Abstract
Background
The COVID-19 pandemic caused by the SARS-CoV-2 coronavirus has currently affected millions of people around the world. To combat the rapid spread of COVID-19 there is an urgent need to implement technological platforms for the production of vaccines, drugs and diagnostic systems by the scientific community and pharmaceutical companies. The SARS-CoV-2 virus enters the cells by the interaction between the receptor-binding domain (RBD) present in the viral surface spike protein and its human receptor ACE2. The RBD protein is therefore considered as the target for potential subunit-based vaccines.
Methods and results
We evaluate the use of Nicotiana benthamiana plants as the host to transiently-producing recombinant RBD (RBDr) protein. The identity of the plant-produced RBDr was confirmed by immune assays and mass spectrometry. Immunogenicity was confirmed through the specific antibodies generated in all of the immunized mice compared to the PBS treated group.
Conclusions
In conclusions, the immunogenicity of the RBDr produced in N. benthamiana was confirmed. These findings support the use of plants as an antigen expression system for the rapid development of vaccine candidates.
Keywords: SARS-CoV-2, RBDr, N. benthamiana, Mice, Immunization, Antibodies
Introduction
The infectious disease COVID-19 produced by the SARS-CoV-2 coronavirus keeps the vast majority of countries on alert, due to the high mortality caused by this disease [1, 2]. Its rapid spread between continents has led the World Health Organization to declare it a pandemic, https://www.who.int. This has resulted not only in the development of therapies and quick affordable diagnostic systems, but also the rapid search for effective vaccines. Several vaccines and vaccine candidates are now being used and evaluated in clinical trials involving platforms based on nucleic acids, viral vectors and subunit vaccines [3].
SARS-CoV-2 is a virus enveloped with RNA genetic material, appearing like a crown, due to the presence of the S surface glycoprotein. This protein has two subunits known as S1 and S2. The amino acid sequence between the 319–591 residues of S1, corresponding to the receptor binding dominion (RBD), has a central role in the viral infection [4]. The RBD has eight cysteines and two N-glycosylation sites (N331 and N343). The inclusion of glycans can play a relevant role in vivo, in the folding, dynamics, stability and accessibility of the solvents by the RBD [5]. The receptor of this virus is the angiotensin converting enzyme 2 (hACE2), found on the external surface of the membrane of several types of human cells, forming the RBD-hACE2 complex. Consequently, the viral particle enters the host cell and produces the release of its genetic material in the cytoplasm of the infected cell [6]. The above reasons have led to consider the RBD as a protein candidate for the development of a subunit vaccine.
There have been diverse biotechnological platforms for drug production, but most of the drugs and recombinant vaccines have been produced in mammal or microbial cells. These productive systems have large advantages, but also disadvantages such as their high production costs [7] leading to alternative production settings that may reduce costs significantly. Plants then arise as production systems for recombinant proteins, offering many advantages over conventional platforms, which include its simple cultivation, low cost, the uncomplicated increase in production scale, among others [8, 9]. It has also been shown that they can assemble simple molecules, such as antibody fragments, to produce more complex ones. The plants, as bioreactors, can also produce drugs in a stable or transitory manner, where the latter is a strong tool for rapidly obtaining heterologous proteins of pharmaceutical importance [10, 11].
In this study, we describe the production of the RBD protein of SARS-CoV-2 in N. benthamiana, as a rapid and scalable alternative for making this antigen. The integrity and identity of the protein was confirmed by immunoblotting and mass spectrometry assays. Finally, we discuss the capacity of the RBD produced in N. benthamiana plants to generate a specific immune response.
Materials and methods
Plant material and growing conditions
N. benthamiana seeds were grown on the TerraPlantR 2 substrate, in floating trays soaked with water under controlled conditions (25 oC with photoperiods of 16 h light/8 h darkness) to obtain plants of approximately 7 weeks of age. Every 15 days we sprinkled the foliar fertilizer Bayfolan® S- Bayer.
RBD recombinant variants as model antigens
For western blot analysis his-RBD (amino acids 331–529 of the Spike protein from SARS-CoV-2) variant produced in Escherichia coli in PET-28 plasmid (donated by Biomedical Research Department, Center for Genetic Engineering and Biotechnology, Havana) was used as positive control. Human ACE2 receptor (hACE2) and chimeric protein hFc-RBD-HRP were supplied by the Center of Molecular Immunology, Havana, Cuba. RBD produced in Pichia pastoris [12] was used as a positive control in the hACE2 inhibition assay.
