Abstract
Candida albicans and Cryptococcus neoformans cause both superficial and disseminated infections in humans. Current antifungal therapies for deep-seated infections are limited to amphotericin B, flucytosine, and azoles. A limitation is that commonly used azoles are fungistatic in vitro and in vivo. Our studies address the mechanisms of antifungal activity of the immunosuppressive drug rapamycin (sirolimus) and its analogs with decreased immunosuppressive activity. C. albicans rbp1/rbp1 mutant strains lacking a homolog of the FK506-rapamycin target protein FKBP12 were found to be viable and resistant to rapamycin and its analogs. Rapamycin and analogs promoted FKBP12 binding to the wild-type Tor1 kinase but not to a rapamycin-resistant Tor1 mutant kinase (S1972R). FKBP12 and TOR mutations conferred resistance to rapamycin and its analogs in C. albicans, C. neoformans, and Saccharomyces cerevisiae. Our findings demonstrate the antifungal activity of rapamycin and rapamycin analogs is mediated via conserved complexes with FKBP12 and Tor kinase homologs in divergent yeasts. Taken together with our observations that rapamycin and its analogs are fungicidal and that spontaneous drug resistance occurs at a low rate, these mechanistic findings support continued investigation of rapamycin analogs as novel antifungal agents.
Cryptococcus neoformans and Candida albicans are two common opportunistic fungal pathogens. Current antifungal agents in clinical use include amphotericin B, fluconazole, and flucytosine (29), which have side effects, lack fungicidal activity, or lack activity against emerging resistant mutants (66). Thus, additional antifungal agents are needed.
Several new antifungal agents are in development. The candins are 1,3-β-glucan synthase inhibitors and are potently active against Candida species and Aspergillus fumigatus (23, 40, 51, 52). The pneumocandin caspofungin acetate–MK-0991 is in phase III clinical trials for candida infections and has been approved for refractory aspergillosis. Broad-spectrum triazoles, including voriconazole, posaconazole, and ravuconazole, are being studied in human trials (50). Combination therapy with different antifungal agents may also improve therapy (20, 28, 62).
Rapamycin (sirolimus) is a natural product of the bacterium Streptomyces hygroscopicus originally discovered in a screen for antimicrobial activity against C. albicans and later found to have potent immunosuppressive activity (4, 63). Rapamycin diffuses into the cell and associates with the peptidyl-prolyl isomerase FKBP12. Rapamycin inhibits FKBP12 enzymatic activity; however, this inhibition is not related to immunosuppressive or antifungal activity. The targets of the FKBP12-rapamycin complex are the TOR kinases. Two TOR proteins, Tor1 and Tor2, have been characterized in the yeasts S. cerevisiae and Schizosaccharomyces pombe, and a single TOR homolog has been identified in C. albicans, C. neoformans, Drosophila melanogaster, and humans (10, 18, 33, 39, 49, 56, 64). The Tor kinase has been functionally conserved from yeast to humans (2). Both immunosuppression in mammalian cells and the toxicity of rapamycin in S. cerevisiae and C. neoformans are mediated via FKBP12-dependent inhibition of Tor kinases (12, 18, 33, 34, 41). The FKBP12-rapamycin complex binds to a small region on Tor (the FKBP12-rapamycin binding domain [FRB domain]) adjacent to the carboxy-terminal kinase domain (17). Mutations in the FRB domain prevent FKBP12-rapamycin binding to TOR and confer rapamycin resistance (12, 15, 18, 34, 41, 59, 69). In addition, TOR proteins have a toxic effector domain that, when overexpressed, arrests cell growth (3).
Recent studies in the model yeasts S. cerevisiae and S. pombe reveal that the Tor kinases function in a nutrient-sensing pathway (reviewed in reference 54). Inhibition of Tor signaling by FKBP12-rapamycin induces autophagy (1, 38, 46), represses ribosomal protein gene expression (14, 53, 68), induces nitrogen catabolite-repressed genes (6, 8, 14, 31), and inhibits translation (5, 7). In S. pombe, rapamycin blocks mating in response to nutrient limitation (65). The Tor2 kinase is essential in S. pombe and S. cerevisiae, whereas in S. pombe the Tor1 kinase is required for mating and stationary-phase entry (64). Thus, the TOR pathway likely serves as a global nutrient-sensing pathway that will be conserved in pathogenic fungi.
Rapamycin inhibits the growth of several fungi, including C. neoformans, C. albicans, Candida stelloidea, A. fumigatus, Aspergillus flavus, Aspergillus niger, Fusarium oxysporum, and Penicillium sp. (18, 24, 47, 67). We are interested in FKBP12 and TOR as targets for novel antifungal agents. The C. albicans RBP1 gene encoding a homolog of the target protein FKBP12 was previously identified (25). We isolated C. albicans rbp1/rbp1 and also TOR1-1/TOR1 mutant strains that were viable and rapamycin resistant. These findings, and results using the yeast two-hybrid assay, demonstrate that rapamycin antifungal activity is exerted via FKBP12 and Tor1 homologs in C. albicans. In addition, we analyzed the antifungal activity of rapamycin analogs with reduced immunosuppressive activity against C. albicans, C. neoformans, and S. cerevisiae (22). Two analogs retain antifungal activity in vitro and promote FKBP12-Tor complexes. Further examination of rapamycin analogs as potential antifungal agents is warranted.
MATERIALS AND METHODS
Media, drugs, and strains
Media were prepared as previously described (55). 5-Fluoroorotic acid (5-FOA) medium was prepared as described previously (9) by using 625 μg of 5-FOA/ml and replacing uracil with 100 μg of uridine/ml. Rapamycin and its analogs (Abbott Labs) were added to media or disks from stock solutions in 90% ethanol–10% Tween 20. Strains used are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| C. albicans | ||
| SC5314 | WT | 27 |
| BWP17 | ura3Δ::imm34/ura3Δ::imm34 his1::hisG/ his1::hisG arg4::hisG/arg4::hisG | 66a |
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 27 |
| YJM126 | ade2/ade2 ura3Δ::ADE2/ura3Δ::ADE2 | This study |
| YAG116 | ura3Δ::imm434/ura3Δ::imm434 rbp1::CaURA3MX3F/RBP1 | This study |
| YAG134 | ura3Δ::imm434/ura3Δ::imm434 rbp1::MX3/RBP1 | This study |
| YAG171 | ura3Δ::imm434/ura3Δ::imm434 rbp1::MX3/rbp1::CaURA3MX3R | This study |
| JRB12 | SC5314 TOR1-1/TOR1 | This study |
| JRB21 | BWP17 TOR1-2/TOR1 | This study |
| C. neoformans | ||
| H99 | Serotype A MATα | 61 |
| M049 | H99 ade2 | 61 |
| MCC1 | M049 frr1::ADE2 ade2 | 18 |
| JEC21 | Serotype D MATα | 43 |
| CN111 | JEC21 TOR1-1 | 18 |
| S. cerevisiae | ||
| JK9-3dα | MATα trp1 his4 leu2-3,112 ura3-52 rme1 GAL+HMLa | 33 |
| JHY3-3B | JK9-3dα fpr1::URA3 | 33 |
| R17 | JK9-3dα TOR2-1 HIS+ | 33 |
| Y190 | MATa trp1-901 his3 leu2-3,112 ura3-52 ade2 gal4 gal80 URA3::GAL-lacZ LYS2::GAL-HIS3 | 32 |
| SMY4-1 | Y190 TOR1-3 fpr1::ADE2 | 16 |
WT, wild type.
