Abstract
Alzheimer’s disease (AD) is a debilitating, chronic neurodegenerative disease. Genetic studies involving genome-wide association studies (GWAS) and meta-analysis have discovered numerous genomic loci associated with AD; however, the causal genes and variants remain unidentified in most loci. Integration of GWAS signals with epigenomic annotations has demonstrated that AD risk variants are enriched in myeloid-specific enhancers, implicating myeloid cells in AD etiology. AD risk variants in these regulatory elements modify disease susceptibility by regulating the expression of genes that play crucial roles in microglial phagocytosis. Several of these AD risk genes are specifically expressed in myeloid cells, whereas others are ubiquitously expressed but are regulated by AD risk variants within myeloid enhancers in a cell type-specific manner. We discuss the impact of established AD risk variants on microglial phagocytosis and debris processing via the endolysosomal system.
Genetic Determinants of Autosomal Dominant Alzheimer’s Disease (ADAD)
AD is the most common cause of dementia, accounting for 60–80% of all dementia cases [1]. It is a chronic, incurable neurodegenerative disease, affecting ∼50 million people worldwide [1]. Hall-mark pathologies in the brain of AD patients include extracellular β-amyloid plaques (Aβ) and intracellular neurofibrillary tangles (NFTs) composed of hyperphosphorylated tau, as well as gliosis and lipidosis (abnormal accumulation of lipids) [2,3]. These changes are associated with widespread neuronal cell death that is one of the causes of memory loss. Although the majority of AD cases have an onset above the age of 65 years (late-onset AD, LOAD), ∼10% of AD cases are associated with earlier onset (early-onset AD, EOAD). Genetic analysis of individuals with earlier onset led to identification of ADAD – a rare heritable form of the disease that represents <1% of AD cases. ADAD is caused by mutations in the amyloid precursor protein (APP) [4], presenilin 1 (PSEN1) [5], or presenilin 2 (PSEN2) [6] genes. Genetic determinants of ADAD led to the amyloid cascade hypothesis (ACH) which proposes that abnormal APP processing causes increased Aβ production leading to spread of NFTs, neurodegeneration, and the clinical manifestations of AD. The C-terminal fragment of APP is recycled through the endosomes where further processing by γ-secretases (whose catalytic subunit is encoded by PSEN1 and PSEN2) leads to the production of Aβ peptides (Aβ37–43) [7]. The longer Aβ peptides are believed to be more prone to aggregation, more toxic to neurons, and exacerbate tau pathology, thus leading to cognitive deficits [8]. In support of the ACH, a mutation in APP that reduces Aβ production strongly reduces the risk of AD. However, therapies targeting Aβ have largely failed, raising the question of whether the ACH is applicable to all forms of AD given that it was proposed based on ADAD, whereas most clinical trials have been performed in sporadic AD cases.
As outlined in later sections, genetic analyses of risk factors for AD have pointed to phagocytic clearance of tissue debris by microglia as a candidate key pathophysiological mechanism. In this review we discuss how AD risk genes converge on this biological process and impact on its different steps. We focus on well-established AD risk genes and their role in microglial phagocytosis, or – in cases where a role in microglia specifically has not been established – their roles in peripheral macrophage function and other immune cells.
Common AD Risk Variants Are Enriched in Microglial and Endolysosomal Genes
Genetic factors contributing to the majority of AD cases (∼90%) remain unclear. Recent GWAS of common genetic variants, frequently called SNPs, have led to the identification of >40 genomic loci associated with LOAD [9]. By far the strongest genetic risk factor for LOAD is the apolipoprotein E (APOE) genotype. There are three isoforms of APOE: APOE2, APOE3, and APOE4. The APOE33 genotype (carriage of two APOE3 alleles) is the most prevalent in all populations and is considered to confer a ‘neutral’ risk of AD. By contrast, having one or two APOE4 alleles markedly increases the risk of AD, whereas carrying one or two APOE2 alleles is markedly protective against AD. APOE is functionally involved in Aβ and apoptotic cell clearance as well as in lipid transport (described later). Aside from APOE, discovering causative genes at genome-wide loci remains challenging because common variants usually map within intergenic or intronic regions that are noncoding and, if located in regulatory elements, they may modulate the expression of one or more genes at some distance from the associated polymorphisms (i.e., not necessarily the closest gene). Interestingly, pathway analyses show that AD common variants mapping within GWAS loci converge on phagocytosis, lipid processing, and the immune response – biological processes exerted mainly by myeloid cells (microglia in the brain) [10]. In addition, several AD risk variants localize within regulatory elements (enhancers or promoters) that are bound by PU.1 – a transcription factor crucial for development and function of myeloid cells and an AD risk gene [11].
Although GWAS have facilitated major achievements in understanding the genetic risks of late-onset AD, further efforts will be necessary to decipher the link between AD genetics and disease pathogenesis. To this end, several groups have integrated multiple scales of functional data, including epigenetic annotations and gene expression data, with network analyses and genetic data from GWAS to elucidate the biological function of AD risk variants. Together, these analyses have found that common AD risk variants are enriched in microglia-specific enhancers, and in active enhancers of monocytes, macrophages, and microglia [12] that affect cell type-specific gene expression, strongly implicating microglia in disease etiology [11,13]. For example, BIN1 is a broadly expressed protein, but AD risk variants are located within a regulatory region downstream from BIN1 that is predicted to be a microglia-specific enhancer. Deletion of this enhancer in human induced pluripotent stem cells (iPSCs) resulted in decreased BIN1 expression in iPSC-derived microglia but not in neurons and astrocytes [13]. In addition, in recent unpublished results, Novikova and colleagues studied the relationship between myeloid active enhancers containing AD risk variants and regulation of target gene expression using summary data-based Mendelian randomization (SMR). They found that the same variants that confer AD risk also regulate myeloid-specific gene expression. Importantly, they nominated candidate causal genes as genome-wide significant loci, and found that several of these genes (including BIN1, RIN3, CD2AP, and ZYX, among others) localize to the endolysosomal network and may regulate endocytosis/phagocytosis [12]. The roles of individual AD risk genes in the endolysosomal network are discussed in detail later.
Polygenic Architecture of AD
The genetic architecture of AD reveals a polygenic component that consists of multiple small-effect loci [14]. One approach developed to determine the combinatorial effects of multiple variants on disease risk, age of onset, and heritability is the polygenic risk score (PRS). This strategy often includes SNPs that did not reach genome-wide significance in GWAS, leading to a more complete understanding of the genetic risk profile of an individual. Despite this, these additive models explain only a modest portion of AD heritability beyond APOE genotype, suggesting either that there are nonadditive effects or that there are many more risk loci that remain to be identified. Pathway analyses demonstrate that AD polygenic risk influences a variety of biological pathways including immune response, cholesterol metabolism, endolysosomal pathways, and APP metabolism [10]. In addition, studies that examined the aggregated effect of risk loci on disease onset indicate that each additional risk variant may lead to earlier AD onset [15]. Interestingly, a polygenic risk score generated from LOAD GWAS actually has a stronger impact on sporadic EOAD risk than LOAD, arguing that sporadic EOAD and LOAD are part of a continuum in which EOAD represents the most genetically loaded sporadic AD cases [16]. Other studies have demonstrated that increased burden of AD risk variants (measured by PRS) is linked to smaller hippocampal volume [17], and that this may be driven by microglia-mediated immune response pathways, possibly driving disease susceptibility [18].
