Abstract
Tissue engineering approaches combine a bioscaffold with stem cells to provide biological substitutes that can repair bone defects and eventually improve tissue functions. The prospective bioscaffold should have good osteoinductivity. Surface chemical and roughness modifications are regarded as valuable strategies for developing bioscaffolds because of their positive effects on enhancing osteogenic differentiation. However, the synergistic combination of the two strategies is currently poorly studied. In this work, a nanoengineered scaffold with surface chemistry (oxygen-containing groups) and roughness (Rq = 74.1 nm) modifications was fabricated by doping nanohydroxyapatite (nHA), chemically crosslinked graphene oxide (GO) and carboxymethyl chitosan (CMC). The biocompatibility and osteoinductivity of the nanoengineered CMC/nHA/GO scaffold was evaluated in vitro and in vivo, and the osteogenic differentiation mechanism of the nanoengineered scaffold was preliminarily investigated. Our data demonstrated that the enhanced osteoinductivity of CMC/nHA/GO may profit from the surface chemistry and roughness, which benefit the β1 integrin interactions with the extracellular matrix and activate the FAK–ERK signaling pathway to upregulate the expression of osteogenic special proteins. This study indicates that the nanocomposite scaffold with surface chemistry and roughness modifications could serve as a novel and promising bone substitute for tissue engineering.
The CMC/nHA/GO scaffold with the surface chemistry and roughness dual effects and the release of phosphate and calcium ions synergistically assist the mineralization and facilitate the bone regeneration.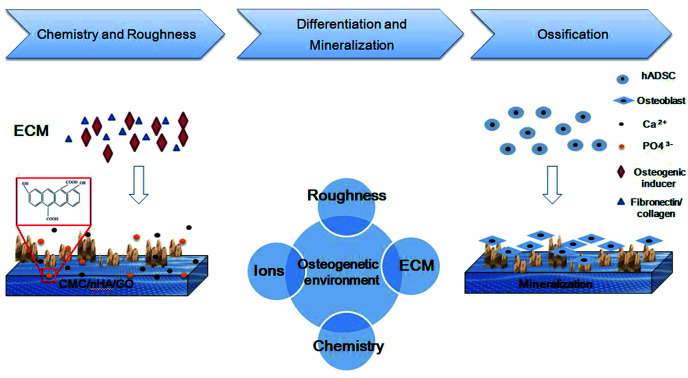
1. Introduction
A potential bone substitute should have good bioactivity, including osteoconductivity and osteoinductivity, in the absence of chemical treatment. Scaffolds made of different types of biomaterials, such as bioceramics, bioglass, titanium alloy, biopolymers and their composites, possess some bioactivity, but these scaffolds also have disadvantages such as bioinertia, tissue rejection, infection and cyst complications. Thus, researchers have paid attention to stem cell epigenetics and growth factor sustained release to improve the bioactivity of scaffolds,1–3 but these methods suffer from disadvantages, such as virus carrier dangers, growth factor inactivation and cumbersome processes.4–6 However, nanoscale bioactivity can be utilized to engineer next generation implants. The physicochemical properties of nanomaterials play a vital role in stem cell biological behaviors, including adhesion, proliferation, differentiation, migration and protein absorption.7–13 A number of studies have shown that carbon nanotubes, graphene nanomaterials and silicon nanomaterials can significantly manipulate osteoblast adhesion and differentiation and promote bone formation via chemical inducing effects.14–19 Although nanomaterials can significantly improve scaffold bioactivity, the limited incorporation of nanomaterials into a scaffold is necessary when considering the biosafety in vivo,20–23 and the bioactivity of nanocomposite scaffolds continues to be explored within the biosafety dose of the nanomaterials.
To further improve the bioactivity of bioscaffold, some studies have reported that topographic substrates can mimic an in vivo microenvironment composed of pores, ridges, and channels to provide physical cues to cells at a nanoscale level.24,25 Many reports have illustrated that the scaffold surface roughness and topographical structure can regulate osteogenesis through cell adhesion effects on the signaling cascades outside of the cytoskeletal signaling.8,24,26–29 Especially, the surface roughness can mimic the nanoscale extracellular matrix properties of tissue and promote both osteoblast and mesenchymal stem cell attachment and proliferation.30 Therefore, enhancing the surface roughness of a scaffold is an effective approach to improve its bioactivity.
We have reviewed some techniques, such as electrospinning, soft lithography, electrodeposition, cold atmospheric plasma and nanomaterial templates, which can benefit fabrication of surface nanostructures.30–33 Based on the principle of a nanomaterial template, we attempted to find a type of nanomaterial to rough scaffold surface. Among the nanomaterials used in regeneration medicine, nanohydroxyapatite (nHA) is an inorganic nanomaterial, which exhibits the highest similarity with the mineral component in natural bone,34 moreover nHA has been proved that it has good osteoinductivity in bone regeneration process.35,36 The enhancing mechanism of nHA was reported that extracellular PO43− could promote osteogenic differentiation in stem cells by inducing phosphate-ATP-adenosine metabolic signaling.37 Therefore, if nHA is used as a template, it not only could improve scaffold surface roughness, but also could enhance the scaffold osteoinductivity due to its chemical effects.
Herein, based on the fabrication of a graphene oxide-based (GO) bioactive scaffold as previously reported,21 we used nHA as a physical and chemical enhancer to further improve the osteoconductivity and osteoinductivity of scaffold (Scheme 1). Briefly, nHA was mixed into the GO and chitosan mixed solution, and the rough, porous, nHA-doped GO (CMC/nHA/GO) scaffold was synthesized via a chemical crosslinking reaction and lyophilization. Then, the roughness and ion release rate were characterized, and the osteoinductivity of the CMC/nHA/GO scaffold was evaluated by in vitro and in vivo experiments. Finally, a preliminary mechanism underlying the synergistically enhanced osteoinductivity of the scaffold was studied. Through a comprehensive evaluation of the CMC/nHA/GO composites scaffold, we hope to develop a promising bioactive bone graft substitute with good osteoinductivity and lay the foundation for further developing tissue engineering.
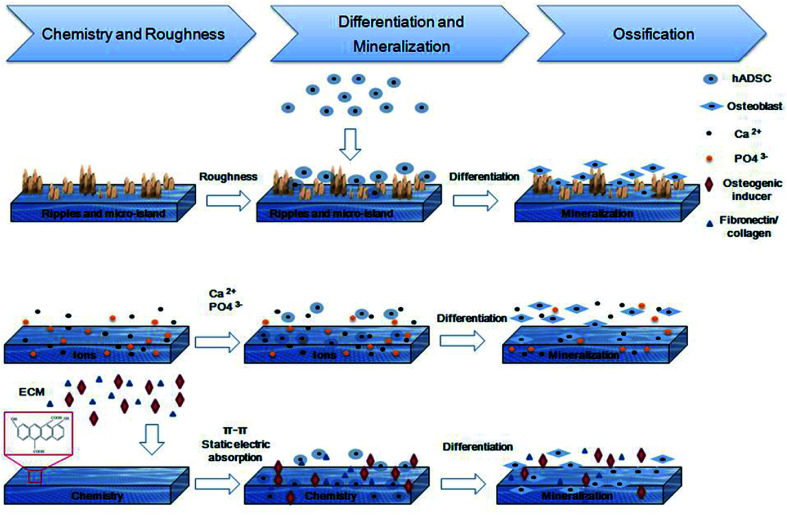
Scheme 1. Introduction of CMC/nHA/GO scaffolds modified surface properties, including, topographies and roughness, ions releasing and chemical composition. The synergistic effects promote the osteogenic differentiation of hADSCs and the formation of new bone.
2. Materials and methods
2.1. Preparation of CMC/nHA/GO composite scaffold
The CMC/nHA/GO composite scaffold was prepared by chemical crosslinking according to our previous report.15 Briefly, 2% carboxymethyl chitosan (CMC) (W/V) was first dissolved in a 0.2% (W/V) GO solution, and subsequently, 2% nHA (W/V) was homogeneously dispersed into the CMC/GO mixed solution under magnetic stirring for 15 min and sonicating for 1 hour. Then, 0.1 M EDC and 0.025 M NHS were added to the CMC/nHA/GO mixture under stirring, and the mixture was held for 1 hour at RT to allow the mixture to completely crosslink. The porous CMC/nHA/GO scaffold was obtained by lyophilization and ultrasonic washing. The control groups of the CMC/GO and CMC scaffolds were prepared simultaneously using a similar method.
2.2. Characterization of the CMC/nHA/GO composite scaffold
The surface morphology and pore size of the scaffolds were observed using scanning electron microscopy (SEM, Zeiss Ultra 55, Germany). Furthermore, the porosities of scaffolds were analyzed by using mercury porosimeter (AutoPore, lv 9510). The surface roughness of the scaffolds was imaged and calculated using atomic force microscopy (AFM, Bruker, USA). The chemical functional groups were confirmed using Fourier transform infrared (FTIR) spectroscopy (EQUINOX 55, Bruker) from 4000 to 400 cm−1 with a resolution of 2 cm−1 over 64 scans using a KBr pellet. The crystalline phases were assayed using an X-ray diffractometer (BRUKER-AXS, D8 ADVANCE) at a scanning rate of 0.033° per second in a 2θ range from 5° to 80° with Cu-Kα radiation (λ = 1.5418 Å). The chemical composition of scaffold was investigated with X-ray photoelectron spectroscopy (Kratos AXIS Ultra DLD) and Raman spectroscopy (Senterra R200-L, Bruker, Germany).
For the Ca2+ and PO43− ion release rate assay, the Ca2+ and PO43− ion concentrations in dd-H2O were measured at 1, 3, 5, 7, 10, 14 and 21 days. Each time, 1 mL of dd-H2O was removed and replaced with fresh solution. The collected solution was analyzed for Ca2+ and PO43− ion concentrations via a spectrophotometric method (DMS-80 UV-visible, Varian, Palo Alto, CA, USA) using known standards and calibration curves based on previous studies.38
The stiffness of the scaffolds were evaluated by a depth-sensing indentation approach using a nanoindenter (MTS Systems, Oak Ridge, TN) of 50 nm radius, calibrated with a fused silica standard. A maximum load of 0.15 mN was set and 15 indents were made at 35 lm intervals on each sample. The maximum indentation depth was set to 200 nm, while the drift rate and Poisson's ratio were set at 2.0 and 0.350, respectively. The load-displacement data were continuously recorded during one complete cycle of loading and unloading. Each type of scaffolds was tested to obtain the average stiffness. Compress loading tests were performed with a tabletop uniaxial testing instrument. The loading was 50 N and crosshead speed was set at 0.5 mm min−1. Samples were sliced into circle with 10 mm diameter and scaffolds were tested (n = 5 for each group). Stress–strain curves and parameters like Young modulus and compress strength were obtained.
