Abstract
Background:
c-Myc has become significantly involved in aggressive B-cell non Hodgkin lymphoma (NHL), but little is known about its importance in T and NK cell NHL (TNKcNHLs) in association with prognostic factors. The study is to investigate the significance of c-Myc expression with clinicopathological features of TNKcNHLs patients.
Methodology:
A cross-sectional study of 32 archived tissue blocks of TNKcNHLs were immunohistochemically stained with c-Myc. The results were microscopically evaluated and statistically analysed to examine the association between the clinicopathological data with the c-Myc expression.
Results:
c-Myc protein expressions were detected in 25/32 (78.1%) cases. The median age was 38-years. Malay ethnicity (92.0%) with 21 males and 11 females. c-Myc expressions were seen in T lymphoblastic lymphoma (20%), ALK-positive ALCL (16%) ,PTCL,NOS (16%), extra nodal NK/T-cell lymphoma, nasal type (12%), extra-nodal involvement (78.1%), elevated serum LDH (83.3%) and high ECOG performance status (82.4%). However, no statistical significant of c-Myc in association with the clinicopathological parameters (p > 0.05).
Conclusion:
There was no statistically significant association of clinicopathological parameters and histological subtypes of TNKcNHLs contributed by small samples tested. However, the attribution of c-Myc in this disease should be further explored.
Key Words: c-Myc protein expression, T and NK cell NHL, immunohistochemistry, clinicopathology
Introduction
Lymphomas encompass malignant diseases that originate from T and B cells in the lymphatic system. It can arise in any tissue, two third are from lymph node and one third from extranodal (Yin et al., 2019). There are Hodgkin lymphoma (HL) and Non-Hodgkin lymphoma (NHL: B-cell, T-cell and natural killer (NK) cell neoplasms). T and NK cell NHLs (TNKcNHLs) are uncommon, constituting about 10-15% of the disease (Satou et al., 2019). They comprised of heterogeneous group of lymphoma entities, classified by morphology, immunophenotype, and genetic characteristics. In the World Health Organisation (WHO) Classification system, TNKcNHLs can be divided into precursor T-cells (T lymphoblastic lymphoma (TLBL), and mature T-cells, termed as peripheral T cell lymphomas (PTCLs). Specifically, 31 entities are listed in the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Swerdlow et al., 2016). Except for a few relatively indolent entities, TNKcNHLs are aggressive tumours with poor prognosis (Armitage, 2012; Zain, 2019).
TNKcNHLs are underrepresented because of lower incidence compared to that of B-cell NHL, however they vary in different geographical regions and racial populations. TNKcNHLs commonly affect adults and the elderly, and are more common in Asia (10.1%) compared to the West (6%) (Au et al., 2005; Zain et al., 2019). Wang and Vose (2013) discovered the incidence rates of most B cell NHL have started to decline in the United States compared to the rising incidence rates of TNKcNHLs, mainly PTCL and AITL. In Malaysian population, lymphoma was generally ranked fourth in males and sixth in females as reported by Azizah et al., (2016). Malay males had the higher rate than Chinese and Indian ethnicities. The same report showed that the lymphoma incidence in 2007-2011 was increasing in trend.
The clinical presentations of TNKcNHLs’ are variable, and associated with poor clinical outcomes (Xu et al., 2012). There are prognostic clinical markers that determine the disease outcome. The commonly used is the International Prognostic Index (IPI) for NHL, which include the age, Ann Harbor staging, Eastern Cooperative Oncology Group (ECOG) performance status and serum LDH (Ramadas et al., 2017). Clinical presentation of B symptoms that carry an adverse outcome, are incorporated as part of Ann Harbor staging.
The c-MYC gene comprised of three exons located on chromosome 8q24 in humans (Huppi et al., 2012). The major product of the c-MYC gene is c-Myc protein, which comprise over 430 amino acids with 150 amino-terminal residues (Dang et al., 2005). Physiologically, c-Myc protein is expressed during embryogenesis and tissues that are highly proliferating like skin epidermis and gastrointestinal mucosa. c-MYC is identified as a regulator of more than 15% of genes that control cell growth, cell cycle, survival and metabolism (Ott et al., 2013).
