Abstract
The bacterial enzyme MurA catalyzes the transfer of enolpyruvate from phosphoenolpyruvate (PEP) to uridine diphospho-N-acetylglucosamine (UNAG), which is the first committed step of bacterial cell wall biosynthesis. From high-throughput screening of a chemical library, three novel inhibitors of the Escherichia coli MurA enzyme were identified: the cyclic disulfide RWJ-3981, the purine analog RWJ-140998, and the pyrazolopyrimidine RWJ-110192. When MurA was preincubated with inhibitor, followed by addition of UNAG and PEP, the 50% inhibitory concentrations (IC50s) were 0.2 to 0.9 μM, compared to 8.8 μM for the known MurA inhibitor, fosfomycin. The three compounds exhibited MICs of 4 to 32 μg/ml against Staphylococcus aureus; however, the inhibition of DNA, RNA, and protein synthesis in addition to peptidoglycan synthesis by all three inhibitors indicated that antibacterial activity was not due specifically to MurA inhibition. The presence of UNAG during the MurA and inhibitor preincubation lowered the IC50 at least fivefold, suggesting that, like fosfomycin, the three compounds may interact with the enzyme in a specific fashion that is enhanced by UNAG. Ultrafiltration and mass spectrometry experiments suggested that the compounds were tightly, but not covalently, associated with MurA. Molecular modeling studies demonstrated that the compounds could fit into the site occupied by fosfomycin; exposure of MurA to each compound reduced the labeling of MurA by tritiated fosfomycin. Taken together, the evidence indicates that these inhibitors may bind noncovalently to the MurA enzyme, at or near the site where fosfomycin binds.
Both gram-positive and gram-negative bacteria are surrounded by a cell wall which protects the cell from destruction by osmotic pressure. It is well established that interference with cell wall biosynthesis is an excellent mechanism for bacterial killing; for example, penicillin and vancomycin specifically interact with the cell wall at different stages of its formation. A major component of the cell wall is a layer of peptidoglycan (murein), which is a polymer of the sugars N-acetylglucosamine (NAG) and N-acetylmuramic acid, and various amino acids. The bacterial MurA enzyme (UDP-NAG enolpyruvyl transferase), in the first committed step of peptidoglycan biosynthesis, catalyzes the transfer of enolpyruvate from phosphoenolpyruvate (PEP) to UDP-NAG (UNAG), releasing inorganic phosphate (6).
MurA is conserved across both gram-positive and gram-negative bacterial species; gram-negative bacteria have one copy of the murA gene (5), and gram-positive bacteria have two copies (9). MurA is an essential enzyme in that its deletion from Escherichia coli and Streptococcus pneumoniae (both copies) is lethal (5, 9), and it has no mammalian homolog. One marketed antibacterial drug, fosfomycin, is a natural product that is a specific inhibitor of the MurA enzyme (13). MurA is thus an attractive target for antibiotic discovery.
Inhibition of MurA by fosfomycin is well characterized. Fosfomycin is a PEP surrogate which forms a covalent adduct with Cys115 of MurA (16). Resistance to fosfomycin can be achieved by several different mechanisms, including enzymatic modification of the antibiotic (3), decreased uptake of the antibiotic (1), and overexpression of MurA (12). In addition, MurA containing a Cys115Asp mutation was enzymatically active but was resistant to fosfomycin (15).
Despite the availability since 1992 of both recombinant MurA and a straightforward assay method to identify inhibitors of the enzyme (17), fosfomycin has remained the only MurA inhibitor of record in the literature to date. In this study, we report the identification of three new inhibitors of the E. coli MurA enzyme. Each compound apparently has a mechanism distinct from that of fosfomycin, in that a noncovalent enzyme-inhibitor complex appears to be formed. The characterization of these inhibitors is presented.
(This study was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy [D. A. Montenegro, L. Licata, J. Melton, I. Turchi, G. C. Webb, B. D. Foleno, E. Wira, W. Jones, J. Masucci, K. Bush, and E. Z. Baum, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2021, 2000].)
MATERIALS AND METHODS
Compounds.
Fosfomycin (disodium salt, catalog no. P-5396) was purchased from Sigma (St. Louis, Mo.) and was dissolved in distilled H20 at 40 mM. Compounds RWJ-3981, RWJ-110192, and RWJ-140998 and their analogs were obtained from The R. W. Johnson Pharmaceutical Research Institute (Raritan, N.J.) and were dissolved in 100% dimethyl sulfoxide at 10 mg/ml for assay purposes.
Expression of MurA.
