Abstract
Objectives:
Paeoniflorin (PF), a compound found in Paeonia lactiflora and Paeonia suffruticosa, has anticancer potential, particularly in inhibiting migration and invasion, the resistant cancer cells hallmarks. To date, the mechanism of overcoming tamoxifen resistance in breast cancer is not yet elucidated. This research aims to explore the potential target of PF as a co-treatment for circumventing breast cancer resistance to tamoxifen with a genomic understanding-bioinformatics.
Methods:
Microarray data originating from GSE67916 and GSE85871 in the NCBI GEO database was analyzed to obtain differentially expressed genes (DEGs). Further analyses were performed on DEGs using the DAVID v6.8, STRING-DB v11.0, the Cytoscape, and cBioportal. Gene expression analysis validation in breast cancer cells and tamoxifen-resistant breast cancer cells was accomplished using GEPIA and ONCOMINE databases. Survival rate analysis of selected genes was conducted using Kaplan–Meier.
Results:
We obtained 175 DEGs from the two samples (tamoxifen-resistant and paeoniflorin-treated). DEG involves in 70 biological processes, 26 cellular components, and 18 molecular functions, and three pathways relevant to breast cancer. The PPI network analysis and hub genes selection obtained 10 genes with the highest degree scores. Genetic changes for selected genes, including IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 were found from 0.5% to 7% of the case population per patient case. Additional analysis using cBioportal revealed FGFR signaling pathway through Ras is important for the PF mechanism in circumventing breast cancer resistance to tamoxifen. ONCOMINE and GEPIA analysis emphasized the importance of selected genes in the tamoxifen-resistance mechanism.
Conclusion:
PF has potential to be used as a co-treatment for circumventing breast cancer resistance to tamoxifen by targeting FGFR2 signaling, but further validation is needed.
Key Words: Breast cancer, bioinformatics, paeoniflorin, tamoxifen resistance
Introduction
Breast cancer is responsible for 2.1 million cancers, the fifth leading cause of cancer related death worldwide in 2018 (Dolatkhah et al., 2020). Approximately 80% of breast cancers are estrogen receptor (ER)-positive type. Thus, endocrine therapy is considered a complement to surgery in most patients with breast cancer (Lumachi et al., 2015). Tamoxifen (Figure 1A) is a central component of ER-positive breast cancer treatment (Chang, 2012). As a selective ER modulator and a triphenylethylene derivative, it acts as an agonist in the uterine but an antagonist in the breast tissue (Hultch, 2018). However, more than half of advanced ER-positive breast cancers are intrinsically resistant to tamoxifen, and about 40% of them develop resistance during treatment (Hultch et al., 2018). Crosstalk between the ER and a set of growth factors receptor, including HER2, EGFR, FGFR, IGFIR, and impaired PI3K/PTEN and NFκB activation are among factors that responsible for tamoxifen resistance (Ali et al., 2016). Therefore, it is necessary to develop compounds that overcome the resistance to tamoxifen.
Figure 1.
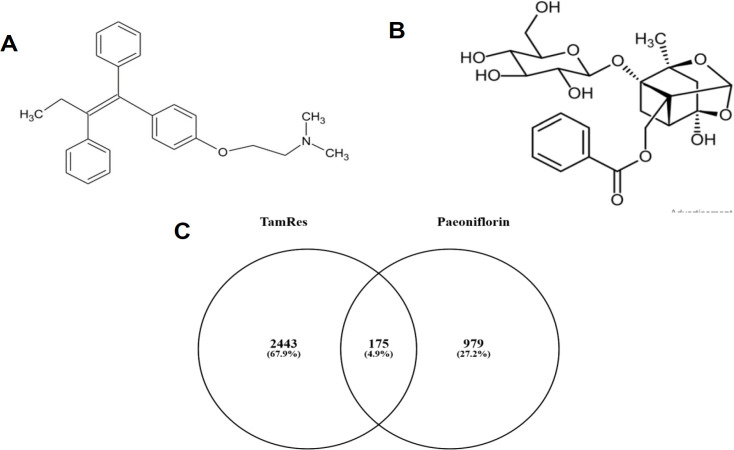
(A) Chemical Structure of Tamoxifen, (B) Chemical Structure of Paeoniflorin, (C) A Venn diagram of the DEGs between tamoxifen-resistant and Paeoniflorin (PF)-treated MCF-7 breast cancer cells
Paeoniflorin (PF, Figure 1B) is a promising adjunct therapy to curb tamoxifen resistance in breast cancer. PF is a pinan monoterpene glycoside with various bioactivities (Xiang et al., 2020). The anticancer activity of PF has been comprehensively studied in many types of cancer, including liver, gastric, breast, lung, pancreatic, colorectal, glioma, bladder, and leukemia (Xiang et al., 2020). The PF remarkably impeded MCF-7 breast cancer cell proliferation in a time- and concentration-dependent manner (Zhang et al., 2017). Besides, PF has been proven to prevent hypoxia-induced migration and invasion in MCF-7 cells (Zhou et al., 2016), two hallmarks of resistant cells (Fares et al., 2020). Therefore, PF has the potential to overcome tamoxifen resistance in breast cancer. Collectedly, it is necessary to conduct further studies on how PF works more broadly in circumventing breast cancer resistance to tamoxifen.
Bioinformatics analysis accelerates the drug targets identification and the drug candidates screening and refinement. This analysis also facilitates the side effects characterization and predicts drug resistance mechanisms (Xia, 2017). This study aims to explore the potential target of PF as a co-treatment for circumventing breast cancer resistance to tamoxifen with a genomic understanding-bioinformatics using several public databases, including GEO2R, DAVID v6.8, STRING-DB v11.0, Cytoscape 3.8.2, CytoHubba, and cBioPortal. The approach would facilitate and fasten the identifying potential target genes process and the PF mechanism in circumventing tamoxifen resistance in breast cancer.
