Abstract
The rapidly spreading Omicron variant is highly resistant to vaccines, convalescent sera, and neutralizing antibodies (nAbs), highlighting the urgent need for potent therapeutic nAbs. Here, a panel of human nAbs from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) convalescent patients show diverse neutralization against Omicron, of which XMA01 and XMA04 maintain nanomolar affinities and excellent neutralization (half maximal inhibitory concentration [IC50]: ∼20 ng/mL). nAb XMA09 shows weak but unattenuated neutralization against all variants of concern (VOCs) as well as SARS-CoV. Structural analysis reveals that the above three antibodies could synergistically bind to the receptor-binding domains (RBDs) of both wild-type and Omicron spikes and defines the critical determinants for nAb-mediated broad neutralizations. Three nAbs confer synergistic neutralization against Omicron, resulting from the inter-antibody interaction between XMA04 and XMA01(or XMA09). Furthermore, the XMA01/XMA04 cocktail provides synergistic protection against Beta and Omicron variant infections in hamsters. In summary, our results provide insights for the rational design of antibody cocktail therapeutics or universal vaccines against Omicron.
Keywords: SARS-CoV-2, COVID-19, Omicron, neutralizing antibodies, coronaviruses
Graphical abstract
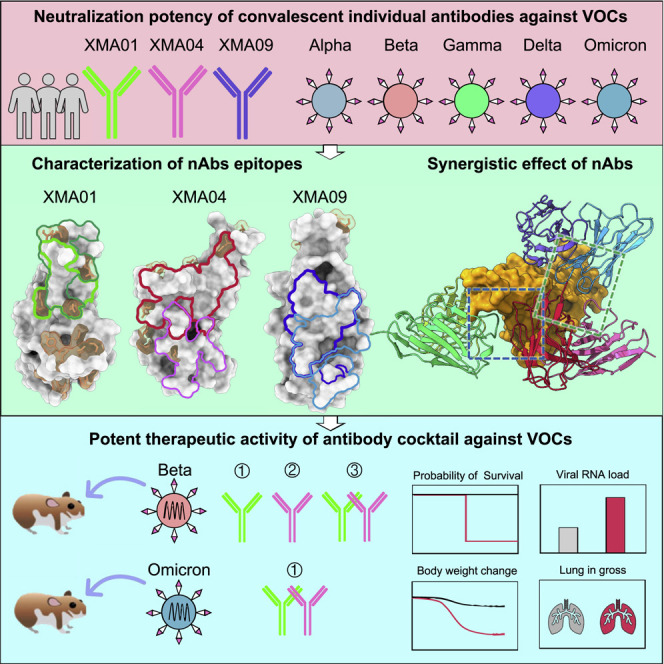
Wang et al. identify three broadly neutralizing antibodies against SARS-CoV-2 variants of concern including Omicron, with structural insight revealing how the three antibodies are resistant to most of the RBD mutations in variants of concern and show that these have synergic neutralization and protection against the Omicron variant.
Introduction
As of February 2022, coronavirus disease 2019 (COVID-19) induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused over 399 million infections and more than 5.7 million deaths (Abu-Raddad et al., 2021). The continuous emergence of multiple SARS-CoV-2 variants, especially variants of concern (VOCs), generate serious threats to existing vaccines and therapeutic antibodies (Abu-Raddad et al., 2021; Madhi et al., 2021; McCallum et al., 2021; Wang et al., 2021), which is exacerbated by the currently rapidly emerging Omicron (B.1.1.529) variant (Hadfield et al., 2018; Scott et al., 2021). More than any previous VOC, the Omicron variant contains 15 mutations in the receptor-binding domain (RBD), 9 of which reside in the receptor-binding motif (RBM), suggesting that the Omicron variant has an unprecedented devastating effect on immune protection established by vaccination and infection. Omicron has been shown to be highly resistant to neutralization by plasma from vaccinated individuals, convalescent sera, and most reported neutralizing antibodies (nAbs), including those authorized under Emergency Use Authorization (EUA) (Cameroni et al., 2022; Cao et al., 2021; Cele et al., 2021; Hoffmann et al., 2021; Liu et al., 2021; Planas et al., 2021; Wang et al., 2022), which highlights the urgent need for broadly nAbs and essential adjustments of current antibody therapeutics. In this study, we generated a panel of human nAbs with diverse broad neutralization against SARS-CoV-2 VOCs, including Omicron. Cryoelectron microscopy (cryo-EM) structures of the combination of three noncompeting nAbs in complex with wild-type spike trimer (WT-S) and Omicron S trimer (Omicron-S) together revealed the critical determinants for nAb-mediated broad neutralization. We also elucidated the synergistic neutralization of three nAbs and potent protection of XMA01/XMA04 cocktail against SARS-CoV-2 Beta and Omicron variants infection in hamsters.
Results
Identification of broadly nAbs against SARS-CoV-2 variants including Omicron
We constructed a panel of eleven nAbs targeting the SARS-CoV-2 RBD that were obtained from convalescent patients infected with the SARS-CoV-2 prototype strain, as previously reported (Tiller et al., 2008). Of the nAbs, XMA09 and XMA18 showed efficient cross-neutralization against SARS-CoV (Figure 1A). Eleven nAbs were classified into 4 clusters by primary identification of their binding sites by cross-blocking assay, with classic antibodies of Classes 1–5 as reference (Barnes et al., 2020; Starr et al., 2021a) (Figure 1B). The cross-neutralizing nAbs XMA09 and XMA18 were evaluated as classes 5 and 4, respectively (Figures 1A and 1C). The VOCs emerged after the Alpha (B.1.1.7) variant, including Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) variants, were refractory to antibody neutralization in varying degrees, indicating the diminished efficacy of vaccines and antibody therapeutics (Abu-Raddad et al., 2021; Madhi et al., 2021; McCallum et al., 2021; Wang et al., 2021). A pseudotyped SARS-CoV-2 neutralization assay revealed that most nAbs from classes 4 and 5 and some nAbs from classes 1 and 2/3 showed broad-neutralizing activities against VOCs as well as variants of interest (VOIs) (Figures 1D, 1E, and S1). Strikingly, two nAbs, XMA01 (from class 1) and XMA03 (from class 2/3) maintained excellent neutralizing potency and breadth against VOCs (Figures 1D and 1E). Especially, XMA01 demonstrated strong and unchanged broad-spectrum neutralization against all tested pseudotyped variants with half maximal inhibitory concentrations (IC50s) less than 10 ng/mL (Figures 1D and S1A). While class 2/3 nAbs XMA04 and XMA55 showed similar neutralizing activities against the SARS-CoV-2 prototype strain and D614G, compared with XMA01, diminished or abolished neutralizing activities against Beta and Gamma variants were observed, respectively, probably resulting from the mutations of E484 residue (Figures 1D, 1E, and S2).
Figure 1.
Identification of antibodies with broadly neutralizing activities against SARS-CoV-2 variants, including Omicron
(A) Heatmap showing the binding activities of nAbs to different types of spike and RBD proteins determined using ELISA and neutralization potency against lentivirus (LV) pseudoviruses carrying spike protein of SARS-CoV and SARS-CoV-2 prototype strain (termed SARS-CoV LV and SARS-CoV-2 LV, respectively).
(B) Antigenic sites of the select 5 representative antibodies (classes 1–5) are shown on the SARS-CoV-2 RBD surface, and the ACE2 footprint is indicated as a black line.
(C) Epitope mapping of our 11 nAbs and 5 representative nAbs by clustering analysis based on blocking ELISA. Blocking ELISA was performed by using naked nAbs (in black font) to block HRP-coupled nAbs (in blue font). The heatmap of blocking ELISA data is shown with blocking rate colored continuously from white to red (blocking rate >75%) in the scale bar.
(D and E) Neutralization potency of 11 nAbs and antibody S309 against SARS-CoV-2 variants. The IC50 values of each nAb against D614G, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Epsilon (B.1.429), Eta (B.1.525), Lota-a (B.1.526a), Lota-b (B.1.526b), Kappa (B.1.617.1), Lambda (C.37), and A.VOL. V2 in order (D) and the fold change of neutralization against variants compared with D614G (E) are shown. Numbers in the box indicate the fold-change value, and blue color indicates less change in neutralizing potency. Neutralization assays were performed by using an LV pseudovirus system.
(F) Pairwise neutralization potency of nAbs against the Omicron variant and D614G. The IC50 values were determined by using a VSV pseudovirus with Omicron spike protein and are presented in the top panel. The dots in brown refer to Omicron, and the dots in gray refer to D614G. The fold changes of neutralization potency against Omicron related to D614G are presented in the bottom panel. The pink stars indicate three optimal nAbs against SARS-CoV-2 variants, including Omicron. The black dashed lines indicate the 10 and 100 ng/mL IC50 values (top panel) or 10- and 100-time fold changes (bottom panel).