Elevated or deregulated expression of c-MYC have been found in many human cancers, and associated with aggressive and poorly differentiated tumours (Beroukhim et al., 2010; Dang, 2012). Activated oncogenic c-MYC is important in the development of Burkitt lymphoma (BL), caused by a translocation between chromosome 8 and one of the three chromosomes that has antibody-encoding genes (Molyneux et al., 2012). Other studies reported the translocation of the MYC gene, contributed to a poorer prognosis in DLBCL (Green et al., 2012; Ott et al., 2013). A recent study on c-myc protein expression and high ki-67 in diffuse large B cell lymphoma (DLBCL) showed the likelihood of disease relapsed (El Hussien et al., 2021)
Many studies on c-MYC focused on aggressive B cell lymphoma such as BL, DLBCL and high-grade B-cell lymphoma, and linked them to poor outcome and survival (Green et al., 2012), and only a few on TNKcNHLs. Chisholm et al., (2015) reported c-Myc protein expression was more than 90% in TNKcNHLs, and correlated with poor survival. However, there is still insu-cient information regarding c-Myc protein expression in TNKcNHLs. This prompted us to conduct a study to examine c-Myc protein expression in association with clinicopathological features of TNKcNHLs diagnosed in our centre, Hospital Universiti Sains Malaysia. We used IHC technique as it was reported to have a high sensitivity (89%-91%) and specificity (88% to 89.4%) in predicting the MYC rearrangement by fluorescent in situ hybridisation (FISH) (Lynnhtun et al., (2014); Nwanze et al., (2017). Furthermore, it is cheaper and less time consuming compared to FISH method.
Materials and Methods
A cross sectional study was conducted in the Department of Pathology, Hospital USM from 2001 until 2018. The histopathological reports were obtained from the laboratory’s databases. 55 cases were reported as TNKcNHLs, and only 32 were selected as they have adequate tissue. The epidemiological data including IPI and information on treatment were retrieved from the patients’ records. Selected FFPE tissue blocks were sectioned at 3μm thickness, then deparaffinized and epitope retrieval using a pH6 Tris-based solution. The slides were incubated overnight with anti-c-MYC primary antibody (clone Y69, ab32072; ABCAM) at dilution 1:500. Localization of the antigen-antibody complex was achieved using the DAKO 3,3’diaminobenzidine tetrahydrochloride (DAB), then visualized with horseradish peroxide (HRP) substrate and DAB chromogen. The positive control tissue used was colorectal carcinoma tissue with adjacent normal colonic mucosa cells for internal negative controls. Microscopic assessment by counting 100 tumour cells in ten representative high-power fields (HPF) (×400), and 5% as a cut-off value (Huang et al., 2014). This was done by two pathologists blinded to the clinicopathological parameters. Nuclear staining for c-Myc was considered positive, and cytoplasmic staining was negative. Data was analysed using software IBM SPSS version 24. Descriptive analysis for the clinicopathological parameters, and Fisher’s exact test to analyse the association of clinicopathological characteristics with c-Myc expression Level of significance in this study was set as p value < 0.05.
Results
Clinicopathological characteristics
55 cases of primary TNKcNHLs from 634 lymphoma cases diagnosed in Department of Pathology ,Hospital USM from 2001 to 2018. Only 32 were selected and had c-Myc IHC staining done. There were 21 (65.6%) males and 11 (34.4%) females. The median age was 38 years old (8 to 81). The ethnicities were Malay (29,90.6%), Chinese (2, 6.3%) and Indian (1, 3.1%). FFPE tissues were from 10 (31.3%) lymph nodes and 22 (68.8%) extra-nodal biopsies. The site of involved organs, the subtypes of TNKcNHLs and clinicopathological data are listed in Table 1. Only 17 (53.2%) patients received chemotherapy. 10 (31.3%) were treated with CHOP (cyclophosphamide, doxorubicin, vincristin, and prednisone) as the first line treatment. Alternative regimens were used, for example RICE (rituximab, ifosfamide, carboplatin and etoposide), ICE, romidepsin and SMILE (etoposide, ifosfomide, methotrexate and dexamethasone), the latter was given to all extra nodal TNKcNHLs patients. Fifteen patients (46.9%) did not received chemotherapy treatment, because of poor performance status, multiorgan insufficiencies and patients’ refusal.
Table 1.