Materials and Methods
Data mining
The microarray data were obtained from the NCBI Gene Expression Omnibus (GEO) database with ID GSE67916 for Tamoxifen-resistant ER+ breast cancer cells and GSE85871 for MCF7 cells treated with paeoniflorin (Lv et al., 2017). The gene expression in both gene series expressions were profiled using the microarray technology with Affymetrix Human Genome U133A 2.0 (Santa Clara, CA). The microarray data of the two GSE codes were further analyzed using the GEO2R to identify differentially expressed genes (DEGs) (Zhang et al., 2020). The adjusted p-value < 0.05 and Iog fold change > 1 were used to select the significant (Hermawan and Putri, 2020a). The two samples derived from the GSE code were then analyzed using Venny 2.1.0 to obtain a Venn diagram (Liu et al., 2019). From the Venn diagram, it is obtained that DEGs are shared by both samples.
Gene ontology and KEGG pathway enrichment analysis
The DEGs from the previous step was analyzed for gene ontology (GO) enrichment analysis consisting of biological processes, molecular functions, and cellular component; and KEGG pathway enrichment analysis using the database for annotation, visualization, and integrated discovery (DAVID) v6.8) (Wulandari et al., 2020). The value of p < 0.05 was selected as the significant value (Hermawan et al., 2021a).
Proten-protein interaction (PPI) network and hub gene selection
The PPI network analysis of the DEGs shared by the two GSE codes was executed using STRING-DB (Karimizadeh et al., 2019) with medium confidence (score = 0.400) and then visualized using Cytoscape software (Hermawan and Putri, 2020b). Comprehensive relationship analysis between nodes in the PPI was carried out using the Maximal Clique Centrality (MCC) function, the most effective method for finding hub genes in the PPI network (Li et al., 2020).
Analysis of genetic alterations of selected target genes
According to the previous study, the genetic alterations of selected target genes were analyzed using cBioPortal (Hermawan et al., 2021b). The breast cancer study with the highest genetic alterations was chosen for further connectivity analysis with the significant value at p < 0.05.
Gene expression analysis validation in non-resistant and tamoxifen-resistant breast cancer cells
Validation analysis of IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 was performed using the GEPIA database (http://gepia.cancer-pku.cn). The GEPIA database analysis was carried out to determine the difference in expression between cancer cells and normal cells based on TCGA and GTEx data (Tang et al., 2017). The default settings were used except for the BRCA with logFC > 1 and p-value < 0.01 dataset.
Validation of gene expression analysis was conducted according to Hermawan et al., (2020). Analysis was carried out on IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 genes in the ONCOMINE database (https://oncomine.org) (Rhodes et al., 2004). The settings used in the ONCOMINE database were cancer type analysis vs. cancer analysis, breast cancer, the clinical outcome of recurrence and metastatic status, and molecular subtype of the estrogen receptor. The analysis was performed to determine the expression level in the recurrence group and the nonrecurrence group after the tamoxifen treatment.
Survival rate analysis
Survival rate analysis of IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 was conducted using Kaplan–Meier survival rate plots in the Kaplan–Meier plotter database (http://kmplot.com) (Gyorffy, 2021). Kaplan–Meier plotter provides a correlation between survival and gene expression, that can be used for clinical cohorts’ evaluation (Gyorffy, 2021). The survival rate analysis was executed using breast cancer mRNA expression datasets. The default settings were used in the analysis with the exception of ER-positive only samples and restricted endocrine therapy only with tamoxifen. The samples number in the analysis is shown in each figure, and the p-value used was < 0.05.
Results
Data collection and processing
The objective of this study was to identify potential target genes and molecular mechanisms of paeoniflorin compounds in circumventing tamoxifen resistance in breast cancer. There was a good GSE67916 and GSE86871 data distribution (Supplementary Figure 1). A total of 3388 DEGs were obtained from the GSE67916 sample of gene expression analysis in Tamoxifen-resistant ER+ breast cancer cell lines, with details: 976 DEGs in the form of genes with increased expression and 2412 DEGs in the form of genes with decreased expression (Supplementary Table 1). Then 1210 DEGs were obtained from the GSE85871 sample of gene expression profiles of MCF-7 cells treated with traditional Chinese medicine components, with details: 857 DEGs of genes with increased expression and 353 DEGs of genes with decreased expression (Supplementary Table 2). Then, 175 DEGs owned by the two samples (tamoxifen-resistant and paeoniflorin-treated) were obtained from the Ven diagram by Venny 2.1.0 (Figure 1C, Supplementary Table 3). The DEGs of these two samples were analyzed further (Figure 1C).
Gene ontology and KEGG pathway enrichment analysis
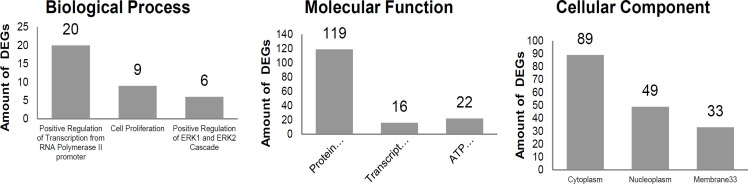
Enrichment analysis of the Ontology Gene (GO) pathway and KEGG pathway was carried out to study DEGs’ biological function and molecular mechanism. The GO and KEGG pathway analyses were performed with DAVID v6.8 using a cut-off p-value <0.05 based on three criteria: biological processes, cellular components, and molecular functions. The results showed that DEG involves in 70 biological processes, 26 cellular components, and 18 molecular functions (Figure 2, Supplementary Table 4). Three KEGG pathway analyses with DAVID revealed three pathways relevant to breast cancer (Table 1).