2.3. Isolation and osteoinductive cultures of hADSCs
Human adipose-derived stem cells (hADSCs) were isolated from the adipose tissue of normal donors as previously reported,39 with informed consent approved according to the procedures of the Ninth People's Hospital affiliated with the Shanghai Jiao Tong University School of Medicine Institutional Review Board (IRB [2014]8). Briefly, adipose tissue was extensively washed with 20 mL of phosphate-buffered saline (PBS) three times, cut into small pieces, and digested with 0.1% collagenase (Sigma Aldrich, St. Louis, MO, USA) solution with shaking at 37 °C for 2 hours. Then, the digested tissues were resuspended in DMEM/F12 (Gibco BRL, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 units per mL of penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA) and incubated at 37 °C with a 5% CO2 humid atmosphere. The hADSCs exhibited fibroblast-like shapes and were used for all the tests.
For the proliferation assays, the proliferation medium (PM) consisted of the DMEM/F12 (Gibco) supplemented with 10% FBS (Gibco), 100 units per mL penicillin and streptomycin (Invitrogen). For osteogenic induction, the differentiation medium (DM) consisted of α-MEM (Gibco) supplemented with 10% FBS (Gibco), 10−8 M dexamethasone, 10 mM β-glycerol phosphate, and 50 mg mL−1l-ascorbic acid (Sigma-Aldrich).
2.4. Proliferation assay
Proliferation of the hADSCs on the substrates was evaluated using cell counting kit-8 (CCK-8) and the Ki-67 staining assay as previously reported.21 The substrates were prepared in a 96 well plate in advance. The hADSCs were seeded at a density of 1 × 104 cells per 100 μL medium per well, and the proliferation of the hADSCs on the substrates was evaluated using the CCK-8 assay (Dojindo, Japan) after 0, 24, 48 and 72 hours. For further evaluating the proliferation of the hADSCs on the CMC/nHA/GO substrates, a Ki-67 staining assay was performed in 24 well culture plates. After 2 days of culture, the hADSCs cells were fixed with 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.3% Triton X-100 (Sigma-Aldrich), and blocked with 10% normal goat serum (Invitrogen). The cells were then subjected to mouse monoclonal anti-Ki-67 (1 : 200, BD Biosciences, USA) at 4 °C for 12 hours and were labeled with fluorescent secondary antibodies (Alexa Fluor 546 goat anti-mouse immunoglobulin G, BD Biosciences). Finally, the cell nuclei were counterstained with Hoechst (Invitrogen) and imaged under a fluorescence microscope (Olympus BX51, Japan). The proliferation rate of the hADSCs was calculated using the proportion of the Ki-67 positive cells out of the Hoechst cells.
2.5. Adhesion assay
For the adhesion assay, the hADSCs were seeded on the substrates at a density of 1 × 104 cells per well, unattached cells were carefully removed from the substrates by washing with PBS after 1, 2 and 8 hours of incubation, and then the attachment of the hADSCs onto the substrates was evaluated by the CCK-8 assay.
2.6. Cell viability assay
To observe the viability of the hADSCs on the substrates, the cells were stained with the LIVE/DEAD viability/cytotoxicity kit (Invitrogen) after they were cultured in the scaffolds for 1, 3 and 5 days, and the cell viability was observed under a fluorescence microscope (Nikon Eclipse E-600 FN). To further evaluate the biocompatibility of the scaffolds, the hADSCs were cultured on the scaffolds for 3 days and stained with the LIVE/DEAD viability/cytotoxicity kit (Invitrogen), then fixed with 4% PFA and counterstained with Hoechst (Invitrogen). The scaffolds were scanned layer by layer using confocal microscopy (Leica SP5), and the reconstructed 3D scaffolds were imaged at 0, 45 and 90 degrees.
2.7. Scanning electron microscope analysis
To observe the cell morphology on the scaffolds, biological SEM was performed after the hADSCs were cultured on the scaffolds for 3 days. The scaffolds were fixed with 2.5% glutaraldehyde, dehydrated with graded ethanol, dried with a supercritical carbon dioxide drying instrument (autosamdri-815, TOUSIMIS, MARYLAND), and sputter coated with gold for the SEM analysis.
2.8. Real-time PCR analysis
The osteoinductivity of the scaffold in vitro was evaluated by analyzing RNA expression of the bone-related genes of the hADSCs. The relative gene expression was analyzed after the hADSCs were cultured on the substrates in PM or DM for 7 and 14 days using a Power SYBR Green PCR Master Mix (Applied Biosystems, USA) along with a 7500 Real-Time PCR Detection System. The primers are shown in ESI Table 1.† Each target gene was tested in triplicate, and the data are expressed as the fold change relative to the CMC substrate after normalization of the expression of GAPDH.
2.9. Western blot analysis
To evaluate the osteogenic differentiation of the hADSCs on the substrates, Western blot was performed. Briefly, the total proteins were extracted from the hADSCs after 7 d of osteoinductive culture or proliferative culture and then the concentration were measured by a BCA Kit (Pierce) according to the manufacturer's protocol. For each sample, 20 mg of protein was loaded onto a 10% SDS/PAGE gel. The gel-separated proteins were then transferred to a PVDF membrane (Millipore, Billerica, MA, USA) at 80 V for 90 min and incubated with the primary antibodies, BSP (1 : 1000, CST), OCN (1 : 1000, Abcam), OPN (1 : 1000, Abcam) and β-actin (1 : 3000, Invitrogen), at 4 °C overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich). The protein expression was visualized using an Odyssey V 3.0 imagescanner (LICOR, Lincoln, NE, USA).
For the cell-material interaction analysis, the expression of β1 integrin was measured by Western blot. The hADSCs were cultured in PM and DM for 7 days and the Western blot were performed as above, the primary antibody β1 integrin was purchased from Abcam. To further investigate the mechanism of the cell-material interactions, the hADSCs were cultured in PM for 24 h and 48 h. Then, Western blot was performed to evaluate the protein expression of ERK1/2 (1 : 1000, CST), p-ERK (1 : 1000, CST), FAK (1 : 1000, Abcam) and p-FAK (1 : 1000, Abcam).
2.10. ALP and ARS staining assay
To further evaluate the osteoinductivity of the scaffold, the alkaline phosphatase (ALP) staining of the hADSCs was detected after they were cultured on CMC/nHA/GO substrates in PM and DM for 7 days. Alizarin red S (ARS) staining was performed after cells were cultured in PM and DM for 14 days. ALP and ARS staining were performed as previously described.40 Briefly, the cells were washed and fixed in 4% polyoxymethylene for 15 min. ALP staining was performed using a BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime) according to the manufacturer's instructions, with the cells were incubated at 37 °C for 30 min.
2.11. Immunofluorescence assay
Moreover, the cellular immunofluorescence was also conducted after the hADSCs were cultured on the substrates in PM and DM for 7 days, as previously described.21 The hADSCs were first fixed in 4% PFA (Sigma) and permeabilized with 1% Triton X-100 (Invitrogen). Then, the cells were incubated with anti-BSP antibody (1 : 200, Abcam) overnight at 4 °C and incubated with anti-rabbit Alexa Fluor 546 secondary antibody (1 : 800, Invitrogen). The nuclei were counterstained with Hoechst (Invitrogen), and the positive ratio was evaluated by calculating the proportion of the BSP positive cells out of the Hoechst positive cells under a fluorescence microscope (Olympus BX51, Japan).
2.12. Ectopic osteogenesis
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Shanghai Animal Experimental Center and approved by the Animal Ethics Committee of the Ninth People's Hospital affiliated with the Shanghai Jiao Tong University School of Medicine (HKDL [2014]54). Four-week-old male nude mice were used in the animal experiment. The hADSCs were seeded onto small patches (approximately 4 mm × 4 mm) of the CMC, CMC/GO and CMC/nHA/GO scaffolds and cultured for a week before implantation. The nude mice were anesthetized with pentobarbital sodium by intraperitoneal injection. Then, the cell-seeded scaffolds were implanted into the subcutaneous tissue. At 8 weeks after surgery, the implants were excised with the peripheral skin tissues and soft tissues. Three animals were used for each group.
2.13. Von Kossa staining and immunohistochemistry
The retrieved implants were fixed with 4% PFA, dehydrated by gradient concentrations of ethanol, embedded in paraffin, and cut into histological slices. The histological slices were stained with hematoxylin and eosin and counterstained with Von Kossa. Briefly, after dewaxing and hydration, Mayer's hematoxylin was applied for nuclear staining, and 5% silvernitrate (Sigma-Aldrich) added to each slice. The slices were then exposed to light for at least 30 min. The histological slices were observed using a light microscope (Olympus BX51, Japan).
Immunohistochemistry was performed using the Histostain-SP Kit (Invitrogen). Two osteogenic marker proteins, BSP and OPN, were detected. The stained slides were observed under a microscope (Olympus BX51, Japan). The positive rates were analyzed by an analysis system (Image-Pro Plus 6.0).
2.14. In situ osteoinductivity of the scaffolds
To assess the ability of the CMC/nHA/GO scaffolds to promote in situ bone repair in a critical-size defect, a 5 mm rat cranial defect model was selected. Adult female SD rats (24 total, 300 g) were used in the study and randomly allocated into four groups: (a) no defect as a negative control (n = 6), (b) CMC scaffold with hADSCs (n = 6), (c) CMC/GO scaffold with hADSCs (n = 6), and (d) CMC/nHA/GO scaffold with hADSCs (n = 6) at eight weeks. Ethical approval was given by the Animal Research Committee of the Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. Anesthesia was induced with an intra-peritoneal injection of pentobarbital (Nembutal, 3.5 mg/100 g). Then, 1.0 cm sagittal incisions were made on both sides of the scalp to expose the calvarium. A 5 mm diameter circular transosseous defect was created on both sides of the rat parietal calvarium using a trephine (Nouvag AG, Goldach, Switzerland), appropriate constructs (5 mm × 1.2 mm) were immediately implanted into the defects, and the incisions were closed. All rats survived the surgeries. At 8 weeks post-operation, the rats were euthanized under general anesthesia, and the calvarias were collected and fixed in 4% PFA.
2.15. Micro-CT measurement
Microcomputed tomography (micro-CT) scans were performed on all the fixed samples using a Scanco Medical 80 micro-CT system (1024 pixel matrix; 20 mm voxel size; 20 mm slice thickness) to assess the morphology of the reconstructed calvarias. After the micro-CT scans, a bone visualization was performed using three-dimensional isosurface renderings. Micro-CT was used to determine the percentage of new bone volume relative to the tissue volume (BV/TV) and the bone mineral density (BMD).