(G and H) Binding affinities of XMA01, XMA04, and XMA09 to Omicron RBD protein (G) and spike protein (H) determined by SPR technology. KD values were calculated using the evaluation software for a Biacore 8K instrument using a 1:1 global fit model.
We next assessed the neutralizing activities of these nAbs against Omicron pseudovirus. Despite an approximately 10-fold decrease in neutralization potency compared with D614G, XMA01 and XMA04 still efficiently neutralized Omicron, with IC50 values of 23.6 and 24.9 ng/mL, respectively; thus, they showed significantly stronger neutralization potency than the widely recognized S309 (sotrovimab) (Cameroni et al., 2022; Pinto et al., 2020), with an IC50 of 284.7 ng/mL against Omicron (Figures 1F, S1B, and S1C). The two nAbs showed effective neutralization against Omicron by blocking ACE2 binding (Figure S2B). Surface plasmon resonance (SPR) assays further confirmed the excellent affinities of XMA01 and XMA04 to Omicron spike or RBD proteins (Figures 1G, 1H, and S3). Of the two SARS-CoV/SARS-CoV-2 cross-neutralizing nAbs, XMA09 was found to bind Omicron and SARS-CoV spikes with excellent picomolar affinity and undiminished neutralizing activity against Omicron pseudovirus when compared with D614G (Figures 1G, 1H, and S3). However, similar to other reported class 5 nAbs, such as S2H97, XMA09 showed poor neutralizing activities against Omicron and other variants (Figures 1F and S1), partially due to its weak ACE2-blocking potential (Figure S2B). Collectively, our findings confirmed that the mutations present in the Omicron RBD mediate substantial resistance to multiple clusters of antibodies and identified XMA01 and XMA04 as potent nAbs against Omicron with excellent affinities. Furthermore, XMA01, XMA04, and XMA09 from class 1, class 2/3, and class 5, respectively, might serve as an antibody cocktail to protect against infection by most SARS-CoV-2 variants including Omicron.
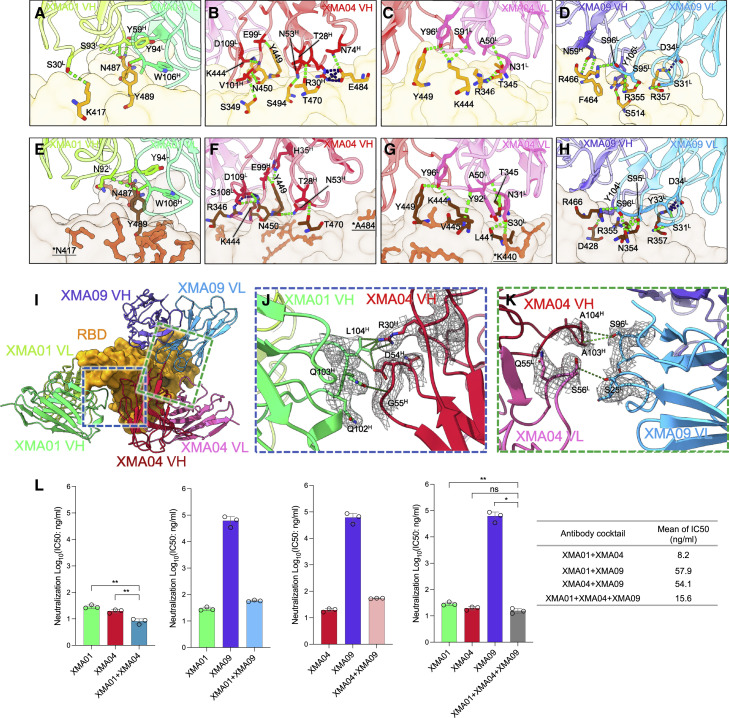
Cryo-EM structures of three antibodies in complex with SARS-CoV-2 WT and Omicron spike proteins
Given the excellent neutralizing breadth and potently noncompeting binding properties of XMA01, XMA04 and XMA09, we then determined the cryo-EM structures of prefusion-stabilized WT-S and Omicron-S in complex with a combination of Fab fragments of three antibodies. Structures of WT-S and Omicron-S simultaneously bound by three antibodies at resolutions of 3.42 and 3.82 Å, respectively, were obtained and showed identical binding modes of the three antibodies (Figures 2A–2D, S4A, S4B, and S5; Table S1; Video S1). Intriguingly, only the spike monomer (but not trimer) bound by the three Fabs was observed and reconstructed, indicating the antibody-induced dissociation of the trimeric spike. To this end, we also determined the cryo-EM structure of trimeric Omicron-S in complex with the mono-antibody XMA01 (Figures 2E, S4C, and S5; Table S1). The medium-resolution structure revealed that three XMA01 bound to three RBDs of the Omicron spike, and the fitting of our three-Fab complex structure revealed that further binding of XMA04 and XMA09, especially the latter, will induce steric clashes with adjacent NTDs on the trimer (Figure 2E). In addition, we also verified the antibody-induced dissociation of spike proteins by negative-staining transmission electronic microscope (TEM). The trimeric WT-S and Omicron-S in complex with XMA09 alone or with the triple-antibody cocktail, respectively, were checked. Both WT-S and Omicron-S were found disrupted by the administration of either XMA09 alone or the triple-antibody cocktail (Figure S6), which is consistent with the observation in cryo-EM analysis. Remarkably, the triple-antibody cocktail aggravated the dissociation potential of spikes compared with XMA09 alone (Figure S6).
Figure 2.
Cryo-EM structures of three antibodies in complex with SARS-CoV-2 WT and Omicron spike proteins
(A and B) Domain-colored cryo-EM map (A) and cartoon representation (B) of the immune complex of WT-S:XMA01:XMA04:XMA09.
(C and D) Domain-colored cryo-EM map (C) and cartoon representation (D) of the immune complex of Omicron-S:XMA01:XMA04:XMA09. WT-RBD and Omicron RBD are colored orange and brown, respectively, and nAbs XMA01, XMA04, and XMA09 are colored spring green, crimson, and dodger blue, respectively.
(E) Cryo-EM density map (transparent gray) of Omicron-S in complex with XMA01 and superimposition of the density map of WT-S:XMA01:XMA04:XMA09 onto one open RBD of Omicron-S:XMA01 shows the steric clashes (orange dashed box) between two adjacent NTDs and two Fabs XMA01 and XMA09.
(F) Structure of WT-S:XMA01:XMA04:XMA09 with highlighted RBD mutations (revealed in the VOCs Alpha, Beta, Gamma, and Delta). WT-RBD and bound nAbs are shown as transparent surfaces and cartoons, respectively.
(G) Structure of Omicron-S:XMA01:XMA04:XMA09 with 15 highlighted mutations in the Omicron RBD (displayed in coral sphere).
(H) Comparisons of the footprints of XMA01 (spring green line), XMA04 (crimson line), and XMA09 (dodger blue line) on WT-RBD (gray surface representation). The ACE2-binding site is marked as the black dotted line, and the mutation sites of Omicron mutations are colored in coral.
(I) Sequence alignment of the spike proteins of SARS-CoV-2, VOCs, and SARS-CoV with strictly conserved residues shown as dots, and the epitopes of three nAbs are highlighted with a color scheme according to (A)–(D).
nAbs targeting the RBD can be categorized into five classes (class 1–5) (Starr et al., 2021a). XMA01, XMA04, and XMA09, which bind to the upper “shoulder”, lower shoulder, and cryptic “waist” regions of RBDs, respectively, belong to class 1, class 3, and class 5 nAbs and resemble AZD8895 (Dong et al., 2021), S309 (Tortorici et al., 2020), and S2H97 (Starr et al., 2021a), respectively (Figure S7). The binding of three nAbs evaded most of the RBD mutations revealed in previous VOCs (K417N, L452R, T478K, E484K, and N501Y) and recent Omicron variants (G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H) (Figures 2F, 2G, and S7A–S7I). The footprints of three nAbs were substantially nonoverlapping except for some common residues involving the binding of two nAbs (Figures 2G and 2H). The footprints of XMA01 and XMA04 contain 2 and 3 Omicron mutations, respectively (K417N and Q493R for XMA01 and N440K, G446S, and E484A for XMA04), which may correspond to their slight decrease in neutralization against Omicron (Figure 2I). In addition, the epitopes of XMA01 and XMA04 overlap with RBD residues involved in ACE2 binding to varying degrees and thereby show potent ACE2-blocking activities (Figure S2B).
