Abstract
Pneumonia, an acute respiratory tract infection, is one of the major causes of mortality worldwide. Depending on the site of acquisition, pneumonia can be community acquired pneumonia (CAP) or nosocomial pneumonia (NP). The risk of pneumonia, is partially driven by host genetics. CYP1A1 is a widely studied pulmonary CYP family gene primarily expressed in peripheral airway epithelium. The CYP1A1 genetic variants, included in this study, alter the gene activity and are known to contribute in lung inflammation, which may cause pneumonia pathogenesis. In this study, we performed a meta-analysis to establish the possible contribution of CYP1A1 gene, and its three variants (rs2606345, rs1048943 and rs4646903) towards the genetic etiology of pneumonia risk. Using PRISMA guidelines, we systematically reviewed and meta-analysed case-control studies, evaluating risk of pneumonia in patients carrying the risk alleles of CYP1A1 variants. Heterogeneity across the studies was evaluated using I2 statistics. Based on heterogeneity, a random-effect (using maximum likelihood) or fixed-effect (using inverse variance) model was applied to estimate the effect size. Pooled odds ratio (OR) was calculated to estimate the overall effect of the risk allele association with pneumonia susceptibility. Egger's regression test and funnel plot were used to assess publication bias. Subgroup analysis was performed based on pneumonia type (CAP and NP), population, as well as age group. A total of ten articles were identified as eligible studies, which included 3049 cases and 2249 healthy controls. The meta-analysis findings revealed CYP1A1 variants, rs2606345 [T vs G; OR = 1.12 (0.75–1.50); p = 0.02; I2 = 84.89%], and rs1048943 [G vs T; OR = 1.19 (0.76–1.61); p = 0.02; I2 = 0.00%] as risk markers whereas rs4646903 showed no statistical significance for susceptibility to pneumonia. On subgroup analysis, both the genetic variants showed significant association with CAP but not with NP. We additionally performed a spatial analysis to identify the key factors possibly explaining the variability across countries in the prevalence of the coronavirus disease 2019 (COVID-19), a viral pneumonia. We observed a significant association between the risk allele of rs2606345 and rs1048943, with a higher COVID-19 prevalence worldwide, providing us important links in understanding the variability in COVID-19 prevalence.
Keywords: Pneumonia, Community acquired pneumonia, Nosocomial pneumonia, CYP1A1, Genetic variants, Meta-analysis, COVID-19
1. Introduction
Pneumonia is an acute inflammatory condition of the lungs usually caused by bacterial, viral or fungal infection (Mackenzie, 2016). According to the site of acquisition, pneumonia is classified as community acquired (CAP) or nosocomial (NP) (Mackenzie, 2016). The global burden of disease study (2015) stated that lower respiratory infections like pneumonia are the third most common cause of death globally (WHO, 2022). Though the biology of the infecting microbe is important, host genetic plays a crucial role in the pathogenesis of pneumonia (Dela Cruz et al., 2018). Pneumonia occurs when pathogens enter the alveoli, infect, multiply and encourage a host immune response. These responses cause inflammation of the lung tissues, marking the pathogenesis of pneumonia (Jain et al., 2021). Host genetic factors that participate in these processes starting from pathogen entry, infection, inflammation, and resolution can all be considered as good candidates in genetic association studies of pneumonia and its complications (Cooke and Hill, 2001; Kumar et al., 2014). The difference in epidemiology, pathogenesis, microbiology, common causative organism, and pathophysiology between CAP and NP depends on the mode of acquisition of pneumonia infection, on host risk factors and other environmental changes (Herold and Sailer, 2004; Torres et al., 2021). Thus, understanding the functional impact of genetic determinants of susceptibility to pneumonia, both CAP and NP, independently is crucial for determining the mechanisms behind pneumonia pathogenesis.
CYP1A1 is a critical enzyme mediating the metabolism of a broad spectrum of xenobiotics and endobiotics [9]. There are several reports highlighting the functionally relevant genetic variants of CYP1A1 to play a clinically important role in several disease phenotypes. A number of studies have investigated the genetic association as well as gene interaction of pneumonia risk with monooxygenase enzyme group, cytochrome P450 (CYP). In a study, a group of researchers identified CYP1A1 gene as a critical regulator of inflammatory responses and phagocytosis in sepsis through signalling pathways that may be promising targets for treating inflammatory diseases (Tian et al., 2020). A study by Fang et al. (2016) demonstrated lower CYP1A1 expression in pigs infected with Mycoplasma hyopneumoniae (M. hyopneumoniae). They further extended their efforts by studying this gene in pulmonary alveolar macrophages (PAM) cell lines suggest CYP1A1 supresses inflammatory response caused by pneumonia infection (Fang et al., 2016). Interestingly, few other studies indicated the role of CYP1A1 genetic polymorphisms in infectious diseases and consequently establishing its role in inflammatory responses. Previously, it was identified that genetic variants of some host genes (CYP1A1, ACE and IL-6) are associated with the diversity in response to CAP (Salnikova et al., 2014; Zhao et al., 2017). The selection of this gene was established based on its role in physiological and pathological processes during pneumonia infection, particularly in the immune and inflammatory responses (Zhao et al., 2017). From the systematic literature search performed for association of CYP1A1 genetic variants and pneumonia, the most widely reported CYP1A1 single nucleotide polymorphisms (SNPs) obtained were rs2606345, rs1048943 and rs4646903. These SNPs had functional consequences which may ultimately be involved with a disease phenotype like pneumonia. The SNP rs2606345 (C > A), is located in the first intron of the gene, has a functional role of lower gene expression in the presence of allele A (Rotunno et al., 2009; Talwar et al., 2017). Another SNP, rs1048943 (T > A, C, G), resulted in a missense amino acid substitution, is characterized by the substrate-specific increased activity for minor allele G (Salnikova et al., 2013c). The presence of minor allele ‘C' of the 3’ untranslated region (UTR) SNP, rs4646903, shows an increased inducibility of CYP1A1 gene expression (Meletiadis et al., 2006; Salnikova et al., 2013c). Thus we can suggest that genetically determined alteration of CYP1A1 expression could contribute to lung inflammation pathogenesis. While there are several evidence of association between CYP1A1 polymorphisms with risk of pneumonia, there are studies which show conflicting results as well (Muñoz et al., 2012; Smith et al., 2001).
In this study we used a meta-analysis approach: 1) to investigate the impact of CYP1A1 risk allele and the risk of pneumonia (including both CAP and NP). This may increase the odds of the incident pneumonia; 2) to determine whether any association between CYP1A1 and pneumonia is generalizable to the coronavirus disease 2019 (COVID-19) as the ongoing COVID-19 pandemic and its consequent prevalence has rarely been examined through the lens of pneumonia. This genetic predisposition with pneumonia may help us in understanding the genetic etiology of COVID-19 infection and its prevalence.
2. Materials and methods
This meta-analysis was conducted as per the recommendations of the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines (Liberati et al., 2009) following the PICOS (Population, intervention, comparison, outcome and study design) strategy. The PICOS outline (Population: pneumonia patients; Intervention/Exposure: individuals carrying the risk allele of CYP1A1 variants, rs2606345, rs1048943 and rs4646903; Comparisons: risk allele carriers in cases and controls vs wild type allele carriers in cases and controls; Outcomes: pneumonia susceptibility in patients carrying the risk allele of CYP1A1 variants; and Study design: case-control studies). The selected studies were those in which the relationship between CYP1A1 gene polymorphisms and risk of pneumonia disease has been evaluated. Bibliographic databases like MEDLINE (PubMed), Web of Science, and Science Direct and worldwidescience.org were searched for all articles published till January 13, 2021. The keywords used to identify relevant studies were “CYP1A1”, “Pneumonia”, “genetic variant”, and “single nucleotide polymorphisms” using AND/OR Boolean operators. Cross references of each study retrieved were also examined for inclusion in case they discuss the effect of CYP1A1 genetic variant and its risk in pneumonia.
Two investigators (DG and SY) independently reviewed each study for its inclusion in the meta-analysis. The inclusion criteria were: (1) studies conducted on human population only, (2) included the effect of CYP1A1 genetic variant with available numeric data, (3) adopted a case-control study design (4) provided a detailed assay method. Studies (1) any other type of lung inflammation apart from pneumonia or pneumonia as a consequence of any exposure or pneumonia existing with comorbid conditions, (2) no defined diagnostic criteria for pneumonia, and (3) genotypic data not in accordance with Hardy Weinberg equilibrium were excluded.