2.16. Statistical analysis
All data presented in this study are shown as the mean ± SD unless specifically indicated otherwise. Each experiment was repeated at least three times. Statistical analyses were performed on the data obtained using a one-way analysis of variance. A value of *P < 0.05 was considered to be statistically significant.
3. Results and discussion
3.1. Characterization of the nanoengineered scaffold and mechanical performance evaluation
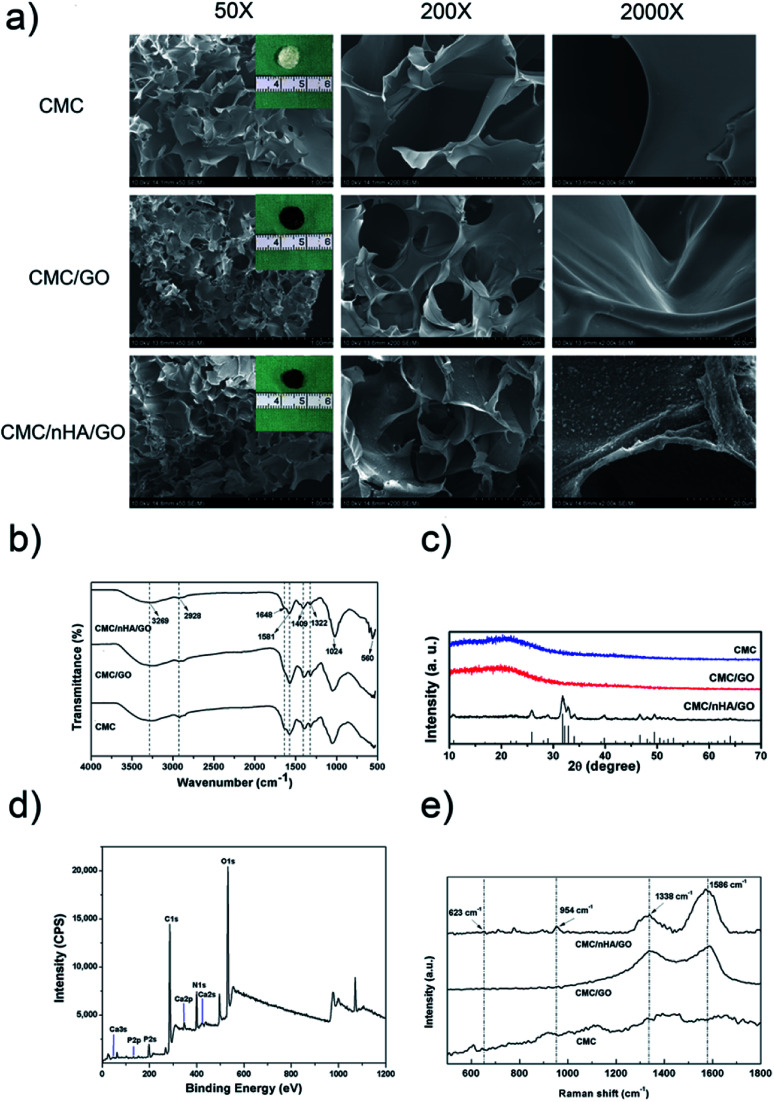
To fabricate a nanocomposite scaffold with the surface chemistry and roughness modifications, a combination of GO and nHA was used to fabricate a chitosan-based scaffold via chemical crosslinking. First, the hydrophilic carboxymethyl chitosan (CMC) was dissolved in a well dispersity of GO solution (ESI Fig. 1†) and formed an electronegative mixture. The nHA was physically absorbed on the surface of GO or CMC to form the nanostructured surface, and after the chemical crosslinking between GO and CMC (ESI Scheme 1†), the porous CMC/nHA/GO scaffold exhibited a rough surface with convex protrusions due to nHA aggregation (Fig. 1a, ESI Fig. 2†). To determine the chemical structure of the scaffold, FTIR was performed, and the spectra showed that the CMC/nHA/GO scaffold had plenty of oxygen-containing groups and phosphate groups (Fig. 1b). The PO43− stretching vibrations at approximately 1024 and 560 cm−1 suggested the presence of an HA phase in the composite scaffolds. The N–H stretching vibration (3269 cm−1) and bending vibration (1581 cm−1), the C O stretching vibration (1648 cm−1), and the C–N stretching vibration (1322 cm−1) proved that the amido bond (–NHCO–) existed in the chemically crosslinked scaffold. The stretching vibrations of the dissociated carboxyl group (–COO–, 1409 cm−1) and methylene (–CH2, 2928 cm−1) indicated that the carboxymethyl groups were in excess in the scaffold. The chemical structure results proved that the scaffold was successfully chemically modified, and the scaffold surface chemical groups play an effective role in inducing cell osteogenic differentiation.
Fig. 1. Characterization of the scaffolds. (a) SEM images of the CMC, CMC/GO and CMC/nHA/GO scaffolds at low (Mag = 50, scale bars: 1 mm), middle (Mag = 200, scale bars: 200 μm) and high (Mag = 2000, scale bars: 20 μm) magnifications and optical images of the CMC, CMC/GO and CMC/nHA/GO scaffolds; (b) FTIR spectra of the scaffolds; (c) XRD spectra of the scaffolds; (d) XPS spectra of the scaffolds; (e) Raman spectra of the scaffolds.
Moreover, the crystallinity degree of the scaffold also influences chemical bioactivity. Cortical bone is a composite material composed of hydroxyapatite (HA) and collagen, and the HA crystals in bone tissue are known to have a low degree of crystallization.37 Research has reported that the low crystallinity in calcium phosphate bone graft substitutes could result in a large amount of poorly crystalline apatite.41 Therefore, we think low crystallinity HA is suitable to be used in scaffolds for bone regeneration. The XRD analysis results demonstrated that the CMC/nHA/GO, CMC/GO and CMC scaffolds all exhibited amorphous structures (Fig. 1c). Notably, the diffraction peak of CMC/nHA/GO at 2θ = 25.9° (002) and 31.9° (211) was consistent with the standard diffraction pattern of nHA (PDF#09-0432), which further confirmed the presence of the crystalline phase nHA in the CMC/nHA/GO scaffold. Additionally, the low significant peaks in the CMC/nHA/GO spectrum also indicated that the incorporated nHA had a low degree of crystallinity. Although highly crystalline nHA facilitates the mechanical performance of scaffolds, nHA with a stable lattice structure lacks bioabsorbability. Low crystallinity nHA is much closer to the nature of primary bone and more easily dissociates into phosphate ions under the physiological conditions to induce the mineralization of new bone.37,42
To further evaluate the chemical composition of CMC/nHA/GO scaffold, XPS spectra was performed, the results in Fig. 1d showed three predominant peaks at 286.1 eV, 399.4 eV, and 532.5 eV, which corresponds to C1s, N1s and O1s respectively. Moreover, the peaks at 132.8 eV and 346.9 eV also revealed the existence of P 2p and Ca 2p. The XPS survey indicates that the synthesized CMC/nHA/GO composite scaffold is consist of C, N, O, Ca and P elements. Meanwhile, Raman spectra was carried out as shown in Fig. 1e, the peaks of 623 and 954 cm−1 represent the stretching and bending vibrations of nHA, and the peaks of 1338 and 1586 cm−1 show the D and G bands of GO. It fully illustrated the composite scaffold is compose of nHA and GO.
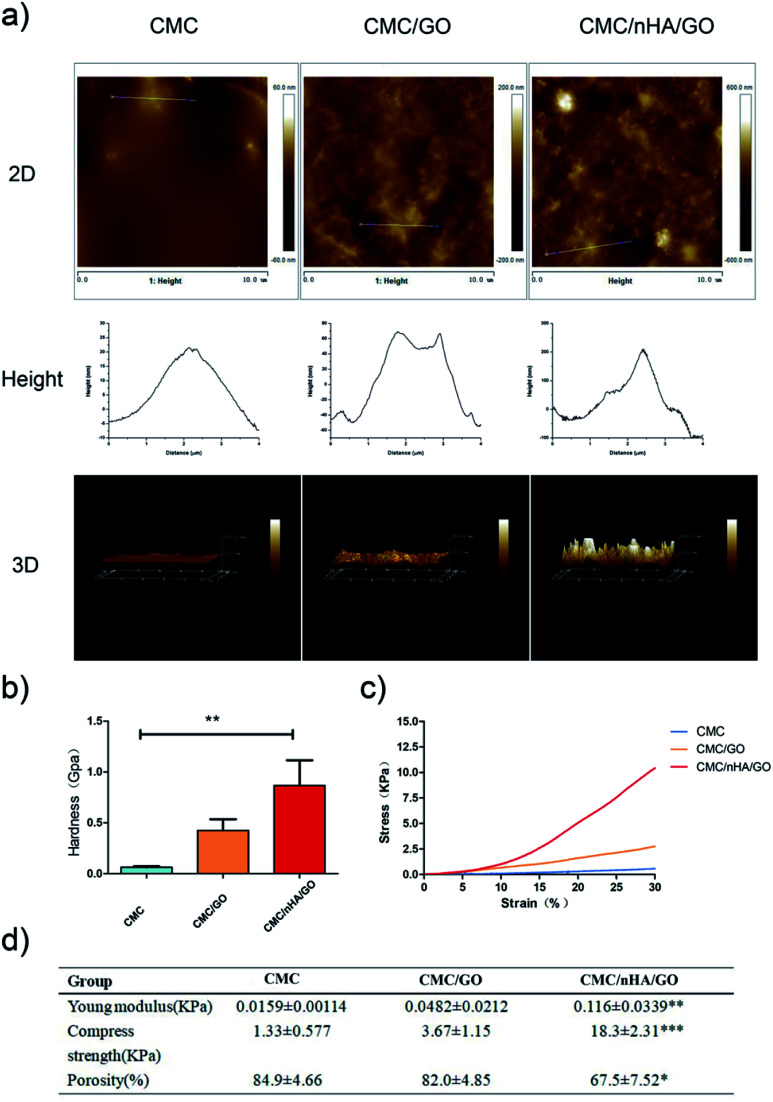
The roughness play an important role on scaffolds physicochemical properties, to quantify the roughness of the scaffold, three types of scaffolds were created and performed atomic force microscopy (AFM). The CMC and CMC/GO scaffolds had relatively smooth surfaces with root mean square (Rq) roughness of 5.7 nm and 25.8 nm. After the scaffold was doped with nHA, the roughness of the CMC/nHA/GO scaffold increased to 74.1 nm (Fig. 2a). To further prove the nHA could enhance the mechanical performance of scaffolds, the stiffness, Young modulus and compress strength of composites scaffold were tested. The hardness of CMC/nHA/GO scaffold was 2.1- and 14.1-fold higher than those of the CMC/GO and CMC scaffolds as shown in Fig. 2b respectively. Analyzing from the strain–stress curves of scaffolds of Fig. 2c, we also concluded that the Young modulus and compress strength of CMC/nHA/GO (0.116 ± 0.0339 kPa Young modulus and 18.3 ± 2.31 kPa compress strength) were higher than those of the CMC/GO (0.0482 ± 0.0212 kPa Young modulus and 3.67 ± 1.15 kPa compress strength) and CMC (0.0159 ± 0.00114 kPa Young modulus and 1.33 ± 0.577 kPa compress strength) scaffolds (Fig. 2d). These results indicated that the CMC/nHA/GO scaffold have significantly higher mechanical strength than CMC/GO and CMC scaffolds. Meanwhile, a certain of porosity is benefit to tissue penetration, we also test the porosity of three kinds of scaffolds, when compare with the porosities of CMC/GO (84.9 ± 4.66%) and CMC scaffolds (82.0 ± 4.85%), the CMC/nHA/GO scaffold have relative low porosity (67.5 ± 7.52%) (Fig. 2d), it mainly due to the nHA's surface occupation and aggregation.