Those best-in-class SARS-CoV-2 nAbs mostly belong to classes 1 and 2, which are able to block the binding of ACE2 to the RBM and always show excellent neutralizing potencies (IC50 <10 ng/mL) (Starr et al., 2021a). However, due to the high mutation rate in the Omicron RBM, the neutralization of those nAbs (including some EUA antibodies, such as REGN10933 [class 1], REGN10987 [class 3], LY-CoV555 [class 2], LY-CoV016 [class 1], and AZD8895 [class 1]) against Omicron is almost lost (Cameroni et al., 2022; Wang et al., 2022). Omicron-resistant antibodies, such as S309 (class 3) and S2H97 (class 5), show moderate neutralizing potencies (IC50 of ∼200–1,000 ng/mL) against the Omicron variant (Cameroni et al., 2022; Liu et al., 2021; Planas et al., 2021). XMA01 is categorized as a class 1 nAb but maintains a high potency of Omicron neutralization (IC50: 23.6 ng/mL). For all 15 RBD mutations in Omicron, only the K417N mutation leads to the loss of one hydrogen bond between XMA01 and RBD (Figures 3A and 3E), which may account for the neutralizing decrease of the Omicron variant. The binding site of XMA01 is similar to that of AZD8895 but farther from the notorious loop 470–490, which enriches several VOC mutations, such as S477N, T478K, E484A, Q493R, and G496S (Figure S7J–S7L). Among these residues, S477, T478, and Q493 are key residues for AZD8895 binding, while only Q493 provides appreciable contacts with XMA01 (Dong et al., 2021) (Figure S7J–S7L). Furthermore, residue F486 in the RBD, which was previously revealed to be critical in mediating the interaction of spike to ACE2 by inserting itself into the hydrophobic pocket of ACE2 (Barnes et al., 2020; Dong et al., 2021; Dussupt et al., 2021; Lan et al., 2020; Yan et al., 2020), also plays a critical role in the binding of XMA01 by mimicking the insertion of its bulky aromatic side chain into the hydrophobic pocket formed by the heavy chain S57, T58, Y59 and light chain Y94, W106 of XMA01 and creates strong interactions with these residues (Figure S7M–S7N).
Figure 3.
Comparisons of three antibodies interacting with WT-RBD and Omicron-RBD and the synergistic neutralization of three nAbs against Omicron
(A–D) Interaction details of the WT RBD bound by XMA01 (A), XMA04 (B and C), and XMA09 (D).
(E–H) Parallel display of interaction details of Omicron-RBD bound by XMA01 (E), XMA04 (F and G), and XMA09 (H). Critical residues involved in interactions are shown as sticks, and hydrogen bonds (lime) and salt bridges (dark blue) are labeled as dashed lines.
(I) Overall view of inter-antibody interactions between XMA01 and XMA04 and between XMA04 and XMA09.
(J and K) Close-up views of interaction details between XMA01 and XMA04 (J) and XMA04 and XMA09 (K) are shown. The key residues involved in inter-antibody interactions are shown as sticks, and their representative density maps are also shown as meshes. Hydrogen bonds (lime dashed lines) and contacts (dark green dashed lines) are labeled.
(L) Neutralization of the Omicron variant by monoantibody, two-antibody cocktail (1:1 mixture), and three-antibody cocktail (1:1:1 mixture) based on VSV pseudotyped with Omicron spike protein. IC50 values for antibody cocktails are listed. The IC50 values were calculated using nonlinear regression (four-parameter) by Prism (v.8.0.1). Data are expressed as the means ± SD for three technical replicates. The significant difference between the groups was analyzed by Student’s unpaired two-tailed t test. ns, no significant difference; ∗p < 0.05; ∗∗p < 0.01.
XMA04, another high potent nAb against Omicron (IC50: 24.9 ng/mL), recognizes an epitope that is near the binding site of the well-known S309 but closer to the RBM and therefore shows excellent ACE2-blocking activity (Figure S2B). XMA04 interacts with the WT RBD by forming an elaborate interaction network containing 15 hydrogen bonds and 6 salt bridges (Figures 3B, 3C, and 3F–3G). Although the mutation of E484A in the Omicron RBD causes the loss of 3 hydrogen bonds and 4 salt bridges, the binding affinities of XMA04 to the Omicron spike or RBD are maintained at ∼10 nM. The XMA09 epitope shows high similarity with the epitopes of the previously reported pan-Sarbecovirus broad antibody S2H97 (Starr et al., 2021a), with the final epitope designated site V (the fifth class of epitopes on the RBD). The XMA09 epitope evades all RBD mutations of concern found in most VOCs and VOIs, including Omicron (Figures 3D, 3H, and S8). Furthermore, both XMA09 and S2H97 as well as some other nAbs belonging to this class (Li et al., 2021) can cross-neutralize against SARS-CoV. The special cryptic neutralizing site recognized by those class 5 nAbs was sequentially highly conserved among Sarbecovirus (Figure 2I). Although with unsatisfactory neutralization against both the WT and Omicron variants, these nAbs may neutralize the virus by destabilizing the viral spike protein, which was previously elucidated for some other nAbs, such as CR3022 (Wrobel et al., 2020) and 7D6/6D6 (Li et al., 2021).
Interaction of three antibodies to WT-RBD and Omicron-RBD and the synergistic neutralization against Omicron
The three antibodies bound to one RBD are close to each other and thereby introduce extra inter-antibody interactions between adjacent nAbs (Figures 3I–3K). XMA04 dramatically interacts with ambilateral XMA01 and XMA09 simultaneously and therefore constructs a three-antibody combinated interaction network. This structural feature indicates the synergistic effect of these antibodies when administered in combination. To test the potential synergistic neutralization of antibody combinations against Omicron, we assessed the neutralization potencies of two-antibody (1:1 mixture) and three-antibody (1:1:1 mixture) combinations compared with a mono-antibody. The results indicated that the XMA01/XMA04 cocktail showed an appreciable synergistic effect, with an improved IC50 of 8.2 ng/mL (Figure 3L). In addition, the three-antibody cocktail also displayed potent neutralizing activity against Omicron, with an IC50 of 15.6 ng/mL (Figure 3L). The present antibody cocktail of AZD8895 and AZD1061 (Dong et al., 2021) has been shown to exhibit enhanced neutralizing activities over individual antibodies because of the synergistic effects. Given that the Omicron can escape most approved antibody cocktails, our two- and three-antibody cocktails provide promising antibody-cocktail candidates that are effective against the Omicron variant.
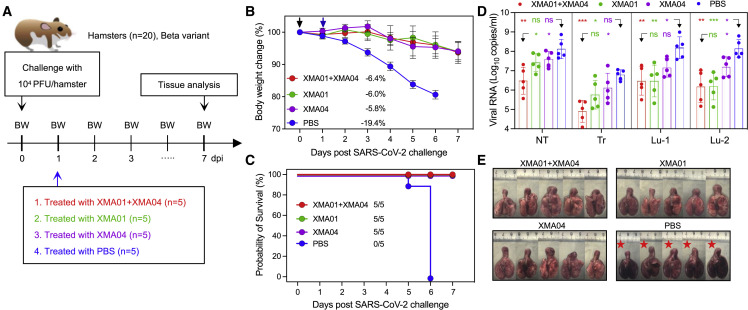
Enhanced therapeutic activities of XMA01/XMA04 cocktail against SARS-CoV-2 Beta and Omicron variants challenge in hamsters
We subsequently evaluated the therapeutic activity of the XMA01, XMA04, and XMA01/XMA04 cocktail against the infection of the Beta variant in a hamster model (Figure 4A). Eight-week-old male hamsters were intranasally challenged with 1 × 104 plaque forming units (PFU) of the Beta variant and treated intraperitoneally with XMA01, XMA04, or XMA01/XMA04 cocktail at a total of 20 mg/kg dose at 1 day post-infection (dpi). Hamsters from the untreated group lost an average of 19.4% body weight, and 100% of the animals were dead within 6 dpi. In contrast, all antibody-treated hamsters survived with slight weight loss: an average decrease of 6.0%, 5.8%, and 6.4% for XMA01-, XMA04-, and XMA01/XMA04-cocktail-treated groups, respectively (Figures 4B and 4C). To determine the viral burden in different groups, we then detected the viral replication in the homogenized tissues of respiratory tract organs including nasal turbinate, trachea, and lung regions proximal and distal to the pulmonary hilum by real-time reverse-transcription polymerase chain reaction (RT-PCR). While the amount of viral RNA in the untreated group surged to approximately 1 × 107–1×109 copies/mL in lung tissues, the antibody-treated groups showed a profound reduction in viral RNA concentrations (1 × 105–1×107 copies/mL) (Figure 4D). Although XMA01 or XMA04 individually could not significantly reduce the amounts of viral RNA in the nonlung respiratory tract, such as nasal turbinate and trachea, the combination of XMA01 and XMA04 succeeded in the significant inhibition of virus replication, especially locally in the nasal turbinate (Figure 4D), indicating the enhanced therapeutic efficacy of antibody combination. The high efficiency of XMA01/XMA04 cocktail for reducing the viral burden in the upper respiratory tract may benefit to decrease the risk of the SARS-CoV-2 transmission within populations. However, the enhancement of XMA01/XMA04 cocktail for the inhibition of virus replication in lung tissues is less conspicuous, possibly due to the high potency and abundant distribution of nAbs in lung tissues. We also assessed the protective efficacies of mono-antibody and XMA01/XMA04 cocktails in viral-infection-related lung damage. For the untreated group, multifocal diffuse hyperemia and consolidation were observed in the gross lung pictures (Figure 4E). In contrast, treatment with XMA01, XMA04, and especially XMA01/XMA04 cocktails effectively inhibited the occurrence of apparent lesions (Figure 4E).
