Abstract
To determine if mutations in the dihydropteroate synthase (DHPS) gene of Pneumocystis carinii f. sp. hominis arose in a single strain that was subsequently widely disseminated, we examined four genomic regions of 22 P. carinii clinical isolates selected based on the absence or presence of mutations in the DHPS gene. By single-strand conformation polymorphism and DNA sequencing, we found varying genotypes for each of the four regions in isolates with DHPS mutations, suggesting that these mutations occurred independently in multiple strains of P. carinii. This suggests that exposure to sulfa will select for these mutations in diverse strains.
Mutations in the human-derived Pneumocystis carinii (P. carinii f. sp. hominis) dihydropteroate synthase (DHPS) gene, the target of sulfa drugs, have been reported with increasing frequency in the United States (1, 4–6, 8, 11–13), Europe (3, 16), and Asia (15) and have been linked to prior exposure to sulfa or dapsone, suggesting the possible emergence of sulfa resistance. Epidemiological studies suggest that these mutations do not represent allelic variants since they were rarely detected in clinical isolates from the early 1980s (3, 16). The localization of these mutations to two sites encoding amino acids in the active site of the enzyme raises the possibility that these mutations arose in single or limited numbers of P. carinii isolates that then became widely disseminated. Alternatively, these mutations may have arisen independently in multiple isolates, and due to their precise location, conferred a survival advantage over wild-type strains or those strains with other random mutations in the DHPS gene. To differentiate between these two mechanisms, we undertook to examine genomic loci with known allelic variation in clinical P. carinii isolates with or without DHPS mutations.
(This work was presented in part at the 7th International Workshops on Opportunistic Protists, Cincinnati, Ohio, 13 to 16 June 2001 [L. Ma and J. A. Kovacs, Abstr. 7th Int. Workshops Opportunistic Protists, abstr. PO16, 2001].)
We used a previously described PCR–single-strand conformation polymorphism (SSCP) technique (2, 12) to analyze four genomic regions, including the intron of the nuclear 26S rRNA gene (26S rRNA), the internal transcribed spacer 1 of the nuclear rRNA gene operon (ITS1), the variable region of the mitochondrial 26S rRNA gene (mt26S), and the intron 6 region of the β-tubulin gene (tubulin). Twenty-two isolates (2 autopsy lung samples, 7 sputum samples, and 13 bronchoalveolar lavage fluid samples) were obtained from patients diagnosed with P. carinii pneumonia between 1986 and 1999. The DHPS gene in these isolates had been typed previously by DNA sequencing and/or SSCP (11, 12). Genomic DNA was extracted either by treatment with proteinase K followed by phenol-chloroform extraction as described previously (11) or by use of the NucliSens isolation kit (Organon Teknika, Durham, N.C.). Two microliters of DNA extract was added to a 20-μl PCR mixture containing 0.25 μM concentrations of each primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 2.5 U of AmpliTaq Gold DNA polymerase (Perkin Elmer, Foster City, Calif.). The primers and the thermal cycling conditions used for amplification of the four variable regions were identical to those described by Hauser et al. (2). After examination by electrophoresis on a 3% NuSieve 3:1 agarose gel (FMC Bioproducts, Rockland, Maine), 2- to 2.5-μl aliquots of PCR products were analyzed by SSCP using the GenePhor Electrophoresis System, a precast GeneGel SSCP gel, and GeneGel SSCP Buffer (Amersham Pharmacia Biotech, San Francisco, Calif.) as described previously (12). The optimal electrophoretic buffer, temperature, and migration time for each genomic locus were as follows: ITS1, buffer B, 12°C, 270 min; 26S rRNA, buffer C, 12°C, 185 min; mt26S, buffer C, 5°C, 260 min; tubulin, buffer A, 12°C, 190 min. These conditions were optimized for our laboratory and varied from those described previously (2). The gels were stained by using the PlusOne DNA silver staining kit (Amersham Pharmacia Biotech).
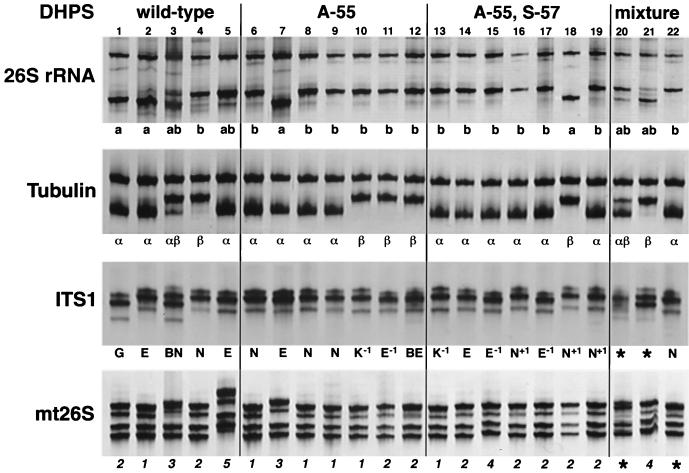
The SSCP patterns for four genomic loci in 22 clinical isolates are shown in Fig. 1. Although the DNA fragments examined in this study are the same as those reported by Hauser et al. (2), the SSCP patterns differ from those in the earlier report, especially for ITS1 and mt26S. This is most likely due to the different electrophoretic conditions. In this study, we found two different SSCP patterns, representing two different genotypes, for the 26S rRNA and β-tubulin loci, respectively. The SSCP patterns for the ITS1 and mt26S loci showed great variability, with at least four different patterns.