E. coli MurA (GenBank accession number AE000399) was expressed as a fusion to the carboxy terminus of the maltose binding protein (MBP) as follows. The entire MurA coding region (nucleotides 6508 to 7768) was amplified from E. coli OC3340 DNA (strain K12) using the primers 5′-AATTCCCGATGGATAAATTTCGTGTTCAG-3′ and 5′-TGCAGATTATTCGCCTTTCACACG-3′ and Pfu polymerase (Stratagene, La Jolla, Calif.). The resultant PCR fragment was flanked by EcoRI and PstI sites which allowed its cloning in frame into the corresponding restriction sites of the pMalC2 polylinker (New England Biolabs, Beverly, Mass.). The 89-kDa MBP-MurA fusion protein (hereafter designated MurA) was expressed and purified by chromatography on amylose resin columns (New England Biolabs) by standard protocols. For mass spectrometry experiments, murA was amplified from E. coli OC3340 DNA using the primers 5′-TATGGATAAATTTCGTGTTCAG-3′ and 5′-TCGAGTTCGCCTTTCACACGCTC-3′ and cloned into the NdeI and XhoI sites of the pET23a(+) polylinker (Novagen). This construct encodes a 46-kDa protein of MurA with LeuGluHis6 at its carboxy terminus (MurA-His). The His-tagged protein was purified on a nickel-nitrilotriacetic acid agarose column (Qiagen, Valencia, Calif.) as recommended by the supplier.
Assay of MurA activity.
MurA (100 nM, 10 μg/ml) was incubated with 20 μM PEP and 75 μM UNAG in 25 mM Tris-HCl (pH 7.8) for 30 min, and inorganic phosphate (Pi) was monitored using malachite green as described previously (17). All MurA incubations were at room temperature. For inhibition studies, MurA and inhibitor were incubated together for 10 min unless otherwise noted, prior to addition of the PEP and UNAG substrates. The IC50 (concentration of inhibitor at which 50% inhibition of MurA is achieved) was determined graphically. In the enzymatic assay, the specific activity of the MurA-His protein and its inhibition by fosfomycin and the compounds were similar to those of the MBP-MurA protein.
Mass spectrometry.
To examine covalent adduct formation, MurA-His (100 μM) was incubated with 1 mM UNAG in 100 μl of 15 mM Tris-HCl (pH 7.8) for 20 min followed by addition of 1 mM inhibitor and continued incubation for 1 h. Samples were desalted, and free compound was removed using MicroSpin G-50 columns (Amersham Pharmacia Biotech, Parsippany, N.J.). Samples were digested with trypsin as described previously (16) except that Triton X-100 was omitted, and peptide masses were determined by matrix-assisted laser desorption ionization mass spectrometry.
Reversibility of inhibition.
MurA (800 nM) and UNAG (1 mM) were incubated alone, or with 63 μM RWJ-3981, RWJ-110192, or RWJ-140998, or 1 mM fosfomycin in 480 μl of 10 mM ammonium acetate (pH 6.8) for 30 min. An aliquot was then set aside for testing as the before-filtration sample. To separate free inhibitor from enzyme-bound complexes, the remainder of each reaction mixture was applied to Ultrafree 0.5-ml centrifugal filtration devices (10K NMWL; Millipore, Bedford, Mass.) that were used as per the manufacturer's instructions. The filtrate should contain free compound and other low-molecular-mass material (<10 kDa); the retentate, containing MurA and MurA inhibitor complexes, becomes concentrated (to 5 to 20 μl) during each centrifugation step. The retentate was washed three times with 0.3 ml of 10 mM ammonium acetate (pH 6.8). The amounts of protein recovered in the washed retentates were determined by bicinchinonic acid protein assay (Pierce, Rockford, Ill.) and were similar for all of the samples (≈50%). Samples were then assayed for MurA activity, with the before-filtration and after-washing samples separately normalized to their respective MurA controls (MurA sample without added inhibitor, 0% inhibition; reaction containing no MurA, 100% inhibition).
Competition of each inhibitor with [3H]fosfomycin for binding to MurA.
MurA (13 μM) and UNAG (225 μM) were incubated in 100 μl of 10 mM Tris-HCl (pH 7.8) with 1 mM RWJ-3981, RWJ-110192, or RWJ-140998 or 4 mM fosfomycin for 10 min. [3H]fosfomycin (1 μCi of >0.5 Ci/mmol, 20 μM final concentration; SibTech, Newington, Conn.) was added, and the incubation was continued. Aliquots (20 μl in duplicate) were removed at 10 and 35 min and added to 100 μl of 0.5-mg/ml bovine serum albumin used as carrier. Cold 10% trichloroacetic acid (TCA; 1 ml) was added to each sample, and labeling of MurA by [3H]fosfomycin was determined by precipitation onto Whatman GF/A filters and scintillation counting as described earlier (11). The extent of labeling of MurA with [3H]fosfomycin in the absence of added inhibitor is defined as the 100% control; counts per minute retained on the filter in the absence of MurA is defined as background (0% control).
MIC determinations.
Broth microdilution MICs were determined according to the National Committee for Clinical Laboratory Standards (18). The MIC is the lowest concentration of compound that inhibited bacterial growth.
Macromolecular synthesis and membrane damage assays.
DNA, RNA, and protein synthesis were monitored by the incorporation of tritiated thymidine, uridine, or amino acids, respectively, into TCA-precipitable material as described previously (11) using S. aureus ATCC 29213. Peptidoglycan synthesis was monitored in a similar fashion by incorporation of [3H]NAG (Amersham catalog no. T-2238, 8.3 Ci/mmol) into TCA-precipitable material, using 0.25 μCi of [3H]NAG per ml of S. aureus culture. Compounds were used at four times the MIC and were administered 10 min before the addition of radiolabel. Membrane damage was assessed by the BacLight assay (Molecular Probes, Eugene, Oreg.), which was performed as described previously (11) on S. aureus 29213 cells at the MICs and at four times the MICs of fosfomycin, RWJ-3981, RWJ-110192, and RWJ-140998.
