Abstract
Objective:
Infection with human tumor viruses is one of the hypothesized causes of cancer. The current investigation aimed to explore the presence and quantitative analysis of a new human tumor virus, Merkel cell polyomavirus (MCPyV) in tissue samples of 114 patients with oral cavity lesions including oral squamous cell carcinoma (OSCC), oral lichen planus (OLP), Dysplasia and oral irritation fibroma (OIF) in Northern Iran.
Methods:
From 114 formalin fixed paraffin embedded samples; 35 with SCC, 29 with OLP, 14 with dysplasia and 36 with OIF were cut, deparaffinized and DNA was extracted. Quantitative detection of MCPyV large T antigen was performed by absolute quantitative Real-Time PCR. Result: MCPyV DNA was detected in 30.6% (n: 11/36) of IF, 24.1% (n; 7/29) of OLP, 21.4% (n:3/14) of dysplasia and 20% (n;7/35) of OSCC samples. The mean MCPyV DNA copy number was 2.32×10−2 ± 3.97 ×10−2, 2.02×10−2 (SD=3.13×10−2), 2.69×10−4 (SD=2.51×10−4), and 2.56×10−4 (SD=6.73×10−4) per cell in OSCC, dysplasia and both of OLP and OIF samples, respectively (P=0.76).
Conclusion:
This study provides the first data from Iran regarding the presence of MCPyV genome in oral cavity lesions and oral cancer. These results also emphasize that MCPyV has an active role in the occurrence of oral lesions and progression to cancer. Further studies should be carried out to clarify the role of MCPyV in oral cavity lesions.
Key Words: Merkel Cell Polyomavirus (MCPyV), Oral Squamous Cell Carcinoma (OSCC), Oral Lichen Planus (OLP)
Introduction
Oral cancer (OC) is a most common malignant tumor in the head and neck tissues and sixth most common cancer with the high mortality at the world (Warnakulasuriya et al., 2009). Oral squamous cell carcinoma (OSCC) is a epigenetic and genetic illness that is the most prevalent type of OC. Mouth and tongue is the most common site for development of OSCC (Bagan et al., 2010; Bello et al., 2010).
Oral oncogenesis is a complex pathway that involves several components such as smoking, alcohol, nutrition and genetic risk factors (Dhull et al., 2018). In addition, many other possible risk factors including infection with certain oncogenic viruses like human papilloma virus (HPV) and Epstein–Barr virus (EBV) have also been suggested (Pezzuto et al., 2015). However, the role of HPV and EBV in the etiology of OC remains controversial. Co-infection with the other tumor viruses has been suggested as a cofactor for HPV and EBV-related carcinogenesis (Moens et al., 2014; Zheng et al., 2010). Merkel cell Polyomavirus (MCPyV) is a new human oncogenic virus that is associated with the Merkel cell carcinoma (MCC), a rare but rapid metastasizing skin neoplasm (Feng et al., 2008). Although, MCC is a rare cancer, but infection caused by this virus is widespread and its can be detected on the skin of ~80% of healthy people who acquired the initial infection early in life (Coursaget et al., 2013). Merkel cells are oval-shaped epithelial neuroendocrine cells with mechanoreceptors properties for light touch sensation and found in the skin of vertebrates. They are found in the basal layers of epidermis, oral mucosa and esophageal epithelium (Harmse et al., 1999). Although, MCPyV transmission is not yet known, but high levels of the viral genome was found in the oral cavity and saliva (Loyo et al., 2010). Previous studies have shown that due to the persistent presence of the MCPyV in oral cells, it can accelerate the carcinogenic process in them. Therefore, based on the previous findings, we designed a study for the first time in our region to determine the presence of MCPyV load in oral cancer cells such as squamous cell carcinoma (OSCC), oral lichen planus (OLP), Dysplasia and oral irritation fibroma (OIF).
Materials and Methods
Oral cancer biopsies
In a during of 2015 to 2019, a total of 114 formalin fixed and paraffin-embedded (FFPE) oral biopsies were collected from the patients referred to the oral pathology department at the school of Dentistry in Babol University of Medical Sciences. These samples included 35 OSCC, 29 OLP, 36 OIF and 14 Dysplasia, respectively. These samples were obtained from 44 women and 70 men with an age range of 44 to 70 years. Histopathological diagnosis was confirmed by oral pathologists and FFPE samples classified in OSCC, OLP, Dysplasia and OIF groups.