Allele frequency data for each case and control were extracted into contingency tables to calculate the odds of pneumonia in patients carrying the risk allele of the associated variants. In case of missing allele frequency data, the corresponding odds ratio (OR) and p value were calculated from genotypic data given. The references of the retrieved articles were manually screened to identify additional studies. In case of studies where genotypic data is given allelic data is calculated to maintain a consensus across studies. The included studies and their characteristics like first author, year of publication, population, disease, genetic variant, odds ratio, genotyping method, risk allele, sample size (cases and control) and male female distribution and their quality assessment score were tabulated. All the included articles described some variant of pneumonia infection, one study discussing Mycoplasma pneumoniae infection (Zhao et al., 2017), CAP (Moroz et al., 2011; Salnikova et al., 2013a; Salnikova et al., 2013c; Salnikova et al., 2010; Smelaya et al., 2011), NP (Salnikova et al., 2014), both CAP and NP (Salnikova et al., 2013b; Salnikova et al., 2008) and relapsing pneumonia (Korytina et al., 2005). The included cases were diagnosed by experienced professionals based on symptoms, medical histories and the clinical, radiology or laboratory results (chest X-ray, spirometry measures, etc.), and physical examination of new lung infiltrates or lower respiratory tract infection. Controls were age and gender matched healthy volunteers with no previous history of relevant infectious diseases.
The methodological assessment of all the selected articles was performed by two reviewers independently using the modified Newcastle-Ottawa Scale (NOS) for non-randomised studies (Wells et al., 2001). The quality score was assigned on the basis of eight categories primarily based on three broad criteria: selection of study groups; comparability of the groups; and ascertainment of either the exposure or outcome of interest for case-control, respectively. A maximum of one star was awarded for each detail present in the study for each parameter except for comparability, where a maximum of two stars can be given. A cumulative score of the number of stars obtained for each study reflected its quality. In case of conflicting scores, a consensus was reached upon discussing with another author (RK). A study was regarded as a high-quality study when it rated six or more stars. Stata 16.0 (Stata Corporation, College Station, TX) (Sterne and Egger, 2001; Support, S. T. StataCorp, 2019) was used to generate pooled ORs between pneumonia patients and healthy controls (DerSimonian and Laird, 1986; Cochran, 1954). Heterogeneity of data was evaluated using the I2 statistics, with I2 greater than 50% considered significant heterogeneity (Higgins et al., 2003). Based on I2 value, a random effect (maximum likelihood) or fixed effect (inverse variance) model was adopted to perform the meta-analysis (Cochran, 1954). Summary ORs were represented as a point estimate and 95% confidence intervals (CIs) on a forest plot (Light et al., 1994), and publication bias was evaluated using regression based Egger's test and Begg's funnel plot (Begg and Mazumdar, 1994; Egger et al., 1997; Sterne et al., 2001). Subgroup analysis was performed based on pneumonia type (CAP and NP), populations (China and Russia) and age group (<12 years and > 12 years). Since M. pneumoniae infection is the most common form of CAP, the data from this article is included with CAP cohort. Patients having frequently recurring (relapsing) pneumonia (J18, according to the ICD-10) with unspecified organism of infection, this has also been included with CAP for easy interpretation. Sensitivity analyses were also performed to assess heterogeneity, to estimate the influence of any individual datasets, by omitting one study at a time and examining their influence on the combined effect.
We additionally conducted a spatial analysis (among countries) of the factors that might account for the variability among COVID-19 prevalence (total cases per million). Here we used country-specific demographic and socio-economic variables (such as population density, GDP, median age, and many others), to discover any association pattern. The COVID-19 dataset used in the study was downloaded from ourworldindata.org on May 24, 2021. The population specific allele frequency data for rs2606345 and rs1048943 were obtained from 1000 genome browser (Consortium, T. G. P, 2015) on March 08, 2021. For this purpose, we initially ran a linear regression fit between COVID-19 prevalence, and the country-wise distribution of risk allele of CYP1A1 SNPs (rs2606345, and rs1048943). To further strengthen the robustness and precision of above association, and to determine the influence of other confounding variables along with the allelic distribution of CYP1A1 variants with COVID-19 prevalence, we considered 20 other predictor variables (socio-economic and demographic factors). To reduce the skewness of the data, all the variables were log-transformed before entering into the regression models. We additionally removed predictor variables with >30% missing data. We next examined the univariate relationships between predictor variables and COVID-19 prevalence to find candidate variables for our final multivariable model. The variables with p < 0.05 were considered for the multivariable models. All the analyses were performed in R 3.6.3.
3. Results
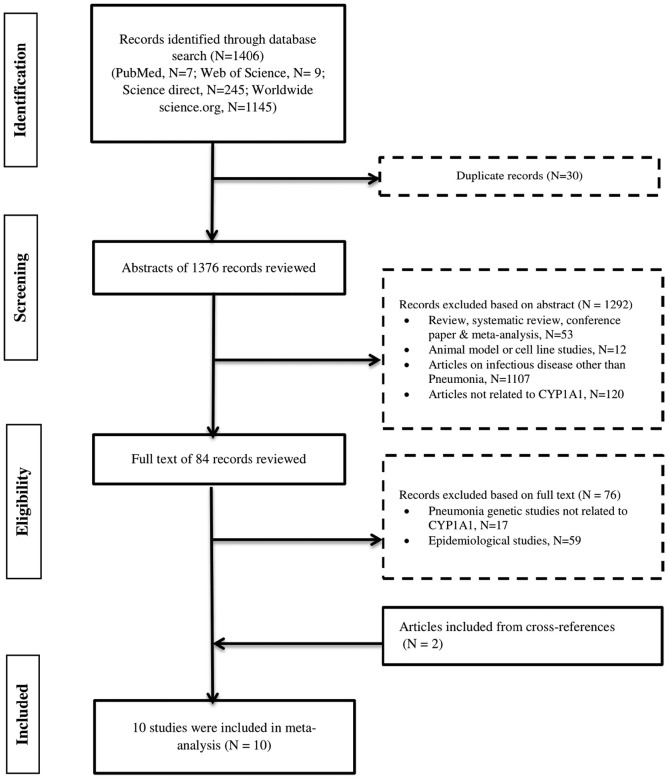
Through the initial search, a total of 1406 articles were identified (7 from PubMed, 9 from Web of science, 245 from Science Direct and 1145 from worldwidescience.org). Based on initial screening of titles and abstract 1292 publications were excluded after removing duplicates (n = 30), leaving 84 articles for full text review. Among them 17 articles were removed as they discussed some other gene but not CYP1A1 gene or its genetic variants and 59 articles were removed as they did not discuss any genetic association. Finally, 8 case control studies and 2 studies from their cross-references that met the pre-defined criteria, were included for the quantitative analysis. The flow chart for the study selection process is represented in Fig. 1 . The current systematic search included ten studies totalling 5298 subjects (3049 cases and 2249 healthy controls). The study population primarily comprised of well-characterized cohorts in Russia (Korytina et al., 2005; Moroz et al., 2011; Salnikova et al., 2013a; Salnikova et al., 2013b, Salnikova et al., 2013c; Salnikova et al., 2014; Salnikova et al., 2008; Salnikova et al., 2010; Smelaya et al., 2011) and one included Chinese cohort (Zhao et al., 2017). The mean age of all the pooled participants was 29.33 ± 5.49 years (31.16 years for cases and 27.49 years for controls). All the studies discuss the association of CYP1A1 genetic variants (rs2606345, rs1048943, rs4646903) with the risk of pneumonia. The demographic characteristics and clinical details of all the included studies are provided in Table 1 . For cumulative quality assessment, three of ten articles were deemed as good quality (cut off score of ≥7), six articles (≥5–6 score) were categorised under moderate, and finally any scores below 5 were judged as poor quality which included one article (Supplementary table 1).
Fig. 1.
Flow chart of study selection in metaanalysis of CYP1A1 polymorphisms with Pneumonia risk.
Study methodology for the inclusion and exclusion of studies exploring the role of CYP1A1 genetic variants in pneumonia patients. The number of studies excluded on each step is represented as N.
Table 1.