Figure 2.
GO Enrichment Analysis on DEGs with DAVID, Consist of Biological Process, Molecular Function, and Cellular Component
Table 1.
KEGG Pathway of DEGs Relevant to Breast Cancer
| Pathway | P-value | DEGs |
|---|---|---|
| hsa04630:Jak-STAT signaling pathway | 0.012149 | IFNB1, STAT2, PTPN11, PIK3CB, IL6ST, PRLR, IRF9 |
| hsa05200:Pathways in cancer | 0.014094 | KITLG, CDK6, COL4A3, TPR, BCL2, CTNNA1, GNAS, LAMB1, PIK3CB, SKP2, FGFR2, PML |
| hsa04151:PI3K-Akt signaling pathway | 0.037994 | RBL2, KITLG, CDK6, IFNB1, COL4A3, BCL2, LAMB1, PIK3CB, PRLR, FGFR |
PPI network and hub gene selection
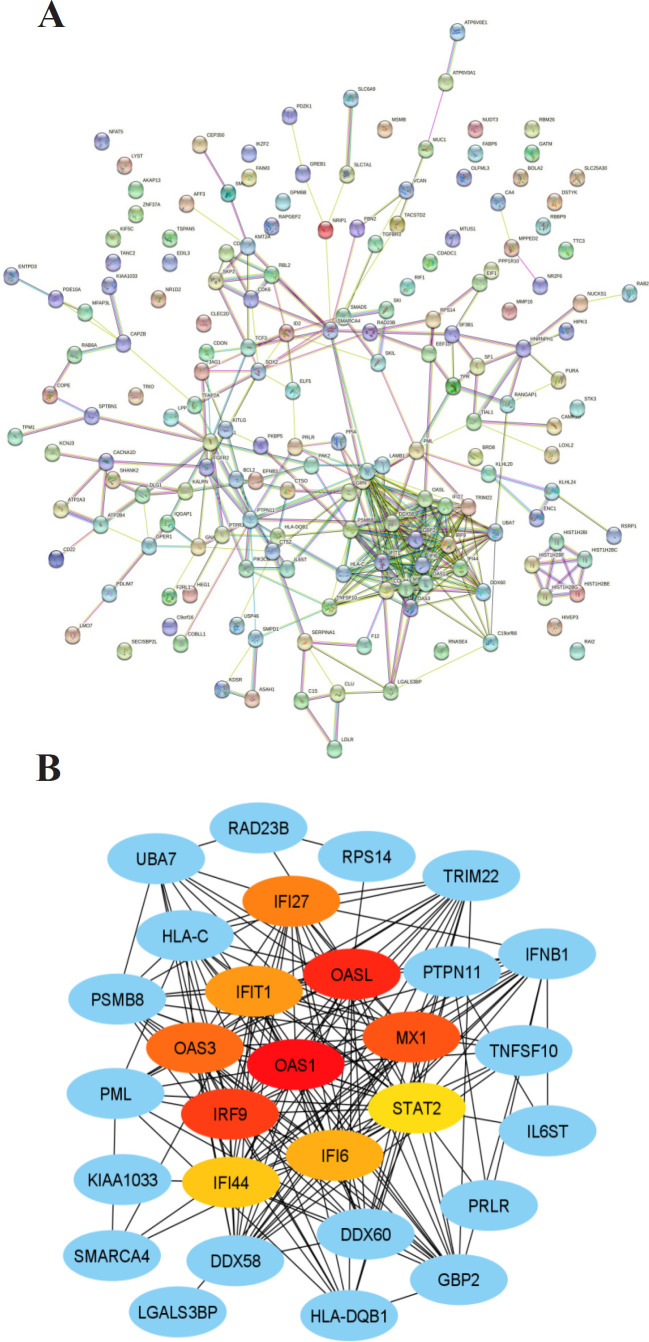
Protein-protein Interaction (PPI) has a central role in running the organisms’ cellular systems. Problems with PPIs can be a cause or indication of disease (Kuzmanov & Emili, 2013). The PPI network analysis was conducted to study the interactions between DEGs. PPI network analysis showed that the PPI was made up from 188 proteins with a confidence level of 0.4 (medium confidence), consisting of 173 nodes, 364 edges, PPI enrichment p-value: < 1.0e-16 and avg. local clustering coefficient: 0.402 (Figure 3A).
Figure 3.
(A). PPI network of the DEGs, as constructed by STRING-DB v11.0 (B). PPI network between hub genes and surrounding DEGs, as analyzed by CytoHubba plugin on Cytoscape 3.8.2
Further analysis with the CytoHubba plugs in to the Cytoscpape 3.8.2 application to select the hub gene, a gene that has a high level of intra-module connectivity (Zu et al., 2019). Present study defined the hub gene as the ten genes with the highest connectivity in the obtained PPI network. A comprehensive analysis of the relationship between nodes in the PPI was carried out using the MCC function, the most effective method for finding hub genes in the PPI network (Li et al., 2020). The top 10 genes with the highest degree scores (Fig. 3B, Table 2), in a particular order, are OAS1, OASL, IRF9, MX1, OAS3, IFI27, IFIT1, IFI6, IFI44, and STAT2. Different colors represent different degree scores of the hub genes. The red color indicated a higher degree score (Xiao et al., 2018).
Table 2.