Fig. 2. Mechanical properties of the scaffolds. (a) AFM of the scaffolds; (b) hardness of the scaffolds as determined by nanoindenter testing (**P < 0.01); (c) stress–strain curve of the scaffolds; (d) Young modulus, compress strength and porosity of the scaffolds (*P < 0.05; **P < 0.01; ***P < 0.001).
3.2. Cell adhesion and proliferation on a nanoengineered scaffold
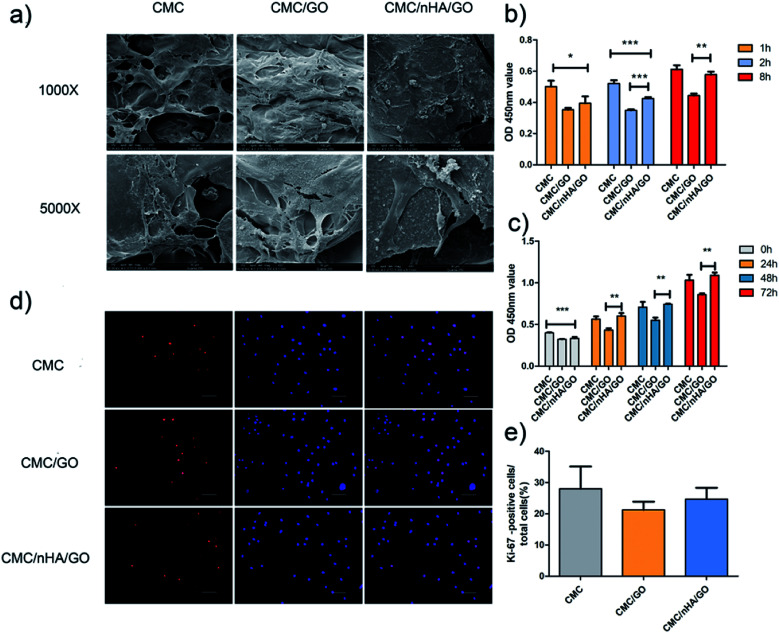
To evaluate the cell morphology on a nanoengineered scaffold, hADSCs were cultured on a scaffold for 7 days in a proliferation medium. The cell composite scaffolds were examined using scanning electric microscopy (SEM) and live/dead staining. The different scaffold roughness all exhibited good cell adhesion ability, and the hADSCs grown on the CMC/nHA/GO scaffold showed more elongated spindle shapes (Fig. 3a). To further confirm the cell viability on the scaffolds, live/dead staining images were obtained, and they demonstrated that there was no difference in the cell viability among three kinds of roughness of the 2D substrates, almost no dead cells, which were indicated by red signals, were observed after 5 days of culture (ESI Fig. 3†). The 3D images of the CMC/nHA/GO scaffold at different degrees (0°, 45° and 90°) were obtained, and the scaffold exhibited a blue color because the CMC biomolecule has strong non-specific absorption ability for the fluorescence dye Hoechst. We clearly saw the live cells with green signals were all attached to the scaffold walls (ESI Fig. 4†), and almost no dead cells were present in the scaffold.
Fig. 3. Effects of the CMC/nHA/GO scaffold on cell adhension and proliferation. (a) Growth and morphology of hADSCs on the CMC, CMC/GO and CMC/nHA/GO scaffolds detected by SEM at low (Mag = 1000, scale bars: 100 μm) and high (Mag = 5000, scale bars: 30 μm) magnification; (b) adhesion rate of hADSCs on substrates for 8 h normalized to the CMC 1 h; (c) proliferation of hADSCs on substrates for 72 h; (d) Ki-67 immunofluorescence staining of cells on substrates for 3 d (scale bars: 20 μm); (e) positive cell ratios of Ki-67 were determined by dividing the number of immune-positive cells to the number of nuclei stained with Hoechst (*P < 0.05, **P < 0.01, ***P < 0.001).
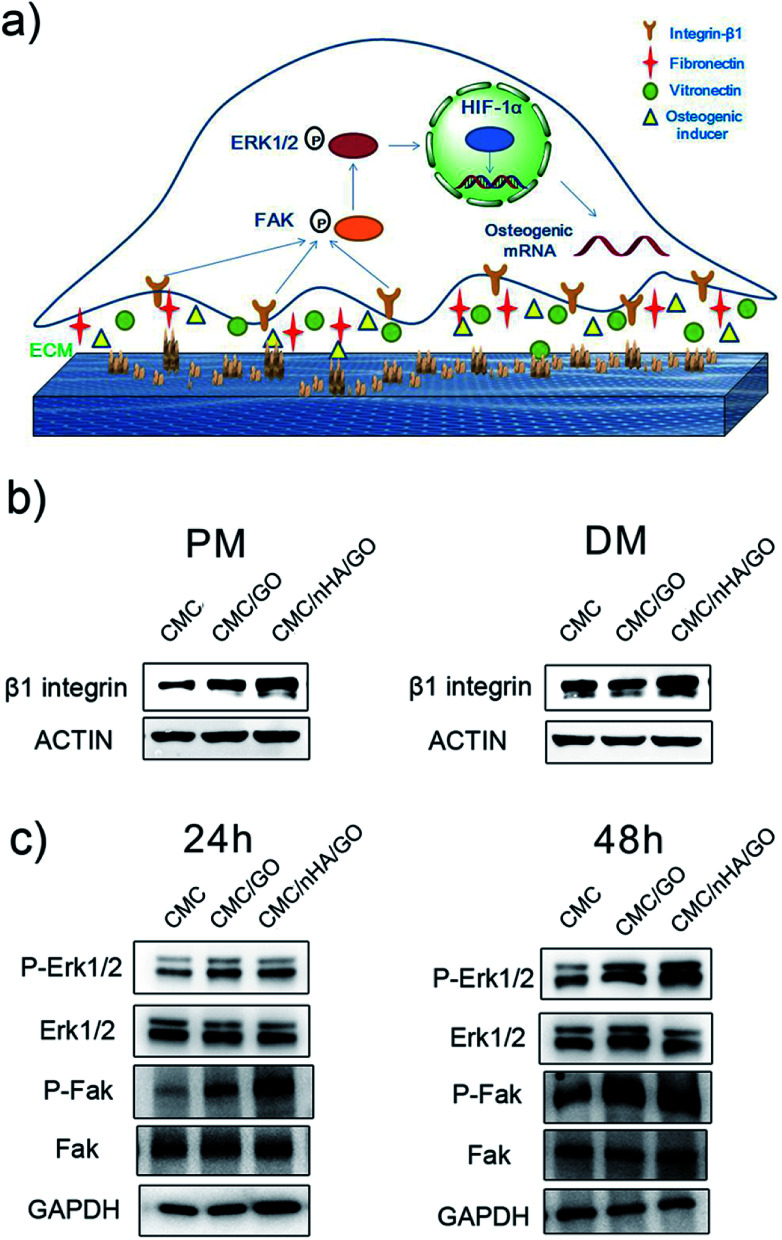
Cell adhesion is an important index to judge the biocompatibility of a scaffold. The cell adhesion results showed that the CMC/nHA/GO groups had a relatively strong adhesive force compare to that of the CMC/GO groups, and the CMC/nHA/GO groups exhibited a high adhesion rate at 8 h (115% of CMC/nHA/GO vs. 80% of CMC/GO) (Fig. 3b). Interestingly, the results from the cell adhesion rate within 8 h showed that the CMC/nHA/GO groups have a relatively good affinity (36% increase from 1 h to 8 h) compared to that of the CMC/GO groups (18% increase from 1 h to 8 h) (Fig. 3b). Based on the above cell adhesion status and morphology, we think that the CMC/nHA/GO scaffold with a rough surface has good adhesion ability. Based on the adhesion results, although the CMC/nHA/GO group has better adhesion than the CMC/GO group, both groups were worse than the CMC group, which mainly contributes to the scaffold surface chemistry. Many studies have reported that GO can cause cell toxicity due to its oxidative stress role.43,44 Although we controlled the amount of GO in the composite scaffold based on our previous experience,21 direct contact with GO will also cause a cell stress reaction and lead to a decrease in the cell adhesion ability.45 After the scaffolds are doped with nHA to modify the surface roughness, the areas of direct contact of the cells with GO is reduced. Therefore, the cell oxidative stress role decreases and the adhesion rate increases. Moreover, recent research has shown that micrometer scale topographical surfaces and nanoscale roughness can directly regulate cellular adhesion through various integrin-mediated signaling mechanisms.46 Therefore, to further confirm the effects of the surface roughness on adhesion, the adhesion protein, β1 integrin, expression level was also determined, as shown in the following (Fig. 6b).
Fig. 6. The molecular mechanism by which CMC/nHA/GO scaffolds promoted osteogenesis. (a) Schematic illustration of the mechanism of CMC/nHA/GO scaffolds promoting osteogenesis; (b) immunoblots showing the up-regulation of β1 integrin in hADSCs on CMC/nHA/GO substrates after incubated in PM and DM for 3 days; (c) immunoblots displaying the phosphorylation of Erk1/2 and FAK in hADSCs on CMC/nHA/GO substrates after incubated in PM for 24 h or 48 h.