Main characteristic of studies included in meta-analysis for CYP1A1 genetic variants associated with risk of pneumonia.
| Study details |
Case |
Control |
Variant details |
Genotypic |
Allelic |
Score |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Study (year) [Ref] | Population (Ethnicity) | Genotyping method | Disease | M | F | Total | Age (in years) |
M | F | Total | Age (in years) | Studied CYP1A1 variants (alt. allele) | Alt. allele frequency | p value | OR (95%CI) | p value | OR (95%CI) |
Quality |
| 1 | Zhao J., et al. (2017) (Zhao et al., 2017) | China (East Asian) |
PCR Sequencing | MPP | 225 | 190 | 415 | 5.13± 2.81 |
154 | 146 | 300 | 5.02 ± 1.63 | rs2606345 (T) | 93.83 | TT (<0.0001) | TT 11.38 (6.29-0.57) |
T(0.764) | 1.07 (0.68–1.66) |
7 |
| 2 | Salnikova, L. E., et al. (2013) (Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c) | Russia (European) | Allele specific tetra-primer PCR | CAP | 307 | 27 | 334 | 26.93± 0.42 |
130 | 11 | 141 (without CAP) | 21.06 ± 0.42 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | 57.83, 12.59, 0.03 | T/T rec (3.9×10-5), TT (0.093), AA (0.188) | TT 2.40 (1.59-3.64), TT 1.54 (0.95-2.50), AA 0.58 (0.26-1.30) |
T(<0.0001), T(0.117), A(0.19) | T 1.90 (1.41–2.55), T 1.43 (0.91–2.25), A 0.59 (0.27–1.3) |
5 |
| 286 | 28 | 314 (Healthy) | 41.65 ± 1.03 | 63.18, 11.25, 0.04 | T/T rec (1.4 × 10-5), TT(0.220), AA(0.0780) | TT 2.00 (1.46-2.74), TT 1.30 (0.87-1.94), AA 0.88 (0.51-1.53) |
T(0.00045), T(0.22), A(0.56) | T 1.5 (1.20–1.9), T 1.2 (0.86–1.83), A 0.68 (0.60–1.3) |
|||||||||||
| 3 | Salnikova, L. E., et al. (2014) (Salnikova et al., 2014) | Russia (European) | PCR-CTPP | NP | 224 | 44 | 268 | 43.1± 1.2 |
116 | 35 | 151 | 42.5 ± 1.5 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | 61.0, 17.33, 0.04 | TT (0.324), TT( 0.377), AA (0.88) | TT 1.23 (0.81-1.86), TT 0.79 (0.47-1.32), AA 1.05 (0.50-2.22) |
T(0.32), T(0.36), A(0.89) | T 1.16 (0.86–1.55), T 0.79 (0.48–1.29), A 1.05 (0.50–2.18) |
7 |
| 4 | Salnikova, L. E., et al. (2008) (Salnikova et al., 2008) | Russia (European) | PCR - Genotyping | CAP | NA | NA | 99 (CAP) | 30.2± 13.1 |
NA | NA | 160 | 21.5 ± 5.5 | rs1048943 (G) | 0.03 | AA (0.035) | AA 0.39 (0.16-0.96) |
A (0.039) | A 0.41 (0.17–0.98) |
4 |
| NP | 57 (NP) | 48.0± 14.7 |
AA (NA) | AA 0.61 (0.20-1.93) |
A (NA) | A 0.63 (0.20–1.92) |
|||||||||||||
| 5 | Korytina, G. F., et al. (2005) (Korytina et al., 2005) | Russia (European) | PCR-RFLP | RP | 33 | 17 | 50 | 11.4± 1.7 |
94 | 133 | 227 | 12.5 ± 1.3 | rs1048943 (G) | 0.02 | AA (0.031) | AA 0.25 (0.08-0.72) |
A(0.009) | A 0.25 (0.099–0.67) |
6 |
| 6 | Salnikova, L. E., et al. (2010) (Salnikova et al., 2010) | Russia (European) | Allele specific PCR genotyping | CAP | NA | NA | 243 | NA | NA | NA | 178 | 21.53 ± 5.49 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | 64.04, 10.95, 0.03 | TT (0.01) | TT 1.61 (1.09-2.38) |
T(0.01) | T 1.43(1.06–1.91) | 5 |
| 7 | Smelaya T.V., et al. (2011) (Smelaya et al., 2011) |
Russia (European) | Comprehensive PCR based | CAP | NA | NA | 277 (CAP) | 25.29± 8.01 |
NA | NA | 178 | NA | rs2606345 (T) | 63.48 | TT (0.011) | TT1.6 (1.11-2.43) |
T(0.0103) | T 1.46 (1.093–1.963) |
5 |
| NP | 158 (NP) | 43.70 ± 17.69 | TT (1 x 10-4) | TT(0) | T(<0.0001) | T 0.48 (0.35–0.65) |
|||||||||||||
| 8 | Moroz, V. V., et al. (2011) (Moroz et al., 2011) | Russia (European) | Allele specific tetra-primer PCR | CAP | 307 | 27 | 334 | 26.9 ± 0.8 | 130 | 11 | 141 | 29.1 ± 0.6 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | 64.04, 11.76, 0.04 | T 1.90 (1.41–2.55), T 1.43 (0.91–2.25), A 0.59 (0.27–1.31) |
6 | |||
| NP | 176 | 40 | 216 | 43.0± 2.0 |
83 | 22 | 105 | 41.0 ± 1.6 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | T 1.31 (0.93–1.85), T 0.91 (0.52–1.59), A 1.32 (0.58–3.00) |
|||||||||
| 9 | Salnikova, L. E., et al. (2013)(Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c) | Russia (European) | Allele specific tetra-primer PCR | CAP | 321 | 29 | 350 | 27.2± 0.8 |
343 | 89 | 432 | 30.0 ± 0.7 | rs2606345 (T), rs4646903 (C), rs1048943 (G) | 69.15, 11.77, 0.04 | T 1.58 (1.27–1.96), T 1.28 (0.91–1.80), A 0.86 (0.53–1.39) |
6 | |||
| 10 | Salnikova, L. E., et al. (2013) (Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c) | Russia (European) | Allele specific tetra-primer PCR | NP | 224 | 44 | 266# | 43.1± 1.2 |
116 | 35 | 150 | 42.5 ± 1.5 | rs2606345 (T), rs4646903 (C), | 62.38, 11.13, 0.04 | T 0.86 (0.64–1.15). T 1.25(0.77–2.04), A 0.94 (0.45–1.96) |
7 | |||
Bold characters highlight important phenotypic groupings and their total counts in the respective study.
M, male; F, female; PCR, polymerase chain reaction; Age of the participants shown in Mean ± Standard deviation. PCR-CTPP, polymerase chain reaction- confronting two-pair primers; RFLP, restriction fragment length polymorphism; MPP, mycoplasma pneumoniae pneumonia; CAP, community acquired pneumonia; NP, nosocomial pneumonia; RP, relapsing pneumonia; alt. Allele, alternate allele for respective SNP; allele frequency (denoted in per cent) of alternate allele calculated from respective study in control population, OR, odds ratio; CI, confidence interval; dom, dominant model; rec, recessive model.
All p values represented are uncorrected. #male/female count not given for 3 samples.
Quality assessment was performed using modified NOS scale (Wells G A, 2001) the detailed scoring can be found in Suppl. Table 1.
All the citations are as in the main manuscript file.