Selection of hub Genes, as Analyzed by CytoHubba plugin of CytoScape Based on MCC
| Rank | Name | Score |
|---|---|---|
| 1 | OAS1 (2'-5'-oligoadenylate synthetase 1) | 1.71E+08 |
| 2 | OASL (2'-5'-Oligoadenylate Synthetase Like) | 1.71E+08 |
| 3 | IRF9 (Interferon regulatory factor 9) | 1.71E+08 |
| 4 | MX1 (Myxovirus resistance 1) | 1.71E+08 |
| 5 | OAS3 (2'-5'-Oligoadenylate Synthetase 3) | 1.71E+08 |
| 6 | IFI27 (Interferon Alpha Inducible Protein 27) | 1.71E+08 |
| 7 | IFIT1 (Interferon Induced Protein With Tetratricopeptide Repeats 1) | 1.71E+08 |
| 8 | IFI6 (Interferon alpha-inducible protein 6) | 1.71E+08 |
| 9 | IFI44 (Interferon Induced Protein 44) | 1.27E+08 |
| 10 | STAT2 (Signal Transducer and Activator of Transcription 2) | 1.23E+08 |
Analysis of genetic alterations of selected genes
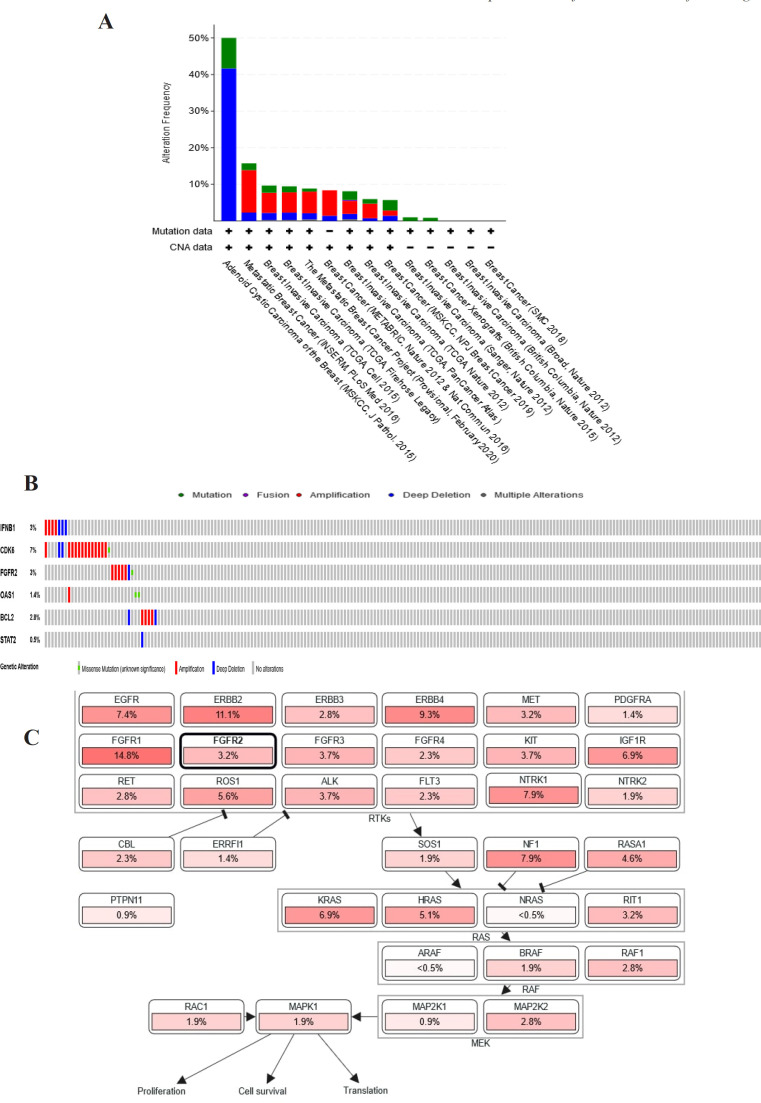
Among the DEGs, IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 were selected to explore their genome changes across breast cancer studies using cBioPortal. FGRF2, BCL2, and CDK6 were preferred among the DEGs of the KEGG pathway enrichment (cancer signaling pathway and PI3K-Akt). IFNB1 was chosen based on Jak-STAT and PI3k-Akt pathways. STAT2 was selected from the hub gene and DEGs from the Jak-STAT pathway, while OAS1 was the hub gene in the first order. A study, BRCA (INSERM) 2016, showed the second-highest genetic alteration among breast cancer studies and had more variation of DEGs-associated genetic changes than the study that ranked first (Figure 4A). Genetic changes for each target gene were found from 0.5% to 7% of the case population per patient case, including STAT2 (0.5%), OAS1(1.4%), BCL2 (2.8%), FGFR2 (3 %), IFNB1 (3%), and CDK6 (7%) (Figure 4B). In addition, most of the gene changes include amplification and deep deletion. Additional analysis using cBioportal revealed FGFR signaling pathway through Ras is important for the PF mechanism in circumventing breast cancer resistance to tamoxifen (Figure 4C).
Figure 4.