Rare AD Risk Variants Implicate Microglial Cells
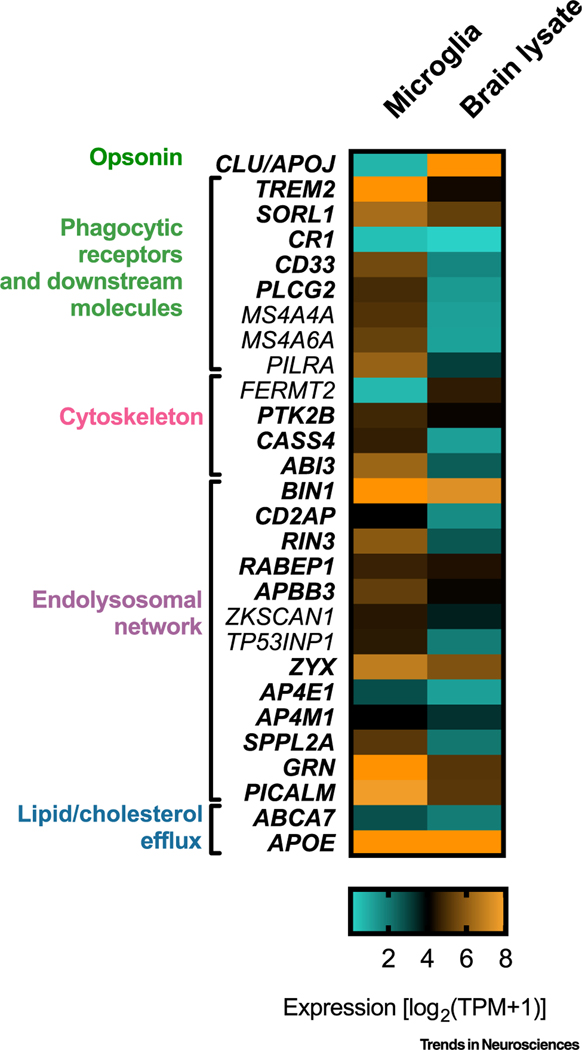
Analyses using whole-exome or whole-genome sequencing led to the discovery of rare variants that typically have a larger effect size than common variants identified by GWAS. A great advantage of these studies is the focus on rare variants in coding regions of the genome, and this links functional variants to causal genes and thus provides better clues of how disease risk is modulated. Rare variants associated with AD have been identified in TREM2, ABCA7, SORL1, ABI3, and PLCG2 [19–21]. In TREM2 (triggering receptor expressed on myeloid cells 2), the partial loss-of-function (LOF) R47H missense mutation has been found to significantly increase AD risk [21]. Of note, homozygous LOF mutation in TREM2 leads to Nasu–Hakola disease (NHD), an autosomal-recessive disorder characterized by early-onset dementia and multifocal bone cysts [22]. PLCG2, a member of phospholipase Cγ family, is another candidate gene in which rare variants have been identified. It colocalizes with TREM2, sharing a common interactome, and is likely a TREM2 downstream signaling effector [23]. A rare PLCG2 variant, P522R, has been associated with decreased risk of AD [19]. Importantly, the P522R polymorphism maps within the PLCγ2 regulatory domain, slightly increasing the activity of the PLCγ2 enzyme, thus acting as a gain-of-function (GOF) mutation [23]. Similarly to TREM2, PLCγ2 is expressed in microglial cells at high levels (Figure 1), and its expression is increased in microglia that surround amyloid plaques in mouse models of AD [23]. Rare LOF variants in ABCA7 (ABC transporter subfamily 7) have been associated with AD and increased disease susceptibility [24]. Similarly, several rare nonsense variants lead to reduced expression of ABCA7 and increased AD risk [25]. Common variants within the ABCA7 locus are also associated with AD risk [26]. ABCA7 may play an important role in phagocytosis of apoptotic cells because its ortholog in Caenorhabditis elegans, ced-7, is indispensable for the efficient detection and removal of apoptotic cells [27]. In mammals, many genes harboring rare variants are highly expressed in microglial cells and modulate the functions of microglia, underscoring the possible involvement of this cell type in the etiology and progression of AD (Figure 1) [19,28].
Figure 1. Expression Pattern of Alzheimer’s Disease (AD) Risk Genes Involved in Phagocytosis.
Heatmap showing expression of selected AD risk genes in ex vivo human microglia and human brain lysates. Values of expression (transcripts per million, TPM) are plotted based on [28]. Genes marked in bold are discussed in this review.
Mechanism of Microglial Phagocytosis
Common and rare AD risk variants converge on microglial cells and the endolysosomal network, an integral part of phagocytosis. Phagocytic clearance of tissue debris is a core function of microglial cells, the brain-resident macrophages, to maintain tissue homeostasis and immune tolerance, as well as to resolve inflammation and promote tissue repair. This is a complex process of recognition, engulfment, digestion, and response [29]. Microglia are professional phagocytes that sense and migrate toward ‘find me’ signals released by myelin debris, apoptotic cells, unwanted synapses, and dystrophic neurites, and they ensure proper recognition of these cues via a variety of phagocytic receptors expressed by microglia [29]. Damaged cells and unwanted synapses additionally expose an ‘eat me’ signal on their surface, phosphatidylserine (PS), that is bound to phagocytic receptors which help to dock extracellular cargo onto the phagocyte membrane (step 1: recognition) [29]. Specific recognition of macromolecules and receptors initiates actin-associated cytoskeleton rearrangement leading to the formation of phagocytic cups and engulfment (step 2: engulfment). Internalized material is then degraded in a process that involves multiple organelles (as illustrated in Figure 2). Material reaches a phagosome that matures and fuses with early endosomes, late endosomes, and lysosomes, forming a phagolysosome [29]. Lysosomes contain batteries of acid hydrolases and cathepsins that ensure degradation (step 3: digestion). This is followed by activation of transcription factors that enhance lipid clearance, reverse cholesterol transport, and release anti-inflammatory mediators (step 4: response) [30]. Early endosomes are involved in the progression of clathrin-mediated endocytosis which employs surface receptors that bind to extracellular cargo concentrated in plasma membranes by clathrin-coated pits [31,32]. Material from the early endosome is recycled back to the plasma membrane or fuses with late endosomes/lysosome. From the latter compartments material can be transported to the trans-Golgi network (TGN) for further sorting. Trafficking between organelles occurs in intracellular vesicles and is regulated by a family of Rab GTPases [33].
Figure 2. Summary of Endolysosomal Processing in Microglial Cells, and Links to Candidate Alzheimer’s Disease (AD) Risk Genes.
Microglial cells constantly probe fragments of plasma membrane by endocytosis (top left) and large macromolecules (i.e., apoptotic bodies, myelin debris, synapses) by phagocytosis (top right). Inside the cell, engulfed material reaches early endosomes or phagosomes which mature and fuse with lysosomes (bottom, middle), forming a phagolysosome (bottom, right). Proteins are sorted to the trans-Golgi network (TGN) via retrograde transport and recycling endosomes. AD risk gene products are overlaid in cellular compartments they are known to regulate. Green indicates AD genes whose decreased expression is associated with increased AD risk; red indicates that increased expression is associated with increased AD risk. Gold indicates an unknown expression effect on AD risk. The expression directionality of AD risk genes is plotted based on [12] CD2AP, BIN1, RIN3, APPB3, ZYX, AP4E1, AP4M1, SPPL2A; [107] GRN; [98] RAB10; [62] CD33C; [64] CD33T; and [57] CR1 (CR1-S, CR1-F).
