Abstract
The pathological and immune response of individuals with COVID-19 display different dynamics in lung and intestine. Here, we depict the single-cell transcriptional atlas of longitudinally collected lung and intestinal tissue samples from SARS-CoV-2-infected monkeys at 3 to 10 dpi. We find that intestinal enterocytes are degraded at 3 days post-infection but recovered rapidly, revealing that infection has mild effects on the intestine. Crucially, we observe suppression of the inflammatory response and tissue damage related to B-cell and Paneth cell accumulation in the intestines, although T cells are activated in the SARS-CoV-2 infection. Compared with that in the lung, the expression of interferon response-related genes is inhibited, and inflammatory factor secretion is reduced in the intestines. Our findings indicate an imbalance of immune dynamic in intestinal mucosa during SARS-CoV-2 infection, which may underlie ongoing rectal viral shedding and mild tissue damage.
Keywords: SARS-CoV-2, rhesus macaque, single-cell transcriptomics, intestinal mucosal immunity
Graphical abstract
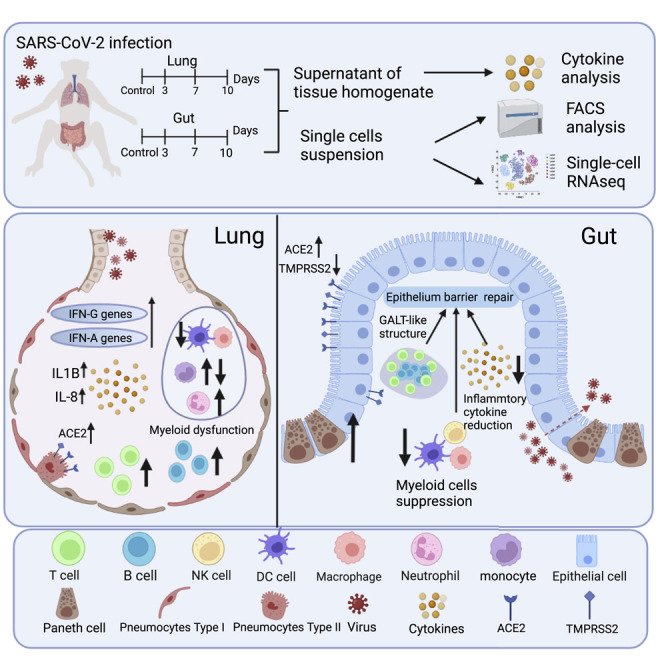
Zheng et al. reveal distinct immune responses in lung and intestine of SARS-CoV-2-infected rhesus monkeys by scRNA-seq, which indicates that the gut-viral ongoing shedding is related to intestinal mucosal immunosuppression. They further demonstrate the roles of T, B, and Paneth cells in epithelial barrier repair and dynamics of inflammatory cytokines of intestines.
Introduction
SARS-CoV-2 has spread rapidly around the world, with the number of confirmed cases globally exceeding 500 million and the number of cumulative deaths reaching 6.2 million (World Health Organization, 2022). The rapid development of vaccines has contributed to a reduction in the number of severe COVID-19 cases (Li et al., 2021; Tenforde et al., 2021), but there remains a lack of specific antiviral therapy for COVID-19 patients and effective control of viral spread (Caracciolo et al., 2021; Simsek Yavuz and Unal, 2020), especially when we are facing an increasing number of waves of breakthrough infections caused by SARS-CoV-2 variants (Ahmad, 2021). In this context, further exploring viral infection patterns and improving our understanding of host immune response mechanisms systematically is necessary to control this pandemic effectively.
Among the clinical manifestations of SARS-CoV-2 infection, fever and cough are the most common symptoms (Kevadiya et al., 2021; Morgan et al., 2020), but 2%–10% of COVID-19 patients exhibit gastrointestinal symptoms such as vomiting, diarrhea, and abdominal pain (Kariyawasam et al., 2021; Zoghi et al., 2021). Notably, a higher SARS-CoV-2 RNA load in rectal swabs was associated with severe COVID-19 outcomes, including death and ICU admission (Li et al., 2020), and the duration of virus in stool samples (22 days) was significantly longer than that in respiratory (18 days) and serum samples (16 days) (Zheng et al., 2020b), indicating that gastrointestinal tract samples showed better maintenance and rapid development of SARS-CoV-2 genetic diversity than respiratory tract samples (Li et al., 2020; Wang et al., 2021; Zheng et al., 2020b). In our previous study in a rhesus macaque model, the observed pattern of ongoing viral shedding in stool for up to 27 days was remarkably similar to the pattern observed in long-haul COVID-19 patients (Zheng et al., 2020a). These results suggest that SARS-CoV-2 replicates and proliferates not only in lung tissues but also in intestinal tissue, where it can elicit ongoing viral shedding.
During the course of SARS-CoV-2 infection, the host antiviral immune response is key for infection clearance but may cause pathological damage. Recent studies revealed that myeloid cells drive lung inflammation in severe COVID-19, and this finding suggests a potential strategy for reducing lung damage by inhibiting the airway-specific inflammatory process (Li et al., 2020; Speranza et al., 2021; Szabo et al., 2021). Additionally, during viral infection, local tissue cells play important roles in viral invasion and histopathologic progression. For instance, angiotensin-converting enzyme 2 (ACE2) and serine protease transmembrane protease 2 (TMPRSS2), which contribute to SARS-CoV-2 entry, are widespread in lung and intestinal tissues, while ACE2 is an interferon-stimulated gene (ISG) in human airway epithelial cells (Ziegler et al., 2020).
Our previous study implied that lung and intestinal tissues show different pathologies during SARS-CoV-2 infection (Zheng et al., 2020a). Although long-term viral shedding was detected in stool swabs, no significant pathological damage was observed in intestinal tissue sections. Here, in biopsies of small intestinal tissue and lung tissue obtained from rhesus macaques at 3, 7, and 10 days post-infection (dpi), we generated a profile of the tissue response to SARS-CoV-2 infection via single-cell RNA sequencing (scRNA-seq). In the lungs, we observed activation of immune molecules at 3–7 dpi and marked inflammatory cell accumulation. Conversely, in the intestine, we observed a suppressed inflammatory response and decreased tissue damage related to B cell and Paneth cell accumulation.
Results
Nasal inoculation with SARS-CoV-2 led to rectal viral shedding but no obvious pathological progression in the intestine in rhesus macaques
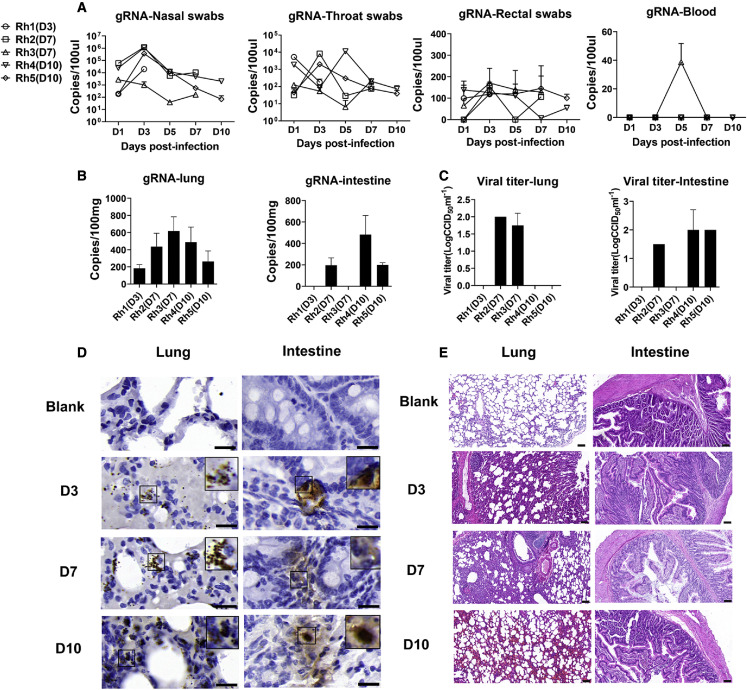
In this study, five rhesus macaques that displayed viral shedding in rectal swabs from 3 to 10 dpi were evaluated (Figure 1A). We observed mild clinical symptoms from 1 to 3 dpi (Table S1). From 4 to 10 dpi, the animals showed less activity and reduced appetite (Table S1). The lung and small intestinal tissues from animals at 3, 7, and 10 dpi were positive for viral nucleic acids (Figure 1B), and viral replication was detected by viral titers (Figure 1C), proving the presence of viral RNA and infectious virions. Meanwhile, the virus was observed in the lungs and intestine by immunohistochemistry (IHC) (Figure 1D). Most importantly, no serious pathological damage was observed in small intestine during this period of SARS-CoV-2 infection (Figure 1E). However, mild to marked interstitial pneumonia was observed in lung tissues (Figure 1E). Together, these results indicate that SARS-CoV-2 can proliferate in intestinal and respiratory tissues and that the associated pathological changes are more serious in lung.
Figure 1.
Viral shedding, viral distribution, viral replication, and histopathological changes in respiratory and intestinal tissues in SARS-CoV-2-infected rhesus macaques
(A and B) The different kinds of swabs, blood (A) and tissue homogenate (B) was collected for 3–10 days post-SARS-COV-2 infection in rhesus macaques. Total RNA was extracted and detected for gRNA. The results are presented as copies per 100 μL swabs or 100-mg tissues. Data are presented as mean ± SD. The data came from two replicate experiments.
(C) In vitro, virus titration was performed to detect infections in lung and intestine tissues at 3, 7, and 10 dpi. The results are presented as CCID50 per 1 mL. Data are presented as mean ± SD. The data came from two replicate experiments.
(D) The presence of SARS-COV-2 N antigen was confirmed by IHC. Necropsy tissue samples at 3, 7, and 10 dpi and normal tissues were analyzed. Scale bar, 20 μm.
(E) Histopathological analyses of small intestines and lungs of rhesus macaque at 3, 7, and 10 dpi. The normal tissues were regarded as blank controls. Scale bar, 100 μm.
See also Table S1.