Construction of the expression vector of the RBD
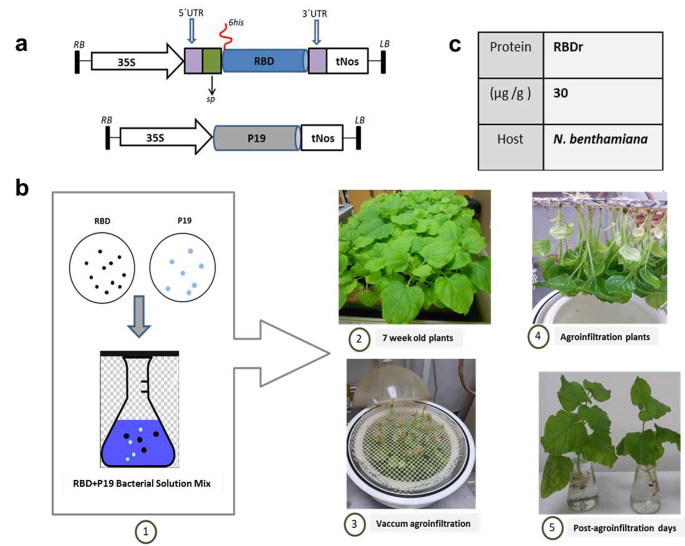
The nucleotide sequence (amino acids 331–530 of the Spike protein from SARS-CoV-2, strain Wuhan-Hu-1 (NCBI Acc. No. YP_009724390)) coding the RBD region that carried six histidine amino acids at the N-terminal end was extracted from the PET-28 plasmid. It was inserted in a plant expression vector, pCambiaHT, using the BamHI restriction sites, and flanked by the 5´ and 3´ non-translatable (UTR) sequences of the Cowpea Mosaic Virus. The 35 S promoter of the Cauliflower Mosaic Virus and the nopaline synthase terminator (tNos) were the signals used for starting and ending the transcription, respectively. The genetic design considered the fusion of the peptide signal of the sporamin from sweet potato (Ipomoea batatas L.), at the N-terminal end of the gene of interest. As a result, we obtained the binary vector pCambiahis-RBDapo (Fig. 1a), used in N.benthamiana transient transformation assays.
Fig. 1.
Schematic representation of transitory transformation in plants of N. benthamiana. a Genetic construction of RBDr. 35 S: promoter of the Cauliflower Mosaic Virus; 5´ and 3´ UTR: non-translational regions of the Cowpea Mosaic Virus; sp: sweet potato sporamin signal peptide; 6his: sequence coding for 6 histidines; tNos: nopaline synthase terminator; RBD: sequence of the receptor binding domain contained in the spike protein of the SARS-CoV-2 virus; P19: gene coding for RNA silencing suppressor p19 from Tomato Bushy Stunt Virus; RB and LB: right and left border, respectively, flanking the transcriptional unit. b Diagram of the vacuum agroinfiltration protocol. c RBDr expression levels
Transitory expression of the N. benthamiana leaves
Agrobacterium tumefaciens strain GV3101 was individually transformed with the expression vector pCambiahis-RBDapo and pCambiaP19, by heat shock (Fig. 1a). The resulting strains were checked by PCR and cultivated in the YEB medium (lab-lemco 4 g/L, sacarose 5 g/L, lacto-pectone 5 g/L, yeast extract 1 g/L, MgSO4 2 mM, pH 7.2 ) supplemented with 50 mg/mL of rifampicin and kanamycin, while stirring at 200 rpm for 16 h at 28 oC. We later transferred them to a YEB medium without antibiotics, under the same conditions where they reached the optical density (OD) of 0.6 to 0.7 at 600 nm. Cell were harvested by centrifugation at 3000 rpm for 30 min and each bacterial pellet was resuspended in the Murashige-Skoog liquid medium (Sigma, USA), with 30 mg/mL of acetosyringone (Sigma-Aldrich, USA) and then incubated at room temperature while slowly stirring in the dark for 4 h.
The vacuum infiltration protocol was performed following the process shown in Fig. 1b, using a final solution, resulting from the mixture of the two bacterial solutions (1:1 proportion). At 7 weeks of growth, we took the plants from the floating tray and submerged them, with their roots upward, in the mixture of the Agrobacterium, within a vacuum chamber. The agro-infiltration procedure had 2 cycles consisting of 1 min of vacuum and 1 min of decompression each. Afterwards, we rinsed the treated plants with water to detach the excess bacteria, and they were incubated in water for 5 days at 23 oC with photoperiods of 16 h of light and 8 h of darkness in glass jars. Agro-infiltrated plants with pCambiaP19 were used as negative control (+ P19).
RBDr expression and extraction
The 5 day-post-infiltrated leaves (5dpi) were harvested and total soluble proteins were extracted [13]. To determine the levels of expression of the RBDr we used an enzyme-linked immunosorbent assay (ELISA). Plates (Nunc MaxiSorp™) were coated with 10 µg/mL of the monoclonal antibody anti-RBD (AcM-RBD) produced at the CIGB, Sancti Spíritus, Cuba, and incubated for 16 h at 4 °C. The plate was blocked with 5% skim milk in PBS-T, for 2 h at 37 °C. In order to quantify the RBDr expression, (serial dilutions starting from 1:100) of N. benthamiana protein extracts containing RBDr were added to plate and kept it for 2 h at 37 °C. As a second antibody was used monoclonal anti-poly-histidine, produced in mice and conjugated to the horseradish peroxidase (mAb-his-HRP, 1:2000, catalog A7058 Sigma-Aldrich), incubated for 1 h at 37 °C. Intermediate washing were established between each step with PBS-T. In the quantification were used a standard curve of RBD produced in P. pastoris. As negatives controls were utilized crude extract + P19 and PBS. The reaction was detected after the addition of 3,3–5,5-tetramethylbenzidine and quantified using a microplate reader at 450 nm using a spectrophotometer (Thermo Scientific UV-Vis, USA).