Modeling studies.
The X-ray crystal structure of MurA with bound fosfomycin was used in modeling studies (23). The docking programs FlexX (19) and Dockvision (10) were used to dock RWJ-3981, RWJ-110192, and RWJ-140998 into the catalytic site of the MurA enzyme. Nonprotein atoms were removed prior to the docking runs. The poses from both programs showed similar orientations for all three inhibitors. The program Hint (14) was used to analyze the more important interactions of the inhibitors with the residues in the catalytic site of the enzyme. The AM1 Hamiltonian (8) was used to calculate the lowest unfilled molecular orbital (LUMO) energies of the analogs of RWJ-3981.
RESULTS AND DISCUSSION
Identification of three inhibitors of MurA.
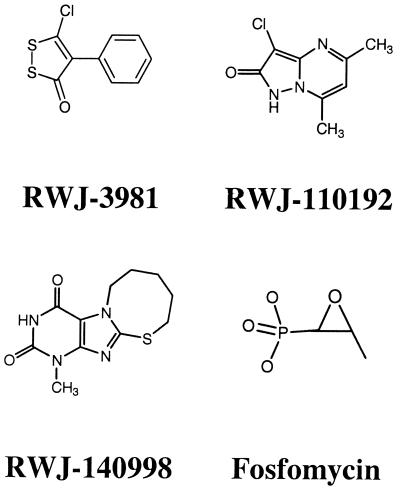
By screening a chemical library, compounds RWJ-3981, RWJ-110192, and RWJ-140998 were identified as inhibitors of the E. coli MurA enzyme (Fig. 1). All three compounds exhibited lower MurA IC50s (0.2 to 0.9 μM) than did the known MurA inhibitor, fosfomycin (IC50 = 8.8 μM) (Table 1). The three compounds have no apparent structural similarity to fosfomycin. Compound RWJ-3981 is a cyclic disulfide. Compound RWJ-110192 is a pyrazolopyrimidine. RWJ-140998 is a purine and contains the 2,4-dioxopyrimidine ring which is also found in the uracil portion of the UNAG substrate. Inhibition of MurA by RWJ-3981, RWJ-110192, or RWJ-140998 was prevented by dithiothreitol (10 mM) added either before or after the enzyme-inhibitor preincubation. In contrast, inhibition of MurA by fosfomycin was unaffected by the presence of dithiothreitol.
FIG. 1.
Structures of MurA inhibitors.
TABLE 1.
Inhibition of MurA as a function of preincubation conditions
| Compounds used:
|
IC50 (μM) of:
|
||||
|---|---|---|---|---|---|
| In 10-min preincubationa | To initiate reaction | Fosfomycin | RWJ- 3981 | RWJ- 110192 | RWJ- 140998 |
| E + Ib | UNAG + PEP | 8.8 | 0.20 | 0.30 | 0.90 |
| E + I + UNAG | PEP | 0.40 | 0.040 | 0.040 | 0.070 |
| E + I + PEP | UNAG | 12.0 | 0.20 | 0.20 | 0.80 |
E, MurA enzyme; I, inhibitor.
Standard conditions unless otherwise noted.
Presence of UNAG during MurA inhibitor preincubation lowers the observed IC50.
Inhibition of MurA by the PEP surrogate fosfomycin requires the presence of the substrate UNAG, which effects a conformation change in the enzyme (16, 22). From structural studies, it has been determined that MurA assumes an open conformation in the absence of ligand (21) and a closed conformation in the presence of UNAG and fosfomycin (23). The binding of UNAG creates an induced fit which renders the enzyme catalytically competent (20, 22). Furthermore, MurA is isolated from E. coli with PEP covalently bound to Cys115; addition of UNAG turns over the bound PEP to product, purging the enzyme (4). Since the new MurA inhibitors might bind to the blocked PEP site and/or require UNAG for binding to the enzyme, the effects of the order of addition of reaction components on IC50s were examined (Table 1).
The presence of PEP during preincubation of MurA with each inhibitor did not change the IC50 observed with preincubation of only MurA and inhibitor. In contrast, the presence of UNAG during the preincubation of MurA with inhibitor decreased the IC50 22-fold for fosfomycin, from 8.8 to 0.4 μM. Similarly, when UNAG was present during the preincubation, each of the RWJ inhibitors exhibited a decrease in its IC50, as follows: 5-fold for RWJ-3981, 7.5-fold for RWJ-110192, and 13-fold for RWJ-140998. The decrease in the IC50s suggests that the presence of UNAG enhances the interaction of the three inhibitors with MurA, possibly in a manner similar to that of the binding of fosfomycin. It is possible that the binding of each compound requires the enzymatically active conformation of MurA, which is achieved in the presence of UNAG (20, 22, 23). In addition, if the binding site of a particular compound overlaps with the PEP binding site, the effect of UNAG may be to purge MurA of PEP and provide access to the binding site.