Differential single-cell profiles in the lungs and intestines during the response to SARS-CoV-2 infection
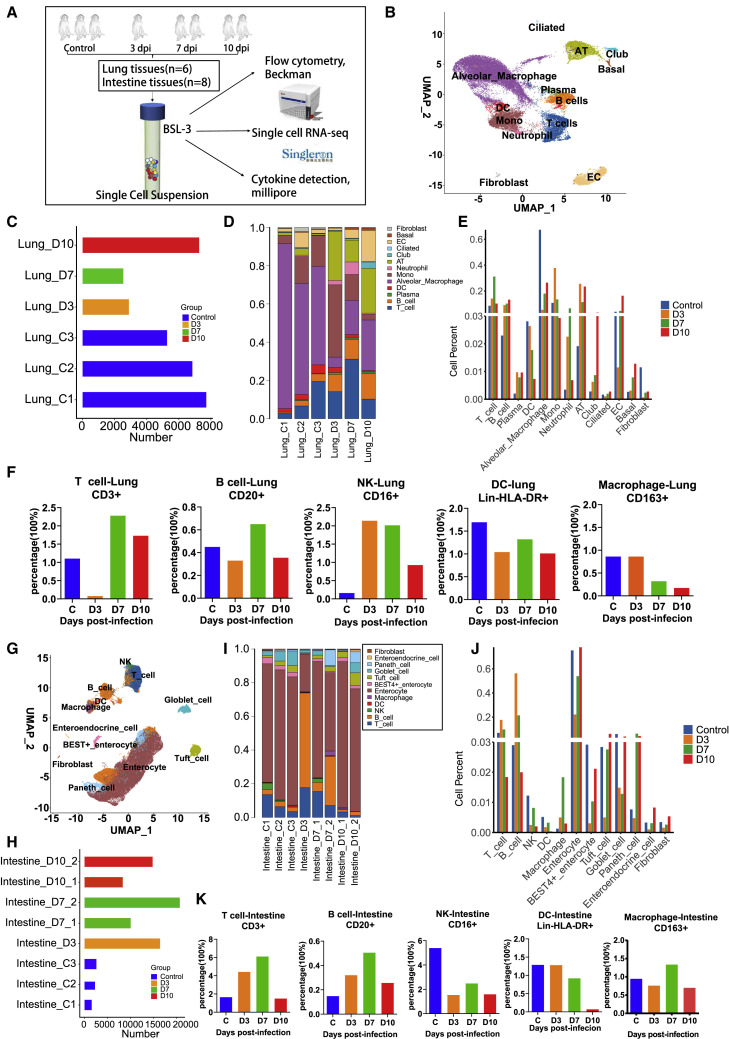
We observed single-cell suspensions from five macaques at different time pointspost-infection (Figure 2A). To investigate the pathogenic mechanism of SARS-CoV-2 in macaques at the single-cell level, we obtained 13 cell populations in the lung (Figures 2B and S1A) and 12 cell populations in the intestine (Figures 2G and S1B) and harvested 32,504 single cells from the lung (Figures 2C) and 76,549 cells from intestinal tissues (Figure 2H), which provided high-quality transcriptomes for all tissue cells and resident lymphoid cells. UMAP of the lung and intestine samples showed distinct separation of the major cell lineages into T cells, B cells, monocytes/macrophages, dendritic cells (DCs), natural killer (NK) cells, and tissue cells (Figures 2B and 2G).
Figure 2.
Global analysis of different immune and tissue cells in SARS-CoV-2-infected rhesus macaques
(A) Experimental workflow showing assays performed on SARS-CoV-2-infected rhesus macaque lung and intestinal tissues at 3, 7, and 10 dpi. Tissue samples were collected from eight rhesus macaques, including three healthy controls and five infected monkeys (one euthanized at 3 dpi, two at 7 dpi, and two at 10 dpi). Two of eight lung tissue samples did not pass pre-machine quality control. scRNA-seq, flow cytometry analysis, and cytokine detection were performed in eight intestinal tissue samples and six lung tissue samples for further analysis.
(B) Identification of different cell clusters across all lung samples (n = 6).
(C) Bar plot shows the cell number of each sample for monkey lung tissues (n = 6).
(D) Cell cluster frequency shown as the fraction of total cells for each infected macaque in lung tissues (n = 6) at different infection time points.
(E) Dynamic changes in each cell cluster frequency on 3 (n = 1),7 (n = 1), and 10 (n = 1) dpi in lung tissues. Three healthy animals were regarded as the control. The average fraction of cells is shown at different infection time points.
(F) Frequency of major immune cell subsets displayed as an average percentage on 3 (n = 1), 7 (n = 1), and 10 (n = 1) dpi in lung samples by flow cytometry. Three healthy animals were regarded as the control.
(G) Identification of different immune and tissue cell clusters across all intestinal samples (n = 8).
(H) Bar plot shows the cell frequency of each sample for intestinal tissues (n = 8).
(I) Cell cluster frequency shown as the fraction of total cells for each infected macaque in intestinal tissues (n = 8)at different infection timepoints.
(J) Dynamic changes in each cell cluster frequency on 3 (n = 1), 7 (n = 2), and 10 (n = 2) dpi in small intestinal tissues. Three healthy animals were regarded as the control. The average fraction of cells is shown at different infection time points.
(K) Frequency of major immune cell subsets displayed as an average percentage on 3 (n = 1), 7 (n = 2), and 10 (n = 2) dpi in gut samples by flow cytometry. Three healthy animals were regarded as the control.
See also Figure S1.
We noted a striking decrease in alveolar macrophage (AM) numbers in the lungs at 3 dpi (Figures 2D and 2E), indicating that AMs in the lungs facilitate SARS-CoV-2 infection, as reported in a clinical case (Wang et al., 2020) and mouse models (Lv et al., 2021). From 3 to 10 dpi, longitudinal increases in alveolar epithelial cells (ATs) and endothelial cells (ECs) in the lungs may contribute to the progression of alveolar or interstitial pathology and be accompanied by neutrophil and monocyte infiltration (Figures 2D and 2E). In contrast, we observed that intestinal enterocytes (IECs) were degraded at 3 dpi but recovered rapidly by 7 and 10 dpi, which revealed that viral infection mildly impacted villus epithelial cells (Figures 2I and 2J). In addition to macrophages, the percentages of NK cells and DCs were decreased in intestinal tissues from 3 to 10 dpi (Figures 2I and 2J). Compared with those in control animals, the proportions of T and B cells in lung tissues continued to increase during the infection period (Figures 2D and 2E), whereas the proportions of T and B cells in intestinal tissues were increased at 3–7 dpi, followed by a gradual decrease at 10 dpi (Figures 2I and 2J). Further FACS analysis of the dynamic characteristics of immune cells also showed significant accumulation of T cells, B cells, NK cells, and DCs in the lung epithelium and reduction of AMs at 3–7 dpi (Figure 2F), which was related to pneumonia. In the intestinal tissue, we found that T cells, B cells, DCs, and macrophage accumulated among intestinal epithelial cells (Figure 2K), but CD16+ NK cell numbers decreased gradually. Therefore, the overall intestinal mucosal immune system maintains activation of the antiviral immune response at 3–7 dpi, which is consistent with the single-cell transcriptomic analysis.
Taken together, our longitudinal sampling of SARS-CoV-2-infected macaques demonstrated distinct scRNA profiles of cell compositions in the lungs and intestines.
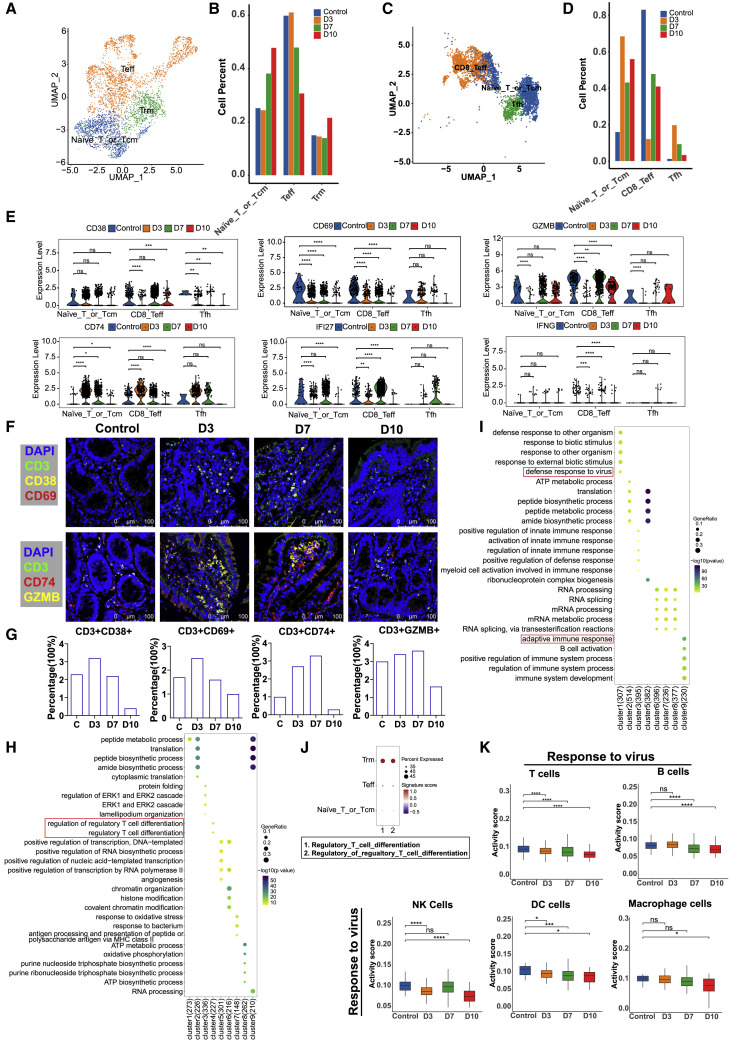
T cells in intestinal tissues indicate an activated antiviral immune response during the early and middle stages of SARS-CoV-2 infection
UMAP embeddings were used to analyze T cell subsets at different time points after infection (3, 7, and 10 dpi). According to the annotation of cell markers, T cells in the lung tissue were divided into three subsets: effector T (Teff) cells, naive T cells or central memory T (TCM) cells, and tissue-resident memory T (TRM) cells (Figures 3A and S2A). Upon SARS-CoV-2 infection, the proportion of TRM cells increased only slightly at 10 dpi in lung tissues; however, we found that the populations of naive T or TCM cells increased in size during the middle (7 dpi) and late (10 dpi) stages of infection, whereas Teff cell numbers began to decrease during the middle stage (7 dpi) and did not recover until 10 dpi (Figure 3B), indicating their involvement in promoting inflammation. Compared with lung tissues, intestinal tissues had three main types of T cells: T follicular helper (Tfh) cells, CD8 Teff cells, and naive T cells or TCM cells (Figures 3C and S2B). We observed naive T or TCM cell and Tfh cell activation in the intestines during the early and middle stages of SARS-CoV-2 infection (Figure 3D), which may be related to the organization of gut-associated lymphoid tissue (GALT) or gut-regional lymph nodes (Macal et al., 2008; Sun et al., 2021). The DEGs identified in T cells, including CD38, CD69, and GZMB, are hallmarks of T cell activation; these markers were expressed in naive T or TCM cells, CD8 Teff cells, and Tfh cells from the intestine at 3–10 dpi (Figure 3E). However, in the lungs, the expression of these genes was mainly activated in Teff cells (Figure S2C). In further immunofluorescence analysis, we found that CD3+CD38+, CD3+CD69+, and CD3+GZMB+ cells were mainly located in the small-intestinal tissue at 3–7 dpi (Figures 3F and 3G), but not at 10 dpi. These data demonstrate that T cells were activated at 3–7 dpi and inhibited at 10 dpi.
Figure 3.
The dynamic characteristics and functional changes in T cells from lung and intestinal tissues during SARS-CoV-2 infection
(A) Subclustering of T cells in all lung tissue samples (n = 6).
(B) Quantification of T cell subsets in 3 (n = 1), 7 (n = 1), and 10 (n = 1) dpi in lung. Three healthy animals were regarded as the control. For each T cell cluster, the average fraction of cells is shown at different infection time points.
(C) Subclustering of T cells in the small intestine (n = 8).
(D) The histogram represents the quantification of T cell cluster frequency at different infection time points (3, 7, and 10 dpi) in the small intestine.
(E) The differential expression levels of the genes CD38, CD69, GZMB, CD74, IFI27, and IFNG at 3 (n = 1), 7 (n = 2), and 10 (n = 2) dpi in the small intestine. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Wilcoxon test.
