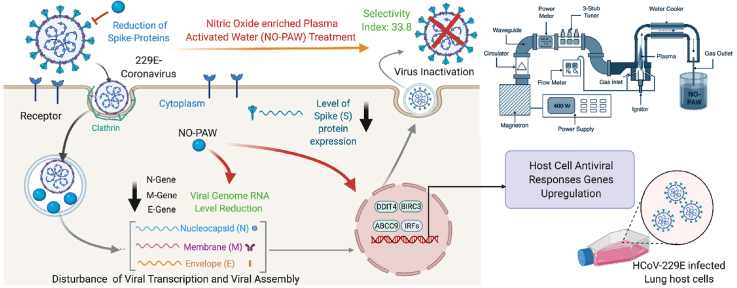
Abstract
The ongoing pandemic caused by the novel coronavirus, SARS-CoV-2, is influencing global health. Moreover, there is a major threat of future coronaviruses affecting the entire world in a similar, or even more dreadful, manner. Therefore, effective and biocompatible therapeutic options against coronaviruses are urgently needed. To address this challenge, medical specialists require a well-informed and safe approach to treating human coronaviruses (HCoVs). Herein, an environmental friendly approach for viral inactivation, based on plasma technology, was considered. A microwave plasma system was employed for the generation of the high amount of gaseous nitric oxide to prepare nitric oxide enriched plasma-activated water (NO-PAW), the effects of which on coronaviruses, have not been reported to date. To determine these effects, alpha-HCoV-229E was used in an experimental model. We found that NO-PAW treatment effectively inhibited coronavirus infection in host lung cells, visualized by evaluating the cytopathic effect and expression level of spike proteins. Interestingly, NO-PAW showed minimal toxicity towards lung host cells, suggesting its potential for therapeutic application. Moreover, this new approach resulted in viral inactivation and greatly improved the gene levels involved in host antiviral responses. Together, our findings provide evidence of an initiation point for further progress toward the clinical development of antiviral treatments, including such coronaviruses.
Keywords: Nitric oxide enriched plasma-activated water, Microwave plasma, Coronavirus, Virus inactivation, Gene expression
Graphical abstract
Highlights
•NO-enriched plasma-activated water (NO-PAW) was prepared using microwave plasma generated gas
•NO-PAW inactivated human coronavirus 229E
•NO-PAW showed no significant toxicity below 2000 μM, against MRC-5 host cells
•NO-PAW reduced viral RNA load and spike protein expression in lung host cells
•Treatment elevated antivirus response gene expression in human host cells
1. Introduction
Coronavirus disease 2019 (COVID‐19) was first reported in 2019 and subsequently categorized as a pandemic by the World Health Organization, in March 2020. Generally, coronaviruses (CoVs) are categorized as positive-stranded RNA viruses belonging to the family Coronaviridae [1]. It is believed that the highly pathogenic human CoVs (HCoVs) belong to the subfamily, Coronavirinae, wherein viruses have been divided into four genera: alpha-, beta-, gamma-, and delta-coronaviruses. To date, seven CoVs that affect humans have been identified, along with the recently discovered, severe acute respiratory syndrome coronavirus‐2 (SARS-CoV-2). These include two alpha-CoVs (HCoV-NL63 and HCoV-229E) and five beta-CoVs, namely HCoV-OC43, HCoV-HKU1, Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2 [2]. CoVs are usually enveloped viruses and have some structural proteins such as membrane (M), spike (S), envelope (E), and nucleocapsid (N) proteins. The S, E, and M proteins are believed to be embedded in the envelope of the virus [3]. COVID-19 can cause severe pneumonia and has resulted in worldwide fatalities during this pandemic. In this concern, the biocompatible treatment approaches dealing with COVID-19 infection are predominantly essential. Therefore, the evaluation of therapeutic options against HCoVs is urgently needed. Even though many antiviral strategies for COVID-19 are under development or have not been sanctioned yet, numerous methodologies have been suggested including repurposed drugs for use against this novel virus. The effectiveness and biocompatibility of many of these approaches are under investigation. Traditional treatment methods have many drawbacks, and innovative strategies for virus inactivation are urgently required. Ritonavir, a protease inhibitor used to treat human immunodeficiency virus (HIV), demonstrated promising in vivo and clinical results against MERS-CoV and SARS-CoV [4,5]. It's in vitro activity against SARS-CoV-2, has been recently proposed [6]. Chloroquine, an antimalarial and immune-modulating drug, has also been proposed as an effective antiviral agent against SARS-CoV [7] and the in vitro activity of hydroxychloroquine against SARS-CoV-2, has been investigated [8]. Nevertheless, evidence of the anti-HCoV activity of hydroxychloroquine at effective doses in clinical settings is limited, because of its toxicity.
Therefore, we aimed to explore the efficacy of nitric oxide enriched plasma-activated water (NO-PAW) against 229-E HCoV, at various concentrations that could be safely administered to infected patients via intraperitoneal, intravenous, intratracheal, or oral administration routes. Non-thermal plasma generates abundant reactive nitrogen and oxygen species [9], consequently stimulating interest in the environmental and biomedical application potential of plasma [[10], [11], [12], [13]]. The role of plasma in microbial inactivation and sterilization [14] has been widely studied. However, plasma-based viral inactivation is a moderately new area of research [15], in which few studies over the past several years have merely described the virucidal properties of plasma along with its mode of action [[16], [17], [18]]. Earlier studies have shown that plasma effectively inactivates λ bacteriophages and feline calicivirus [19,20]. Notably, the biological influence of plasma is dose-dependent, which is essential in plasma-based biomedical applications [21]. Plasma-activated water (PAW) produced by plasma successfully inactivates ɸ174, T4, and MS2 bacteriophages and has revealed an effect similar to that of direct plasma exposure [22]. PAW-induced structural modification and S protein damage in a SARS-CoV-2 pseudovirus was recently demonstrated, providing a new disinfection strategy to combat the pandemic [23]. This report specifically shows the effect of PAW on the receptor-binding domain (RBD) of the S protein of pseudovirus particles. However, the inactivation effects and therapeutic potential of PAW on real corona viruses or SARS-CoV-2, have not yet been reported.
It is well recognized that nitric oxide (NO) plays a role in the pathogenesis of several human viral contaminations [24], and antiviral activity [25]. One study shows that NO can be utilized for antiviral activity on the virions and/or impede the intracellular viral replication process [26]. While the mentioned studies have presented the antiviral effects of NO produced by donor drugs, the present study is presumably the first to exhibit the antiviral effects of plasma-generated, gaseous NO. Based on the literature, improvement of NO-based treatments for viral inactivation, with enhanced outcomes and lacking side effects, is required straightway. Our plasma research center developed the so-called microwave plasma system, which generates a major component of plasma-based gaseous NO. Earlier reports produced from the use of this system have suggested that plasma-generated NO-treated water could be useful as fertilizer, enhancing plant vitality while offering sanitation effects [27,28], as well as extending potential anticancer activity [29]. It has also been shown that NO-PAW potentially modulates immune cells and several immune factors (cytokines and chemokines), which could represent a promising treatment modality against cancer or inflammatory diseases. Since NO-PAW has shown therapeutic and biocompatibility potential, the present work was undertaken to investigate its effect on viral inactivation for disease-treatment purposes. SARS-CoV-2 experiments are limited to performance in a Biosafety Level 3 laboratory, which greatly restricts relevant research [30]. Consequently, utilization of an HCoV-229E model system could alleviate this limitation by enabling study of the coronavirus under Biosafety Level 2 facility conditions. Herein, a 229-E alpha-HCoV was employed as model to evaluate the inactivation of SARS-CoV-2 by NO-PAW, after infection of human lung fibroblast host cells. We aimed to establish the antiviral potential of NO-PAW against HCoVs, thus offering a new inactivation and treatment approach to combatting the pandemic.
2. Materials and methods
2.1. Research material and reagents
For virus culture, HCoV-229E was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) while MRC-5 human lung fibroblast host cells were purchased from the Korean Cell Line Bank (KCLB, Seoul, Korea). The QuantiChrom Nitric Oxide Assay Kit and QuantiChrom Peroxide Assay Kit (BioAssay Systems, Hayward, CA, USA) were used to measuring total NO and H2O2 levels, respectively, and the alamarBlue Assay Kit (Invitrogen, Waltham, MA, USA) was used to measure cell cytotoxicity. All procedures were performed according to the manufacturers' instructions. Chloroquine diphosphate and ritonavir were purchased from Sigma-Aldrich (Yongin, Korea). Stocks of these drugs were prepared in dimethyl sulfoxide (DMSO) and stored at −20 °C until further use. An antibody specific to the HCoV S protein was obtained from Sino Biological (Chesterbrook, PA, USA). All cell culture plasticware was purchased from SPL Life Sciences (Pocheon, Korea). Nitrogen (99.99%) and oxygen (99.99%) gases were purchased from Dong-A Scientific (Seoul, Korea). Other analytical reagent-grade solvents and chemicals were purchased from Sigma-Aldrich (Merck, St. Louis, MO, USA). Distilled water with a resistivity of 18.2 MΩ.Cm, obtained from a Milli-Q® water purification system (Millipore, Bedford, MA, USA) was used to prepare all aqueous solutions in this study.