RBP1 insertion-deletion constructs.
The RBP1 disruption plasmids pAG70 and pAG71 contain the CaURA3MX3 (30) disruption cassette replacing nucleotides (nt) 40 to 189 of the RBP1 open reading frame (ORF). These plasmids are identical except that the CaURA3MX3 cassettes are in an opposite orientation. The RBP1 disruption plasmids were constructed in three steps. First, the genomic RBP1 locus and ∼900 bp of flanking sequence was PCR amplified from the uracil auxotrophic strain YJM126 and subcloned into pGEM-T Easy (Promega), resulting in plasmid pAG54. Next, the NotI insert was subcloned into the NotI site of the yeast-E. coli shuttle vector pRS314 (CEN ARS TRP1 [58]), resulting in plasmid pAG67. Finally, the CaURA3MX3 disruption cassette was subcloned into plasmid pAG67 by in vivo PCR-directed recombination.
Primers.
The primers used were as follows: JM37, CCTCGACATCATCTGCCC; PR80, CAAGGAGTCAACCACCACTAAG; PR83, TCTGAGTCTGGGTGTGGGTC; PR92, CAGCACTGGAAGGTATGAGTG; PR103, ATGTCTG AAGAACTTCCACAAATTGAAATTGTTCAAGAAGCAGCTGAAGCTTC GTACGC; PR104, GCACCACCTTTACCATAATTGTTAGTTAAAGAAATATGCATAGGCCACTAGTGGATCTG; PR130, ATGTCTGAAGAACTT CCACAAATTGAAATTGTTCAAGGCATAGGCCACTAGTGGATCTG; PR131, TTAGCACCACCTTTACCATAATTGTTAGTTAAAGAAATATCAGCTGAAGCTTCGTACGC; JOHE2920, GTACGAGAATTCATGTCTGAAGAACTTCCACAA; JOHE2921, GCAACGGGATCCTTATTGACCATTAACACCAAG; JOHE6244, TTTATGGCACGAACAATGGCACGATGCTTTGG AAGATGCTAGCAGGTTTTTCTTTGGTGAACACAACACAGAAAAGAT GTTT; JOHE6245, TTTATGGCACGAACAATGGCACGATGCTTTGGAA GATGCTCGCAGGTTTTTCTTTGGTGAACACAACACAGAAAAGATGT TT; JOHE6246, TTTATGGCACGAACAATGGCACGATGCTTTGGAAGA TGCTATCAGGTTTTTCTTTGGTGAACACAACACAGAAAAGATGTTT; JOHE6247, GGCAAGGTGTTTCTTGAAGC; and JOHE6248, TACTTCTTGATTCGCGATAGC.
PCR conditions.
The RBP1 ORF and ∼900 nt of flanking genomic DNA was PCR amplified by using primers PR80 and PR83. The 50-μl reaction consisted of 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, 100 μg of genomic DNA, 0.5 μM PR80, 0.5 μM PR83, 0.2 mM concentrations of each deoxynucleoside triphosphate (dNTP), and 0.5 U of Vent polymerase (New England Biolabs). DNA amplification was initiated with a 4-min denaturation at 94°C followed by 30 amplification cycles (94°C for 1 min, 55°C for 15 s, and 72°C for 3 min) and was terminated with a 7-min 72°C extension.
The CaURA3MX3 cassettes were PCR amplified for in vivo PCR-directed recombination by using the primer pairs PR103-PR104 (for the forward orientation of the disruption cassette) and PR130-PR131 (for the reverse orientation). These primers amplify the CaURA3MX3 cassette (from plasmid pAG61 [30]) by adding 5′-terminal extensions homologous to nt 1 to 39 (PR103 and PR131) and 190 to 226 (PR102 and PR130) of the RBP1 ORF. Each 100-μl reaction consisted of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.1 mM MgCl2, 0.1% gelatin, 100 ng of pAG61, 0.5 μM concentrations of each primer, 0.2 μM concentrations of each dNTP, and 2.5 U of REDTAQ DNA Polymerase (Sigma). DNA amplification was initiated with a 3-min 94°C denaturation followed by 30 cycles (94°C for 30 s, 55°C for 10 s, and 72°C for 3 min) and was terminated with a 7-min 72°C extension.
RBP1 disruption.
The gene disruption cassettes from plasmids pAG70 and pAG71 were PCR amplified as described for the RBP1 locus except that the denaturation time was reduced to 1 min. To disrupt the first RBP1 ORF, the ∼3.5-kb PCR product was transformed into C. albicans CAI4 (27) by electroporation (60). Prototrophic transformants were selected on minimal medium (synthetic dextrose [SD]). Homologous disruptants were verified by colony PCR (45) using primers PR92 and JM37 and Southern blotting. A resulting rbp1::CaURA3MX3F/RBP1 strain, YAG116, was chosen.