(F) Analysis of the expression of T cell activation-related factors by a confocal immunofluorescence assay. Top: T cells were detected by an anti-CD3 antibody followed by Alexa Fluor 488 dye-conjugated secondary antibody. The T cell activation-related molecules CD38 and CD69 were detected with anti-CD38 and anti-CD69 primary antibodies, followed by secondary antibodies conjugated with Alexa Fluor 555 and Alexa Fluor 647 dyes, respectively. Bottom: T cells were detected by an anti-CD3 antibody followed by Alexa Fluor 488 dye-conjugated secondary antibody. The T cell activation-related granzyme B and molecules CD74 were detected with anti-granzyme B and anti-CD74 primary antibodies, followed by secondary antibodies conjugated with Alexa Fluor 555 and Alexa Fluor 647 dyes, respectively. Nuclei were stained with DAPI (magnification, 630×). Scale bar, 100 μm. Control, normal tissues.
(G) Percentage of CD3+CD38+, CD3+CD69+, CD3+CD74+, and CD3+GZMB+ cells were computed in total intestine cells (number of cells from three randomly selected fields captured by confocal fluorescence microscope) for normal controls and COVID-19 intestine samples.
(H) Genes clustered by their expression pattern during the progression of SARS-CoV-2 infection (3, 7, 10 dpi). GO term analysis revealed the top five enriched biological processes in nine clusters from the lung.
(I) Genes clustered by their expression pattern during the progression of SARS-CoV-2 infection (3, 7, 10 dpi). GO term analysis revealed the top five enriched biological processes in nine clusters from the small intestine. Cluster 4 does not contained a significantly enriched term of biological process, so it is not shown.
(J) Signature scores of the GO term “regulatory T cell differentiation” genes in T cell subsets (Trm, Teff, and Naive_T or _Tcm) in the lung by Ucell package.
(K) Signature scores of the GO term “response to virus” gene module in major immune cells, including B cells, DC cells, macrophages, NK cells, and T cells of the small intestine by Ucell package.∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Wilcoxon test.
See also Figure S2.
Further analysis of the DEG profiles of lung and intestinal tissues revealed that the genes IFN-G, IFI27, and CD74, which are related to the “IFN-γ signaling response,” were inhibited at 7–10 dpi in the lungs (Figure S2C). However, in intestinal tissues, the levels of the genes CD74, IFI27, and IFN-G were elevated during the early and middle stages of infection, and there was no enrichment or expression of IFN-γ-related genes during the late stage of infection (10 dpi) (Figure 3E). Immunofluorescence analysis of small intestinal tissue sections also demonstrated that the percentage of CD74 was downregulated in CD3+ T cells during the last stage of infection (10 dpi) (Figures 3F and 3G).
Finally, we analyzed the transcription dynamics of T cell-related genes from 3 to 10 dpi during SARS-CoV-2 infection. Nine time-dependent expression patterns were identified in the lung (Figure S2D) and small intestine (Figure S2E). Their biological significance was investigated in the lung (Figure 3H) and intestine (Figure 3I). In lung tissues, cluster 4 (227 genes) including “regulatory T cell differentiation” genes showed a decreasing trend at 7–10 dpi (Figures 3H and S2D). Further analysis showed that the genes related to “regulatory T cell differentiation” were mainly from Trm cells (Figure 3J). In GO analysis of small intestine, clusters 1 and 9 genes were significantly enriched in “defense response to virus” and “adaptive immune response” (Figure 3I), and these genes associated with the antiviral immune system are activated during the early and middle stages of infection (Figures S2E and S2F). Additionally, the gene module related with “response to virus” was significantly downregulated in T, B, NK, DC, and macrophage cells, especially at 10 dpi, which demonstrated widespread immune suppression in the gut (Figure 3K). Therefore, we concluded that T cells were mainly activated during the early and middle stages (3–7 dpi) of infection in the small intestine.
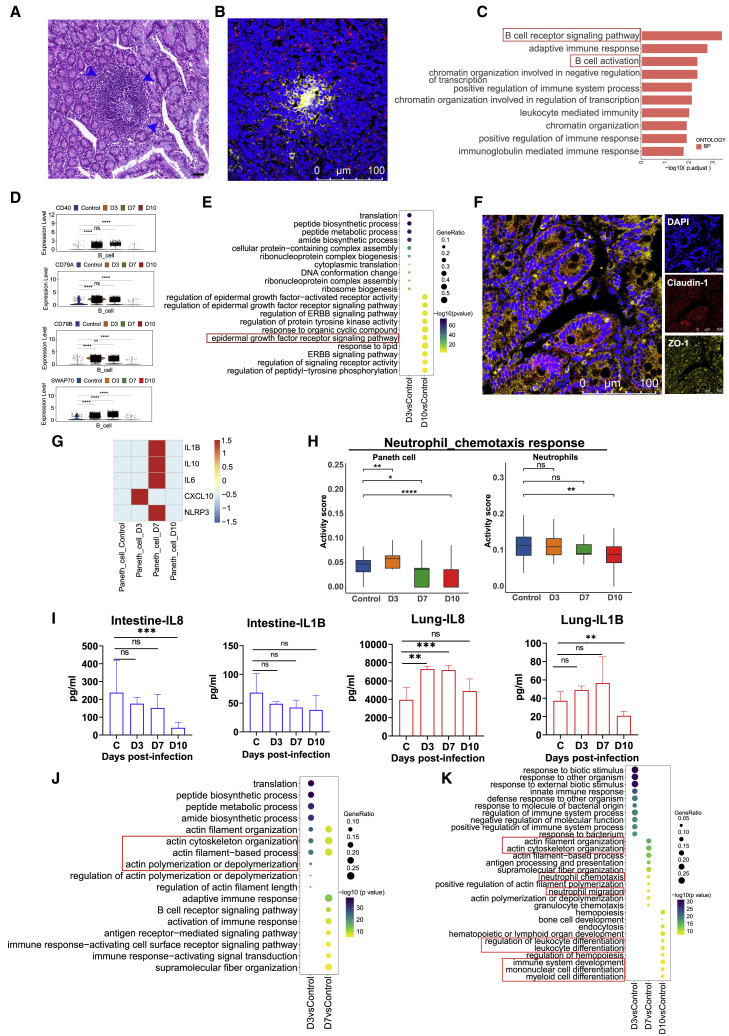
SARS-CoV-2 infection induced B cell and Paneth cell accumulation in the intestine, which was related to a weakened inflammatory response and tissue damage
In the intestinal samples, we observed increases in B cell and Paneth cell numbers after infection (Figures 2I and 2J). As Paneth cells contribute to the epithelial niche during pathogen invasion, they play key roles in mucosal immunity (Goga et al., 2021). We observed that Paneth cells increased at 7–10 dpi (Figures 2I and 2J). Similarly to T cells, B cells also exhibit an activated antiviral immune response during the early and middle stages of SARS-CoV-2 infection. B and T cell migrating to the intestines is associated with the formation of structure of lymphoid tissue (Cabrita et al., 2020) (Figures 2I–2K). HE analysis revealed that lymphocyte accumulation was similar to that of the GALT structure at 7 dpi (Figure 4A). Furthermore, immunofluorescence analysis indicated that intestinal GALT-like was present, with central B cell accumulation and surrounding marginal T cells (Morbe et al., 2021) (Figure 4B). According to the GO analysis, “B cell receptor signaling pathway” and “B cell activation” pathways were enriched in B cells of intestinal tissues at 3 dpi (Figure 4C). The expression levels of the representative B cell-activation-related genes CD40, CD79A, CD79B, and SWA70 were increased at 3–7 dpi (Figure 4D) in intestine. However, except for CD79A, CD79B, SWA70, and CD40, all exhibited ongoing high expression levels in lung tissues at 3–7 dpi (Figure S3A). We found that “epidermal growth factor receptor signaling-related pathways” were enriched in enterocyte cells at 10 dpi (Figure 4E). Moreover, immunofluorescence analysis revealed that tight-junction (TJ)-related proteins, including ZO-1 and claudin-1, showed high expression at 10 dpi, which indicates that intestinal epithelial barrier repair occurs during the last stage of infection (Figure 4F). The findings above suggested that enterocytes, Paneth cells, and B cells may be involved in sustaining intestinal epithelial homeostasis.
Figure 4.
The dynamic characteristics and functional changes of B and Paneth cells from lung and intestinal tissues during SARS-CoV-2 infection
(A) HE analysis of lymphocyte accumulation in the small intestine of rhesus macaques infected with SARS-CoV-2. The blue arrows indicate the accumulation of lymphocytes. Scale bar, 20 μm.
(B) Immunofluorescence analysis of T and B cell distributions in the small intestine of rhesus macaques infected with SARS-CoV-2. CD20+ B cells, yellow; CD3+ T cells, red. Scale bar, 100 μm.
(C) Enriched top 10 GO terms for upregulated genes in SARS-CoV-2-infected macaques at 3 dpi compared with normal controls in B cells from the intestine.
(D) Expression of the CD40, CD79A, CD79B, and SWAP70 genes compared with the control at 3, 7, and 10 dpi in B cells from the intestine. ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Wilcoxon test.
(E) The top 10 up-GO-enriched biological processes in D3 versus Control, D10 versus Control samples in the enterocyte population from the intestine. None of the top 10 enriched biological processes from GO enrichment analysis were observed in day 7 samples compared with normal controls in enterocytes, so it is not shown.
(F) Confocal imaging shows the junctional protein localization of ZO-1 and claudin-1 in the small intestine at 10 dpi. The ZO-1 protein (yellow), Claudin-1 protein (red), and cell nucleus (blue) were stained with Alexa Fluor 555, Alexa Fluor 647, and DAPI dye, respectively. Scale bar, 100 μm.
(G) The averaged expression of the IL-1B, CXCL10, IL-10, IL-6, TNF, and NLRP3 genes at 3, 7, and 10 dpi in Paneth cells from the intestine.
(H) Evaluation of the GO term “neutrophil chemotaxis” in Paneth cells from the small intestine and neutrophils from the lung during the whole infection progression.∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Wilcoxon test.
(I) Protein expression levels of IL1B and IL8 (CXCL8) in the small intestine and lung during the SARS-COV-2 infection process. Cytokine cytometric bead arrays were performed to measure the IL1B and IL8 concentrations. The average and SD are shown for the protein concentrations (pg/mL). ∗∗p < 0.01, ∗∗∗p < 0.001; Mann-Whitney U test was used to compare control and infected groups.
(J) The top 10 up-GO-enriched biological processes in day 3 samples compared with normal controls, and day 7 samples compared with controls in the Paneth cell population from the intestine. None of the top 10 enriched biological processes from GO enrichment analysis were observed in day 10 samples compared with normal controls in Paneth cells, so it is not shown. Dot color indicates the significance of the enrichment (p value), and dot size indicates the gene ratio annotated to each GO term.
(K) The top 10 up-enriched biological processes by GO enrichment analysis in day 3 samples compared with normal controls, day 7 samples compared with normal controls, and day 10 samples compared with normal controls in neutrophils from lung. Dot color indicates the significance of the enrichment (p value), and dot size indicates the gene ratio annotated to each GO term.
See also Figure S3.