For a better understanding of the proliferation of the nanoengineered scaffolds, a cck-8 assay and Ki-67 immunofluorescence staining were completed. The cck-8 assay directly reflects the cell proliferation status on the scaffold at different time points, and the results showed that the proliferation rate on the scaffold with CMC/nHA/GO group was higher than that on the scaffold with CMC/GO group and similar with that of the CMC group scaffold (Fig. 3c). Ki-67 is a cellular proliferation marker that is present during all the active phases of the cell cycle (G1, S, G2, and mitosis),47 and it markedly increases during the S phase of the cell cycle.48 Ki-67 immunofluorescence staining is a gold standard to evaluate cellular proliferation. The Ki-67 staining results after the cells were cultured on a 2D substrate for 48 h showed that the CMC/nHA/GO group exhibited a higher proliferation rate (24.7%) than that of the CMC/GO group (21.2%) (Fig. 3d and e). This result is consistent with the CCK-8 assay results. Referring to the effects of the surface chemistry and roughness on proliferation, the chemistry and roughness are suspected to alter the initial protein interactions and, consequently, mediate the cell proliferation. Some studies have reported that the chemistry and roughness could mimic the nanoscale extracellular matrix properties of the tissue and promote cell attachment and proliferation.49,50 Therefore, based on our proliferation results, we think the nanoengineered CMC/nHA/GO scaffold with surface chemistry and a rough surface possesses good effects for accelerating cell proliferation.
3.3. Synergistic enhancement of osteoinductivity in a scaffold by nanoengineering the surface chemistry and roughness
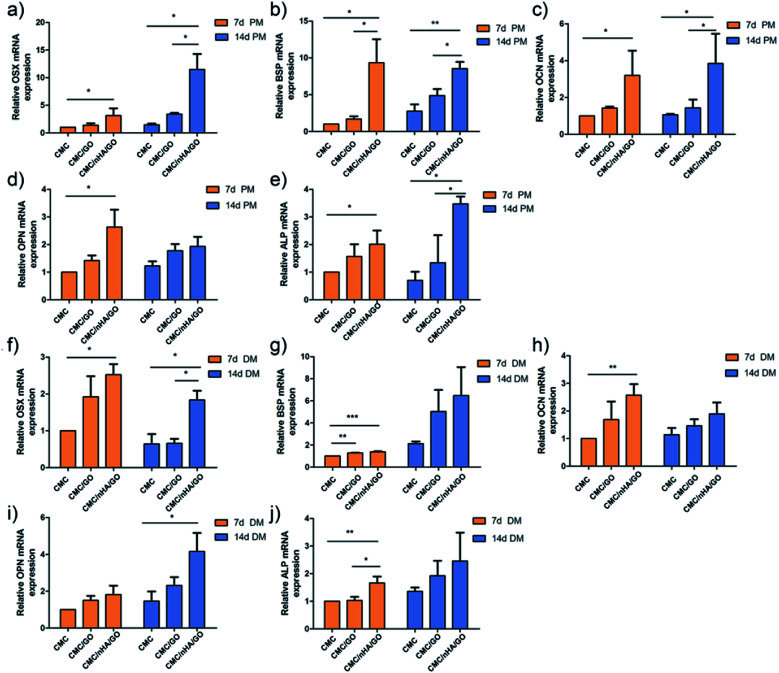
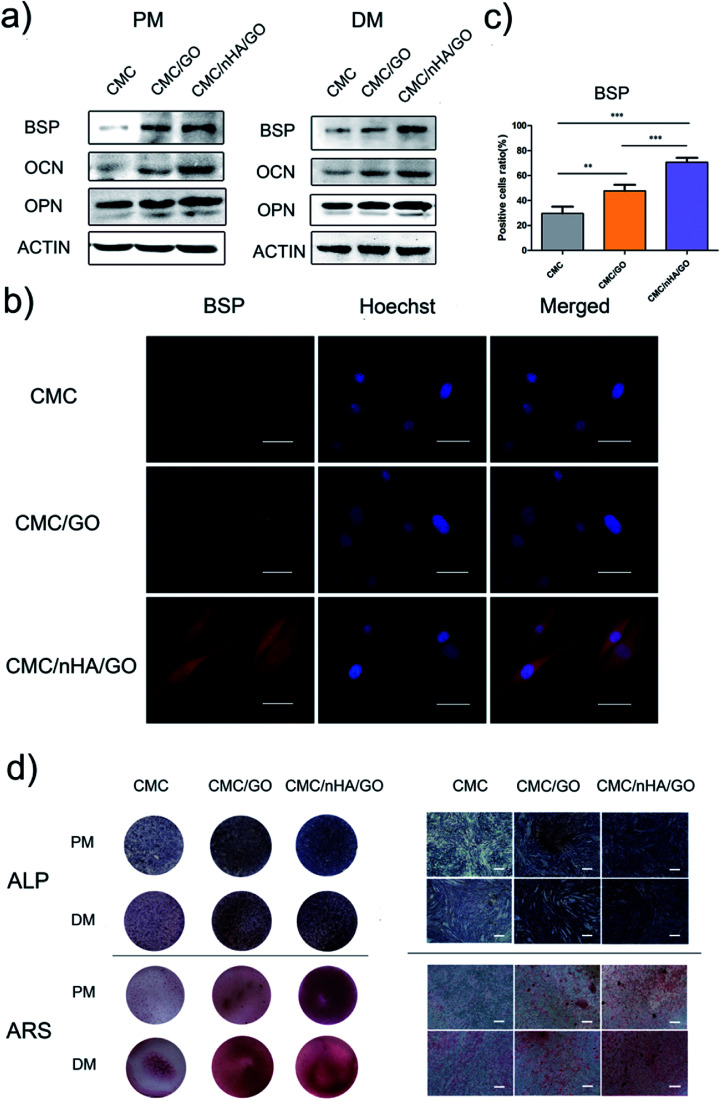
To evaluate the osteoinductivity of the nanoengineered scaffolds, we observed the osteogenic differentiation of hADSCs under different culture conditions, including a proliferation medium (PM) and differentiation medium (DM). The DM was a standard osteogenic-inducing medium containing dexamethasone, β-glycerol phosphate and l-ascorbic acid, and it can regulate stem cell osteogenic differentiation via chemical signal-mediated signaling pathways. To exclude the effects of ectogenic chemicals on cell differentiation, we verified the nanoengineered scaffold osteoinductivity under normal PM culture conditions and compared the results to those from the DM groups. First, using the mRNA level, we detected osteogenic-specific markers, such as osterix (OSX), bonesialoprotein (BSP), osteocalcin (OCN), osteopontin (OPN) and alkaline phosphatase (ALP), after the cells were incubated with the 2D substrates for 7 and 14 days. The gene expression results indicated that the CMC/nHA/GO group scaffold significantly augmented the stem cell osteogenic differentiation, and the gene upregulated levels under the PM conditions were mostly higher than those under the DM conditions (Fig. 4). To further confirm the osteogenic differentiation, the protein levels of BSP, OCN and OPN were determined after 7 days of PM and DM culture. The Western blot results showed that the CMC/nHA/GO group exhibited high protein expression levels under PM and DM conditions (Fig. 5a and b). The CMC/nHA/GO scaffold-induced osteogenic protein relative expression levels in the PM group were better than those in the DM group, and similar expression profiles were obtained for their respective mRNA levels. These results indicated that the CMC/nHA/GO scaffold with surface chemistry and roughness significantly enhanced cell osteogenic differentiation and had a higher inducing level when compare with that of the chemical inducing effects.
Fig. 4. Evaluation of the expression of osteogenesis-related genes of hADSCs on CMC/nHA/GO substrates. The expression of OSX (a), BSP (b), OPN (c), CON (d) and ALP (e) by qPCR after hADSCs were incubated in PM for 7 d or 14 d; the expression of OSX (f), BSP (g), OPN (h), CON (i) and ALP (j) by qPCR after hADSCs were cultured in DM for 7 d or 14 d (*P < 0.05, **P < 0.01, ***P < 0.001).
Fig. 5. Evaluation of the osteogenic differentiation of hADSCs on CMC/nHA/GO substrates. (a) The protein expression of osteogenic markers of hADSCs incubated in PM or DM for 7 d; (b) BSP immunofluorescence staining of hADSCs on substrates incubated in PM for 7 d (scale bars: 50 μm); (c) positive cell ratios of BSP were determined by dividing the number of immune-positive cells to the number of nuclei stained with Hoechst (*P < 0.05, **P < 0.01, ***P < 0.001); (d) light microscopy and optical images of ALP staining of hADSCs on substrates incubated in PM or DM for 7 d and ARS staining of hADSCs on substrates incubated in PM or DM for 14 d (scale bars: 200 μm).
We think the surface chemistry and physical effects synergistically enhance the scaffold osteoinductivity. For the CMC/nHA/GO scaffold, its surface was modified with oxygen-containing functional groups by incorporating GO and roughened with some micro-island structures via doping with nHA. In regards to the chemistry-inducing effects, some researchers have reported that a GO-based scaffold can induce stem cell differentiation via the π–π stacking interactions of the sp2-bonded carbon atoms in a single two-dimensional layer with the aromatic rings of the inducer molecules.21,51 Additionally, nHA has ion inducing effects. Low crystallinity nHA can release phosphate and calcium ions in a physiological environment, and these ions will take part in ossification. The mechanism has two parts. In one, phosphate ions induce the ATP pathway, causing adenosine metabolism and promoting cell osteogenic differentiation.52 In the other, dissociated phosphate ions absorb calcium ions and aggregate to accelerate inorganic mineral in situ deposition and the formation of new bone tissue.53 Recently, a new study reported that nHA could significantly influence stem cells osteogenic differentiation by stimulating DNA methylation.54 Considering the above studies, we think the properties of nHA can significantly influence cell osteogenic differentiation. From the nHA crystallinity degree analysis results (Fig. 1c) and the released ions curve results (ESI Fig. 5†), we can conclude that the doped nHA in the CMC/nHA/GO scaffold has a low crystallinity, and the phosphate ions had a sustained release (accumulated 2% release rate within 21 days) and participate in the ossification period. The fast calcium ion release rate (accumulated 12% release rate within 21 days) implied an early ossification process improved with the amount of calcium ion participation. The early ossification with the CMC/nHA/GO scaffold was evaluated by performing ALP and ARS staining after culturing hADSCs for 7 or 14 days, and the results showed that the number of calcium mineral nodules deposited by the hADSCs on the CMC/nHA/GO scaffold was more than that on the CMC/GO and CMC scaffold. The CMC/nHA/GO group exhibited greater mineral formation under the PM conditions than under the DM conditions (Fig. 5d). These staining results fully imply that the CMC/nHA/GO scaffold can improve the stem cell osteogenic differentiation under non-chemical inducer effects. We think that the scaffold surface chemistry effects play a main role in inducing stem cell differentiation.