3.1. Meta-Analysis Results
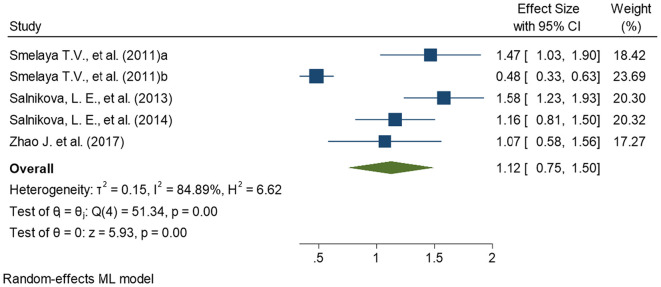
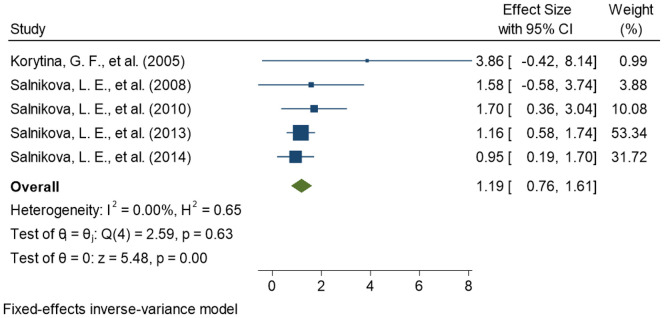
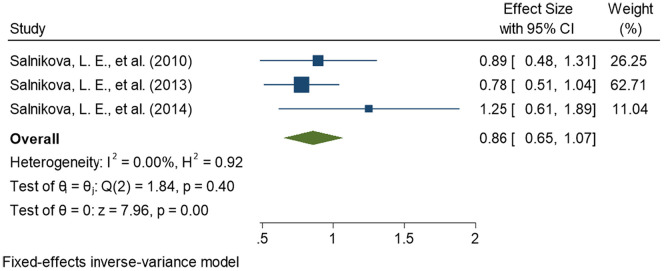
This meta-analysis compares pneumonia patients as cases, comprising CAP and NP subjects both, with healthy controls for association of CYP1A1 genetic variants with pneumonia susceptibility To maintain the precision in assessing the effect size in each meta-analyses performed, we removed all the studies (n = 3) with overlapping samples before meta-analysis (Moroz et al., 2011; Salnikova et al., 2013b, Salnikova et al., 2013c). Finally, a total of seven studies were included for the meta-analyses. Among the seven included studies, four provided the data on rs2606345 (Salnikova et al., 2013a; Salnikova et al., 2014; Smelaya et al., 2011; Zhao et al., 2017), five provided data on rs1048943 (Korytina et al., 2005; Salnikova et al., 2013a; Salnikova et al., 2014; Salnikova et al., 2008; Salnikova et al., 2010) and three provided the data on rs4646903 (Salnikova et al., 2013a; Salnikova et al., 2014; Salnikova LE, 2010). The details of included studies, their sample size, population, risk allele, and allele distribution in cases and controls for each SNP are represented in Table 2 . Of all the CYP1A1 variants studied, we observed the most significant association of rs2606345, and rs1048943 but no significant association was established for rs4646903. Our meta-analysis demonstrated that CYP1A1 genetic polymorphisms significantly correlated with the increased risk of pneumonia under the allelic model for rs2606345 [T vs G; OR = 1.12 (0.75–1.50); p < 0.02; I2 = 84.89%], and rs1048943 [G vs T; OR = 1.19 (0.76–1.61); p < 0.02; I2 = 0.00%] as risk markers (Fig. 2, Fig. 3 ) however no statistical significance was achieved for rs4646903 [C vs T; OR = 0.86 (0.65–1.07); p = 0.4; I2 = 0.00%] (Fig. 4 ), when compared with healthy controls.
Table 2.
Pooled odds ratio for allelic comparisons for studies exploring association of CYP1A1 variants- rs2606345, rs4646903, rs1048943 in patients with risk of pneumonia.
| Gene (SNP) |
Risk allele | No. of studies | Population | Total samples | All patients |
Total | Control |
Total | OR (95%CI) |
p value | I2 | Model | Test of publication bias |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk allele present | Risk allele absent | Risk allele present | Risk allele absent | Egger's test | |||||||||||
| CYP1A1 (rs2606345) | T | 4 | Chinese, Russian | 2663 | 2115 | 735 | 2850 | 1511 | 609 | 2120 | 1.12 (0.75-1.50) |
0.02 | 84.89 | R | 0.0029 |
| CYP1A1 (rs1048943) | G | 5 | Russian | 2107 | 91 | 1827 | 1918 | 78 | 2218 | 2296 | 1.19 (0.76-1.61) |
0.02 | 0.00 | F | 0.1719 |
| CYP1A1 (rs4646903) | C | 3 | Russian | 1609 | 165 | 1533 | 1698 | 161 | 1357 | 1518 | 0.86 (0.65-1.07) |
0.40 | 0.00 | F | 0.1828 |
Bold characters highlight significantly associated alleles with respective P values.
All patients include patients of CAP and NP both; OR, odds ratio; CI, confidence interval; F, Fixed effect model.
All p values calculated using chi-square test.
Fig. 2.
Forest plot determining association of CYP1A1 variant, rs2606345, with pneumonia.
The square and horizontal lines correspond to the study- specific odds ratio(OR) and 95% confidence interval (CI). The area of the square refers to the study specific weight (random model; maximum likelihood).The diamond represents the summary of OR and 95% CI. Smelaya et al., 2011 includes CAP patients; Smelaya et al. (2011) includes NP patients.
Fig. 3.
Forest plot determining association of CYP1A1 variant, rs1048943, with pneumonia.
The square and horizontal lines correspond to the study- specific odds ratio(OR) and 95% confidence interval (CI). The area of the square refers to the study specific weight (Fixed effect; inverse of variance).The diamond represents the summary of OR and 95% CI.
Fig. 4.
Forest plot determining association of CYP1A1 variant, rs4646903, with pneumonia.
The square and horizontal lines correspond to the study- specific odds ratio(OR) and 95% confidence interval (CI). The area of the square refers to the study specific weight (fixed effect; inverse of variance).The diamond represents the summary of OR and 95% CI.
3.2. Test for publication bias
As shown in Supplementary fig. 1, visualization of the Begg's funnel plot suggested that Egger's linear regression test yielded evidence of publication bias among the included studies, therefore, we further performed subgroup analysis and sensitivity analysis to assess the robustness and consistency of our meta-analysis findings.
3.3. Subgroup and sensitivity analysis
Due to heterogeneity in the meta-analysis, we attempted to subgroup the studies. Subgroup analysis was performed based on pneumonia type (CAP and NP), different population (China and Russia), and age group (<12 years and > 12 years) (Table 3 , Supplementary fig. 5). Significant association was observed for CAP subgroup for rs2606345 [OR = 1.43 (1.19–1.66); p < 0.0001] and rs1048943 [OR = 1.29(0.76–1.18); p = 0.02]. Sensitivity analysis results are as represented in Table 4 , showing no change from overall effect size after removing one study at a time.
Table 3.
Sub-group analysis for included studies comparing pneumonia patients with healthy controls for association of CYP1A1 genetic variants with pneumonia risk based on different subgroups like pneumonia type, population and age.
| S.No. | SNP | Subgroup characteristics | Number of studies | Heterogeneity |
OR (95% CI) | p value | ||
|---|---|---|---|---|---|---|---|---|
| I2 | p value | |||||||
| 1 | rs2606345 | Pneumonia subtype | CAP | 3 | 0.00 | 0.24 | 1.43 (1.19–1.66) | <0.0001 |
| NP | 2 | 83.87 | 0.00 | 0.78 (0.32–1.25) | 0.06 | |||
| Population | China | 1 | – | – | 1.07 (0.58–1.56) | 0.76 | ||
| Russia | 4 | 88.63 | 0.00 | 1.14 (0.70–1.59) | 0.03 | |||
| Age | <12 years | 1 | – | – | 1.07 (0.58–1.56) | 0.76 | ||
| >12 years | 4 | 88.63 | 0.00 | 1.14 (0.70–1.59) | 0.03 | |||
| 2 | rs1048943 | Pneumonia subtype | CAP | 3 | 0.00 | 0.38 | 1.29(0.76–1.18) | 0.02 |
| NP | 2 | 0.00 | 0.59 | 1.02(0.30–1.73) | 0.63 | |||
| Age | <12 years | 1 | – | – | 3.86 (−0.42–8.14) | 0.007* | ||
| >12 years | 4 | 0.00 | 0.78 | 1.16(0.74–1.59) | 0.18 | |||
| 3 | rs4646903 | Pneumonia subtype | CAP | 2 | 0.00 | 0.64 | 0.81(0.59–1.04) | 0.15 |
| NP | 1 | – | – | 1.25 (0.61–1.89) | 0.36 | |||
Bold characters highlight significantly associated alleles with respective p values.
CAP, Community acquired pneumonia; NP, nosocomial pneumonia;
All p values calculated using chi square test, except * where p value calculated using Fischer exact test.
Table 4.
