Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most prevalent types of cancer and is responsible for close to one million annual deaths globally. In Pakistan, HCC accounts for 10.7% of cancer incidence. Prior studies indicated an association between interleukin 4 (IL-4) and cytotoxic T lymphocyte protein 4 (CTLA-4) gene polymorphisms in many types of cancers, including HCC that are either hepatitis B virus (HBV)- or hepatitis C Virus (HCV)-induced. The association of IL-4 and CTLA-4 genetic polymorphisms with HCV-induced HCC is not yet determined in the Pakistani population. Therefore, this research is designed to investigate the implication of IL-4 and CTLA-4 gene polymorphisms by determining the association of IL-4 -590 C/T (rs2243250) and CTLA-4 + 49 A/G (rs231775) with HCC in Pakistan.
Methods
Different bioinformatics tools were employed to determine the pathogenicity of these polymorphisms. Samples were collected from HCV-induced HCC patients, followed by DNA extraction and ARMS-PCR analysis.
Results
The SNP analysis results indicated a positive association of IL-4 -590C/T and CTLA-4 + 49A/G gene polymorphisms with HCV-induced HCC in Pakistan. The CTLA-4 polymorphism might enhance therapeutic efficiency of HCC chemotherapy medicines. The IL-4 polymorphism might introduce new transcription factor binding site in IL-4 promoter region.
Conclusion
This study delineated risk factor alleles in CTLA-4 and IL-4 genes associated with HCV-mediated HCC among Pakistani patients that may have application to serve as genetic markers for pre- and early diagnosis and prognosis of HCC in HCV patients.
Keywords: HCC, CTLA-4, Gene polymorphism, IL-4, HCV
Background
Hepatocellular carcinoma (HCC) is a multifactorial primary liver malignancy caused by excessive alcohol intake, obesity and viral infections [1]. Globally, hepatitis B and C virus infections are a major cause of HCC. Chronic HBV and HCV infections have been responsible for 44% and 22% of HCC cases, respectively [2]. HCC is responsible for almost 830,180 deaths around the globe [3]. In Pakistan, about 70% of HCC is imputed to HCV, whereas HBV is the major etiological contributor of HCC disease in Asian and Pacific countries [4].
HCV promotes generation of cytokines which contribute to HCC progression. Cancer-associated genetic polymorphisms have been identified in cytokines that might facilitate their targeting for anti-cancer therapies [5]. Cytokine IL-4 is involved in regulation of various important biological functions including B-cell mitogenesis, class switching to IgE, cell homeostasis, and tissue repair [6]. During T-cell signalling, IL-4 promotes T-cell differentiation. In cancer, IL-4 functions in the tumor microenvironment to mediate pro-tumor activity by activating tumor-associated or myeloid-derived suppressor cell (MDSC) associated macrophages [7]. Compared to IL-4, CTLA-4CTLA-4 acts as a negative regulator of T-cell signalling. It is present on T-cells and binds B7-1 (CD-80) and B7-2 ligands (CD-86). It induces changes that directly stop TCR immune synapse and block CD28 signaling, thus downregulating interaction of T-cells with antigen-presenting cells [8]. Antibodies that target CTLA-4CTLA-4 are used clinically as anti-cancer immunotherapeutic agents [9].
Genetic variants of IL-4 and CTLA-4 are reported to have association with several cancers including HCC, colorectal cancer, and head and neck cancer [10–14]. A recent publication indicated an IL-4IL-4 variant (rs2243250) with relation to HCV-induced HCC in an Egyptian population [15]. Previously, its association was delineated in a Chinese population [16]. Similarly, association between CTLA-4 rs231775 and HCV-induced HCC was found in the Chinese Han population [17] and an Egyptian population [18]. Previous studies have delineated the contribution of pathogenic single nucleotide polymorphisms (SNPs) on clinical outcomes in cancer patients. The presence of certain SNPs boosts the efficiency of some treatment drug’s efficacy, leading to increased patient survival rate. For example, the genetic variant rs9582036 was identified in the VEGFR1 receptor that increased patient survival after treatment with bevacizumab [19]. Likewise, HCC patients having the KDR gene rs1870377 AA genotype were reported to show better response to first-line therapy sorafenib [19].
In cancer, CTLA-4 mediated modulation of IL-4IL-4 is responsible for immune dysregulation [20]. During HBV infection, up-regulated expression of CTLA-4 results in the increased activity of IL-4 that functions to generate anti-infection response [21]. During HIV infection, CTLA-4 and IL-4 polymorphism influenced the treatment response in patients [22]. Similarly, CTLA-4 and IL-4 polymorphism is also reported to induce HCC progression [23]. The association of genetic polymorphisms of IL-4 and CTLA-4 with HCV-mediated HCC is not investigated in the Pakistani population. The objectives of the current research include the prediction of pathogenicity of IL-4 -590 C/T (rs2243250) and CTLA-4 + 49 A/G (rs231775) and the study of association between IL-4 -590 C/T (rs2243250) and CTLA-4 + 49 A/G (rs231775) polymorphisms with HCV-induced HCC. The IL-4 polymorphism lies in the promoter region; therefore, the potential influence of IL-4 -590 C/T polymorphism on the regulation of IL-4 expression was also estimated. Structural change in CTLA-4 protein due to the polymorphism was predicted and investigation for CTLA-4 structural changes on the therapeutic efficiency of HCC first and second line therapeutics was carried out. This study determined putative pre-diagnostic genetic marker for HCV-induced HCC and provided insight on functional impact of IL-4 and CTLA-4 genetic variants in HCV-mediated HCC.