Another reason for the improvement in the osteoinductivity of scaffold is that the scaffold physical characteristics, including the roughness, topography and nanostructure, play an important role in influencing the stem cell differentiation status. For instance, single-crystal apatite nanowires sheathed in graphitic shells can lead to excellent osteogenic differentiation and bony fusion through a programmed smart behavior.55 A surface with nanostructured strontium carbonate biomorphic crystals can enhance osteoblast adhesion and differentiation by mimicking nanotopographical features similar to cellular filopodia.56 We speculated that the nHA-doped CMC/nHA/GO scaffold with a surface roughness and micro-island structures could activate cytoskeletal signaling cascades effects and induce stem cell differentiation. The hADSCs immunocytochemistry results shown that the BSP positive rate for the CMC/nHA/GO group (70.7% positive rate) was higher than that of the CMC/GO (47.7% positive rate) and CMC (29.6% positive rate) groups under the PM conditions (Fig. 5b and c). Moreover, when compared with the CMC/GO group, the CMC/nHA/GO group under the PM conditions had more remarkable statistical significance (***P < 0.001) than that under the DM conditions (*P < 0.05) (Fig. 5c, ESI Fig. 6†). We also observed that the expression levels of β1 integrin in the CMC/nHA/GO substrate were higher than those in the CMC/GO and CMC substrates (Fig. 6b). Based on these results, we proposed that the rough surface structures could modulate the adhesion protein expression and induce the downstream signal molecule cascade reaction, enhancing the stem cell osteogenic differentiation.
3.4. The nanoengineered scaffold mechanism for enhancing cell osteogenic differentiation
To interpret the nanoengineering scaffold mechanism of osteoinductivity, efforts have focused on focal adhesion interactions and the resulting changes in intracellular tension.57,58 Herein, to investigate the osteogenic induction mechanism of the CMC/nHA/GO scaffold, we first performed a Western blot to detect the expression levels of β1 integrin, which serves as the predominant mediator of cell adhesion and plays a crucial role in osteogenesis.59–61 The results revealed that the expression of β1 integrin in the CMC/nHA/GO group was higher than that in the CMC/GO and CMC groups under the PM conditions, and similar results were observed under the DM conditions (Fig. 5b). These results suggested that the hADSCs have a better communication with the CMC/nHA/GO substrate, which means better interaction with the biophysical microenvironment such as more absorption of osteogenic inducer embedded in the ECM and tighter contact with the surface roughness and varying nanotopographical features.
As studies have reported, many β1 integrin-mediated cellular activities are involved in the activation of the intracellular signaling pathways associated with nonreceptor tyrosine kinases,62 most notably focal adhesion kinase (FAK), which is associated with the β-integrin subunit.63,64 FAK localizes at focal adhesions and can influence cellular transcriptional events by means of adhesion-dependent phosphorylation of downstream signaling molecules to regulate essential cellular processes, such as survival, growth, migration, and differentiation.65,66 Many reports have shown that FAK is activated in response to both the ECM and soluble signaling factors, suggesting that the FAK family maybe at the crossroads of multiple signaling pathways that affect pivotal cell processes. Therefore, the transmission of adhesion-dependent signaling by FAK is of particular importance on topographic microsurfaces.67 In our study, the Western blot results revealed that the CMC/nHA/GO scaffold with surface roughness and chemistry facilitated the phosphorylation of FAK in the hADSCs at 24 h, and a similar trend was observed at 48 h (Fig. 6c).
The FAK-mediated signaling pathways involve several downstream molecules, including extracellular signal-regulated kinase (ERK) in the mitogen-activated protein kinase (MAPK) pathway.68 Recently, GO was reported to promote VEGF expression in stem cells by phosphorylating ERK1/2 and then upregulating HIF-1α via the β1integrin/FAK signaling pathway.69 Enhanced phosphorylation of ERK1/2 in the hADSCs was observed in the CMC/nHA/GO group scaffold at both 24 h and 48 h (Fig. 6c), which is in agreement with related studies.70 On the basis of the above results, the osteoinductivity of the CMC/nHA/GO scaffold was concluded to be beneficial to cell-surface roughness interaction and β1 integrin combination with the bioactive constituents such as fibronectin, laminin, collagen embedded in the ECM, osteogenic inducer adsorbed by the graphene oxide and Ca2+, PO43− ions released by the nanohydroxyapatite, and then the activation of the phosphorylation of FAK. This leads to the phosphorylation and activation of ERK1/2, then increasing the related transcription factor levels, at last upregulating the expression of osteogenic special genes, including OPN, BSP and OCN (Fig. 4). Above all, β1 integrin provides a key connection between the cultured hADSCs and the nanoengineered scaffold, and its activation of the FAK–ERK signaling pathway demonstrate that integrins can serve as a master regulator to influence the osteogenesis of hADSCs on CMC/nHA/GO scaffold (Fig. 6a).
3.5. In vivo evaluation of the nanoengineered scaffold on new bone formation
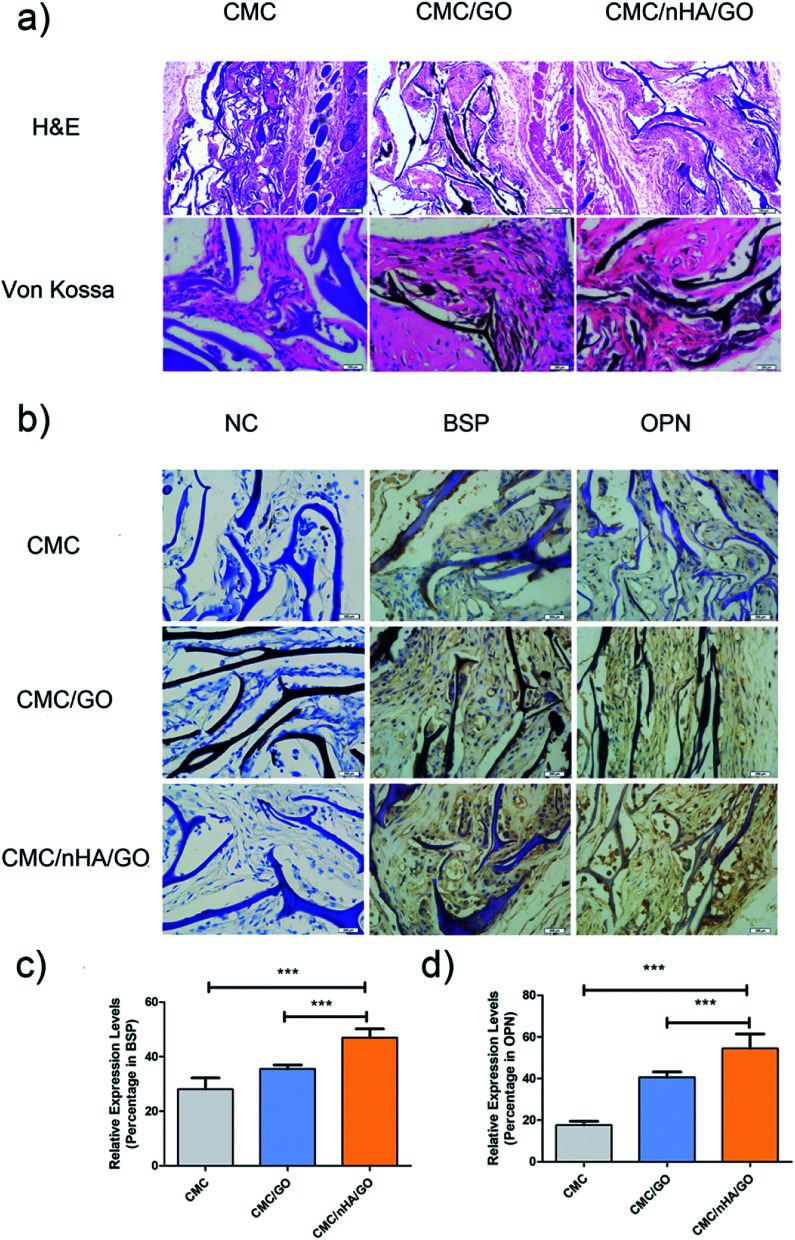
To evaluate the bone regenerative capacity of the nanoengineered scaffold, ectopic bone formation in nude mice was analyzed. Significant osseous features after 8 weeks of implantation were observed in the Von Kossa staining, which reflected the ossification ability of the scaffold. The CMC/nHA/GO and CMC/GO scaffold showed abundant deposits of mineralized bone matrix diffusely distributed while the CMC scaffold had tiny black deposits (Fig. 7a). Moreover, from the histological analysis results, the CMC/nHA/GO implant exhibited no sign of infection or complication, and it had more new tissue mass and fibrous encapsulation across the implant architecture than the CMC/GO and CMC scaffold. The results implied that the CMC/nHA/GO scaffold had a good biocompatibility. Furthermore, to further evaluate the expression levels of osteogenic proteins, such as BSP and OPN in the implants, immunohistochemistry was performed. The brown areas, which indicated the positive expression of BSP and OPN, were observed in the CMC/nHA/GO and CMC/GO group, and no obvious positive staining was observed in the CMC group (Fig. 7b). The quantitative analysis of the BSP expression showed that the CMC/nHA/GO group (46.9%) has a significantly higher positive ratio than the CMC/GO (35.5%) and CMC groups (28.0%) (Fig. 7c). Similar results were observed for the OPN expression of the CMC/nHA/GO (54.5%), CMC/GO (40.5%) and CMC groups (17.7%) after 8 weeks of implantation (Fig. 7d). These results clearly illustrate that the CMC/nHA/GO scaffold further enhanced the osteoinductivity of the CMC/GO scaffold.
Fig. 7. Histological analysis and immunohistochemical analysis of CMC/nHA/GO scaffolds postimplantation. (a) HE staining images of hADSCs/scaffold complexes after implantation for 8 weeks; Von Kossa staining of cells/scaffold complexes at 8 weeks postimplantation; dark deposits indicated the calcium deposition (scale bars: 100 μm); (b) immunohistochemical analysis of osteogenic differentiation in cells/scaffold complexes after being implanted for 8 weeks. BSP and OPN staining demonstrated positive brown staining in the tissue (scale bars: 200 μm); positive expression ratios of BSP (c) and OPN (d) in CMC/nHA/GO, CMC/GO and CMC groups at 8 weeks postimplantation (*P < 0.05, **P < 0.01, ***P < 0.001).
To better evaluate the osteoinductivity of the CMC/nHA/GO scaffold, critical-size calvarial defect repairing experiments were performed in rats and were characterized by micro-CT. hADSCs were seeded onto the scaffolds in proliferation medium for days, before we implanted the scaffold, the viability of the hADSCs on the CMC/nHA/GO scaffold was evaluated by performing live/dead staining (ESI Fig. 3†). Confocal microscope imaging showed that hADSCs attached well to the surface and grew vigorously on the CMC/nHA/GO scaffold, thereby suggesting the CMC/nHA/GO scaffold provided a suitable microenvironment for hADSCs. Bone growth was evaluated 8 weeks after implantation of the three types of cell composite scaffolds, and defects that were left untreated without scaffolds were used as the negative controls. Representative images of each group are shown in Fig. 8a.