2.2. Microwave plasma torch system and nitric oxide enriched plasma-activated water
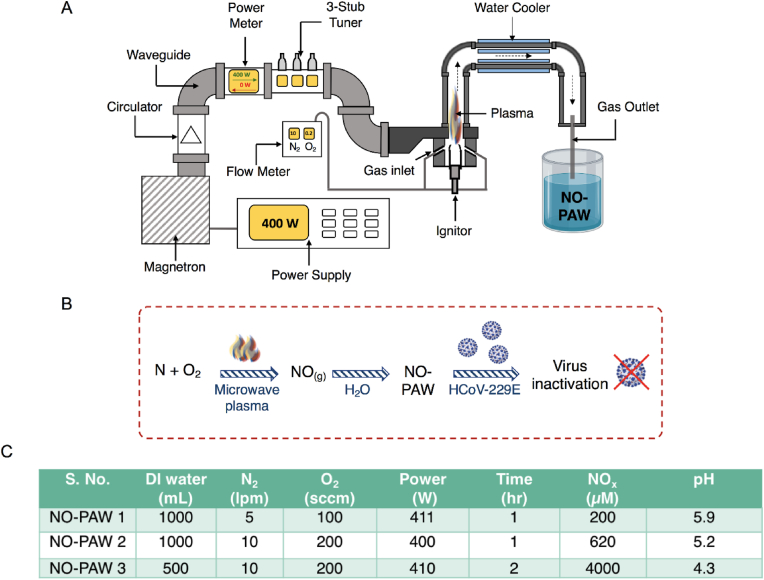
Fig. 1A schematically represents the generation of NO and the treatment of liquids with the gas generated by the microwave plasma system, the configuration of which has been well described earlier [27,29]. Microwaves (2.45 GHz) radiated from a magnetron passed through a circulator, power monitor, and three-stub tuner, before being guided through a tapered waveguide and entering a discharge tube made of quartz. To generate a microwave plasma torch, nitrogen (5 or 10 lpm) and oxygen (100 or 200 sccm) gases, controlled by a mass flow meter, were mixed and injected into the system, and approximately 400–410 W microwave power, applied. Resultantly, a plasma torch (temperature: 6000 K; plasma density: 1013/cm3) was generated in the discharge tube [31]. The gas generated by the torch flame was cooled by passage through a metal pipe wrapped with water tubing (Fig. 1A) and then injected into either 1 L or 0.5 L of deionized (DI) water (pH 6.7). DI was preferable to other solvents—such as cell culture media—because the solution contained many components and its chemical composition would be even more complicated after plasma-generated gas exposure. Therefore, we chose to start with a simple background liquid, to simplify the system. NO-PAW has been added to the cells right after the preparation throughout the experiments for consistency.
Fig. 1.
Generation of NO-PAW using gaseous NO by the microwave plasma system. (A) Schematic diagram of the experimental setup for the preparation of NO-PAW using gaseous NO. (B) Scheme for preparation of NO-PAW using plasma-generated gaseous NO. (C) Optimization of high NOx generation from gaseous NO exposed DI water, by changing various parameters as indicated.
2.3. Nitric oxide enriched plasma-activated water characterization
Since NO is slowly oxidized to nitrite and nitrate within 20% in the liquid phase after 1 h of NO-PAW generation [27], quantitating total NO/NO2−/NO3− is a feasible method to estimate the NO level. By this way, the total level of NO, NO2− and NO3− are presented as NOx. The NOx produced in the NO-PAW was analyzed by their respective detector or using a commercial QuantiChrom Nitric Oxide Assay Kit based on the improved Griess method. The concentration of H2O2 in NO-PAW was measured by using a commercial QuantiChrom Peroxide Assay Kit based on the Fe2+/Fe3+−xylenol orange oxidation reaction induced by H2O2. All measurements were performed following the instruction of the manufacturers. The measurement of •OH was measured using 3′-(p-aminophenyl) fluorescein (APF, Thermo fisher). Briefly, after 20 min of incubation, the APF fluorescence intensity in PAW was detected by a plate reader (Synergy HT, Biotek). The oxidation-reduction potential [32] and conductivity of the water samples were analyzed by an ORP30 Tester and CON30 Tester (Clean Instruments, Shanghai, China), respectively. The pH value of the NO-PAW samples was obtained using a pH spear (Eutech Instruments, Paisley, United Kingdom). All measurements were carried out right after the plasma treatment in triplicates. The gas-phase Fourier transform infrared spectroscopy (FTIR) of the plasma was analyzed by a MATRIX-MG5 Gas analyzer (Bruker, MA, USA). This is an automated real-time FT-IR gas analyzer with OPUS GA software. Also, this software simulates these parameters by the specific temperature as it can be varied at room temperature. In this machine, the gas cell has a heating system and we have used a 25 °C temperature in our setup. The gas outlet of the microwave plasma system was connected to the gas analyzer for the in-situ measurement of the plasma composition.
2.4. Cell and virus culture (inoculation)
MRC-5 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, Amarillo, TX, USA) supplemented with 10% fetal bovine serum (FBS, RDTech, Palatine, IL, USA) and 1% antibiotics (Welgene Inc., Gyeongsan, Korea), and maintained in 5% CO2. Cells were passaged every 2–3 d, to maintain a healthy state, and routinely tested for mycoplasma contamination, using the MycoAlert™ Mycoplasma Detection Kit (Lonza, Basel, Switzerland). The viral strain was propagated by inoculation of flasks containing 24 h-old seeded host cells, which were approximately 80–90% confluent. After 6–7 days of incubation in a fresh infection medium at 37 °C, the supernatant, including the working viral stock, was harvested by centrifugation (2000 rpm, 15 min). The viral titer was calculated using the 50% tissue culture infective dose (TCID50) by evaluating the cytopathic effect (CPE), in the form of cell rounding and sloughing and cytoplasmic vacuolization recorded using a bright-field microscope (10X). Additional virus stock was prepared after propagation and kept frozen at −80 °C, for future use. Virus inactivation experiments were performed in serum-free DMEM to prevent restriction by any factor that exists in serum. The second passage of HCoV-229E in susceptible MRC-5 host cells, at a titer of 1 × 10−6.8 TCID50/mL from laboratory stocks kept at −80 °C, was utilized for all experiments. As mentioned, all infection experiments were performed in a Biosafety Level 2 laboratory.
2.5. Infectivity titer determination using median tissue culture infectious dose (TCID50)
The viral titer was determined using an end-point titration assay. First, a 96-well culture plate (sample dilution plate) was inoculated with serial two-fold dilutions of defrosted HCoV-229E. In total, nine dilutions were performed, and eight replicates of each dilution were allocated per column. Thereafter, an additional 96-well culture plate with MRC-5 host cells was cultured in the culture medium described above. MRC-5 cells were rinsed with phosphate-buffered saline (PBS) after 24 h of seeding before each well was inoculated with virus from the sample plate and incubated to allow viral infection and replication within the MRC-5 cells. HCoV-229E was grown and propagated in MRC-5 cells cultured in serum-free DMEM. For the NO-PAW treatment studies, MRC-5 cells were seeded the day before HCoV-229E infection, in 96-well plates, in DMEM containing 10% FBS. Briefly, experimental HCoV-229E virus aliquots were thawed under running water at room temperature (RT) for 5 min and treated with NO-PAW or market-available drugs for 2 h at room temperature, inside a cell culture hood, before inoculation of seeded MRC-5 host cells. The virus culture and controls were subjected to CPE determination using a Nikon bright-field microscope for up to 7–10 days of culture, and TCID50 was calculated as previously described [33].
2.6. Cytotoxicity assay
Cytotoxicity of NO-PAW and positive control drugs was assessed in MRC-5 cells using the alamarBlue™ assay. MRC-5 cells were seeded in 96-well cell culture plates at a density of 104 cells/well and incubated for 24 h in a cell culture incubator. The next day, MRC-5 host cells were treated with various concentrations of NO-PAW or drugs and incubated for an additional 4 days. Thereafter, the culture medium was discarded, and cells were incubated with 10% alamarBlue solution in DMEM, for 2 h. Mean fluorescence intensity was measured using a Synergy HT spectrophotometer (BioTek, Winooski, VT, USA) at 540 nm excitation and 600 nm emission. Viability was calculated as a percentage or fraction by comparing the fluorescence of untreated control cells with that of treated cells. The 50% cytotoxic concentration (CC50) is the concentration of NO-PAW or drugs that reduced cell viability by 50%. Herein, the CC50 was calculated using the inbuilt curve fitting tool of GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, USA).
