Abstract
Klebsiella pneumoniae 5064, isolated in New York, carried plasmid-mediated resistance to multiple β-lactams and was unresponsive to clavulanic acid. The β-lactamase gene responsible for cephalosporin resistance encoded FOX-5, with 96 to 97% amino acid identities to other members of the FOX family of β-lactamases. The blaFOX-5 coding region was located next to a transposase gene from the Aeromonas salmonicida insertion element ISAS2.
AmpC β-lactamases in functional group 1 are characterized by their hydrolysis of cephalosporins and resistance to clavulanic acid inhibition (6). These β-lactamases were originally described as chromosomal, inducible enzymes in the Enterobacteriaceae, with clinically problematic cephalosporin resistance occurring by stable derepression of the chromosomal ampC gene (15). In the late 1980s, ceftazidime resistance caused by a plasmid-borne AmpC enzyme was discovered in a Klebsiella pneumoniae clinical isolate (20). Since this time, there have been increasing reports of plasmid-encoded AmpC enzymes throughout the world (1, 5, 10, 13, 19, 23).
The FOX family of AmpC-type β-lactamases has 76% amino acid homology to the Aeromonas sobria β-lactamase CepS (24) and 72% homology to the K. pneumoniae CMY-1 AmpC enzyme (2). FOX-1 was discovered in a K. pneumoniae isolate from Argentina, and FOX-2 was identified in an E. coli strain from Guatemala (3, 11). Two additional members of the FOX family were recently isolated in Italy and Spain (4, 16). In the United States, K. pneumoniae and Escherichia coli strains producing FOX-type β-lactamases in isolates from several southern states have been described (G. Jacoby, J. Tran, and M. Alvarez, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1481, 1999; N. D. Hanson, P. Coudron, E. S. Moland, C. C. Sanders, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1481, 1999).
In this paper, we present the sequence and biochemical characterization of FOX-5, a plasmid-borne AmpC enzyme that was identified in a K. pneumoniae isolate from The Mount Sinai Hospital of New York City.
(This work has been presented in part previously [A. M. Queenan, K. Bush, and S. Jenkins, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1470, 2000].)
K. pneumoniae 5064 was a urine culture isolate from a 90-year-old female nursing-home patient (Table 1). The MICs shown in Table 2 were determined by the NCCLS broth microdilution method (18). K. pneumoniae 5064 was resistant to penicillins and cephalosporins, including cephamycins, and refractory to inhibition by clavulanic acid. This clinical isolate was susceptible to imipenem, aztreonam, and gentamicin, intermediate to tetracycline, and resistant to ciprofloxacin and trimethoprim-sulfamethoxazole (Vitek Biomerieux, Durham, N.C.). K. pneumoniae 5064 contained a plasmid of approximately 125 kb, which was isolated with the Qiagen Miniprep kit, electroporated into E. coli DH5-α, and selected on Luria-Bertani agar containing 2 μg of ceftazidime per ml. The β-lactam resistance profile of the E. coli DH5-α/p5064 transformant was similar to that of the clinical isolate, but ciprofloxacin, trimethoprim-sulfamethoxazole, and tetracycline resistance were not transferred with this plasmid.
TABLE 1.
Bacterial strains
| Strain | Description | pI of β-lactamase(s) | Source |
|---|---|---|---|
| K. pneumoniae 5064 | Clinical isolate | 5.6, 7.2, 7.6 | This study |
| E. coli DH5-α | Recipient strain for plasmid transformation | None | Invitrogen |
| E. coli DH5-α/p5064 | Carries ∼125-kb plasmid p5064 from K. pneumoniae 5064 | 5.6, 7.2 | This study |
| E. coli DH5-α/pFOX-5 | Carries plasmid pFOX-5, 1.7-kb subclone of p5064 in pACYC184 | 7.2 | This study |
TABLE 2.