Microglial Phagocytosis – A Process Affected by AD Risk Variants?
Genes that promote amyloidogenesis in ADAD (APP, PSEN1, PSEN2) primarily impact on endolysosomal functions [34]. Most amyloidogenic processing of APP occurs during endocytosis in the endolysosomal system. In particular, early endosomes are relevant in AD pathogenesis as the first compartments along the endocytic pathway that carry out the initial processing of APP. In AD, endosomal structures display several morphological and functional abnormalities, including impaired transport to endosomes, enlarged endosomes, and accelerated endocytosis, leading to excessive production of Aβ [34]. Such endosomal dysfunction has been reported to disrupt cholinergic neurons and lead to neurodegeneration [35]. Furthermore, in AD the lysosomal system is also activated, as evidenced by increased lysosomal gene expression. Novel analysis and interpretation of GWAS studies that implicate microglia in the etiology of AD encourage moving beyond neuronal cells to understand how AD risk variants impact on microglial phagocytic clearance of tissue debris and Aβ plaques. We describe later AD risk genes and how they are involved in the control of phagocytosis, with particular emphasis on the endolysosomal network (illustrated in Figure 3). Each subheading introduces an important phagocytic component.
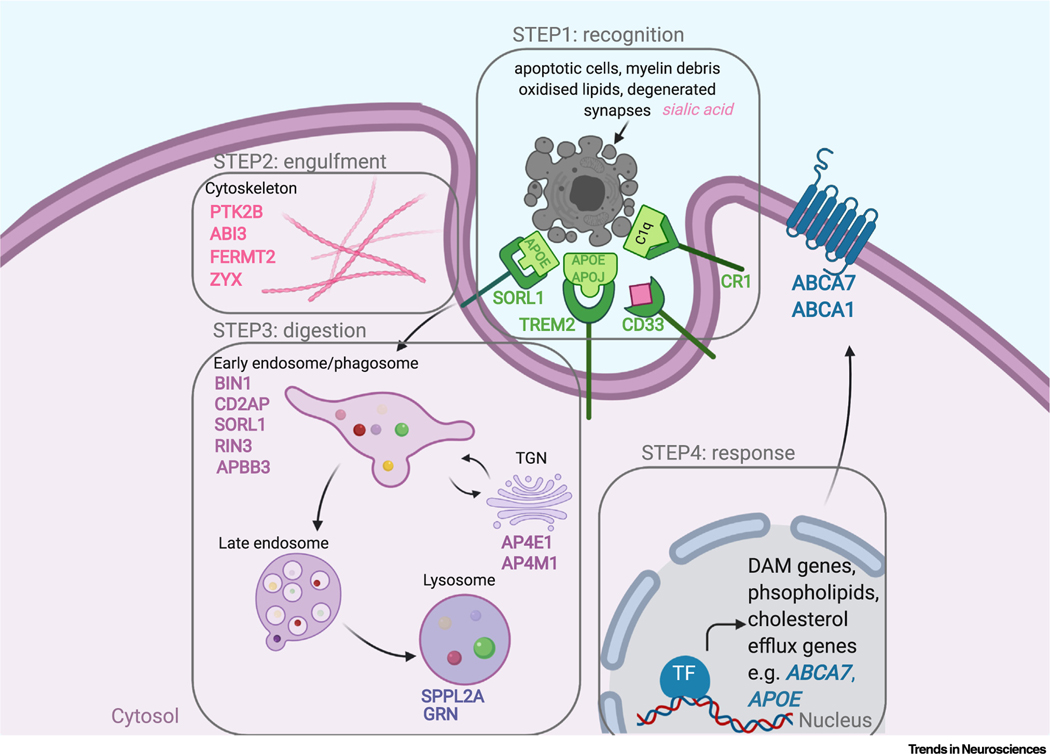
Figure 3. Candidate Alzheimer’s Disease (AD) Risk Genes in Microglial Phagocytosis.
Phagocytosis of apoptotic cells, myelin debris, synapses, and degenerated neurites is completed in four steps. Step 1: recognition [achieved by ‘find-me’ signals opsonized with apolipoprotein (APOE and APOJ, that are recognized by phagocytic receptors including TREM2 (triggering receptor expressed on myeloid cells 2), CR1 (complement receptor 1), and SORL1 (sortilin 1)]. Step 2: engulfment [achieved by cytoskeleton rearrangements regulated by e.g., PTK2B (protein tyrosine kinase 2β), ABI3]. Step 3: digestion [performed in the endolysosomal network and controlled by e.g., BIN1 (bridging integrator 1), CD2AP CD2-associated protein), RIN3 (Ras and Rab interactor 3), and SPPL2a (signal peptide peptidase-like 2a)]. Step 4: response [comprising activation of transcriptional program of clearance, i.e., DAM (disease/damage-associated microglia) genes and genes involved in phospholipids and cholesterol efflux, e.g., ABCA7, APOE]. AD risk genes are listed in each step and are overlaid in the cellular compartment whose function they control. Abbreviations: TF, transcription factor; TGN, trans-Golgi network.
Phagocytic Substrates
Daily, many apoptotic cells, degenerating synapses, and myelin debris need to be effectively cleared to ensure brain homeostasis and prevent debris accumulation and exaggerated inflammation. Phagocytic substrates are usually coated with opsonins that facilitate their recognition and docking to the phagocyte membrane via opsonin–receptor interaction. Apolipoproteins and complement proteins are well-known opsonins that coat many extracellular cargoes dedicated for degradation. One of the main apolipoproteins that coats Aβ and apoptotic cells is APOE, the major genetic risk factor for AD and major lipid transporter in the brain. Its capacity as an opsonin depends on its lipidation status. APOE2 is highly lipidated and binds more substrate, whereas APOE4 is poorly lipidated, thus compromising recognition and phagocytosis [36]. APOE is highly expressed in astrocytes, but it is upregulated in microglial cells following tissue damage, and has been identified as a key marker of disease/damage-associated microglia (DAM) [37,38]. In recent years more attention has been paid to microglial APOE. It has been shown that iPSC-derived microglia from homozygous APOE44 individuals display decoupled lipid metabolism, as evidenced by increased cholesterol biosynthesis and decreased cholesterol efflux leading to the formation of foamy glia [39]. APOE44 microglia also upregulate genes involved in inflammatory response while downregulating those responsible for cell movement. As a consequence, APOE44 microglia display reduced phagocytosis of Aβ in culture [40].
Another important apolipoprotein in the brain is clusterin (CLU) that encodes APOJ. SNPs have been identified in the CLU locus by two independent GWAS analyses [41,42]. APOJ opsonizes apoptotic cells and assists docking to phagocyte membranes [43]. APOJ is frequently found in injured tissue, suggesting that it participates in tissue repair [44].