This study was approved by the local ethics committee with the ethical number: IR.MUBABOL.HRI.REC.1399.035 in Babol University of Medical Sciences, Babol, Iran.
DNA extraction
Deparaffinization of FFPE samples was performed according to a previously described procedure (Yahyapour et al., 2012). Genomic DNA was extracted from FFPE samples using the DNA Extraction Mini Kit from Tissue (Yekta TajhizAzma, Tehran, Iran) according to the manufacturer’s instructions. The quality and quantity of purified DNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Quantitative Real-Time PCR
A QReal-time PCR was performed to detect and measure the amount of MCPyV large T-antigen (LTag) DNA load using a Rotor-Gene® Q (Qiagen GmbH, Hilden, Germany) real-time PCR system according to a previously described procedure (Sadeghi et al., 2015). A cellular gene (human RNase P) was used in order to normalize the number of viral copies per cells. Pairs of primer and specific TaqMan probe for the human RNase P gene and MCPyV LTag gene are listed in Table 1.
Table 1.
Primer and Specific Probes Used in This Study
| Target Gene | Sequences (5'-3') | Primer and Probe |
|---|---|---|
| Human RNase P | 5'-AGATTTGGACCTGCGAGCG-3' | RNP F-Primer |
| 5'-GAGCGGCTGTCTCCACAAGT-3' | RNP R-Primer | |
| FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ1 | RNP Probe | |
| MCPyV LTag | 5'-CCAAACCAAAGAATAAAGCACTGA-3' | LTag F-Primer |
| 5'-TCGCCAGCATTGTAGTCTAAAAAC-3' | LTag R-Primer | |
| HEX- AGCAAAAACACTCTCCCCACGTCAGACAG-BHQ1 | LTag Probe |
Statistical analysis
Statistical analysis was performed using SPSS version 22 software. The chi-square test and one-way analysis of variance (ANOVA) was used to compare differences between the study groups for categorical and continuous variables, respectively. Heterogeneity of variance was tested via Levene method, and in case of heterogeneous variation among the study groups, we performed robust test of equality of mean by Brown-Forsythe. An ordinal logistic regression model was used to estimate the incidence of cancer in MCPyV-related subjects. A P -value of less than 0.05 (<0.05) was considered statistically significant.
Results
Demographic and histopathological characteristics of all 114 enrolled subjects are shown in Table 2. In total, 24.6% (n; 28/114) were positive for the presence of MCPyV genome. MCPyV DNA was detected in the 20.4% (n; 7/49) of malignant and premalignant and 27.7% (n; 18/65) of non-malignant samples. There was no significant difference in the presence of MCPyV genome in the malignant and non-malignant groups (P=0.471). Regarding the site of tumor in malignant and pre-malignant group, MCPyV infection was detected in 25.0% (n; 6/24) of tongue.
Table 2.