MICs of antimicrobial agents for K. pneumoniae 5064 and E. coli transformants
| Agenta | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| K. pneumoniae 5064 (TEM, SHV, FOX-5) | E. coli DH5-α/p5064 (TEM, FOX-5) | E. coli DH5-α/pFOX-5 (FOX-5) | E. coli DH5-α (none) | |
| Benzylpenicillin | >512 | >512 | >512 | 32 |
| Benzylpenicillin + clavulanate | >512 | >512 | >512 | 64 |
| Amoxicillin | >512 | >512 | 256 | 16 |
| Amoxicillin + clavulanate | 256 | 256 | 256 | 8 |
| Piperacillin | >512 | 512 | 32 | ≤0.5 |
| Piperacillin + tazobactam | 128 | 8 | 32 | ≤0.5 |
| Cefoxitin | 512 | 256 | 512 | 32 |
| Cefoxitin + clavulanate | 512 | 128 | 256 | 32 |
| Cefoxitin + tazobactam | 256 | 128 | 256 | 8 |
| Ceftazidime | 64 | 64 | 128 | 0.5 |
| Ceftazidime + clavulanate | 128 | 64 | >128 | 0.5 |
| Cefotaxime | 16 | 8 | 32 | ≤0.12 |
| Cefotaxime + clavulanate | 16 | 8 | 32 | ≤0.12 |
| Cefotetan | 64 | 32 | 128 | 0.5 |
| Cefotetan + clavulanate | 64 | 32 | >128 | 0.5 |
| Cefepime | 0.5 | ≤0.25 | 0.5 | ≤0.25 |
| Cefepime + clavulanate | ≤0.25 | ≤0.25 | 0.5 | ≤0.25 |
| Cefpodoxime | 128 | 512 | >512 | 0.5 |
| Cefpodoxime + clavulanate | 256 | 512 | >512 | 0.25 |
| Imipenem | 0.25 | 0.25 | 0.5 | 0.25 |
| Imipenem + clavulanate | 0.5 | 0.25 | 0.25 | 0.25 |
| Aztreonam | 8 | 2 | 16 | 0.25 |
| Aztreonam + clavulanate | 8 | 1 | 8 | ≤0.12 |
| Ciprofloxacin | 4 | ≤0.25 | ≤0.25 | ≤0.25 |
| Gentamicin | 8 | 1 | 0.5 | 0.5 |
| Tobramycin | 4 | 1 | ≤0.25 | ≤0.25 |
| Tetracycline | 8 | 0.5 | 1 | 1 |
Clavulanate, 2 μg/ml; tazobactam, 4 μg/ml.
β-lactamases in K. pneumoniae 5064 and the E. coli transformant were examined by isoelectric focusing (IEF) of freeze-thaw lysates (7, 17) separated on Ampholine PAGplates (pH 3.5 to 9.5; Amersham Pharmacia, Piscataway, N.J.), with visualization using nitrocefin. To test inhibition, filter paper saturated with a single inhibitor (10 μg of aztreonam per ml, 100 μM tazobactam, or 50 mM EDTA) was applied to the IEF gel for 10 min before development with nitrocefin. IEF analysis revealed that K. pneumoniae 5064 contained three β-lactamases, with pI values of 5.6, 7.2, and 7.6 (Table 1). The E. coli transformant carried the pI 5.6 and pI 7.2 enzymes. The enzymes with pI values of 5.6 and 7.6 were inhibited by tazobactam, consistent with TEM-2-like and SHV-type β-lactamases. The β-lactamase with pI 7.2 was inhibited by aztreonam but not by tazobactam or EDTA.
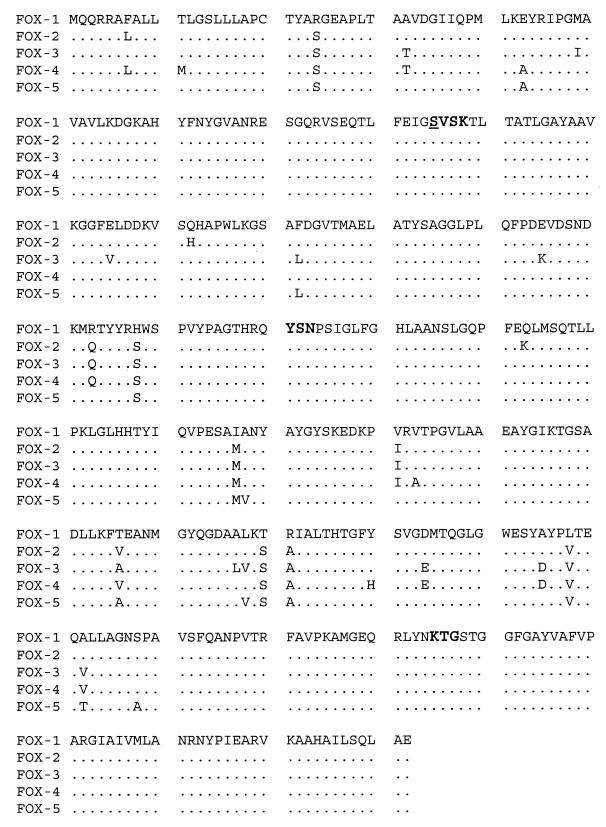
The isoelectric point and inhibition profile data of the pI 7.2 enzyme suggested an AmpC β-lactamase, possibly of the FOX family. PCR of DNA from K. pneumoniae 5064 with FOX-specific primers FOX1F and FOX1H produced a band of approximately 1 kb, which was predicted to encompass the entire FOX-coding region (3). The PCR product was cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.), and 13 clones were sequenced with M13rev and T7 primers by ACGT, Inc. (Northbrook, Ill.). The amino acid sequence and comparison to other members of the FOX family are shown in Fig. 1. The FOX-5 β-lactamase was similar to all members of the FOX family, with amino acid identities of 96.1% to FOX-4, 96.6% to FOX-1 and FOX-3, and 96.9% to FOX-2.