(A) Genetic Alterations of DEGs (IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2) presented on a chart of 14 breast cancer studies (B) Oncoprint cBioportal in the 2016 BRCA study (INSERM) : Genetic alteration of selected DEGs (IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2) in the patient population. (C). Results of DEGs analysis by cBioportal in the pathway section related to gene alteration. The results showed that DEGs are involved in the FGFR2 signaling pathway through Ras
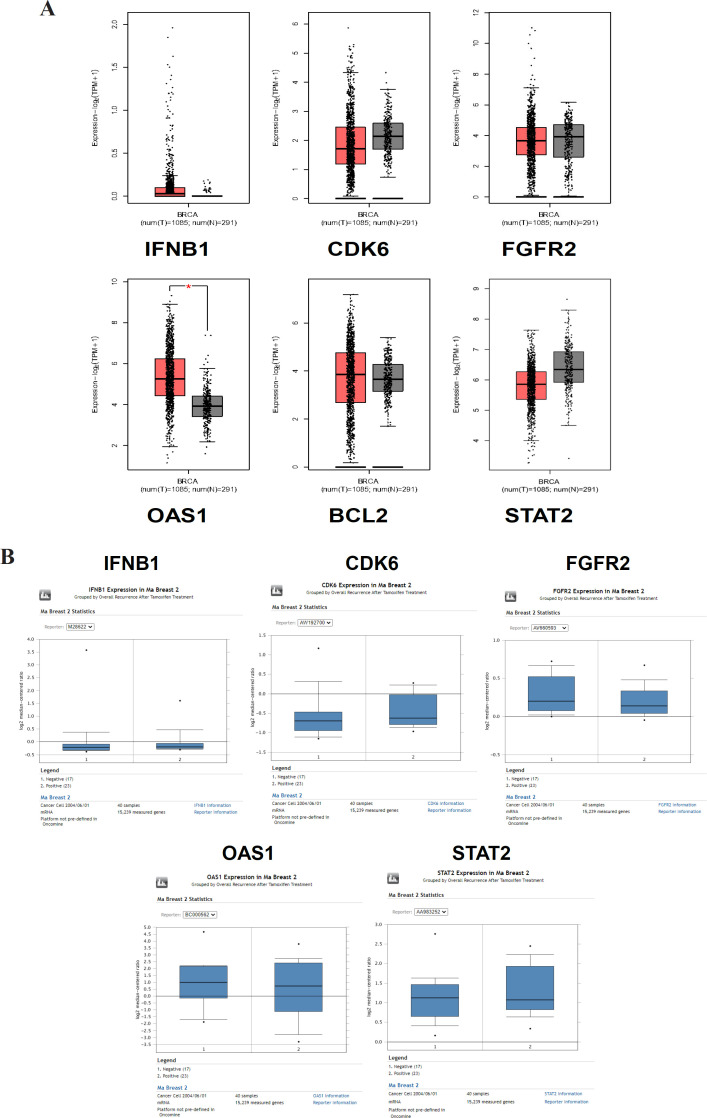
Validation of gene expression analysis in non-resistant and tamoxifen-resistant breast cancer cells
Validation analysis with GEPIA showed that the mRNA expression in non-resistant breast cancer cells and normal cells were not significantly different except that OAS1 was expressed significantly higher in tumor cells than in normal cells (p-value < 0.01) (Figure 5A). ONCOMINE analysis showed that CDK6 and STAT were overexpressed in the tamoxifen-resistant group or recurrent group (Figure 5B). However, the expression of IFNB1 did not differ between recurrent and nonrecurrent groups. Furthermore, FGFR2 and OAS1 were expressed slightly lower than nonrecurrent, but it is not apparent. In this analysis using ONCOMINE, the BCL2 was excluded as it is not available in the dataset used.
Figure 5.
(A). Validation analysis of gene expression of IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 in non-resistant breast cancer cells and normal cells using GEPIA. (B). Gene expression analysis of IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 in recurrent and non-recurrent group after treated with tamoxifen using ONCOMINE
Survival rate analysis
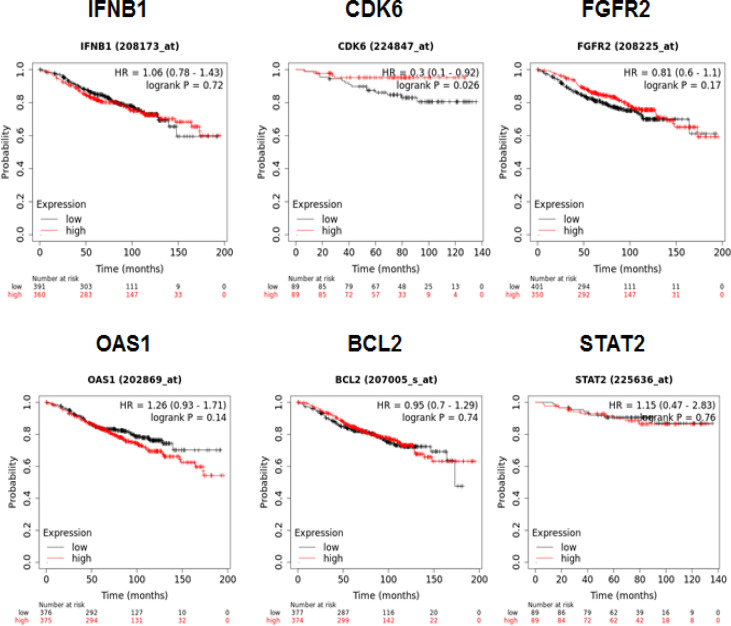
Kaplan–Meier curves showed that the survival rate of low mRNA expression of CDK6 was significantly lower than that of the high mRNA expression group (p = 0.026) (Figure 6). The survival rate of STAT2 between the low expression and high expression groups was not significantly different, but both had a fairly high survival rate, above 80%. Overall survival rates of the high-expressed mRNA of IFNB1 (p = 0.72), FGFR2 (p = 0.17), OAS1 (p = 0.14), and BCL2 (p = 0.74) were worse than those of the lower mRNA expression, but the results were not significant.
Figure 6.
