bacteroidetes and Firmicutes phyla. The relative abundance of Succiniclasticum that belonges to Firmicutes was 2.06-folds higher in LS cows than that in HS group. However, Ruminococcaceae UCG-014, Ruminococcaceae NK4A214 group, Ruminococcus 1, Ruminococcus 2, Saccharofermentans and Ruminococcaceae UCG-001 in Ruminococcaceae, Prevotellaceae UCG-001 and Prevotellaceae UCG-003 in Prevotellaceae, Butyrivibrio 2, Eubacterium ruminantium group, Lachnospiraceae XPB1014 group, Lachnospiraceae AC2044 group, Pseudobutyrivibrio, probable genus 10, and Eubacterium ventriosum group in Lachnospiraceae were all significantly greater in HS cows than LS cows (Fig. 4, Appendix Table 2). The real-time qPCR results showed the copy number of Prevotella bryantii was significantly greater in LS than those in HS group. There was no significant difference in the other bacterial genera between HS and LS cows (Fig. 5).
Fig. 4.
Comparison of rumen bacteria at genus level in lactating cows of HS and LS. HS = high total milk solid group; LS = low total milk solid group.
Fig. 5.
The copy numbers of rumen bacteria by real-time qPCR in lactating cows of HS and LS. ∗ indicates the correlation is statistically significant (P < 0.05). HS = high total milk solid group; LS = low total milk solid group.
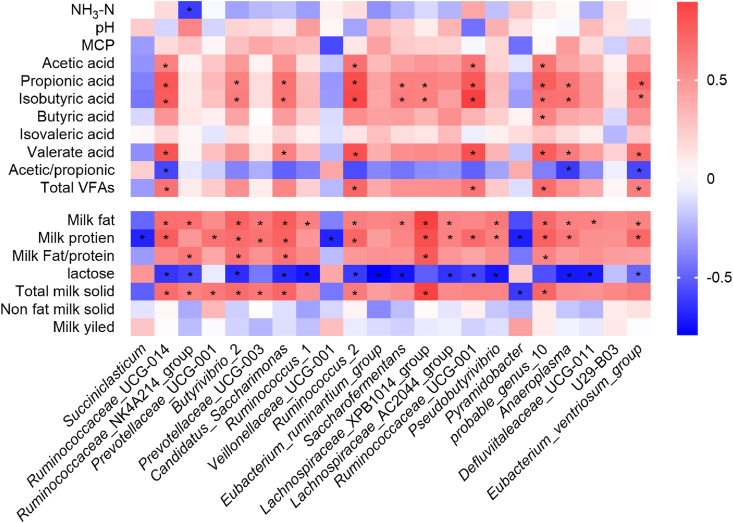
3.5. Correlation of rumen bacterial differentiation with milk composition and fermentation parameters
The Spearman’s rank correlation analysis was used to explore the correlations between milk composition (%), ruminal fermentation parameters and the relative abundances of those bacterial genera that differed significantly between HS and LS cows. The results were shown in Fig. 6. The TMS content was positively correlated with the abundances of Ruminococcaceae UCG-014, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, Butyrivibrio 2, Prevotellaceae UCG-003, Candidatus Saccharimonas, Ruminococcus 2, Lachnospiraceae XPB1014 group, probable genus 10 and Eubacterium ventriosum group, yet Pyramidobacter had negative correlations with TMS content. The relative abundance of these bacterial genera was negatively correlated with milk lactose concentration, but positively correlated with milk protein and fat contents. In contrast, the abundances of Succiniclasticum and Veillonellaceae UCG-001 correlated negatively with milk protein concentration, and Pyramidobacter correlated negatively with milk protein concentration and TMS content.
Fig. 6.
The correlations between the differential bacteria in HS and LS cows and the milk composition and rumen fermentation parameters. Blue color indicates negative correlations, and red color represents positive correlations. Color darkness stands for the value of correlation coefficients, the darker the color, the greater the coefficient. ∗ indicates the correlation is statistically significant (P < 0.05). HS = high total milk solid group; LS = low total milk solid group. NH3-N = ammonia nitrogen; MCP = microbial crude protein; VFA = volatile fatty acid.
As for the rumen fermentation parameters, Ruminococcaceae NK4A214 group abundance correlated negatively with NH3-N concentration. Ruminococcaceae UCG-014, Candidatus Saccharimonas, Anaeroplasma, and Eubacterium ventriosum group negatively correlated with the acetate-to-propionate ratio. Ruminococcaceae UCG-014, Ruminococcus 2, Ruminococcaceae UCG-001, and probable genus 10 correlated positively with acetate, propionate, isobutyrate and total VFA concentrations.