FIG. 1.
Alignment of FOX-1 through FOX-5 deduced protein sequences, containing 382 amino acids. Bold type indicates the conserved elements for the serine β-lactamases. The underlined serine corresponds to amino acid position 80 for E. coli AmpC in the work of Jaurin and Grundström (14).
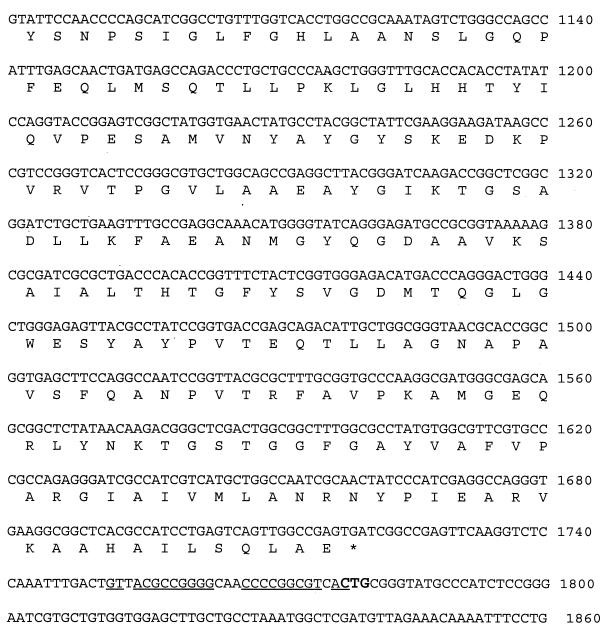
A subclone of the FOX-5 region of p5064 was obtained by ligating EagI fragments of p5064 into pACYC184 cut by EagI (New England BioLabs, Beverly, Mass.) to yield pFOX-5. Transformants selected on agar containing 2 μg of ceftazidime per ml contained a single pI 7.2 β-lactamase and displayed a β-lactam resistance profile similar to that of K. pneumoniae 5064, except for the tazobactam-inhibited piperacillin resistance, which was probably contributed by the pI 5.6 TEM-2-type β-lactamase on the original plasmid (Tables 1 and 2). The nucleotide sequence of the p5064 fragment containing blaFOX-5 is shown in Fig. 2. The transcriptional start site was mapped by primer extension (Promega, Madison, Wis.), and putative −10 and −35 sequences are underlined (21). Immediately 5′ to the blaFOX-5 gene is a region with 98% DNA homology to the ISAS2 insertion element transposase from Aeromonas salmonicida (12), a member of the IS30 family. The DNA sequence downstream from the FOX-5 coding region contains an imperfect inverted repeat of 27 bp, followed by a CTG, two other features of ISAS2. These results suggest that the blaFOX-5 gene is part of an insertion sequence element. CMY-4 and several CTX-M β-lactamases are also reported to be associated with insertion elements (P. D. Stapleton, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 1999; GenBank AF286192 and GenBank AF252622).
FIG. 2.