RBDr antigen purification
Before starting the purification, TSP was clarified by centrifugation at 9000 rpm for 20 min and the supernatant was filtrated with a membrane of 0.22 μm (Sartorius Minisart, Germany). Recombinant His-tagged proteins were then purified by Immobilized Metal Affinity Chromatography (IMAC) using the procedure [12] with modifications described in the caption of Fig. 2. This fraction was concentrated through centricon 3 kDa (Millipore. USA). The protein concentration of the all fractions were determined by the Bradford method [14].
Fig. 2.
Purification of RBDr from N. benthamiana leaves agroinfiltrated. a Schematic representation of recombinant antigen purification process. b Stained 12% SDS-PAGE loaded with 10 µg in line 1,2,3,6 ; 200 ng in 4,7 and 1 µg in 5,8. Bottom, Western blot using the anti-histidine monoclonal antibody conjugated with peroxidase of the fractions obtained during purification; 1: TSP extract containing the RBDr; 2: matrix unbound fraction; 3: washing of the process at 70mM of imidazole; 4 and 5: Elution of the RBDr at 500mM; 6: + P19 leaves extract; 7: RBD produced and purified from E.coli; 8: eluate of concentrated RBDr in centricon (3.0 MWCO, Sartorius); MW: Prestained SDS-PAGE standards, Broad Range, cat # 161–0318 (BIO-RAD, USA); simple arrow indicates the band corresponding to the RBDr protein and the double arrow is E. coli his-RBD
Samples from the fractions of the RBDr purification process were assayed in a 12% acrylamide SDS-PAGE stained with coomassie Blue G25 (Applichem, Germany). For Western blotting, the mAb-his-HRP was used to detect the his-tag of RBDr. A variant of RBD produced in E.coli, his-RBD, was used as a positive control. The substrate used for the colorimetric detection of the assay was diaminobenzidine (Sigma, USA).
Electrospray ionization mass spectrometry (ESI-MS) analysis
The RBDr purified by IMAC was concentrated and separated using SDS-PAGE in polyacrylamide gel at 12.5% under reducing conditions. The bands of protein corresponding to RBDr were cut from the gel and divided into small cubes of approximately 1 mm3. The gel cubes were treated to obtain tryptic peptides. The tryptic peptides were analyzed with a hybrid orthogonal QToF-2 tandem mass spectrometer (Micromass, UK). The sample was sprayed using 1200 V for the capillary and 35 V for the cone. The electrospray ionization-mass spectrometry (ESI-MS) was set to the 200–2000 m/z range and multiply-charged ions were selected for collision induced dissociation based fragmentation. For sequencing peptides, multiply-charged ions were selected for collision induced dissociation (CID) based fragmentation. The ESI-MS/MS of tryptic peptides with z ≥ 2 + were deconvoluted by MaxEnt 3.0 and the collision energies between 15 and 55 eV were used to obtain sequence information in the MS/MS spectrum. The aminoacids were manually assigned and some of them identified by MASCOT MS/MS Ion Search using SARS-CoV-2 and SwissProt databases with propionamide (C) as fixed modification, deamidated (NQ) and oxidation (M) as variable modifications, with a peptide tolerance and MS/MS tolerance of 0.6 Da and 0.3 Da respectively. The collision gas used was Argon, and the mass spectra were processed through the MassLynx v4.1 software (Micromass, UK).
Immunogenicity of the RBDr in mice
According to the Institutional Committee for Animal Care and Use of the CIGB, Cuba, the regulations for the use of laboratory animals were followed to study the immunogenicity of RBDr in mice. Balb/c mice 6 weeks old were used in the experiment. They were maintained under controlled temperature and lighting, with feed and water ad libitum. Two experimental groups were designed, which consisted of nine animals per group for the subcutaneous injection. The first one was the group of interest in which we inoculated 5 µg RBDr. A second group identified as the negative control (-RBD) to which we administered the same volume of PBS. The inoculum was mixed with aluminum hydroxide in a 6:1 ratio (antigen/adjuvant). Three doses were assayed, corresponding to 0, 21 and 42 days, while mice bleeding were developed at 0, 21, 42 and 57 days in order to evaluate specific antibody production and their ability to inhibit the binding of RBD to hACE2.
Anti RBDr titers of immunized mice
To evaluate the immune response against RBDr, the described protocol [12] was followed with the adaptations described below and only up to the plate reading step. Coating with 5 µg/mL of RBDr produced in P. pastoris and only the SS-1 antibody was used. In addition, the RBDr and -RBD sera used in the assay were diluted in PBS-T (dilutions from 1:25 to 1:10000).