Sensitivity analysis after each study was excluded by turns.
| S.No. | SNP | No. Studies | Study Omitted | Pooled OR (95% CI) for remainders | p value | Heterogeneity |
|
|---|---|---|---|---|---|---|---|
| I2 | p value | ||||||
| 1 | rs2606345 | 4 | Zhao et al. (2017) | 1.14 (0.70–1.59) | 0.025 | 88.63 | 0.00 |
| Salnikova et al. (2014) | 1.12 (0.66–1.58) | 0.01 | 87.63 | 0.00 | |||
| Smelaya TV et al. (2011)a | 1.05 (0.63–1.47) | 0.18 | 86.36 | 0.00 | |||
| Smelaya TV et al. (2011)b | 1.34 (1.13–1.55) | 0.0002 | 12.48 | 0.22 | |||
| Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c | 1.00 (0.62–1.39) | 0.09 | 81.53 | 0.00 | |||
| 2 | rs1048943 | 5 | Salnikova et al. (2014) | 1.30 (0.70–1.82) | 0.008 | 0.00 | 0.57 |
| Salnikova et al. (2008) | 1.17(0.74–1.61) | 0.05 | 0.00 | 0.48 | |||
| Korytina et al. (2005) | 1.16 (0.74–1.59) | 0.18 | 0.00 | 0.78 | |||
| Salnikova et al. (2010) | 1.13 (0.68–1.58) | 0.09 | 0.00 | 0.58 | |||
| Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c | 1.22 (0.60–1.85) | 0.02 | 0.00 | 0.46 | |||
| 3 | rs4646903 | 3 | Salnikova et al. (2014) | 0.81 (0.59–1.04) | 0.15 | 0.00 | 0.64 |
| Salnikova et al. (2010) | 0.85 (0.60–1.09) | 0.48 | 44.64 | 0.18 | |||
| Salnikova et al., 2013a, Salnikova et al., 2013b, Salnikova et al., 2013c | 1.00 (0.65–1.35) | 0.79 | 0.00 | 0.36 | |||
Smelaya TV et al. (2011)a includes CAP patients; Smelaya TV et al. (2011)b includes NP patients.
Significant p values are represented with bold characters.
3.4. Genetic variability of CYP1A1 genetic variants in global populations and a possible link to COVID-19
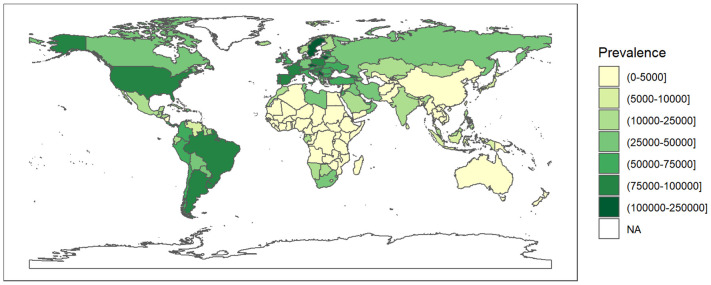
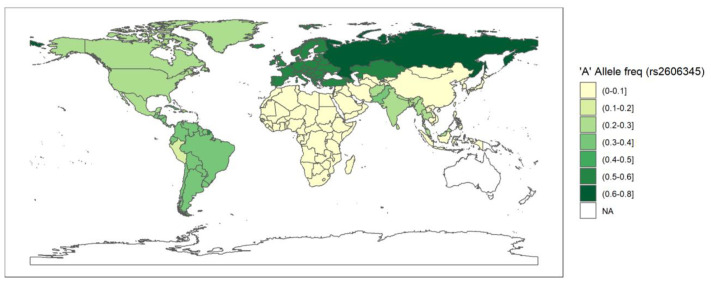
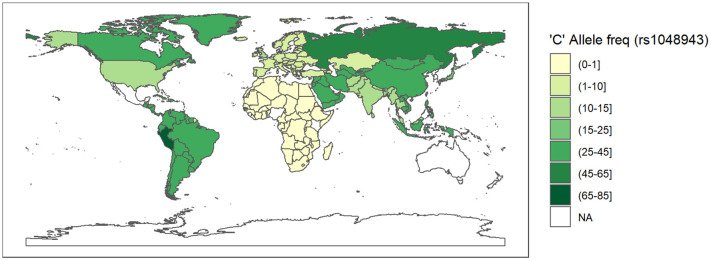
Since rs2606345 (A) is the major allele in Europeans (66.6%) but not in other populations (African 5%, Asian 5–30%) (Consortium, T. G. P, 2015), we explored its possible association, and the risk allele (C) of rs1048943, with high variability in regional prevalence of the ongoing COVID-19 pandemic, through a spatial analysis. The COVID-19 prevalence varied widely across countries (Fig. 5 ). To assess the pattern of association between country-specific prevalence with all the candidate predictor variables (as shown in Supplementary table 2) from a univariate regression analysis for each individual predictor. We found that all the variables except population, population density, diabetes prevalence, and male smokers were significantly associated with prevalence in these univariate models. Excluding these non-significant variables and variables with >30% missing data (ICU patients per million, hospital patients per million, total tests per thousand, poverty, and female smokers), all the remaining variables were included the multivariable linear regression model where we sequentially added them in one-by-one and evaluated the model fit. The variable producing the best fit was retained in the model. Our final model included total death per million (p < 2 × 10–16), percentage of population with age > 65 older (p = 1.78 × 10–5), human development index (p = 1.73 × 10–10), stringency index (p = 0.0286), SNP frequency for rs2606345A (p = 0.0005) and SNP frequency for rs1048943G (p = 0.01). This model had good agreement between these variables with total cases per million across countries (r2 = 0.8983, p < 2.2 × 10–16). The linear model regression exploring the variation in COVID-19 prevalence with the allele frequency data (for rs2606345 and rs1048943) for each country are depicted in Fig. 6, Fig. 7 . This model showed a significant association between COVID-19 prevalence and risk allele frequency of rs2606345 and rs1048943 along with other covariates (age > 65 older, human development index and stringency index.
Fig. 5.
A geospatial distribution map of prevalence (the number of cases per million) as on 24 May 2021 worldwide due to COVID-19.
COVID-19 prevalence is extracted from ourworldindata.org till 24 May 2021. The white colored areas in the map show the absence of data. A half open intervals includes only one of its end-points and is denoted by mixing notations for open and closed intervals. For e.g., (0–1] means greater than 0 and less than or equal to 1 and [0, 1) means greater than or equal to 0 and less than 1.
Fig. 6.
A geospatial frequency maps depicting the distribution of risk allele ‘A' frequency of rs2606345 (CYP1A1).
Fig. 7.
A geospatial frequency maps depicting the distribution of risk allele ‘C' frequency of rs1048943 (CYP1A1).
The data is obtained from population frequency data of the 1000genome browser on 8 March 2021. The white coloured areas in the map show the absence of data. A half open intervals includes only one of its end-points and is denoted by mixing notations for open and closed intervals. For e.g., (0–1] means greater than 0 and less than or equal to 1 and [0,1) means greater than or equal to 0 and less than 1.
Since the CYP1A1 variants (rs2606345, and rs1048943) showed similar association with pneumonia as well as its relationship with COVID-19 prevalence, we additionally checked if these SNPs are in linkage disequilibrium (LD). The results from SNiPA tool revealed these SNPs are not in LD (Supplementary table 3).
4. Discussion
This study is an attempt to investigate the role of host genetic factors in pneumonia susceptibility. Most infectious diseases, like pneumonia, are a consequence of a complex network of host genetics and pathogen genetic factors that may be inducible by several non-genetic factors. The host immune-mediated response determines the course of the disease, its susceptibility, progression, and severity. Such immune response related genes may serve as good candidates in establishing a genetic association in infectious diseases. A few genetic studies investigated the role of CYP1A1 gene and its variants with development and outcome of CAP and NP (Salnikova et al., 2013b; Salnikova et al., 2008; Smelaya et al., 2011). CYP1A are a family of monooxygenase enzymes involved in biotransformation reactions (Danielson, 2002). Presence of CYP1A1 polymorphic variants changes the gene expression and/ activity resulting in the altered redox balance (Stading et al., 2020). This imbalance can cause chronic inflammation in the lungs aggravating the disease pathogenesis (Hussain et al., 2014; Stading et al., 2020). This study provides the first meta-analysis results reiterating the prior experimental observations that suggest a genetic contribution of CYP1A1 variants (rs2606345, rs1048943, rs4646903) establishing a more precise risk estimate of pneumonia. We additionally noted, the genetic variant (rs2606345 and rs1048943) that are crucial for CYP1A1 activity, is statistically associated with COVID-19 prevalence. This study may provide us insights towards understanding the role of CYP1A1 genetics, with inflammation, pneumonia as well as COVID-19 susceptibility.