Figure 4.
Therapeutic activity of XMA01/XMA04 cocktail against SARS-CoV-2 Beta variant challenge in hamsters
(A) Schematic design. A total of 20 male hamsters, divided equally into 4 groups, were infected intranasally with the 1 × 104 PFU SARS-CoV-2 Beta variant and then treated intraperitoneally with XMA01, XMA04, and XMA01/XMA04 cocktails at a total dose of 20 mg/kg or PBS as a control at 1 dpi. The body weight was monitored daily, and examination of hamster lung samples was collected at 7 dpi.
(B) Changes in body weight induced by virus challenge were plotted. Data are expressed by means ± SEM. The significant difference between groups was analyzed by Student’s unpaired two-tailed t test.
(C) Survival analysis. The number of surviving hamsters is indicated.
(D) Viral RNA concentrations in lysates from the nasal turbinate (NT), trachea (TR), and lung regions proximal (Lu-1) and distal (Lu-2) to the pulmonary hilum. Data are expressed by means ± SEM. The significant difference between the groups was analyzed by Student’s unpaired two-tailed t test. ns, no significant difference; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(E) Gross observations of lung tissues from the therapeutic and control groups. Red stars indicate hamster death due to virus infection.
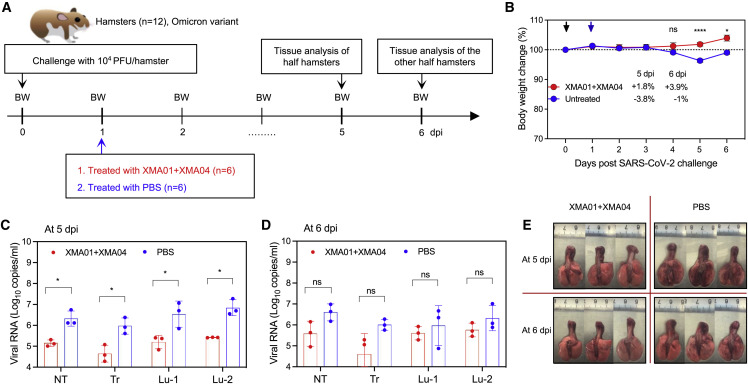
To further determine the therapeutic efficacy of the XMA01/XMA04 cocktail against the Omicron variant, we conducted a similar virus-challenge experiment as that of the Beta variant and evaluated the therapeutic activity of the XMA01/XMA04 cocktail (Figure 5A). Both untreated and treated groups were individually separated into two subgroups, hamsters of which were sacrificed at 5 and 6 dpi for viral RNA load quantification and gross lung observation, respectively. Although no animal death was observed for both the untreated and the antibody-cocktail-treated groups compared with the Beta variant, putatively due to the relatively weaker pathogenicity of the Omicron variant (Figure 5B), the Omicron variant caused slight body weight decrease (3.8%) for the untreated group at 5 dpi but nearly no decrease for the treated group (Figure 5B). Also, the significant decrease of viral RNA loads in the tissues of nasal turbinate, trachea, and lung regions proximal and distal to the pulmonary hilum were achieved by the XMA01/XMA04 cocktail at 5 dpi (Figure 5C). The hamsters in the untreated group recovered from virus infection (increased body weight) to some extent at 6 dpi (Figure 5B); however, the tissue viral RNA loads still maintained at high levels (1 × 106–1 × 107) (Figure 5D). The antibody-cocktail-treated group showed relatively lower tissue viral RNA loads at 6 dpi compared with that of the untreated group (Figure 5D). We noted that the viral RNA loads of the cocktail-treated group at 6 dpi slightly increased (while no statistical difference) when compared with that at 5 dpi, putatively resulted from the decreased antibody concentration in hamsters. Nevertheless, the relatively lower viral RNA loads at 5 and 6 dpi (1 × 105–1 × 106) indicated that antibody cocktail efficiently inhibits viral replication to maintain concentrations of the Omicron RNA at a relatively low level since 5 dpi. Given the reduction of viral loads caused by the XMA01/XMA04 cocktail, no viral-infection-related lung damage was observed in the cocktail-treated group at 5 dpi compared with the untreated group (Figure 5E). Taken together, these results determined that the potently synergistic neutralization of XMA01 and XMA04 effectively protected hamsters from infection and infection-related lung damage of two notorious VOCs, Beta and Omicron.
Figure 5.
Therapeutic activity of XMA01/XMA04 cocktail against SARS-CoV-2 Omicron variant challenge in hamsters
(A) Experimental schedule. Two groups (n = 6) were intraperitoneally treated with PBS and XMA01/XMA04 cocktail (20 mg/kg) at 1 day post intranasally challenge with the 1 × 104 PFU SARS-CoV-2 Omicron variant then were divided into two subgroups (n = 3) that were sacrificed at 5 and 6 dpi, respectively.
(B) Daily body-weight changes were observed and plotted by means ± SEM. The significant differences of body-weight changes at 4, 5, and 6 dpi were analyzed using Student’s unpaired two-tailed t test. ns, no significant difference; ∗p < 0.05; ∗∗∗∗p < 0.0001.
(C and D) Viral RNA concentrations in different tissues as in Figure 4D from subgroups that were separately sacrificed at 5 (C) and 6 dpi (D). Data are expressed by means ± SEM. The significant difference between the groups was analyzed by Student’s unpaired two-tailed t test. ns, no significant difference; ∗p < 0.05.
(E) Gross observations of lung tissues from different subgroups.
Discussion
The ongoing coronavirus 2019 (COVID-19) pandemic causes significant emergence of SARS-CoV-2 VOCs showing increased transmissibility and obvious resistance to the existing vaccines and therapeutic antibodies (Abu-Raddad et al., 2021; Madhi et al., 2021; McCallum et al., 2021; Wang et al., 2021). Currently, the concern is further exacerbated by the rapidly emerging Omicron variant that has an alarming 15 mutations almost accumulating in the key neutralizing epitopes in the RBD (Hadfield et al., 2018; Scott et al., 2021). Many studies have reported that the Omicron variant can abolish the neutralization potency of FDA-approved therapeutic antibodies, just with the exception of S309 (Cameroni et al., 2022; Cao et al., 2021; Cele et al., 2021; Hoffmann et al., 2021; Liu et al., 2021; Planas et al., 2021; Wang et al., 2022). In this study, a panel of human nAbs were obtained from SARS-CoV-2 convalescent patients and showed diverse broad neutralization against SARS-CoV-2 VOCs, of which XMA01 and XMA04 classified into classes 1 and 3, respectively, were identified as more potent nAbs against the Omicron variant by about 10 times higher compared with S309 (Figure 1E). XMA01 derived from the paired germlines of IGHV 3–64 and IGKV15 mainly binds with the epitope focusing on residue F486 by forming several strong interactions. The high conservation of residue F486 that is farther from the notorious loop 470–490 confers XMA01 broad-neutralization potency against SARS-CoV-2 VOCs, including the Omicron variant. Although many AZD8895-like public nAbs with a public clonotype encoded by the same germline sequences (the heavy chain: IGHV1-58 and IGHJ3, and the light chain: IGKV3-20 and IGKJ1), such as S2E12, target the similar antigenic site as XMA01 (Dong et al., 2021; Tortorici et al., 2020), their relatively weaker affinities with conserved F486 and higher overlapping with Omicron mutation sites together mediate the largely decreased neutralization against Omicron variant (Cameroni et al., 2022). Besides, based on the structural analysis, XMA01 exhibits a different binding orientation to the left shoulder compared with IGHV1-58-derived nAbs (Figures S9A–S9C), resulting in contacting an epitope lower than the common site recognized by IGHV1-58-derived nAbs and evading the three VOC mutation residues S477, T478, and E484 (Figures S9D and S9E) (Reincke et al., 2022). Thus, XMA01 maintains more potent and broad neutralization against SARS-CoV-2 variants, and its epitope site focusing on the F486 residue may provide important inspiration for universal vaccine design in the future.