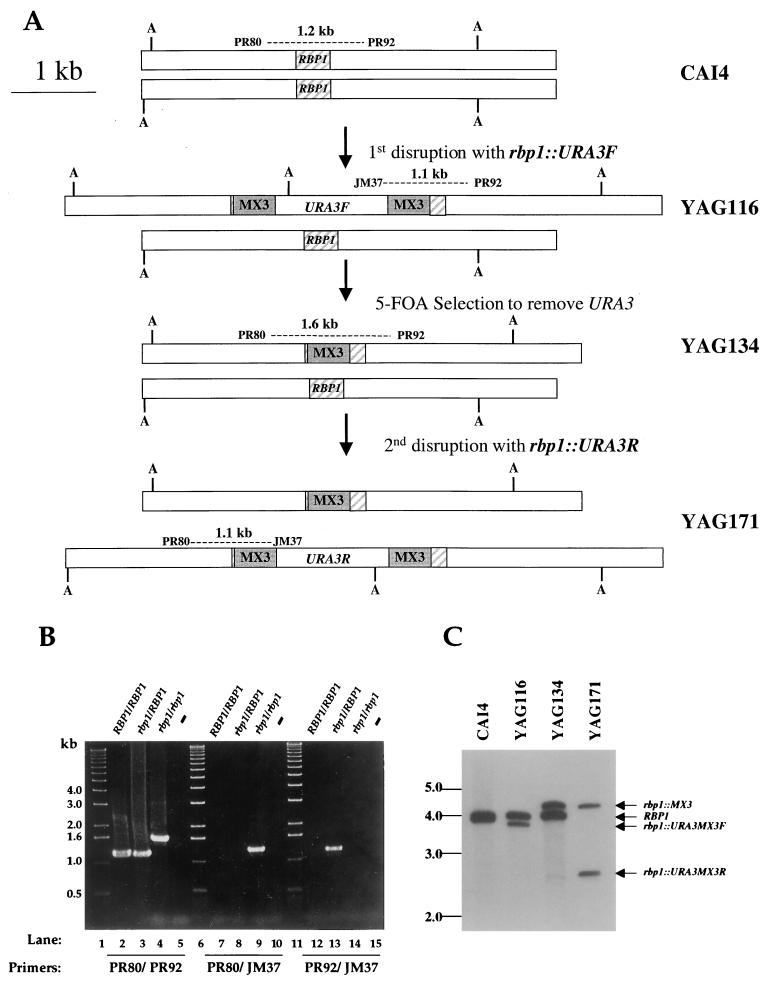
To select for loss of the CaURA3F ORF in strain YAG116, these cells were grown overnight in rich medium plus uridine, ∼105 cells were plated on 5-FOA medium, and 5-FOA-resistant cells were assayed by Southern blotting and colony PCR with primers PR80 and PR92 to verify the loss of the CaURA3 ORF. A resulting rbp1::MX3/RBP1 strain, YAG134, was used for a second round of gene disruption using the RBP1 disruption construct from plasmid pAG71. As above, homologous disruptants were verified by colony PCR with primers PR80 and JM37 and Southern blotting. Strain YAG171 was shown to have the genotype rbp1::MX3/rbp1::CaURA3MX3R (Fig. 1B and C)
FIG. 1.
Disruption of the C. albicans RBP1 genes encoding FKBP12. (A) Disruption alleles of the RBP1 gene. The RBP1 gene is depicted on a 5-kb genomic fragment including the 375-bp RBP1 ORF. One RBP1 wild-type allele was replaced with the CaURA3MX3 forward cassette, followed by treatment with 5-FOA to counterselect URA3. A second transformation replaced the second RBP1 allele with the CaURA3MX3 reverse cassette. Dashed lines represent PCR products used to verify homologous replacement of RBP1. Gray boxes indicate MX3 direct repeats. (B) PCR verification of homologous integration of insertion. Primers PR80 and PR92 amplified an ∼1.2-kb product containing the RBP1 ORF. Primers PR80 and JM37 amplified the rbp1::CaURA3MX3R allele. Primers PR92 and JM37 amplified the rbp1::CaURA3MX3F allele. Lanes: 1, 6, and 11, 1-kb markers; 2, 7, and 12, RBP1/RBP1 strain CAI4; 3, 8, and 13, rbp1/RBP1 strain YAG116; 4, 9, and 14, rbp1/rbp1 strain YAG171; 5, 10, and 15, no-DNA controls. (C) Southern analysis of RBP1 and rbp1 alleles. Genomic DNA from the wild-type RBP1/RBP1 strain CAI4, heterozygous rbp1/RBP1 strains YAG116 and YAG134, and homozygous rbp1/rbp1 strain YAG171 was cleaved with AflIII (A), electrophoresed in a 0.8% gel, transferred to a nylon membrane, and hybridized to a random-primed 32P-labeled 375-bp gel-purified fragment spanning the RBP1 gene. The probe hybridizes to the RBP1 wild-type alleles in strain CAI4 (4.0 and 3.9 kb), to rbp1::URA3MX3F and RBP1 alleles in strain YAG116 (3.6 and 4.0 kb), to rbp1::MX3 and RBP1 alleles in strain YAG134 (4.2 and 4.0 kb), and to rbp1::MX3 and rbp1::URA3MX3R alleles in strain YAG171 (4.2 and 2.7 kb). The positions of DNA markers are shown on the left in kilobases.
Mutagenesis of C. albicans TOR1.
C. albicans TOR1-1 and also TOR1-2 mutant strains (JRB12 and JRB21) were created by transformation of strain SC5314 or BWP17 with 81-mer oligonucleotides designed to carry the wild-type TOR1 sequence or a single base pair change (A→C or G→T) in the Tor1 FRB domain which result in single amino acid substitutions (S→R or S→I) and also disrupt an NheI restriction site. The wild-type partial sequence of C. albicans Tor1 was found in the Genome Project at Stanford University. Transformants were selected on yeast extract-peptone-dextrose (YPD) plates containing 0.1 μg of rapamycin/ml. Multiple resistant isolates were obtained with the mutagenic oligonucleotides, whereas none were obtained with the wild-type oligonucleotide. These mutants were not cross-resistant to FK506 plus fluconazole and are therefore not rbp1/rbp1 mutants. Homologous recombination was verified by colony PCR with primers JOHE 6247 and JOHE 6248, followed by digestion of the 1.5-kb PCR product with the restriction enzyme NheI. The presence of a 1.5-kb uncleaved fragment indicated the presence of the mutant TOR1-1 allele. Genomic DNA flanking the TOR1 and TOR1-1 alleles was isolated by PCR from genomic DNA from strain JRB12 and cloned, and restriction digestion with NheI and sequence analysis of multiple clones revealed five wild-type and six mutant alleles of the eleven analyzed. A similar analysis of the wild-type TOR1 strain SC5314 yielded six wild-type and no mutant TOR1 sequences.
Two-hybrid assays.