The CYP P450 enzymes are primarily known to metabolise xenobiotics (Danielson, 2002), and also endobiotics that are derived from different physiological or pathological processes such as inflammation (Bui et al., 2011). There have been studies that tried to deduce the interplay between CYP1A1 gene and lung infections. Some studies attempted to establish an association of the CYP1A1 genetic variants with pneumonia (Korytina et al., 2005; Salnikova et al., 2013c; Salnikova et al., 2014; Zhao et al., 2017), others related the changes in its expression and activity with the infection (Fang et al., 2016; Fang et al., 2015). On the basis of such evidence, CYP1A1 seems to be an important risk factor for pneumonia. Several investigations reported the change in CYP1A1 activity by infection or inflammatory stimuli; where the activity is induced in some cases or downregulated in other (Morgan, 2001; Santes-Palacios et al., 2016). Tian et al. (2020) in their study found upregulation of CYP1A1 in peritoneal macrophages activated by bacterial lipopolysaccharides (LPS) (Tian et al., 2020). Additionally, an increased production of pro-inflammatory cytokines like TNF-α and IL-6 was observed in CYP1A1 overexpressed macrophage cells after LPS stimulation. Moreover, CYP1A1 upregulation reduced the bacterial phagocytosis by decreasing the expression of macrophage channel, SR-A. The authors concluded CYP1A1 as an important driver of inflammation and sepsis (Tian et al., 2020). On the contrary, in LPS-stimulated bovine mammary epithelial cells, CYP1A1 expression was drastically suppressed as compared to controls along with the elevated expression of TNF-α and IL-6. This effect was attenuated upon CYP1A1 overexpression (Hussain et al., 2014; Zhang et al., 2018). Further, Fang et al. (2015) also reported CYP1A1 downregulation in pig infected with M. hyopneumoniae (46). This study was extended in pulmonary alveolar macrophage (PAM) cells, where inflammatory response, caused by M. hyopneumoniae infection, was supressed upon CYP1A1 overexpression, via PPAR-γ signalling pathway (8). Comparably, the genetic polymorphism included in our study, rs2606345, determining lower CYP1A1 expression, exacerbate similar role in lung inflammation. Such evidence demonstrated that inflammatory stimuli or infection regulate CYP1A1 expression, in turn affecting cytokine production. These responses are species and tissue specific (Morgan et al., 1994.; Ryan and Levin, 1993). Thus, the above findings depict immense potential of CYP1A1 in inflammation and may serve as a useful target in mitigating infections like pneumonia.
Our study revealed the association of rs2606345 and rs1048943 with risk of pneumonia, particularly CAP. The alternate allele (A in plus-strand or T in minus-strand) of rs2606345 increased pneumonia susceptibility. Despite following a stringent criterion for inclusion of studies in the present study, the possibility of observer bias in diagnosing pneumonia cannot be ruled out when dealing with retrospective records. We, therefore advice readers caution, when interpreting the results. There is a need to conduct large scale cohort studies in future to validate our findings. We also observed, this allele to be the major allele in European (66.6%) and Russian (~80%) population unlike in the other populations (African 5%, Asian 5–30%, American 39%) (Consortium, T. G. P, 2015). Interestingly, on spatial analysis, we noted significant association of this allele frequency distribution with total cases per million due to the recent outbreak of COVID-19 (Fig. 6). Likewise, for rs1048943, higher the risk allele ‘C' frequency also showed association with COVID-19 prevalence, across countries worldwide (Fig. 7). We would also like to warn that since this is an ongoing pandemic, the numbers are changing with time and this is a circumstantial evidence.
The COVID-19, a viral pneumonia (Berlin et al., 2020; Gandhi et al., 2020 ), is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Kim et al., 2020; Wu et al., 2020; Yao et al., 2020; Zhang and Holmes, 2020). This activates the host immune system to release several inflammatory cytokines (IL-1, IL-6, TNFα, and IFNγ) (Li and Fan, 2020). There is a very fine balance between the immune response produced by the host cells on encountering SARS-CoV-2 and lung damage. The immune cells accelerate the production of cytokines to kill the pathogen. In case of cytokine rush, such accelerated immune response may in turn, damage the host cells. Several studies correlated the downregulation of CYP1A1 with increasing pro-inflammatory cytokines levels (IL-6, TNF-α, IL-1, IL-1β) (Wang et al., 2022). In this study we speculate that CYP1A1 could be crucial in COVID-19 prevalence. Since this study establishes the association of CYP1A1 genetic variants with pneumonia susceptibility and one of the major symptoms in patients dying with COVID-19 infection is pneumonia (Guan et al., 2020; Surendra et al., 2021). Pneumonia may be considered a proxy phenotype for studying association with CYP1A1 with COVID-19. We also observed in this study, there is a positive correlation of ‘A' allele frequency of CYP1A1 SNP (rs2606345) with COVID-19 prevalence among populations worldwide. Since CYP1A1 plays a vital role in innate immune response (inflammatory responses) against any kind of infection (Stading et al., 2020), specifically in lungs (Fang et al., 2016). According to the GTEx database (https://gtexportal.org/home/gene/CYP1A1), the expression of this gene is the highest in lungs with >1500 transcript per million (TPM) expression. This is almost 3 times higher than the average expression of CYP1A1 in other tissues like adipose, breast, liver, and skin (where the TPM is ~500) (GTEx Consortium, 2013, Lonsdale et al., 2013). Also, interestingly, the biological role of the allele ‘A' or ‘T' of rs2606345 is known to have ~70–80% reduced CYP1A1 promoter activity thereby reducing the enzyme activity as well (Talwar et al., 2017). Therefore, we speculate that due to the reduced enzyme activity in individuals carrying A or T allele, they are at a higher risk of pneumonia. A recent in vitro study demonstrated, CYP1A1 gene expression to be inversely proportional to the angiotensin-converting enzyme 2 (ACE2) gene expression. Decrease in ACE2 expression results in suppression of SARS-CoV-2 infection in mammalian cells (Tanimoto et al., 2021). This finding from Tanimoto et al. study are concurrent with our speculations establishing the role of CYP1A1 with COVID-19 prevalence. Hence, we believe exploring the role of theseCYP1A1 variants (rs2606345 and rs1048943) may provide us clues toward understanding COVID-19 pathophysiology. We agree that our data could be a preliminary information which warrants further validation. As observed, factors like demographic, socio-economic and environmental also account for variation in prevalence of COVID-19 across countries (Miller et al., 2020; Sorci et al., 2020).
There are several limitations to such interpretation of this study. As, meta-analysis findings often have several limitations. A common problem posed to the validity of a meta-analysis finding is publication bias. Due to the absence of studies reporting negative results, meta-analysis could produce skewed findings towards a positive result. Though unpublished material should be included in any meta-analysis performed, identification or sourcing such studies is difficult. Meta-analysis performed on individual patient data often demands extraordinary cooperation of all the investigating groups and meticulous integrity in data reporting to largely avoid publication, which is highly unlikely (Lyman and Kuderer, 2005). Further, meta-analyses outcomes statistically estimate the biological significance and their range of their variability based on available literature. Such studies are critical for hypothesis generation or quantitative evidence-based designing of translational studies. Evidence for heterogeneity within the primary studies affect the summary estimate. Heterogeneity due to confounding factors in the participants, intervention and outcome is a consequence of methodological or clinical variability and the true intervention effect will differ for different studies. Due to limited data availability, a limitations of this meta-analysis is that the summary estimates may not be robust due to high heterogeneity in age distribution, population and disease type of included cohort. We attempted to minimise the effect of these variables addressed in the subgroup analysis. However, more primary studies are required for true estimation of the summary effect. Another major limitation of our meta-analysis was lack of genetic association studies between CYP1A1 genetic variants and pneumonia from different populations our findings were limited to Russian population, primarily. Therefore, experimental studies are warranted to establish the functional significance. Similarly, our meta-analysis findings are limited, therefore further experimental validation in population specific pneumonia patient cohorts to establish the role of CYP1A1 variants in pneumonia infection. Next generation sequencing data of this region is inevitable to rule out the presence of other functional variants in the CYP1A1 gene region and its association with pneumonia, both CAP and NP. Due to limited data available, other factors which may influence the COVID-19 prevalence (like epigenetic, other comorbidities like cancer, other respiratory diseases, etc.) could not be included. Further, the uncertainty in estimating its prevalence while the pandemic is still on, and the limited population data and sample size in this meta-analysis are some major constraints.