Survival Rate Analysis of IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2 in ER+ Breast Cancer and Tamoxifen-Treated Only, Using Kaplan–Meier Curve in KMPlotter
Discussion
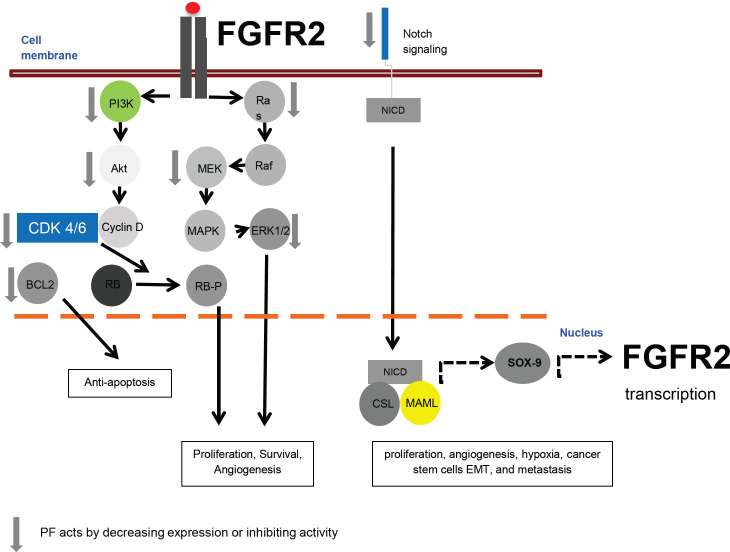
The results of this study using genomic understanding-based bioinformatics analysis highlighted the potential target genes, namely: IFNB1, CDK6, FGFR2, OAS1, BCL2, and STAT2. Furthermore, by using cBioportal, it was found that the PF mechanism in circumventing breast cancer resistance to tamoxifen is via the FGFR signaling pathway through Ras. PF circumvent breast cancer resistance to tamoxifen by targeting FGFR2 via inhibition of its downstream signaling or inhibition of its transcriptional activity through Notch signaling (Figure 7). The FGFR pathway is initiated when the FGFR receptor or Fibroblast Growth Factor Receptor binds to its ligand, Fibroblast Growth Factor, changing the FGFR conformation, resulting in dimerization and activation of FGFR. Activated FGFR phosphorylates FRS2a and FRS2a, which subsequently bind to the adapter containing the SH2 domain, Grb2. Grb2 further bind to SOS, GAB1, and Cbl via its SH3 domain to activate Ras/Raf/MAPK, including ERK MAPK, p38 MAPK, and JNK MAPK (Xie et al., 2020) and its downstream protein, including cyclin-dependent kinases (CDKs) and BCL2 (Chambard et al., 2007; Modi et al., 2012; Wang et al., 2015).
Figure 7.
Proposed Molecular Mechanisms of PF in Circumventing Breast Cancer Resistance to Tamoxifen. PF inhibits the FGFR2 pathway through its downstream signaling. PF also inhibits the transcriptional activity of FGFR2 through inhibition of Notch signaling
The RAS protein serves as the center of a primary intracellular signaling pathway that regulates cell growth, motility, angiogenesis, and immune evasion (Sever and Brugge, 2015). Oncogenic RAS mutations constitutively increase the interaction of breast cells with the basement membrane, induce a phenotype independent of anchorage, invasion, tumorigenic potential, TGF-β and IGF-1 secretion, EGFR activation, mitogen-activated protein kinase (MAPK), and estrogen insensitivity. (Galiè, 2019). Studies have also shown that the MAPK pathway facilitates tamoxifen resistance (Eanes and Patel, 2016). ERK phosphorylation and increased activity are associated with endocrine resistance and decreased survival in breast cancer patients. One possible mechanism is that ERK phosphorylates Ser 118 in the ER, leading to ligand-independent ER activation (Haagenson and Wu, 2014).
In a study conducted by Guo et al., (2017), PF treatment hindered VSMC proliferation by decreasing the expression of four proteins involved in the G0/G1 and G1/S transitions: cyclin D1, cyclin E, CDK4 and CDK2. Furthermore, PF increases the expression of p21, which is known to form a heterotrimetric complex with the cyclin-CDK complex, thereby inhibiting its activity (Guo et al., 2017). PF also exhibited antitumor activity in gastric cancer cells by inhibiting cell proliferation and stimulating apoptosis by reducing the expression of PI3K, p-Akt and Akt (Zheng et al., 2015). Besides, the effect of PF in inhibiting the Akt pathway is also known to occur in primary human liver sinusoidal endothelial cells (HHSECs) (Gong et al., 2015). Additionally, PF is known to have effects on proteins associated with apoptosis. Research conducted by Hu et al. (2018) on mouse chondrocytes shows that PF reduces the expression of mRNA and protein Bcl-2, a protein involved in the apoptosis. Future studies on the FGFR2 and its downstream, including PI3K/Akt, Ras, and MAPK upon PF treatment in tamoxifen-resistant breast cancer cells are needed to the present study’s results.
The second proposed mechanism of PF in circumventing breast cancer resistance to tamoxifen is via inhibition of FGFR2 transcriptional activity through Notch signaling. In invasive breast cancer, increased expression of the Notch signaling pathway components has been reported, including Jagged1-2, Dll1, Dll3, and Dll4, Notch receptors, Hes and Hey target genes, and accumulation of NICD (Wang et al., 2010). Notch signaling is known to regulate many cellular processes, including proliferation, apoptosis, angiogenesis, hypoxia, cancer stem cell activity, EMT, and metastasis (Acar et al., 2016). Crosstalk between estradiol and Notch signaling has a significant role in human breast carcinogenesis and angiogenesis (Kontomanolis et al., 2018). NOTCH-1 activates ERα-dependent transcription in the presence or absence of estradiol. Therefore, Notch signaling regulates estrogen signaling in breast cancer cells, and the combination of antiestrogens and Notch inhibitors may be more effective in ERα(+) breast cancer (Kontomanolis et al., 2018). The Notch-1 target gene, SOX9, is expressed in multi-potent cells in the liver. Overexpression of SOX9 is frequently detected in patients with liver cancer (Huang et al., 2019). SOX9 is a transcription factor (Lefebvre and Ginzberg, 2018) and its overexpression is frequent in breast cancer subtypes (Jana et al., 2020).