Since MurA was present in the assay at 100 nM, the IC50s for the compounds (40 to 70 nM) in the presence of UNAG are close to the theoretical lower limit of 50 nM (half the enzyme concentration) for a 1:1 stoichiometric addition of inhibitor to enzyme. These data suggested that the possibility of formation of a covalent or tightly bound complex between MurA and each inhibitor should be investigated.
Lack of covalent adduct formation between RWJ-3981, RWJ-110192, or RWJ-140998 and MurA.
The mechanism of inhibition of MurA by fosfomycin is well established. Fosfomycin forms a covalent adduct with the active site nucleophile Cys115 of MurA (15, 16, 23). Using conditions that detected the covalent adduct between fosfomycin and MurA-Cys115 in mass spectrometry of tryptic digests, none of the three RWJ inhibitors appeared to form a covalent adduct with MurA, either at Cys115 or elsewhere on the MurA protein (data not shown).
Determination of reversibility of inhibition.
To determine whether the inhibition by the compounds was reversible, each compound was incubated with MurA and UNAG, followed by filtration and washing to separate free compound and other low-molecular-weight components from free enzyme and from enzyme-inhibitor complex. An irreversible complex is expected to remain inhibited after washing.
Incubation of MurA with fosfomycin, RWJ-3981, RWJ-110192, or RWJ-140998 prior to filtration and washing substantially inhibited each sample (77 to 116% inhibition) (Table 2). After washing, the fosfomycin sample, which is expected to covalently and irreversibly modify the MurA protein, remained inhibited. Similarly, the RWJ-3981 and RWJ-140998 samples remained inhibited after washing. In contrast, the RWJ-110192 sample, which was fully inhibited before washing, recovered 64% of its activity, displaying only a 36% inhibition after washing.
TABLE 2.
Effects of filtration and washing on the reversibility of the putative MurA-inhibitor complex
| Compound | % Inhibitiona
|
Reversible | |
|---|---|---|---|
| Before filtration | After washing | ||
| Fosfomycin | 111 ± 10 | 132 ± 10 | No |
| RWJ-3981 | 116 ± 17 | 101 ± 3 | No |
| RWJ-110192 | 101 ± 13 | 36 ± 24 | Yes |
| RWJ-140998 | 77 ± 27 | 102 ± 10 | No |
Average of two (RWJ-3981) or three experiments. MurA and each inhibitor were subjected to three cycles of ultrafiltration and dilution to remove free inhibitor and then assayed for activity as described in Materials and Methods.
Based on these data, inhibition of MurA by RWJ-140998 and RWJ-3981 appeared to be irreversible; inhibition by RWJ-110192 appeared to be reversible. The lack of evidence for covalent adducts between MurA and each compound from the tryptic peptide mass spectrometry experiment discussed above would suggest that the basis for inhibition is complex formation via strong noncovalent binding of compound to MurA. The binding of RWJ-110192 would appear to be weaker than that of RWJ-3981 or RWJ-140998 since inhibited enzyme recovered partial activity during washing.
Competition with [3H]fosfomycin for binding to MurA. To determine whether interaction with each inhibitor prevented the binding of fosfomycin to MurA, MurA (with UNAG) was incubated with each inhibitor, followed by incubation with [3H]fosfomycin and determination of radioactivity bound to MurA. As expected (Table 3), the presence of unlabeled fosfomycin reduced the labeling of MurA to background levels.
TABLE 3.
Labeling of MurA with [3H]fosfomycin after preincubation with compounds
| Samplea | [3H]fosfomycin labeling in cpm (% of control)b after:
|
|
|---|---|---|
| 10 min | 35 min | |
| Background (no MurA) | 127 ± 73 (0) | 97 ± 34 (0) |
| MurA | 896 ± 25 (100) | 1091 ± 59 (100) |
| MurA + unlabeled fosfomycin | 143 ± 34 (2) | 145 ± 23 (5) |
| MurA + RWJ-3981 | 120 ± 28 (−1) | 90 ± 7 (−1) |
| MurA + RWJ-110192 | 354 ± 16 (29) | 582 ± 114 (49) |
| MurA + RWJ-140998 | 140 ± 35 (2) | 112 ± 9 (1) |
Reaction mixtures containing MurA, UNAG, and the indicated compounds were preincubated for 30 min followed by addition of [3H]fosfomycin and TCA precipitation as described in Materials and Methods.
% of control = [(sample cpm − background)/(MurA cpm − background)] × 100.