Nucleotide and deduced amino acid sequence for the p5064 FOX-5 region. Nucleotides 1 to 520 correspond to the sequence of the ISAS2 transposase, which is shown in italic type. Bold type indicates the transposase nucleotide and amino acid differences from the data reported by Gustafson et al. The FOX-5 transcription start is marked with an arrow, and putative −10 and −35 regions are underlined. The sequence used for the primer extension experiment is underlined. The FOX-5 putative ribosome-binding site is double underlined. The sequence downstream of the FOX-5 translational stop is from a second FOX-5 clone that extended past the EagI site. Elements of ISAS2, i.e., the imperfect inverted repeat and CTG, are underlined and bold, respectively (12).
The FOX-5 β-lactamase was purified from the DH5-α/pFOX-5 transformant grown in tryptic soy broth with 2 μg of ceftazidime per ml. A freeze-thaw lysate (7) was passed through a Sephadex S-75 column in 50 mM phosphate buffer (pH 7.0). Active fractions were further purified using cation exchange in 25 mM phosphate buffer (pH 6.2) through HiTrap S (Amersham-Pharmacia), followed by anion exchange through HiTrap Q (Amersham-Pharmacia) in 25 mM Tris (pH 8.5). FOX-5 purity (98%) was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels stained with Coomassie blue; the molecular mass calculated from the gel was 37 kDa.
Hydrolysis data were obtained by measuring initial rates at 25°C on a Shimadzu 1600UV spectrophotometer (22). All substrates were prepared fresh in 50 mM phosphate buffer, pH 7.0. Km and Vmax were determined by averaging results of Eadie-Hofstee, Hanes, Cornish Bowden direct linear plots and a least-squares fit to the Michaelis-Menten equation. The concentration of inhibitor that decreased enzymatic activity by 50% (IC50) was determined by graphing percent control activity against concentration of inhibitor, using initial rates obtained after a 5-min preincubation of enzyme and inhibitor at 25°C. Cephaloridine at 100 μM was used as the substrate for the IC50 studies. The Ki of aztreonam was determined under steady-state conditions using the Dixon plot for inhibitors of high affinity, with cephaloridine as a substrate at 100 and 300 μM (9).
FOX-5 had a hydrolysis profile similar to those of the other FOX enzymes (Table 3) (4, 11, 16). Cephaloridine and cephalothin were hydrolyzed 70 to 80 times faster than penicillin. Hydrolysis of cefoxitin was 1,000-fold slower than cephaloridine hydrolysis. Notable among the FOX family of β-lactamases is a low Km value of 0.9 μM for cefoxitin, which increased the catalytic efficiency (kcat/Km) for this substrate. Cefepime and cefpodoxime had low kcat values, but the catalytic efficiency of cefpodoxime was 10-fold higher than that for cefepime, primarily due to the low Km for cefpodoxime. Cefotaxime, ceftazidime, and aztreonam had hydrolysis rates too slow for reliable determination of kinetic parameters. Clavulanic acid and tazobactam were poor inhibitors, while aztreonam inhibited FOX-5 with a Ki of 1.6 nM (Table 3).
TABLE 3.
Substrate and inhibitor profiles of purified FOX-5 β-lactamase
| Compound | kcat (S−1) | Relative kcat | Km (μM) | kcat/Km (mM−1 s−1) | IC50 (μM) |
|---|---|---|---|---|---|
| Substrates | |||||
| Cephaloridine | 790 ± 52 | 100 | 1,300 ± 58 | 610 | |
| Cephalothin | 870 ± 15 | 110 | 71 ± 5.3 | 12,000 | |
| Benzylpenicillin | 11 ± 0.3 | 1.4 | 9.2 ± 0.6 | 1,200 | |
| Cefoxitin | 0.7 ± 0.04 | 0.09 | 0.85 ± 0.09 | 820 | |
| Cefepime | 0.6 ± 0.1 | 0.08 | 86 ± 2 | 7.0 | |
| Cefpodoxime | 1.5 ± 0.04 | 0.19 | 2.3 ± 0.5 | 650 | |
| Ceftazidime | <0.08 | <0.01 | NDa | ND | |
| Cefotaxime | 0.08 | 0.01 | ND | ND | |
| Aztreonam | <0.08 | <0.01 | ND | ND | |
| Inhibitors | |||||
| Clavulanic acid | >45 | ||||
| Tazobactam | 28 ± 3 | ||||
| Aztreonam | 0.0002b |
ND, not determined.
Ki was 1.6 nM.