SOX9 is elevated in breast cancer patients after endocrine therapy failure (Domeneci et al., 2018). Recently, mouse genetic studies have revealed a feed-forward loop on early pancreatic FGF10/FGFR2b/SOX9 progenitors to sustain their proliferation. SOX9 acts as a link between FGF10 and Notch in progenitor proliferation. (Li et al., 2015). The SOX9 ablation resulted in significant downregulation of FGFR2 and FGFR4, which indicates that SOX9 is required to maintain the expression of FGFR2 and FGFR4 (Seymor et al., 2012). A previous study by Zhang et al., (2018) showed that using different concentrations of PF significantly decreased NOTCH-1 and HES-1 mRNA and protein expression in MCF-7 breast cancer cells. Overall, the axis between Notch signaling-FGFR2 and PF treatment in circumventing breast cancer to tamoxifen are warranted for future studies.
The advantage of using the genomic understanding-based bioinformatics in this study is that it can accelerate the identifying the target of a compound for a drug candidate of a specific disease. In addition, in terms of duration, time needed is relatively shorter than other techniques such as screening in wet laboratory experiments. However, the results of this study, in which targets and molecular mechanisms of PF in circumventing breast cancer resistance to tamoxifen still need to be validated within vitro, in vivo, toxicity test, and clinical trials.
In Conclusion, PF circumvent breast cancer resistance to tamoxifen by targeting FGFR2 via inhibition of its downstream signaling or inhibition of its transcriptional activity through Notch signaling. The results of this study need further confirmation for the development of PF as drugs for circumventing breast cancer resistance to tamoxifen.
Author Contribution Statement
Data collection, analysis and interpretation of data, and writing draft: Moh. Arikus Wibowo and Eri Prasetyo Nugroho. Study concept and design, supervision, funding acquisition, editing, and final approval of manuscript: Adam Hermawan.
Funding statement
This study was supported by Rekognisi Tugas Akhir (RTA) year 2021 Universitas Gadjah Mada, SK Rektor UGM No.1185/UN1.P.III/SK/HUKOR/2021.
Availability of data
The data that support the findings of this study are available in the supplementary files and available in http://ugm.id/PFRTA
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
We thank Ms. Herwandhani Putri for her writing assistance.
References
- Acar A, Simões BM, Clarke RB, Brennan K. A role for notch signalling in breast cancer and endocrine resistance. Stem Cells Int. 2016;2016:2498764. doi: 10.1155/2016/2498764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Rasool M, Chaoudhry H, et al. Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation. 2016;12:135–9. doi: 10.6026/97320630012135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambard JC, Lefloch R, Pouysségur J, Lenormand P. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Chang M. Tamoxifen resistance in breast cancer. Biomol Ther. 2012;20:256–67. doi: 10.4062/biomolther.2012.20.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenici G, Aurrekoetxea-Rodríguez I, Simões BM, et al. A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene. 2019;38:3151–69. doi: 10.1038/s41388-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes L, Patel YM. Inhibition of the MAPK pathway alone is insufficient to account for all of the cytotoxic effects of naringenin in MCF-7 breast cancer cells. Biochim Open. 2016;3:64–71. doi: 10.1016/j.biopen.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct Targeted Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiè M. RAS as supporting actor in breast cancer. Front Oncol. 2019;9:1–9. doi: 10.3389/fonc.2019.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WG, Lin JL, Niu QX, et al. Paeoniflorin diminishes ConA-Induced IL-8 production in primary human hepatic sinusoidal endothelial cells in the involvement of ERK1/2 and akt phosphorylation. Int J Biochem Cell Biol. 2015;62:93–100. doi: 10.1016/j.biocel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhao Y, Li L, et al. Concentration-dependent effects of paeoniflorin on proliferation and apoptosis of vascular smooth muscle cells. Mol Med Rep. 2017;16:9567–72. doi: 10.3892/mmr.2017.7776. [DOI] [PubMed] [Google Scholar]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol. 2021;19:4101–9. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagenson KK, Wu GS. The role of MAP kinases and MAP kinase phosphatase-1 in resistance to breast cancer treatment. Cancer Metastasis Rev. 2010;29:143–9. doi: 10.1007/s10555-010-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermawan A, Ikawati M, Jenie RI, et al. Identification of Potential Therapeutic target of naringenin in breast cancer stem cells inhibition by bioinformatics and in vitro studies. Saudi Pharm J. 2021a;29:12–26. doi: 10.1016/j.jsps.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermawan A, Ikawati M, Khumaira A, et al. Bioinformatics and in vitro studies reveal the importance of p53, PPARG and notch signaling pathway in inhibition of breast cancer stem cells by hesperetin. Adv Pharm Bull. 2021b;11:351–60. doi: 10.34172/apb.2021.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermawan A, Putri H. Integrative bioinformatics analysis reveals potential target genes and TNFα signaling inhibition by Brazilin in metastatic breast cancer cells. Asian Pac J Cancer Prev. 2020a;21:2751–62. doi: 10.31557/APJCP.2020.21.9.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermawan A, Putri H. Bioinformatics studies provide insight into possible target and mechanisms of action of nobiletin against cancer stem cells. Asian Pac J Cancer Prev. 2020b;21:611–20. doi: 10.31557/APJCP.2020.21.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermawan A, Putri H, Ikawati M. Bioinformatic analysis reveals the molecular targets of tangeretin in overcoming the resistance of breast cancer to tamoxifen. Gene Rep. 2020;21:100884. [Google Scholar]