Phagocytic Receptors
As noted earlier, genetic analyses implicate TREM2 in AD risk and suggest a crucial role for microglia in AD pathology because TREM2 is highly expressed in this cell type [45]. TREM2 recognizes several tissue damage-associated lipids (e.g., PS) that are present in degraded myelin, on the surface of apoptotic cells, or in unwanted synapses [46]. Importantly, TREM2 also binds to apolipoproteins including APOE and APOJ that opsonize tissue debris, thus facilitating their uptake [47]. Microglia lacking TREM2 fail to recognize chemotactic signals or respond properly to an injury, and show reduced migration toward apoptotic cells injected into the brain [48]. It has been shown in mouse models of demyelination induced by cuprizone, and also in an AD mouse model (5×FAD), that a lipid-rich environment (e.g., degenerated myelin, Aβ plaques, apoptotic cells) triggers the clearance response, including activation of endolysosomal pathways and immune reaction, leading to a switch from homeostatic microglia to DAM [37,38,49]. Consistent with this notion, lack of TREM2 in microglia results in reduced phagocytosis of apoptotic cells and myelin debris, as well as a reduced clearance response, leading to debris accumulation outside the cell and secondary events such as necrosis and sustained inflammation [38,49]. Similarly, microglia carrying the TREM2 R47H LOF variant show reduced proliferation, decreased activation, and weaker recruitment to Aβ plaques in two mouse models of AD [50,51]. Importantly, in AD patients expressing the R47H variant of TREM2, fewer microglia were associated with plaques, and this may explain the higher neuritic dystrophy around amyloid plaques [52]. This reduction in microglial recruitment to plaques may be due to an impaired capacity to recognize lipids – the R47H mutation resides in the ligand-binding domain of TREM2 [46]. Other TREM2 variants associated with NHD, T66M and W50C, markedly reduce phagocytosis of apoptotic cells in human iPSC-derived microglia [53]. TREM2 has been extensively studied in microglia in mouse models of AD, including amyloidosis models (APP/PS1 and 5×FAD) and a tauopathy model (PS19) [46,54,55]. Strikingly, studies on the effect of TREM2 on Aβ load, tau spreading, microglial recruitment, and neuroinflammation in these mouse models are conflicting. Overall, these studies suggest a protective role of TREM2 in the early stages of disease progression, but a detrimental role in late stages of AD when NFTs are present [46,54,55].
Another phagocytic receptor is complement receptor 1 (CR1). CR1 recognizes complement proteins such as C1q and C3b/C4b. A copy-number variant of CR1 that elevates the number of C3b/C4b binding sites (CR1-S isoform) is reported to decrease CR1 expression, and this contributes to reduced Aβ clearance and a 30% increased risk of AD in those individuals [56,57]. Conversely, a low-copy repeat variant of CR1 produces the CR1-F isoform that has fewer C3b/C4b binding sites. One may speculate that the CR1-F isoform might have higher phagocytic capacity [58]. Importantly, CR1 is a GWAS-significant locus [41], whereas the opsonin C4b maps within the HLA-DRB1 locus, which is also an AD risk locus [59]. CR1 is highly expressed in erythrocytes where it removes immune complexes, apoptotic cells, microbes, and Aβ [60]. In the brain CR1 is expressed by reactive astrocytes and microglia, which might suggest that it plays a similar role in the brain and in peripheral cells.
CD33 is a phagocytic receptor involved in the control of the immune response. It has been identified in several GWAS as an AD susceptibility locus [61]. A common risk variant (C) upstream of the CD33 gene is associated with higher expression of full-length CD33 in monocytes and microglia, increased microglial activation in the brain, and reduced phagocytosis of Aβ associated with worsening of cognitive functions [61–63]. Conversely, the protective allele (T) at this site decreases expression of CD33 and leads to enhanced phagocytosis of Aβ [64]. Consistently, loss of CD33 in human monocytic cell lines leads to increased phagocytosis of several different substrates [65]. Another variant has been discovered in exon 2, which encodes the sialic acid ligand-binding domain. This variant results in expression of a shorter form of CD33 that lacks exon 2 and decreased expression of the full-length isoform [66]. This variant is protective, resulting in increased phagocytosis and reduced amyloid burden [64,66]. Phagocytic receptors carrying AD risk variants are listed in Table 1 (Key Table).
Table 1. Key Table.
Phagocytic Receptors Identified as AD Risk Genes, and Mutations Associated with AD That Impact on Phagocytosisa
| Receptor | Species | Model/studies | Mutation | Alterations in phagocytosis | Refs |
|---|---|---|---|---|---|
| TREM2 | Human | Postmortem AD brains | R47H mutation | Reduced recruitment and phagocytosis of Aβ plaques | [52] |
| TREM2 | Human | NHD patient-derived microglia | T66M and W50C mutation | Reduced phagocytosis of apoptotic cells | [53] |
| TREM2 | Mouse | 5×FAD and APP/PS1 | R47H mutation | Reduced recruitment and phagocytosis of Aβ plaques | [50,51] |
| TREM2 | Mouse | 5×FAD and APP/PS1 | TREM2 knockout | Reduced recruitment and phagocytosis of Aβ plaques | [46] |
| CR1 | Human | Patient CSF | C3b/C4 increased repeatsb | Decreased Aβ clearance in CSF | [57] |
| CD33 | Human | Patient-derived monocytes | Full-length with risk allele (C)c | Decreased uptake of dextran and Aβ | [62] |
| CD33 | Human | Post-mortem AD brains | Full-length with protective allele (T)b | Decreased insoluble Aβ in the brain – indicator of enhanced Aβ uptake and clearance | [64] |
| CD33 | Human | Human monocytic cell lines | CD33 knockout | Increased phagocytosis of dextran, polystyrene beads, myelin, and Aβ | [65] |
| CD33 | Mouse | APP/PS1 | CD33 knockout | Decreased insoluble Aβ and reduced Aβ plaque burden – indicator of enhanced Aβ uptake and clearance | [64] |
Abbreviations: CSF, cerebrospinal fluid; NHD, Nasu–Hakola disease.
Variant associated with decreased expression.
Variant associated with increased expression.
Cytoskeleton
Macrophages are able to engulf extracellular cargo via a phagocytic cup as a result of rearrangements in the actin cytoskeleton. These rearrangements enable focal adhesion (i.e., direct contact) between phagocytic substrates and integrins on the surface of the phagocyte. Several genes participating in focal adhesion have been implicated in AD through the association of common (PTK2B, CASS4) and rare (ABI3) variants with AD. PTK2B (protein tyrosine kinase 2β) and CASS4 (Crk-associated substrate) both lie within reported AD-associated GWAS loci, although specific causal variants have not been linked to either gene. PTK2B encodes PYK2 and belongs to the focal adhesion kinase (FAK) family. Both PTK2B and CASS4 are involved in macrophage migration through modulation of F-actin dynamics [67,68]. Another example is ABI3, a member of the ABI (Abselson-interactor) family, that regulates the organization of the actin cytoskeleton. ABI3 is expressed in microglia (Figure 1) and its expression is elevated in aging individuals [19] and AD brains, as well as in microglia associated with Aβ plaques, suggesting that it controls migration of microglia to plaques [69]. Two other AD risk genes, CD2AP (CD2-associated protein) and ZYX (zyxin) may play a dual role in phagocytosis because they control both the actin cytoskeleton and endosome function depending on which cell type they are expressed in. CD2AP is located within a GWAS locus for LOAD [70]. In non-neuronal cells, CD2AP plays a role in the organization of the actin cytoskeleton. For example, in macrophages and dendritic cells, CD2AP colocalizes with F-actin, underscoring the importance of CD2AP in actin remodeling that is essential for proper engulfment of extracellular material and phagocytosis [71]. CD2AP-deficient leukocytes display defects in actin dynamics that affect their migration to lymph nodes [72]. CD2AP has also been found in early endosomes where it participates in APP sorting to lysosomes. Neuronal Cd2ap knockout results in endosomal accumulation of APP and promotes Aβ generation [73]. ZYX (zyxin) has been identified as a putatively causal gene within the GWAS locus previously called the EPHA1 locus. ZYX is primarily associated with actin filaments, suggesting that it participates in actin remodeling and active engulfment. ZYX is required for clathrin-mediated endocytosis and immune synapse formation [74], and may increase phagocyte motility [75]. In addition, Aβ has been shown to repress ZYX expression in HEK293T cells, demonstrating that ZYX is a direct target of Aβ [76]. ZYX is also enriched in microglia (Figure 1).