4. Discussion
In this study, a large individual variation in TMS content was noted in the selected 45 Holstein cows, and the content ranged from 10.4% to 14.1% (Appendix Table 1) when the cows were fed the same diet. This result was not reported in the previous research, but the individual variations in feed efficiency, utilization efficiency of nitrogen, and milk protein yield had been reported (Shabat et al., 2016; Wang et al., 2019; Wu et al., 2018). The selected HS cows had greater TMS than LS cows, which attributed to the greater content of milk fat and milk protein in the present study. However, on the basis of TMS, there was more milk fat, but less lactose, and no change in milk protein in TMS in HS cows when compared with those in LS cows. Our results were consistent with a previous study by Nichols et al. (2018). We also noted that HS cows had greater feed intake, 10.8% higher than that in LS cows. When the milk yield was similar between 2 groups, the increase of the feed intake was coincident with the change in TMS content and the TMS yield (increased by 12.5% and 13.0% respectively). Moreover, the CP, NDF, and ADF intakes were all found higher in HS than that in LS group. The results suggest that TMS content seems apparently to be associated with feed and the nutrient intakes in those lactating cows. Mostly current research showed that the DMI of different diets was associated with TMS (Jenkins and McGuire, 2006; Nichols et al., 2018), the results of this present study warrant a further study.
The TMS content was mainly affected by the interaction between feed intake and rumen fermentation (Schären et al., 2018; Pitta et al., 2018). In the present study, we found the individual VFA and total VFA concentrations were relatively greater in HS cows compared with LS cows, although some were not significant statistically. The higher TMS in HS group was likely due to the differences in the VFA concentrations which were attributed to the differentiation of the fermentation rate caused by the change in DMI (including the CP, NDF, and ADF intakes). VFA are the major precursors for TMS, so the greater VFA concentration could explain the greater content of milk fat in HS cows. In addition, there was no significant change in the VFA profile, the molar percentages of individual VFA, which suggested that the rumen fermentation pattern did not change. The similarity of pH and NH3-N indicated the stable acid-base environment of the cow fed the same composite TMR. The similarity of MCP between 2 groups explained the reason why there was no difference in the milk protein to TMS ratio in both groups.
The roles of rumen bacteria in feed and nitrogen utilization efficiency were reported recently (Shabat et al., 2016; Wang et al., 2019). Some studies have found that rumen bacteria were tightly related to the milk components (Zou et al., 2019; Zeng et al., 2019). In this study, the results of 16s rRNA gene sequencing showed that there were differences in the rumen bacterial richness and diversity (Sobs, Simpson and Shannon indices) between HS and LS cows. It had been reported that the rumen bacteria richness was negatively related to milk fat in cows (Xue et al., 2018), and a lower bacteria richness was found in cows with a higher protein yield when compared with cows with a lower protein yield (Xue et al., 2019). As for bacterial diversity, the alpha index was not associated with milk yield (Tong et al., 2018). We assumed the greater the bacterial diversity, the more fermentation products can be produced, such as VFA, AA and glucose, which might be beneficial to milk production.
The present study also showed that the variation in TMS was associated with the changes in some bacterial genera in the rumen of cows. The higher abundance of predominant genus Succiniclasticum in LS cows than HS cows indicated that Succiniclasticum could decrease TMS by affecting milk protein content. Possibly, Succiniclasticum restrains the nitrogen metabolism and absorption of the host. A similar result was reported in our previous research in goats (Wang et al., 2019). Succinivibrio dextrinisolvens was related to the feed protein degradation (Wang et al., 2017). Selenomonas that is at the same order (Selenomonadales) as Succiniclasticum, was a proteolytic bacterium (Zhou et al., 2019). On the contrary, Xue et al. (2019) showed that Succinimonas (Proteobacteria phylum), which has the same function as Succiniclasticum in the rumen, was positively associated with milk protein content.
A previous study showed Succiniclasticum could promote the use of fiber by converting succinate produced by rumen fiber decomposition to propionate which is the major precursor to gluconeogenesis in ruminants (Wang et al., 2019). A greater propionate concentration in the rumen fluid in HS cows was noted, which was not correlated with the lactose concentration in the present study. Possibly, the effect of Succiniclasticum might be compromised by the great complexity of interactions among different bacteria species. The specific role of Succiniclasticum on protein degradation and nitrogen metabolism in the rumen needs to be further explored.