RBD-hACE2 Inhibition Assay
The capacity of the polyclonal antibodies generated by RBDr to inhibit the binding of the hACE2 receptor was evaluated through a competitive ELISA previously reported [12]. Mice sera after the last immunization with RBDr (T57) were used for the analysis. As positive control was considered the sera of nine volunteers (90% inhibition) immunized with RBD of P. pastoris and mice sera immunized with PBS at T57 were used as a negative control.
Statistics analysis
The mean of the experimental values was represented and the standard deviation indicated as error bars. In the analysis of the data, we used the GraphPad Prism software version 8.0.2, with a Kruskal-Wallis test and Dunn’s test.
Results
Transient transformation of Nicotiana benthamiana, expression and purification of RBDr
The strains of A. tumefaciens transformed with the genetic constructions represented in the Fig. 1a were used during the transitory co-transformation of N. benthamiana plants. Five days after vacuum infiltration (5dpi), no visible damages (necrosis and change of coloration) were observed in the treated plants (Fig. 1b). The antigen concentration in the total soluble protein extract was estimated by ELISA, quantifying an expression of 30 µg RBDr per gram of leaves (Fig. 1c).
The designed RBDr included a polyhistidine peptide, so the purification included an immobilized metal chelate affinity chromatography step (Fig. 2a). The IMAC fractions were analyzed by SDS-PAGE and Western blot using mAb-his-HRP. In the eluates fractions (lanes 4 and 5) the identity of the recombinant protein was confirmed, as well as in the sample resulting from the final concentration (lane 8, several subpopulations with size between 25 and 41 kDa approximately, presumably glycoforms thereof, coexisted). Additionally, bands above 41 kDa were observed in the SDS-PAGE, not detected in the Western blot. No signals were detected in lanes 2 and 3 corresponding to unbound and washed fraction, either in lane 6 corresponding to the initial extract of agroinfiltrated plants only with pCambiaP19 vector (Fig. 2b).
ESI-MS analysis of the RBDr protein
Mass spectrometry (MS) is the analytical tool of choice to confirm the amino acid sequence, to evaluate the integrity of the N- and C-terminal ends, and to detect posttranslational modifications in natural or recombinant proteins. We analyzed the RBDr through ESI-MS with the goal to confirm the amino acid sequence and to verify the correct assembling of disulfide bonds of RBDr. Sample processing, including an efficient proteolytic digestion, the recovery of the proteolytic peptides and a desalting step prior to ESI-MS analysis, plays a key role to obtain the highest sequence coverage. The ESI-MS spectrum through the RBDr protein digestion is shown in Fig. 3a. Some of the signals with stronger relative intensities, designed by the blue arrows, were chosen for sequencing. The sequence assignment based on the agreement between the expected and experimental m/z of tryptic peptides are summarized in Table 1. Two of the four-disulfide bonds present in the RBDr, C379-C432 and C480-C488, were identified. This study confirmed coverage of the sequence of RBDr of 51.3%, thereby indicating the identity of the antigen (Fig. 3b).
Fig. 3.
RBDr characterization through mass spectrometry. a ESI-MS spectrum through the digestion of the RBDr protein in polyacrylamide gel. The identified disulfide bonds between the cysteines (C379-C432 and C480-C488) are highlighted in red as -S-S-. The signs marked with blue arrows were the peptides that were analyzed with the MASCOT software as the criterion for the identity of the RBDr protein in the databases of the sequences. b Amino acid sequence of the RBDr protein. The residues highlighted in brown, correspond to the 4 amino acids contributed by the sub-cloning vector. The green colour highlights the six residues of the histidine for their purification by affinity chromatography using immobilized metal chelates. Those highlighted in blue are the residues of the spacer arm, and those in black correspond to the RBD theoretical sequence that includes the identified amino acids sequence in violet. Red letters are the cysteine residues. The red lines represent the disulfide bonds that link to cysteines, C336-C361, C379-C432, C480-C488 and C391-C525. The asterisks represent the identified bonds
Table 1.