The yeast S. cerevisiae two-hybrid strain used was SMY4 (Y190 TOR1-3 fpr1::ADE2) (16). The two-hybrid fusion plasmids used were pGAD424 and pGBT9 (26). The C. albicans RBP1 gene was fused to the GAL4 activation domain [GAL4(AD)] in plasmid pGAD424. First, the RBP1 gene was amplified with primers JOHE2920 and JOHE2921 by using plasmid pAG54 as a template. The PCR product was cleaved with EcoRI and BamHI and cloned in the corresponding sites in plasmid pGAD424, yielding plasmid pMCC4. Plasmids pML80 (TOR1) and pML82 (TOR1-1) expressing the S. cerevisiae TOR1 gene or the TOR1-1 mutant allele, respectively, fused to the GAL4 DNA-binding domain [GAL4(BD)] in plasmid pGBT9 were as described previously (41). Plasmids expressing the GAL4(BD)-C. neoformans TOR1 FRB domain, the GAL4(BD)-C. neoformans TOR1-1 mutant FRB domain, and the GAL4(AD)-C. neoformans FKBP12 were as described previously (18). The two-hybrid strain SMY4 was cotransformed with the two-hybrid fusion plasmids, cells were grown on SD-Leu-Trp synthetic medium, and the β-galactosidase activity was assayed by using the chromogenic substrate chlorophenol-β-d-galactopyranoside (CPRG) (16). For two-hybrid growth assays, cells were resuspended in top agar (0.7% Bacto agar in water) on the surface of synthetic SD-Leu-Trp-His medium with 10 mM 3-aminotriazole. Disks without or with 1 μg of rapamycin or an analog were placed on the surface, and cells were incubated for 2 to 3 days at 30°C. Colony formation indicated GAL4-dependent expression of the GAL-HIS3 reporter gene fusion by FKBP12-ligand-TOR interactions.
Growth inhibition assays.
C. albicans, C. neoformans, or S. cerevisiae cells were cultured in YPD liquid medium overnight, and ∼106 cells were resuspended in 3 ml of top agar (0.7% Bacto agar in water), which was poured onto the surface of YPD medium (90-mm plates) and allowed to solidify. Disks without or with 1 μg of rapamycin or an analog were placed on the surface. Cultures were incubated for 24 to 36 h at 30°C. Experiments to determine MICs and minimal fungicidal concentrations (MFCs) were performed by the broth microdilution method according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) (44). The optical density at 600 nm (OD600) was measured with a Beckman spectrophotometer after incubation for 48 h (C. albicans) or 72 h (C. neoformans) at 30°C. The MIC was defined as the lowest drug concentration in which a visual turbidity of ≤80% inhibition was observed compared to that produced by the growth control. The MFC was determined as previously described (21). Briefly, 100-μl aliquots from wells with growth inhibition were plated onto Sabouraud agar plates. The lowest concentration that yielded three or fewer colonies was recorded as the MFC.
RESULTS
Disruption of the C. albicans RBP1 genes by homologous recombination.
S. cerevisiae cells treated with rapamycin irreversibly arrest growth, and the effect of rapamycin is mediated by the prolyl-isomerase FKBP12 (33). The FKBP12-rapamycin complex binds to and inhibits the TOR kinases, and strains that lack FKBP12 are viable and rapamycin resistant (33). The human pathogenic yeast C. albicans is markedly sensitive to rapamycin (63). We found that rapamycin has fungicidal activity (Table 2), and in a standard Luria-Delbrück fluctuation test the spontaneous rate of rapamycin resistance was found to be low (∼1 in 108 cells). The presumed target of rapamycin in this fungus is the FKBP12 homolog encoded by the RBP1 gene (25). To test this hypothesis, a homozygous rbp1/rbp1 mutant strain was constructed.
TABLE 2.
FKBP12 and TOR mutations confer resistance to rapamycin and analogsa
| Straina | MICs and MFCs (μg/ml)b of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rapamycin
|
Analog
|
|||||||||
| 2
|
18
|
19
|
23
|
|||||||
| MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | |
| C. neoformans WT | 0.19 | 0.39 | 3.12 | 3.12 | >100 | >100 | >100 | >100 | 1.56 | 3.12 |
| C. neoformans frr1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| C. neoformans TOR1-1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| C. albicans WT | <0.09 | 0.39 | 6.25 | 12.5 | >100 | >100 | >100 | >100 | 0.78 | 0.78 |
| C. albicans rbp1/rbp1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| C. albicans TOR1-1/TOR1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
WT, wild type.
Drug dilutions were from 100 to 0.09 μg/ml.
C. albicans is diploid, and both RBP1 alleles were mutated by replacing 150 nt within the RBP1 ORF (i.e., nt 40 to 189) with the URA3-containing disruption cassette CaURA3MX3 (30). To ensure that gene disruption was the result of two independent events, two different RBP1 disruption plasmids were constructed in which nt 40 through 189 of the RBP1 ORF were replaced with the CaURA3MX3 cassette in opposite orientations (Fig. 1A). Ca. 900 bp of genomic sequence flanking the RBP1 ORF were present to facilitate homologous recombination. The rbp1Δ::CaURA3MX3 allele in the forward orientation (Fig. 1A) was PCR amplified and electroporated into the ura3/ura3 strain CAI4 to generate the rbp1::CaURA3MXF/RBP1 heterozygous mutant strain, YAG116. To reuse the CaURA3MX3 cassette for the second round of gene deletion, the rbp1/RBP1 YAG116 cells were treated with 5-FOA to select for loss of the URA3 ORF by recombination of the flanking direct repeats (designated as MX3). A 5-FOA resistant, uracil auxotrophic rbp1::MX3/RBP1 strain, YAG134, was transformed with the rbp1::CaURA3MX3 allele in the reverse orientation (Fig. 1A), resulting in the homozygous rbp1/rbp1 mutant YAG171. The genotypes of wild-type (CAI4), rbp1/RBP1 heterozygous (YAG116 and YAG134), and rbp1/rbp1 homozygous (YAG171) strains were verified by both colony PCR (Fig. 1B) and Southern blotting (Fig. 1C).
C. albicans FKBP12 homolog Rbp1 is required for rapamycin antifungal action.
The FKBP12 homolog Rbp1 was found to mediate rapamycin action in C. albicans. The wild-type RBP1/RBP1 parental strain and the rbp1/RBP1 heterozygous mutant were sensitive to inhibition by rapamycin, whereas the rbp1/rbp1 homozygous mutant was rapamycin resistant (Fig. 2A). The rbp1/RBP1 heterozygous strain readily gave rise to rapamycin-resistant colonies. Consistent with the interpretation that these isolates arise from spontaneous homozygosis that produces rbp1/rbp1 mutants by mitotic recombination, these isolates were prototrophic, cross-resistant to another drug that targets Rbp1 (FK506 plus fluconazole), and the majority lacked the wild-type RBP1 gene by PCR analysis. In summary, the FKBP12 homolog Rbp1 mediates rapamycin antifungal activity against C. albicans.