In most cases, the closer genetic distance in animals, the more similar functions of ruminal bacteria display. The present study showed the positive correlations between the Prevotellaceae UCG-003, Prevotellaceae UCG-001 abundances and the TMS content. Xue et al., (2020) showed that Prevotella genus contributed to the metabolism of glutathione, phenylalanine, starch, sucrose, and galactose, and was involved in AA and carbohydrate metabolism by ruminal microbes. Moreover, Prevotella appeared to be a major bacterium that can reduce nitrogen loss through effectively utilizing forages to produce succinate and acetate as the end-product of glucose catabolism (Liu et al., 2019). That might partly explain the reason why the VFA and TMS in HS group compared with LS cows were greater in the present study.
Lachnospiraceae and Ruminococcaceae are deemed to be highly specialized in degradation of complex plant materials to VFA in the rumen (Biddle et al., 2013). Lachnospiraceae plays a predominant role in biohydrogenation in the rumen, especially Butyrivibrio species has been identified as the most important biohydrogenation bacterium (Huws et al., 2011; Dewanckele et al., 2018; Zhang et al., 2019). However, our research showed an interesting result that the abundances of Butyrivibrio 2, Eubacterium ventriosum group Lachnospiraceae XPB1014 group and probable genus 10 in Lachnospiraceae positively correlated with the TMS content, milk protein and milk fat. In addition, these bacteria had positive correlations with the propionate and isobutyrate concentrations, which explained the greater proportion of milk protein, fat and TMS in HS cows. We also found in this study that HS cows harbored the greater abundance of Ruminococcaceae in the rumen, such as Ruminococcaceae UCG-014, Ruminococcaceae NK4A214 group, Ruminococcus 1, Ruminococcus 2, Saccharofermentans, and Ruminococcaceae UCG-001, within which Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-014, and Ruminococcus 2 correlated positively with the TMS content. Acetate is the main substrate produced in the rumen for the milk fat synthesis, and an energy source for the microbial protein synthesis (Xue et al., 2020). The present study showed that probable genus 10, Ruminococcaceae UCG-001, Ruminococcus 2, and Ruminococcaceae UCG-014 boosted milk protein, fat and TMS, which might be due to the positive correlation with the acetate concentration. Thus, we deemed that these bacteria genera from Lachnospiraceae and Ruminococcaceae might be dominant bacteria linked to TMS (Biddle et al., 2013).
5. Conclusion
TMS was greater with the higher DMI, and milk fat was the most changeful component in milk. Propionic acid and valeric acid concentrations were also greater in the high TMS group cows. Moreover, the significant correlations between some rumen bacterial genera and TMS content were found in this study. The positive roles of Ruminococcaceae UCG-014, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, Butyrivibrio 2, Prevotellaceae UCG-003, Candidatus Saccharimonas, Ruminococcus 2, Lachnospiraceae XPB1014 group, probable genus 10, and Eubacterium ventriosum group, and the negative roles of Succiniclasticum in TMS content were noted. And, Ruminococcaceae UCG-014, Ruminococcus 2, Ruminococcaceae UCG001, probable genus 10 and Eubacterium ventriosum group might boost the total VFA production in the rumen of lactating cows. In conclusion, the DMI of dairy cows and some special bacteria in the rumen were significantly varied with TMS content, which suggests the potential function of rumen bacteria contributing to TMS content of dairy cows.
Author contributions
Kaizhen Liu: Investigation, Data curation, Writing - original draft. Yangdong Zhang: Conceptualization, Writing – review. Guoxin Huang: Methodology, Validation. Shengguo Zhao: Conceptualization, Project administration. Nan Zheng: Investigation, Writing - review & editing. Jiaqi Wang: Supervision, Conceptualization, Writing - review & editing.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This study was supported by the Scientific Research Project for Major Achievements of the Agricultural Science and Technology Innovation Program (CAAS ZDXT2019004), the Agricultural Science and Technology Innovation Program (ASTIP-IAS12) and Modern Agro-Industry Technology Research System of the PR China (CARS-36).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2021.12.005.
Contributor Information
Yangdong Zhang, Email: zhangyangdong@caas.cn.
Jiaqi Wang, Email: jiaqiwang@vip.163.com.
Appendix.