Representation of theoretical RBD peptides. Those marked with blue letters are the sequenced fragments that belonged to RBDr
| Frg # | Res # | Sequence | Theor (Bo) | (M + H) | (M + 2 H) | (M + 3 H) | (M + 4 H) | (M + 5 H) |
|---|---|---|---|---|---|---|---|---|
| T1 | 1–17 | MGSSHHHHHHSSGLVPR(G) | 1898.88 | 1899.89 | 950.45 | 633.97 | 475.73 | 380.78 |
| T2 | 18–39 | (R)GSHMASNITNLCPFGEVFNATR(F) | 4678.16 | 4679.16 | 2340.09 | 1560.39 | 1170.55 | 936.64 |
| T6 | 51–71 | (R)ISNCVADYSVLYNSASF | ||||||
| T3 | 40–48 | (R)FASVYAWNR(K) | 1112.54 | 1113.55 | 557.28 | 371.85 | 279.14 | 223.52 |
| T4 | 49–49 | (R)K(R) | 146.11 | 147.11 | 74.06 | 49.71 | 37.53 | 30.23 |
| T5 | 50–50 | (K)R(I) | 174.11 | 175.12 | 88.06 | 59.05 | 44.54 | 35.83 |
| T7 | 72–79 | (K)CYGVSPTK(L) | 3059.40 | 3060.41 | 1530.71 | 1020.81 | 765.86 | 612.89 |
| T12 | 118–137 | (K)LPDDETGCVLAWNSNNLDSK(V) | ||||||
| T8 | 80–96 | (K)LNDLCFTNVYADSFVIR(G) | 3966.04 | 3967.05 | 1984.03 | 1323.02 | 992.52 | 794.22 |
| T18 | 203–221 | (R)VVVLSFELLHAPATVCGPK(K) | ||||||
| T9 | 97–101 | (R)GDEVR(Q) | 574.27 | 575.28 | 288.14 | 192.43 | 144.58 | 115.86 |
| T10 | 102–110 | (R)QIAPGQTGK(I) | 898.49 | 899.50 | 450.25 | 300.50 | 225.63 | 180.71 |
| T11 | 111–117 | (K)IADYNYK(L) | 885.42 | 886.43 | 443.42 | 296.15 | 222.36 | 178.09 |
| T13 | 138–147 | (K)VGGNYNYLYR(L) | 1217.58 | 1218.59 | 609.80 | 406.87 | 305.40 | 244.52 |
| T14 | 148–150 | (R)LFR(K) | 434.26 | 435.27 | 218.14 | 145.76 | 109.57 | 87.86 |
| T15 | 151–151 | (R)K(S) | 146.11 | 147.11 | 74.06 | 49.71 | 37.53 | 30.23 |
| T16 | 152–159 | (K)SNLKPFER(D) | 989.53 | 990.54 | 495.77 | 330.85 | 248.39 | 198.91 |
| T17 | 160–202 | (R)DISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYR(V) | 4765.12 | 4766.12 | 2383.57 | 1589.38 | 1192.29 | 954.03 |
| T19 | 222–222 | (K)K(-) | 146.11 | 147.11 | 74.06 | 49.71 | 37.53 | 30.23 |
Murine immunization and serum analysis
Animals subcutaneous administration was carried out with three doses (Fig. 4a) of protein containing 5 µg of RBDr mixed with aluminum hydroxide adjuvant. Through the duration of the experiment, mice showed no signs of disease or adverse effects following vaccination. Sera from mice immunized at times 0, 21, 42 and 57 days were evaluated by ELISA as described in materials and methods. Specific antibody titers were observed in all inoculated animals after the third dose compared to the control sera -RBD. At 57 days, the highest antibody titers were detected in two of nine immunized mice. Antibody titers were statistically significant on T57 compared to earlier times (indicated with different letters) (Fig. 4b).
Fig. 4.
Titration of antibodies in mice immunized with RBDr produced in N. benthamiana. a Immunization schedule. From T0 to T42 we performed the administration of the recombinant antigen. The syringes (filled in red) throughout the entire period, show the time of the blood extraction of the inoculated animals. b Response in immunized mice in each trial period. The values represent the antibodies titters expressed as the base 10 logarithm. +RBDr: experimental group inoculated with the recombinant antigen produced in plants; –RBD: mice immunized with PBS; Graph show mean ± standard deviation (SD) for n = 9 mice/group. c Binding inhibition capacity of polyclonal antibodies RBDr to hACE2. +RBDr: sera of immunized mice with RBDr, C + sera of persons immunized with RBD produced in P. pastoris, that showed high percentages of inhibition and taken as the positive control; -RBD: mice immunized with PBS. For the evaluation, we chose all sera at time T57. We observed two sera with high inhibition capacity, similar to those used as the positive control. Data are mean ± SD. from n = 9 repetitions/groups. The different letters represent significant differences for P < 0.0001
The inhibition of the binding of hFc-RBD-HRP to hACE2 mediated by sera from animals immunized with RBDr, suggests a probable decrease of the infective capacity of the virus, and it therefore offers information of the possible usefulness of a vaccine candidate against SARS-CoV-2. Figure 4c represents inhibition of RBD binding to hACE2 in the presence of sera extracted at T57. Of all the sera tested, two of them showed an inhibition of approximately 90%, similar to the sera referred to as positive control. In the rest of the animals, the mean percentage of inhibition was between 40 and 50%. In contrast, at T0 the sera did not show any inhibitory capacity in the binding to hACE2, thus confirming that this inhibition is due to the anti-RBDr antibodies generated in the mice immunized with the antigen produced in N. benthamiana.
Discussion
The propagation of the novel coronavirus SARS-CoV-2, the cause of COVID-19, has become the center of attention worldwide because of the levels of infection and mortality [15]. This virus is genetically related to SARS-CoV, and it is characterized by a more rapid spread than the rest of the coronaviruses. It can infect cells of the lung, kidney, heart and intestine, among other tissues, producing the multiple organ dysfunction syndrome (MODS) [16]. Among the treatment alternatives to survive and control the virus infection, SARS-CoV-2 specific antiviral drugs, redirected drugs and specific vaccines against the SARS-CoV-2 virus have been assayed [17].