Methods
In silico method
Data retrieval
SNPs of IL-4 and CTLA-4 were selected by identifying their integral role in several diseases through literature review [15, 24, 25]. Information regarding the variants of IL-4 and CTLA-4 was retrieved from ENSEMBL having genome assembly “GRCh38:CM000667.2” & “GRCh38:CM000664.2”, respectively. ENSEMBL and RegulomeDB were employed to get co-ordinates of IL-4 (+ 49 A/G) and CTLA-4 (-590 C/T) SNP and mapped on genome assembly GRCH38.p13. Protein data bank was accessed for downloading protein structures of CTLA-4 and IL-4.
SNP Analysis
Pathogenicity of IL-4 SNP (Variant ID: rs2243250) and CTLA-4 SNP (ID: rs231775) was predicted using the following six tools: SIFT, PolyPhen2.0, REVEL, MetaLR, MutationAssessor and CADD [26]. SIFT and PolyPhen2.0 predicts protein function changes based on homology in the primary sequence and physiochemical relationship between neighboring amino acids. REVEL integrates prediction scores of SIFT, PolyPhen, MutationAssessor, MetaLR, REVEL, and CADD to make a pathogenicity prediction for a variant. MetaLR uses allele frequency and logistic regression integrating variant deleteriousness scores to predict impact of missense variants. MutationAssessor employs evolutionary conservation knowledge for predicting functionality alteration of the protein on the basis of homology. CADD is used for the prediction of SNP variants deleteriousness via functional or evolutionary conservation data. Regulome DB was used to determine the coordinates, rank, and score of the SNPs [27].
Transcription regulation prediction
Alibaba 2.0 was employed to predict the alteration in transcription factor binding sites (www.gene-regulation.com). HOPE project analysis was used to determine protein structural alteration and predict resultant pathogenicity (www.cmbi.umcn.nl/hope/).
In situ mutagenesis
Protein tertiary structure for CTLA-4 was retrieved from Alphafold database (AF-P16410-F1). Mutation in wildtype CTLA-4 structure was introduced through the mutagenesis tool of PyMol v4.0.2. Both mutated and wildtype structures of CTLA-4 were superimposed to highlight differences.
Molecular docking
Chemical structures for Avastin (PubChem 349,985,080) and Sorafinib (PubChem 216,239) were downloaded from PubChem database. Docking of the drugs against CTLA-4 (AF-P16410-F1) was done in CB Dock and the vina scores and cavity sizes were retrieved. PyMOL was used for docked structure visualization (http://clab.labshare.cn/cb-dock/php/). LigPlot (version v.1.4.5) was used to visualize hydrogen bonds and hydrophobic interactions between protein and ligand [28].
In vitro method
Study design and patient selection
A retrospective case–control study was conducted in which patients were recruited from the Combined Military Hospital Rawalpindi, Pakistan. The sample size (N) was 429, out of which 213 were HCC patients and 216 were control patients. The inclusion criterion was set as HCV-infected HCC patients and all other HCC patients were excluded. The confirmation of HCV infection in patients was achieved by quantifying the viral load through quantitative-reverse transcriptase polymerase chain reaction (qRT-PCR). Isolation of HCV RNA from the patients’ plasma was performed through Instant virus RNA kit CE IVD ªAnalytik Jena. TaqMan probes were employed for quantifying the HCV in iQ5 real-time PCR detection system ªBio-Rad Laboratories. Healthy subjects were used as control. Sample size was estimated through the procedure explained by Suresh and Chandrashekara (2015) [29] and validated using the G*Power Software version 3.1.9.2 for Windows. Sample collection was performed after written consent was obtained from the subjects. The study was approved by the Institutional Review Board committee, National University of Sciences and Technology, Islamabad, Pakistan (IRB No. 04–2019-03/06). The study was performed in accordance to the principles of the Declaration of Helsinki [30].
Design of primers
Primer1 (http://primer1.soton.ac.uk/primer1.html) was used for designing the allele-specific primer sets for IL4 -590 C/T (rs2243250) and CTLA4 + 49 A/G genes. For CTLA4, forward and reverse primer sequences used as internal controls were CACAAGGCTCAGCTGAACCTGGATG (for Allele A) and ACAGGAGAGTGCAGGGCCAGGTCCTAGT (for Allele B), respectively. Forward and reverse outer primers were GTGGGTTCAAACACATTTCAAAGCTTCAGG and TCCATCTTCATGCTCCAAAAGTCTCACTC, respectively. For IL4, forward and reverse primer sequences used as internal controls were TCACGGATTTCTGTTGTGTTTC and GCCTCCCAACCATTCCCTTA, respectively. While ACACTAAACTTGGGAGAACATTGTC for detection of C allele and ACACTAAACTTGGGAGAACATTGTT for T allele detection for used.
DNA extraction
Total DNA was isolated from blood using standardized phenol–chloroform protocol [31].
Tetra amplification refractory mutation system polymerase chain reaction (Tetra ARMS-PCR)
Tetra ARMS-PCR was used for identification of CTLA-4 + 49 A/G and IL-4 -590 C/T polymorphism in genes. Reaction mixture (total volume 20 µl) was prepared by adding 5X buffer solution (4μL), 2 mM dNTPs (2μL), Taq polymerase (0.5 U), primer (10 pmol each), distilled water (10.9 μL) and DNA template (1 μL). The conditions used in PCR were: Denaturation (95˚C for 4 min), 25 cycles of denaturation (95 °C for 45 s), annealing (58 °C for 45 s), and extension (72 °C for 45 s), and a final extension (72 °C for 5 min). Two percent agarose gel was used to analyze the PCR product of ARMS-PCR. A 100 bp ladder was also loaded alongside the PCR products for comparison of size. The results were analyzed by Wealtec dolphin-doc gel analysis systems.