Recently, focal adhesion proteins (encoded by genes emerging from GWAS) have been proposed to be a key component of AD pathogenesis and to drive synapse loss in AD [77]. Other studies propose that focal adhesion regulates neuronal survival, and can lead to cell death if compromised [78]. Although it is worth exploring these hypotheses in neuronal cells, such studies should be extended to microglial cells because the focal adhesion candidate AD genes nominated by GWAS are expressed in microglia at higher levels than in other cell types (Figure 1), and importantly regulate actin dynamics and the engulfment of phagocytic substrates.
Early Endosomes
Genetic studies of sporadic AD also implicate genes that regulate early endosome function, trafficking, and maturation, including BIN1, CD2AP, PICALM, and SORL1 [71,79,80]. The BIN1 (bridging integrator 1) locus has been identified by GWAS as the most significant susceptibility locus in AD after APOE [81]. Integrative bioinformatic approaches revealed that BIN1 is the strongest causal candidate gene within the BIN1 locus and linked lower expression of BIN1 with increased AD risk [12]. Other studies found an insertion allele that increases the expression of BIN1 in AD brain and contributes to tau spreading [82]. Conflicting results suggest that BIN1, like TREM2, may have opposing effects on different stages of the disease, or that BIN1 has distinct underlying roles in neuronal and microglial cells. Whereas the roles of BIN1 in neuronal cells have been extensively studied ([83] for review), its roles in microglia are relatively understudied. It has been shown in the PS19 mouse model that genetic deletion of Bin1 from microglia reduces tau secretion through extracellular vesicles that are shed from endosomal compartments in a sex-dependent manner [84]. Specifically, BIN1-depleted microglia from males but not from females show lower levels of heat-shock proteins involved in tau spreading [84]. Interestingly, in mouse hippocampus, loss of Bin1 in neurons affects the transcriptome of surrounding microglia (that express Bin1). Pathway analysis of these microglia revealed that the LXR/RXR pathway is the top inhibited pathway, suggesting that neuronal BIN1 may enhance microglial clearance processes [85].
One candidate AD risk gene is PICALM (phosphatidylinositol-binding clathrin assembly protein) that has pleiotropic effects on endocytosis and regulates both clathrin-mediated endocytic uptake and autophagy [86]. The PICALM locus has been identified by GWAS [87]; however, it is unclear whether PICALM itself, or other genes in the locus, impacts on AD pathogenesis. In non-neuronal cells, knockdown of PICALM causes enlargement of endosomes and disruption of the TGN [88]. However, none of these studies directly addressed the role of PICALM in microglia despite the observation that PICALM is highly expressed in these cells (Figure 1).
Another candidate AD risk gene that participates in vesicle trafficking to early endosomes is RIN3 (Ras and Rab interactor 3). RIN3 is located within the SLC24A4 GWAS locus and has been independently identified as a potentially causal gene that has deleterious rare variants [59,89]. RIN3 is highly expressed in microglia (Figure 1); however, its role in these cells has been poorly explored. In HEK293T cells, RIN3 stabilizes the active form of Rab5 that promotes trafficking to endosomes [90]. Sustained activation of Rab5 mediated by RIN3 may underlie AD pathogenesis because abnormal overactivation of Rab5 has been observed in post-mortem brain samples from AD patients [91]. Interestingly, RIN3 localizes in vesicles shed from plasma membrane to early endosomes, together with BIN1 and CD2AP, suggesting that these three risk factors cooperate in endocytosis [92].
SORL1 (sortilin 1) has a dual role in phagocytosis: it both regulates the function of sorting endosomes and is present in the plasma membrane where it acts as a surface receptor for APOE [93]. The SORL1 locus has been consistently identified by GWAS as being associated with AD [94,95]. In addition, rare deleterious coding variants in this gene have been found in AD patients [20]. In neurons, decreased expression of SORL1 in AD patients leads to reduced sorting and increased processing of APP, contributing to elevated Aβ production [96]. In immune cells, the role of SORL1 may be different than in neurons because SORL1 regulates phagocyte migration toward chemoattractants and the immune response [97] (see Outstanding Questions). One may speculate that the dual functions of neuronal SORL1 in APP processing and in microglial phagocytic clearance of Aβ might result in an increased load of senile plaques, a major hallmark of AD pathology.
Outstanding Questions.
How do AD risk factors impact on the function of microglia? AD risk variants are enriched in myeloid-specific enhancers including microglial enhancers, implicating this cell type in AD pathogenesis. However, how AD risk variants impact on microglial function in AD remains unclear. Is this phenomenon specific to LOAD GWAS risk variants, or are ADAD mutations associated with APP also important for microglia? Is dysfunction of microglia sufficient to drive the disease?
Is endolysosomal processing compromised in microglia during AD, and how is this process related to the main pathological AD hallmarks (β-amyloidosis and tau spreading)? GWAS signals enriched in microglial enhancers are predicted to affect genes involved in endolysosomal processing, including BIN1, CD2AP, RIN3, and others. How do these genes affect the morphology and function of endocytic compartments in microglia? Are early endosomes particularly affected, given that the vast majority of AD risk gene products are localized here? Is the role of these genes in microglial endolysosomal processes similar to that observed in neuronal cells, or does it differ depending on cell type? How does myeloid cell dysfunction in the endolysosomal pathway contribute to plaque deposition and tau spreading at different stages of the disease?
What would be the most suitablemodel to address efferocytosis in myeloid cells? Differences in the immune system and microglia between mouse and human, and preclinical models that only partially model human disease, highlight the need for better models. One may need to think about a human system that allows the immune response to be modeled without species bias. Organoids composed of neurons and mixed glial populations may be of potential interest for understanding microglial phagocytosis of tissue debris in AD. Transplantation of hiPSC-derived hematopoietic progenitor cells (genetically edited to modify disease risk) back into mouse brain may allow assessment of human microglial function in vivo in a disease-relevant context.