2.7. Effective antiviral concentration determination
The inactivation effect of NO-PAW and positive control drugs against HCoV-229E was evaluated using microscopic observation of CPE. MRC-5 cells (104 cells/mL) were seeded in 96-well plates, and a volume of 100 μL/well media was used. After 24 h incubation, NO-PAW and the positive control drug solutions were added to 100 TCID50 viruses and incubated for 2 h. Thereafter, treated 100 TCID50 viruses were added to MRC-5 host cells in 96-well plates. Three controls were used for these experiments: 1) cell control (cells without viruses), 2) virus control (cells with viruses), and 3) positive controls (cells with market-available drugs). Cytotoxicity measurements were performed as described, using the alamarBlue assay. The percentage of CPE was observed after 4 days of infection and treatment and calculated using the controls. The antiviral concentration of 50% effectiveness (EC50) was expressed as the drug concentration that achieved a 50% inhibition rate of virus-induced CPE. The resultant graph presented treatment doses and %CPE on the X- and Y-axes, respectively. The EC50 was calculated using GraphPad Prism's inbuilt curve fitting function.
2.8. Selectivity index analysis
The selectivity index (SI)—an indicator that measures the interface between cytotoxicity and antiviral activity—was analyzed by dividing CC50 by EC50 for each tested treatment candidate and virus.
A high SI suggests low toxicity at concentrations that display efficient antiviral activity. It also specifies that a significant range of concentrations displays an effective antiviral effect with nominal toxicity. In contrast, a low SI would indicate critical toxicity at concentrations with effective antiviral activity.
2.9. Quantitative real-time PCR analysis - viral RNA load detection and host cell response
For the isolation of viral RNA, 20 μL of cell suspension containing infected and non-infected cells was collected and mixed with 80 μL of PBS, centrifuged at 5000 rpm for 10–12 min and resuspended in 150 μL of 0.1% diethylpyrocarbonate (DEPC, Sigma-Aldrich, Korea) according to an established protocol [34]. Primer pairs used for qRT-PCR were designed and purchased from DNA Macrogen (Seoul, Korea) and are listed in Table 1. Cellular RNA from MRC-5 cells was extracted using the TRIzol method [35,36]. The effects of NO-PAW treatment on viral RNA load and early activated antiviral response gene expression in MRC-5 host cells after HCoV-229E infection were analyzed.
Table 1.
List of PCR primers used in this study.
| Gene Name | Sequence (5′-3′) |
|---|---|
| ACTIN-forward | GGC ATC CTC ACC CTG AAG TA |
| ACTIN-reverse | AGG TGT GGT GCC AGA TTT TC |
| 18sRNA-forward | CAGGTCTGTGATGCCCTTAGA |
| 18sRNA-reverse | GCTTATGACCCGCACTTACTG |
| 229E (E-Gene)-forward | ATGTTCCTTAAGCTAGTGGATGA |
| 229E (E-Gene)-reverse | TTAGAAATCAATAACTCGTTTAG |
| 229E (M-Gene)-forward | ATTGGCTTCAGGTGTTCAGG |
| 229E (M-Gene)-reverse | TCATGTTGCTCATGGGAGAG |
| 229E (N-Gene)-sense | AGGCGCAAGAATTCAGAACCAGAG |
| 229E (N-Gene)-antisense | AGCAGGACTCTGATTACGAGAAG |
| ABCC9-forward | CTGGCAGTGGGAAATCATCG |
| ABCC9-reverse | GGCTTCCCAGAGTCTGTCAT |
| BIRC3-forward | CTGTGATGGTGGACTCAGGT |
| BIRC3-reverse | TTCATCTCCTGGGCTGTCTG |
| DDIT4-forward | CAGCTGGATGTGTGTGTAGC |
| DDIT4-reverse | TACACAAACCACCTCCACGA |
| FOSL1-forward | CCGCCCTGTACCTTGTATCT |
| FOSL1-reverse | CTGCTGCTACTCTTGCGATG |
| IRF1-forward | GCCTTCTTCCCTCTTCCACT |
| IRF1-reverse | TTAATCCAGATGAGCCCCGG |
| CD13-forward | CTCACTGCAGCCTCAACTTC |
| CD13-reverse | CAAGGGACCCAGAATAGCCA |
| IRF2-forward | TTAACTCAGGACTCCAGCCC |
| IRF2-reverse | TATCTCGTCCGTTCTGAGGC |
2.10. Immunocytochemistry
To evaluate whether NO-PAW and the positive control drugs reduced the number of viral particles in HCoV-infected cells, we followed a standard fluorescent immunostaining protocol to detect viral antigens in the human host cells [37]. Briefly, 2 × 105 MRC–5 host cells for HCoV–229E were plated in each well of 12-well plates, the day before the experiment. After treatment, 200 μL of virus suspension was overlaid on a monolayer of MRC-5 host cells. The cells were rinsed twice with 1X PBS and incubated with the virus for 24 h in a fresh infection medium. Infected cells were then fixed in 100% ice-cold methanol at 4 °C for 5–6 min and labeled with anti-HCoV spike glycoprotein (1:200) in PBS containing 1% bovine serum albumin (BSA), at room temperature for 2 h. Thereafter, cells were washed thrice in 1X PBS and labeled with secondary antibody-tagged Alexa-fluor 647 (1:400) in PBS containing 1% BSA, for 30 min in the dark at room temperature. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, Korea) to achieve nuclear staining and observed using an Olympus confocal fluorescence microscope. Representative results were determined twice for each NO-PAW dose-, drug-, and virus subgroups. Up to five fields of view of the combined DAPI and Alexa Fluor images were obtained for each sample.
2.11. Flow cytometric analysis
Anti-HCoV spike glycoprotein antibody was used to analyze the expression of spike protein using a flow cytometric analysis. Briefly, NO-PAW treated and untreated virus-infected cells were harvested and stained with the desired antibody with secondary antibody-tagged Alexa-fluor 488. Samples were incubated for 45 min in ice and analyzed using a BD FACSVerse system equipped with the FACS suite software.
2.12. Statistics
Experimental data are expressed as the mean ± SD of three biological replicates. Statistical comparison was performed using either using a Two-tailed unpaired parametric t-test among two groups or a one-way ANOVA with Dunnett corrections for multiple comparisons to untreated control using PRISM9.3 software. Differences were designated statistically significant if * p < 0.05, **p < 0.01, ***p < 0.001.
3. Results
3.1. Preparation of nitric oxide enriched plasma-activated water (NO-PAW)
Herein, we used a microwave plasma torch source to prepare NO-PAW, as shown in Fig. 1A. Microwave plasma was generated using a mixture of oxygen and nitrogen gas flowing at 100 or 200 sccm and 5 or l0 lpm, respectively. Gaseous NO in water acts as a signaling molecule that can affect the virus inactivation process (Fig. 1B). The level of NOx generated by microwave plasma increased, when the amount of DI water and exposure time were reduced and increased, respectively. Approximately 4000 μM of NOx was detected in the in-water after exposure to plasma-generated gas formed using 10 lpm nitrogen and 200 sccm oxygen, within 2 h of treatment (Fig. 1C). Moreover, the pH remained in the 6.5–4.5 range from the lowest to highest level of NOx generation in the liquid phase by plasma-generated gaseous NO, under different treatment conditions.