With the exception of the XMA01 epitope, broad nAb XMA09 is also worth investigating. By binding to the special cryptic neutralizing site V that is highly conserved among Sarbecoviruses and distant from ACE2 footprint, XMA09, as well as the previously reported nAb S2H97 (Starr et al., 2021a), broadly inhibit infection of SASR-CoV-2 variants, including the Omicron variant, and even SARS-CoV. The sequence conservation of cryptic neutralizing site V may be necessary for the requirement of structure and function of Sarbecoviruses spike protein. Furthermore, our structural information indicated that XMA09 may destabilize the viral spike protein and further inhibit SARS-CoV-2 infection, as did previously reported CR3022 (Wrobel et al., 2020) and 7D6/6D6 (Li et al., 2021). However, the class 5 nAbs showed less potent neutralization against SARS-CoV-2 when compared with those ACE2-competing nAbs such as XMA01 and XMA04. Thus, further efforts are needed for enhancing neutralization potency, e.g., expansion of antibody volume by modifying immunoglobulin G (IgG) as IgMs (Ku et al., 2021) or particles forms (Ma et al., 2020) to enhance the blocking of the RBD attachment to the receptor.
The Omicron variant, with accumulating mutations, could affect nAbs targeting different antigenic sites. Nevertheless, some class 3 (represented by S309) and 4 (represented by CR3022) nAbs that show broad-neutralization potency against Sarbecovirus are impaired by Omicron mutations (Cao et al., 2021; Liu et al., 2021). The neutralization potency of nAbs BRII-198 (class 3) approved by National Medical Products Administration (NMPA) (Ju et al., 2020) and ADG2 (class 4) under evaluation in clinical trials (Rappazzo et al., 2021) are relatively less impaired by the Omicron variant, with IC50 values ranging from 100 ng/mL to 1 μg/mL (Liu et al., 2021). Because of their epitopes located distantly from the ACE2 footprint, nAbs BRII-198 and ADG2 exhibit lower neutralizing activities against SARS-CoV-2 variants compared with XMA01 and XMA04, which strongly block ACE2 attachment (Figure S2B). Notably, nAb BRII-198 has been reported not invincible; especially, R346 mutation on RBD, which is contained in approximately 10% of Omicron viruses in GISAID, was reported to diminish the activity of BRII-198 (Liu et al., 2021). In contrast, XMA01 could directly escape the adverse influence of R346 mutation by spatial exclusion from interacting with this residue. Another class of public nAbs using IGHV 3–53/66 targeting class 1 antigenic sites (e.g., RENG10933) are substantially escaped by SARS-CoV-2 Beta, Gamma, and Omicron variants due to the abolished interaction of nAb with the negatively charged lysine on residue 417. However, nAb DXP-604, in a clinical trial from this class, shows moderate neutralizing activity against the Omicron variant, with an IC50 value of 287 ng/mL (Cao et al., 2021). XMA01 and XMA04 showed at least 5- to 10-fold higher neutralizing efficacies against Omicron when compared with all the above nAbs, implying their clinical applicability against SARS-CoV-2 and variants.
In addition, many studies have demonstrated that the combination of two noncompeting antibodies can efficiently protect against SARS-CoV-2 infection, and antibody cocktails have been approved under EUA (Baum et al., 2020; Dong et al., 2021; Starr et al., 2021b; Weisblum et al., 2020). Further, we speculate that antibody cocktails composed of three antibodies targeting different epitopes on the RBD are also reasonable and might provide stronger synergistic protection due to their adequate cover of ACE2 footprints as well as multiple neutralization mechanisms. In this study, structure analyses revealed that XMA01, XMA04, and XMA09 recognize three noncompeting epitopes then nearly shield the receptor-binding site to efficiently hinder spike protein attachment with ACE2. Excitingly, XMA04 is conformationally close to the other antibodies, mediating simultaneous interaction with XMA01 and XMA09. Thereby, an appreciable synergistic effect with an improved IC50 of 8.2 ng/mL was observed for the XMA01 and XMA04 cocktail, and the enhanced therapeutic activity of the XMA01 and XMA04 mixture was further confirmed in a hamster model. Previously reported antibodies, AZD8895 and AZD1061, recognized similar epitopes as XMA01 and XMA04, respectively, and also showed synergistic effect to promote the neutralizing activity (Dong et al., 2021). Unfortunately, unlike XMA01 and XMA04, these two antibodies showed a significant loss in neutralization potency against the Omicron variant (IC50s: ∼1–10 μg/mL), which may limit the usage of their combination (Planas et al., 2021).
In summary, our studies generated a panel of human nAbs with diverse broad neutralization against SARS-CoV-2 VOCs. Potent and synergistic neutralization of XMA01, XMA04, and XMA09 against the Omicron variant were determined by both structural and functional analyses. The XMA01 and XMA04 cocktail further provided potent therapeutic activities against the infection of the Beta and Omicron variants in hamsters. Collectively, our results provide insights for the rational development of therapeutic antibody cocktails and universal vaccines against circulating SARS-CoV-2 VOCs and emerging variants in the future.
Limitations of the study
Some limitations are present in this study. The neutralization assay was performed using pseudoviruses instead of authentic variants. While the efficacy of the SARS-CoV-2-related pseudoviruses system has been validated, neutralization assay using the authentic virus could measure a variety of neutralizing mechanisms of nAbs including the ACE2-dependent manner. Due to the limitation of authentic SARS-CoV-2 variants, we just performed therapeutic efficacy of the antibody cocktail against the Beta and Omicron variants in hamster models. Inclusion of the other authentic variants in future studies will illuminate the broad therapeutic efficacy of this antibody cocktail.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| XMA01 | This paper | N/A |
| XMA03 | This paper | N/A |
| XMA04 | This paper | N/A |
| XMA08 | This paper | N/A |
| XMA09 | This paper | N/A |
| XMA12 | This paper | N/A |
| XMA18 | This paper | N/A |
| XMA26 | This paper | N/A |
| XMA47 | This paper | N/A |
| XMA55 | This paper | N/A |
| XMA99 | This paper | N/A |
| C102 | (Barnes et al., 2020) | N/A |
| P2B-2F6 | (Ju et al., 2020) | N/A |
| S309 | (Pinto et al., 2020) | N/A |
| CR3022 | (Wrobel et al., 2020) | N/A |
| S2H97 | (Starr et al., 2021a) | N/A |
| Biological samples | ||
| PBMCs of donor | This paper | N/A |
| Recombinant proteins and peptides | ||
| SARS-CoV S2P protein | This paper | N/A |
| SARS-CoV-2 S2P protein | This paper | N/A |
| SARS-CoV-2 RBD protein | This paper | N/A |
| SARS-CoV-2 Omicron RBD | Sino Biological | Cat#40592-V08H121 |
| SARS-CoV-2 WT-S | Acro Biosystem | Cat#SPN-C52H9-500ug |
| SARS-CoV-2 Omicron-S | Sino Biological | Cat#40589-V08H26 |
| Bacterial and virus strains | ||
| Authentic SARS-CoV-2 Beta (B.1.351) virus (hCoV-19/China/AP100/2021) | This paper | GISAID access number: EPI_ISL_2,779,639 |
| Authentic SARS-CoV-2 Omicron (B.1.1.529) virus | This paper | GISAID access number: EPI_ISL_8,182,026 |
| SARS-CoV wildtype pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 wildtype pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 D614G pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Alpha (B.1.1.7) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Gamma (P.1) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Beta (B.1.351) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Epsilon (B.1429) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Eta (B.1.525) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Lota-a (B.1.526a) variant (S477N) pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Lota-b (B.1.526b) variant (E484K) pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Kappa (B.1.617.1) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Delta (B.1.617.2) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 A.VOI.V2 variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 Lambda (C.37) variant pseudovirus (LV) | (Zhang et al., 2022) | N/A |
| SARS-CoV-2 D614G pseudovirus (VSV) | This paper | N/A |
| SARS-CoV-2 Omicron (B.1.1.529) variant pseudovirus (VSV) | This paper | N/A |
| Critical commercial assays or kits | ||