To conclude, our meta-analysis demonstrates the association of CYP1A1 polymorphisms with CAP, where rs2606345 and rs1048943 are risk-associated and rs4646903 is protective. However, the lack of data in other ethnic groups prevented us from drawing conclusion of causality of the variant as presence of association in different ethnicities would have provided evidence in this direction. Though this is a statistical analysis, our findings along with the previous reports revealing the role of CYP1A1 in inflammatory response to infection, may help us deduce the importance of this gene in infectious disease like pneumonia. The direct evidences of the molecular mechanism by which CYP1A1 contributes to pneumonia pathogenesis is still yet to be explored. We also observed that genetic variant, rs2606345 and rs1048943, governing CYP1A1 expression may give us clues towards the genetic basis of COVID-19 prevalence across populations worldwide.
Author contributions
RK devised the concept of the review, reviewed the manuscript and supervised the overall study till final manuscript preparation. DG and SY performed the literature search independently and, reviewed articles and, extracted the data. DG generated the forest and funnel plots. PS (Priyanka Singh) performed the linear regression and calculated Pearson's correlation and prepared all the spatial analysis images. SG helped us through the meta-analysis and running the statistical analysis through STATA. DG and PS (Pooja Singh) performed the quality assessment of the included articles and prepared the supplementary material. Upon discrepancies, RK resolved the conflict. DG and SY wrote the manuscript. ST cross-checked all the data, re-calculated the numerical values and statistical analysis. NK, SK, PRP helped in manuscript writing, reference management, and preparation of tables. RK and AA reviewed the manuscript, figures and tables. DG, SY and NK edited the manuscript. SY and NK prepared the tables. RK, YH, and BP finally revised the figures, tables and the whole manuscript. VS improved the writing of the overall manuscript. RK supervised the whole study. AA gave valuable inputs regarding the COVID-19 prevalence and directed us to estimate the inter-ethnic difference for COVID-19 infection. AA and LS reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding information
Financial support for this research work has been provided by the Council of Scientific and Industrial Research (CSIR), grant number OLP1154
The data is obtained from population frequency data of the 1000genome browser on 8 March 2021. The white colored areas in the map show the absence of data. A half open intervals includes only one of its end-points and is denoted by mixing notations for open and closed intervals. For e.g., (0–1] means greater than 0 and less than or equal to 1 and [0,1) means greater than or equal to 0 and less than 1.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
We are highly indebted to our founder director, Prof. S.K. Brahmachari, whose vision in the field of genomics paved us a path. SY, PS (Pooja Singh), ST, NK, and PRP acknowledge CSIR, Government of India for providing their fellowships. DG, PS (Priyanka Singh) and SK acknowledges ICMR, UGC and DBT, Govt. of India, for their financial assistance, respectively. We thank the anonymous reviewers for their helpful suggestions in improving the manuscript. The authors are thankful to Dr. L.E. Salnikova, V. A. Negovsky Research Institute of General Reanimatology, Russian Academy of Medical Sciences, Moscow Neurology Division, for sharing genotypic and allelic data from their patient population.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2022.105299.
Appendix A. Supplementary data
Supplementary material
References
- Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Bui P., Imaizumi S., Beedanagari S.R., Reddy S.T., Hankinson O. Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase derived eicosanoids. Drug Metab. Dispos. 2011;39(2):180–190. doi: 10.1124/dmd.110.035121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran W.G. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. doi: 10.2307/3001666. [DOI] [Google Scholar]
- Consortium, T. G. P A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke G.S., Hill A.V. Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- Danielson P.B. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 2002;3(6):561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- Dela Cruz C.S., Christiani D.C., Cormier S.A., Crothers K., Doerschuk C.M., Evans S.E., Goldstein D.R., Khatri P., Kobzik L., Kolls J.K., Levy B.D., Metersky M.L., Niederman M.S., Nusrat R., Orihuela C.J., Peyrani P., Prince A.S., Ramírez J.A., Ridge K.M., Sethi S., Suratt B.T., Sznajder J.I., Tsalik E.L., Walkey A.J., Yende S., Aggarwal N.R., Caler E.V., Mizgerd J.P. Future research directions in Pneumonia. NHLBI Working Group Report. Am. J. Respir. Crit. Care Med. 2018;198:256–263. doi: 10.1164/rccm.201801-0139WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.M., Zhao W.M., Fu Y.-F., Tu F., Li B.-X., Wang X.M., Zhao F., Ren S.W. Difference in susceptibility to mycoplasma pneumonia among various pig breeds and its molecular genetic basis. Sci. Agric. Sin. 2015;48(14):2839–2847. doi: 10.3864/j.issn.0578-1752.2015.14.015. [DOI] [Google Scholar]
- Fang X., Zhao W., Xu J., Tu F., Wang X., Li B., Fu Y., Ren S. CYP1A1 mediates the suppression of major inflammatory cytokines in pulmonary alveolar macrophage. Dev. Comp. Immunol. 2016;65:132–138. doi: 10.1016/j.dci.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Gandhi R.T., Lynch J.B., Del Rio C. Mild or moderate Covid-19. N. Engl. J. Med. 2020;383:1757–1766. doi: 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., DSC Hui, Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold C.J., Sailer J.G. Community-acquired and nosocomial pneumonia. Eur. Radiol. Suppl. 2004;3:E2–20. doi: 10.1007/s00330-003-2162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T., Al-Attas O.S., Al-Daghri N.M., Mohammed A.A., De Rosas E., Ibrahim S., Vinodson B., Ansari M.G., El-Din K.I. Induction of CYP1A1, CYP1A2, CYP1B1, increased oxidative stress and inflammation in the lung and liver tissues of rats exposed to incense smoke. Mol. Cell. Biochem. 2014;391(1–2):127–136. doi: 10.1007/s11010-014-1995-5. [DOI] [PubMed] [Google Scholar]
- Jain V., Yilmaz G., Bhardwaj A. StatPearls Publishing; Treasure Island (FL): 2021. Pneumonia Pathology. StatPearls [Internet]https://www.ncbi.nlm.nih.gov/books/NBK526116/ 2021. Available online. [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korytina G.F., Yanbaeva D.G., Babenkova L.I., Etkina E.I., Victorova T.V. Genetic polymorphisms in the cytochromes P-450 (1A1, 2E1), microsomal epoxide hydrolase and glutathione S-transferase M1, T1, and P1 genes, and their relationship with chronic bronchitis and relapsing pneumonia in children. J. Mol. Med. (Berl) 2005;83(9):700–710. doi: 10.1007/s00109-005-0660-6. [DOI] [PubMed] [Google Scholar]
- Kumar V., Wijmenga C., Xavier R.J. Genetics of immune-mediated disorders: from genome-wide association to molecular mechanism. Curr. Opin. Immunol. 2014;31:51–57. doi: 10.1016/j.coi.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Fan J.G. Characteristics and mechanism of liver injury in 2019 coronavirus disease. J. Clin. Transl. Hepatol. 2020;8(1):13–17. doi: 10.14218/JCTH.2020.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339 doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light R.J., Singer J.D., Willett J.B. In: The handbook of research synthesis. Cooper H., Hedges L.V., editors. Russell Sage Foundation; New York: 1994. The visual presentation and interpretation of meta-analysis; pp. 439–454. [Google Scholar]
- Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman G.H., Kuderer N.M. The strengths and limitations of meta-analyses based on aggregate data. BMC Med. Res. Methodol. 2005;5:14. doi: 10.1186/1471-2288-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie G. The definition and classification of pneumonia. Pneumonia. 2016;8:14. doi: 10.1186/s41479-016-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J., Chanock S., Walsh T.J. Human pharmacogenomic variations and their implications for antifungal efficacy. Clin. Microbiol. Rev. 2006;19(4):763–787. doi: 10.1128/CMR.00059-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.E., Bhattacharyya R., Miller A.L. Data regarding country-specific variability in Covid-19 prevalence, incidence, and case fatality rate. Data Brief. 2020;32 doi: 10.1016/j.dib.2020.106276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E.T. Regulation of cytochrome p450 by inflammatory mediators: why and how? Drug Metabolism. 2001;29:207–212. [PubMed] [Google Scholar]