3.2. Physical characterization of nitric oxide enriched plasma-activated water
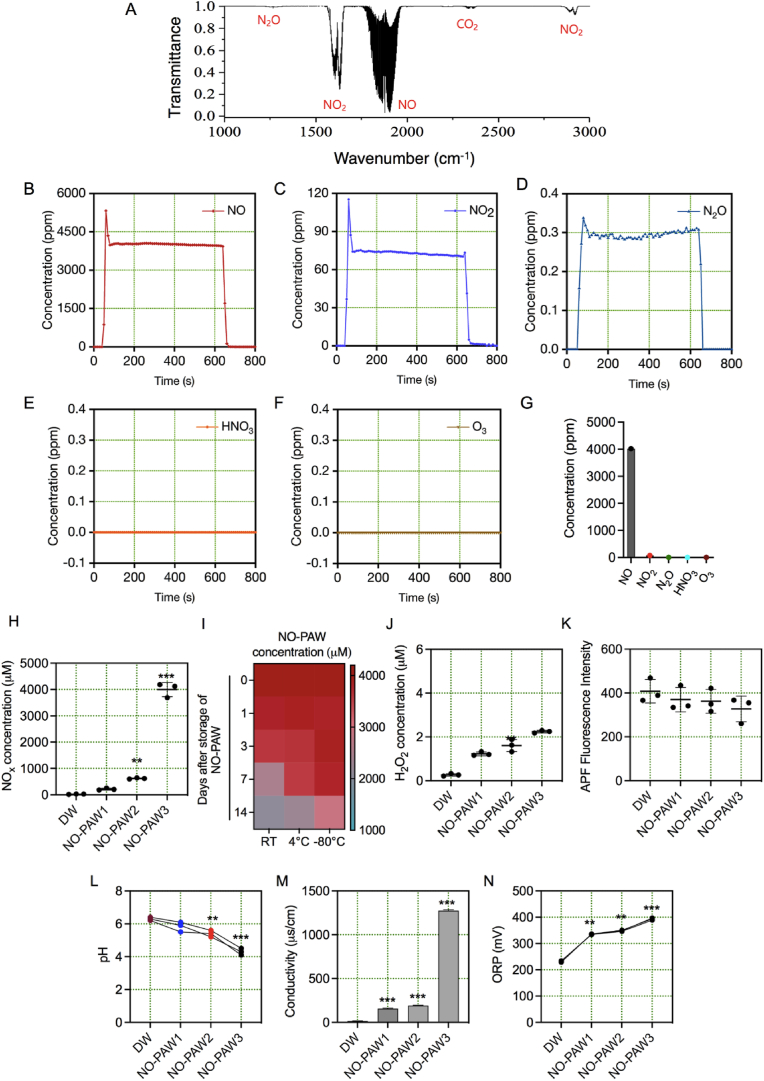
The physicochemical properties of microwave plasma-generated nitric oxide water are shown in Fig. 2. First, we analyzed the composition of the plasma by using real-time gas phase FTIR analysis. The FTIR spectrum reveals the existence of gaseous NO at 1700-2000 cm−1, NO2 at 1550-1650 cm−1, and N2O 2800-2900 cm−1 (Fig. 2A). We have noticed that NO is the dominant species with a high concentration of about 4000 ppm (Fig. 2B). On the other hand, the concentration of NO2 and N2O are significantly smaller, about 75 ppm and 0.3 ppm, respectively (Fig. 2C–D). Other common plasma-generated gases such as HNO3 and O3 were not detected in our system (Fig. 2E–G). The gas composition analyzed by FTIR proved that NO is the main product of the microwave plasma, all absolute values were calculated using inbuild OPUS GA software in Bruker Matrix-MG5 gas FTIR instrument. In this analyzer, the optical path length is 5 m of the optical cavity between a pair of mirrors and the gas cell size is 20 cm. The OPUS GA software establishes an easy-to-use graphical user interface to control MATRIX-MG gas analyzers. All concentration values are given by OPUS GA software in real-time. The software utilizes the extinction coefficient parameter automatically and shows the final concentration as per gas and temperature. After that, we measure the concentration of some dissolved reactive species that are commonly produced in PAW as well as several important physical parameters. Fig. 2H shows the NOx concentration in water at different treatment conditions as mentioned in this report. In the case of NO -PAW3, a maximum of 4000 ppm of NO in the gas phase and approximately 4000 μM NOx were measured in the liquid phase, but with NO-PAW1 and NO-PAW2, it was only about 200 μM and 620 μM, respectively in the liquid phase. The high amount of NOx produced in the PAW is in agreement with our previous report, where NO was detected by using an electrochemical sensor [27]. We emphasize that the concentration of NOx in the PAW depends on several parameters, such as the microwave power, feeding gas flow rate, treatment time, and volume of the sample. Since our experiments were mainly based on NOx concentration, therefore we have additionally checked the aging/decay effect of NOx concentration in our prepared NO-PAW using different conditions when stored at RT (∼25 °C), 4 °C, and −80 °C until 14 days. We observed that the concentration of NOx in PAW was decreased at RT, while it was more stable at lower temperatures (Fig. 2I). To validate next, whether NO is the main product as observed by FTIR analysis, we have detected H2O2 and OH radical in NO-PAW which shows that these species exist in less or negligible amounts (Fig. 2J–K). These findings indicated that these short-lived reactive species were slightly present in prepared PAW. The plasma generated gaseous NO gets immediately dissolved into the liquid phase and get converted into NO2− and NO3− and further to nitrous and nitric acid (HNO2 and HNO3). The pH (-log [H+]) is a measurement of hydrogen ion concentration in a sample. The pH value of different treatment conditions is also shown in Fig. 2L. The pH value of the plasma-generated water is lowered in accordance with the NOx concentration. Corresponding electrical conductivity and ORP are also measured and presented in Fig. 2M − N, respectively. The calculation of electrical conductivity involves the ability of a substance to transmit an electrical current over a defined area. On the other hand, electrical conductivity is a non-specific measurement of the concentration of both positively and negatively charged ions within a sample. Similarly, ORP reflects electron transfer ability through a chemical reaction. Measurements of ORP show the redox status of water. Regardless of the oxidant type or concentration, ORP rises as pH falls. The presence of any hydrogen ions present in a substance will impact the pH level and most probably influence conductivity levels and ORP which is seen in Fig. 2. NO-PAW3 has the lowest 4 pH, the highest conductivity (1300 μs cm−1), and the highest ORP (400 mV) in this experiment. It can be explained as previously described that the dissolved plasma generated gaseous NO can be converted into NO2− and NO3− anions that can raise the solution conductivity, especially for NO-PAW3, which was subjected to the highest amount of NOx. Also, the highest decrease in the pH can be attributed to the high concentration of the NOx species in the NO-PAW3. The increased ORP value also suggests that the NO-PAW3 has the strongest oxidizing capability.
Fig. 2.
Physical characteristics of NO-PAW prepared by microwave plasma system. (A) Gas-phase FTIR spectrum of the microwave plasma system. The concentration of microwave plasma system compositions was determined from FTIR analysis (B) NO (C) NO2 (D) N2O (E) HNO3 and (F) O3. (G) Summary of the gaseous species composition concentration of the microwave plasma. (H) NOx concentration in prepared NO-PAW. (I) Influence of various storage conditions on NOx concentrations in NO-PAW. (J–N). H2O2, •OH, pH, Conductivity, and ORP of prepared NO-PAW using microwave plasma system, respectively.
3.3. Nitric oxide enriched plasma-activated water reduced the cytopathogenic effect
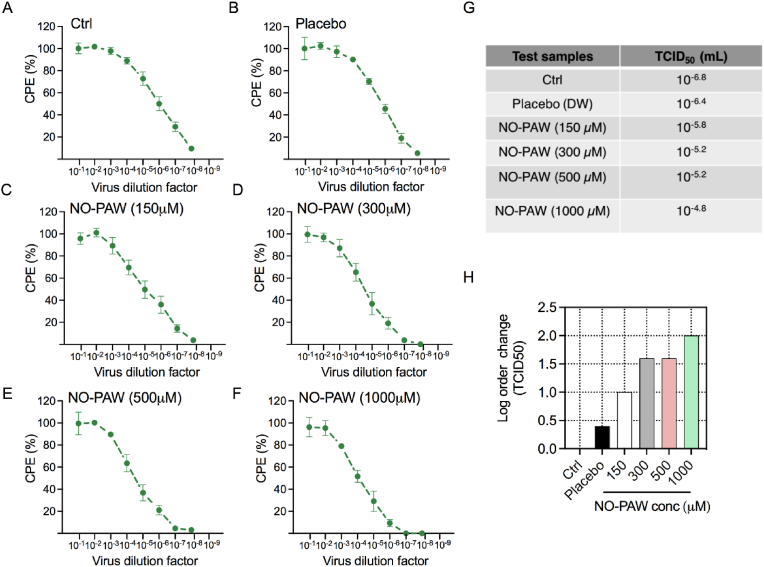
First, we tested the effect of NO-PAW on HCoV-229E infectivity. Accordingly, it was added directly after 229E viral adsorption in the cells, and viral growth was evaluated. Preliminary testing indicated that HCoV-229E-exposed MRC-5 cells started to exhibit signs of cytolytic infection-depicted by rounded cell morphology and clumping—approximately 4 days after infection. To compare the effect of NO-PAW at various concentrations (low to high), we have utilized NO-PAW3 due to the presence of high NOx concentration as compared to NO-PAW1 and NO-PAW2 in this experiment. In the presence of NO-PAW (NO-PAW3), these indications of CPE were repressed; NO-PAW inhibited cell detachment, which was followed by clumping (data not shown). The degree of CPE inhibition by NO-PAW was concentration-dependent in the HCoV-229-exposed MRC-5 cells (Fig. 3A–F). A concentration of 1000 μM NO-PAW provided near-complete defense against a detachment of the cell monolayer caused by HCoV-229E, during the seven-day investigational period. Viral infection was observed in exposed cells after 4 days of infection at higher dilutions; however, it was detected nearly 7 days after infection, in case of lower dilutions. Therefore, the effects of NO-PAW at various concentrations (150, 300, 500, and 1000 μM) were examined at 7 days—based on the CPE observed—using the TCID50 assay, which reflects the number of viral particles identified in the sample. The decrease in HCoV-229E virus growth correlated well with the variations in TCID50, in a concentration-dependent manner (Fig. 3G). The addition of 150, 300, and 500 μM NO-PAW, respectively, for 7 days decreased viral infectivity by approximately 1.0, 1.6, and 1.6 logs, respectively (Fig. 3H). Importantly, NO-PAW strongly suppressed viral growth by 2-log order of change, at concentrations of 1000 μM or higher. In sum, the results obtained from the CPE inhibition assay of anti-HCoV-229E activity indicated that NO-PAW inhibits CPE in HCoV-229E-infected MRC-5 cells.