Each of the three compounds (RWJ-3981, RWJ-110192, and RWJ-140998) reduced the labeling of MurA by [3H]fosfomycin compared to that of the MurA control, suggesting that these compounds may bind at or near the active site of the enzyme and prevent access of fosfomycin. Compared to RWJ-3981 and RWJ-140998, RWJ-110192 was less effective at preventing labeling of MurA with [3H]fosfomycin. Whereas labeling of MurA by [3H]fosfomycin was essentially abolished by the presence of either RWJ-3981 or RWJ-140998, labeling in the presence of RWJ-110192 was 29% at 10 min and 49% at 35 min, suggesting either weaker interactions with MurA than fosfomycin or less overlap in the binding sites of RWJ-110192 and fosfomycin. Moreover, the increase in labeling of MurA at 35 min compared to 10 min suggested that the MurA-RWJ-110192 interaction was reversible, consistent with the partial recovery of MurA activity observed after filtration and washing of MurA with RWJ-110192 (Table 2).
Thus, all three inhibitors reduced the labeling of MurA by [3H]fosfomycin, suggesting that the compounds may bind at or near the active site. However, we cannot exclude the possibility that the binding of the compounds may cause a conformation change in MurA which prevents fosfomycin from binding.
Modeling studies: docking of inhibitors into the active site of MurA.
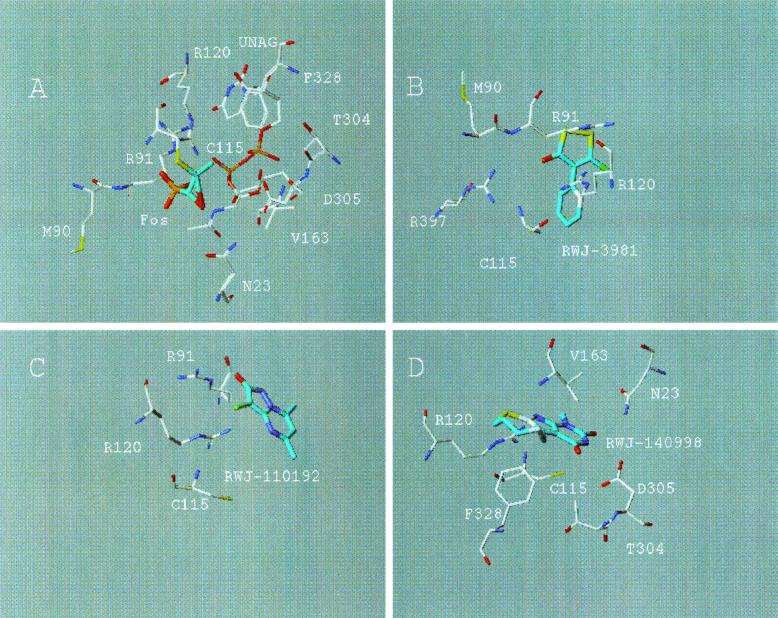
The crystal structure of the MurA enzyme complexed with the substrate UNAG and with fosfomycin, a PEP surrogate which binds at the PEP site, consists of two globular domains with the active site located between them (23). This structure was used as the starting point for modeling studies, to dock each of the compounds into the active site of MurA. The predicted binding of each compound is shown in Fig. 2.
FIG. 2.
DockVision poses of the crystal structure of MurA with bound fosfomycin (A), RWJ-3981 (B), RWJ-110192 (C), and RWJ-140998 (D). The carbons of the inhibitors are shown in cyan.
For RWJ-3981 (Fig. 2B), the centroid of the sulfur-containing ring was predicted to bind 7.1 Å from the sulfur of the catalytic Cys115. The carbonyl oxygen of the inhibitor may form strong hydrogen bonds with Arg120 and Arg397. The sulfur atoms can make hydrophobic contacts with the side chain carbon atoms of Met90 and Arg91.
For RWJ-110192 (Fig. 2C), the centroid of the rings was predicted to bind farther from Cys115 than for RWJ-3981. The centroid of the bicyclic ring system was 8.8 Å from the sulfur of Cys115. This inhibitor may overlap with the NAG of UNAG. The carbonyl oxygen can form a strong hydrogen bond with Arg91. Also, there may be a relatively strong electrostatic attraction between the guanidinium group of Arg91 and the negatively charged pyrazolone ring of RWJ-110192.
RWJ-140998 (Fig. 2D) was also predicted to bind farther away than RWJ-3981 from the catalytic Cys115. The centroid of the rings was 8.7 Å from the sulfur of Cys115. Although RWJ-140998 contains the 2,4-dioxopyrimidine ring also found in the uracil portion of the UNAG substrate, the proposed binding of the dioxopyrimidine of RWJ-140998 was relatively far (>10 Å) from the UNAG site. The NH of the pyrimidindione ring can form a strong hydrogen bond to Asp305, while the carbonyl oxygen may form a hydrogen bond to Asn23. Arg120 may form a hydrogen bond with one of the nitrogen atoms of the imidazole. The phenyl ring of Phe328 can form a hydrophobic contact with the sulfur atom of the thiazocine ring, while the side chains of Thr304 and Val163 may form hydrophobic contacts with the methylene carbons of this ring.
Confirmation of these hypotheses about the interactions between MurA and RWJ-3981, RWJ-110192, and RWJ-140998 would require cocrystallization studies of MurA with each inhibitor.
SARs of RWJ-3981 and RWJ-140998.