Although GO enrichment analysis of intestinal B and Paneth cells revealed that neutrophil-related pathways were enriched in the gut at 10 dpi compared with 7 dpi (Figures S3B and S3C), we did not find neutrophil genes in the intestine (Figure 2J). However, Paneth cells are functionally similar to neutrophils when exposed to a variety of stimuli, including bacterial or viral antigens (van Es and Clevers, 2014). We found that major inflammatory genes, including IL1B, IL6, CXCL10, IL10, and NLRP3, were activated in Paneth cells from the intestine at 3 and 7 dpi but not at 10 dpi (Figure 4G). Therefore, we compared the gene transcriptome of the GO term “neutrophil chemotaxis” between Paneth cells from the intestine and neutrophils from the lung. The results showed that the levels of neutrophil-related genes were increased at 3 dpi and decreased at 7–10 dpi in the intestine and were substantially reduced in the lung (Figure 4H). Neutrophil chemotactic cytokine analysis confirmed that CXCL8 and IL1B showed a decreasing trend at 3–10 dpi in the small intestine; however, the opposite tendency was observed at 3–7 dpi in lung tissues (Figure 4I). As the gene clustering pattern suggested, the upregulated genes in Paneth cells mostly encode factors related to cell cytoskeleton organization and epithelial cell differentiation (Figures 4J and S3C) associated with the epithelium barriers at 3-7 dpi. Although we found “cell cytoskeleton organization” genes upregulated in neutrophils of lung on 7 dpi, most highly expressed genes were related to neutrophil chemotaxis and immune cell differentiation (Figure 4K) at 7–10 dpi.
Therefore, Paneth cells and B cells play important roles in the normal damage repair mechanism in the intestinal epithelium and in intestinal inflammation (Lueschow and McElroy, 2020). We concluded that the potential pathological changes induced by SARS-CoV-2 were suppressed by the above-mentioned responses in the intestine during the late stage of infection.
Myeloid cells function in the degradation of maintaining intestinal mucosal immunity in the late stage of infection
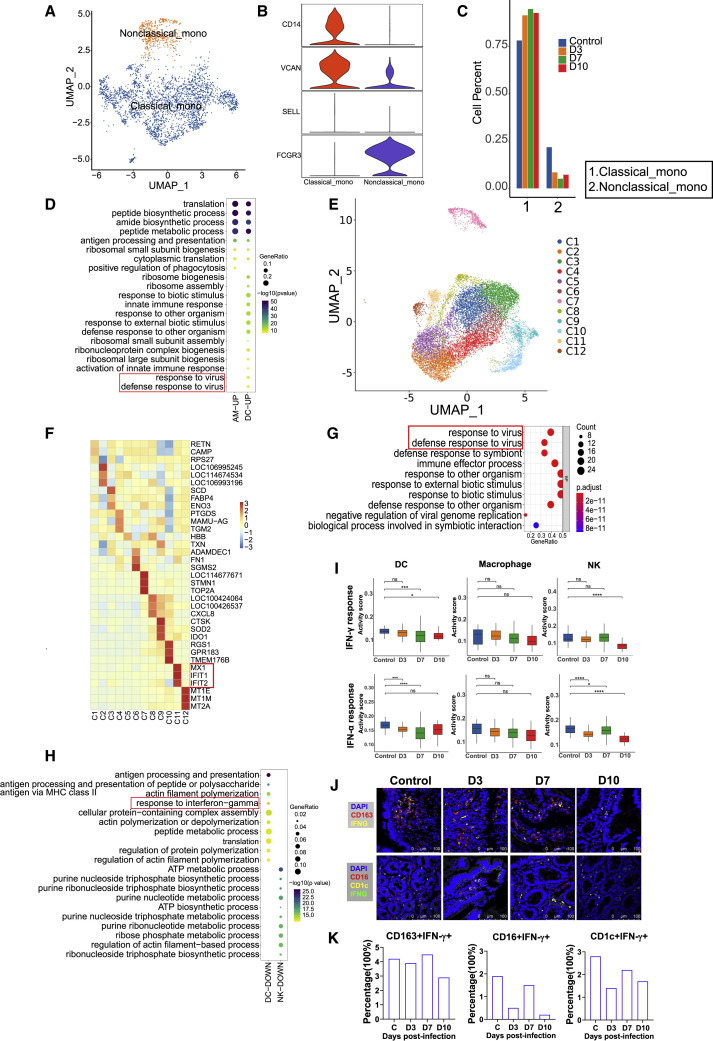
Another family of main immune cells, myeloid cells, in the lungs were mainly classified into four types: monocytes, AMs, DCs, and neutrophils (Figure 2B). The nonclassical monocytes showed substantially decreasing numbers (Figures 5A–5C), whereas the numbers of classical monocytes increased at 3–10 dpi (Figures 5A–5C). Additionally, other myeloid cells, including AM, DC, and NK, exhibited a different changing trend during infection progress (Figure 2B). These results indicate that SARS-CoV-2 infection evokes disordered changes in myeloid cells in lung tissues. GO analysis revealed that genes of “response to virus” were especially enriched in DCs at 3 dpi (Figure 5D) but not at 7–10 dpi (Figure S4A) in lung. Subclustering of AMs revealed 12 populations (Figures 5E and 5F). The genes MX1, IFIT1 and IFIT2, related to IFN induction, was enriched in cluster 11 (Figure 5F). The cluster 11 genes were enriched in the “response to virus” signaling pathways, indicating activation of antiviral immune response (Figure 5G). The “response to virus” pathways were not enriched in the monocytes and neutrophils (Figure S4B). In contrast to the lung, the “response to IFN-G” was downregulated in intestinal DCs at 3 dpi (Figure 5H).
Figure 5.
Changes in myeloid cell subsets in the lung and intestine of SARS-CoV-2-infected rhesus macaques
(A) Subclustering of monocytes in all lung tissue samples (n = 6).
(B) Violin plots showing marker genes of monocyte clusters in the lung. The y axis represents the normalized gene expression value.
(C) The histogram represents the quantification of classical and nonclassical monocyte cluster frequencies among lung tissue samples on 3 (n = 1), 7 (n = 1), and 10 (n = 1) dpi. There are three control samples. The average is shown for each infection time point group.
(D) The GO analysis for top 20 enriched biological processes are revealed in SARS-CoV-2-infected macaques at 3 dpi compared with normal controls across AMs and DCs in lung. The dot color depth indicates the statistical significance of the enrichment (p value), and the dot size is equal to the gene ratio annotated to each term.
(E) UMAP embedding of 12 AM clusters in total lung tissue samples (n = 6).
(F) Heatmap showing the top three differentially expressed genes across 12 AM subclusters from the total lung tissue samples (n = 6).
(G) The top 10 in AM cluster 11 of the lungs, as determined by GO enrichment analysis. The dot color depth indicates the significance of the enrichment (p value), and the dot size is equal to the gene ratio annotated to each term.
(H) The top 10 enriched biological processes by GO analysis for downregulated genes are revealed in SARS-CoV-2-infected macaques at 3 dpi and compared with normal controls in DCs and NK cells from the small intestine. The dot color depth indicates the significance of the enrichment (p value), and the dot size is equal to the gene ratio annotated to each term.
(I) Hallmark gene set scores for “IFN-γ response” (top) and “IFN-α response” (bottom), evaluated for all immune cell clusters across different infection timepoints (3, 7,and 10 dpi) and compared with the normal control in small intestine.∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Wilcoxon test.
(J) Immunofluorescence imaging revealed protein expression of IFN-γ on CD163+ macrophage cells (top) or IFN-γ expression on CD1c+ DCs and CD16+ NK cells (bottom) at different infection time points (3, 7,and 10 dpi) and compared with the normal control. Top: IFN-γ protein (yellow), CD163 protein (red), and cell nucleus (blue). Bottom: IFN-γ(green), CD16 (red), and CD1c (yellow).
(K) Percentage of CD163+IFN-γ+, CD16+IFN-γ+, and CD1c+IFN-γ+ cells were computed in total intestine cells (number of cells from three randomly selected fields captured by confocal fluorescence microscope) for normal controls and COVID-19 intestine samples.
See also Figure S4.
The DEGs associated with antiviral genes were compared between the lungs and intestines. We found that the IF127, IFIT2, IFIT3, MX1, MX2, and ISG15 genes associated with the IFN response were mainly expressed in monocytes, macrophages, DCs, and neutrophils in the lungs at 3 dpi (Figure S4C). We also observed IFN-γ-related gene expression in the intestines at 3–7 dpi (Figure S4D). The IFI27 gene was inhibited at 10 dpi in NK, DC, and macrophage cells (Figure S4E), whereas the IFIT2, IFIT3, MX1, MX2, and ISG15 genes showed nearly no expression in intestinal immune cells at 10 dpi (Figure S4D). Additionally, we found significant concomitant downregulation of IFN-γ- and IFN-α-stimulated signatures within DCs, macrophages, and NK cells during the whole infection process, especially at 10 dpi (Figure 5I). Furthermore, compared with control, IFNγ expression on CD163+ macrophages, CD1c+ DC, or CD16+ NK cells exhibited a decreasing trend at 10 dpi (Figures 5J and 5K).
Taken together, the above observations revealed broad suppression of DCs, NK cells, and macrophages during the antiviral immune response in intestinal tissues, especially the “IFN response” in the late stage of SARS-CoV-2 infection.
ACE2 and TMPRSS2 were enriched in intestinal epithelial cells, in which IFN-related genes were activated
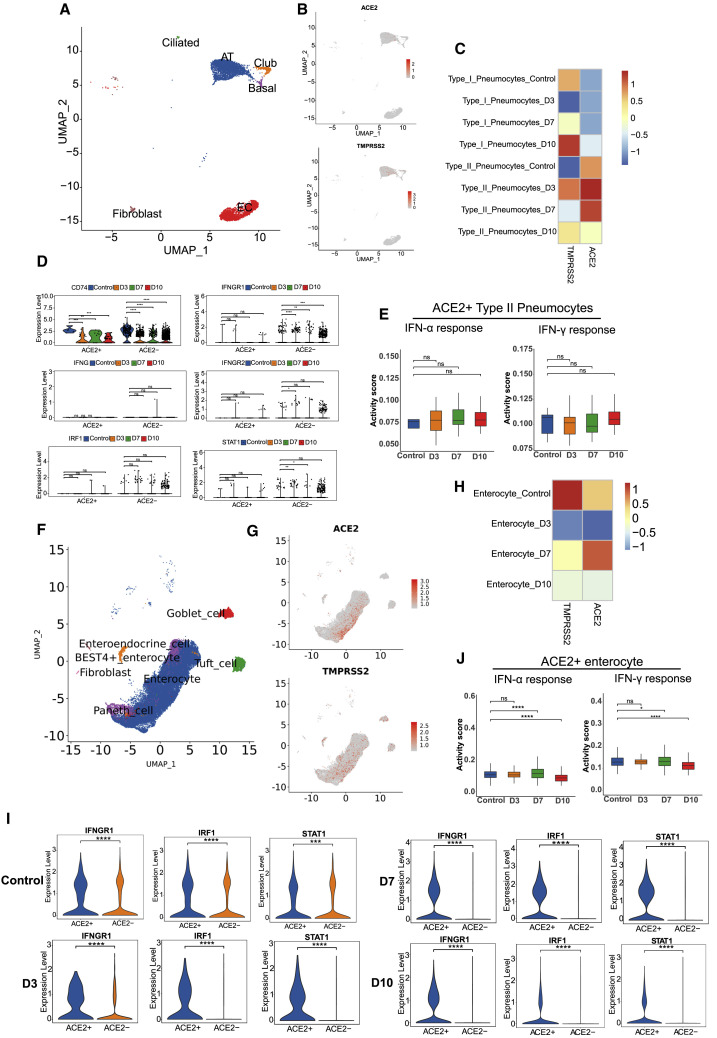
In addition to immune cells, tissue cells also play important roles in the immune response upon SARS-CoV-2 infection. We investigated ACE2 and TMPRSS2 expression in different tissue cells, and ACE2 expression was mainly identified in type II ATs (Figures 6A–6C). Moreover, ACE2 and TMPRSS2 co-expression was observed predominantly in type II ATs (Figures 6B and 6C). ACE2 expression in type II ATs peaked at 3–7 dpi (Figure 6C). Some studies have shown that SARS-CoV-2 receptor ACE2 is an ISG in epithelial cells (Ziegler et al., 2020). Compared with other type II ATs, in which CD74, IFNG, IFNGR1, IFNGR2, STAT1, and IRF1 are regulated as IFN-γ response genes, ACE2+ type II ATs showed only CD74 activation, indicating the limited IFN-γ antiviral immune response in lung (Figure 6D). Although, the difference was not statistically significant, the IFN-α- and IFN-γ-induced signatures showed an increasing tendency from 3 to 10 dpi in ACE2+ type II pneumocytes (Figure 6E).