MCPyV Detection in Censorious and Non-Censorious from Oral Biopsies Using qReal-Time PCR
| Oral Cavity Cases (n=114) Number (percentages) |
MCPyV Number (percentages) |
MCPyV DNA Load (Copies/Cell) |
MCPyV DNA Load (Copies/Cell) |
|||
|---|---|---|---|---|---|---|
| Variable | Positive | Negative | Range | Mean | P-Value | |
| Gender | ||||||
| Male | 44 (38.6) | 9 (20.5) | 35 (79.5) | 9.0×10-6-5.77×10-3 | 7.14×10-4 | 0.42 |
| Female | 70 (61.4) | 19 (27.1) | 51 (72.9) | 3.0×10-6-1.01×10-1 | 1.17×10-4 | |
| Age | ||||||
| < 65 yr | 86 | 19 (22.1) | 67 (77.9) | 9.0×10-6-5.64×10-2 | 6.35×10-3 | |
| ≥ 65 yr | 28 | 9 (32.1) | 19 (67.9) | 3.0×10-6-1.01×10-1 | 1.52×10-2 | 0.28 |
| Average (yr) | 52.65 (79/16±23/53) | - | - | - | - | |
| Range (yr) | 12-87 | - | - | - | - | |
| Histopathology | ||||||
| OSCC | 35 (30.7) | 7 (20.0) | 28 (80.0) | 3.0×10-6-1.01×10-1 | 2.32×10-2 | |
| OLP | 29 (25.4) | 7 (24.1) | 22 (75.9) | 6.9×10-5-7.86×10-4 | 2.7×10-4 | 0.76 |
| Dysplasia | 14 (12.3) | 3 (21.4) | 11 (78.6) | 8.61×10-4-5.64×10-2 | 2.02×10-2 | |
| OIF | 36 (31.6) | 11 (30.6) | 25 (75.9) | 9.0×10-6-2.28×10-3 | 2.57×10-4 | |
| Site of lesion | ||||||
| Buccal Mucosa | 47 (41.2) | 7 (14.9) | 40 (85.1) | 2.2×10-5-1.01×10-3 | 2.71×10-4 | |
| Floor of Mouth | 5 (4.4) | 3 (60.0) | 2 (40.0) | 5.7×10-5-1.01×10-1 | 3.39×10-2 | |
| Gingiva & alveolar mucosa | 20 (17.5) | 3 (15.0) | 17 (85.0) | 8.1×10-5-5.41×10-2 | 1.83×10-2 | 0.013 |
| Labial mucosa | 10 (8.8) | 6 (60.0) | 4 (40.0) | 1.3×10-5-2.28×10-3 | 4.51×10-4 | |
| Tongue | 24 (21.1) | 8 (33.3) | 16 (66.7) | 3.0×10-6-8.3110-3 | 8.31×10-3 | |
| Unknown | 8 (7.0) | 1 (12.5) | 7 (87.5) | 9.0×10-6 | 9.0×10-6 | |
| Type of Samples | ||||||
| Malignant | 35 | 7 (20) | 28 (80) | 3.0×10-6-1.01×10-1 | 2.32×10-2 | 0.66 |
| Pre-malignant | 14 | 3 (21.4) | 11 (78.6) | 8.61×10-4-5.64×10-2 | 2.02×10-2 | |
| Non-malignant | 65 | 18 (27.7) | 47 (72.3) | 9.0×10-7-2.28×10-3 | 7.51×10-4 | |
| Total | 114 | 28 (24.6) | 86 (75.4) | 3.0×10-6-1.01×10-1 | 8.14×10-3 | - |
In non- malignant group, 24.1% (n; 7/29) of OLP, and 30.6% (n; 11/36) of OIF samples were positive for MCPyV DNA genome. In tumoral group, 42.9% (n; 3/7) of tongue biopsy specimens carried the MCPyV DNA. In non- tumoral oral lesions, 28.6% (n; 6/21) of buccal mucosa, 23.8% (5/21) for both lip and tongue were positive for MCPyV DNA.
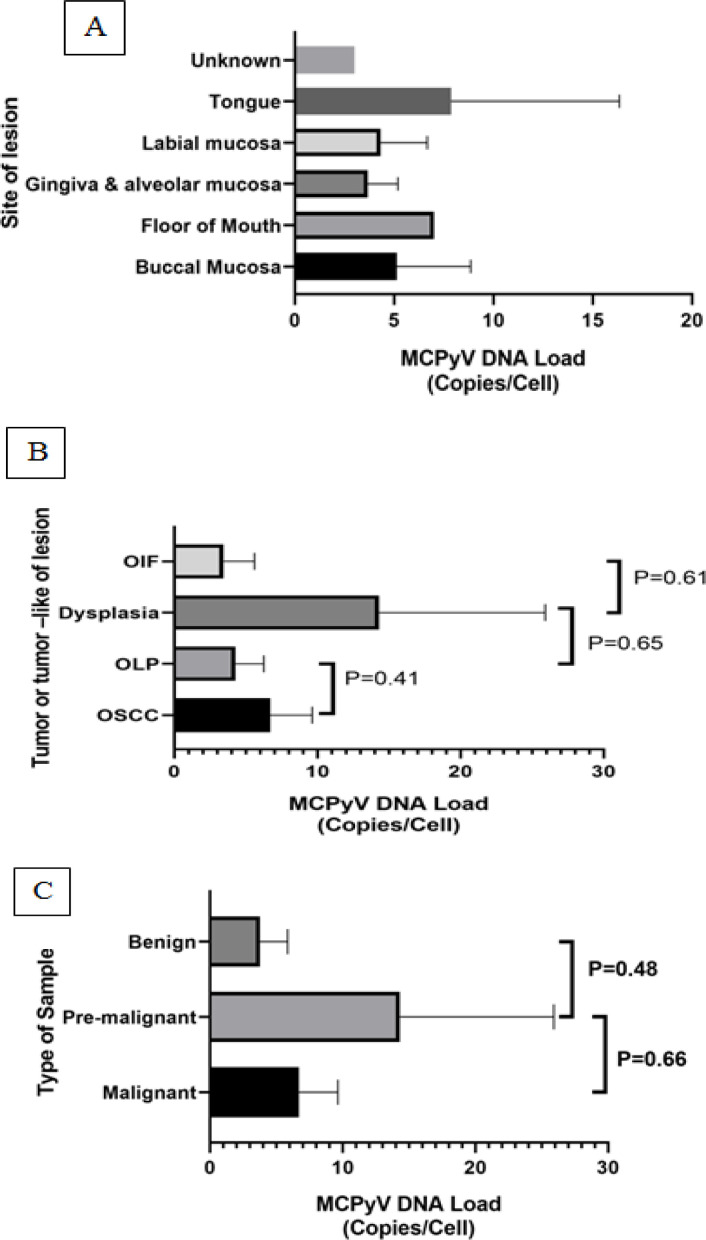