In recent years, plants have been considered as a powerful expression system for the production of therapeutic proteins [18, 19] that show similar, or even greater biological activity than the homologous proteins expressed in cultured mammalian cells [20]. In this respect, we used in this study the transitory transformation of N. benthamiana plants due to the advantages it can offer in terms of speed, versatility and ease of agroinfiltration. Previous studies have demonstrated the production of SARS-CoV-2 related proteins in N. benthamiana. Recent studies estimated between 2 and 4 µg RBD/g fresh weight using the same host as us [21] Similarly, Rattanapisit obtained 8 µg of such antigen/g fresh weight of leaf from the same plant [22]. Also other studies confirm 10 µg [5] and 25 µg [23] of RBD in N. benthamiana. The RBDr expression level reported here is in the same order than those were previously informed, in spite of we assayed a platform involving a non-replicative methods by the construction of a vector that carried 5´and 3´ UTR, of the Cowpea Mosaic virus, a successful design for the accumulation of other heterologous proteins [24]. However, this does not imply that plant made vaccines against SARS-CoV-2 are not a possible alternative, taking into account the performance of clinical trials with vaccine candidates produced in N. benthamiana plants [25].
The preparation of the sample for the analysis of mass spectrometry plays a determinant role in the quality of the result. The confirmation of the main part of the amino acid sequence depends on the efficiency of the digestion and the recovery of the tryptic peptides [26]. In our case, the digestion in gel under native conditions, made it possible to recover the necessary fragments to confirm 51.3% of the RBDr. Under our conditions, it was possible to identify the links of C379-C432 and C480-C488. It was impossible to identify the first C336-C361 because these Cys residues are included in the T2 and T6 fragments that could not be sequenced because of the presence of N-glycans, which make the peptides more hydrophilic. Therefore, recovery of the glycopeptides by RP-HPLC is difficult. C391-C525 was not identified because we were unable to sequence the C terminal end. Taking into account that the analysis was performed through a digestion in gel, we observed a greater coverage of sequences compared to other studies using fragments of the gel for the peptide analyses. In this sense, a study of the analysis of the protein extract of HeLa, optimizing six different gel digestion methods, revealed a sequencing coverage of less than 20% [27]. In another case, was reached peptide sequencing of 44% by also using gel digestion [28]. In contrast, when the variable region of an antibody was purified for its analysis by mass spectrometry, using digestion in solution, the coverage confirmed was of more than 80% of the sequence [29].
Both the humoral and cellular immune response developed with vaccines, have had great relevance in different viral disease protection [30, 31]. The doses used in the immunization have a determinant role in immunogenicity [32]. In our study, the administration of 5 µg of RBDr brought about the appearance of specific antibodies in all the animals, as of the third injection, compared to animals -RBD. In addition, approximately 22% of the animals immunized with RBD of plant origin developed IgG titers 1:10000 (log10 4), very similar to the average the titers shown by Siriwattananon [23] in the first immunization but lower with respect to the second immunization evaluated by them with 10 µg of RBD-Fc. It is presumed that the delay in the immunological response could be influenced by the low dose used. In that sense, when was used 10 µg of RBD, produced in humans cell line, compared to the 5.0 and 2.5 µg doses higher antibodies titers were obtained [33]. Also, the dose trials related to a vaccine candidate based on the mRNA of RBD in mice, showed that the highest dose (15 µg) produced the highest titers when compared to 2 µg in the immunized mice [34]. Another study showed that, mice injected with hemagglutinin (HA) from the Influenza virus, stimulated lower HA titers when using 3 µg of the inoculum, compared to the highest dose vaccine, which induced higher HA titers [35]. These results could explain the heterogeneity of immune response observed in mice immunized with the dose evaluated of RBDr, when two of nine mice were high responders. It is possible that immunization with higher dose of RBDr could increase the immunogenecity. In addition, another factor that could influence the generation of antibodies is the combination of antigens with potent adjuvants that increase the immune response regardless of the dose used [36]. This is the case of the virus-like particle of protein S, produced in N. benthamiana, as a vaccine candidate that showed high anti-S antibody titers with one of the adjuvants employed (oily emulsion of DL-α-tocopherol and squalene), even when using low dosages of the antigen [25]. Taking into account this report, RBDr could be combined with another adjuvant to increase the immunogenicity.