Plasmid-mediated AmpC enzymes have been reported with increasing frequency in several U.S. medical centers (8;G. A. Jacoby, P. Han, M. Alvarez, and F. Tenover, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C40, 1995). In one study, the occurrence of AmpC-type β-lactamases in clinical isolates of E coli, K. pneumoniae, and Proteus mirabilis was 1.2%, with half of these strains carrying the AmpC on a plasmid (8). This report is the first biochemical characterization of a FOX family plasmid-mediated β-lactamase in the United States, indicating the expansion of this family of enzymes into North America. The association of FOX-5 with an insertion element suggests a mechanism for its incorporation into the K. pneumoniae p5064 plasmid. As a result, further transmission of FOX β-lactamases may be expected.
Nucleotide sequence accession number.
The GenBank accession number for the FOX-5 sequence is AY007369.
Acknowledgments
We thank Nancy Hanson for sharing the amino acid sequence of her plasmid-mediated FOX β-lactamase before publication.
REFERENCES
- 1.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43:1924–1931. doi: 10.1128/aac.43.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Wagner S, Jungwirth R, Schneider I, Meyer D. A novel class C β-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob Agents Chemother. 1997;41:2041–2046. doi: 10.1128/aac.41.9.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou G, Oliver A, Ojeda M, Monzon C, Martinez-Beltran J. Molecular characterization of FOX-4, a new AmpC-type plasmid-mediated β-lactamase from an Escherichia coli strain isolated in Spain. Antimicrob Agents Chemother. 2000;44:2549–2553. doi: 10.1128/aac.44.9.2549-2553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Singer S B. Effective cooling allows sonication to be used for liberation of β-lactamases from Gram-negative bacteria. J Antimicrob Chemother. 1989;24:82–84. doi: 10.1093/jac/24.1.82. [DOI] [PubMed] [Google Scholar]
- 8.Coudron P E, Moland E S, Thomson K S. Occurrence and detection of AmpC β-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791–1796. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon M. The graphical determination of Km and Ki. Biochem J. 1972;129:197–202. doi: 10.1042/bj1290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fosberry A P, Payne D J, Lawlor E J, Hodgson J E. Cloning and sequence analysis of blaBIL-1, a plasmid-mediated class C β-lactamase gene in Escherichia coli BS. Antimicrob Agents Chemother. 1994;38:1182–1185. doi: 10.1128/aac.38.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez Leiza M, Perez-Diaz J C, Ayala J, Casellas J M, Martinez-Beltran J, Bush K, Baquero F. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother. 1994;38:2150–2157. doi: 10.1128/aac.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustafson C E, Chu S, Trust T J. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 13.Horii T, Arakawa Y, Ohta M, Ichiyama S, Wacharotayankun R, Kato N. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob Agents Chemother. 1993;37:984–990. doi: 10.1128/aac.37.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaurin B, Grundstrom T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc Natl Acad Sci. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones R N. Important and emerging β-lactamase-mediated resistances in hospital-based pathogens: the AmpC enzymes. Diagn Microbiol Infect Dis. 1998;31:461–466. doi: 10.1016/s0732-8893(98)00029-7. [DOI] [PubMed] [Google Scholar]
- 16.Marchese A, Arlet G, Schito G C, Lagrange P H, Philippon A. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother. 1998;42:464–467. doi: 10.1128/aac.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthew M, Harris A M. Identification of β-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol. 1976;94:55–67. doi: 10.1099/00221287-94-1-55. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5, 5th ed. NCCLS Standards. Vol. 20. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 19.Nordmann P. Trends in β-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27:S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 20.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhart F C, et al., editors. Escherichia coli and Salmonella, cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 792–821. [Google Scholar]
- 22.Sykes R B, Bonner D P, Bush K, Georgopapadakou N H. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob Agents Chemother. 1982;21:85–92. doi: 10.1128/aac.21.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzouvelekis L S, Tzelepi E, Mentis A F, Tsakris A. Identification of a novel plasmid-mediated β-lactamase with chromosomal cephalosporinase characteristics from Klebsiella pneumoniae. J Antimicrob Chemother. 1993;31:645–654. doi: 10.1093/jac/31.5.645. [DOI] [PubMed] [Google Scholar]
- 24.Walsh T R, Hall L, MacGowan A P, Bennett P M. Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J Antimicrob Chemother. 1995;36:41–52. doi: 10.1093/jac/36.1.41. [DOI] [PubMed] [Google Scholar]