Figure 6.
Altered features of ACE2 and TMPRSS2 in lung and intestinal tissues across the stages of SARS-CoV-2 infection
(A) UMAP projection reveals the ACE2 or TMPRSS2 distribution in various types of tissue cell populations in the lung samples (n = 6).
(B) ACE2 and TMEPRSS2 distribution in various types of tissue cell populations in the lung samples (n = 6).
(C) Heatmap showing the relative expression level of ACE2 and TMPRSS2 genes across type I and type II pneumocytes in infected samples at 3 (n = 1), 7 (n = 1), and 10 (n = 1) dpi in lung.
(D) Interferon stimulation-related genes expressed among ACE2+ and ACE2– type II pneumocytes in infected samples at 3 (n = 1), 7 (n = 1), and 10 (n = 1) dpi in lung.∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Wilcoxon test.
(E) Hallmark gene set scores for “IFN-γ response” (left) and “IFN-α response” (right), evaluated for ACE2+ type II pneumocytes across different infection timepoints (3, 7, and 10 dpi) and compared with the normal control in lung tissues.Wilcoxon test.
(F) UMAP projection shows the ACE2 or TMPRSS2 distribution in various types of tissue cell populations in the intestine (n = 8).
(G) ACE2 and TMPRSS2 distributions in various types of tissue cell populations in the intestine (n = 8).
(H) Heatmap showing the relative expression level of the ACE2 and TMPRSS2 genes across enterocytes at different infection time points (3, 7, and 10 dpi) and compared with the normal control in intestinal tissues.
(I) Interferon stimulation-related genes IFNGR1, IRF1, and STAT1 expressed among ACE2+ and ACE2–enterocytes (Seurat package) across clusters at 3, 7, and 10 dpi in the small intestine.∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Wilcoxon test.
(J) Hallmark genes set scores for “IFN-γ response” (left) and “IFN-α response” (right), evaluated for ACE2+ enterocytes across different infection time points (3, 7,and 10 dpi) and compared with the normal control in small intestinal tissues.∗p < 0.05, ∗∗∗∗p < 0.0001, Wilcoxon test.
In contrast to the findings in lung tissue, the ACE2 and TMPRSS2 genes were widely expressed in a variety of intestinal tissue cells, including enterocytes, tuft cells, goblet cells, Paneth cells, BEST4+ enterocytes, and enteroendocrine cells (Figures 6F and 6G). At different time points after infection (3–10 dpi), we found that ACE2 and TMPRSS2 were mainly expressed in enterocytes (Figure 6G), and ACE2 expression exhibited an increasing trend from 3 to 7 dpi, whereas TMPRSS2 levels decreased during the infection period (Figure 6H). Moreover, IFN-γ-related genes of enterocytes, including IFNGR1, STAT1, and IRF1, were activated in ACE2+ enterocytes at 3–7 dpi, which implied that the immune response to IFN-γ was activated during the early and middle stages of infection (Figure 6I). We found that IFN-α- and IFN-γ-induced gene modules all showed significant upregulation at 7 dpi and marked downregulation at 10 dpi in ACE2+ enterocytes (Figure 6J). Consequently, we concluded that ACE2+ enterocytes are the main target of SARS-CoV-2. The inhibition of IFN pathways in the last stage may help explain the symptoms of late gut-viral shedding.
Characteristics of inflammatory molecules in lung and intestinal tissues
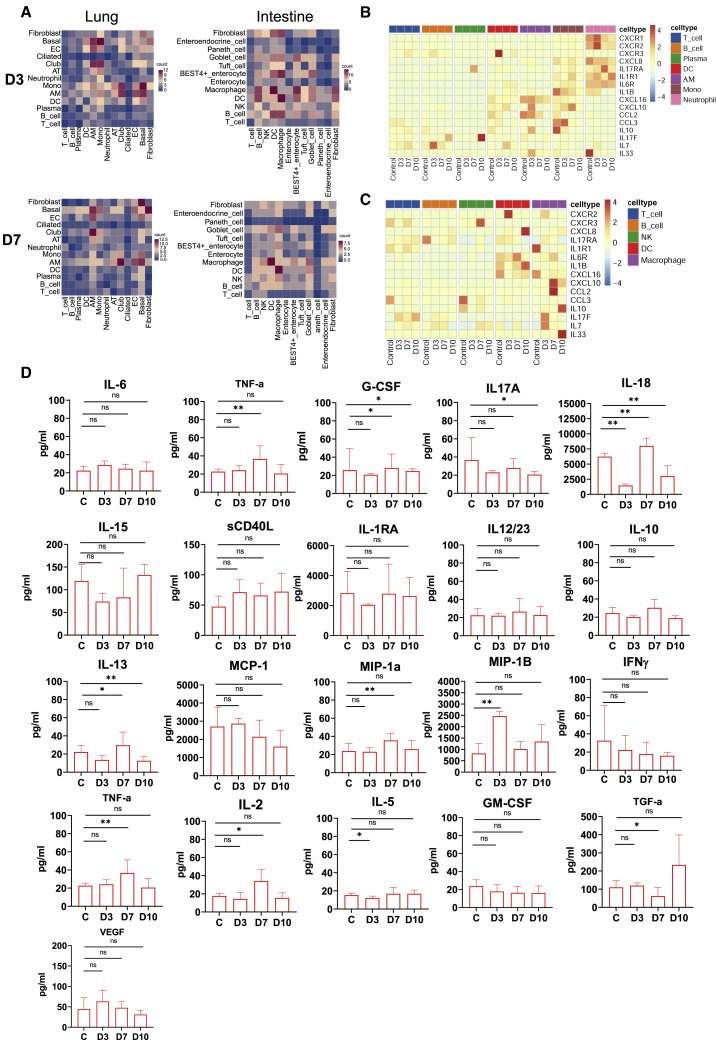
We use CellPhone DB to investigate the cell-cell interaction (CCI) network among the cell types identified in this study. Remarkably, AMs exhibited strong interactions with ECs and AT cells throughout the infection period (3–10 dpi) (Figures 7A and S5A), indicating interaction between these cells was associated with the antiviral immune response. However, the macrophages strongly interacted with DCs at 3 and 7 dpi in gut (Figures 7A and S5A). Further analysis revealed obvious enrichment in the ligand-receptor pairs CD74_MIF, CD74_COPA, and CD74_APP between macrophages and other cells at 7 dpi (Figure S5B), suggesting that these pairs might be associated with the production of epithelial-mesenchymal transition (ECM) components, as reported by previous research (Wu et al., 2020).
Figure 7.
The dynamic characteristics of inflammatory molecules in lung and intestinal tissues
(A) Cell-cell communication was identified by CellPhone DB in day 3 samples and day 7 samples from lung and small intestinal tissue. The darker the color intensity is, the more pairs of receptor-ligand interactions there are between cells.
(B and C) Heatmap showing the normalized average expression of the indicated chemokines and cytokines at 3, 7, and 10 dpi in the tissue cells of the lungs (B) and small intestine (C).
(D) Protein expression levels of multiple cytokines in the intestine during the SARS-COV-2 infection process (3, 7,and 10 dpi). The average and SD are shown for the protein concentrations (pg/mL). ∗p < 0.05, ∗∗p < 0.01; Mann-Whitney U test was used to compare control and infected groups.
See also Figure S5.
Here, we also compared the expression of chemokines and cytokines in the lungs and intestines. Genes encoding chemokines and inflammatory cytokines appeared to be upregulated in lung tissue from 3 to 10 dpi (Figures 7B and S5C). The genes, including CXCR1, CXCR2, IL17RA, ILR1, and IL6R, were mainly observed on neutrophils in the lung. In addition, CXCL10 and CXCL16 were mainly expressed in AMs and monocytes at 3–7dpi (Figure 7B). Compared with the lungs, inflammatory factor genes were mainly from DCs, macrophages, and fibroblasts in intestine (Figures 7C and S5D). The cytokine cytometric bead assay demonstrated that the protein expression levels of proinflammatory cytokines (IL6, TNF-α, G-CSF, IL17A, IL18, IL15, sCD40 L, IL1RA, and IL12/23), anti-inflammatory factors (IL10), chemokines (IL13, MCP-1 [CCL2], MIP-1a[CCL3], and MIP-1B[CCL4]), and adaptive immune response cytokines (IFN-γ, TNF-α, IL-2, IL-5, GM-CSF) and the cytokines related to intestinal epithelial and endothelial homeostasis (VEGF, TGF-a) were significantly decreased or not obviously changed compared with the control at the last stage of infection (10 dpi) (Figure 7D). As stated above, the intestinal mucosa exhibited a less inflammatory state at the late stage of infection.
Discussion
In this study, on the basis of the evidence of SARS-CoV-2 duplicating in intestinal and respiratory tissues of monkeys with different pathological changes, we revealed a spatial scRNA profile of these two tissues on the level of cell composition and activation during 3–10 dpi. Overall, we find longitudinal increases in ATs and ECs in the lungs with neutrophil and monocyte infiltration, contributing to the constant progression of interstitial pathology. In intestine, we observed that IECs were degraded at 3 dpi but recovered rapidly and may result in mild infection in villus epithelial cells. Lymphocyte infiltration was basically noticed in tissue damage and inflammation (Sakai and Kobayashi, 2015). We found that the activation of lymphocytes in the intestines was limited at 10 dpi, although T and B cells were increased at 3–7 dpi. In comparison, significant accumulations of T cells, B cells, NK cells, AMs, and neutrophils were detected in lung through 3 to 10 dpi. We conclude that activation of intestinal mucosal antiviral response occurs within 3–7 dpi, the early and middle stages of infection.
Patients with severe COVID-19 display features of impairment and exhaustion of SARS-CoV-2-reactive CD8+ T cells (Chen and John Wherry, 2020; Kusnadi et al., 2021), and these impaired T cell responses can result from deficient IFN production, as IFN promotes the survival and effector functions of T cells (Acharya et al., 2020). Our findings also revealed that DEGs associated with antiviral genes were upregulated in immune cells at 3–7 dpi, which may lead to proinflammatory cytokine production, which results in lung injury during SARS-CoV-2 infection. In our study, hallmark genes of T cell activation were widely expressed in T cells of the intestines at 3 dpi, and the IFN-related genes, including IF127, CD74, and IFNG, were enriched in the intestines from 3 to 7 dpi, which indicates that the antiviral immune response was activated in the intestines only during the early and middle stage of infection. These results suggest that intestinal IFN-related antiviral immunity may help control rectal viral shedding during the early stage of SARS-CoV-2 infection.