Fig. 3.
NO-PAW inhibited the cytopathic effect in lung epithelial cells. (A–F) Cytopathic effect (CPE) was detected in MRC-5 cells, 7 days after exposure to HCoV-229E virus, at the indicated infectious viral titers. (G) The average log TCID50/mL value of three independent experiments is presented. (H) The graph indicates the log order of change in viral TCID50, at indicated NO-PAW concentrations.
3.4. Nitric oxide enriched plasma-activated water exhibits antiviral activity against HCoV-229E
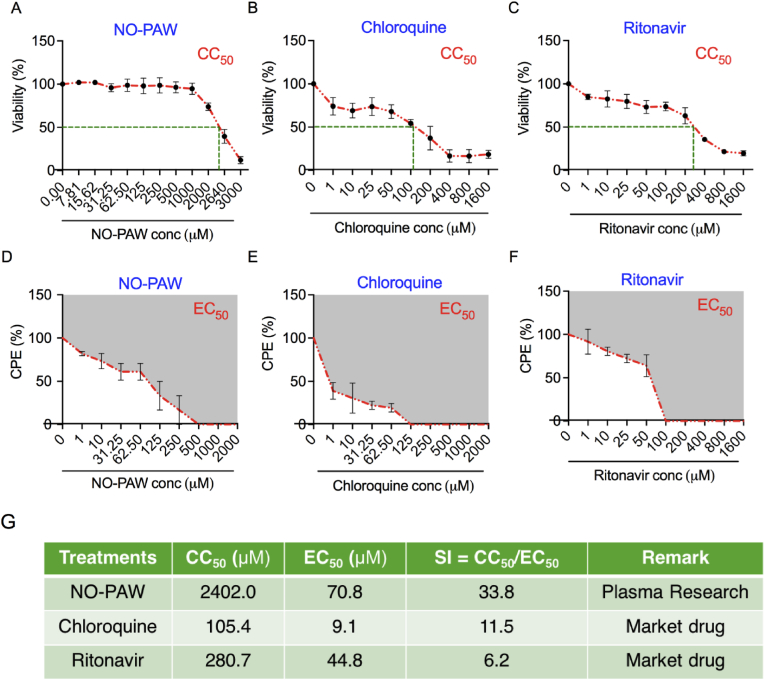
Standard assays were used to measure the cytotoxicity of NO-PAW and its effects on HCoV-229E infection rates. Initially, cytotoxicity of NO-PAW in MRC-5 cells was determined using the alamarBlue assay along with market-available antiviral drugs such as chloroquine and ritonavir. To conduct this analysis on all three groups, cells were treated with each treatment candidate at various concentrations, and the cells were incubated for 4 days. Our results indicated a CC50 value of 2402.0 μM for NO-PAW in MRC-5 cells, suggesting that its working concentration was not toxic to these cells. The corresponding values for chloroquine and ritonavir were 105.4 and 280.7 μM, respectively (Fig. 4A–C). Subsequently, we determined the effective concentration that inhibited HCoV-229E propagation by 50% (EC50). To determine EC50, MRC-5 cells were infected with 100 TCID50 HCoV-229E viruses in the presence of variable concentrations of NO-PAW, chloroquine, and ritonavir. Data showed that NO-PAW inhibits HCoV-229E infection in MRC5 cells, with an EC50 of 70.8 μM, whereas chloroquine and ritonavir showed this effect at 9.1 μM and 44.8 μM with an EC50, respectively. In addition, NO-PAW exhibited better SI (33.8) whereas chloroquine and ritonavir display a narrowed SI of 11.5 and 6.2, respectively (Fig. 4D–F). The higher SI ratio demonstrates the theoretically more efficient and safer drug would be at the time of infection for a given viral infection. These results indicated that NO-PAW—reflected the highest SI value and had the strongest anti-HCoV-229E activity among the tested drugs.
Fig. 4.
NO-PAW shows effective anti-viral activity. (A–C) Viability of MRC-5 cells after treatment with NO-PAW, chloroquine, and ritonavir at indicated concentrations, after 4 days. (D–F) Cytopathic effect (CPE) was detected in MRC-5 cells after 4 days of exposure to 100 TCID50 HCoV-229E viruses. (G) Anti-viral activity of NO-PAW, chloroquine, and ritonavir against HCoV-229E in MRC-5 cells.
3.5. Nitric oxide enriched plasma-activated water upregulates antiviral host genes response following HCoV-229E infection
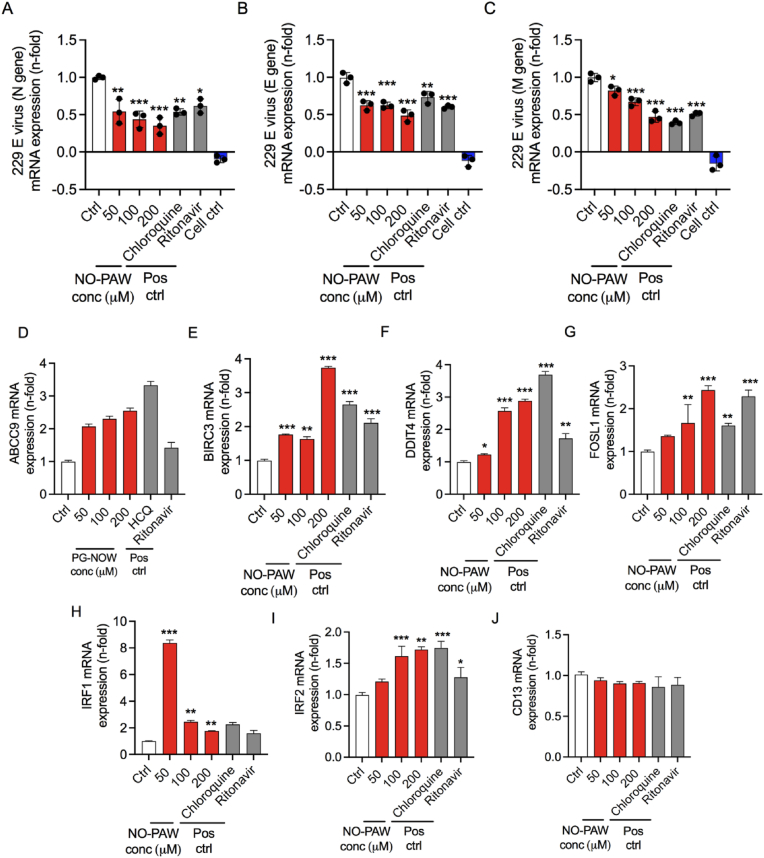
Since above results indicate that NO-PAW could effectively inhibit HCoV-229E infection in a concentration-dependent manner. Further investigation was conducted to determine whether NO-PAW could affect the expression of the 229E virus at the molecular level in the evaluated samples. It is well-known that the coronaviral genome encodes mainly four structural proteins: N, E, M, and the S protein, which are essential to construct a whole viral particle [38,39]. Few methods for coronavirus detection are available; recently described techniques for screening for human coronavirus include PCR amplification, with primers stated for the N protein gene [40,41] and E protein gene [42] in earlier reports. The coronavirus N protein is required for coronavirus RNA synthesis, binds to viral RNA, and is associated with viral genome replication [43,44] including host cellular response to viral infection [45], whereas M is considered as the vital organizer of coronavirus assembly, cooperating with other major structural proteins [39]. Also, E proteins have participated in virus assembly and growing [46]. Our data showed that NO-PAW treatment reduced the expression of N, M, and E gene messenger RNA (mRNA) levels more noticeably than chloroquine and ritonavir in HCV-229E virus (Fig. 5A–C). Following the construction of virus assembly, epithelial cells are primary targets for viral replication and represent the first line of defense against virus entry and infection. Friedman et al. reported a database of approximately 319 genes involved in the antiviral response, some of which seem to be activated in MRC-5 cells during HCoV-229E infection [47]. Based on this literature, the expression of genes involved in the antiviral response was examined in HCV-229E-infected MRC-5 host cells. Herein, RNA was extracted following infection of MRC-5 cells with HCoV-229E, and related antiviral host response genes were quantified using qPCR analysis. We evaluated the expression of interferon regulatory factors (IRF1, IRF2), ATP-binding cassette, sub-family C member 9 (ABCC9), DNA damage-inducible transcript 4 (DDIT4), FOS-like 1, AP-1 transcription factor (FOSL1), and baculoviral IAP repeat-containing 3 (BIRC3). Interestingly, NO-PAW enhanced the mRNA levels of ABCC9, BIRC3, DDIT4, IRF2, and FOSL1 antiviral genes involved in host defense (Fig. 5D–I). However, IRF1 mRNA expression was significantly elevated at lower concentrations (50 μM) as compared to control and higher NO-PAW concentrations (Fig. 5H). Notably, chloroquine and ritonavir also improved the expression of these antiviral host response genes in MRC-5 cells; however, they presented with high toxicity compared to NO-PAW, as previously indicated. Moreover, we have checked the expression of CD13, a common receptor on human cells for several molecular functions and biological processes including viral receptor activity. Though, our results indicated that NO-PAW did not affect the CD13 expression in host lung cells following 229E infection, suggesting that the normal expression of this receptor is maintained after treatment (Fig. 5J).