The mean MCPyV DNA load according to histopathologic diagnosis in patients with tumoral and non-tumoral oral lesions shown in Figure 1.
Figure 1.
The Mean MCPyV DNA Load According to Histopathologic Diagnosis in Patients with Neoplastic and Non-Neoplastic Oral Lesions. The p-value was determined by ANOVA. [A. by site of lesion; B. by type of lesion (OSCC, OLP, Dysplasia and OIF); C. by type of samples (Benign, pre-malignant and malignant)]
The mean MCPyV DNA copy number was 2.32×10−2 ±3.97×10−2, 2.02×10−2 ±3.13×10−2, 2.69×10−4 ±2.51×10−4, and 2.56×10−4 ±6.73×10−4 per cell in OSCC cases, dysplasia and both of OLP and OIF samples, respectively (P=0.28). The mean of cancerous and non-cancerous groups was 2.19×10-3 and 7.5×10-4, respectively. There was statistically significant difference between these two studied groups (P=0.01). Furthermore, mean of MCPyV viral load was 2.23×10−2 and 0.02×10−4 in the cancerous and non-cancerous samples, respectively, which was not significant (P=0.082).
Discussion
MCPyV, is the most recently discovered human cancer virus has been known as a etiological factor in the majority of MCC, a rare and very aggressive skin cancer (Feng et al., 2008). In spite of the fact that MCPyV transmission route has not been defined, the fragments of MCPyV genome have been shown in the head and neck malignant tissue including oral cancer (Loyo et al., 2010; Wieland et al., 2009l; Kantola et al., 2009). If the head and neck tissue is exposed constantly with this oncogenic virus, it is possible that malignancy may develop in the region.
To investigate whether this oncogenic virus could have a role in the etiology of OSCC a cross-sectional study was designed in northern Iranian city of Babol and a total of 114 oral specimens including 35 OSCC and 79 non-neoplastic oral specimens were examined for MCPyV genome.
In the present study, both neoplastic and non-neoplastic oral specimens were found to harbor MCPyV DNA. However, MCPyV infection in non-neoplastic lesions (30.6%) was considerably higher than OSCC cases (20.0%). The higher rate of MCPyV infection in our non-neoplastic group might be explained by histopathological distribution of samples (82.3% of non-neoplastic samples were diagnosed with irritation fibroma and only 17.7% had dysplasia histology). MCPyV infection was frequently detected in oral samples by other investigators (Loyo et al., 2010; Tanio et al., 2015; Hamiter et al., 2017).