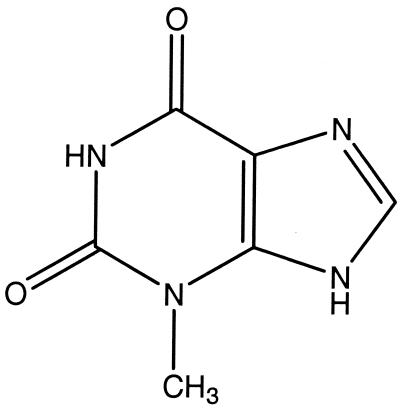
To investigate the structure-activity relationships (SARs) of RWJ-3981 and RWJ-140998, several structural analogs of these two compounds were tested for inhibitory activity in the MurA assay. The RWJ-140998 analog, 1-methylxanthine (Fig. 3), did not inhibit the MurA enzyme at a concentration of 25 μM. Thus, some features of the thiazocine ring appear to be necessary for inhibitory activity, consistent with the prediction of interactions between MurA and this moiety in the modeling study (Fig. 2).
FIG. 3.
Structure of 1-methylxanthine.
For the RWJ-3981 analogs (Table 4), the presence of the chlorine on the ring containing the disulfide appeared to be essential for activity, as Compound 3, which lacked the chlorine, was inactive (IC50 > 25 μM). Replacing the chlorine with a dimethylamino group (Compound 2) decreased activity about 100-fold. Replacing it with 4-methoxyanilino, 2-hydroxyethylthio, or N-morpholino (Compounds 4, 5, and 6, respectively) decreased activity >100-fold. Compound 1, with a dichlorinated phenyl ring, was approximately as active as RWJ-3981.
TABLE 4.
Structure-activity relationship (SAR) of analogs of RWJ-3981
| Compound name or no. | Substituent at position:
|
IC50 (μM) | LUMO energy (kcal/mol) | |
|---|---|---|---|---|
| R1 | R2 | |||
| RWJ-3981 | Cl | Phenyl | 0.2 | −59.5 |
| 1 | Cl | 3,4-Dichlorinated phenyl | 0.4 | −63.6 |
| 2 | Dimethylamino | Phenyl | 22 | −50.0 |
| 3 | H | Phenyl | >25 | −55.8 |
| 4 | 4-Methoxyanilino | Phenyl | >25 | −50.7 |
| 5 | 2-Hydroxyethylthio | Phenyl | >25 | −56.0 |
| 6 | N-Morpholino | Phenyl | >25 | −52.1 |
The SAR of the dithiolanones (Table 4) demonstrated that the chlorine atom is necessary for activity, yet Hint calculations showed no significant interaction of the chlorine with any of the neighboring residues in the active site of MurA in the modeling studies. The LUMO energies of the dithiolane derivatives were calculated (Table 4). The LUMO is centered on the disulfide moiety in the ring, suggesting that attack of a nucleophile (i.e., Cys115) would occur at one of the sulfur atoms. Replacing the chlorine at position 5 of the dithiolanone ring lowered the LUMO energy of RWJ-3981 by 3.7 kcal/mole relative to Compound 3, which had no substitutions. All of the other substitutions (dimethylamino, 4-methoxyanilino, 2-hydroxyethylthiol, or N-morpholino [Compounds 2, 4, 5, and 6, respectively]) increased the LUMO energy relative to that of RWJ-3981. These results suggest that RWJ-3981 should be more reactive than the other analogs toward nucleophilic attack, thus rationalizing the observed SAR.
The presence of a disulfide in RWJ-3981 does indeed raise the possibility that Cys115 of MurA could perform nucleophilic attack, with the opening of the ring of RWJ-3981 and concomitant formation of a disulfide bond between the compound and Cys115. This mechanism would be analogous to that observed for fosfomycin, with nucleophilic attack by Cys115 at C-2 of the antibiotic, opening of the ring of the epoxide, and formation of a covalent bond between C-2 and Cys115 (13, 16). However, repeated attempts to detect a putative covalent adduct between MurA and RWJ-3981 by mass spectrometry were unsuccessful, under conditions that did detect the adduct between MurA and fosfomycin. If the covalent adduct did form in the case of RWJ-3981, it did not survive the conditions of mass spectrometry. The docking programs used in the modeling study cannot address covalent binding between proteins and ligands. Thus, it is possible that the irreversible complex detected between MurA and RWJ-3981 (Table 2) was the result of either tight binding or covalent adduct formation.
Antibacterial activity of the MurA inhibitors.
Each of the MurA inhibitors was tested for antibacterial activity using both gram-negative and gram-positive bacteria (Table 5). Each of the compounds had modest gram-positive antibacterial activity, with MICs of 4 to 32 μg/ml against the strains of Staphylococcus aureus, Enterococcus faecalis, and Enterococcus faecium tested. Like fosfomycin, RWJ-3981 inhibited a lipopolysaccharide-deficient E. coli strain approximately equally as well as it inhibited S. aureus. Gram-positive bacteria contain two copies of murA, encoding MurA1 and MurA2. Both MurA1 and MurA2 from S. pneumoniae were inhibited by fosfomycin in enzyme assays (9). Our enzyme assays used E. coli MurA, and it is not known whether RWJ-3981, RWJ-110192, and RWJ-140998 can inhibit either of the MurA enzymes from gram-positive bacteria.