Fig. 5.
Expression of genes involved in antiviral response after NO-PAW treatment. (A–C) q-PCR analysis of HCoV-229E virus N, E, and M gene expression, after 7 days of relevant treatment [48]. q-PCR analysis of ABCC9, BIRC3, DDIT4, FOSL1, IRF1, IRF2, and CD13 in HCoV-229E-infected MRC-5 cells, after 7 days of relevant treatment.
3.6. Nitric oxide enriched plasma-activated water reduces the expression of spike protein following HCoV-229E infection
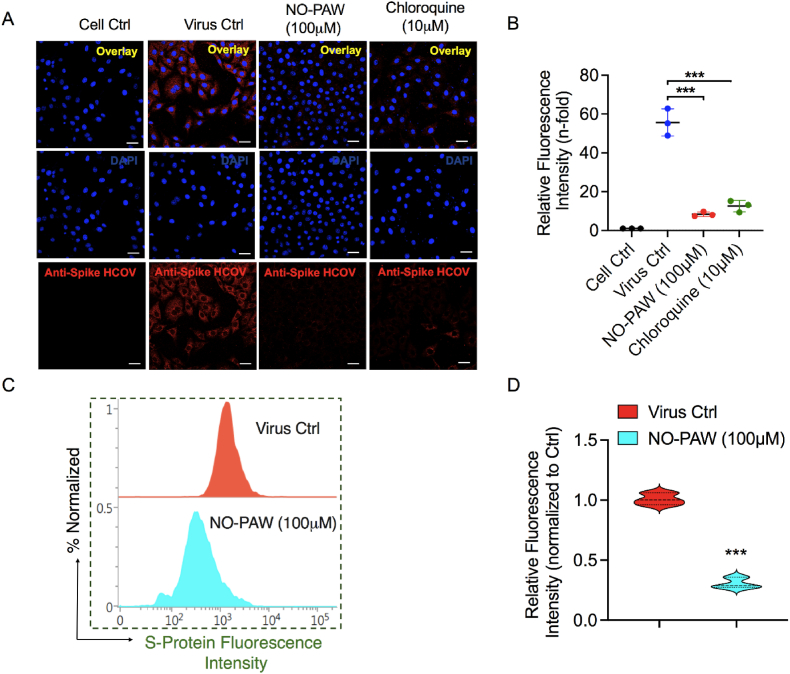
Finally, we investigated the integration of HCoV-229E in MRC-5 host cells after exposure to NO-PAW and relevant positive control drugs. The spike protein is a glycosylated protein-making spike on the viral surface that mediates the virus entry into the host cells. Fig. 6 shows representative spike protein fluorescent images of HCoV-229E-infected MRC-5 cells treated with 100 μM NO-PAW and 10 μM chloroquine. In Fig. 6A, the red fluorescence intensity qualitatively indicates infection of MRC-5 host cells with live HCoV-229E virus. Treatment with NO-PAW noticeably reduced the expression of viral spike glycoprotein, as indicated by a reduction in red fluorescence in HCoV-229E-infected MRC-5 cells, when compared to the virus control (Fig. 6B). To examine further to quantify the expression of the spike protein of viral particles in HCoV-infected MRC5, we performed flow cytometry analysis after NO-PAW treatment in those cells. Results revealed that expression of spike protein was significantly reduced after NO-PAW treatment in infected MRC-5 host lung cells (Fig. 6C–D). Taken together, NO-PAW has the potential to inactivate HCoV-229E virus through the diminution of spike protein levels.
Fig. 6.
NO-PAW reduces the expression of spike protein in HCoV-229E-infected MRC-5 cells. (A) Representative immunofluorescence images of MRC5 human lung fibroblasts infected with HCoV-229E virus. (B) A quantitative graph has been shown calculated from panel A. (C, D) Flow cytometry analysis of spike protein in HCoV-229 E infected MRC-5 cells after 100 μM NO-PAW treatment after 4 days.
4. Discussion
The public health crisis caused by SARS-CoV-2 has become a worldwide pandemic. The severity of the COVID‐19 pandemic demands the rapid development and implementation of successful countermeasures to lower person-to-person transmission. Although the disinfection of viruses in the environment can effectively prevent their transmission, efficient treatment strategies for viral inactivation need to be established. We have developed a promising antiviral treatment approach using gaseous NO based on plasma technology. The entire plasma NO-generating system used in this study was separated from the atmospheric environment; hence, the NO-PAW components were uniquely controlled by the inlet gases, N2 and O2. First, we confirmed whether our system generated a significant amount of NO among other species. FTIR analysis revealed the dominance of NO concentration over other species which proved the high amount of NO that can be transported to the PAW (Fig. 2). The liquid phase analysis also confirmed the high NOx level in the prepared NO-PAW, at standardized parameters. The effects of reactive nitrogen species, such as NOx, on biological systems, are known to depend on their concentrations. Low concentrations can stimulate cell function and initiate many cell signaling pathways, but at high concentrations, it can provoke cell death [[49], [50], [51]]. Moreover, the formation of endogenous NO appears to have an encouraging antiviral effect against human respiratory viruses. It might be postulated that the administration of added NO could have a beneficial therapeutic effect against these infections when endogenous NO concentrations are not sufficiently abundant to attain significant antiviral effects. Various strategies for administering additional NO in the body have been recognized, some of which are in the early stages of development, whereas others have been applied at the clinic level [52]. Many exogenous NO donors have been manufactured and translated at the clinical level (not as antiviral treatments) [50,53]. In most in vitro studies, the donor is supplied to the cell culture media and its biological effects (e.g., inhibition of viral replication) were examined. However, researchers usually do not measure the concentration of generated NO; therefore, the correlation between the therapeutic level of the donor and inhibition of the virus is not very clear. A facile, potent substitute was discovered in the pulmonary administration of gaseous NO through inhalation. This is necessary from the perspective of the main topic of this discussion, namely the treatment of HCoVs. Inhalation of gaseous NO has been successfully used in the treatment of respiratory conditions, including pulmonary infections and tuberculosis [54]. More recently, portable devices that can generate NO have been established for clinical applications [55].