FIG. 1.
SSCP analysis of four genomic loci (indicated at the left) for P. carinii f. sp. hominis isolates. Mixture represents either wild-type and double mutation (A/T-55 and S/P-57) or single A-55 mutation and double mutation (A-55 and S/P-57). The DHPS genotype is indicated at the top. Patient numbers (1 to 22) are shown under the DHPS genotypes. Below each lane is the genotype for each locus. The genotypes for 26S rRNA and tubulin were named based on SSCP patterns alone, and the genotypes for ITS1 and mt26S were determined by SSCP analysis combined with DNA sequencing. Undetermined genotypes are indicated by “★.” The genotypes for ITS1 correspond to those described previously (9), with the variable numbers of T at positions 62 to 71 given in superscript (“−1” and “+1” represent 9 and 11 T's, respectively, compared to the commonly seen 10 T's). For mt26S, all five genotypes contain a G-to-A change at position 288 compared to the published sequenced (14), with an additional C-to-A change at position 85, a C-to-T change at position 248, a C-to-T change at position 85, or a C-to-T change at position 80 in types 2, 3, 4, and 5, respectively.
To more accurately determine the genotypes for ITS1 and mt26S, for selected isolates we performed direct sequencing and/or sequencing of individual clones after subcloning of PCR products. Sequencing of the ITS1 region of 20 isolates revealed nucleotide variations at seven positions (6, 14, 15, 21, 28, 80, and 81) which were identical to those reported by Lee at al. (9). In addition, for 10 isolates we observed variable numbers of the nucleotide T in a poly(T) tract at positions 62 to 71, as previously described (9). By sequencing of individual clones after subcloning, two isolates (no. 3 and 12) were found to contain two types of ITS1 sequence. For two isolates (no. 20 and 21), direct sequencing was unsuccessful and the ITS1 genotypes could not be accurately determined by SSCP alone. Among the 20 isolates sequenced, there were five types of ITS1 sequences, excluding variation in the T's in positions 62 to 71. The ITS1 genotype for each isolate was assigned as described previously (9). Sequencing of the mt26S sequences of 12 isolates revealed four genotypes with nucleotide variations at positions 85, 248, and 288 as described previously (7, 10, 14). A novel genotype with a C-to-T change at position 80 was identified for one isolate (no. 5) by three separate PCRs followed by direct sequencing. The genotypes of unsequenced isolates were determined by comparing the SSCP patterns to those of the sequenced isolates. For two isolates (no. 20 and 22), the mt26S genotypes could not be accurately determined.
Based on the DHPS sequences, the 22 isolates were divided into four groups, including 5 wild-type isolates (Thr-55 and Pro-57), 7 single Ala-55 mutants, 7 double mutants (Ala-55 and Ser-57), and 3 mixtures of either wild-type and double mutation (Ala/Thr-55 and Ser/Pro-57) or single Ala-55 mutation and double mutation (Ala-55 and Ser/Pro-57). As shown in Fig. 1, each group had a mixture of genotypes for each of the four genomic loci. The variability in ITS1 and mt26S was greater than that in 26S rRNA and tubulin. Among the 14 isolates containing either single Ala-55 or double mutations, there were 10 unique types based on analysis of these four loci (Table 1). When taking into account the variable numbers of T at positions 62 to 71, there was one additional type. These observations strongly suggest that DHPS mutations did not arise from a single strain of P. carinii, but rather that they arose independently in multiple different strains. Thus, while mutations may randomly occur in the DHPS gene (presumably at a low frequency), mutations at codons 55 and 57 are likely to confer a selective survival advantage to P. carinii during exposure to sulfa drugs. Nevertheless, an alternative explanation is that these mutations arose in single or limited numbers of P. carinii isolates that then became widely disseminated, with the complexity of genetic types resulting from horizontal transfer of DNA during sexual reproduction or another mechanism.
TABLE 1.