Statistical analysis
Microsoft office 2016 Excel (Rehmond, WA, USA) was used for organization of the genotyping results. Statistical significance of results was tested using GraphPad Prism software ver8.0.1 (GraphPad Software Inc., San Diego, CA, USA). Genotype distribution of gene polymorphism and their associated strength (odds ratio, OR and relative risk, RR) in patients and healthy control was calculated by two-way Fisher’s exact test. Koopman asymptotic score and Baptista-Pike method was computed for the calculation of OR and RR, respectively. A probability of less than 0.05 was taken as significant.
Results
In silico prediction of pathogenicity for CTLA-4 SNP + 49A/G
RegulomeDB provided the coordinates, score, and rank of the variant (Variant ID: rs231775). Rank of CTLA-4 + 49A/G polymorphism was 5, depicting transcription factor binding sites and DNase peak in CTLA-4. The score of this SNP was 0.13454, showing less regulatory functional role of the SNP. ENSEMBLE indicated CTLA-4 + 49A/G SNP as a missense variant. Table 1 includes RegulomeDB annotation score and information regarding the chromosomal location (Chr:bp), allelic mutation (A), amino acid alteration (AA) and coordinates (AA coord.) of CTLA-4 SNP. In silico pathogenicity prediction results obtained for CTLA + 49A/G polymorphism (Variant ID: rs231775) were: SIFT “Tolerated” (score: 0.2), PolyPhen “Benign” (score: 0.009), CADD “Likely Benign” (Score: 0), REVEL “Likely Benign” (Score: 0.007), MetaLR “Tolerated” (Score: 0) and MutationAssessor “low” (Score: 0.28). Overall, CTLA-4 SNP rs231775 was predicted as “benign or non-pathogenic”.
Table 1.
Annotation scores and chromosomal location (Chr: bp), allelic mutation (A), amino acid alteration (AA) and coordinates (AA coord.) of CTLA4 and IL4 SNP
| CTLA4 | IL4 | ||
|---|---|---|---|
| ENSEMBLE | Variant ID | rs231775 | rs2243250 |
| Chr:bp | 2:203,867,991 | 5:132,009,153 | |
| Alleles | A/G | C/T | |
| Variant type | Missense | Promoter region | |
| AA | T/A | – | |
| AA coord | 17 | – | |
| Regulome Annotation | SCORE | 0.13454 | 0.60906 |
| RANK | 5 | 4 |
HOPE structural alteration analysis for CTLA-4
HOPE analysis was employed to predict the potential influence of this SNP on protein structure and function. HOPE predicted that CTLA-4 SNP (Variant ID: rs231775) mutation (threonine converted to alanine) leads to substitution of a small sized amino acid residue that is more hydrophobic than the wild type residue. The substitution of alanine may affect the interaction of CTLA-4 with other proteins, and increased hydrophobicity may alter folding of the protein due to loss of H-bonds. The altered amino acid was not located in the conserved region, suggesting that this mutation may not be highly damaging.
CTLA-4 variant influence on therapeutic efficiency of chemotherapeutic drugs
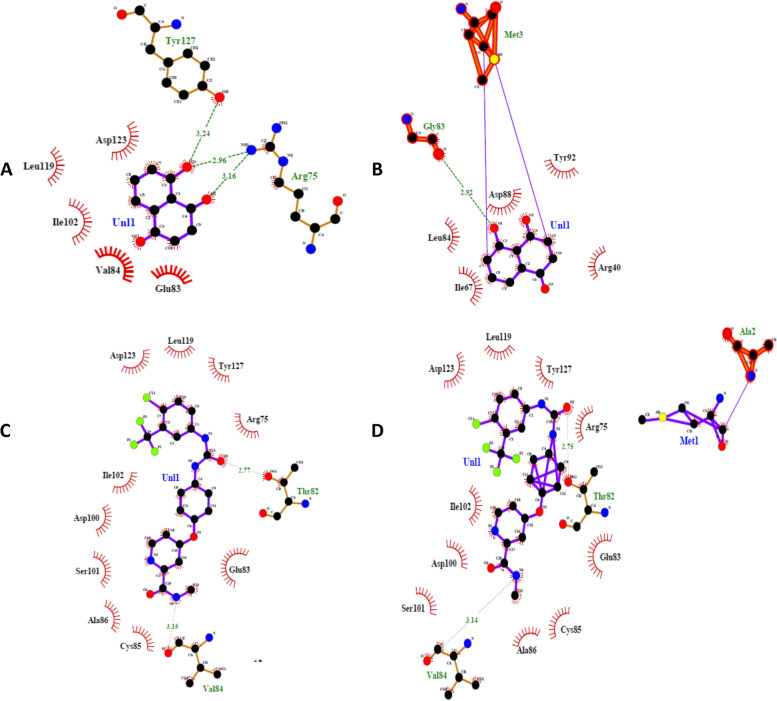
FDA approved Sorafenib is a first line treatment drug, whereas Avastin is a second line treatment drug for HCC. Both drugs were docked through CB-Dock with wildtype and mutated CTLA-4 protein to analyse the influence of SNP on binding energy and interaction of CTLA-4 with Avastin and Sorafenib. Table 2 depicts results obtained via CB-Dock with the Vina score and Cavity size of different docked structures. Molecular docking outcomes indicated that introduction of alanine in place of threonine at position 17 in CTLA-4 resulted in the addition of a covalent bond in both CTLA-4-Avastin and CTLA-4-Sorfanib complexes. Avastin binding with CTLA-4 was possible due to generation of five hydrophobic and two hydrogen bond interactions. Amino acids Glu83, Val84, Ile102, Leu119, and Asp123 participated in non-electrostatic interactions, whereas amino acids Arg75 and Tyr127 hydrogen bonded with Avastin. Residue Arg75 made two hydrogen bonds with the oxygen atoms of Avastin with estimated distance 2.96 Å and 3.19 Å (Fig. 1a). CTLA-4 + 49A/G polymorphism reduced the number of hydrogen bonds and introduced a covalent bond (Met3) (Fig. 1b). Similar influence of CTLA-4 polymorphism was found for the CTLA-4 and Sorafenib model complex. Sorafenib interacted with CTLA-4 by making two hydrogen bonds (Thr82 and Val84) and ten hydrophobic interactions (Cys85, Glu83, Ala86, Ser101, Asp100, Ile102, Asp123, Leu119, Tyr127 and Arg75). Allele G substitution in CTLA-4 resulted in the addition of four hydrophobic interactions along with a covalent bond (Fig. 1c and 1d).