TABLE 5.
MICs of MurA inhibitors against various bacterial strains
| Bacterial strain | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|
| RWJ- 3981 | RWJ- 110192 | RWJ- 140998 | Fosfomycin | |
| E. coli OC2530a | 8 | >32 | >32 | 16 |
| E. faecalis ATCC29212 | 8 | 16 | 16 | >32 |
| E. faecium OC3312 | 8 | 16 | 16 | >32 |
| S. aureus ATCC29213 | 4 | 32 | 32 | 16 |
| MRSA OC2878 | 4 | 16 | 16 | 16 |
| S. aureus OC4172 | 4 | 16 | 16 | 1 |
Hypersusceptible, LPS deficient.
Because fosfomycin uptake is enhanced by the presence of glucose-6-phosphate due to the induction of the hexose phosphate transporter system (2, 7, 13), broth microdilution MICs were also determined in the presence of this sugar (25 μg/ml) (data not shown). The MICs of fosfomycin did decline as much as eightfold for S. aureus 29213, OC2878, and the hypersensitive E. coli but were unchanged for the other strains tested. However, for RWJ-3981, RWJ-110192, and RWJ-140998, glucose-6-phosphate had no effect on MICs for any of the strains, consistent with the lack of structural similarity of the compounds to either fosfomycin or to hexose phosphate.
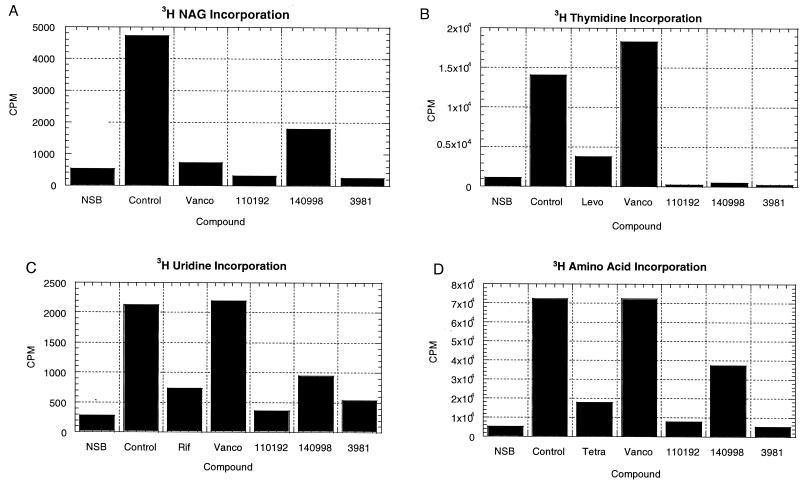
To determine whether the three compounds inhibited peptidoglycan synthesis in S. aureus, the incorporation of [3H]NAG was examined (Fig. 4A). In addition, the effects of these compounds on DNA, RNA, and protein synthesis were determined (Fig. 4B, C, and D). All three compounds inhibited peptidoglycan synthesis in S. aureus, as determined by reduced incorporation of [3H]NAG compared to that of the control (Fig. 4A). However, all three compounds also inhibited DNA, RNA, and protein synthesis within 10 min at four times the MIC (Fig. 4B, C, and D show [3H]thymidine incorporation, [3H]uridine incorporation, and [3H]-amino acid incorporation, respectively). The rapid inhibition of multiple bacterial functions suggested that the antibacterial activities of these compounds against S. aureus in the MIC assay was not due to specific inhibition of MurA. In contrast, inhibition by vancomycin was specific to peptidoglycan synthesis, without inhibiting DNA, RNA, or protein synthesis (Fig. 4B, C, and D). In an attempt to ascertain whether cell wall synthesis might be specifically inhibited at a lower concentration of the compounds, the labeling experiment was repeated at the MIC (data not shown). RWJ-3981 and RWJ-110192 inhibited DNA, RNA, protein, and cell wall synthesis from 78 to 99%, whereas RWJ-140998 did not inhibit any of these processes by more than 22% under these conditions. Thus, for all three compounds, it was not possible to detect specific inhibition of cell wall biosynthesis.
FIG. 4.
Effects of MurA inhibitors on peptidoglycan (A), DNA (B), RNA (C), and protein synthesis (D) in S. aureus 29213 cells. Vancomycin, levofloxacin, rifampin, and tetracycline were positive controls for inhibition of peptidoglycan, DNA, RNA, and protein synthesis, respectively. NSB, nonspecific binding (cpm bound to filter in the absence of cells).
Because of our previous experience with membrane-damaging agents which inhibited bacterial DNA, RNA, and protein synthesis within 10 min of exposure, these compounds were tested in a propidium iodide uptake assay to measure cellular integrity (11). None of the compounds appeared to damage the bacterial cell membrane (data not shown). RWJ-3981 did not protect mice from death in an S. aureus lethal infection model (data not shown), nor did it cause overt toxicity when dosed at 80 mg/kg of body weight.