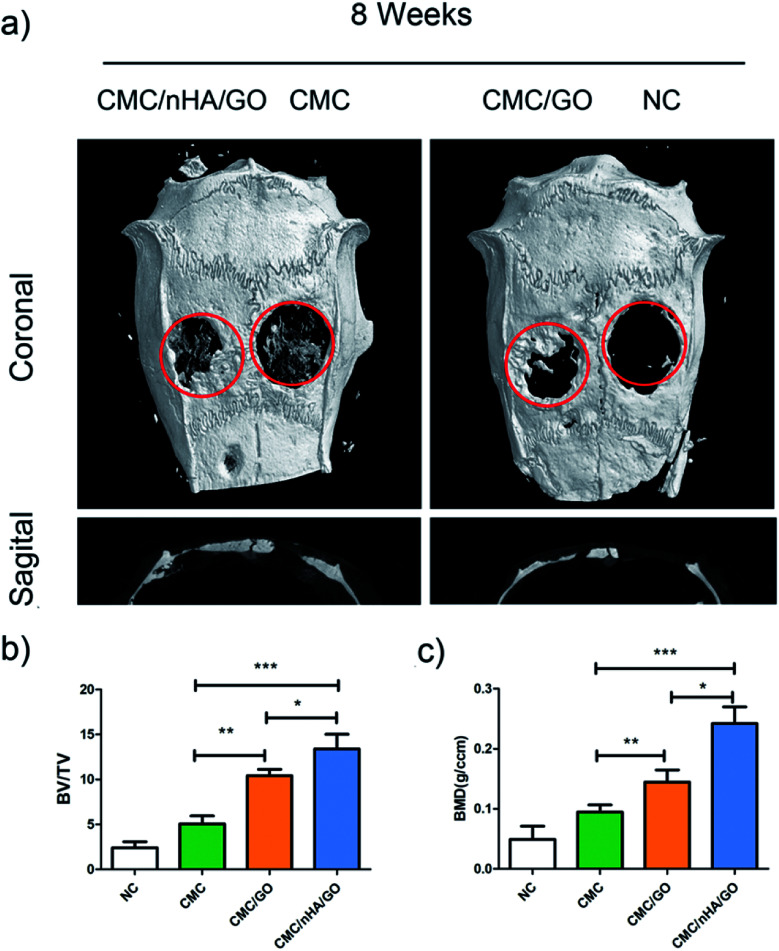
Fig. 8. Micro-CT imaging analysis. (a) Representative coronal and sagittal images of calvarial bone defects after 8 weeks implantation; (b) bone volume to tissue volume (BV/TV); (c) bone mineral density (BMD) morphometric analysis (*P < 0.05, **P < 0.01, ***P < 0.001).
The micro-CT images showed that new bone tissue formed in the margins of the defects, and extended across the scaffolds in the CMC/nHA/GO and CMC/GO groups. There was also a little sporadic bone tissue in the defect in the CMC/nHA/GO group. In the negative control group, there was almost no new bone tissue formation, and only a small amount of new bone tissue showed a diffuse distribution in the center of the defect in the CMC group. The quantitative micro-CT analysis results showed that the CMC/nHA/GO group (13.4%) exhibit a significantly higher ratio of new bone volume to tissue volume (BV/TV) compared to the ratio of the CMC/GO (10.4%), CMC (5.0%) and NC (2.4%) groups (Fig. 8b). Similar results were found in the bone mineral density (BMD) analysis, the BMD in the CMC/nHA/GO group (0.242 g cm−3) was markedly greater than that in the CMC/GO (0.144 g cm−3), CMC (0.0945 g cm−3) and NC (0.0490 g cm−3) groups (Fig. 8c). These results indicated that the CMC/nHA/GO scaffold exhibited a stronger in vivo bone regenerationability than that of the CMC/GO and CMC scaffold. Above all, our data demonstrated that the CMC/nHA/GO scaffold significantly enhanced in vivo bone regeneration by hADSCs in accordance with our in vitro analysis.
4. Conclusions
To summarize, we fabricated a bioactive scaffold through modification of the surface chemistry and roughness by doping nanomaterials of GO and nHA. The nanoengineered CMC/nHA/GO scaffold was enriched in oxygen-containing groups and micro-island structures; under the dual effects of the surface chemistry and roughness, the nanoengineered scaffold had improved cell adhesion and proliferation abilities and enhanced osteoinductivity compared with that of the unique chemistry effect of the CMC/GO scaffold. Moreover, the bioscaffold sustained the release of phosphate and calcium ions, which synergistically assists the ECM ossification and promotes new bone regeneration. Furthermore, through investigating the primary osteogenic mechanism, we speculated that the nanoengineered scaffold could activate the cytoskeletal FAK–ERK signaling cascade effects and induce osteogenesis. Together with the advantages of the surface chemistry and roughness, this might provide an innovative strategy for developing bioactive bone substitutes. The nanoengineered scaffold has promising potential for applications in bone tissue engineering.
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
Z. Y. and C. X. contributed equally to this work. We are gratefully to Prof. Daxiang Cui, Guangxia Shen and Chunlei Zhang for providing testing instruments and assisting us to perform animal experiments. The work was supported by the National High Technology Research and Development Program (863 Program) (2015AA020311), National Natural Science Foundation of China (Grant No. 81601623, 81320108010, 81470663, 81470662, 81500757); The Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2016MS03); Shanghai Rising-Star Program (17QA1402000) and the Science and Technology Commission of Shanghai (17DZ2260100). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00383a
References
- Lv L. Liu Y. Zhang P. Zhang X. Liu J. Chen T. Su P. Li H. Zhou Y. Biomaterials. 2015;39:193–205. doi: 10.1016/j.biomaterials.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Pan T. Song W. Gao H. Li T. Cao X. Zhong S. Wang Y. ACS Appl. Mater. Interfaces. 2016;8:19217–19227. doi: 10.1021/acsami.6b02969. [DOI] [PubMed] [Google Scholar]
- Tang W. Lin D. Yu Y. Niu H. Guo H. Yuan Y. Liu C. Acta Biomater. 2016;32:309–323. doi: 10.1016/j.actbio.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Shah N. J. Hyder M. N. Quadir M. A. Dorval Courchesne N. M. Seeherman H. J. Nevins M. Spector M. Hammond P. T. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12847–12852. doi: 10.1073/pnas.1408035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. Zhou H. Zou D. Xie Q. Bi X. Gu P. Fan X. Biomaterials. 2013;34:6717–6728. doi: 10.1016/j.biomaterials.2013.05.042. [DOI] [PubMed] [Google Scholar]
- Liao Y. H. Chang Y. H. Sung L. Y. Li K. C. Yeh C. L. Yen T. C. Hwang S. M. Lin K. J. Hu Y. C. Biomaterials. 2014;35:4901–4910. doi: 10.1016/j.biomaterials.2014.02.055. [DOI] [PubMed] [Google Scholar]
- Genchi G. G. Marino A. Grillone A. Pezzini I. Ciofani G. Adv. Healthcare Mater. 2017;6(9):1700002. doi: 10.1002/adhm.201700002. [DOI] [PubMed] [Google Scholar]
- Krishna L. Dhamodaran K. Jayadev C. Chatterjee K. Shetty R. Khora S. S. Das D. Stem Cell Res. Ther. 2016;7:188. doi: 10.1186/s13287-016-0440-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini F. Prasetyanto E. A. Taraballi F. Pandolfi L. Monroy F. Lopez-Montero I. Tasciotti E. De Cola L. Small. 2016;12:4881–4893. doi: 10.1002/smll.201601017. [DOI] [PubMed] [Google Scholar]
- Gong T. Xie J. Liao J. Zhang T. Lin S. Lin Y. Bone Res. 2015;3:15029. doi: 10.1038/boneres.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. Lin S. Li Y. Zhang T. Shao X. Tian T. Zhou T. Li Q. Lin Y. Chem. Commun. 2018;54:1327–1330. doi: 10.1039/C7CC09397G. [DOI] [PubMed] [Google Scholar]
- Liao J. Tian T. Shi S. Xie X. Ma Q. Li G. Lin Y. Bone Res. 2017;5:17018. doi: 10.1038/boneres.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T. Gong T. Xie J. Lin S. Liu Y. Zhou T. Lin Y. ACS Appl. Mater. Interfaces. 2016;8:22884–22891. doi: 10.1021/acsami.6b07097. [DOI] [PubMed] [Google Scholar]
- Dong M. Jiao G. Liu H. Wu W. Li S. Wang Q. Xu D. Li X. Liu H. Chen Y. Biol. Trace Elem. Res. 2016;173:306–315. doi: 10.1007/s12011-016-0686-3. [DOI] [PubMed] [Google Scholar]
- Nardecchia S. Serrano M. C. Gutiérrez M. C. Portolés M. T. Ferrer M. L. Monte F. D. Adv. Funct. Mater. 2012;22:4411–4420. doi: 10.1002/adfm.201200684. [DOI] [Google Scholar]
- Jakus A. E. Secor E. B. Rutz A. L. Jordan S. W. Hersam M. C. Shah R. N. ACS Nano. 2015;9:4636–4648. doi: 10.1021/acsnano.5b01179. [DOI] [PubMed] [Google Scholar]
- Tian T. Liao J. Zhou T. Lin S. Zhang T. Shi S. R. Cai X. Lin Y. ACS Appl. Mater. Interfaces. 2017;9:30437–30447. doi: 10.1021/acsami.7b09176. [DOI] [PubMed] [Google Scholar]
- Yu P. Bao R.-Y. Shi X.-J. Yang W. Yang M.-B. Carbohydr. Polym. 2017;155:507–515. doi: 10.1016/j.carbpol.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Zhou T. Li G. Lin S. Tian T. Ma Q. Zhang Q. Shi S. Xue C. Ma W. Cai X. Lin Y. ACS Appl. Mater. Interfaces. 2017;9:42589–42600. doi: 10.1021/acsami.7b14267. [DOI] [PubMed] [Google Scholar]
- Singh A. V. Mehta K. K. Worley K. Dordick J. S. Kane R. S. Wan L. Q. ACS Nano. 2014;8:2196–2205. doi: 10.1021/nn405253d. [DOI] [PubMed] [Google Scholar]
- Ruan J. Wang X. Yu Z. Wang Z. Xie Q. Zhang D. Huang Y. Zhou H. Bi X. Xiao C. Adv. Funct. Mater. 2016;26:1085–1097. doi: 10.1002/adfm.201504141. [DOI] [Google Scholar]
- Zhang Q. Lin S. Shi S. Zhang T. Ma Q. Tian T. Zhou T. Cai X. Lin Y. ACS Appl. Mater. Interfaces. 2018;10:3421–3430. doi: 10.1021/acsami.7b17928. [DOI] [PubMed] [Google Scholar]
- Jeong K. J. Song Y. Shin H. R. Kim J. E. Kim J. Sun F. Hwang D. Y. Lee J. J. Biomed. Mater. Res., Part A. 2017;105:1637–1645. doi: 10.1002/jbm.a.35993. [DOI] [PubMed] [Google Scholar]
- Das R. K. Zouani O. F. Labrugere C. Oda R. Durrieu M. C. ACS Nano. 2013;7:3351–3361. doi: 10.1021/nn4001325. [DOI] [PubMed] [Google Scholar]