The capacity of viral RBD to bind itself to the hACE2 determines the success of the infectivity of SARS-CoV-2. In the protection against COVID-19 the formation of neutralizing antibodies that can inhibit the binding of the viral receptor to hACE2 is crucial [37, 38]. In this sense, some reports have shown a correlation between the percentage of inhibition of ACE2 binding, mediated by RBD antibodies, and the generation of neutralizing antibody responses [39]. Although the inhibition percentage of the RBDr sera to reduce the protein binding to its receptor hACE2 average 40–50% compared with the mice treated with PBS, in this study was confirmed that those animals with the higher RBD-specific IgG titers had inhibition percentage near to 90% (coinciding with the highest IgG titers), indicating that the antigen herein obtained is functional. Similar results were obtained on evaluating a group of monoclonal antibodies with a high affinity against RBD, where they exceptionally demonstrated that five candidates showed more than 90% of inhibition, higher than the rest, with 50% of inhibition [40]. Another study, evaluated the immunogenicity of RBD bound to Fc (2.5, 5 and 10 µg) produced in human cell lines using Freud’s adjuvant; with an immunization schedule similar to ours they obtained 66% of inhibition of the binding to ACE2 [33].
Conclusions
In this study, we have confirmed the transient production of RBDr in N. benthamiana. By ESI-MS the identity of this protein and the correct assembling of two of their four disulfide bonds were demonstrated. While this paper does not describe dose studies necessary, the production of specific anti-RBDr antibody in mice immunized showed inhibition of binding of RBD to the hACE2 receptor, confirming the immunogenicity of the antigen produced in N. benthamiana, in spite of the low dosage assayed. This study suggests the transient expression N. benthamiana as a tool to assemble and present the correctly folded antigens to the immune system that could be used as vaccines candidates against SARS-CoV-2 virus which could be particularly useful when new variants of the virus have emerged.
Author contributions
The authors are grateful to colleagues from the Animal Biotechnology Department of the Center for Genetic Engineering and Biotechnology for their collaboration in the immunization of the animals used in the trial. Y.C, A.L, A.H, C.E.G. design of the genetic construction, expression and purification of the protein. I A. mass spectrometric analysis. Y.C, R.U.M. RBDr immunogenicity assays. Y.C. writing the manuscript. All authors contributed to data processing.
Funding
This work was supported with funds from the BioCubaFarma and the Center for Genetic Engineering and Biotechnology, La Habana, Cuba.
Availability of data and material
The data obtained during the investigation were included in the manuscript.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
The approval for the experiments in animals were granted by the Institutional Committee for Animal Use and Care of the CIGB, Cuba.
Compliance with Ethical Standards
All animals were cared for following the guidelines of the American Association for the Accreditation of Laboratory Animal Care (AAALAC), and studies were conducted under the approval of the CIGB Animal Care and Use Committee, protocol number, protocol number: CICUAL/CIGB/20098.
Consent for publication
The authors have given their consent to the disclosure of this study in the journal.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueiredo ADA, Moreira DC. Letalidad de la COVID-19: ausencia de patrón epidemiológico. Gac Sanit. 2022;35(4):355–357. doi: 10.1016/j.gaceta.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belete TM. Review on up-to-date status of candidate vaccines for COVID-19 disease. Infect drug Resist. 2021;14:151–161. doi: 10.2147/IDR.S288877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han P, Li L, Liu S, et al. Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2. Cell. 2022;185(4):630–640. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin Y-J, König-Beihammer J, Vavra U, et al. N-glycosylation of the SARS-CoV-2 receptor binding domain is important for functional expression in plants. Front Plant Sci. 2021;12:1–14. doi: 10.3389/fpls.2021.689104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. cell 181(2):271 – 80.10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed]
- 7.O’Flaherty R, Bergin A, Flampouri E, et al. Mammalian cell culture for production of recombinant proteins: A review of the critical steps in their biomanufacturing. Biotechnol Adv. 2020;43:1–17. doi: 10.1016/j.biotechadv.2020.107552. [DOI] [PubMed] [Google Scholar]
- 8.Ortega-Berlanga B, Pniewski T. Plant-Based Vaccines in Combat against Coronavirus Diseases. Vaccines. 2022;10(2):138. doi: 10.3390/vaccines10020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdiani A, Hansen OK, Nielsen UB, et al. Bioreactor-based advances in plant tissue and cell culture: challenges and prospects. Crit Rev Biotechnol. 2019;39(1):20–34. doi: 10.1080/07388551.2018.1489778. [DOI] [PubMed] [Google Scholar]
- 10.Kowalczyk T, Merecz-Sadowska A, Picot L, et al. Genetic Manipulation and Bioreactor Culture of Plants as a Tool for Industry and Its Applications. Molecules. 2022;27(3):795. doi: 10.3390/molecules27030795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar M, Tomar M, Punia S, et al. Plant-based proteins and their multifaceted industrial applications. LWT. 2022;154:2–15. doi: 10.1016/j.lwt.2021.112620. [DOI] [Google Scholar]
- 12.Limonta-Fernández M, Chinea-Santiago G, Martín-Dunn AM et al (2021) The SARS-CoV-2 receptor-binding domain expressed in Pichia pastoris as a candidate vaccine antigen. medRxiv [DOI] [PMC free article] [PubMed]