| Ni Sepharose 6 Fast Flow | Cytiva | Cat#17-5318-03 |
| MabSelect SuRe resin | Cytiva | Cat#17-5474-02 |
| ExpiFectamine™ CHO Transfection Kit | Thermo Scientific | Cat#A29129 |
| SARS-CoV-2 RT-PCR Kit | Wantai | Cat#WS-1248 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat#52906 |
| live/dead-Aqua | Molecular Probes | Cat#L34957 |
| CD3-PE-Cy7 | BD Biosciences | Cat#557851; RRID: AB_396896 |
| CD19-BV786 | BD Biosciences | Cat#563325; RRID: AB_2744314 |
| CD27-BV650 | BD Biosciences | Cat#563228; RRID: AB_2744352 |
| anti-human IgM-PerCP-Cy5.5 | BD Biosciences | Cat#561285; RRID: AB_10611998 |
| anti-human IgG-BV421 | BD Biosciences | Cat#562581; RRID: AB_2737665 |
| Streptavidin-APC | Molecular Probes | Cat#S32362 |
| DyLight 488 NHS Ester | ThermoFisher Scientific | Cat#46402 |
| EZ-Link Sulfo-NHS-LC-Biotin | ThermoFisher Scientific | Cat#21335 |
| Deposited data | ||
| Cryo-EM structure of WT-S:XMA01:XMA04:XMA09 | This paper | EMDB: EMD-32516 PDB: 7WHZ |
| Cryo-EM structure of Omicron-S:XMA01:XMA04:XMA09 | This paper | EMDB: EMD-32517 PDB: 7WI0 |
| Cryo-EM structure of Omicron-S:XMA01 | This paper | EMDB: EMD-32518 |
| Experimental models: Cell lines | ||
| ExpiCHO cells | ThermoFisher Scientific | Cat# A29127 |
| H1299-ACE2hR | (Zhang et al., 2022) | N/A |
| HBK21-hACE2 | (Xiong et al., 2020) | N/A |
| 293T | (Zhang et al., 2022) | N/A |
| Vero cells | ATCC | Cat# CCL-81 |
| Experimental models: Animals | ||
| LVG Syrian hamsters | Charles River | Cat#501 |
| Recombinant DNA | ||
| Plasmid encoding SARS-CoV S2P | This paper | N/A |
| Plasmid encoding SARS-CoV-2 S2P | This paper | N/A |
| Plasmid encoding SARS-CoV-2 RBD | This paper | N/A |
| Software and algorithms | ||
| Columbus Analysis system (version 2.5.0) | PerkinElmer | https://www.perkinelmer.com/ |
| GraphPad Prism (version 8.0.1) | Graphpad | https://www.graphpad.com/ |
| MotionCor2 | (Zheng et al., 2017) | http://msg.ucsf.edu/em/software/motioncor2.html |
| Gctf | (Zhang, 2016) | https://en.wikibooks.org/w/index.php?title=Software_Tools_For_Molecular_Microscopy&stable=0#Gctf |
| Cryosparc V3 | (Punjani et al., 2017) | https://cryosparc.com |
| ResMap | (Kucukelbir et al., 2014) | http://resmap.sourceforge.net |
| Chimera | (Pettersen et al., 2004) | http://www.cgl.ucsf.edu/chimera |
| Coot | (Emsley and Cowtan, 2004) | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot%20 |
| PHENIX | (Adams et al., 2010) | http://phenix-online.org |
| Molprobity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu |
| ChimeraX | (Goddard et al., 2018) | https://www.cgl.ucsf.edu/chimerax/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Ningshao Xia (nsxia@xmu.edu.cn).
Materials availability
All requests for resources and reagents should be directed to the Lead Contact author. All reagents, which includes antibodies, proteins, plasmids, and virus, will be made available on request after completion of a Materials Transfer Agreement for non-commercial usage.
Experimental model and subject details
Cell lines
Cell lines used in this study were obtained from the ATCC (H1299, BHK21, 293T and Vero) or Thermo Fisher Scientific Inc. (ExpiCHO cells). All cell lines used in this study were routinely tested for mycoplasma and found to be mycoplasma-free.
Sample donors and collection
Ten convalescent individuals infected with the SARS-CoV-2 prototype strain (aged 33–71 years) were recruited with informed consent. Then, sera and PBMCs were collected. This study was designed in accordance with the Declaration of Helsinki and subsequently approved by the medical ethics committee of the School of Public Health, Xiamen University.
Method details
Protein expression and purification
The spike proteins and/or RBDs of SARS-CoV, SARS-CoV-2 were obtained as previously reported (Wrapp et al., 2020). In brief, a gene encoding the ectodomain of a prefusion conformation-stabilized spike proteins (GenBank: MN908947, GenBank: MN908947 for SARS-CoV and SARS-CoV-2 spike genes, respectively) with the proline substitutions at 986 and 987 and the ‘GSAS’ substitutions at the S1/S2 furin cleavage site (residues 682–685), a C-terminal T4 fibritin trimerization motif, an HRV3C protease and 8xHisTag were synthesized and individually cloned into pTT5 vector. To determine the blocking capacity of mAbs, we also synthesized gene of SARS-CoV-2 spike fluorescin probe comprising SARS-COV-2 gene sequence, a C-terminal T4 fibritin trimerization motif, an HRV3C protease, 8xHisTag and a C-terminal green fluorescent protein (mGamillus).
Recombinant expression of these proteins was performed by the ExpiCHO expression system (Thermo Scientific, A29133). Briefly, plasmids encoding targeted proteins were transiently transfected into ExpiCHO cells using ExpiFectamine CHO transfection kit (Thermo Scientific, A29129). The cell-free supernatants were obtained 7 days after transfection by centrifugation and filtration with a 0.22 μm filter. Subsequently, the proteins were purified by Ni Sepharose 6 Fast Flow (Cytiva) column, and stored in the PBS buffer.
Specific memory B cell sorting and antibody gene amplification
The SARS-CoV-2 RBD specific B cells were obtained in the same way as previously reported (Tiller et al., 2008). In brief, PBMCs collected from SARS-CoV-2 convalescent individuals were incubated with a cocktail containing live/dead-Aqua (Molecular Probes, dilution: 1 per 100), CD3-PE-Cy7 (BD Biosciences, dilution: 1 per 200), CD19-BV786 (BD Biosciences, dilution: 1 per 200), CD27-BV650 (BD Biosciences, dilution: 1 per 100), anti-human IgM-PerCP-Cy5.5 (BD Biosciences, dilution: 1 per 100), anti-human IgG-BV421 (BD Biosciences, dilution: 1 per 100), RBD-FITC and biotinylated RBD, followed with Streptavidin-APC (Molecular Probes) binding to biotinylated RBD. IgG + memory B cells (CD3-CD19 + CD27+IgM-IgG+) that bind to RBD were single cell sorted from PBMC samples from each donor. Single cells were sorted by fluorescence-activated cell sorting on an Aria III sorter (BD Biosciences) into 96-well PCR plates containing 20 μL per well of lysis buffer (5 μL of 5 × first strand buffer (Invitrogen), 1.25 μL dithiothreitol (Invitrogen), 0.5 μL RNase Out (Invitrogen) and 0.0625 μL Igepal (Sigma)). Then, the antibody variable genes (IgH, Igλ and Igκ) were amplified by real-time (RT-) PCR and nested PCR reactions and were sequenced. Antibody genes were analyzed for the variable regions of IgG heavy and light chains using the IMGT V-quest webserver (http://www.imgt.org/IMGT/vquest).
Expression of antibodies and Fabs
Variable region genes of antibody heavy chain and light chain were codon optimized and cloned into pTT5 vectors (Youbio, VT2202) containing the constant region of human IgG1 heavy chain and light chain, respectively. For the expression of Fabs, the variable region genes of heavy chain were cloned into pTT5 vector just carrying heavy chain constant CH1 region. The paired heavy and light chain expression cassettes were then transiently co-transfected into ExpiCHO cells with equal amounts of plasmids according to the manufacturer’s instructions (Life Technologies), and antibodies were purified from culture supernatant 5–7 days after transfection, using a protein-A column (Cytiva).
Binding assay for nAbs by indirect ELISA
The binding activities of nAbs against SARS-CoV-2 spike protein were determined using an indirect ELISA. The nAbs were added to antigen-coated microwell plates, and incubated at 37°C for 30 min. Then, incubation of HRP-conjugated anti-human antibody at 37°C for 30 min to detect the bound mAbs, followed by washing five times. Finally, substrate solution was incubated for 15 min at 37°C, and stopped by 50 μL of 2 M H2SO4. OD was determined at 450 nm with a reference wavelength of 630 nm. The binding activities of nAbs against SARS-CoV-2 RBD and SARS-CoV spike protein were determined by same method.
Blocking assay by ELISA and cluster analysis
Briefly, the unlabeled nAbs (50 μg per well) or 20mM phosphate-buffered saline (PBS) were added to SARS-CoV-2 RBD-coated 96-well microplates and then incubated for 30 min at 37°C. Next, HRP-conjugated nAbs were added at selected dilutions, at which OD readings was ∼1.5 present. After incubation for 30 min at 37°C, the microplates were rinsed and the color was developed. The blocking rate was measured quantitatively by comparing OD in the presence and absence of competitor mAbs, and transformed using the formula [1- (ODinhibited/ODoriginal)] x100%.