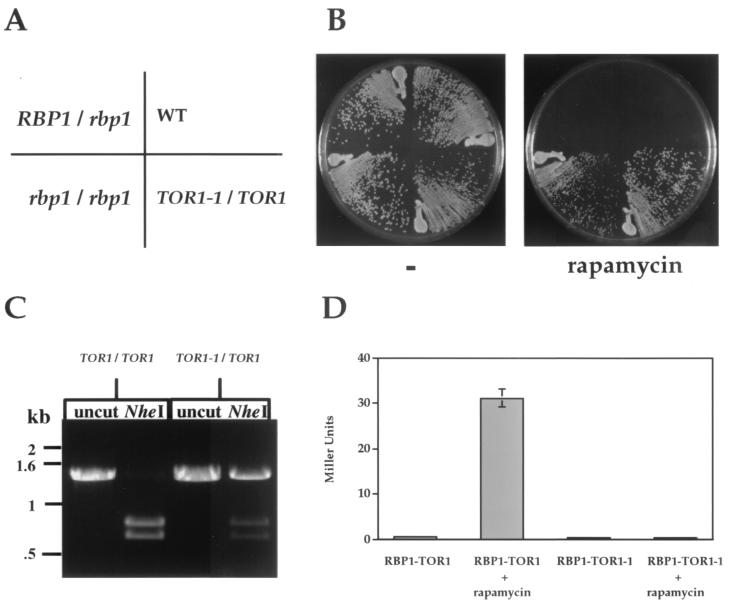
FIG. 2.
C. albicans rbp1/rbp1 and TOR1-1/TOR1 mutants are rapamycin resistant. (A and B) The wild-type (SC5314), rbp1/RBP1 heterozygous (YAG116), rbp1/rbp1 homozygous (YAG171), and TOR1-1/TOR1 heterozygous (JRB12) mutants were grown on YPD medium without or with 1 μg of rapamycin/ml for 24 h at 30°C. (C) Genomic DNA from the wild type (SC5314) and the TOR1-1/TOR1 mutant strain (JRB12) was PCR amplified, cleaved with NheI, and electrophoresed, demonstrating that strain JRB12 is heterozygous for the TOR1-1 and TOR1 alleles. (D) C. albicans FKBP12 homolog binds rapamycin and forms a complex with Tor1. The S. cerevisiae GAL4(BD)-TOR1 wild-type and S1972R mutant fusions were coexpressed with the C. albicans GAL4(AD)-FKBP12 fusion protein (Rbp1) in the two-hybrid host strain SMY4 (fpr1 TOR1-3), with or without rapamycin (1 μg/ml). β-Galactosidase activity was measured by CPRG assay in duplicate.
The rbp1/rbp1 mutant strain grew more slowly than the isogenic wild-type strain on rich medium. However, this growth defect was corrected when the medium was supplemented with uridine (400 μM) and cytosine (3 μg/ml), suggesting that this is the result of incomplete Ura3 expression and not the rbp1 mutation. rbp1/rbp1 mutant strains had no defect in filamentous or invasive growth or growth at different temperatures or on minimal medium (not shown).
C. albicans TOR1-1 mutation confers rapamycin resistance.
To test if the Tor1 kinase is the target of the FKBP12-rapamycin complex, two different point mutations were introduced into the FRB domain of C. albicans Tor1 to result in an amino acid substitution (serine to isoleucine or arginine) at a serine residue implicated in rapamycin-FKBP12 binding to fungal and mammalian Tor proteins (11, 12, 15, 17, 18, 34, 41, 59, 69). These amino acid substitutions were introduced into the C. albicans TOR1 gene by transformation with a single-stranded mutagenic 80-mer oligonucleotide and selection on rapamycin-containing medium. PCR amplification with primers flanking the site of mutation and digestion with NheI (a site present in wild-type TOR1 is destroyed by the introduced mutation) revealed that both wild-type TOR1 and the TOR1-1 or TOR1-2 mutant allele were present (Fig. 2C and data not shown). The PCR products for strain JRB12 (TOR1-1/TOR1) were cloned; of 11 clones, 5 were digested by NheI and contained the wild-type TOR1 sequence, whereas 6 were NheI resistant and contained the mutant sequence. The resulting heterozygous TOR1-1/TOR1 (JRB12) and TOR1-2/TOR1 (JRB21) cells were viable and rapamycin resistant, indicating that the Tor1 kinase is the target of the Rbp1-rapamycin complex and that the TOR1-1 and TOR1-2 mutations confer dominant drug resistance (Fig. 2A).
C. albicans FKBP12 homolog binds to wild-type Tor1 but not to a Tor1-1 mutant.
The yeast two-hybrid system was used to test whether the C. albicans FKBP12 homolog Rbp1 interacts with the Tor1 protein. Protein-drug-protein interactions were monitored by measuring β-galactosidase expression from a GAL-lacZ reporter gene (Fig. 2D). Rapamycin-dependent interactions were detected between wild-type S. cerevisiae Tor1 and the C. albicans FKBP12 homolog Rbp1. In contrast, the S. cerevisiae rapamycin-resistant Tor1-1 S1972R mutant protein failed to interact with FKBP12-rapamycin (Fig. 2D). The S1972R mutation does not affect the stability of the Tor1-1 mutant as previously reported (41). Thus, the S1972R mutation prevents the formation of the FKBP12-rapamycin-TOR complex. These findings indicate that the C. albicans FKBP12 homolog Rbp1 forms a complex with rapamycin that interacts with the TOR kinase at the FRB domain in vivo.
Nonimmunosuppressive rapamycin analogs are toxic to C. albicans, C. neoformans, and S. cerevisiae via FKBP12-dependent inhibition of TOR.