Table 2.
CB Dock scores as a result of docking between CTLA4/IL4 (Proteins) & Tetrahydroxyflavanone (ligand)
| Protein | Drug | Vina score |
|---|---|---|
| CTLA4 (wild) | Avastin | -5.1 |
| Sorafinib | -7.3 | |
| CTLA4 (Mutated) | Avastin | -5.0 |
| Sorafinib | -7.3 |
Fig. 1.
Molecular interaction between CTLA-4 wildtype/mutated and Avastin and Sorafenib predicted through LigPlot. A CTLA-4 (wildtype) and Avastin complex. B CTLA-4 (mutated) and Avastin complex. C CTLA-4 (wildtype) and Sorafebin complex and. D CTLA-4 (mutated) and Sorafebin complex. Purple lines represent interaction between ligand atoms. Orange line depicts interaction between protein atoms. Hydrogen bonding is shown in green dotted lines. Red spiked semi circles denote hydrophobic interactions. Purple line between ligand and amino acid represents covalent bonds
In silico prediction of SNP pathogenic impact on IL-4
RegulomeDB ranked the IL-4 SNP at 4, indicating transcription factor binding sites and DNase peak, and a score of 0.60906 suggested high regulatory function of the SNP (Table 1). Alibaba 2.0 transcription site analysis estimated the generation of two additional transcription binding sites for MIG1 and SP1 due to IL-4 rs2243250. SP1 is located within the E1 region that is responsible for enhancer activity. This SP1 site-generating mutation is thought to have no effect on normal functioning of the E1 region, but more research is needed to draw any conclusion [32].
Viral load of HCV in HCV-induced HCC patients
Viral load of HCV was determined through qRT-PCR analysis in the plasma of HCV-induced HCC patients to ensure the HCV infection. Viral load more than 800,000 IU/L is considered high. In this study, high viral load of HCV was observed and mean viral load along with standard deviation found was 339,840,893.2 ± 1,092,261,683 IU/L.
Comparison of CTLA-4 + 49A/G and IL-4 -590 C/T genotypes in HCV-associated HCC patients
Association of CTLA-4 and IL-4 polymorphism with pathogenicity of HCV-induced HCC was evaluated through ARMS-PCR in 213 patient samples. Outcomes indicated that the prevalence of genotype AG (62.9%) in CTLA-4 was more than GG genotype (37.1%) (Table 3). The OR and RR scores indicated that individuals with AG genotype are more at risk of developing HCV-induced HCC (< 0.0001). IL-4 genotypes CC and CT had association with HCV-induced HCC (< 0.0001). However, values obtained of RR and OR indicated CC as risk factor genotype and CT as protective genotype for HCV-mediated HCC (Table 3).
Table 3.
Genotype frequencies of CTLA4 + 49A/G and IL4 -590C/T polymorphism in HCV induced HCC patients and Control groups
| Gene | Genetype | Patient (n = 213) | Control (n = 216 | Odds ratio | 95% CI –odds ratio | Relative risk | 95% CI—relative risk | P value |
|---|---|---|---|---|---|---|---|---|
| n, (%) | n, (%) | |||||||
| CTLA4 | AG | 134 (62.9%) | 55 (25.5%) | 4.96 | 3.29 -7.52 | 2.154 | 1.76—2.64 | < 0.0001 |
| GG | 79 (37.1%) | 161 (74.5%) | 0.20 | 0.13—0.30 | 0.4643 | 0.37—0.56 | < 0.0001 | |
| IL4 | CC | 138 (64.8%) | 60 (27.8%) | 4.78 | 3.19 -7.17 | 2.14 | 1.75—2.65 | < 0.0001 |
| CT | 46 (21.6%) | 136 (62.9%) | 0.16 | 0.11—0.25 | 0.37 | 0.28—0.48 | < 0.0001 | |
| TT | 29 (13.6%) | 20 (9.3%) | 1.54 | 0.84—2.84 | 1.22 | 0.91—1.53 | 0.17 |
Gender-based comparison of CTLA-4 + 49A/G and IL-4 -590 C/T genotypes in HCV-associated HCC patients
Gender based analysis of CTLA-4 + 49A/G and IL-4 -590 C/T genotypes revealed both AG and GG genotypes of CTLA-4 were significantly associated with the disease. AG genotype in both genders was found as pathogenic while GG genotype was indicated as protective (Table 4). Similarly, genotypes CC and CT have significant association with HCV-induced HCC in both females and males. CC genotype in both genders eas indicated as risk factor while CT was shown to be protective (Table 4).
Table 4.