NO in NO-PAW is dissolved from the gaseous phase, after being produced by the plasma-induced breakdown of molecular oxygen and nitrogen. The most used treatments in the SARS-CoV-2 pandemic include various repurposed drugs and traditional methods that may have potential safety, biocompatibility, and therapeutic efficiency issues. In contrast, NO-PAW does not exhibit side effects or treatment efficacy concerns. Based on the investigation of the HCoV-229E model, NO-PAW drastically inactivated viruses and inhibited their infection rate in lung host cells, reflecting the inactivation ability of the present plasma-based strategy. Data from TCID50 assays on MRC-5 cells suggest that NO-PAW exerts therapeutic effects against the HCoV-229E virus by inhibiting viral replication. In the cultured cells, virus particles decreased and TCID50 was altered by 2-log order of change at a NO-PAW concentration of 1000 μM, suggesting the practicality of NO-PAW in therapeutic applications. NO-PAW presented antiviral effects in a dose-dependent manner at specific 100 TCID50 virus dilution factors, but its cytotoxicity in lung host cells was not dose-dependent, up to very high concentrations. The antiviral compounds inhibited cell growth, with the SI depending on experimental design and the methods used to measure cytotoxicity [56]. Thus, the SIs of antiviral drugs, as well as CC50 and EC50 (by visualization of CPE), could be easily determined. We also confirmed the antiviral activity of two compounds (chloroquine and ritonavir) that have been previously reported to inhibit HCoV-229E replication in vitro, along with NO-PAW. Of the evaluated alternatives, NO-PAW was the most powerful candidate with an EC50 of 70.8 μM, a CC50 of 2402 μM, and an SI of 33.8, indicating that it could efficiently fight HCoV-229E infection. Both chloroquine and ritonavir had lower SI values than NO-PAW (Fig. 4). However, most studies showed an antiviral effect in vitro for these two control drugs, with potential cytotoxicity that warrants further investigation. In agreement with prior studies [6,57], we also observed the high cytotoxicity of chloroquine and ritonavir in MRC-5 host cells, which precluded the study of their anti-HCoV effect. In contrast, NO-PAW showed no or minimal toxicity at concentrations up to 2000 μM in lung host cells (Fig. 4). On the other hand, storage conditions are critical factors affecting the composition and antiviral potential of PAW. In this regard, we revealed that NOx concentration was comparably more stable when stored at lower temperatures after the generation of the NO-PAW as shown in Fig. 2.
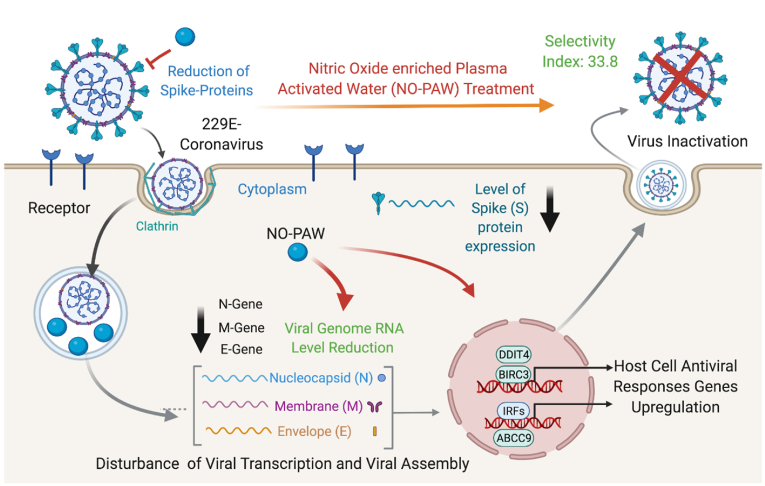
Generally, viral infections initiate a broad range of defense mechanisms inside host cells. Upon infection, viral particles are present in the cytosol and are recognizable by intracellular sensors. These sensors initiate signaling pathways that activate IFN regulatory factors (IRFs). Other members of the IRF family have been shown to play a vital role in the regulation of viral replication and apoptosis [58]. The inhibitor of apoptosis (IAP) family member, BIRC3 has been shown to trigger IRF to endorse an interleukin-1-mediated inflammatory response [59]. Largely, IFNs cannot directly display antiviral functions; however, they activate the downstream JAK-STAT signaling pathway by binding to the receptors, IFN alpha and beta receptor subunit 1 (IFNAR1) and IFNAR2, to induce the transcription of IFN-stimulated genes (ISGs) for antiviral activity [60,61]. It is also stated that an adaptor protein named mitochondrial antiviral signaling adaptor, localized in the mitochondria, is recruited and activates IRFs to trigger the expression of IFNs and pro-inflammatory cytokines [62]. These all genes through various pathways collectively enhance the IRFs expression in host cell antiviral responses. Besides, DDIT4, which is DNA damage-inducible transcript 4, stimulated by IFN was upregulated after treatment of NO-PAW in infected MRC-5 cells. Earlier literature suggests that DDIT4 protein levels enhance at the time of viral entry or/and primary viral transcription and are then down-regulated at late stages of infection. Mata et al. demonstrated that the transient early induction in DDIT4 levels probably signifies an antiviral response, where DDIT4 serves as a host defense factor [63] and prevented induction of mTORC1 signaling by the virus [64]. One another critical factor in antiviral immunity seems to be mammalian ABCC9 protein, which was recognized to be essential for antiviral immunity in human cells [65]. We observed that NO-PAW efficiently upregulated the expression of the above discussed antiviral gene response in MRC-5 host cells, upon HCoV-229E infection (Fig. 5). Considering its efficacy in host defense system enhancement activity, NO-PAW can act as an anti-viral agent after more preclinical studies for the treatment of coronavirus infections. Interestingly, NO-PAW suppressed the detectable level of viral N, M and E gene mRNA, suggesting its potential for viral inactivation itself. Confocal microscopy revealed a wide distribution of spikes in the cytoplasm of MRC-5 cells transfected with HCoV-229E. Spike staining is prominent in virus-infected MRC-5 cells, which may be partly explained by their adherence to membrane receptors and synthesis of spike protein, prior to them assembling to form new virus particles during the replication process. Flow cytometric analysis revealed the remarkable reduction of spike protein levels after NO-PAW treatment (Fig. 6). The results indicated that NO-PAW have the potential to suppress the spike protein expression in 229E-infected MRC-5 cells, suggesting possible viral inactivation in host cells. These findings propose that NO-PAW efficiently reducing the construction of viral genome assembly and inducing the expression of host cell antiviral response genes to lead biocompatible and nontoxic candidate for therapeutic purposes (Fig. 7). Further studies should be extended to pseudoviruses to explore the specificity of NO-PAW against novel and future coronaviruses.
Fig. 7.
Possible mechanism of virus inactivation by NO-PAW in host cells.
5. Conclusion
Herein, we confirmed the anti-coronavirus effect of plasma generated gaseous NO based NO-PAW by comparing it to two well-known antiviral compounds, hydroxychloroquine, and ritonavir, which have been previously reported to inhibit HCoV replication in vitro. Our findings indicated that, among all tested compounds, plasma generated gaseous NO offered the most powerful means to fight HCoV infection in lung host cells. NO-PAW reduced viral RNA load and decreased the expression of spike proteins, thereby improving the host antiviral response. The outcomes of the present investigation, conducted in our plasma bioscience center, could reflect future treatment strategies against coronavirus infection that are affordable and could be used in a clinical setting. Administration routes could include oral, intratracheal, or intraperitoneal delivery, or form part of a combined treatment regimen against coronavirus diseases. Future clinical trials concerning this topic are required to further improve understanding of the antiviral activity of NO-PAW.
CRediT authorship contribution statement
Nagendra Kumar Kaushik: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing, Writing – original draft. Pradeep Bhartiya: Data curation, Formal analysis. Neha Kaushik: Data curation, Formal analysis, Software, Writing – original draft. Yungoh Shin: Investigation, Methodology. Linh Nhat Nguyen: Data curation, Formal analysis, Writing – original draft. Jang Sick Park: Investigation, Methodology. Doyoung Kim: Investigation, Methodology. Eun Ha Choi: Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing, Conceptualization, All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the National Research Foundation(NRF) of Korea, funded by the Korean government (2021R1A6A1A03038785, 2021R1F1A1055694), and by Kwangwoon University in 2021.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Nagendra Kumar Kaushik, Email: kaushik.nagendra@kw.ac.kr.
Eun Ha Choi, Email: ehchoi@kw.ac.kr.