Upon scRNA analysis of intestinal tissue, we observed that B cell and Paneth cell numbers increased significantly after SARS-COV-2 infection. We proposed that both B cells and T cells accumulated in the mucous layer of the intestine, indicating the development of local GALT-like structure (Farstad et al., 1997; Spencer and Sollid, 2016). It was reported that Paneth cell and GALT dysfunction leads to dysbiosis, a compromised epithelial barrier, and susceptibility to enteric bacterial and viral infection (Holly and Smith, 2018). Paneth cells of the intestine are functionally similar to neutrophils (van Es and Clevers, 2014). In our study, neutrophil-related pathways were activated during the early stage of infection in Paneth cells, and the neutrophil-chemotactic cytokines CXCL8 and IL-1B were inhibited during the last stage of infection in the gut. Therefore, Paneth cells and B cells may help maintain gastrointestinal barrier homeostasis and participate in suppressing inflammation and tissue damage induced by SARS-CoV-2. In contrast to neutrophils from the lung, we found that the activated genes of intestinal Paneth cells were related to tissue barrier repair during the middle and last stages of infection (7–10 dpi), whereas the activated genes of lung neutrophils mainly functioned in immune and inflammatory responses during the last stage of infection. The different functional states of Paneth cells and neutrophils may be one factor in the different pathological changes between intestinal and respiratory tissues in COVID-19.
SARS-CoV-2 can spread and replicate in tissues, subsequently triggering immune dysfunction and inflammation (Lowery et al., 2021; Weatherhead et al., 2020). From the viewpoint of the cell tropism of SARS-CoV-2 infection in the lung and intestine, we found that ACE2 and TMPRSS2 were expressed in type II pneumocytes, IECs, tuft cells, goblet cells, and enteroendocrine cells, similar to the results of previous studies (Jackson et al., 2022; Ziegler et al., 2020). Additionally, previous in vitro research showed that ACE2 is a human ISG in airway epithelial cells (Ziegler et al., 2020). Our data indicate that ACE2 is abundant in the intestines and that the antiviral immune response of ACE2+ intestinal cells is related to inflammatory state and outcome of virus control. In our previous study, lung tissue with COVID-19 was found to be in a state of ongoing high inflammation at 3–9 dpi infection (Zheng et al., 2020a). However, the secretion level of inflammatory factors in intestinal tissue infected with SARS-CoV-2 was limited at the last stage of infection (10 dpi) in the gut. Therefore, lung and gut are in different inflammatory states in the SARS-CoV-2 infected monkeys.
In summary, our study demonstrates that intestinal and lung tissues exhibit different cellular immune response characteristics and inflammatory state during SARS-CoV-2 infection. The maintenance of the intestinal mucosal barrier is related to B cells and Paneth cells. The IFN-related antiviral functions of T and B lymphocytes and myeloid cells were inhibited at 10 dpi, which may lead to ongoing viral shedding from intestinal tissue. Notably, the small intestine appears to be in a reduced inflammatory state during SARS-CoV-2 infection, which may explain the mild intestinal pathological damage. However, it is not yet clear whether this inflammation is entirely resolved.
Limitations of the study
During the COVID-19 epidemic, we had limited numbers of macaques to perform our experiments, so this study mainly relied on five SARS-CoV-2-infected monkeys. At every infection time point, we compared the immune response in only one or two animals. Therefore, statistical analysis could not be performed for some data, such as the cell frequency, which was just represented by means. Furthermore, we mainly focused on the immune response in the early intestinal tract, which was limited to a short sampling window (up to 10 dpi). Moreover, due to limitations of the intranasal rhesus macaque infection model, the animals showed only mild clinical symptoms. Therefore, the conclusions of this study should be used carefully in the context of intestinal immune response in severe COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD20 Monoclonal Antibody | ThermoFisher Scientific | Cat#MA1-10135; RRID: AB_11152399 |
| CD3-εPolyclonal Antibody | ImmunoWay | Cat#YT0761 |
| CD3 epsilon antibody | Genetex | Cat#GTX42110; RRID: AB_11178619 |
| Recombinant Anti-CD16 Antibody | Abcam | Cat#ab246222; |
| Purified anti-human CD1c Antibody | Biolegend | Cat#331502; RRID: AB_1088995 |
| Recombinant Anti-CD163 Antibody | Abcam | Cat#ab182422; RRID: AB_2753196 |
| CD74 (D5N3I) XP® Rabbit mAb | Cell Signaling | Cat#77274S; RRID: AB_2799893 |
| Granzyme B mouse mAb(PT0564) | ImmunoWay | Cat#YM4028 |
| CD38 Monoclonal Antibody | Proteintech | Cat#60006-1-Ig; RRID: AB_716994 |
| CD69 Polyclonal Antibody | Proteintech | Cat#10803-1-AP; RRID: AB_2074965 |
| ZO-1 Monoclonal Antibody | ThermoFisher Scientific | Cat#33–9100; RRID: AB_2533147 |
| Recombinant Anti- Claudin 1 Antibody | Abcam | Cat#ab211737 |
| IFN gamma Monoclonal Antibody | R&D systems | Ca#AF-285-NA; RRID: AB_354445 |
| IFN gamma Monoclonal Antibody | ThermoFisher Scientific | Cat#MA5-23718; RRID: AB_2609805 |
| Alexa Fluor 488 Donkey Anti-Goat IgG (H + L) | ThermoFisher Scientific | Cat#A11055; RRID: AB_2534102 |
| Alexa Fluor® 488 Goat Anti-rat IgG H&L | Abcam | Cat#ab150157; RRID: AB_2722511 |
| Alexa Fluor® 555 Goat Anti-Mouse IgG-H&L | Abcam | Cat#ab150114; RRID: AB_2687594 |
| Alexa Fluor 647 donkey Anti-Rabbit IgG (H + L) | Abcam | Cat#ab150075; RRID: AB_2752244 |
| NHP T/B/NK Cell Cocktail | BD Biosciences | Cat#558639; RRID: AB_1645290 |
| PerCP-Cy™5.5 Mouse Anti-Human CD3 | BD Biosciences | Cat#552852; RRID: AB_394493 |
| Brilliant Violet 421™ Anti-human CD19 Antibody | BioLegend | Cat#302234; RRID: AB_11142678 |
| FITC Anti-human CD14 Antibody | BioLegend | Cat#301804; RRID: AB_314186 |
| PE Mouse Anti-Human CD56 | BD Biosciences | Cat#556647; RRID: AB_396511 |
| PE/Cyanine7 Anti-human HLA-DR Antibody | BioLegend | Cat#307616; RRID: AB_493588 |
| PE/Cyanine7 Anti-human CD163 Antibody | BioLegend | Cat#333614; RRID: AB_2562641 |
| SARS-CoV-2 Nucleocapsid Antibody, Rabbit Mab | Sino Biological | Cat#40143-R001; RRID: AB_2827974 |
| Bacterial and virus strains | ||
| SARS-CoV-2-KMS1/2020 | (Zheng et al., 2020a) | https://pubmed.ncbi.nlm.nih.gov/33180882/ |
| Chemicals, peptides, and recombinant proteins | ||
| Lymphocyte Separation Medium (Human) | Solarbio | Cat#P8610 |
| Trypsin-EDTA Solution | SIGMA | Cat#T4049 |
| RNase-free Water | TAKARA | Cat#9012 |
| Dulbecco’s Modified Eagle’s Medium (DMEM) | Sigma | Cat#d8371 |
| DAPI | Abcam | Cat#ab104139 |
| Phosphate Buffered Saline (PBS) | Corning | Cat#21-040-CVC |
| Fetal bovine serum (FBS) | Gibco | Cat#10099–141 |
| TRNzol Universal Reagent | TianGen | Cat#DP424 |
| Albumin bovine serum | Sigma-Aldrich | Cat#A1933 |
| Sodium Citrate buffer | Solarbio® | Cat#C1010 |
| Triton®X-100 | BioFrox | Cat#1139ML500 |
| Tween 20 | BioFrox | Cat#1247ML500 |
| Critical commercial assays | ||
| GEXSCOPE® Single Cell RNA Amplification & Library Reagents Kit | Singleron | Cat1110011LA1 |
| GEXSCOPE® Single Cell Barcoding Beads Kit | Singleron | Cat#1110011SB2 |
| GEXSCOPE® Single Cell Microchip & Wash Buffer Set Kit | Singleron | Cat#1110011MC3 |
| Hematoxylin-Eosin/HE Staining Kit | Solarbio | Cat#G1120 |
| Metal Enhanced DAB Substrate Kit | Solarbio | Cat#DA1015 |
| Universal Two-Step Detection Kit (Mouse/Rabbit Enhanced Polymer Detection System) | ZSGB-BIO | Cat#PV-9000 |
| One Step PrimScript RT-PCR Kit | TaKaRa | Cat#RR064A |
| Non-human Primate Cytokine Magnetic Bead Panel | Millipore | PRCYTOMAG-40K, PCYTMG-40K-PX23, PRCYMAG40PMX23BK |
| GEXSCOPE® Single Cell RNA Amplification & Library Reagents Tissue | Singleron | Cat#1110011LA1 |
| GEXSCOPE® Tissue Preservation Solution | Singleron | Cat#SN154211 |
| GEXSCOPE® Tissue Dissociation Mix | Singleron | Cat#SN421201 |
| Oligonucleotides | ||
| Probe: ORF1ab-P: 5′-CCGTCTGCGGTAT GTGGAAAGGTTATGG-3′ |
(Zheng et al., 2020a) | https://pubmed.ncbi.nlm.nih.gov/33180882/ |
|
Primer: ORF1ab-F: 5′-CCCTGTGGGTTT TACACTTA-3′ |
(Zheng et al., 2020a) | https://pubmed.ncbi.nlm.nih.gov/33180882/ |
|
Primer: ORF1ab-R: 5′-ACGATTGTGCA TCAGCTG-3′ |
(Zheng et al., 2020a) | https://pubmed.ncbi.nlm.nih.gov/33180882/ |
| Software and algorithms | ||
| Prism 8.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator | Adobe Illustrator | https://www.adobe.com/ |
| Adobe Photoshop | Adobe Photoshop | https://www.adobe.com/ |
| BioRender | BioRender | https://biorender.com/ |
| Python version 3.7 | Python (2020) | https://www.python.org/downloads/release/python-370/ |
| R version 3.5 | R Foundation for Statistical Computing (2017) | https://www.R-project.org |
| Seurat (v3.1.2, R package) | (Stuart et al., 2019) | https://github.com/satijalab/seurat |
| ClusterProfiler (v3.16.1, R package) | (Yu et al., 2012) | http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| CellPhoneDB(v2.1.7) | (Efremova et al., 2020) | https://github.com/Teichlab/cellphonedb |
| UCell (v1.1.0, R package) | (Andreatta and Carmona, 2021) | https://github.com/carmonalab/UCell |
| Mfuzz (2.46.0, R package) | (Kumar and Futschik, 2007) | http://www.bioconductor.org/packages/release |
| Deposited data | ||
| Raw and analyzed data | This paper | CNSA:CNP0002884 |
| Other | ||
| Iron beads | Solarbio | Cat#YA3031 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by lead author, Longding Liu (longdingl@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Biosafety and animal ethics
All experiments associated with the SARS-CoV-2 virus were performed in BSL-3 and ABSL-3 laboratories, and these facilities were approved by the Institutional Biosafety Committee of Institute of Medical Biology (IMB) & Chinese Academy of Medical Sciences (CAMS). The animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of the IMB & CAMS.