Distribution of Clinicopathological Features, Sites of the Lesion, and Subtypes of T and NK Cell Lymphoma Patients (n = 32).
| Variables | Mean (SD) | n (%) |
|---|---|---|
| Age | 38.2 (22.7) | |
| Age group | ||
| Less than 60 | 24 (75.0) | |
| More than 60 | 8 (25.0) | |
| Race | ||
| Malay | 29 (90.6) | |
| Chinese | 2 (6.3) | |
| Indian | 1 (3.1) | |
| Gender | ||
| Male | 21 (65.6) | |
| Female | 11 (34.4) | |
| Site | ||
| Nodal | 10 (31.3) | |
| Extra nodal | 22 (68.8) | |
| Extra nodal | ||
| Less than 1 | 25 (78.1) | |
| More than 1 | 7 (21.9) | |
| Ann-Arbor staging | ||
| I-II | 16 (50.0) | |
| III-IV | 16 (50.0) | |
| LDH value | ||
| Normal | 14 (43.8) | |
| Elevated | 18 (56.3) | |
| ECOG | ||
| Less than 2 | 15 (46.9) | |
| More than 2 | 17 (53.1) | |
| B symptoms | ||
| Yes | 12 (37.5) | |
| No | 20 (62.5) | |
| IPI score | ||
| 0 to 2 | 18 (56.3) | |
| 3 to 5 | 14 (43.8) | |
| Sites of lesion | ||
| Nodal | 10 (31.3) | |
| Extranodal | 22 (68.7) | |
| Bone | 4 (12.5) | |
| Nasopharynx | 6 (18.8) | |
| Skin | 3 (9.4) | |
| Soft tissue | 3 (9.4) | |
| Tonsils | 2 (6.3) | |
| Trephine | 4 (12.5) | |
| Subtypes | ||
| ALK-not done, ALCL | 1 (3.1) | |
| ALK-negative, ALCL | 1 (3.1) | |
| ALK-positive, ALCL | 6 (18.8) | |
| AITL | 1 (3.1) | |
| ENTKL, nasal type | 3 (9.4) | |
| PTCL, NOS | 6 (18.8) | |
| Variables | n (%) | |
| T lymphoblastic lymphoma | 6 (18.8) | |
| T-cell lymphoma (not being subtyped) | 8 (25.0) | |
ALCL, anaplastic large cell lymphoma; ALK, anaplastic lymphoma kinase; AITL, angioimmunoblastic T cell lymphoma; ENTKL, extranodal T and NK cell lymphoma; PTCL, peripheral T cell lymphoma; NOS, not otherwise specified.
c-Myc Immnunohistochemistry Assessment
Microscopic assessment of the c-Myc stain on the tumour cell nucleus are done on the selected 32 cases. Stained nucleus more than 5% were considered as positive (Huang et al., 2014). Figure 1 shows positive nuclear staining of the tumour cells. The study observed that c-Myc immunoreactivity was seen in 78.1% of the cases. Table 2 shows c-Myc expressions according to the clinicopathology parameters and histopathology subtypes. However in this study, we found no statistical significant association between c-Myc expressions and clinicopathological factors or with the subtypes (p>0.05). There were 11 (34.4%) patients who succumbed to death and among them, c-Myc expressions were seen in eight cases. The main cause of demise were complications of infection. Three patients are still alive, whereas the rest of the cases in 18/32 (56%) remain unknown of their status. Two out of three survivors exhibited c-Myc expression and, have been disease-free for more than ten years.
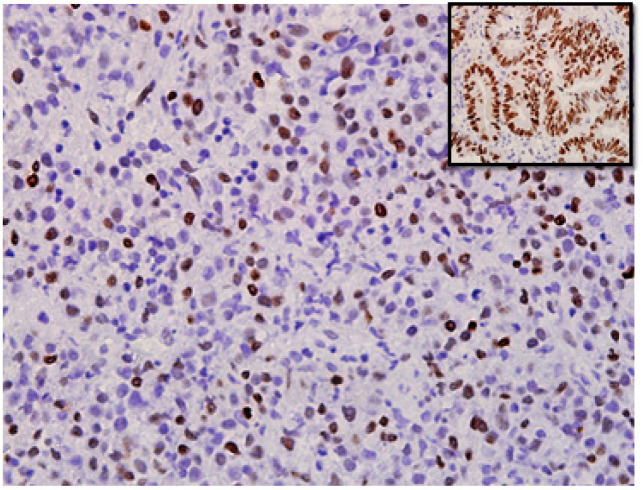
Figure 1.