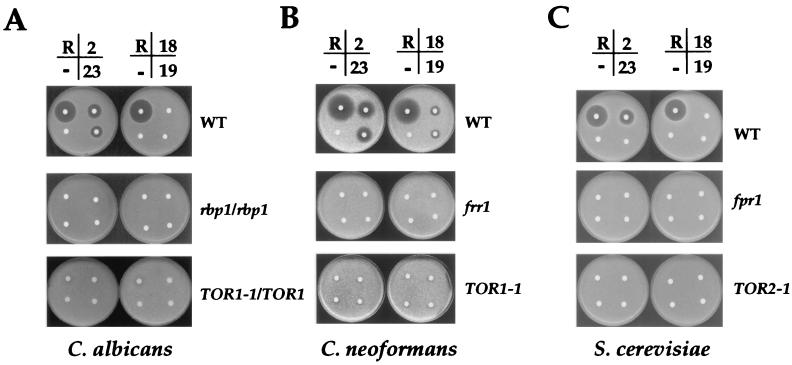
Previously, several rapamycin derivatives were prepared, and their immunosuppressive and antifungal activities were determined (22). Analog 2 is 1,2,3,4- tetrahydro-rapamycin. Analogs 18 [(S)-2-methyl-thienyl], 19 [(S)-NOHCOOibu], and 23 [(S)-NOHCON-piperidyl] are 7-substituted rapamycin analogs. We assessed the fungal activities of these rapamycin analogs with diminished immunosuppressive activities by determining MICs and MFCs using the NCCLS criteria, as well as a drug diffusion assay. Analog 2 was fungicidal to wild-type C. albicans, C. neoformans, and S. cerevisiae but not to the isogenic FKBP12 or TOR mutants (Fig. 3, Table 2). Analog 23 was also toxic and fungicidal to C. albicans and C. neoformans via FKBP12-dependent inhibition of Tor but showed no activity against S. cerevisiae (Fig. 3). Analogs 18 and 19 were weakly toxic to C. neoformans in the drug diffusion assay (Fig. 3B); however, the MIC was >100 μg/ml (Table 2). Analogs 18 and 19 had no activity against either C. albicans or S. cerevisiae (Fig. 3A and C; Table 2). In summary, analogs 2 and 23 are the most toxic to C. neoformans and C. albicans, and they share a common mechanism of antifungal action with rapamycin involving FKBP12-dependent inhibition of Tor kinases.
FIG. 3.
The antifungal activity of rapamycin and its analogs is mediated via FKBP12 and TOR proteins. C. albicans (A), C. neoformans (B), and S. cerevisiae (C) wild-type and FKBP12 (rbp1/rbp1, frr1, or fpr1) and TOR (TOR1-1/TOR1, TOR1-1, and TOR2-1) mutant strains were grown in YPD liquid medium overnight, and ∼106 cells were resuspended in the top agar on the surface of YPD medium. Disks without (−) or with 1 μg of rapamycin (R) or analogs 2, 18, 19, and 23 were placed on the surface of the medium and incubated for 24 to 36 h at 30°C.
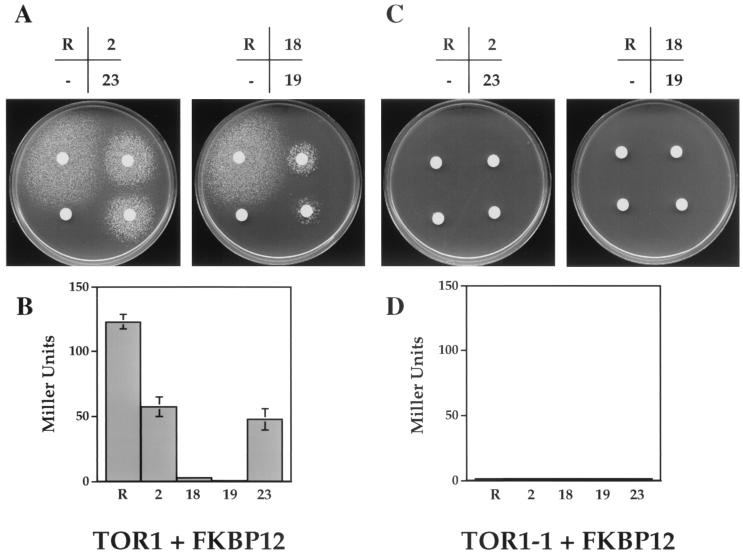
The yeast two-hybrid system was used to test whether rapamycin analogs mediate cryptococcal FKBP12-TOR interactions similar to the way rapamycin does. A two-hybrid host strain whose growth is rendered resistant to rapamycin by a TOR1-3 mutation and which lacks endogenous yeast FKBP12 was employed for these studies (SMY4-1). This specialized two-hybrid reporter strain was transformed with plasmids expressing the C. neoformans FKBP12 fused to GAL4(BD) [GAL4(BD)-FKBP12] and the C. neoformans Tor1 wild-type protein or the Tor1-1 mutant protein fused to the protein GAL4(AD) [GAL4(AD)-TOR1]. A large halo of colonies was observed surrounding the disks containing rapamycin (Fig. 4A), which is indicative of GAL4-dependent expression of the GAL-HIS3 reporter gene fusion as a consequence of FKBP12-rapamycin-Tor1 interactions. Rapamycin analogs 2 and 23, and to a lesser extent analogs 18 and 19, also promoted this interaction (Fig. 4A). These observations were quantitated by monitoring expression of the GAL-lacZ reporter gene (Fig. 4B). A similar concentration of analogs 2 and 23 promoted ∼50% the level of reporter gene expression compared to rapamycin itself, whereas analogs 18 and 19 had significantly less activity. No growth on medium lacking histidine or the GAL-lacZ reporter gene was observed when a mutation that prevents FKBP12-rapamycin binding to TOR was introduced into the GAL4-TOR1 fusion protein (Fig. 4C and D). In summary, rapamycin analogs 2 and 23 promote FKBP12 binding to the wild-type Tor1 kinase but not to a Tor1-1 mutant.
FIG. 4.
Rapamycin and analogs promote FKBP12-TOR interactions. The wild-type GAL4(BD)-TOR1 FRB (A and B) and the mutant GAL4(BD)–TOR1-1 FRB (Ser1862Leu) domains (C and D) were coexpressed with the GAL4(AD)-FKBP12 fusion protein in the two-hybrid host SMY4 (fpr1::hisG TOR1-3). (A and C) Cells were resuspended in top agar on the surface of medium lacking histidine and disks bearing rapamycin, the analogs 2, 28, 19, or 23, or no drug were placed on the surface. Growth indicates GAL4-dependent expression of the GAL-HIS3 reporter gene and results from the formation of FKBP12-drug-Tor complexes. (B and D) β-Galactosidase activity was measured by the CPRG assay and was determined in duplicate.
DISCUSSION
The identification of novel drug targets is of great importance in the field of medical mycology. We have previously demonstrated that rapamycin is toxic to C. neoformans via FKBP12-rapamycin inhibition of the Tor1 kinase (18). Here we assessed the antifungal activities and mechanisms of action of rapamycin and its analogs with diminished immunosuppressive activities against C. neoformans and C. albicans.