The following is the supplementary data to this article:
References
- Bauman D.E., Griinari J.M. Nutritional regulation of milk fat synthesis. Annu Rev Nutr. 2003;23:203–227. doi: 10.1146/annurev.nutr.23.011702.073408. [DOI] [PubMed] [Google Scholar]
- Bekele A.Z., Koike S., Kobayashi Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett. 2010;305:49–57. doi: 10.1111/j.1574-6968.2010.01911.x. [DOI] [PubMed] [Google Scholar]
- Bernabucci U., Basirico L., Morera P., Dipasquale D., Vitali A., Piccioli Cappelli F., et al. Effect of summer season on milk protein fractions in Holstein cows. J Dairy Sci. 2015;98:1815–1827. doi: 10.3168/jds.2014-8788. [DOI] [PubMed] [Google Scholar]
- Biddle A., Stewart L., Blanchard J., Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. [Google Scholar]
- Chen Y., Penner G.B., Li M., Oba M., Guan L. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl Environ Microbiol. 2011;77(16):5770–5781. doi: 10.1128/AEM.00375-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechow C.D., Rogers G.W., Cooper J.B., Phelps M.I., Mosholder A.L. Milk, fat, protein, somatic cell score, and days open among Holstein, Brown Swiss, and their crosses. J Dairy Sci. 2007;90:3542–3549. doi: 10.3168/jds.2006-889. [DOI] [PubMed] [Google Scholar]
- Dewanckele L., Vlaeminck B., Hernandez-Sanabria E., Ruiz-Gonzalez A., Debruyne S., Jeyanathan J., et al. Rumen biohydrogenation and microbial community changes upon early life supplementation of 22:6n-3 enriched microalgae to goats. Front Microbiol. 2018;9:573. doi: 10.3389/fmicb.2018.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Wu Z.Y., Zhang W.X. Effects of garlic addition on bacterial communities and the conversions of nitrate and nitrite in a simulated pickle fermentation system. Food Control. 2020;113:107215. [Google Scholar]
- Hurtaud C., Rulquin H., Vérité R. Effects of level and type of energy source (volatile fatty acids or glucose) on milk yield, composition and coagulating properties in dairy cows. Reprod Nutr Dev. 1998;38:315–330. doi: 10.1051/rnd:19980312. [DOI] [PubMed] [Google Scholar]
- Huws S.A., Kim E.J., Lee M.R., Scott M.B., Tweed J.K., Pinloche E., et al. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ Microbiol. 2011;13:1500–1512. doi: 10.1111/j.1462-2920.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- Jenkins T.C., McGuire M.A. Major advances in nutrition: impact on milk composition. J Dairy Sci. 2006;89:1302–1310. doi: 10.3168/jds.S0022-0302(06)72198-1. [DOI] [PubMed] [Google Scholar]
- Kmicikewycz A.D., Heinrichs A.J. Effect of corn silage particle size and supplemental hay on rumen pH and feed preference by dairy cows fed high-starch diets. J Dairy Sci. 2015;98:373–385. doi: 10.3168/jds.2014-8103. [DOI] [PubMed] [Google Scholar]
- Li D., Wang J.W., Bu D.P., Yang S.L., Wei H.U., Zhou L.Y. Determination of the effects of soybean oil and linseed oil in diets on the quantities of rumen cellulolytic bacteria in beef cattle by real-time PCR. Chin J Anim Nutr. 2008;20:256–260. [Google Scholar]
- Liu C., Wu H., Liu S., Chai S., Meng Q., Zhou Z. Dynamic alterations in Yak rumen bacteria community and metabolome characteristics in response to feed type. Front Microbiol. 2019;10:1116. doi: 10.3389/fmicb.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Xu Q., Wang L., Wang J., Guo W., Zhou M. The impact of diet on the composition and relative abundance of rumen microbes in goat. Asian-Australas J Anim Sci. 2017;30:531–537. doi: 10.5713/ajas.16.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols K., Bannink A., Pacheco S., Van Valenberg H.J., Dijkstra J., Van Laar H. Feed and nitrogen efficiency are affected differently but milk lactose production is stimulated equally when isoenergetic protein and fat is supplemented in lactating dairy cow diets. J Dairy Sci. 2018;101:7857–7870. doi: 10.3168/jds.2017-14276. [DOI] [PubMed] [Google Scholar]
- Nkrumah J.D., Okine E.K., Mathison G.W., Schmid K., Li C., Basarab J.A. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 2006;84:145–153. doi: 10.2527/2006.841145x. [DOI] [PubMed] [Google Scholar]
- Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J., et al. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol. 2013;79:3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D.W., Indugu N., Baker L., Vecchiarelli B., Attwood G. Symposium review: understanding diet-microbe interactions to enhance productivity of dairy cows. J Dairy Sci. 2018;101:7661–7679. doi: 10.3168/jds.2017-13858. [DOI] [PubMed] [Google Scholar]