Extra Nodal NK/T-Cell Lymphoma, Nasal Type Tissue Stained for c-MYC Immunohistochemistry. The c-MYC staining is mainly nuclear (photographed at x400 magnification). Inset picture shows positive control with nuclear staining using tissue from colorectal carcinoma
Table 2.
Association between Clinicopathological Factors with c-MYC Expression in T- and NK- NHL Patients (n=32).
| Variables | C-MYC | p valuea | |
|---|---|---|---|
| Negative n (%) | Positive n (%) | ||
| Age | |||
| Less than 60 | 7 (29.2) | 17 (70.8) | 0.15 |
| More than 60 | 0 (0.0) | 8 (100.0) | |
| Race | |||
| Malay | 6 (20.7) | 23 (79.3) | 0.541 |
| Chinese | 1 (50.0) | 1 (50.0) | |
| Indian | 0 (0.0) | 1 (100.0) | |
| Gender | |||
| Male | 4 (19.0) | 17 (81.0) | 0.667 |
| Female | 3 (27.3) | 8 (72.7) | |
| Site | |||
| Nodal | 2 (20.0) | 8 (80.0) | >0.950 |
| Extra nodal | 5 (22.7) | 17 (77.3) | |
| Extra nodal | |||
| Less than 1 | 5 (20.0) | 20 (80.0) | 0.653 |
| More than 1 | 2 (28.6) | 5 (71.4) | |
| Ann-Arbor staging | |||
| I-II | 1 (6.3) | 15 (93.8) | 0.083 |
| III-IV | 6 (37.5) | 10 (62.5) | |
| LDH value | |||
| Normal | 4 (28.6) | 10 (71.4) | 0.669 |
| Elevated | 3 (16.7) | 15 (83.3) | |
| ECOG | |||
| Less than 2 | 4 (26.7) | 11 (73.3) | 0.678 |
| More than 2 | 3 (17.6) | 14 (82.4) | |
| B symptoms | |||
| Yes | 2 (16.7) | 10 (83.3) | 0.683 |
| No | 5 (25.0) | 15 (75.0) | |
| IPI score | |||
| 0 to 2 | 4 (25.0) | 14 (77.8) | >0.950 |
| 3 to 4 | 3 (18.8) | 11 (78.6) | |
| T/NK-cell NHL subtypes | |||
| ALCL, ALK not done | 1 (100.0) | 0 (0.0) | 0.43 |
| ALCL, ALK-negative | 0 (0.0) | 1 (100) | |
| ALCL, ALK-positive | 2 (33.3) | 4 (66.7) | |
| AITL | 0 (0.0) | 1 (100.0) | |
| ENK/T-cell, nasal type | 0 (0.0) | 3 (100.0) | |
| PTCL, NOS | 2 (33.3) | 4 (66.7) | |
| TLL | 1 (16.7) | 5 (83.3) | |
| T-cell lymphoma | 1 (12.5) | 7 (87.5) | |
| Mortality | |||
| Alive | 1 (33.3) | 2 (66.7) | 0.85 |
| Dead | 3 (27.3) | 8 (72.7) | |
| Unknown | 3 (16.7) | 15 (83.3) | |
P values were calculated by Fisher’s exact test (two-sided)a and were considered to be statistically significant if p < 0.05
Discussion
Mature TNKcNHLs comprise approximately 10-15% of all NHLs (De Leval and Jaffe, 2020). In contrast to B cell NHL, most TNKcNHLs are lack defining genetic alterations, and their classification relies on a combination of morphological, immunophenotyping and genetic characteristics (Quintanilla-Martinez, 2017). Within 17 years, only 55 (8.6%) TNKcNHLs cases from 634 lymphoma were diagnosed in our centre. This is similar to the low incidence of TNKcNHLs during the ten year period recorded in the United States (US) Surveillance, Epidemiology and End Result (SEER) stating only 6% incidence, compared to B-NHLs (Ward et al., 2019). Our study exhibited uneven ethnic distributions as reflected by the local population of Kelantan (Department of Statistic Malaysia, 2015), which showed predominantly Malay (95.7%), Chinese (3.4%) and Indian (0.3%) populations. However, the epidemiological data in this study did not portray the true TNKcNHLs incidence in Kelantan population, because lymphoma cases are also diagnose and manage in another hospital in this state.