- Jeon O. Alsberg E. Adv. Funct. Mater. 2013;23:4765–4775. doi: 10.1002/adfm.201300529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C. Piemontese M. Lumetti S. Ravanetti F. Macaluso G. M. Passeri G. Acta Biomater. 2012;8:2963–2968. doi: 10.1016/j.actbio.2012.04.043. [DOI] [PubMed] [Google Scholar]
- Ozdemir T. Xu L. C. Siedlecki C. Brown J. L. Integr. Biol. 2013;5:1407–1416. doi: 10.1039/C3IB40068A. [DOI] [PubMed] [Google Scholar]
- Shi M. Chen Z. Farnaghi S. Friis T. Mao X. Xiao Y. Wu C. Acta Biomater. 2016;30:334–344. doi: 10.1016/j.actbio.2015.11.033. [DOI] [PubMed] [Google Scholar]
- Cai X. Xie J. Yao Y. Cun X. Lin S. Tian T. Zhu B. Lin Y. Bone Res. 2017;5:17048. doi: 10.1038/boneres.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Favi P. Cheng X. Golshan N. H. Ziemer K. S. Keidar M. Webster T. J. Acta Biomater. 2016;46:256–265. doi: 10.1016/j.actbio.2016.09.030. [DOI] [PubMed] [Google Scholar]
- Jung H. Hwang D. Kim E. Kim B. J. Lee W. B. Poelma J. E. Kim J. Hawker C. J. Huh J. Ryu du Y. Bang J. ACS Nano. 2011;5:6164–6173. doi: 10.1021/nn2006943. [DOI] [PubMed] [Google Scholar]
- Liu X. Carbonell C. Braunschweig A. B. Chem. Soc. Rev. 2016;45:6289–6310. doi: 10.1039/C6CS00349D. [DOI] [PubMed] [Google Scholar]
- Surwade S. P. Zhou F. Li Z. Powell A. O'Donnell C. Liu H. Chem. Commun. 2016;52:1677–1680. doi: 10.1039/C5CC08183A. [DOI] [PubMed] [Google Scholar]
- Ohji T. Fukushima M. Int. Mater. Rev. 2013;57:115–131. doi: 10.1179/1743280411Y.0000000006. [DOI] [Google Scholar]
- Shakir M. Jolly R. Khan M. S. Rauf A. Kazmi S. Int. J. Biol. Macromol. 2016;93:276–289. doi: 10.1016/j.ijbiomac.2016.08.046. [DOI] [PubMed] [Google Scholar]
- Cunniffe G. M. Curtin C. M. Thompson E. M. Dickson G. R. O'Brien F. J. ACS Appl. Mater. Interfaces. 2016;8:23477–23488. doi: 10.1021/acsami.6b06596. [DOI] [PubMed] [Google Scholar]
- Fujisaki K. Tadano S. Journal of biomechanics. 2007;40:1832–1838. doi: 10.1016/j.jbiomech.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Regnault W. F. Icenogle T. B. Antonucci J. M. Skrtic D. J. Mater. Sci.: Mater. Med. 2008;19:507–515. doi: 10.1007/s10856-007-3178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes B. T. Diekman B. O. Gimble J. M. Guilak F. Nat. Protoc. 2010;5:1294–1311. doi: 10.1038/nprot.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. Wang Z. Bi X. Zhou H. Wang Y. Gu P. Fan X. Biochem. Biophys. Res. Commun. 2014;446:98–104. doi: 10.1016/j.bbrc.2014.02.058. [DOI] [PubMed] [Google Scholar]
- Hu M. H. Lee P. Y. Chen W. C. Hu J. J. Mater. Sci. Eng., C. 2014;45:82–88. doi: 10.1016/j.msec.2014.08.065. [DOI] [PubMed] [Google Scholar]
- Fan C. Li J. Xu G. He H. Ye X. Chen Y. Sheng X. Fu J. He D. J. Mater. Sci. 2010;45:5814–5819. doi: 10.1007/s10853-010-4656-4. [DOI] [Google Scholar]
- Zhang W. Yan L. Li M. Zhao R. Yang X. Ji T. Gu Z. Yin J. J. Gao X. Nie G. Toxicol. Lett. 2015;237:61–71. doi: 10.1016/j.toxlet.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Wang K. Ruan J. Song H. Zhang J. Wo Y. Guo S. Cui D. Nanoscale Res. Lett. 2011;6:8. doi: 10.1007/s11671-010-9751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C. Wakefield D. L. Han Y. Muller D. A. Holowka D. A. Baird B. A. Dichtel W. R. Chem. 2016;1:273–286. doi: 10.1016/j.chempr.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Wang C. Li Z. Lu Z. Li Y. Yin J. J. Zhou Y. T. Gao X. Fang Y. Nie G. Zhao Y. Adv. Mater. 2012;24:5391–5397. doi: 10.1002/adma.201202678. [DOI] [PubMed] [Google Scholar]
- Bruno S. Darzynkiewicz Z. Cell Proliferation. 1992;25:31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z. Zhao H. Zhang S. Lee M. Y. Lee E. Y. Zhang Z. Oncotarget. 2015;6:11735–11750. doi: 10.18632/oncotarget.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Bachhuka A. Han S. Wei F. Lu S. Visalakshan R. M. Vasilev K. Xiao Y. ACS Nano. 2017;11:4494–4506. doi: 10.1021/acsnano.6b07808. [DOI] [PubMed] [Google Scholar]
- Liu H. W. Huang W. C. Chiang C. S. Hu S. H. Liao C. H. Chen Y. Y. Chen S. Y. Adv. Funct. Mater. 2014;24:3715–3724. doi: 10.1002/adfm.201303853. [DOI] [Google Scholar]
- Luo Y. Shen H. Fang Y. Cao Y. Huang J. Zhang M. Dai J. Shi X. Zhang Z. ACS Appl. Mater. Interfaces. 2015;7:6331–6339. doi: 10.1021/acsami.5b00862. [DOI] [PubMed] [Google Scholar]
- Shih Y. R. Hwang Y. Phadke A. Kang H. Hwang N. S. Caro E. J. Nguyen S. Siu M. Theodorakis E. A. Gianneschi N. C. Vecchio K. S. Chien S. Lee O. K. Varghese S. Proc. Natl. Acad. Sci. U. S. A. 2014;111:990–995. doi: 10.1073/pnas.1321717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn D. ChemPhysChem. 2015;16:2069–2075. doi: 10.1002/cphc.201500231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S. W. Jang H. L. Nam K. T. Beck, Jr G. R. Biomaterials. 2015;65:32–42. doi: 10.1016/j.biomaterials.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong N. Cha M. Park Y. C. Lee K. M. Lee J. H. Park B. C. Lee J. ACS Nano. 2013;7:5711–5723. doi: 10.1021/nn305767t. [DOI] [PubMed] [Google Scholar]
- Wang G. Zhao X. Moller M. Moya S. E. ACS Appl. Mater. Interfaces. 2015;7:23412–23417. doi: 10.1021/acsami.5b08493. [DOI] [PubMed] [Google Scholar]
- Biggs M. J. Richards R. G. Dalby M. J. Nanomedicine. 2010;6:619–633. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam L. R. Massich M. D. Hao L. Shin Wong L. Mader C. C. Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4377–4382. doi: 10.1073/pnas.1201086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. Aslani A. Sambito M. A. Kalkhoran N. M. Slamovich E. B. Webster T. J. J. Biomed. Mater. Res., Part A. 2008;84:265–272. doi: 10.1002/jbm.a.31469. [DOI] [PubMed] [Google Scholar]
- Saidak Z. Le Henaff C. Azzi S. Marty C. Da Nascimento S. Sonnet P. Marie P. J. J. Biol. Chem. 2015;290:6903–6912. doi: 10.1074/jbc.M114.621219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris F. Hammar E. Bosco D. Pilloud C. Maedler K. Donath M. Y. Oberholzer J. Zeender E. Morel P. Rouiller D. Halban P. A. Diabetologia. 2002;45:841–850. doi: 10.1007/s00125-002-0840-7. [DOI] [PubMed] [Google Scholar]
- Schaller M. D. Borgman C. A. Cobb B. S. Vines R. R. Reynolds A. B. Parsons J. T. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5192–5196. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch S. M. Vuori K. Ruoslahti E. Chan-Hui P. Y. J. Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. C. Guan J. L. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10148–10152. doi: 10.1073/pnas.91.21.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurenova E. Xu L. H. Yang X. Baldwin Jr. A. S. Craven R. J. Hanks S. K. Liu Z. G. Cance W. G. Mol. Cell. Biol. 2004;24:4361–4371. doi: 10.1128/MCB.24.10.4361-4371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem S. Li J. Yee S. P. Fellows G. F. Goodyer C. G. Wang R. J. Pathol. 2009;219:182–192. doi: 10.1002/path.2577. [DOI] [PubMed] [Google Scholar]
- Reddig P. J. Juliano R. L. Cancer Metastasis Rev. 2005;24:425–439. doi: 10.1007/s10555-005-5134-3. [DOI] [PubMed] [Google Scholar]
- Westhoff M. A. Serrels B. Fincham V. J. Frame M. C. Carragher N. O. Mol. Cell. Biol. 2004;24:8113–8133. doi: 10.1128/MCB.24.18.8113-8133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Chang Q. Xu L. Li G. Yang G. Ding X. Wang X. Cui D. Jiang X. Adv. Healthcare Mater. 2016;5:1299–1309. doi: 10.1002/adhm.201500824. [DOI] [PubMed] [Google Scholar]
- Zhou C. Zhang X. Xu L. Wu T. Cui L. Xu D. Amino Acids. 2014;46:1673–1680. doi: 10.1007/s00726-014-1729-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.