We established that rapamycin and two less immunosuppressive analogs are toxic to C. neoformans and C. albicans via FKBP12-dependent inhibition of Tor1. First, we constructed a C. albicans rbp1/rbp1 mutant by a sequential two-step method by using two different rbp1::URA3 disruption alleles in which the marker gene was in opposite orientations. Second, point mutations were introduced into the FRB domain of the C. albicans Tor1 homolog by transformation with mutagenic 80-mer oligonucleotides. Both classes of mutants are resistant to rapamycin and its analogs, indicating that antifungal activity is mediated via binding to the C. albicans FKBP12 homolog Rbp1 and Tor1. Third, rapamycin promoted interactions between the C. albicans FKBP12 homolog and wild-type S. cerevisiae Tor1 in the two-hybrid assay. Rapamycin analogs 2 and 23 also promoted strong interactions between the cryptococcal FKBP12 and Tor1 homologs. Our studies extend the understanding of the conserved antifungal mechanism of action of rapamycin and its analogs from the basidiomycete C. neoformans to the ascomycetous human pathogen C. albicans.
Although analogs 2 and 23, with the highest toxicity to C. neoformans and C. albicans, can only achieve <50% of the antifungal activity of rapamycin itself, they were previously found to be ∼1,000-fold less immunosuppressive than rapamycin in a human T-cell proliferation assay (22). Additional work is warranted to develop or to test existing rapamycin analogs (42) with a greater selective toxicity for lower eukaryotes. There is a clear advantage for the use of analogs of known immunosuppressive drugs, such as rapamycin, since there is a wealth of structural, molecular, and pharmacological information on the activity of these compounds (13, 37, 57). Rapamycin was approved by the Food and Drug Administration as an immunosuppressant for renal transplant recipients in August 1999. New studies are investigating the efficacy of using rapamycin alone or in combination with FK506 and cyclosporine A in other transplant conditions. Interestingly, we have found previously that cyclosporine A and FK506, and also analogs of these compounds with reduced or no immunosuppressive activity, are toxic to C. neoformans (19, 48). Additional advantages of rapamycin and its analogs are their oral activity, fungicidal activity, and the fact that spontaneous drug resistance is infrequent in vitro.
The use of rapamycin as an antifungal agent in an in vivo setting was reported by earlier studies wherein rapamycin was found to protect 50% of mice from an otherwise-lethal infection with C. albicans (4) and to improve the survival of mice with invasive aspergillosis (35, 36). These studies and our findings suggest that patients receiving rapamycin may benefit from both the immunosuppressive and the antifungal activities of rapamycin. It remains to be tested if rapamycin or its analogs with decreased immunosuppressive activity confer a beneficial effect in animal models of candidiasis and cryptococcal meningitis or in transplant recipients currently receiving rapamycin.
ACKNOWLEDGMENTS
We thank Christina Hull for comments. Strain SGY243 (YJM126) was obtained from Squibb and strain BWP17 was obtained from Aaron Mitchell.
These studies were supported by R01 grant AI41937 (to J.H., M.E.C., and J.R.P.) and a supplement (to M.C.C.) from the NIAID, by P01 grant AI44975 from the NIAID to the Duke University Mycology Unit, and in part by MUCU grant 21363 (to M.D.). Alan Goldstein is supported by the DUMC interdisciplinary training program in AIDS-NIAID T32-AI07392-10. Maria E. Cardenas is supported by K01 award CA77075 from the NCI. Joseph Heitman is a Burroughs-Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abeliovich H, Dunn J W A, Kim J, Klionsky D J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcon C M, Cardenas M E, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive. TOR function in yeast. Genes Dev. 1996;10:279–288. doi: 10.1101/gad.10.3.279. [DOI] [PubMed] [Google Scholar]
- 3.Alarcon C M, Heitman J, Cardenas M E. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell. 1999;10:2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker H, Sidorowicz A, Sehgal S N, Venzina C. Rapamycin (AY-22,989), a new antifungal antibiotic. III. In vitro and in vivo evaluation. J Antibiot. 1978;31:539–545. doi: 10.7164/antibiotics.31.539. [DOI] [PubMed] [Google Scholar]
- 5.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck T, Hall M N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 7.Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram P G, Choi J H, Carvalho J, Ai W, Zeng C, Chan T F, Zheng X F. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 9.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 10.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–759. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 11.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 12.Cafferkey R, Young P R, McLaughlin M M, Bergsma D J, Koltin Y, Sathe G M, Faucette L, Eng W-K, Johnson R K, Livi G P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardenas M E, Cruz M C, Poeta M D, Chung N, Perfect J R, Heitman J. Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin Microbiol Rev. 1999;12:583–611. doi: 10.1128/cmr.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardenas M E, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardenas M E, Hemenway C, Muir R S, Ye R, Fiorentino D, Heitman J. Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J. 1994;13:5944–5957. doi: 10.1002/j.1460-2075.1994.tb06940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J, Chen J, Schreiber S L, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 18.Cruz M C, Cavallo L M, Görlach J M, Cox G, Perfect J R, Cardenas M E, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz M C, Poeta M D, Wang P, Wenger R, Zenke G, Quesniaux V F J, Movva N R, Perfect J R, Cardenas M E, Heitman J. Immunosuppressive and nonimmunosuppressive cyclosporin analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin dependent inhibition of calcineurin. Antimicrob Agents Chemother. 2000;44:143–149. doi: 10.1128/aac.44.1.143-149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Poeta M, Cruz M C, Cardenas M E, Perfect J R, Heitman J. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob Agents Chemother. 2000;44:739–746. doi: 10.1128/aac.44.3.739-746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Poeta M, Schell W A, Dykstra C C, Jones S, Tidwell R R, Czarny A, Bajic M, Bajic M, Kumar A, Boykin D, Perfect J R. Structure: in vitro activity relationships of pentamidine analogues and dication-substituted bis-benzimidazoles as new antifungal agents. Antimicrob Agents Chemother. 1998;42:2495–2502. doi: 10.1128/aac.42.10.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickman D A, Ding H, Li Q, Nilius A M, Balli D J, Ballaron S J, Trevillyan J M, Smith M L, Seif L S, Kim K, Sarthy A, Goldman R C, Plattner J J, Bennani Y L. Antifungal rapamycin analogues with reduced immunosuppressive activity. Bioorg Med Chem Lett. 2000;10:1405–1408. doi: 10.1016/s0960-894x(00)00184-0. [DOI] [PubMed] [Google Scholar]