Lymphoid neoplasm incidence increases with age, and is found higher in males than females (Wang and Vose, 2013). The median age is between fifth to seventh decades, but it may vary according to the subtypes (Wang and Vose, 2013). In our retrospective study, the median age of patients with TNKcNHLs at diagnosis was 38 years (8 to 81 years) which is almost two decades younger than the previous reports from Western countries (Vose et al., 2008). This finding is comparable with three studies from China identified that the median age were 49 (3 to 82 years) (Lee et al., 2008), 44 years (8 to 77 years) (Huang et al., 2014) and 48 years (10 to 73 years) ( Wang et al., 2017). The present study showed male to female ratio of 1.9:1, and male predilection was reported in previous studies (Lee et al., 2008; Xu et al., 2012; Huang et al., 2014).
MYC protein regulates about 15% of human genes involved in the control of cell cycle, metabolism, protein biosynthesis, chromatin remodelling, and stem cell self-renewal (Chisholm et al., 2015). An elevated or deregulated expression of c-MYC have been detected in many human cancers and correlated with aggressive, poorly differentiated tumours (Pelengaris et al., 2002). Many of previous c-Myc studies were done using molecular tests such as FISH. However, Mohammed et al., (2019) showed that, c-Myc and BCL2 IHCs protein over expressions were comparable with RT-PCR c-Myc and BCL2 genes arrangements in understanding the DLBCL prognostication. Another study on c-Myc protein expression in bladder carcinoma showed it could predict the clinical behaviour of the disease (Elwy et al., 2019). IHC of c-Myc are now emerging as a favourable alternative as it is a cheaper and easier technique which can be used to study the effects of this protein in disease behaviour.
The role of c-Myc in TNKcNHLs has not been discussed as intensively as in B NHL, therefore, little is known about its importance in association with the prognostic value and survival index (Huang et al., 2014; Chisholm et al., 2015). In the present study, we conducted c-Myc IHC test on FFPE tissue and examine its expression in association with patients’ clinicopathological parameters of TNKcNHLs cases diagnosed in our centre. Our result showed 25 (78.1%) cases expressed c-Myc protein, with expression of each subtypes as shown in Table 2. A study by Chisholm et al., (2015) reported c-Myc protein was expressed in 94% of TNKcNHLs cases, with predominantly in TLBL and ENKTL-NT. Also, in a report by Huang et al., (2014) demonstrated c-Myc expression in 64.2% of 53 ENKTL cases. Similar findings were also observed in TLBL and ENKTL-NT cases in our centre. Huang et al,. (2014) and another two studies by Chen et al., (2017) and Wang et al., (2017) reported that overexpression of c-Myc protein in ENKTL cases are related to unfavourable clinical outcomes. However, in our study, we could not proceed with survival analysis as complete patients’ data were not available.
ALK-positive ALCL is usually associated with a favourable prognosis, but in published case reports (Moritake et al., 2011; Liang et al., 2013), described unfavourable clinical outcomes in those with concurrent ALK and MYC translocations. Liang et al., (2013) summarized three reported cases of confirmed ALK and MYC translocations, with, one of the patients was in complete remission for nearly two years after subjected to aggressive chemotherapy, radiotherapy and hematopoietic stem cell transplantation (HSCT). The other two cases however, only received aggressive chemotherapy, resulting in death within six months from the diagnosis. In present study, there were 4/6 (66.7%) cases of ALK-positive ALCL that expressed c-Myc, with one case displayed aggressive clinical course that the patient died a few months after diagnosis. Haemato-oncologists need to consider an intensified therapy if c-Myc expression is identified in such patient. Manso et al., (2016) reported 15/74 (20.3%) AITL and 13/54 (24.1%) PTCL,NOS cases were positive for c-Myc IHC, with c-MYC protein was significantly (p<0.05) expressed in the AITL. We observed that 4/5 (80%) PTCL,NOS and 1/1 (100%) AITL cases expressed c-Myc in our study. The proportion of cases with c-Myc expression in our study varied according to TNKcNHLs subtypes, however, no association was significant (p>0.05).