Typing of 14 P. carinii f. sp. hominis isolates with either single or double DHPS mutations, based on analysis of four genetic loci
| No. of isolates sharing a genotype | Genotype at each locusa
|
|||
|---|---|---|---|---|
| 26S rRNA | Tubulin | ITS1 | mt26S | |
| 3 | b | α | N | 1 |
| 2 | b | α | N | 2 |
| 2 | b | α | E | 2 |
| 1 | b | α | E | 4 |
| 1 | b | α | K | 1 |
| 1 | b | β | E | 2 |
| 1 | b | β | K | 1 |
| 1 | b | β | BE | 2 |
| 1 | a | α | E | 3 |
| 1 | a | β | N | 2 |
For further details, see the legend for Fig. 1.
Acknowledgments
We thank Steven R. Meshnick, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Mich., for his suggestions to initiate this study.
REFERENCES
- 1.Beard C B, Carter J L, Keely S P, Huang L, Pieniazek N J, Moura I N, Roberts J M, Hightower A W, Bens M S, Freeman A R, Lee S, Stringer J R, Duchin J S, del Rio C, Rimland D, Baughman R P, Levy D A, Dietz V J, Simon P, Navin T R. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg Infect Dis. 2000;6:265–272. doi: 10.3201/eid0603.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser P M, Francioli P, Bille J, Telenti A, Blanc D S. Typing of Pneumocystis carinii f. sp. hominis by single-strand conformation polymorphism of four genomic regions. J Clin Microbiol. 1997;35:3086–3091. doi: 10.1128/jcm.35.12.3086-3091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helweg-Larsen J, Benfield T L, Eugen-Olsen J, Lundgren J D, Lundgren B. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet. 1999;354:1347–1351. doi: 10.1016/S0140-6736(99)03320-6. [DOI] [PubMed] [Google Scholar]
- 4.Huang L, Beard C B, Creasman J, Levy D, Duchin J S, Lee S, Pieniazek N, Carter J L, del Rio C, Rimland D, Navin T R. Sulfa or sulfone prophylaxis and geographic region predict mutations in the Pneumocystis carinii dihydropteroate synthase gene. J Infect Dis. 2000;182:1192–1198. doi: 10.1086/315824. [DOI] [PubMed] [Google Scholar]
- 5.Kazanjian P, Armstrong W, Hossler P A, Burman W, Richardson J, Lee C H, Crane L, Katz J, Meshnick S R. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J Infect Dis. 2000;182:551–557. doi: 10.1086/315719. [DOI] [PubMed] [Google Scholar]
- 6.Kazanjian P, Locke A B, Hossler P A, Lane B R, Bartlett M S, Smith J W, Cannon M, Meshnick S R. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in AIDS patients. AIDS. 1998;12:873–878. doi: 10.1097/00002030-199808000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane B R, Ast J C, Hossler P A, Mindell D P, Bartlett M S, Smith J W, Meshnick S R. Dihydropteroate synthase polymorphisms in Pneumocystis carinii. J Infect Dis. 1997;175:482–485. doi: 10.1093/infdis/175.2.482. [DOI] [PubMed] [Google Scholar]
- 9.Lee C H, Helweg-Larsen J, Tang X, Jin S, Li B, Bartlett M S, Lu J J, Lundgren B, Lundgren J D, Olsson M, Lucas S B, Roux P, Cargnel A, Atzori C, Matos O, Smith J W. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C H, Lu J J, Bartlett M S, Durkin M M, Liu T H, Wang J, Jiang B, Smith J W. Nucleotide sequence variation in Pneumocystis carinii strains that infect humans. J Clin Microbiol. 1993;31:754–757. doi: 10.1128/jcm.31.3.754-757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L, Borio L, Masur H, Kovacs J A. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J Infect Dis. 1999;180:1969–1978. doi: 10.1086/315148. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Kovacs J A. Rapid detection of mutations in the human-derived Pneumocystis carinii dihydropteroate synthase gene associated with sulfa resistance. Antimicrob Agents Chemother. 2001;45:776–780. doi: 10.1128/AAC.45.3.776-780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei Q, Gurunathan S, Masur H, Kovacs J A. Failure of co-trimoxazole in Pneumocystis carinii infection and mutations in dihydropteroate synthase gene. Lancet. 1998;351:1631–1632. doi: 10.1016/S0140-6736(05)77687-X. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair K, Wakefield A E, Banerji S, Hopkin J M. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi T, Hosoya N, Endo T, Nakamura T, Sakashita H, Kimura K, Ohnishi K, Nakamura Y, Iwamoto A. Relationship between mutations in dihydropteroate synthase of Pneumocystis carinii f. sp. hominis isolates in Japan and resistance to sulfonamide therapy. J Clin Microbiol. 2000;38:3161–3164. doi: 10.1128/jcm.38.9.3161-3164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visconti E, Ortona E, Margutti P, Marinaci S, Zolfo M, Mencarini P, Celentano L P, Siracusano A, Tamburrini E. Identification of dihydropteroate (DHPS) gene mutant in Pneumocystis carinii in respiratory samples of HIV positive patients from 1992 to 1997. J Eukaryot Microbiol. 1999;46:132S. [PubMed] [Google Scholar]