Gender based genotype frequencies comparison of CTLA4 + 49A/G and IL4 -590C/T polymorphism in HCV induced HCC patients and Control groups
| Gene | Gender based genotypes | HCV induced HCC % | Control % | Oddsratio | 95% CI-odds ratio | Relativerisk | 95% CI- relative risk | P value |
|---|---|---|---|---|---|---|---|---|
| Present | Present | |||||||
| CTLA4 | Male AG | 28.50% | 18.65% | 4.46 | 2.43—8.26 | 2.11 | 1.66–2.87 | < 0.0001 |
| Male GG | 18.65% | 28.50% | 0.22 | 0.12—0.41 | 0.47 | 0.34 -0.63 | < 0.0001 | |
| Female AG | 33.62% | 18.30% | 5.32 | 3.06—9.35 | 2.16 | 1.66—2.85 | < 0.0001 | |
| Female GG | 35.25% | 64.75% | 0.18 | 0.11—0.33 | 0.46 | 0.35 -0.60 | < 0.0001 | |
| IL4 | Male CC | 32.64% | 16.06% | 5.15 | 2.80 -9.56 | 2.37 | 1.70—3.37 | < 0.0001 |
| Male CT | 9.84% | 31.61% | 0.17 | 0.09—0.34 | 0.37 | 0.24—0.55 | < 0.0001 | |
| Male TT | 4.66% | 5.18% | 1.01 | 0.41—2.70 | 1.01 | 0.56 -1.51 | > 0.99 | |
| Female CC | 31.91% | 12.34% | 4.62 | 2.68—8.04 | 2.01 | 1.56—2.62 | < 0.0001 | |
| Female CT | 11.49% | 31.91% | 0.14 | 0.083 -0.25 | 0.37 | 0.26—0.51 | < 0.0001 | |
| Female TT | 8.51% | 3.83% | 2.26 | 0.99—5.42 | 1.39 | 1.00—1.77 | 0.07 |
Association of co-existence of CTLA-4 + 49A/G and IL-4 -590 C/T genotypes in HCV-associated HCC patients
In the present study, the influence of the co-existence of CTLA-4 and IL-4 genotypes in HCV-mediated HCC patients was also evaluated. Genotypes GG (CTLA-4) and TC (IL-4) co-existed in 11.74% of patients and demonstrated significant association with HCV-mediated HCC. However, OR and RR indicated their co-existence as protective. Likewise, genotypes CA (CTLA-4) and TT (IL-4) were found to have co-existence in 8.45% patients with significant association with disease. Contrary to genotypes GG (CTLA-4) and TC (IL-4), genotypes CA (CTLA-4) and TT (IL-4) were determined as risk factor. Similarly, genotypes GA (CTLA-4) and CC (IL-4) were also identified as risk factor and co-existed in around 45% of the patients (Table 5).
Table 5.
CTLA-4 and IL-6 co-existing genotypes, along with their relative risk, odds ratio and p-value in HCV-induced HCC patients
| Genotype (CTLA4 + IL6) | Number | Relative Risk (95% CI) | Odds Ratio (95% CI) | P-value | |
|---|---|---|---|---|---|
| Patients (%) | Control (%) | ||||
| GG + TT | 11 (5.16%) | 14 (6.48%) | 0.88 (0.52—1.28) | 0.78 (0.35—1.71) | 0.68 |
| GG + TC | 25 (11.74%) | 107 (48.61%) | 0.31 (0.21—0.43) | 0.140 (0.085—0.23) | < 0.0001 |
| GG + CC | 43 (20.19%) | 40 (18.52%) | 1.05 (0.82—1.31) | 1.113 (0.68—1.81) | 0.71 |
| GA + TT | 18 (8.45%) | 6 (2.91%) | 1.51 (1.10—1.85) | 3.077 (1.26—7.56) | 0.01 |
| GA + TC | 21 (9.86%) | 29 (13.43%) | 0.82 (0.57—1.12) | 0.7053 (0.39—1.27) | 0.29 |
| GA + CC | 95 (44.60%) | 20 (9.26%) | 2.19 (1.86—2.59) | 7.890 (4.63—13.72) | < 0.0001 |
Discussion
HCC incidence is decreasing in several high-risk countries, but its incidence is increasing in low-risk countries [33, 34]. There are many modified risk factors linked with HCC that increase association of an individual with HCC, such as alcohol intake, smoking or toxin exposure. Apart from extrinsic factors, genetic polymorphism is also identified as a major contributor. HCV infection is among the prominent causes of HCC [35]. The present study aimed to identify whether CTLA-4 + 49 A/G and IL-4–590 C > T polymorphisms associate in HCV-induced HCC Pakistani patients. A further aim was to evaluate potential pathogenic contributions of these polymorphisms to HCC.
The IL-4 and CTLA-4 SNPs were localized to the promoter region and protein-coding region, respectively. RegulomeDB analysis indicated likely regulatory roles and transcription factor binding sites for the IL-4 SNP, whereas gene regulatory roles of the CTLA-4 polymorphism were estimated as low. The CTLA-4 SNP represents as missense variant, and pathogenicity of this SNP was estimated through in silico SNP analysis tools: SIFT, PolyPhen2.0, CADD, REVEL, metaLR and MutationAssessor. Scores from each tool classified both CTLA-4 + 49 A/G and IL-4–590 C/T SNPs in tolerant or benign class.
Evidence from literature indicated the involvement of the CTLA-4 + 49 A/G variant in a number of diseases such as rheumatoid arthritis [36], Behcet’s disease [37], Graves’ disease [38], diabetes mellitus, thyroid diseases and colorectal cancer [39]. However, information is lacking on the relationship of the CTLA-4 + 49 A/G polymorphism and HCV induced HCC in the Pakistani population. In a study conducted among Chinese population, CTLA-4 + 49 A/G association with HBV induced HCC was concluded [40]. CTLA-4 polymorphism rs3087243 G > A also showed association with HBV induced HCC among eastern Chinese Han population [17]. CTLA-4 polymorphism along with IL-10 and TNF-alpha also reported associated with HCC [23]. In the present study, CTLA-4 + 49 A/G polymorphism was identified as a potential risk factor for HCV induced HCC in Pakistan. Genotype AG was found to be in high frequency in HCV induced HCC patients while the dominant genotype in control samples was GG. The role of genotype GG was found as protective in HCV-induced HCC patient.