In the current study, out of the 114 examined specimens, the MCPyV DNA genome was detected in 28 (24.6%), and MCPyV positivity was not associated with cancerous or non-cancerous lesions (p= 0.66). Also similar to our results, in the study of Loyo et al (Loyo et al., 2010), Which evaluated a wide range of diverse human tissues for the presence of this virus, there was no significant relationship between virus levels and cancerous or non-cancerous tissues. Out of 47 SCC samples, 19 (40.4%) were MCPyV positive, which was higher than the positive rate observed in the SCC group in our study (7/35, 20%).
Few studies have examined the presence of the MCPyV virus genome in cancerous and noncancerous oral specimens. However, there is partial and incomplete information on the frequency of MCPyV infection in oral neoplasms and its association with cancer induction in the oral cavity which requires further and more detailed studies. In different studies, the prevalence of MCPyV virus genome in oral cancer samples varied from 7% to 28.6 (Tanio et al., 2015; Hamiter et al., 2017).
The existence of this discrepancy between different research groups may be due to the nature of the samples (paraffin blocks or fresh tumor samples), the efficiency of DNA extraction procedure, the sensitivity of diagnostic methods such as PCR tests and the presence of possible contamination in molecular tests, sample size and differences in virus prevalence in different racial populations and geographical areas. For example, in the Windon study (2020), immunohistochemistry (IHC) was used to detect the virus in oral cavity cancers, and in contrast to the present study, which was performed by PCR, young people were more likely to develop cancer (Windon et al., 2020). Also, in the present study, no association was found between MCPyV infection and sex, age, type and sites of lesions.
In our study, the prevalence of MCPyV in OSCC of the tongue was 12.5% (3/24). Tanio et al found the MCPyV prevalence rate of 3.33% (2/60) in tongue tissue samples (Tanio et al., 2015).
In a study directed by Hamiter et al, the prevalence of MCPyV genome in tongue biopsies were 28.6% (Hamiter et al., 2017). Our study showed a much higher MCPyV positivity rate of 33.3% (8/24). The prevalence of MCPyV in our study may have been even higher if we had access to additional fresh frozen tissue, as was the case in Loyo’s study (Loyo et al., 2010). Also, Tanio et al reported the prevalence of MCPyV in 7.7% (4/52), 5.3% (1/19), 4% (7/176), 1% (1/10) and 0.0% (0/11) and 0.0% (0/4) in gingiva, floor of mouth, buccal mucosa and lip; respectively (Tanio et al., 2015). In the present study, the prevalence of MCPyV were found in 33.3% (8/24), 15% (3/20), 60% (3/5), 14.9% (7/47) and 60% (6/10) in tongue, gingiva, floor of mouth, buccal mucosa and lip; respectively and there was statistically significant difference between the sites of lesion (P=0.013). We detected 24.1% (n; 7/29) MCPyV genome in the OLP samples. These data are contrast with Regnault et al (Masson Regnault et al., 2017). As shown in Fig.1, the difference between MCPyV DNA loads (as a copy per cell) in tumor and non-tumor groups was not statistically significant. Quantitative detection of MCPyV and normalization of viral copy numbers to cell numbers could be informative and valuable marker when evaluating viral carcinogenic process and tumorigenic mechanism. Relative quantitation studies showed that MCC samples are generally positive for the virus at higher than 1 copy per tumor cell ((Loyo et al., 2010; Harmse et al., 1999). However the viral copy numbers in a large set of non-MCC malignancies were 2-4 log lower than that seen in MCC (Imajoh et al., 2012; Mohebbi et al., 2018; Behdarvand et al., 2017; Yahyapour., 2016; Sadeghi et al., 2015; Salehi-Vaziri., 2015). Despite widespread presence of MCPyV in diverse human malignant and non- malignant tissue samples, it appears that only high level of virus supporting a direct tumorigenic mechanism and tumor viral clonality. Low copy numbers of MCPyV genome in both tumor and non-tumor oral tissues might be explained by simple persistent viral replication in oral as a passenger virus or viral shedding from another organ (e.g., respiratory tract) to mouth without any pathological outcome. The low prevalence of MCPyV has been demonstrated in some natural human tissue such as liver biopsy or respiratory secretions, and Two studies by Loyo et al and Baez et al, with detecting higher levels of the virus in salivary specimens than other oral tissues, confirm that the virus originated in the underlying tissues and was not correlated with oral SCC (Loyo et al., 2010). Usually, if MCPyV stimulated tumor in humans arose as a monoclonal development of a viral transformed cell (e.g., MCC tumors); more than one DNA copy per tumor cell would be present (Feng et al., 2008). In comparison with previous publications in other region, present study was conducted in a more diversity of neoplastic and non-neoplastic oral samples and to our knowledge, this is the first study in the Caspian Sea region to examine the role of this oncogenic virus (MCPyV) in OSCC using highly sensitive real-time PCR method. The findings of the present study should be interpreted cautiously, due to some limitations including, the small percentage of samples with normal histology as a control and lack of fresh biopsy samples for analysis. Evaluation of fresh biopsy specimens using quantitative real-time PCR may help clarify our viewpoint to the role of aforesaid oncogenic viruses in the development of oral cancer.