In this study, there is no significant association between c-Myc protein expression and the IPI (patient’s age, gender, Ann Arbor staging, LDH levels, B symptoms, extra-nodal involvement, and ECOG). In contrast to a report by Huang et al., (2014), 53 ENKTL patients, the age and clinical stage were significantly associated with c-Myc expression. Chen et al., (2017) and Wang et al., (2017) also reported no significant association between c-Myc expression and clinicopathological features, although they were able to demonstrate low three year OS rate and poor outcome of survival analysis in their c-Myc expressed TNKcNHLs.
We concluded, there is a role of c-Myc in TNKcNHLs that can predict the disease behaviour, as observed by the protein expression by the tumour cells. However, we were not able to demonstrate its significant association with clinicopathological parameters or histological subtypes, due to the small sample size. Future investigations by recruiting more samples and including molecular testing in the study c-Myc in TNKcNHLs should be explored.
Author Contribution Statement
None.
Acknowledgements
The authors declare there is no conflict of interest regarding publishing the article. This article is a part of an approved dissertation of Master of Pathology. This research was approved by the Human Ethical Committee of the School of Medical Sciences, Universiti Sains Malaysia (USM/JEPeM/18010081). We thank Mrs Ummi Atikah Ayub for the technical assistance. Authors contributions: NH conducted the experiment, microscopic analysis, analysed data and draft write up; SAMI editing draft; FAH microscopic analysis and finalising draft editing. This research was supported by Universiti Sains Malaysia -Research University Individual (RUI) Grant (1001/PPSP/8012247) and University Bridging Grant (304/PPSP/6316134).
References
- Armitage JO. The aggressive peripheral T-cell lymphomas: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87:511–9. doi: 10.1002/ajh.23144. [DOI] [PubMed] [Google Scholar]
- Au WY, Ma SY, Chim CS, et al. Clinicopathologic features and treatment outcome of mature T-cell and natural killer-cell lymphomas diagnosed according to the World Health Organization classification scheme: a single center experience of 10 years. Ann Oncol. 2005;16:206–14. doi: 10.1093/annonc/mdi037. [DOI] [PubMed] [Google Scholar]
- Azizah AM, Nor Saleha IT, Noor Hashimah A, Asmah ZA, Mastulu W. Malaysian National Cancer Registry Report 2007-2011, In ‘Lymphoma (ICD10: C81-C85; C96) National Cancer Institute. 2016: 42–3. [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YP, Chen BZ, Zhu WF, et al. The clinical significance of c-MYC expression, rearrangement, and copy number gain in extranodal NK/T-cell lymphoma: A retrospective study in China. Exp Mol Pathol. 2017;103:38–43. [Google Scholar]
- Chisholm KM, Bangs CD, Bacchi CE. Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am J Surg Pathol. 2015;39:294–303. doi: 10.1097/PAS.0000000000000365. [DOI] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8:177–8. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leval, L Jaffe, E.S Lymphoma classification. Cancer J. 2020;26:176–85. doi: 10.1097/PPO.0000000000000451. [DOI] [PubMed] [Google Scholar]
- El-Hussien M, Mokhtar N, Khorshed E. C-MYC protein expression and high ki-67 proliferative index are predictives of disease relapse in diffuse large b cell lymphoma. Asian Pac J of Cancer Biol. 2021;6:15–20. [Google Scholar]
- Elwy A, Elsaba T, Abd Elzaher A, Nassar M. Prognostic value of c-myc immunohistochemical expression in muscle invasive urothelial carcinoma of the urinary bladder: a retrospective study. Asian Pac J Cancer Prev. 2019;20:3735–46. doi: 10.31557/APJCP.2019.20.12.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–7. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- Huang X, Sun Q, Fu H, et al. Both c-Myc and Ki-67 expression are predictive markers in patients with extranodal NK/T-cell lymphoma, nasal type: A retrospective study in China. Pathol Res Pract. 2014;210:351–6. doi: 10.1016/j.prp.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Huppi K, Pitt JJ, Wahlberg BM, et al. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet. 2012;3:69. doi: 10.3389/fgene.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Uhm JE, Lee HY, et al. Clinical features and prognostic factors of patients with “peripheral T cell lymphoma, unspecified”. Ann of Hematol. 2008;88:111–9. doi: 10.1007/s00277-008-0544-2. [DOI] [PubMed] [Google Scholar]