Allele G in CTLA-4 + 49 causes threonine replacement with alanine. Molecular docking of CTLA-4 wildtype and mutated protein with Sorafenib and Avastin indicated that alanine substitution caused an additional covalent bond between CTLA-4 and the drugs. Formation of a covalent bond between ligand and protein is irreversible and a very strong interaction [41]. In most natural biological systems non-covalent interactions are typical [42], and these interactions in comparison to covalent bonding are weak and reversible [41, 42]. The presence of a covalent bond between mutated CTLA-4 protein and Avastin and Sorafenib dictates a strong interaction. This further hints towards the protective influence of CTLA-4 GG genotype that might enhance therapy influence.
Studies conducted earlier provided insight into the possible role of IL-4 polymorphism with HCV-induced HCC. The outcomes of the current study pointed towards the association of -590 C > T mutation of IL-4 with increased risk of HCV-induced HCC, and align with a previous study including IL-4 [43]. Annotation scores from RegulomeDB for -590 SNP indicated the regulatory role of this SNP and the presence of transcription factor binding sites. This finding was further confirmed by Alibaba 2.0 analysis. The C > T mutation created 2 additional binding sites, MIG1 and SP1. SP1 is located within the E1 region that is responsible for activity of enhancers in IL-4. Such SP1 mutation was thought to have no effect on normal functioning of the E1 region, but more research is needed to draw any conclusion [44]. These mutations can alter the normal cellular functions, which may lead to cancer formation. These findings support -590 C > T (rs2243250) involvement in hepatocellular carcinoma.
IL-4 -590 C > T polymorphism has a direct association with cancers [24], and is linked with certain diseases such as atopic asthma & allergic rhinitis [25] and smoking linked cancer [36]; however, many studies report that this polymorphism does not have a significant association with disease incidence [45]. A study linked with the Egyptian population showed that IL-4 -590 C > T polymorphism was associated with HCV induced HCC. IL-4 can promote macrophage and Th cell production, which in turn supports progression of cancers like HCC [43]. IL-4 -590 C > T polymorphism is associated with both HBV and HCV induced HCC in a Caucasian population [46]. In our study cohort, genotype “CC” was found to be in high frequency in HCV induced HCC patients and is indicated as risk factor allele.
Conclusion
In the present study, CTLA-4 + 49 genotype AG and IL-4 -590 genotype CC were found to be associated with the pathogenicity of HCV-mediated HCC. Age-based and gender-based analysis revealed the association of CTLA-4 AG genotype with adults (age 20–39) and both genders. Similarly, IL-4 association with age groups 20-39yrs and 40-59yrs and with both genders was revealed. Computation analysis estimated that CTLA-4 polymorphism might have influence on the hydrophobicity and structure of the protein while IL-4 polymorphism might lead to altered transcription factor binding sites in its promoter region. The CTLA-4 polymorphism might augment efficacy of clinically available chemotherapy drugs. Thus CTLA-4 & IL-4 SNP polymorphisms have strong association with HCV induced HCC among Pakistani patients that may have application to serve as genetic markers for pre- and early diagnosis and prognosis of HCC in HCV patients.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting project number (RSP2022R502), King Saud University, Riyadh Saudi Arabia for funding this project.
Authors’ contributions
MS, YB, KK, AR, MF, FF, NWA, SR, MQ, and TA designed the study, conceived the study and analyzed the results. KK, NMA, SR, TA, AA, and NWA conceived an initial part of the study, performed the experiment, and helped in compiling the results. MS and YB experimented. KK, MF, SR, FF, AA, and TA helped in writing the results. SR, TA, and AA wrote the paper with input from all other authors MS, SR, YB, KK, FF, TA, NWA, MQ, JHT and AA made a substantial contribution in the interpretation of data and revising the manuscript for intellectual content. NMA performed bioinformatics. All authors read and approved the final manuscript.
Funding
The authors extend their appreciation to the Researchers Supporting project number (RSP2022R502), King Saud University, Riyadh Saudi Arabia for funding this project. We are grateful to the Department of Healthcare Biotechnology, Atta-ur-Rahman School of Applied Biosciences, National University of Sciences and Technology, Islamabad, Capital, Pakistan and Higher Education Commission for their funding through grant number 10067. Funding body has no role in designing the study.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declarations
Ethics approval and consent to participate
The experimental protocol for the use of human blood samples was approved (Ref: No: IRB No. 04–2019-03/06) by the ethical committee of Combined Military Hospital and ASAB, NUST. Informed consent was obtained from all subjects and/or their legal guardian(s).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Shabbir, Email: mshabbir@asab.nust.edu.pk.
Yasmin Badshah, Email: yasmeenb@asab.nust.edu.pk.
Khushbukhat Khan, Email: kkhan.phdabs18asab@nust.edu.pk.
Janeen H. Trembley, Email: trem0005@umn.edu
Areeb Rizwan, Email: areebrizwan03@gmail.com.
Fatima Faraz, Email: fatimafaraz1997@gmail.com.
Syeda Alveena Shah, Email: syeda.alveena10@gmail.com.
Mahrukh Farooqi, Email: farooqui96@hotmail.com.
Naeem Mahmood Ashraf, Email: naeem.mahmood@uog.edu.pk.
Tayyaba Afsar, Email: Tayyaba_sona@yahoo.com.
Ali Almajwal, Email: aalmajwal@ksu.edu.sa.
Nawaf W. Alruwaili, Email: nawaf.alwanas@gmail.com
Suhail Razak, Email: smarazi@ksu.edu.sa.