Blocking capacity of nAbs against ACE2 binding
For SARS-CoV-2 spike protein-blocking assay, nAbs were pre-made as 2-fold serial dilutions using DMEM containing 10% FBS. Aliquots (44 μL per well) of diluted samples and spike protein probes (11 μL per well) were mixed in a 96-well plate with U shaped bottom. Half of the culture medium (50 μL) of 293T-ACE2iRb3 cell plate were gently removed, and 50 μL of sample/probe mixtures were added to each well. Cell image acquisitions performed with Opera Phenix (green, red and near-infrared channels in confocal mode) using a 20× water immersion objective at 1-h after probe incubation in wash-free and live-cell conditions.
All quantitative image analyses were based on images that acquired by Opera Phenix (PerkinElmer). All image data were transfer to Columbus system (version 2.5.0, PerkinElmer Inc.) for analysis. Multiparametric image analysis was performed as described in the following. The signals of blue channel or near-infrared channel were used to detect the nucleus. As the ACE2 is a membrane protein, the signals of ACE2-mRuby3 (red channel) were used to determine the cell boundary. Then, the cells were further segmented into the regions of membrane (outer border: 0%, inner border: 15%), cytoplasm (outer border: 20%, inner border: 45%), and nucleus (outer border: 55%, inner border: 100%). The median fluorescence intensity (MFI) of probe channel (Ex488/Em525) in the cytoplasmic region (cMFI) and the MFI of ACE2-mRuby3 (Ex561/Em590) on the membrane were calculated for inter-well normalization. The cMFI inhibition ratio (%) of the test sample was calculated using the following equation: [(cMFIpc-cMFItst)/(cMFIpc-cMFIblk)]×100%. In this formula, the cMFIpc is the cMFI value of probe-only well (as positive control), the cMFItst is the cMFI value of test well and the cMFIblk is the cMFI value of cell-only well. For each plate, 5 replicates of probe-only well and 1 cell-only well were included. The blocking capacities of nAbs were expressed as means of IC50.
Pseudovirus neutralization assay based on lentiviral pseudovirus
Neutralizing capacities of nAbs against SARS-CoV, SARS-CoV-2 ancestral strain and related variant strains were test based on lentiviral (LV) pseudotyping particles bearing spike protein, according to previous report (Chang et al., 2021). In briefly, lentiviral pseudovirions carrying spike protein were produced by co-transfection of a lentiviral packaging plasmid (psPAX2, Addgene), spike plasmids of SARS-CoV-2 variants and a green fluorescent protein (mNeonGreen) reporter vector (pLvEF1α-mNG, carrying EF1α promoter-driven mNeonGreen expressing cassette) in 293 T cells. The mixtures of serially diluted nAbs and LV pseudotyping particles inoculum (0.5 TU/cell), incubated for 1 h, were transferred into 96-well cell culture plates with an optically clear bottom that were pre-seeded with H1299-ACE2hR cells (H1299 cells that stably over-expressing human ACE2 and nuclear-localized H2B-mRuby3) for 36 h incubation. Then, the fluorescence images were collected by Opera Phenix or Operetta CLS high-content equipment (PerkinElmer) and quantitatively determined using Columbus Software 2.5.0 (PerkinElmer). To determine the antibody neutralizing activity, the reduction percentage of mNeonGreen (+) cells in the nAbs treated wells compared to the control wells was calculated. The IC50 value was determined by the 4-parameter logistic (4PL) regression using GraphPad Prism (version 8.0.1) (https://www.graphpad.com).
Pseudovirus neutralization assay based on VSV pseudovirus
Omicron variant and D614G pseudovirus using Vesicular Stomatitis Virus (VSV) carrying the spike protein were produced as our previous study (Xiong et al., 2020). Briefly, mAbs with 2-fold serial dilutions with 10% FBS-DMEM (GIBCO, 12,100,061) from 2 μg/mL were mixed with diluted pseudovirus (MOI = 0.05), incubated at 37°C for 1 h. A mixture of 80 μL was added to the precoated BHK21-hACE2 cells (BHK21 cells stably expressing hACE2). After incubation for 12 h, post-infection cells were fluorescently imaged using Opera phenix or Operetta CLS (PerkinElmer), and quantitatively analyzed by Columbus image management analysis software to detect the number of green fluorescent positive cells. The inhibition rate was calculated by reduction of GFP positive cells with presence of nAbs compared with the untreated control wells. The IC50 value identified by the maximum dilution concentration required to achieve infection inhibition by 50% was determined by the 4-parameter logistic (4PL) regression using GraphPad Prism (version 8.0.1).
Affinity assay
Antibody affinity to spike and RBD proteins were tested using surface plasmon resonance technology (SPR) by a Biacore 8K instrument (GE Healthcare). Firstly, the S2P or RBD was linked to a CM-5 sensor chip (GE Healthcare) by amine-coupled method. Then, serially diluted antibodies (800, 400, 200, 100, 50, 25, 12.5 and 6.25 nM) flow through the sensor surface at a flow rate of 30 μL/min in PBS-P + buffer (0.2 M phosphate buffer with 27 mM KCl, 1.37 M NaCl, and 0.5% Surfactant P20 (Tween 20)). The flow durations were 120 s for the association stage and 300 s for dissociation. Finally, association rates (ka), dissociation rates (kd), and affinity constants (KD) were calculated using evaluation software equipped for Biacore 8K instrument.
Negative-staining electronic microscopy
The SARS-CoV-2 WT-S and Omicron-S (Sino Biological Inc.) were incubated with XMA09-Fab alone and three-nAb Fabs, respectively, for 45 min. Next, the spike proteins (WT-S and Omicron-S) and immune complexes (WT-S:XMA09, WT-S:XMA01:XMA04:XMA09, Omicron-S:XMA09 and Omicron-S:XMA01:XMA04:XMA09) were diluted in PBS (pH 7.4) and then absorbed onto 200 mesh carbon-coated copper grids (Quantifoil Micro Tools) for 1 min. The grids were washed twice with double-distilled water and negatively stained with 2% phosphotungstic acid (pH 6.4) for 30 s. Specimens were evaluated and imaged with the FEI Tecnai T12 TEM at 1,500,00× magnification.
Cryo-EM sample preparation and data collection
Aliquots (3 μL) of 3.5 mg/mL mixtures of purified SARS-CoV-2 WT-S (ACRO Biosystems) or Omicron-S proteins (Sino Biological Inc.) in complex with excess Fab fragments of nAbs were incubated in 0.01% (v/v) Digitonin (Sigma) and then loaded onto glow-discharged (60 s at 20 mA) holey carbon Quantifoil grids (R1.2/1.3, 200 mesh, Quantifoil Micro Tools) using a Vitrobot Mark IV (ThermoFisher Scientific) at 100% humidity and 4°C. Data were acquired using the SerialEM software on an FEI Tecnai F30 transmission electron microscope (ThermoFisher Scientific) operated at 300 kV and equipped with a Gatan K3 direct detector. Images were recorded in the 36-frame movie mode at a nominal 390,00× magnification at super-resolution mode with a pixel size of 0.389 Å. The total electron dose was set to 60 e− Å−2 and the exposure time was 4.5 s.
Image processing and 3D reconstruction
Drift and beam-induced motion correction was performed with MotionCor2(Zheng et al., 2017) to produce a micrograph from each movie. Contrast transfer function (CTF) fitting and phase-shift estimation were conducted with Gctf (Zhang, 2016). Micrographs with astigmatism, obvious drift, or contamination were discarded before reconstruction. The following reconstruction procedures were performed by using Cryosparc V3 (Punjani et al., 2017). In brief, particles were automatically picked by using the “Blob picker” or “Template picker”. Several rounds of reference-free 2D classifications were performed and the selected good particles were then subjected to ab-initio reconstruction, heterogeneous refinement and final non-uniform refinement. The resolution of all density maps was determined by the gold-standard Fourier shell correlation curve, with a cutoff of 0.143 (Scheres and Chen, 2012). Local map resolution was estimated with ResMap (Kucukelbir et al., 2014).
Atomic model building, refinement, and 3D visualization
The initial model of nAbs were generated from homology modeling by Accelrys Discovery Studio software (available from: URL: https://www.3dsbiovia.com). The structure of RBD from the structure of WT trimeric spike (pdb no. 6VSB (Wrapp et al., 2020)) were used as the initial modes of our WT-RBD and Omicron RBD. We initially fitted the templates into the corresponding final cryo-EM maps using Chimera (Pettersen et al., 2004), and further corrected and adjusted them manually by real-space refinement in Coot (Emsley and Cowtan, 2004). The resulting models were then refined with phenix.real_space_refine in PHENIX (Adams et al., 2010). These operations were executed iteratively until the problematic regions, Ramachandran outliers, and poor rotamers were either eliminated or moved to favored regions. The final atomic models were validated with Molprobity (Chen et al., 2010; Robert and Gouet, 2014). All figures were generated with Chimera or ChimeraX (Goddard et al., 2018; Pettersen et al., 2021).