Virus and cells
The viral strain SARS-CoV-2-KMS1/2020 (GenBank accession number: MT226610.1) was stocked by the IMB & CAMS. The virus was propagated in Vero cells cultured in DMEM (Sigma) supplemented with 2% fetal calf serum (FBS, Gibco), 50 U/mL streptomycin, and 50μg/mL penicillin at 37°C in 5% CO2. Virus titers were measured by cytopathic efficiency (CPE) assay, and virus samples were stored at 106.0 TCID50/ml at −80°C for subsequent experiments.
Animal study
Five male Chinese rhesus macaques aged 8–12 months (weight: 2–3 kg) were nasally inoculated with the SARS-CoV-2-KMS1/2020 virus at a dose of 2×105 TCID50. Three healthy male Chinese rhesus macaques aged 8–12 months (weight: 2–3 kg) were used as controls in this study. Throat swabs, nasal swabs, fecal swabs, and blood samples were collected at 1, 3, 5, 7, and 10 dpi. Macaques at 3, 7, and 10 dpi were euthanized after anesthetization. The lungs and small intestines were collected for tissue section preparation. In addition, single-cell suspensions of tissue pieces were used for single-cell RNA-seq and flow cytometry.
Method details
SARS-CoV-2 genomic RNA detection by qRT–PCR
The total RNA was extracted from swabs or tissues using TRIzol Universal Reagent (TianGen Biotech). Tissues were mechanically homogenized with iron beads prior to RNA extraction. The SARS-CoV-2 RNA standard was generated by vitro transcription of the ORF1a regions. The primers and probes (ORF1ab-F:5′-CCCTGTGGGTTTTACACTTA-3′; ORF1ab-R:5′-ACGATTGTGCATCAGCTG-3′; ORF1ab-P: 5′-CCGTCTGCGGTATGTGGAAAGGTTATGG-3′) were used to measure the number of viral ORF1ab gene copies. The serially diluted SARS-CoV-2 gRNA standards were used to establish a standard curve for the quantification of viral gRNA levels. The qRT–PCR was performed with the one-step PrimeScript™ RT–PCR kit (Takara).
Viral titer detection
Vero cells were cultured in MEM supplemented with 50 units/mL penicillin, 50 μg/mL streptomycin, and 10% FBS (Gibco) at 37°C in 5% CO2. The supernatants of tissue homogenates were collected by homogenizing the tissue in 500 μL of PBS, freezing at −80°C and thawing 3 times, and centrifuging for 10 min at 12000 rpm. Then, the viral titers in the supernatants of the tissue homogenates were measured. Tenfold serial dilutions of the tissue supernatants were plated in eight replicates in Vero cells and cultured for 6 days at 37°C in DMEM added with 2% FBS (Gibco), 50 units/mL penicillin, and 50 μg/mL streptomycin. Six days after inoculation, the 50% endpoint titer using serial dilutions was calculated by the Spearman-Karber method.
Histopathologic analyses
For histopathological examination, lung and small intestine tissue sections were fixed in 10% paraformaldehyde. Then, the tissues embedded with paraffin were sliced at thicknesses of 4 μm. After that, the tissue sections were stained with hematoxylin and eosin (H&E, Solarbio). Briefly, the tissue sections were deparaffinized, dehydrated, and washed with tap water for 2 min. The nucleus were stained with hematoxylin dye for 15 min, followed by washing twice with tap water. Then, the sections were differentiated with differentiation solution for 3 min. After washing twice, the slices were stained with Eosin Y Aqueous Solution for 2 min. Finally, the sections were dehydrated in a series of alcohol, cleared with xylene and sealed with resin.
Immunohistochemistry (IHC)
Tissue slices were incubated at 65°C for 30 min followed by xylene dewaxing and ethanol series hydration. For antigen retrieval, the tissue sections were microwaved for 15 min in glass beaker filled with 0.01 M citrate buffer (pH = 6.0). After heating, the glass beaker with slides were cooling for 20 min at room temperature. Then, the tissue sections were incubated with hydrogen peroxide with a final concentration of 3% for 10 min at room temperature to inhibit the endogenous peroxidase activity and then washed with 1x TBS with 0.05% Tween 20. Then, the slices were incubated with blocking solution with 5% BSA for 1 h at room temperature. SARS-CoV-2 antigen was detected using a Rabbit anti-SARS-CoV-2 N protein antibody(Sino Biological) at a 1/500 dilution. After overnight incubation at 4°C, antibody labeling was visualized by a rabbit-specific HRP/DAB IHC polymer detection kit (Solarbio) followed by hematoxylin counterstaining. Subsequently, a histological section scanner was used to capture and evaluate the digital images.
Immunofluorescence (IF)
For immunofluorescence examination, paraffin-embedded lung and small intestine tissue sections were pretreated by dewaxing, hydration, heat antigen retrieval and blocking. According to the detailed antibody panels provided by the Key resources table, the tissue sections were stained with the following primary antibodies: CD3 (at 1/100 dilution), CD1c (at 1/500 dilution), CD163 (at dilution 1/50), CD38 (at 1/50 dilution), CD69 (at 1/200 dilution), CD74 (at 1/200 dilution), CD20 (at 1/200 dilution), ZO-1 (at 1/50 dilution), Claudin 1 (at 1/1000 dilution), and CD16 (at 1/100 dilution). Then, the sections were rinsed five times in PBST for removing the unbound antibodies. To detect IFN-γ and granzyme protein, the tissue sections were permeabilized with 0.1% Triton X-100 for 10 min, blocked with 5% BSA for 1 h. Then, the tissue sections were stained with a primary antibody against IFN-γ (at 1/500 dilution) or granzyme B (at 1/200 dilution) overnight for 2 h at room temperature and then labeled with the fluorescent dye-conjugated secondary detection antibodies for 1 h at room temperature. Finally, sections were rinsed five times with PBST. The nuclei is stained with DAPI for 5 min. After washing three times with PBST, a Leica TCS SP8 laser confocal microscope was used to capture and evaluate the immunofluorescence images.
Multiplex cytokine analysis
The cytokine and chemokine concentrations in the supernatants of the tissue homogenates were measured by the Non-human Primate Cytokine Magnetic Bead Panel (Millipore) according to the manufactures’ kit instructions, followed by plate reading on Luminex 200. The analyte concentrations in each sample were calculated by the 5-parameter logistic method.
Flow cytometry analysis
To prepare single-cell suspensions, the minced tissue sections were passed through a 70-μm nylon mesh filter. Then, single-cell suspensions from tissues were incubated with antibody panels as described in the Key resources table. The CD3+T, CD20 + B, CD16 + NK cells were labeled by NHP T/B/NK Cell cocktail (BD Bioscience). And the DC was stained by the lin and HLA-DR (Biolegend) moleculars. The lineage cocktail including CD56 (BD Bioscience), CD14 (Biolegend), CD3 (BD Bioscience), CD19 (Biolegend) surface molecular. In addition, the anti-CD163 antibody molecular was used to recognize macrophage cells. Briefly, the single-cell suspension was firstly washed twice with PBS. Then, the cells were incubated with a anti-cell-surface protein antibody cocktail at RT for 0.5 h. Then, the single-cell samples were rinsed twice with PBS for removing redundant antibodies. Finally, the stained samples were analyzed by a CytoFlex flow cytometer (Beckman Coulter). The data was analyzed by FlowJo v10 (BD Biosciences).
Preparation of single-cell suspensions
Firstly, the fresh tissue samples were rinsed five times with PBS. The clean samples were cut into small pieces, and then digested in 2 mL of Tissue Dissociation Solution (Singleron Biotechnologies) at 37°C for 15 min with ongoing agitation. After filtering through a 40-μm sterile strainer (Becton Dickinson), the cell suspensions were centrifuged at 500 g for 5 min at 4°C and resuspended in 1 mL of PBS. 2 mL of Red Blood Cell Lysis Buffer (Singleron Biotechnologies) was added in cell samples at 18–20°C for 10 min in order to remove red blood cells. Finally, the single-cell suspensions were centrifuged at 500 g for 5 min at 18–20°C and subsequently resuspended in PBS. A hemocytometer was used to count the cells. All the procedures were performed in a BSL3 facility.
Cell preparation for 3′ barcoded single-cell RNA-seq (scRNA-seq)
The prepared single-cell suspension was loaded onto a single-cell microfluidic chip according to the manufactures’ protocols (Singleron GEXSCOPE). Then, 60 μL of resuspended barcoded beads was slowly injected onto the chip over 30 s to allow the beads to slowly spread inside the chip. After slowly adding 100 μL of Lysis Buffer Mix to the chip, the cells were lysed, and mRNA was released during incubation at room temperature for 20 min. Excess liquid from the inlet and outlet reservoirs was immediately removed. Emulsions were immediately acquired from the chip and incubated in a thermal cycler (Bio-Rad) for reverse transcription.
Single-cell library preparation and sequencing
scRNA-seq library preparations was constructed according to manufactures’ protocols. The captured mRNA was incubated in a preheated ThermoMixer with the reverse transcription mix at 42°C at 1000 rpm for 90 min. Using PCR to amplify ten or fifty nanograms of barcoded full-length cDNA was amplified, the products were purified and fragmented, and adapters were added. The purified cDNA was used for library construction. Finally, the cDNA and library quality was measured by a Qubit and Agilent Fragment Analyzer and sequenced on an Illumina NovaSeq platform with 150 bp paired-end reads.
scRNA-seq data processing and quality control
Illumina bcl2fastq (version 2.19.1) was used to convert the raw sequencing data to FASTQ format, and STAR (version 3.1.0) was used to align the sequenced reads to the Macaca mulatta reference genome sequence (https://www.ncbi.nlm.nih.gov/genome/?term=Macaca+mulatta). We used the CeleScope pipeline (https://github.com/singleron-RD/CeleScopeanalysis) to acquire gene-cell expression matix. Then, the Seurat package was used to analyze and process the gene expression matrix. Cells with <200 genes and >50% mitochondrial genes were filtered out.
Dimensionality reduction, clustering and visualization
Dimensionality reduction, clustering and visualization were performed by Seurat v3. A filtered expression matrix for each sample dataset was used to identify cell subsets. NormalizeData function with ‘scale.factor’ = 10000 was used to normalize gene expression. After data normalization, Highly variable genes were used to perform principal component analysis (PCA). Subsequently, graph-based clustering with 20 PCs at a resolution of 1.2 was performed and visualized using uniform manifold approximation and projection (UMAP) with the Seurat function RunUMAP. The cell type identity of each cluster was determined using canonical markers from the literature and the reference database SynEcoSysTM, which contains collections of canonical cell type markers from single-cell seq data from CellMakerDB, PanglaoDB and recently published literature. The canonical markers and their corresponding cell types are shown in Table S2. After cell type identification, immune cells and other cells were extracted and clustered separately for further detailed analysis. Further clustering was performed by Seurat with 20 PCs at a resolution of 0.8.