References
- 1.London W, McGlynn K. Liver cancer Cancer epidemiology and prevention. 2006;3:763–786. doi: 10.1093/acprof:oso/9780195149616.003.0039. [DOI] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel R: Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. rtdCA Cancer J Clin, 71: 209–49. In.; 2021. [DOI] [PubMed]
- 4.Munaf A, Memon MS, Kumar P, Ahmed S, Kumar MB. Comparison of viral hepatitis-associated hepatocellular carcinoma due to HBV and HCV-cohort from liver clinics in Pakistan. Asian Pac J Cancer Prev. 2014;15(18):7563–7567. doi: 10.7314/APJCP.2014.15.18.7563. [DOI] [PubMed] [Google Scholar]
- 5.Waldmann TA. Cytokines in cancer immunotherapy. Cold Spring Harb Perspect Biol. 2018;10(12):a028472. doi: 10.1101/cshperspect.a028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond Jak/Stat. J Allergy Clin Immunol. 2000;105(6):1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki A, Leland P, Joshi BH, Puri RK. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine. 2015;75(1):79–88. doi: 10.1016/j.cyto.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Agdashian D, ElGindi M, Xie C, Sandhu M, Pratt D, Kleiner DE, Figg WD, Rytlewski JA, Sanders C, Yusko EC. The effect of anti-CTLA4 treatment on peripheral and intra-tumoral T cells in patients with hepatocellular carcinoma. Cancer Immunol, Immunother. 2019;68(4):599–608. doi: 10.1007/s00262-019-02299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Yang W, Huang Y, Cui R, Li X, Li B. Evolving roles for targeting CTLA-4 in cancer immunotherapy. Cell Physiol Biochem. 2018;47(2):721–734. doi: 10.1159/000490025. [DOI] [PubMed] [Google Scholar]
- 10.Hu L, Liu J, Chen X, Zhang Y, Liu L, Zhu J, Chen J, Shen H, Qiang F, Hu Z. CTLA-4 gene polymorphism+ 49 A/G contributes to genetic susceptibility to two infection-related cancers—hepatocellular carcinoma and cervical cancer. Hum Immunol. 2010;71(9):888–891. doi: 10.1016/j.humimm.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 11.Zou C, Qiu H, Tang W, Wang Y, Lan B, Chen Y. CTLA4 tagging polymorphisms and risk of colorectal cancer: a case–control study involving 2,306 subjects. Onco Targets Ther. 2018;11:4609. doi: 10.2147/OTT.S173421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erfani N, Haghshenas MR, Hoseini MA, Hashemi SB, Khademi B, Ghaderi A. Strong association of CTLA-4 variation (CT60A/G) and CTLA-4 haplotypes with predisposition of Iranians to head and neck cancer. Iranian J Immunol. 2012;9(3):188–198. [PubMed] [Google Scholar]
- 13.Li H, Duan N, Zhang Q, Shao Y: Impact of IL-4 polymorphisms on head and neck cancer susceptibility in the Chinese Han population. 2019.
- 14.Shamoun L, Skarstedt M, Andersson RE, Wågsäter D, Dimberg J. Association study on IL-4, IL-4Rα and IL-13 genetic polymorphisms in Swedish patients with colorectal cancer. Clin Chim Acta. 2018;487:101–106. doi: 10.1016/j.cca.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 15.Abd-Elfattah M-E, Naguib M, Elkheer M, Abdelsameea E, Nada A. The role of IL-4 gene polymorphism in HCV-related hepatocellular carcinoma in Egyptian patients. Egyptian Liver Journal. 2021;11(1):1–6. doi: 10.1186/s43066-021-00081-z. [DOI] [Google Scholar]
- 16.Lu Y, Wu Z, Peng Q, Ma L, Zhang X, Zhao J, Qin X, Li S. Role of IL-4 gene polymorphisms in HBV-related hepatocellular carcinoma in a Chinese population. PLoS ONE. 2014;9(10):e110061. doi: 10.1371/journal.pone.0110061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Liu J, Chen Y, Tang W, Liu C, Sun Y, Chen J. Association of CTLA-4 tagging polymorphisms and haplotypes with hepatocellular carcinoma risk: A case-control study. Medicine. 2019;98(29):e16266. [DOI] [PMC free article] [PubMed]
- 18.El-Said HH, Ghanayem NM, Badr EA, El-Fert AY, Gaballah AK. Cytotoxic T-lymphocyte antigen-4 gene polymorphisms in hepatocellular carcinoma patients in Egypt. Menoufia Medical Journal. 2014;27(2):372. doi: 10.4103/1110-2098.141711. [DOI] [Google Scholar]
- 19.Lambrechts D, Claes B, Delmar P, Reumers J, Mazzone M, Yesilyurt BT, Devlieger R, Verslype C, Tejpar S, Wildiers H. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol. 2012;13(7):724–733. doi: 10.1016/S1470-2045(12)70231-0. [DOI] [PubMed] [Google Scholar]
- 20.Miska J, Lui JB, Toomer KH, Devarajan P, Cai X, Houghton J, Lopez DM, Abreu MT, Wang G, Chen Z. Initiation of inflammatory tumorigenesis by CTLA4 insufficiency due to type 2 cytokines. J Exp Med. 2018;215(3):841–858. doi: 10.1084/jem.20171971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H, Zhang R, Zhang W. CTLA-4 interferes with the HBV-specific T cell immune response. Int J Mol Med. 2018;42(2):703–712. doi: 10.3892/ijmm.2018.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Oliveira Rodrigues R, Rabenhorst SHB, de Carvalho PG, Sasahara GL, Vasconcelos LMF, de Arruda ÉAG, da Silva SFR, Ribeiro IF: Association of IL10, IL4, IFNG and CTLA4 gene polymorphisms with efavirenz hypersensitivity reaction in patients infected with human immunodeficiency virus. Japanese Journal of Infectious Diseases 2017:JJID. 2016.2075. [DOI] [PubMed]