- Hu PF, Chen WP, Bao JP, Wu LD. Paeoniflorin inhibits IL-1β-induced chondrocyte apoptosis by regulating the Bax/Bcl-2/caspase-3 signaling pathway. Mol Med Rep. 2018;17:6194–200. doi: 10.3892/mmr.2018.8631. [DOI] [PubMed] [Google Scholar]
- Huang Q, Li J, Zheng J, Wei A. The carcinogenic role of the notch signaling pathway in the development of hepatocellular carcinoma. J Cancer. 2019;10:1570–9. doi: 10.7150/jca.26847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultsch S, Kankainen M, Paavolainen L, et al. Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC Cancer. 2018;18:850. doi: 10.1186/s12885-018-4757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Madhu Krishna B, Singhal J, et al. SOX9: The master regulator of cell fate in breast cancer. Biochem Pharmacol. 2020;174:113789. doi: 10.1016/j.bcp.2019.113789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontomanolis EN, Kalagasidou S, Pouliliou S, et al. The Notch pathway in breast cancer progression. Sci World J. 2018;2018:2415489. doi: 10.1155/2018/2415489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmanov U, Emili A. Protein-protein interaction networks: Probing disease mechanisms using model systems. Genome Med. 2013;5:37. doi: 10.1186/gm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Cai JH, Tsai JJP, Wang CCN. Identification of hub genes associated with development of head and neck squamous cell carcinoma by integrated bioinformatics analysis. Front Oncol. 2020;10:681. doi: 10.3389/fonc.2020.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Zhai WJ, Teng CB. Notch signaling in pancreatic development. Int J Mol Sci. 2015;17:48. doi: 10.3390/ijms17010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumachi F, Santeufemia DA, Basso SM. Current medical treatment of estrogen receptor-positive breast cancer. World J Biol Chem. 2015;6:231–9. doi: 10.4331/wjbc.v6.i3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi PK, Komaravelli N, Singh N, Sharma P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol Biol Cell. 2012;23:3722–30. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: A Cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5:1–21. doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour PA, Shih HP, Patel NA, et al. A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development. 2012;139:3363–72. doi: 10.1242/dev.078733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani N, Fan CO, Flores-Villanueva P, Generali D, Li Y. The fibroblast growth factor receptors in breast cancer: From Oncogenesis to Better Treatments. Int J Mol Sci. 2020;21:2011. doi: 10.3390/ijms21062011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Chenwei L, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lu X, Dong X, et al. PERK1/2 silencing sensitizes pancreatic cancer BXPC-3 cell to gemcitabine-induced apoptosis via regulating Bax and Bcl-2 expression. World J Surg Onc. 2015;13:66. doi: 10.1186/s12957-015-0451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z Li Y, Sarkar FH. 2010) Notch signaling proteins: legitimate targets for cancer therapy. Curr Protein Pept Sci. 11:398–408. doi: 10.2174/138920310791824039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulandari F, Ikawati M, Meiyanto E, et al. Bioinformatic analysis of CCA-1 1 a novel curcumin analog, uncovers furthermost noticeable target genes in colon cancer. Gene Rep. 2020;21:100917. [Google Scholar]
- Xia X. Bioinformatics and drug discovery. Curr Top Med Chem. 2017;17:1709–26. doi: 10.2174/1568026617666161116143440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang Q, Wei S, et al. Paeoniflorin: A monoterpene glycoside from plants of paeoniaceae family with diverse anticancer activities. J Pharm Pharmacol. 2020;72:483–95. doi: 10.1111/jphp.13204. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Feng M, Ran H, Han X, Li X. Identification of key differentially expressed genes associated with nonsmall cell lung cancer by bioinformatics analyses. Mol Med Rep. 2018;17:6379–86. doi: 10.3892/mmr.2018.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Su N, Yang J, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther. 2020;5:181. doi: 10.1038/s41392-020-00222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yu K, Han X, et al. Paeoniflorin influences breast cancer cell proliferation and invasion via inhibition of the notch-1 signaling pathway. Mol Med Rep. 2018;17:1321–5. doi: 10.3892/mmr.2017.8002. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zuo J, Wang L, et al. Identification of differentially expressed genes between mucinous adenocarcinoma and other adenocarcinoma of colorectal cancer using bioinformatics analysis. J Int Med Res. 2020;48:030006052094903. doi: 10.1177/0300060520949036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YB, Xiao GC, Tong SL, et al. Paeoniflorin inhibits human gastric carcinoma cell proliferation through up-regulation of microRNA-124 and suppression of PI3K/Akt and STAT3 signaling. World J Gastroenterol. 2015;21:7197–207. doi: 10.3748/wjg.v21.i23.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang S, Song C, Hu Z. Paeoniflorin prevents hypoxia-induced epithelial–mesenchymal transition in human breast cancer cells. Onco Targets Ther. 2016;9:2511–18. doi: 10.2147/OTT.S102422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Fei Q, Qian L, et al. Identification of key pathways and genes in nasopharyngeal carcinoma using bioinformatics analysis. Oncol Lett. 2019;17:4683–94. doi: 10.3892/ol.2019.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Jin Z, Deng Y, et al. Co-expression network analysis identifies four hub genes associated with prognosis in soft tissue sarcoma. Front Genet. 2019;10:37. doi: 10.3389/fgene.2019.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in the supplementary files and available in http://ugm.id/PFRTA