In summary, we have identified and characterized three new inhibitors of the MurA enzyme which are chemically different from fosfomycin and appear to bind to the enzyme. These compounds represent three new scaffolds available for further chemical modification to develop MurA inhibitors with increased specificity and antibacterial activity.
ACKNOWLEDGMENTS
We thank Raul Goldschmidt and Yuan Chang for cloning of the MurA enzymes, Jeffrey Fernandez and Haiyong Jin for enzyme purification and characterization, and John Masucci and Bill Jones for mass spectrometry experiments. We thank John Melton, Mike Loeloff, and Ellyn Wira for assistance with screening, and Jamese Hilliard for performing the BacLight and some macromolecular synthesis assays. We thank Raul Goldschmidt, Anne Marie Queenan, and Mark Macielag for critical reading of the manuscript.
REFERENCES
- 1.Arca P, Reguera G, Hardisson C. Plasmid-encoded fosfomycin resistance in bacteria isolated from the urinary tract in a multicenter survey. J Antimicrob Chemother. 1997;40:393–399. doi: 10.1093/jac/40.3.393. [DOI] [PubMed] [Google Scholar]
- 2.Barry A L, Fuchs P C. In vitro susceptibility testing procedures for fosfomycin tromethamine. Antimicrob Agents Chemother. 1991;35:1235–1238. doi: 10.1128/aac.35.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernat B A, Laughlin L T, Armstrong R N. Fosfomycin resistance protein (fosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry. 1997;36:3050–3055. doi: 10.1021/bi963172a. [DOI] [PubMed] [Google Scholar]
- 4.Brown E D, Marquardt J L, Lee J P, Walsh C T, Anderson K S. Detection and characterization of a phospholactoyl-enzyme adduct in the reaction catalyzed by UDP-N-acetylglucosamine enolpyruvoyl transferase, MurZ. Biochemistry. 1994;33:10638–10645. doi: 10.1021/bi00201a010. [DOI] [PubMed] [Google Scholar]
- 5.Brown E D, Vivas E I, Walsh C T, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugg T D H, Walsh C T. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- 7.Dette G A, Knothe H, Schoenenbach B, Plage G. Comparative study of fosfomycin activity in Mueller-Hinton media and in tissues. J Antimicrob Chemother. 1983;11:517–524. doi: 10.1093/jac/11.6.517. [DOI] [PubMed] [Google Scholar]
- 8.Dewar M J S, Zoebisch E G, Healy E F, Stewart J J P. Development and use of quantum mechanical molecular models. 76. AM1: a new general purpose quantum mechanical molecular model. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- 9.Du W, Brown J R, Sylvester D R, Huang J, Chalker A F, So C Y, Holmes D J, Payne D J, Wallis N G. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J Bacteriol. 2000;182:4146–4152. doi: 10.1128/jb.182.15.4146-4152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart T N, Ness S R, Read R J. Critical evaluation of the research docking program for the CASP2 challenge. Proteins Suppl. 1997;1:205–209. doi: 10.1002/(sici)1097-0134(1997)1+<205::aid-prot27>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Hilliard J J, Goldschmidt R M, Licata L, Baum E Z, Bush K. Multiple mechanisms of action for inhibitors of histidine protein kinases from bacterial two-component systems. Antimicrob Agents Chemother. 1999;43:1693–1699. doi: 10.1128/aac.43.7.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horii T, Kimura T, Sato K, Shibayama K, Ohta M. Emergence of fosfomycin-resistant isolates of Shiga-like toxin-producing Escherichia coli O26. Antimicrob Agents Chemother. 1999;43:789–793. doi: 10.1128/aac.43.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahan F M, Kahan J S, Cassidy P J, Kropp H. Mechanism of action of fosfomycin (phosphonomycin) Ann N Y Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 14.Kellogg G E, Joshi G S, Abraham D J. New tools for modeling and understanding hydrophobicity and hydrophobic interactions. Med Chem Res. 1991;1:444–453. [Google Scholar]
- 15.Kim D H, Lees W J, Kempsell K E, Lane W S, Duncan K, Walsh C T. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 16.Marquardt J L, Brown E D, Lane W S, Haley T M, Ichikawa Y, Wong C H, Walsh C T. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 17.Marquardt J L, Siegele D A, Kolter R, Walsh C T. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J Bacteriol. 1992;174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 19.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 20.Schonbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- 21.Schonbrunn E, Sack S, Eschenburg S, Perrakis A, Krekel F, Amrhein N, Mandelkow E. Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin. Structure (London) 1996;4:1065–1075. doi: 10.1016/s0969-2126(96)00113-x. [DOI] [PubMed] [Google Scholar]
- 22.Schonbrunn E, Svergun D I, Amrhein N, Koch M H J. Studies on the conformational changes in the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine enolpyruvyltransferase (MurA) Eur J Biochem. 1998;253:406–412. doi: 10.1046/j.1432-1327.1998.2530406.x. [DOI] [PubMed] [Google Scholar]
- 23.Skarzynski T, Mistry A, Wonacott A, Hutchinson S E, Kelly V A, Duncan K. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, and enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure (London) 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]