Another candidate AD gene product that localizes to the early endosome is RAB10. RAB10 is a putative protective factor for AD [98]. Whole-genome sequencing identified variants in the RAB10 locus in individuals who did not develop AD despite carrying APOE4 allele/alleles and a strong family history of AD [99]. RAB10 is involved in the transport of proteins from early endosomes to recycling endosomes or alternatively to the TGN [98]. RAB10 is also involved in APP processing and the production of Aβ. Several studies indicate that decreased expression of RAB10 reduces the Aβ42/Aβ40 ratio, whereas overexpression of RAB10 increased this ratio [99].
Recent unpublished computational analyses have nominated APBB3 (amyloid β precursor protein binding family B member 3) as an additional AD candidate gene [12]. APBB3 interacts with APP in early endosomes; however, its detailed role in myeloid cells or endosomal system remains unknown [100]. An overview of nominated AD risk genes and their roles in the control of the endolysosomal network is depicted in Figure 2.
Late Endosomes and Lysosomes
SPPL2A (signal peptide peptidase-like 2a) maps to an AD risk locus identified by GWAS [101]. Integrative genomic analysis further nominated SPPL2A as an AD risk gene. SPPL2A encodes an endosomal/lysosomal protease and presenilin homolog that localizes to late endosomes/lysosomes [102]. SPPL2A is enriched in aged human microglia; however, its function in these cells remains undetermined. In other immune cells, SPPL2A has been shown to regulate B cell homeostasis and development in vivo, and is responsible for proteolytic cleavage of tumor necrosis factor (TNF)-α and CD74 [103]. Another lysosomal marker is progranulin (encoded by GRN). GRN is highly expressed by microglia but is also present in neurons and astrocytes [104] (Figure 1). Several studies have identified disease-associated GRN polymorphisms. For example, haploinsufficiency of GRN causes frontotemporal dementia [105], whereas complete GRN deficiency leads to a lysosomal storage disease with severe neurodegeneration [106]. In addition, one GRN variant has been associated with both reduced expression of GRN and increased AD risk [107]. GRN localizes in LAMP1-positive lysosomes and controls lysosome function, and both decreased and excess levels of GRN negatively regulate lysosomal biogenesis [104]. GRN is regulated by transcription factor TFEB, a master regulator of lysosome biogenesis that maps within the TREM2 locus [59,108]. Lack of GRN in microglia leads to defective maturation of cathepsins and decreased lysosomal activity [109]. In a mouse model of AD, PDGF–APPSw,Ind (line J9) mice that have GRN-deficient microglia display reduced phagocytosis, increased amyloid plaque burden, and exacerbated cognitive symptoms compared with mice with microglia that express GRN [110]. Importantly, lentiviral overexpression of GRN can rescue these molecular and cognitive deficits, and protects against Aβ toxicity [110]. Similarly, lack of GRN leads to the accumulation of toxic lipid droplets in microglia in the hippocampus that correlates with cognitive decline [111]. GRN may also be an important modulator of anti-inflammatory response in immune cells because it inhibits TNF-α downstream signaling [112].
The Trans-Golgi network
The TGN serves as a major sorting platform that receives input from endocytic compartments. Recent unpublished work has nominated two genes that control TGN functions, AP4M1 and AP4E1, as AD risk genes based on the identification of AD risk variants within enhancers that regulate their expression [12]. In addition, whole-exome sequencing identified an AD-linked rare variant in AP4M1 [113], whereas AP4E1 maps to the SPPL2A locus [59]. AP4E1 and AP4M1 encode two subunits of adaptor-related protein complex 4 (AP-4), ε1 and μ1, respectively. They play an important role in the endocytic pathway by sorting integral membrane proteins. AP4E1 is reported to control the maturation and formation of autophagosomes [114]. AP4M1 is involved in functional recognition of APP [115] and its transport from the TGN to endosomes. If AP-4–APP interaction is impaired, APP is cleaved by γ-secretase to generate Aβ peptide [116]. LOF mutations in AP-4 subunits impair autophagy [117]. Given the fact that autophagy in AD is involved in Aβ and tau degradation, one may speculate that deficiency in the AP-4 complex (in particular, reduced AP4E1 expression is associated with increased AD risk; Figure 2) could lead to abnormal biogenesis of autophagosomes [117] and impaired clearance of Aβ plaques or NFTs.
Concluding Remarks and Future Perspectives
AD-associated genetic variants affect a variety of biological pathways that have an impact on disease susceptibility. Although the identity of most causal genes remains elusive, recent GWAS and post-GWAS bioinformatic analyses implicate microglia and phagocytic clearance of tissue debris, also known as efferocytosis, as key players in AD pathogenesis [118]. This broad biological process includes recognition of released and membrane-tethered chemotactic signals recognized by phagocytic receptors; rearrangements of actin cytoskeleton leading to formation of a phagocytic cup; digestion of engulfed material in endolysosomal compartments; and finally activation of transcription factors that enhance the clearance transcriptional program. Many AD risk genes control the functions of one or more efferocytotic steps (Figure 3). Elimination of apoptotic tissue debris triggers a ‘response’ phase of phagocytosis leading to activation of clearance pathways controlled by transcription factors [liver X receptors (LXRs) and retinoid X receptors (RXRs)] that help to deal with the metabolic load derived from engulfed membranes, and also enhances the expression of anti-inflammatory signals [30]. The latter is a part of the DAM response in microglia that is regulated by several AD risk genes including APOE and TREM2 [37,38]. Importantly, this hypothesis does not exclude APP metabolism from being a part of AD pathology, but instead suggests that APP processing may be a part of broader cellular process – microglial efferocytosis – that, we argue, may be a central mechanism of AD.
Existing evidence arising from GWAS implicates dysregulation of phagocytic clearance in myeloid cells in the etiology of AD; however, extrapolating these findings from preclinical models of AD to humans carries several caveats and challenges. First, the immune systems in mice and humans differ widely. Second, preclinical models of AD are limited in various ways, and do not recapitulate the multifactorial nature of sporadic AD. Importantly, most animal models of AD do not accurately mimic the human disease and manifest incomplete neuropathology and symptomatology at a young age. All these concerns highlight the need for human cell-based models such as human induced pluripotent stem cells (hiPSCs) differentiated into CNS cell types (see Outstanding Questions). Although 2D cultures may not be ideal owing to abnormal cell behavior and activation, one may implement more complex models including cocultures and organoids. 3D culture techniques have been developed successfully and represent a more physiological environment that can include neurons, astrocytes, microglia, and oligodendrocytes. This system allows AD genetic risk factors to be studied in a human cellular context. iPSC technology together with advances in gene editing provide a convenient platform to study human AD risk variants in the correct genomic context, allowing better understanding of disease mechanisms. Recent studies show that cortical organoids produced from ADAD patient-derived hiPSCs spontaneously develop Aβ plaques and NFTs [119]. Importantly, organoids derived from AD patient cells show alterations in the endolysosomal system (i.e., enlarged early endosomes), which suggests that molecular features of AD are preserved [119,120]. So far, studies in AD cerebral organoids have focused on APP processing in neuronal cells. Future organoid models investigating AD risk factors should also include microglia. However, a limitation of these 2D and 3D cultures is the absence of the physiological context of the brain. An alternative approach is to transplant hiPSC-derived hematopoietic progenitor cells (especially those genetically edited to induce or correct AD mutations) into mouse brain to study the microglial response in more physiological conditions [121].