- 13.Ruiz Y, Ramos PL, Soto J, et al. The M4 insulator, the TM2 matrix attachment region, and the double copy of the heavy chain gene contribute to the enhanced accumulation of the PHB-01 antibody in tobacco plants. Transgenic Res. 2020;29(2):171–186. doi: 10.1007/s11248-019-00187-6. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Vestergaard LS, Nielsen J, Richter L, et al. Excess all-cause mortality during the COVID-19 pandemic in Europe–preliminary pooled estimates from the EuroMOMO network, March to April 2020. Eurosurveillance. 2020;25(26):1–6. doi: 10.2807/1560-7917.ES.2020.25.26.2001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Wang T, Cai D, et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38–42. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De P, Chakraborty I, Karna B, et al. Brief review on repurposed drugs and vaccines for possible treatment of COVID-19. Eur J Pharmacol. 2021;898:173977. doi: 10.1016/j.ejphar.2021.173977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniell H, Jin S, Zhu XG, et al. Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol J. 2021;19(3):430–447. doi: 10.1111/pbi.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirzaee M, Osmani Z, Frébortová J, et al. Recent advances in molecular farming using monocot plants. Biotechnol Adv 107913. 2022 doi: 10.1016/j.biotechadv.2022.107913. [DOI] [PubMed] [Google Scholar]
- 20.Aviezer D, Brill-Almon E, Shaaltiel Y, et al. A plant-derived recombinant human glucocerebrosidase enzyme—a preclinical and phase I investigation. PLoS ONE. 2009;4(3):2–6. doi: 10.1371/journal.pone.0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diego-Martin B, González B, Vazquez-Vilar M, et al. Pilot Production of SARS-CoV-2 Related Proteins in Plants: A Proof of Concept for Rapid Repurposing of Indoor Farms Into Biomanufacturing Facilities. Front Plant Sci. 2020;11:1–28. doi: 10.3389/fpls.2020.612781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattanapisit K, Shanmugaraj B, Manopwisedjaroen S, et al. Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci Rep. 2022;10(1):1–11. doi: 10.1038/s41598-020-74904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siriwattananon K, Manopwisedjaroen S, Shanmugaraj B, et al. Plant-Produced Receptor-Binding Domain of SARS-CoV-2 Elicits Potent Neutralizing Responses in Mice and Non-human Primates. Front Plant Sci. 2021;12:1–15. doi: 10.3389/fpls.2021.682953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sainsbury F, Lomonossoff GP. Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 2008;148(3):1212–1218. doi: 10.1104/pp.108.126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward BJ, Gobeil P, Séguin A, et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat Med. 2021;27:1–8. doi: 10.1038/s41591-021-01370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikulášek K, Konečná H, Potěšil D, et al. SP3 Protocol for Proteomic Plant Sample Preparation Prior LC-MS/MS. Front Plant Sci. 2021;12:369. doi: 10.3389/fpls.2021.635550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodman JK, Zampronio CG, Jones AM, et al. Updates of the In-Gel Digestion Method for Protein Analysis by Mass Spectrometry. Proteomics. 2018;18(23):1–5. doi: 10.1002/pmic.201800236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borchers C, Peter JF, Hall MC, et al. Identification of in-gel digested proteins by complementary peptide mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal Chem. 2000;72(6):1163–1168. doi: 10.1021/ac990937m. [DOI] [PubMed] [Google Scholar]
- 29.Babrak L, McGarvey JA, Stanker LH, et al. Identification and verification of hybridoma-derived monoclonal antibody variable region sequences using recombinant DNA technology and mass spectrometry. Mol Immunol. 2017;90:287–294. doi: 10.1016/j.molimm.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havervall S, Ng H, Jernbom Falk A, et al. Robust humoral and cellular immune responses and low risk for reinfection at least 8 months following asymptomatic to mild COVID-19. J Intern Med. 2022;291(1):72–80. doi: 10.1111/joim.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Menezes Martins R, Maria de Lourdes SM, de Lima SMB, et al. Duration of post-vaccination immunity to yellow fever in volunteers eight years after a dose-response study. Vaccine. 2018;36(28):4112–4117. doi: 10.1016/j.vaccine.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Xu W, Xia S, et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target therapy. 2020;5(1):1–10. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q, Ji K, Tian S, et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat Commun. 2021;12(1):1–10. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardi N, Parkhouse K, Kirkpatrick E, et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKee AS, Marrack P. Old and new adjuvants. Curr Opin Immunol. 2017;47:44–51. doi: 10.1016/j.coi.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1):1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Gao C, Das T, et al. The spike-ACE2 binding assay: An in vitro platform for evaluating vaccination efficacy and for screening SARS-CoV-2 inhibitors and neutralizing antibodies. J Immunol Methods. 2022;503:1–10. doi: 10.1016/j.jim.2022.113244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goel RR, Apostolidis SA, Painter MM, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6(58):1–19. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter JD, Hutter CAJ, Zimmermann I et al (2020) Sybodies targeting the SARS-CoV-2 receptor-binding domain. 10.1101/2020.04.16.045419. Biochemistry 1–12.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data obtained during the investigation were included in the manuscript.
Not applicable.