References
- 1.Wolf Y.I., et al. Origins and evolution of the global RNA virome. mBio. 2018;9(6) doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Hoek L. Human coronaviruses: what do they cause? Antivir. Ther. 2007;12(4 Pt B):651–658. [PubMed] [Google Scholar]
- 3.Ashour H.M., et al. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9(3) doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F.-W., et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. JID (J. Infect. Dis.) 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu C.M. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choy K.-T., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020:178. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent M.J., et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2(1) doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell. Discov. 2020;6(1) doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Sanctis V., et al. Preliminary data on COVID-19 in patients with hemoglobinopathies: a multicentre ICET-A study. Mediterr. J. Hematol. Infect. Dis. 2020;12(1) doi: 10.4084/MJHID.2020.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaushik N., et al. The inactivation and destruction of viruses by reactive oxygen species generated through physical and cold atmospheric plasma techniques: current status and perspectives. J. Adv. Res. 2022 doi: 10.1016/j.jare.2022.03.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duarte S., Panariello B.H.D. Comprehensive biomedical applications of low temperature plasmas. Arch. Biochem. Biophys. 2020:693. doi: 10.1016/j.abb.2020.108560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau M., Orange N., Feuilloley M.G.J. Non-thermal plasma technologies: new tools for bio-decontamination. Biotechnol. Adv. 2008;26(6):610–617. doi: 10.1016/j.biotechadv.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno A. Industrial applications of atmospheric non-thermal plasma in environmental remediation. Plasma Phys. Contr. Fusion. 2007;49(5A):A1–A15. [Google Scholar]
- 14.Nicol M.J., et al. Antibacterial effects of low-temperature plasma generated by atmospheric-pressure plasma jet are mediated by reactive oxygen species. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-59652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss M., et al. Virucide properties of cold atmospheric plasma for future clinical applications. J. Med. Virol. 2017;89(6):952–959. doi: 10.1002/jmv.24701. [DOI] [PubMed] [Google Scholar]
- 16.Xia T., et al. Inactivation of airborne porcine reproductive and respiratory syndrome virus (PRRSv) by a packed bed dielectric barrier discharge non-thermal plasma. J. Hazard Mater. 2020;393 doi: 10.1016/j.jhazmat.2020.122266. [DOI] [PubMed] [Google Scholar]
- 17.Su X., et al. Inactivation efficacy of nonthermal plasma-activated solutions against newcastle disease virus. Appl. Environ. Microbiol. 2018;84(9) doi: 10.1128/AEM.02836-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipić A., et al. Inactivation of pepper mild mottle virus in water by cold atmospheric plasma. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.618209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aboubakr H.A., et al. Virucidal effect of cold atmospheric gaseous plasma on feline calicivirus, a surrogate for human norovirus. Appl. Environ. Microbiol. 2015;81(11):3612–3622. doi: 10.1128/AEM.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasuda H., et al. Biological evaluation of DNA damage in bacteriophages inactivated by atmospheric pressure cold plasma. Plasma Process. Polym. 2010;7(3–4):301–308. [Google Scholar]
- 21.Dai X., et al. Dosing: the key to precision plasma oncology. Plasma Process. Polym. 2020;17(10) [Google Scholar]
- 22.Guo L., et al. Mechanism of virus inactivation by cold atmospheric-pressure plasma and plasma-activated water. Appl. Environ. Microbiol. 2018;84(17) doi: 10.1128/AEM.00726-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L., et al. Plasma-activated water: an alternative disinfectant for S protein inactivation to prevent SARS-CoV-2 infection. Chem. Eng. J. 2021:421. doi: 10.1016/j.cej.2020.127742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akaike T. Role of free radicals in viral pathogenesis and mutation. Rev. Med. Virol. 2001;11(2):87–101. doi: 10.1002/rmv.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majano P.L., Garcia-Monzon C. Does nitric oxide play a pathogenic role in hepatitis C virus infection? Cell Death Differ. 2003;10(S1):S13–S15. doi: 10.1038/sj.cdd.4401115. [DOI] [PubMed] [Google Scholar]
- 26.Klingström J., et al. Nitric oxide and peroxynitrite have different antiviral effects against hantavirus replication and free mature virions. Eur. J. Immunol. 2006;36(10):2649–2657. doi: 10.1002/eji.200535587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang M.H., et al. Dynamics of nitric oxide level in liquids treated with microwave plasma-generated gas and their effects on spinach development. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-018-37711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji S.H., et al. Assessment of the effects of nitrogen plasma and plasma-generated nitric oxide on early development of Coriandum sativum. Plasma Process. Polym. 2015;12(10):1164–1173. [Google Scholar]
- 29.Lee C.B., et al. Anticancer activity of liquid treated with microwave plasma-generated gas through macrophage activation. Oxid. Med. Cell. Longev. 2020;2020:1–13. doi: 10.1155/2020/2946820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Na Y.H., et al. Production of nitric oxide using a microwave plasma torch and its application to fungal cell differentiation. J. Phys. Appl. Phys. 2015;48(19) [Google Scholar]
- 32.Michel V., et al. A complex eIF4E locus impacts the durability of va resistance to Potato virus Y in tobacco. Mol. Plant Pathol. 2019;20(8):1051–1066. doi: 10.1111/mpp.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murata T., et al. Reduction of severe acute respiratory syndrome coronavirus‐2 infectivity by admissible concentration of ozone gas and water. Microbiol. Immunol. 2020;65(1):10–16. doi: 10.1111/1348-0421.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sizun J., Arbour N., Talbot P.J. Comparison of immunofluorescence with monoclonal antibodies and RT-PCR for the detection of human coronaviruses 229E and OC43 in cell culture. J. Virol Methods. 1998;72(2):145–152. doi: 10.1016/S0166-0934(98)00013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaushik N., et al. Blockade of cellular energy metabolism through 6-aminonicotinamide reduces proliferation of non-small lung cancer cells by inducing endoplasmic reticulum stress. Biology. 2021;10(11) doi: 10.3390/biology10111088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen L.N., et al. Plasma-synthesized mussel-inspired gold nanoparticles promote autophagy-dependent damage-associated molecular pattern release to potentiate immunogenic cancer cell death. J. Ind. Eng. Chem. 2021;100:99–111. [Google Scholar]
- 37.Buonanno M., et al. Far-UVC light (222 nm) efficiently and safely inactivates airborne human coronaviruses. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-67211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2004;576(1–2):174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters P.S. 2006. The Molecular Biology of Coronaviruses; pp. 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myint S., et al. Evaluation of nested polymerase chain methods for the detection of human coronaviruses 229E and OC43. Mol. Cell. Probes. 1994;8(5):357–364. doi: 10.1006/mcpr.1994.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart J.N., Mounir S., Talbot P.J. Diagnosis of Human Viruses by Polymerase Chain Reaction Technology. 1992. Detection of coronaviruses by the polymerase chain reaction; pp. 316–327. [Google Scholar]
- 42.Nomura R., et al. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 2004;78(16):8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bisht H., et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Boheemen S., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3(6) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride R., van Zyl M., Fielding B. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieto-Torres J.L., et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar B., et al. Transcriptomic profiling and genomic mutational analysis of Human coronavirus (HCoV)-229E -infected human cells. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies H., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 49.Napoli C., et al. Effects of nitric oxide on cell proliferation. J. Am. Coll. Cardiol. 2013;62(2):89–95. doi: 10.1016/j.jacc.2013.03.070. [DOI] [PubMed] [Google Scholar]
- 50.Carpenter A.W., Schoenfisch M.H. Nitric oxide release: Part II. Therapeutic applications. Chem. Soc. Rev. 2012;41(10) doi: 10.1039/c2cs15273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saura M., et al. An antiviral mechanism of nitric oxide. Immunity. 1999;10(1):21–28. doi: 10.1016/S1074-7613(00)80003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang T., Zelikin A.N., Chandrawati R. Progress and promise of nitric oxide‐releasing platforms. Adv. Sci. 2018;5(6) doi: 10.1002/advs.201701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P.G., et al. Nitric oxide donors: chemical activities and biological applications. Chem. Rev. 2002;102(4):1091–1134. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- 54.Bhatraju P., et al. Inhaled nitric oxide: current clinical concepts. Nitric Oxide. 2015;50:114–128. doi: 10.1016/j.niox.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Yu B., et al. Producing nitric oxide by pulsed electrical discharge in air for portable inhalation therapy. Sci. Transl. Med. 2015;7(294) doi: 10.1126/scitranslmed.aaa3097. [DOI] [PubMed] [Google Scholar]
- 56.Yang C.-W., et al. Identification of anti-viral activity of the cardenolides, Na +/K + -ATPase inhibitors, against porcine transmissible gastroenteritis virus. Toxicol. Appl. Pharmacol. 2017;332:129–137. doi: 10.1016/j.taap.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taniguchi T., Tanaka N., Taki S. Regulation of the interferon system, immune response and oncogenesis by the transcription factor interferon regulatory factor-1. Eur. Cytokine Netw. 1998;9(3 Suppl):43–48. [PubMed] [Google Scholar]
- 59.Harikumar K.B., et al. K63-linked polyubiquitination of transcription factor IRF1 is essential for IL-1-induced production of chemokines CXCL10 and CCL5. Nat. Immunol. 2014;15(3):231–238. doi: 10.1038/ni.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moraga I., et al. Receptor density is key to the alpha2/beta interferon differential activities. Mol. Cell Biol. 2009;29(17):4778–4787. doi: 10.1128/MCB.01808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Secombes C.J., Zou J. Evolution of interferons and interferon receptors. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gack M.U., Pfeiffer J. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J. Virol. 2014;88(10):5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mata M.A., et al. Chemical inhibition of RNA viruses reveals REDD1 as a host defense factor. Nat. Chem. Biol. 2011;7(10):712–719. doi: 10.1038/nchembio.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawaoka Y., et al. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog. 2017;13(9) doi: 10.1371/journal.ppat.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Croker B., et al. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nat. Genet. 2007;39(12):1453–1460. doi: 10.1038/ng.2007.25. [DOI] [PubMed] [Google Scholar]