Genetic studies have been extremely important in revealing the genes and pathways that underlie AD risk. Early studies on ADAD focused attention on APP processing, whereas more recent GWAS analyses of common and rare variants strongly suggest that microglial function, in particular phagocytosis of tissue debris (efferocytosis), may be involved in AD development and progression [12,13]. It will be important to determine whether and how APP processing intersects with efferocytosis, or whether these two disease mechanisms represent parallel ways of developing AD. With this review we hope to stimulate further research aimed at understanding efferocytosis in immune cells in the context of AD risk factors, with the goal of identifying novel therapeutic targets and effective therapeutic interventions.
Highlights.
Genetic variants associated with Alzheimer’s disease (AD) are enriched in myeloid-specific enhancers, including microglial enhancers, implicating this cell type in the etiology of AD.
AD risk variants in these enhancers modulate disease susceptibility by regulating enhancer activity and in turn the expression of genes involved in microglial phagocytosis and the endolysosomal pathway.
Common risk variants are enriched for proteins expressed in early endosomes.
Pathway analysis of AD genetics combined with myeloid genomics strongly implicates dysregulation of phagocytosis of tissue debris in the etiology of AD.
Acknowledgements
This work was supported by grants from the National Institute on Aging (U01 AG052411, RF1AG054011, U01AG058635), the JPB Foundation, and the Neurodegeneration Consortium (to A.M.G.). The authors thank Dr Sarah Neuner, Anastasia Efthymiou, and Gloriia Novikova for commenting on the manuscript.
Footnotes
Disclaimer Statement
A.M.G. has consulted for Eisai, Biogen, Pfizer, AbbVie, Cognition Therapeutics, and GSK; she also served on the Scientific Advisory Board of Denali Therapeutics (2015–2018).
References
- 1.Alzheimer’s Association (2020) 2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 16, 391–460 [DOI] [PubMed] [Google Scholar]
- 2.Foley P (2010) Lipids in Alzheimer’s disease: a century-old story. Biochim. Biophys. Acta 1801, 750–753 [DOI] [PubMed] [Google Scholar]
- 3.Holtzman DM et al. (2011) Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 3, 77sr1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goate A et al. (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349, 704–706 [DOI] [PubMed] [Google Scholar]
- 5.Clark RF et al. (1995) The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat. Genet. 11, 219–222 [DOI] [PubMed] [Google Scholar]
- 6.Rogaev EI et al. (1995) Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376, 775–778 [DOI] [PubMed] [Google Scholar]
- 7.Hardy JA and Higgins GA (1992) Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185 [DOI] [PubMed] [Google Scholar]
- 8.Clavaguera F et al. (2009) Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews SJ et al. (2020) Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 19, 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naj AC et al. (2017) Genomic variants, genes, and pathways of Alzheimer’s disease: an overview. Am. J. Med. Genet. B Neuropsychiatr. Genet. 174, 5–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K-L et al. (2017) A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novikova G et al. (2019) Integration of Alzheimer’s disease genetics and myeloid genomics reveals novel disease risk mechanisms. BioRxiv Published online August 12, 2019. 10.1101/694281 [DOI]
- 13.Nott A et al. (2019) Brain cell type-specific enhancer-promoter interactome maps and disease risk association. Science 366, 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escott-Price V et al. (2015) Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 138, 3673–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naj AC et al. (2014) Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 71, 1394–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruchaga C et al. (2018) Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement. 14, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley SF et al. (2017) Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol. Psychiatry 81, 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancaster TM et al. (2019) Microglia-mediated immunity partly contributes to the genetic association between Alzheimer’s disease and hippocampal volume. Brain Behav. Immun. 79, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sims R et al. (2017) Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vardarajan BN et al. (2015) Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 77, 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson T et al. (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerreiro RJ et al. (2013) Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 70, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magno L et al. (2019) Alzheimer’s disease phospholipase C-gamma-2 (PLCG2) protective variant is a functional hypermorph. Alzheimers Res. Ther. 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg S et al. (2015) Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat. Genet. 47, 445–447 [DOI] [PubMed] [Google Scholar]
- 25.De Roeck A et al. (2019) The role of ABCA7 in Alzheimer’s disease: evidence from genomics, transcriptomics and methylomics. Acta Neuropathol. 138, 201–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JC et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinchen JM et al. (2005) Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434, 93–99 [DOI] [PubMed] [Google Scholar]
- 28.Gosselin D et al. (2017) An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sierra A et al. (2013) Janus-faced microglia: beneficial and detrimental consequences of microglial phagocytosis. Front. Cell. Neurosci. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doran AC et al. (2020) Efferocytosis in health and disease. Nat. Rev. Immunol. 20, 254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solé-Domènech S et al. (2016) The endocytic pathway in microglia during health, aging and Alzheimer’s disease. Ageing Res. Rev. 32, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mettlen M et al. (2018) Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxfield FR and McGraw TE (2004) Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 34.Nixon RA (2017) Amyloid precursor protein and endosomal–lysosomal dysfunction in Alzheimer’s disease: inseparable partners in a multifactorial disease. FASEB J. 31, 2729–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S et al. (2016) Evidence that the rab5 effector APPL1 mediates APP-βCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer’s disease. Mol. Psychiatry 21, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J et al. (2015) Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Aβ metabolism in apoE4-targeted replacement mice. Mol. Neurodegener. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasemann S et al. (2017) The TREM2–APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keren-Shaul H et al. (2017) A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 [DOI] [PubMed] [Google Scholar]
- 39.TCW J et al. (2019) Cholesterol and matrisome pathways dysregulated in human APOE ε4 glia. BioRxiv Published online July 25, 2019. 10.1101/713362 [DOI]
- 40.Lin Y-T et al. (2018) APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambert J-C et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 [DOI] [PubMed] [Google Scholar]
- 42.Harold D et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunin P et al. (2016) Clusterin facilitates apoptotic cell clearance and prevents apoptotic cell-induced autoimmune responses. Cell Death Dis. 7, e2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster EM et al. (2019) Clusterin in Alzheimer’s disease: mechanisms, genetics, and lessons from other pathologies. Front. Neurosci. 13, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y et al. (2018) TREM2-dependent effects on microglia in Alzheimer’s disease. Front. Aging Neurosci. 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y et al. (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh FL et al. (2016) TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91, 328–340 [DOI] [PubMed] [Google Scholar]
- 48.Mazaheri F et al. (2017) TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 18, 1186–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantoni C et al. (2015) TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol. 129, 429–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song WM et al. (2018) Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215, 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng-Hathaway PJ et al. (2018) The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 13, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan P et al. (2016) TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 92, 252–264 [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Reitboeck P et al. (2018) Human induced pluripotent stem cell-derived microglia-like cells harboring TREM2 missense mutations show specific deficits in phagocytosis. Cell Rep. 24, 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leyns CEG et al. (2017) TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. U. S. A. 114, 11524–11529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jay TR et al. (2015) TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villegas-Llerena C et al. (2016) Microglial genes regulating neuroinflammation in the progression of Alzheimer’s disease. Curr. Opin. Neurobiol. 36, 74–81 [DOI] [PubMed] [Google Scholar]