Therapeutic activity against Beta and Omicron variants in hamsters
The therapeutic activity of antibodies against SARS-CoV-2 Beta variant strain (GISAID: EPI_ISL_2,779,639) and Omicron variant strain (share an identical sequence with EPI_ISL_8,182,026) that were passaged on Vero cells (#CCL-81, ATCC) in vivo were performed in a Syrian hamster model (Wu et al., 2021). For Beta variant, groups of 8-week-old male hamsters were intranasally challenged with 1 × 104 PFU of SARS-CoV-2 Beta variant. After 24 h, the infected hamsters were treated intraperitoneally with XMA01, XMA04 and XMA01/XMA04 cocktail (1:1) at 20 mg/kg dose or PBS. The health status of the hamsters was observed and the changes in body weight were recorded daily. The lung tissues of hamsters were collected at 5 days post-infection (dpi). The therapeutic efficacy of XMA04 was determined depending on the indicators including body weight, tissue viral RNA load, and lung pathological examination in gross. For Omicron variant, we conducted similar virus challenge experiment as that of Beta variant. Three hamsters in XMA01/XMA04 cocktail (1:1) and PBS treated groups were sacrificed at 5 dpi and at 6 dpi for viral RNA load quantification and gross lung observation, respectively.
SARS-CoV-2 RNA quantification
The tissue samples including lung, trachea and nasal turbinate were separated from infected hamsters and homogenized with TissueLyser II (Qiagen), and SARS-CoV-2 RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen). Then, the viral RNA concentration was quantified using a SARS-CoV-2 RT-PCR Kit (WS-1248, Wantai BioPharm) according to the manufacturer’s instruction.
Quantification and statistical analysis
GraphPad Prism (version 8.0.1) was used for all statistical calculations. To compare continuous variables, Student’s unpaired two-tailed t test was performed between groups. For statistical difference analysis, p values less than 0.05 were considered statistically significant. ns: no significant difference; ∗: p < 0.05; ∗∗: p < 0.01; ∗∗∗: p < 0.001; ∗∗∗∗: p < 0.0001. IC50 values were calculated by non-linear regression analysis (log(agonist) vs response - Variable slope (four parameters)).
Additional resources
None.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 81991491, 81991495, and 82001756), CAMS Innovation Fund for Medical Sciences (grant no. 2019RU022), and Science and Technology Major Projects of Xiamen (grant no. 3502Z20203023).
Author contributions
T.C., Z.Z., S.W., Q.Z., S.L., J.Z., and N.X. designed the study; S.W., Y.W., and T.L. prepared the recombinant spike proteins; Y.Z., T.Z., J.Z., and Y.W. participated in the neutralization assay; L.Y., T.C., and Y.G. designed the therapy experiment in the hamster model; Z.T., D.Y., X.C., Z.C., and C.L. tested the binding activity and affinity; Q.Z., H.S., and L.L., prepared the cryo-EM grids and recorded the cryo-EM movies; Q.Z., Y.H., and H.S. processed the data and obtained all 3D reconstructions; Z.Z., S.W., H.S., S.L., Z.Z., Q.Z., and N.X. analyzed data; S.W., Z.Z., Q.Z., and S.L. wrote the manuscript; and Q.Y., T.C., Z.Z., S.W., H.S., Q.Z., S.L., J.Z., and N.X. participated in the discussion and interpretation of the results. All authors reviewed and approved the paper.
Declaration of interests
The authors declare that they have no conflicts of interest.
Published: May 24, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110862.
Supplemental information
Data and code availability
Structure coordinates are deposited in the Protein Data Bank under accession codes 7WHZ (WT-S:XMA01:XMA04:XMA09) and 7WI0 (Omicron-S:XMA01:XMA04:XMA09). The corresponding EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-32516 (WT-S:XMA01:XMA04:XMA09), EMD-32517 (Omicron-S:XMA01:XMA04:XMA09) and EMD-32518 (Omicron-S:XMA01). Reagents will be made available to the scientific community by contacting Zizheng Zheng or Ningshao Xia and completing a materials transfer agreement. Video S1 has been deposited on Mendeley at https://doi.org/10.17632/kmg9s6t684.1. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for C.-V. Effectiveness of the BNT162b2 covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021;602:657–663. doi: 10.1038/d41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/d41586-021-03824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Hou W.H., Zhao L., Zhang Y.L., Wang Y.B., Wu L.F., Xu T.T., Wang L.L., Wang J., Ma J., et al. The prevalence of antibodies to SARS-CoV-2 among blood donors in China. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zost S.J., Greaney A.J., Starr T.N., Dingens A.S., Chen E.C., Chen R.E., Case J.B., Sutton R.E., Gilchuk P., et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol. 2021;6:1233–1244. doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V., Sankhala R.S., Mendez-Rivera L., Townsley S.M., Schmidt F., Wieczorek L., Lal K.G., Donofrio G.C., Tran U., Jackson N.D., et al. Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations. Nat. Immunol. 2021;22:1503–1514. doi: 10.1038/s41590-021-01068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein. Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., Lier M., Dopfer-Jablonka A., Jack H.M., Behrens G.M., Pohlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization – implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J.W., Wang R.K., Sun J., Ge X.Y., Yu J.Z., Shan S.S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ku Z., Xie X., Hinton P.R., Liu X., Ye X., Muruato A.E., Ng D.C., Biswas S., Zou J., Liu Y., et al. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 2021;595:718–723. doi: 10.1038/s41586-021-03673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li T., Xue W., Zheng Q., Song S., Yang C., Xiong H., Zhang S., Hong M., Zhang Y., Yu H., et al. Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants. Nat. Commun. 2021;12:5652. doi: 10.1038/s41467-021-25997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021;602:676–681. doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Izu A., Pollard A.J. ChAdOx1 nCoV-19 vaccine efficacy against the B.1.351 variant. N. Engl. J. Med. 2021;385:571–572. doi: 10.1056/NEJMc2110093. [DOI] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., De Marco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein. Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021;602:671–675. doi: 10.1038/d41586-021-03827-2. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D.L., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reincke S.M., Yuan M., Kornau H.C., Corman V.M., van Hoof S., Sanchez-Sendin E., Ramberger M., Yu W., Hua Y., Tien H., et al. SARS-CoV-2 Beta variant infection elicits potent lineage-specific and cross-reactive antibodies. Science. 2022;375:782–787. doi: 10.1126/science.abm5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., Hsiao N.Y., Moyo S., Singh L., Tegally H., Dor G., Maes P., Pybus O.G., Kraemer M.U.G., Semenova E., et al. Track Omicron's spread with molecular data. Science. 2021;374:1454–1455. doi: 10.1126/science.abn4543. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z.M., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell. Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Zhang L., Li Q.Q., Liang Z.T., Li T., Liu S., Cui Q.Q., Nie J.H., Wu Q., Qu X.W., Huang W.J. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes. Infec. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F.W., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Hussain S., Harvey R., Martin S.R., Roustan C., Rosenthal P.B., Skehel J.J., Gamblin S.J. Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein. Nat. Commun. 2020;11:5337. doi: 10.1038/s41467-020-19146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.T., Huang X.F., Yuan L.Z., Wang S.J., Zhang Y.L., Xiong H.L., Chen R.R., Ma J., Qi R.Y., Nie M.F., et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci. Transl Med. 2021;13 doi: 10.1126/scitranslmed.abg1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes. Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei M., Wu Y., Wang J., Hong Y., Huang Y., Yuan L., Ma J., Wang K., Wang S., et al. Cross-species tropism and antigenic landscapes of circulating SARS-CoV-2 variants. Cell. Rep. 2022;38:110558. doi: 10.1016/j.celrep.2022.110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates are deposited in the Protein Data Bank under accession codes 7WHZ (WT-S:XMA01:XMA04:XMA09) and 7WI0 (Omicron-S:XMA01:XMA04:XMA09). The corresponding EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-32516 (WT-S:XMA01:XMA04:XMA09), EMD-32517 (Omicron-S:XMA01:XMA04:XMA09) and EMD-32518 (Omicron-S:XMA01). Reagents will be made available to the scientific community by contacting Zizheng Zheng or Ningshao Xia and completing a materials transfer agreement. Video S1 has been deposited on Mendeley at https://doi.org/10.17632/kmg9s6t684.1. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.