- 23.Douglas C M, D'Ippolito J A, Shei G J, Meinz M, Onishi J, Marrinan J A, Li W, Abruzzo G, Flattery A, Bartizal K, Mitchell A, Kurtz M B. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-β-d-glucan synthase inhibitors. Antimicrob Agents Chemother. 1997;41:2471–2479. doi: 10.1128/aac.41.11.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang A, Wong G K, Demain A L. Enhancement of the antifungal activity of rapamycin by the coproduced elaiophylin and nigericin. J Antibiot. 2000;53:158–162. doi: 10.7164/antibiotics.53.158. [DOI] [PubMed] [Google Scholar]
- 25.Ferrara A, Cafferkey R, Livi G P. Cloning and sequence analysis of a rapamycin-binding protein-encoding gene (RBP1) from Candida albicans. Gene. 1992;113:125–127. doi: 10.1016/0378-1119(92)90679-j. [DOI] [PubMed] [Google Scholar]
- 26.Fields S. The two-hybrid system to detect protein-protein interactions. Methods Enzymol. 1993;5:116–124. [Google Scholar]
- 27.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franzot S P, Casadevall A. Pneumocandin L-743,872 enhances the activities of amphotericin B and fluconazole against Cryptococcus neoformans in vitro. Antimicrob Agents Chemother. 1997;41:331–336. doi: 10.1128/aac.41.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgopapadakou N H, Walsh T J. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob Agents Chemother. 1996;40:279–291. doi: 10.1128/aac.40.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein A L, Pan X, McCusker J H. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 33.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 34.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.High K P. The antimicrobial activities of cyclosporine, FK506, and rapamycin. Transplantation. 1994;57:1689–1700. [PubMed] [Google Scholar]
- 36.High K P, Washburn R G. Invasive aspergillosis in mice immunosuppressed with cyclosporin A, tacrolimus (FK506), or sirolimus (rapamycin) J Infect Dis. 1997;175:222–225. doi: 10.1093/infdis/175.1.222. [DOI] [PubMed] [Google Scholar]
- 37.Ingle G R, Sievers T M, Holt C D. Sirolimus: continuing the evolution of transplant immunosuppression. Ann Pharmacother. 2000;34:1044–1055. doi: 10.1345/aph.19380. [DOI] [PubMed] [Google Scholar]
- 38.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 40.Kurtz M B, Bernard E M, Edwards F F, Douglas C M, Marrinan J A, Dropinski J, Armstrong D. Aerosol and parenteral pneumocandins are effective in a rat model of pulmonary Aspergillosis. Antimicrob Agents Chemother. 1995;39:1784–1789. doi: 10.1128/aac.39.8.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz M C, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 42.Luengo J I, Yamashita D S, Dunnington D, Beck A K, Rozamus L W, Yen H-K, Bossard M J, Levy M A, Hand A, Newman-Tarr T, Badger A, Faucette L, Johnson R K, D'Alessio K, Porter T, Shu A Y L, Heys R, Choi J, Kongsaeree P, Clardy J, Holt D A. Structure-activity studies of rapamycin analogs: evidence that the C-7 methoxy group is part of the effector domain and positioned at the FKBP12-FRAP interface. Chem Biol. 1995;2:471–481. doi: 10.1016/1074-5521(95)90264-3. [DOI] [PubMed] [Google Scholar]
- 43.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 45.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. TOR, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 47.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odom A, Poeta M D, Perfect J, Heitman J. The immunosuppressant FK506 and its nonimmunosuppressive analog L-685,818 are toxic to Cryptococcus neoformans by inhibition of a common target protein. Antimicrob Agents Chemother. 1997;41:156–161. doi: 10.1128/aac.41.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson T F. Role of newer azoles in surgical patients. J Chemother. 1999;11:504–512. doi: 10.1179/joc.1999.11.6.504. [DOI] [PubMed] [Google Scholar]
- 51.Petraitiene R, Petraitis V, Groll A H, Candelario M, Sein T, Bell A, Lyman C A, McMillian C L, Bacher J, Walsh T J. Antifungal activity of LY303366, a novel echinocandin B, in experimental disseminated candidiasis in rabbits. Antimicrob Agents Chemother. 1999;43:2148–2155. doi: 10.1128/aac.43.9.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petraitis V, Petraitiene R, Groll A H, Bell A, Callender D P, Sein T, Schaufele R L, McMillian C L, Bacher J, Walsh T J. Antifungal efficacy, safety, and single-dose pharmacokinetics of LY303366, a novel echinocandin B, in experimental pulmonary aspergillosis in persistently neutropenic rabbits. Antimicrob Agents Chemother. 1998;42:2898–2905. doi: 10.1128/aac.42.11.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohde J, Heitman J, Cardenas M E. The Tor kinases link nutrient sensing to cell growth. J Biol Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 55.Rose M D, Winston F, Hieter P. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 56.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 57.Saunders R N, Metcalfe M S, Nicholson M L. Rapamycin in transplantation: a review of the evidence. Kidney Int. 2001;59:3–16. doi: 10.1046/j.1523-1755.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 58.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stan R, McLaughlin M M, Cafferkey R T, Johnson R K, Rosenberg M, Livi G P. 1994. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 2030;269:32027–32033. [PubMed] [Google Scholar]
- 60.Thompson J R, Register E, Curotto J, Kurtz M, Kelly R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast. 1998;14:565–571. doi: 10.1002/(SICI)1097-0061(19980430)14:6<565::AID-YEA251>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 61.Toffaletti D L, Rude T H, Johnston S A, Durack D T, Perfect J R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Horst C M, Saag M S, Cloud G A, Hamill R J, Graybill J R, Sobel J D, Johnson P C, Tuazon C U, Kerkering T, Moskovitz B L, Powderly W G, Dismukes W E. Treatment of Cryptococcal meningitis associated with the acquired immunodeficiency syndrome. N Engl J Med. 1997;337:15–21. doi: 10.1056/NEJM199707033370103. [DOI] [PubMed] [Google Scholar]
- 63.Vezina C, Kudelski A, Sehgal S N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing Streptomycete and isolation of the active principle. J Antibiot. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 64.Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J Biol Chem. 2001;276:7027–7032. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
- 65.Weisman R, Choder M, Koltin Y. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J Bacteriol. 1997;179:6325–6334. doi: 10.1128/jb.179.20.6325-6334.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong G K, Griffith S, Kojima I, Demain A L. Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J Antibiot. 1998;51:487–491. doi: 10.7164/antibiotics.51.487. [DOI] [PubMed] [Google Scholar]
- 68.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng X-F, Fiorentino D, Chen J, Crabtree G R, Schreiber S L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]