In conclusion, our finding showed that MCPyV can be detected in both malignant and non-malignant oral tissues and the lack of higher viral load in malignant tissues than other ones weakened the hypothesis of the pathogenic role of these carcinogenic viruses in the development of oral malignancy yet, to determine our results, more world-wide epidemiological studies with larger sample size and if possible, on fresh biopsy samples should be done.
Author Contribution Statement
Study concept and design: Yousef Yahyapour, and Farzin Sadeghi; Analysis and interpretation of data: Mohammad Chehrazi, Farzin Sadeghi, Yousef Yahyapour and Maryam Seyed Majidi; Drafting of the manuscript: Mina Hasani Estalkhi, Ali Hasanzadeh and Yousef Yahyapour; Critical revision of the manuscript for important intellectual content: Mina Hasani Estalkhi , Maryam Seyed Majidi, Farzin Sadeghi, Ali Hasanzadeh, Mohammad Chehrazi and Yousef Yahyapour; Statistical analysis: Mohammad Chehrazi.
Funding statement
This study was financially supported by a grant from Babol University of Medical Sciences.
This project was approved by 4 scientific committee
A. scientific committee in Faculty of Dental Medicine for student thesis (Mina Hasani Estalkhi); B. scientific committee in Infectious Diseases & Tropical Medicine Research; C. and scientific committee in Oral Health Research Center and D. Research Committee in Vice chancellor of Babol University of Medical Sciences (Project code: 9809350).
Availability of data (if apply to your research)
All data is reserved by the co-responding author and will be provided to applicants if required.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncology. 2010;246:414–7. doi: 10.1016/j.oraloncology.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Behdarvand A, Zamani M, Sadeghi F, et al. Evaluation of Merkel cell polyomavirus in non-small cell lung cancer and adjacent normal cells. Microbial Pathogenesis. 2017;108:21–6. doi: 10.1016/j.micpath.2017.04.033. [DOI] [PubMed] [Google Scholar]
- Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: means, markers and perspectives (II) Oral Oncol. 2010;46:636–43. doi: 10.1016/j.oraloncology.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Coursaget P, Samimi M, Nicol JT, Gardair C, Touzé A. Human Merkel cell polyomavirus: virological background and clinical implications. Apmis. 2013;121:755–69. doi: 10.1111/apm.12122. [DOI] [PubMed] [Google Scholar]
- Dhull AK, Atri R, Dhankhar R, Chauhan AK, Kaushal V. Major risk factors in head and neck Cancer: a retrospective analysis of 12-year experiences. World J Oncol. 2018;9:80. doi: 10.14740/wjon1104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiter M, Asarkar A, Rogers D, et al. A pilot study of Merkel cell polyomavirus in squamous cell carcinoma of the tongue. Oral Oncol. 2017;74:111–4. doi: 10.1016/j.oraloncology.2017.09.021. [DOI] [PubMed] [Google Scholar]
- Harmse JL, Carey FA, Baird AR, et al. Merkel cells in the human oesophagus. J Pathol. 1999;189:176–9. doi: 10.1002/(SICI)1096-9896(199910)189:2<176::AID-PATH416>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Imajoh M, Hashida Y, Nemoto Y, et al. Detection of Merkel cell polyomavirus in cervical squamous cell carcinomas and adenocarcinomas from Japanese patients. Virol J. 2012;9:1–9. doi: 10.1186/1743-422X-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantola K, Sadeghi M, Lahtinen A, et al. Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: implications for respiratory transmission and latency. J Clin Virol. 2009;45:292–5. doi: 10.1016/j.jcv.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyo M, Guerrero-Preston R, Brait M, et al. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. Int J Cancer. 2010;126:2991–6. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson Regnault M, Vigarios E, Projetti F, et al. No detection of Merkel cell polyomavirus in oral lichen planus: Results of a preliminary study in a French cohort of patients. J Med Virol. 2017;89:2055–7. doi: 10.1002/jmv.24887. [DOI] [PubMed] [Google Scholar]
- Moens U, Van Ghelue M, Ehlers B. Are human polyomaviruses co-factors for cancers induced by other oncoviruses? Rev Med Virol. 2014;24:343–60. doi: 10.1002/rmv.1798. [DOI] [PubMed] [Google Scholar]
- Mohebbi E, Noormohamadi Z, Sadeghi-Rad H, et al. Low viral load of Merkel cell polyomavirus in Iranian patients with head and neck squamous cell carcinoma: Is it clinically important? J Med Virol. 2018;90:344–50. doi: 10.1002/jmv.24953. [DOI] [PubMed] [Google Scholar]
- Pezzuto F, Buonaguro L, Caponigro F, et al. Update on head and neck cancer: Current Knowledge on Epidemiology, Risk Factors, Molecular Features and Novel Therapies. Oncology. 2015;89:125–36. doi: 10.1159/000381717. [DOI] [PubMed] [Google Scholar]
- Sadeghi F, Salehi-Vaziri M, Alizadeh A, et al. Detection of Merkel cell polyomavirus large T-antigen sequences in human central nervous system tumors. J Med Virol. 2015;87:1241–7. doi: 10.1002/jmv.24178. [DOI] [PubMed] [Google Scholar]
- Salehi-Vaziri M, Sadeghi F, Alamsi-Hashiani A, et al. Merkel cell polyomavirus and human papillomavirus infections in cervical disease in Iranian women. Arch Virol. 2015;160:1181–7. doi: 10.1007/s00705-015-2368-4. [DOI] [PubMed] [Google Scholar]
- Tanio S, Matsushita M, Kuwamoto S. Low prevalence of Merkel cell polyomavirus with low viral loads in oral and maxillofacial tumours or tumour-like lesions from immunocompetent patients: Absence of Merkel cell polyomavirus-associated neoplasms. Mol Clin Oncol. 2015;3:1301–6. doi: 10.3892/mco.2015.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Wieland U, Mauch C, Kreuter A. Merkel cell polyomavirus DNA in persons without merkel cell carcinoma. Emerg Infect Dis. 2009;15:1496. doi: 10.3201/eid1509.081575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windon M, Fakhry C, Rooper L, et al. The role of age and merkel cell polyomavirus in oral cavity cancers. Otolaryngol Head Neck Surg. 2020;163:1194–7. doi: 10.1177/0194599820932553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyapour Y, Sadeghi F, Alizadeh A, et al. Detection of merkel cell polyomavirus and human papillomavirus in esophageal squamous cell carcinomas and non-cancerous esophageal samples in northern Iran. Pathol Oncol Res. 2016;22:667–72. doi: 10.1007/s12253-016-0048-7. [DOI] [PubMed] [Google Scholar]
- Yahyapour Y, Shamsi-Shahrabadi M, Mahmoudi M, et al. Evaluation of human papilloma virus infection in patients with esophageal squamous cell carcinoma from the Caspian Sea area, north of Iran. Asian Pac J Cancer Prev. 2012;13:1261–6. doi: 10.7314/apjcp.2012.13.4.1261. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Xia P, Zheng HC, et al. The screening of viral risk factors in tongue and pharyngolaryngeal squamous carcinoma. Anticancer Res. 2010;30:1233–8. [PubMed] [Google Scholar]