- Morgan E.T., Thomas K.B., Swanson R., Vales T., Hwang J., Wright K. Selective suppression of cytochrome P-450 gene expression by interleukins 1 and 6 in rat liver. Biochim. Biophys. Acta. 1994;1219(475) doi: 10.1016/0167-4781(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Moroz V.V., Smelaya T.V., Salnikova L.E., Golubev A.M., Rubanovich A.V. Genetic study of predisposition to community-acquired pneumonia. Vestn. Ross. Akad. Med. Nauk. 2011;11:12–16. [PubMed] [Google Scholar]
- Muñoz B., Magaña J.J., Romero-Toledo I., Juárez-Pérez E., López-Moya A., Leyva-García N., López-Campos C., Dávila-Borja V.M., Albores A. The relationship among IL-13, GSTP1, and CYP1A1 polymorphisms and environmental tobacco smoke in a population of children with asthma in Northern Mexico. Environ. Toxicol. Pharmacol. 2012;33(2):226–232. doi: 10.1016/j.etap.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Rotunno M., Yu K., Lubin J.H., Consonni D., Pesatori A.C., Goldstein A.M., Goldin L.R., Wacholder S., Welch R., Burdette L., et al. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 2009;4, (5) doi: 10.1371/journal.pone.0005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D., Levin W. Handbook of Experimental Pharmacology. Springer; 1993. Age-and gender-related expression of rat liver cytochrome P450, Cytochrome P450; p. 461e476. [DOI] [Google Scholar]
- Salnikova L.Y., Smelaya T.V., Moroz V.V., Golubev A.M., Ponasenkov N.Kh., Khomenko R.V., Lapteva N.S., Kuznetsov G.I., Poroshenko G., Rubanovich A.V. Xenobiotic detoxification genes and their role in the development of pneumonia. Oboire. Animatologi. 2008;4:6. doi: 10.15360/1813-9779-2008-6-9. [DOI] [Google Scholar]
- Salnikova L.Y., Smelaya T.V., Moroz V.V., Golubev A.M., Lapteva N.S., Poroshenko G.G., Rubanovich A.V. Genetic predisposition to the development of acute community acquired pneumonia. Oboire. Animatologi. 2010;6, (1):5. doi: 10.15360/1813-9779-2010-1-5. [DOI] [Google Scholar]
- Salnikova L.E., Smelaya T.V., Golubev A.M., Rubanovich A.V., Moroz V.V. CYP1A1, GCLC, AGT, AGTR1 gene-gene interactions in community-acquired pneumonia pulmonary complications. Mol. Biol. Rep. 2013;40(11):6163–6176. doi: 10.1007/s11033-013-2727-8. [DOI] [PubMed] [Google Scholar]
- Salnikova L.E., Smelaya T.V., Moroz V.V., Golubev A.M., Rubanovich A.V. Functional polymorphisms in the CYP1A1, ACE, and IL-6 genes contribute to susceptibility to community-acquired and nosocomial pneumonia. Int. J. Infect. Dis. 2013;17(6):e433–e442. doi: 10.1016/j.ijid.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Salnikova L.E., Smelaya T.V., Moroz V.V., Golubev A.M., Rubanovich A.V. Host genetic risk factors for community-acquired pneumonia. Gene. 2013;518(2):449–456. doi: 10.1016/j.gene.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Salnikova L.E., Smelaya T.V., Vesnina I.N., Golubev A.M., Moroz V.V. Genetic susceptibility to nosocomial pneumonia, acute respiratory distress syndrome and poor outcome in patients at risk of critical illness. Inflammation. 2014;37(2):295–305. doi: 10.1007/s10753-013-9740-x. [DOI] [PubMed] [Google Scholar]
- Santes-Palacios R., Ornelas-Ayala D., Cabañas N., Marroquín-Pérez A., Hernández-Magaña A., Del Rosario Olguín-Reyes S., Camacho-Carranza R., Espinosa-Aguirre J.J. Regulation of human cytochrome P4501A1 (hCYP1A1): A plausible target for chemoprevention? Biomed. Res. Int. 2016;5341081 doi: 10.1155/2016/5341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelaya T.V., Salnikova L.E., Moroz V.V., Golubev A.M., Zarzecki V., Rubanovich A.V. Genetic polymorphism and the incidence of complication of pneumonia of various genesis. Oboire. Animatologi. 2011;7:2. [Google Scholar]
- Smith G.B., Harper P.A., Wong J.M., Lam M.S., Reid K.R., Petsikas D., Massey T.E. Human lung microsomal cytochrome P4501A1 (CYP1A1) activities: impact of smoking status and CYP1A1, aryl hydrocarbon receptor, and glutathione S-transferase M1 genetic polymorphisms. Cancer Epidemiol. Biomark. Prev. 2001;10(8):839–853. [PubMed] [Google Scholar]
- Sorci G., Faivre B., Morand S. Explaining among-country variation in COVID-19 case fatality rate. Sci. Rep. 2020;10(1):18909. doi: 10.1038/s41598-020-75848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stading R., Chu C., Couroucli X., Lingappan K., Moorthy B. Molecular role of cytochrome P4501A enzymes in oxidative stress. Curr. Opin. Toxicol. 2020;20-21:77–84. doi: 10.1016/j.cotox.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne J.A., Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J. Clin. Epidemiol. 2001;54(10):1046–1055. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- Sterne J.A.C., Bradburn M.J., Egger M. 2001. Meta-Analysis in Stata. Systematic Reviews In Health Care: Meta-Analysis in Context; pp. 347–372. [Google Scholar]
- Support, S. T. StataCorp . StataCorp LLC; College Station, TX: 2019. Citing Stata software, documentation, and FAQs. Stata Statistical Software: 2019, Release 16.https://www.stata.com/support/faqs/resources/citing-software-documentation-faqs/ Available online. [Google Scholar]
- Surendra H., Elyazar I.R., Djaafara B.A., Ekawati L.L., Saraswati K., Adrian V., Widyastuti, Oktavia D., Salama N., Lina R.N., et al. Clinical characteristics and mortality associated with COVID-19 in Jakarta, Indonesia: A hospital-based retrospective cohort study. Lancet Reg. Health West Pac. 2021;9 doi: 10.1016/j.lanwpc.2021.100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwar P., Kanojia N., Mahendru S., Baghel R., Grover S., Arora G., Grewal G.K., Parween S., Srivastava A., Singh M., et al. Genetic contribution of CYP1A1 variant on treatment outcome in epilepsy patients: a functional and interethnic perspective. Pharmacogenomics J. 2017;17(3):242–251. doi: 10.1038/tpj.2016.1. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Hirota K., Fukazawa T., Matsuo Y., Nomura T., Tanuza N., Hirohashi N., Bono H., Sakaguchi T. Inhibiting SARS-CoV-2 infection in vitro by suppressing its receptor, angiotensin-converting enzyme 2, via aryl-hydrocarbon receptor signal. Sci. Rep. 2021 Aug 17;11(1):16629. doi: 10.1038/s41598-021-96109-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.X., Tang X., Zhu J.Y., Luo L., Ma X.Y., Cheng S.W., Zhang W., Tang W.Q., Ma W., Yang X., et al. Cytochrome P450 1A1 enhances inflammatory responses and impedes phagocytosis of bacteria in macrophages during sepsis. Cell Commun Signal. 2020;18(1):70. doi: 10.1186/s12964-020-0523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A., Cilloniz C., Niederman M.S., Menéndez R., Chalmers J.D., Wunderink R.G., van der Poll T. Pneumonia. Nat Rev Dis Primers. 2021;7:25. doi: 10.1038/s41572-021-00259-0. [DOI] [PubMed] [Google Scholar]
- Wang G., Xiao B., Deng J., Gong L., Li Y., Li J., Zhong Y. The role of cytochrome P450 enzymes in COVID-19 pathogenesis and therapy. Front. Pharmacol. 2022 Feb 2;(13) doi: 10.3389/fphar.2022.791922. 791922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G.A., Shea B., O’Connell D., Peterson J., Welch V., Losos M., Tugwell P. University of Ottawa; 2001. The Newcastle–Ottawa Scale (NOS) for Assessing the Quality of Non randomised Studies in Meta-Analyses. [Google Scholar]
- WHO The top 10 Causes of Death. 2022. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death Available online. (accessed on 20 October 2021)
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183(3):730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.Y., Wang H., Qi S., Wang X., Li X., Zhou K., Zhang Y., Gao M.Q. CYP1A1 relieves lipopolysaccharide-induced inflammatory responses in bovine mammary epithelial cells. Mediat. Inflamm. 2018;4093285 doi: 10.1155/2018/4093285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang W., Shen L., Yang X., Liu Y., Gai Z. Association of the ACE, GSTM1, IL-6, NOS3, and CYP1A1 polymorphisms with susceptibility of mycoplasma pneumoniae pneumonia in Chinese children. Medicine. 2017;96:15. doi: 10.1097/md.0000000000006642. e6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material