Differential gene expression analysis
The FindMarkers function in the Seurat package (Wilcoxon rank-sum test) was used to identify the DEGs. For a given cell type, all other cell types serve as controls. To identify the DEGs, we set ‘min.pct’ = 0.1 and ‘logfc.threshold’ = 0.25. Genes were defined as significantly upregulated if |logFC| > 0.25 and FDR <0.05. The statistical significance differences shown in the dynamic violin plots were derived from the results of DEGs.
Pathway enrichment analysis
For the DEGs in different cell types or conditions, genes with p val <0.05 and |logFC| > 0.25 were selected for Gene ontology (GO) functional enrichment analysis which performed by clusterProfiler (3.16.1) (Yu et al., 2012). The upregulated genes and downregulated genes were analyzed separately.
Identifying time-dependent transcriptional programs in Macaca mulatta
The time-dependent transcriptional program in Macaca mulatta was identified by Mfuzz (Kumar and Futschik, 2007). Based on the stage of infection progression, the samples from rhesus monkeys were analyzed at 3, 7, and 10 dpi and compared with the samples from the control rhesus monkeys. First, the average expression function in Seurat was used to calculate the average expression of each gene in each viral-infection stage of infection. Second, the expression matrix for each gene was added to 0.000001 to avoid 0, which is unacceptable for Mfuzz. Next, the data were preprocessed by the fifilter.std (min.std = 0), standardize() and mestimate() functions according to the tutorial. Then, genes were clustered into 9 different expression patterns. Genes with a maximum expression of less than 1 were excluded prior to GO analysis of the 9 programs. Only GO terms with adjusted p < 0.05 are shown in the figure.
Cell–cell interaction (CCI) analysis
Cellphonedb function which was curated by the CellPhoneDB v.2.0 (Efremova et al., 2020) database was applied to predict CCIs. Counts and celltype annotation files were input data. The permutation number for evaluating the null distribution of average ligand–receptor pair expression in randomized cell identities was set to 1000. Receptor and ligand expressed in at least 10% of cells were used to perform further statistical analysis. The significant ligand-receptor pairs were predicted with a p value threshold of <0.05. We used the R circlize package (v0.4.10) to generate a chordal graph visualizing the mean expression levels of these significant ligand–receptor pairs with a rank value <0.3 in different celltypes.
Calculation of IFN-related gene set activity score
The enrichment of IFN-related hallmark gene sets (IFN-α and IFN-γ) was evaluated in different kinds of immune cells and tissue cells. UCell was applied to calculate these gene signatures in single-cell datasets based on the Mann–Whitney U statistical analysis (R package, version 1.1.0), which evaluates the rank of each query gene’s expression level in individual cells. We used the UCell score to evaluate the degree to which individual cells expressed a certain gene set. IFN-related gene sets used in the analysis included “HALLMARK_INTERFERON_ALPHA_RESPONSE” and “HALLMARK_INTERFERON _ GAMMA_RESPONSE” from MsigDB GO terms. The Wilcoxon rank-sum test was used to analysis the statistical significance.
Quantification and statistical analysis
The statistical analysis related with single-cell sequencing was performed using R (version 3.5.1). The Wilcoxon rank-sum test was used in this study. The following values indicated statistical significance: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82041017, 32070923, 32000133), Basic Research Project of Science and Technology of Yunnan Province (202001AT070143), and Major Science and Technology Special Projects of Yunnan Province (202002AA100009). The funders had no role in the study, design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank the Kunming National High-Level Biosafety Primate Research Center for its support. We would further like to thank Singleron Biotechnologies for sequencing of scRNA-seq libraries. We would also like to thank BioRender, as the Graphic Abstract was created with BioRender.com.
Author contributions
L.D.L., Q.H.L., H.W.Z., Y.L.C., H.J.S., and Z.L.H. conceived and designed the experiments, analyzed data, made figures, and wrote the manuscript; H.L., J.L., Z.L.H., and X.Z. conducted the rhesus macaque animal studies and performed the viral titer assays; H.W.Z. and Y.L.C. processed samples for scRNA-seq profiling and performed flow cytometry studies; F.M.Y. and Y.Y.L. led the clinical care of the rhesus macaques; J.L.L., C.K.L., L.Q., Y.Y.Z., and Q.Z. conducted the H&E and IHC experiments; Y.Y.L. and Z.X. performed the qRT-PCR assays.
Declaration of interests
The authors declare no conflict of interest.
Published: May 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110864.
Supplemental information
Data and code availability
Raw and processed data are available on CNGB Nucleotide Sequence Archive (CNSA:https://db.cngb.org/cnsa) with accession number CNP0002884. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Front. Immunol. 2021;12:742167. doi: 10.3389/fimmu.2021.742167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M., Carmona S.J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., Johansson I., Phung B., Harbst K., Vallon-Christersson J., et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- Caracciolo M., Correale P., Mangano C., Foti G., Falcone C., Macheda S., Cuzzola M., Conte M., Falzea A.C., Iuliano E., et al. Efficacy and effect of inhaled adenosine treatment in hospitalized COVID-19 patients. Front. Immunol. 2021;12:613070. doi: 10.3389/fimmu.2021.613070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Farstad I.N., Norstein J., Brandtzaeg P. Phenotypes of B and T cells in human intestinal and mesenteric lymph. Gastroenterology. 1997;112:163–173. doi: 10.1016/s0016-5085(97)70231-2. [DOI] [PubMed] [Google Scholar]
- Goga A., Yagabasan B., Herrmanns K., Godbersen S., Silva P.N., Denzler R., Zund M., Furter M., Schwank G., Sunagawa S., et al. miR-802 regulates Paneth cell function and enterocyte differentiation in the mouse small intestine. Nat. Commun. 2021;12:3339. doi: 10.1038/s41467-021-23298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly M.K., Smith J.G. Paneth cells during viral infection and pathogenesis. Viruses. 2018;10:225. doi: 10.3390/v10050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyawasam J.C., Jayarajah U., Riza R., Abeysuriya V., Seneviratne S.L. Gastrointestinal manifestations in COVID-19. Trans. R. Soc. Trop. Med. Hyg. 2021;115:1362–1388. doi: 10.1093/trstmh/trab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnadi A., Ramirez-Suastegui C., Fajardo V., Chee S.J., Meckiff B.J., Simon H., Pelosi E., Seumois G., Ay F., Vijayanand P., et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6:eabe4782. doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ren L., Zhang L., Wang Y., Guo L., Wang C., Xiao Y., Wang Y., Rao J., Wang X., et al. High anal swab viral load predisposes adverse clinical outcomes in severe COVID-19 patients. Emerg. Microbes Infect. 2020;9:2707–2714. doi: 10.1080/22221751.2020.1858700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.N., Huang Y., Wang W., Jing Q.L., Zhang C.H., Qin P.Z., Guan W.J., Gan L., Li Y.L., Liu W.H., et al. Effectiveness of inactivated SARS-CoV-2 vaccines against the Delta variant infection in Guangzhou: a test-negative case-control real-world study. Emerg. Microbes Infect. 2021;10:1751–1759. doi: 10.1080/22221751.2021.1969291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery S.A., Sariol A., Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: implications for COVID-19. Cell Host Microbe. 2021;29:1052–1062. doi: 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueschow S.R., McElroy S.J. The Paneth cell: the curator and defender of the immature small intestine. Front. Immunol. 2020;11:587. doi: 10.3389/fimmu.2020.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Wang Z., Qu Y., Zhu H., Zhu Q., Tong W., Bao L., Lv Q., Cong J., Li D., et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 2021;7:24. doi: 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Sankaran S., Chun T.W., Reay E., Flamm J., Prindiville T.J., Dandekar S. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 2008;1:475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- Morbe U.M., Jorgensen P.B., Fenton T.M., von Burg N., Riis L.B., Spencer J., Agace W.W. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021;14:793–802. doi: 10.1038/s41385-021-00389-4. [DOI] [PubMed] [Google Scholar]
- Morgan K., Samuel K., Vandeputte M., Hayes P.C., Plevris J.N. SARS-CoV-2 infection and the liver. Pathogens. 2020;9:430. doi: 10.3390/pathogens9060430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Kobayashi M. Lymphocyte 'homing' and chronic inflammation. Pathol. Int. 2015;65:344–354. doi: 10.1111/pin.12294. [DOI] [PubMed] [Google Scholar]
- Simsek Yavuz S., Unal S. Antiviral treatment of COVID-19. Turk. J. Med. Sci. 2020;50:611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J., Sollid L.M. The human intestinal B-cell response. Mucosal Immunol. 2016;9:1113–1124. doi: 10.1038/mi.2016.59. [DOI] [PubMed] [Google Scholar]
- Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Perez-Perez L., Meade-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J., et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci. Transl. Med. 2021;13:eabe8146. doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Lagarrigue F., Wang H., Fan Z., Lopez-Ramirez M.A., Chang J.T., Ginsberg M.H. Distinct integrin activation pathways for effector and regulatory T cell trafficking and function. J. Exp. Med. 2021;218:e20201524. doi: 10.1084/jem.20201524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Dogra P., Gray J.I., Wells S.B., Connors T.J., Weisberg S.P., Krupska I., Matsumoto R., Poon M.M.L., Idzikowski E., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814.e6. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenforde M.W., Patel M.M., Ginde A.A., Douin D.J., Talbot H.K., Casey J.D., Mohr N.M., Zepeski A., Gaglani M., McNeal T., et al. Effectiveness of SARS-CoV-2 mRNA vaccines for preventing Covid-19 hospitalizations in the United States. Clin. Infect. Dis. 2021;74:1515–1524. doi: 10.1093/cid/ciab687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es J.H., Clevers H. Paneth cells. Curr. Biol. 2014;24:R547–R548. doi: 10.1016/j.cub.2014.04.049. [DOI] [PubMed] [Google Scholar]
- Wang C., Xie J., Zhao L., Fei X., Zhang H., Tan Y., Nie X., Zhou L., Liu Z., Ren Y., et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang D., Zhang L., Sun W., Zhang Z., Chen W., Zhu A., Huang Y., Xiao F., Yao J., et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021;13:30. doi: 10.1186/s13073-021-00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherhead J.E., Clark E., Vogel T.P., Atmar R.L., Kulkarni P.A. Inflammatory syndromes associated with SARS-CoV-2 infection: dysregulation of the immune response across the age spectrum. J. Clin. Invest. 2020;130:6194–6197. doi: 10.1172/JCI145301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2022. WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. [Google Scholar]
- Wu X., Chen S., Lu C. Amyloid precursor protein promotes the migration and invasion of breast cancer cells by regulating the MAPK signaling pathway. Int. J. Mol. Med. 2020;45:162–174. doi: 10.3892/ijmm.2019.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Li H., Guo L., Liang Y., Li J., Wang X., Hu Y., Wang L., Liao Y., Yang F., et al. Virulence and pathogenesis of SARS-CoV-2 infection in rhesus macaques: a nonhuman primate model of COVID-19 progression. PLoS Pathog. 2020;16:e1008949. doi: 10.1371/journal.ppat.1008949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e9. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghi G., Moosavy S.H., Yavarian S., HasaniAzad M., Khorrami F., Sharegi Brojeni M., Kheirandish M. Gastrointestinal implications in COVID-19. BMC Infect. Dis. 2021;21:1135. doi: 10.1186/s12879-021-06824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data are available on CNGB Nucleotide Sequence Archive (CNSA:https://db.cngb.org/cnsa) with accession number CNP0002884. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.