- Liang X, Branchford B, Greffe B, et al. Dual ALK and MYC rearrangements leading to an aggressive variant of anaplastic large cell lymphoma. J Pediatr Hematol Oncol. 2013;35:e209–13. doi: 10.1097/MPH.0b013e3182815046. [DOI] [PubMed] [Google Scholar]
- Lynnhtun K, Renthawa J, Varikatt W. Detection of MYC rearrangement in high grade B cell lymphomas: Correlation of MYC immunohistochemistry and FISH analysis. Pathology. 2014;46:211–5. doi: 10.1097/PAT.0000000000000076. [DOI] [PubMed] [Google Scholar]
- Manso R, Bellas C, Martín-Acosta P, et al. C-MYC is related to GATA3 expression and associated with poor prognosis in nodal peripheral T-cell lymphomas. Haematologica. 2016;101:e336–8. doi: 10.3324/haematol.2016.143768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed A, Rashed H, Abdelrahman A, et al. C-MYC and BCL2: Correlation between protein over-expression and gene translocation and impact on outcome in diffuse large b cell lymphoma. Asian Pac J Cancer Prev. 2019;20:1463–70. doi: 10.31557/APJCP.2019.20.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux EM, Rochford R, Griffin B, et al. Burkitt’s lymphoma. Lancet. 2012;379:1234–44. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- Moritake H, Shimonodan H, Marutsuka K, et al. C-MYC rearrangement may induce an aggressive phenotype in anaplastic lymphoma kinase positive anaplastic large cell lymphoma: Identification of a novel fusion gene ALO17/C-MYC. Am J Hematol. 2011;86:75–8. doi: 10.1002/ajh.21887. [DOI] [PubMed] [Google Scholar]
- Nwanze J, Siddiqui MT, Stevens KA, Saxe D, Cohen C. MYC immunohistochemistry predicts MYC rearrangements by FISH. Front Oncol. 2017;7:209. doi: 10.3389/fonc.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122:3884–91. doi: 10.1182/blood-2013-05-498329. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Quintanilla-Martinez L. The 2016 updated WHO classification of lymphoid neoplasias. Hematol Oncol. 2017;35:37–45. doi: 10.1002/hon.2399. [DOI] [PubMed] [Google Scholar]
- Ramadas AA, Jose R, Varma B, Chandy ML. Cervical lymphadenopathy: Unwinding the hidden truth. Dent Res J. 2017;14:73–8. doi: 10.4103/1735-3327.201136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou A, Bennani NN, Feldman AL. 2019) Update on the classification of T-cell lymphomas, Hodgkin lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev Hematol. 12:833–43. doi: 10.1080/17474086.2019.1647777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose J, Armitage J, Weisenburger D. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–30. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- Wang JH, Bi XW, Li PF, et al. Overexpression of MYC and BCL2 predicts poor prognosis in patients with extranodal NK/T-cell lymphoma, nasal type. J Cancer. 2017;8:793–800. doi: 10.7150/jca.17766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EM, Sherman RL, Henley SJ, et al. Annual report to the nation on the status of cancer, featuring cancer in men and women age 20–49 years. J Natl Cancer Inst. 2019;12:1279–97. doi: 10.1093/jnci/djz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PP, Wang Y, Shen Y, et al. Prognostic factors of Chinese patients with T/NK-cell lymphoma: a single institution study of 170 patients. Med Oncol. 2012;29:2176–82. doi: 10.1007/s12032-011-0011-0. [DOI] [PubMed] [Google Scholar]
- Yin X, Xu A, Fan F, et al. Incidence and mortality trends and risk prediction nomogram for extranodal diffuse large B-cell lymphoma: An Analysis of the Surveillance, Epidemiology, and End Results Database. Front Oncol. 2019;9:1198. doi: 10.3389/fonc.2019.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zain JM. Aggressive T-cell lymphomas: 2019 updates on diagnosis, risk stratification, and management. Am J Hematol. 2019;94:929–46. doi: 10.1002/ajh.25513. [DOI] [PubMed] [Google Scholar]