- 23.Jj W, Zb W, Tc T. Association of CTLA-4, TNF alpha and IL 10 polymorphisms with susceptibility to hepatocellular carcinoma. Scand J Immunol. 2019;90(6):e12819. doi: 10.1111/sji.12819. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Zhang TL, Wang X. Association between polymorphisms in interleukins 4 and 13 genes and chronic periodontitis in a Han Chinese population. BioMed Res Int. 2016;(7):2016. [DOI] [PMC free article] [PubMed]
- 25.Chen G, Hu C, Song Y, Zhang H, Li S, Lai P, Huang P. Effects of IL-4-590C/T (rs2243250) Polymorphism on the Susceptibility of Smoking-Related Cancer: A Meta-Analysis Involving 11,407 Subjects. Biomed Res Int. 2019;(I);13–26. [DOI] [PMC free article] [PubMed]
- 26.Falahi S, Karaji AG, Koohyanizadeh F, Rezaiemanesh A, Salari F. A comprehensive in Silico analysis of the functional and structural impact of single nucleotide polymorphisms (SNPs) in the human IL-33 gene. Comput Biol Chem. 2021;94:107560. [DOI] [PubMed]
- 27.Dong S, Boyle AP. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum Mutat. 2019;40(9):1292–1298. doi: 10.1002/humu.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, Swindells MB. "LigPlot+: multiple ligand–protein interaction diagrams for drug discovery." 2011. p. 2778–86. [DOI] [PubMed]
- 29.Suresh K, Chandrashekara S. Sample size estimation and power analysis for clinical research studies. Journal of human reproductive sciences. 2012;5(1):7. doi: 10.4103/0974-1208.97779. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Dentists GAotWMAJTJotACo: World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. 2014, 81(3):14–18. [PubMed]
- 31.Sambrook J, Russell DW: Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harbor Protocols 2006, 2006(1):pdb. prot4455. [DOI] [PubMed]
- 32.Hural JA, Kwan M, Henkel G, Hock MB, Brown MA. An intron transcriptional enhancer element regulates IL-4 gene locus accessibility in mast cells. J Immunol. 2000;165(6):3239–3249. doi: 10.4049/jimmunol.165.6.3239. [DOI] [PubMed] [Google Scholar]
- 33.Velazquez ER, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77(14):3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73:4–13. doi: 10.1002/hep.31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Cao H, Xu Z, Wang D, Zeng Y. SNP rs2596542G> A in MICA is associated with risk of hepatocellular carcinoma: a meta-analysis. Bioscience reports. 2019;39(5);121–34. [DOI] [PMC free article] [PubMed]
- 36.Mochizuki M, Amemiya S, Kobayashi K, Kobayashi K, Shimura Y, Ishihara T, Nakagomi Y, Onigata K, Tamai S, Kasuga A. Association of the CTLA-4 gene 49 A/G polymorphism with type 1 diabetes and autoimmune thyroid disease in Japanese children. Diabetes Care. 2003;26(3):843–7. [DOI] [PubMed]
- 37.Abdel Galil SM, Hagrass HA. The role of CTLA-4 exon-1 49 A/G polymorphism and soluble CTLA-4 protein level in Egyptian patients with Behcet's disease. Biomed Res Int. 2014(1);123–34. [DOI] [PMC free article] [PubMed]
- 38.Kouki T, Sawai Y, Gardine CA, Fisfalen M-E, Alegre M-L, DeGroot LJ. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves’ disease. J Immunol. 2000;165(11):6606–6611. doi: 10.4049/jimmunol.165.11.6606. [DOI] [PubMed] [Google Scholar]
- 39.Du X, Tang F, Liu M, Su J, Zhang Y, Wu W, Devenport M, Lazarski CA, Zhang P, Wang X. A reappraisal of CTLA-4 checkpoint blockade in cancer immunotherapy. Cell Res. 2018;28(4):416–432. doi: 10.1038/s41422-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu X, Qi P, Zhou F, Ji Q, Wang H, Dou T, Zhao Y, Gao C. + 49G> A polymorphism in the cytotoxic T-lymphocyte antigen-4 gene increases susceptibility to hepatitis B-related hepatocellular carcinoma in a male Chinese population. Hum Immunol. 2010;71(1):83–87. doi: 10.1016/j.humimm.2009.09.353. [DOI] [PubMed] [Google Scholar]
- 41.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10(9):885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulusu G, Desiraju GR. Strong and weak hydrogen bonds in protein–ligand recognition. J Indian Inst Sci. 2020;100(1):31–41. doi: 10.1007/s41745-019-00141-9. [DOI] [Google Scholar]
- 43.Aboushousha T, Emad M, Rizk G, Ragab K, Hammam O, Fouad R, Helal NS. IL-4, IL-17 and CD163 Immunoexpression and IL-6 Gene Polymorphism in Chronic Hepatitis C Patients and Associated Hepatocellular Carcinoma. Asian Pac J Cancer Prev. 2021;22(4):1105–1113. doi: 10.31557/APJCP.2021.22.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henkel G, Brown MA: PU. 1 and GATA: components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci 1994, 91(16):7737–7741. [DOI] [PMC free article] [PubMed]
- 45.Tindall E, Severi G, Hoang H, Ma C, Fernandez P, Southey M, English D, Hopper J, Heyns C, Tangye S: Comprehensive analysis of the cytokine-rich chromosome 5q31. 1 region suggests a role for IL-4 gene variants in prostate cancer risk. Carcinogenesis 2010, 31(10):1748–1754. [DOI] [PubMed]
- 46.Zheng Z, Li X, Li Z, Ma X-C. IL-4− 590C/T polymorphism and susceptibility to liver disease: a meta-analysis and meta-regression. DNA Cell Biol. 2013;32(8):443–450. doi: 10.1089/dna.2013.2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.