- Ramos M.H., Kerley M.S. Mitochondrial complex i protein differs among residual feed intake phenotype in beef cattle. J. Anim. Sci. 2013;91:3299–3304. doi: 10.2527/jas.2012-5589. [DOI] [PubMed] [Google Scholar]
- Satoshi K., Yasuo K. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. Fems Microbiol Lett. 2001;204:361–366. doi: 10.1111/j.1574-6968.2001.tb10911.x. [DOI] [PubMed] [Google Scholar]
- Schären M., Frahm J., Kersten S., Meyer U., Hummel J., Breves G., et al. Interrelations between the rumen microbiota and production, behavioral, rumen fermentation, metabolic, and immunological attributes of dairy cows. J Dairy Sci. 2018;101:4615–4637. doi: 10.3168/jds.2017-13736. [DOI] [PubMed] [Google Scholar]
- Shabat S.K., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Berg Miller M.E., et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016;10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.S., Chai Z., Song L.J., Liu J.X., Wu Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J Dairy Sci. 2012;95:5978–5984. doi: 10.3168/jds.2012-5499. [DOI] [PubMed] [Google Scholar]
- Stevenson D.M., Weimer P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- Stiverson J., Morrison M., Yu Z. Populations of select cultured and uncultured bacteria in the rumen of sheep and the effect of diets and ruminal fractions. Int J Microbiol. 2011:750613. doi: 10.1155/2011/750613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubner S. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J Microbiol Meth. 2002;50:155–164. doi: 10.1016/s0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- Tong J., Zhang H., Yang D., Zhang Y., Xiong B., Jiang L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS One. 2018;13 doi: 10.1371/journal.pone.0198225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu K., Wang Z., Bai X., Peng Q., Jin L. Bacterial community diversity associated with different utilization efficiencies of nitrogen in the gastrointestinal tract of goats. Front Microbiol. 2019;10:239. doi: 10.3389/fmicb.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cao P., Wang L., Zhao Z., Chen Y., Yang Y. Bacterial community diversity associated with different levels of dietary nutrition in the rumen of sheep. Appl Microbiol Biotechnol. 2017;101:3717–3728. doi: 10.1007/s00253-017-8144-5. [DOI] [PubMed] [Google Scholar]
- Wu X., Sun H., Xue M., Wang D., Guan L.L., Liu J. Serum metabolome profiling revealed potential biomarkers for milk protein yield in dairy cows. J Proteomics. 2018;184:54–61. doi: 10.1016/j.jprot.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Xue M., Sun H., Wu X., Guan L.L., Liu J. Assessment of rumen microbiota from a large dairy cattle cohort reveals the pan and core bacteriomes contributing to varied phenotypes. Appl Environ Microbiol. 2018;84:1403–1414. doi: 10.1128/AEM.00970-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M.Y., Sun H.Z., Wu X.H., Guan L.L., Liu J.X. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J Dairy Sci. 2019;102:5031–5041. doi: 10.3168/jds.2018-15974. [DOI] [PubMed] [Google Scholar]
- Xue M.Y., Sun H.Z., Wu X.H., Liu J.X., Guan L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome. 2020;8:64–83. doi: 10.1186/s40168-020-00819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Guo C., Sun D., Seddik H.E., Mao S. The ruminal microbiome and metabolome alterations associated with diet-induced milk fat depression in dairy cows. Metabolites. 2019;9:154–172. doi: 10.3390/metabo9070154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Medrano R.F., Wang M., Beauchemin K.A., Ma Z.Y., Wang R., et al. Corn oil supplementation enhances hydrogen use for biohydrogenation, inhibits methanogenesis, and alters fermentation pathways and the microbial community in the rumen of goats. J Anim Sci. 2019;97:4999–5008. doi: 10.1093/jas/skz352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Min L., Zheng N., Wang J. Effect of heat stress on bacterial composition and metabolism in the rumen of lactating dairy cows. Animals. 2019;9(11):925. doi: 10.3390/ani9110925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Bao Y., Zhao G. Effects of dietary crude protein and tannic acid on rumen fermentation, rumen microbiota and nutrient digestion in beef cattle. Arch Anim Nutr. 2019;73:30–43. doi: 10.1080/1745039X.2018.1545502. [DOI] [PubMed] [Google Scholar]
- Zou C., Gu Q., Zhou X., Xia Z., Muhammad W.I., Tang Q., et al. Ruminal microbiota composition associated with ruminal fermentation parameters and milk yield in lactating buffalo in Guangxi, China-A preliminary study. J Anim Physiol Anim Nutr. 2019;103:1374–1379. doi: 10.1111/jpn.13154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.