- 57.Brouwers N et al. (2012) Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol. Psychiatry 17, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fonseca MI et al. (2016) Analysis of the putative role of CR1 in Alzheimer’s disease: genetic association, expression and function. PLoS ONE 11, e0149792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunkle BW et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johansson JU et al. (2018) Peripheral complement interactions with amyloid β peptide in Alzheimer’s disease: polymorphisms, structure, and function of complement receptor 1. Alzheimers Dement. 14, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naj AC et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradshaw EM et al. (2013) CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16, 848–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raj T et al. (2014) CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum. Mol. Genet. 23, 2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griciuc A et al. (2013) Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacherjee A et al. (2019) Repression of phagocytosis by human CD33 is not conserved with mouse CD33. Commun. Biol. 2, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Estus S et al. (2019) Evaluation of CD33 as a genetic risk factor for Alzheimer’s disease. Acta Neuropathol. 138, 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salazar SV et al. (2019) Alzheimer’s disease risk factor Pyk2 mediates amyloid-β-induced synaptic dysfunction and loss. J. Neurosci. 39, 758–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh MK et al. (2008) A novel Cas family member, HEPL, regulates FAK and cell spreading. Mol. Biol. Cell 19, 1627–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satoh J-I et al. (2017) Microglia express ABI3 in the brains of Alzheimer’s disease and Nasu–Hakola disease. Intractable Rare Dis. Res. 6, 262–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naj A et al. (2011) Genome-wide association study of late-onset Alzheimer disease identifies disease-associated variants in MS4A4/MS4A6E, CD2AP, CD33, and EPHA1. Alzheimers Dement. 7, S191 [Google Scholar]
- 71.Cormont M et al. (2003) CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic 4, 97–112 [DOI] [PubMed] [Google Scholar]
- 72.Srivatsan S et al. (2013) CD2-associated protein regulates plasmacytoid dendritic cell migration, but is dispensable for their development and cytokine production. J. Immunol. 191, 5933–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ubelmann F et al. (2017) Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep. 18, 102–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joosten B et al. (2018) Super-resolution correlative light and electron microscopy (SR-CLEM) reveals novel ultrastructural insights into dendritic cell podosomes. Front. Immunol. 9, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wen X-M et al. (2020) Zyxin (ZYX) promotes invasion and acts as a biomarker for aggressive phenotypes of human glioblastoma multiforme. Lab. Investig. 100, 812–823 [DOI] [PubMed] [Google Scholar]
- 76.Lanni C et al. (2013) Zyxin is a novel target for beta-amyloid peptide: characterization of its role in Alzheimer’s pathogenesis. J. Neurochem. 125, 790–799 [DOI] [PubMed] [Google Scholar]
- 77.Dourlen P et al. (2019) The new genetic landscape of Alzheimer’s disease: from amyloid cascade to genetically driven synaptic failure hypothesis? Acta Neuropathol. 138, 221–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caltagarone J et al. (2007) Focal adhesions regulate Aβ signaling and cell death in Alzheimer’s disease. Biochim. Biophys. Acta 1772, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pant S et al. (2009) AMPH-1/amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat. Cell Biol. 11, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tebar F et al. (1999) Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell 10, 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seshadri S et al. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chapuis J et al. (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Acker ZP et al. (2019) Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: impact of genetic risk factors. Mol. Neurodegener. 14, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crotti A et al. (2019) BIN1 favors the spreading of Tau via extracellular vesicles. Sci. Rep. 9, 9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McAvoy KM et al. (2019) Cell-autonomous and non-cell autonomous effects of neuronal BIN1 loss in vivo. PLoS One 14, e0220125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tooze SA et al. (2014) Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 6, a018358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambert J-C. et al. (2011) Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol. Aging 32, 756. [DOI] [PubMed] [Google Scholar]
- 88.Meyerholz A et al. (2005) Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 89.Schwartzentruber J et al. (2020) Genome-wide meta-analysis, fine-mapping, and integrative prioritization identify new Alzheimer’s disease risk genes. MedRxiv Published online January 27, 2020. 10.1101/2020.01.22.20018424 [DOI]
- 90.Kajiho H et al. (2011) Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J. Biol. Chem. 286, 24364–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu W et al. (2018) Dysregulation of Rab5-mediated endocytic pathways in Alzheimer’s disease. Traffic 19, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Juul Rasmussen I et al. (2019) Blood–brain barrier transcytosis genes, risk of dementia and stroke: a prospective cohort study of 74,754 individuals. Eur. J. Epidemiol. 34, 579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao N et al. (2018) Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol. Psychiatry 83, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rogaeva E et al. (2007) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reitz C et al. (2011) Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch. Neurol. 68, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Talbot H et al. (2018) Regulatory roles of sortilin and SorLA in immune-related processes. Front. Pharmacol. 9, 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCarthy C et al. (2010) SorLA modulates atheroprotective properties of CLA by regulating monocyte migration. Atherosclerosis 213, 400–407 [DOI] [PubMed] [Google Scholar]
- 98.Tavana JP et al. (2019) RAB10: an Alzheimer’s disease resilience locus and potential drug target. Clin. Interv. Aging 14, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ridge PG et al. (2017) Linkage, whole genome sequence, and biological data implicate variants in RAB10 in Alzheimer’s disease resilience. Genome Med. 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanahashi H and Tabira T (1999) Molecular cloning of human Fe65L2 and its interaction with the Alzheimer’s beta-amyloid precursor protein. Neurosci. Lett. 261, 143–146 [DOI] [PubMed] [Google Scholar]
- 101.Marioni RE et al. (2018) GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Behnke J et al. (2011) Signal-peptide-peptidase-like 2a (SPPL2a) is targeted to lysosomes/late endosomes by a tyrosine motif in its C-terminal tail. FEBS Lett. 585, 2951–2957 [DOI] [PubMed] [Google Scholar]
- 103.Hüttl S et al. (2015) Processing of CD74 by the intramembrane protease SPPL2a is critical for B cell receptor signaling in transitional B cells. J. Immunol. 195, 1548–1563 [DOI] [PubMed] [Google Scholar]
- 104.Kao AW et al. (2017) Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baker M et al. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 106.Smith KR et al. (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 90, 1102–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sheng J et al. (2014) Progranulin polymorphism rs5848 is associated with increased risk of Alzheimer’s disease. Gene 542, 141–145 [DOI] [PubMed] [Google Scholar]
- 108.Sardiello M et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 109.Götzl JK et al. (2018) Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol. Neurodegener. 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Minami SS et al. (2014) Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 20, 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marschallinger J et al. (2020) Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mendsaikhan A et al. (2019) Microglial progranulin: involvement in Alzheimer’s disease and neurodegenerative diseases. Cells 8, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bis JC et al. (2018) Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol. Psychiatry 25, 1859–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mattera R et al. (2017) AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. U. S. A. 114, E10697–E10706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ross BH et al. (2014) Structural and functional characterization of cargo-binding sites on the μ4-subunit of adaptor protein complex 4. PLoS One 9, e88147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burgos PV et al. (2010) Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell 18, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Davies AK et al. (2018) AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat. Commun. 9, 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Márquez-Ropero M et al. (2020) Microglial corpse clearance: lessons from macrophages. Front. Immunol. 11, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gonzalez C et al. (2018) Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 23, 2363–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Israel MA et al. (2012) Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hasselmann J et al. (2019) Development of a chimeric model to study and manipulate human microglia in vivo. Neuron 103, 1016–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]