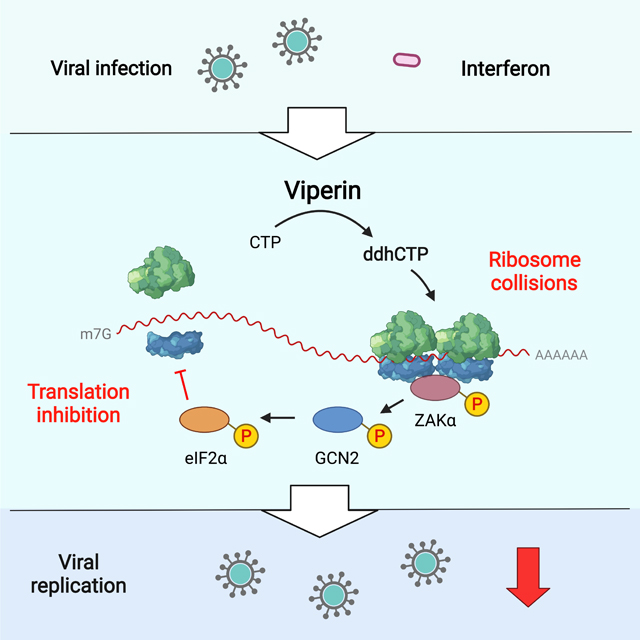
Summary
Innate immune responses induce hundreds of interferon-stimulated genes (ISGs). Viperin, a member of the radical S-adenosylmethionine (SAM) superfamily of enzymes, is the product of one such ISG that restricts the replication of a broad spectrum of viruses. Here we report a previously unknown antiviral mechanism in which viperin activates a ribosome collision-dependent pathway that inhibits both cellular and viral RNA translation. We found that the radical SAM activity of viperin is required for translation inhibition and that this is mediated by viperin’s enzymatic product, 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP). Viperin triggers ribosome collisions and activates the MAPKKK ZAK pathway that in turn activates the GCN2 arm of the integrated stress response pathway to inhibit translation. The study illustrates the importance of translational repression in the antiviral response and identifies viperin as a translation regulator in innate immunity.
Keywords: Viperin, innate immunity, antiviral response, interferon-stimulated gene, ribosome collision, translational regulation, integrated stress response pathway, Zika virus, Kunjin virus
eTOC Blurb
Hsu et al. show that viperin, a broad-spectrum antiviral protein, inhibits viral replication of two medically relevant flaviviruses by limiting viral protein production. Viperin synthesizes a nucleoside derivative, ddhCTP which induces ribosome collisions, leading to activation of the integrated stress response and consequently translation inhibition.
Graphical Abstract
Introduction
The recent Zika virus (ZIKV) epidemic and the current COVID-19 pandemic, which has caused more than 395 million confirmed cases, provides an impetus for the generation of broad-spectrum antiviral therapeutics. The innate immune response serves as a robust first line of defense against virus infections and type I interferon (IFN-I) release during infection plays a critical role in antiviral immunity. IFN secretion is induced when host pattern recognition receptors (PRRs) sense pathogen-associated molecular patterns (PAMPs), leading to the transcription of multiple interferon-stimulated genes (ISGs). The proteins encoded by some of these ISGs restrict viral infection by inhibiting different stages of the virus life cycle. Despite the clear importance of the IFN-mediated response, the cellular/antiviral functions of most ISGs are unknown and understanding how they coordinate with cellular networks during the antiviral response remains a critical goal.
Viperin is the product of Rsad2, one of the most highly induced ISGs upon viral infection (Fink et al., 2007). It inhibits the replication of a broad spectrum of viruses, including dsDNA viruses, positive- and negative-sense single strand RNA viruses, and retroviruses in humans and other species. Examples include ZIKV, West Nile virus (WNV), influenza A virus (IAV), human immunodeficiency virus-1 (HIV-1) and human cytomegalovirus (HCMV) (Chin and Cresswell, 2001; Nasr et al., 2012; Szretter et al., 2011; Van der Hoek et al., 2017; Wang et al., 2007). Viperin contains a radical SAM domain, which contain a [4Fe-4S] cluster responsible for generating the catalytically essential 5’-deoxyadenosyl radical from S-adenosyl methionine (SAM). Mutation of the [4Fe-4S] cluster-binding motif diminish the antiviral activity of viperin against WNV, dengue virus, tick-borne encephalitis virus, hepatitis C virus, HIV and Bunyamwera virus, indicating that the radical SAM activity is critical for restricting viral replication (Carlton-Smith and Elliott, 2012; Jiang et al., 2008; Jiang et al., 2010; Nasr et al., 2012; Upadhyay et al., 2014). Given the remarkably broad and complex catalytic activities mediated by the radical SAM superfamily, the enzymatic function of viperin took a long time to emerge. It has now been shown that recombinant rat, human and bovine viperin proteins convert cytidine triphosphate (CTP) to 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP), which inhibits RNA synthesis by flavivirus RNA-dependent RNA polymerases in vitro (Gizzi et al., 2018).
A common cellular response to viral infection is translation repression. Infection activates the integrated stress response (ISR), which phosphorylates the translation initiation factor, eIF2α, to inhibit initiation. Four eIF2α kinases (GCN2, PKR, PERK, HRI) serve as cellular stress sensors that detect distinct stress stimuli and phosphorylate eIF2α (Pakos-Zebrucka et al., 2016). Although PKR and PERK are well-known to be activated by viral double-stranded RNA and viral proteins respectively (Balachandran et al., 2000; Smith, 2014), GCN2 has been identified as a potent antiviral factor against HIV, Sindbis virus and vesicular stomatitis virus (Berlanga et al., 2006; Jiang et al., 2017; Krishnamoorthy et al., 2008). GCN2 is considered as a critical sensor which monitors global translation (Wu et al., 2020), and can be activated by UV irradiation, uncharged tRNAs, or ribosome stalling and collisions (Deng et al., 2002; Dong et al., 2000; Ishimura et al., 2016; Wu et al., 2020). Recently, it has been shown that ribosome collisions activate the GCN2 arm of the ISR, which specifically depends on the mitogen-activated protein kinase kinase kinase (MAPKKK) ZAK (Wu et al., 2020). This signaling pathway mediates inhibition of global translation induced by UV irradiation and amino acid starvation (Wu et al., 2020). Although ribosome collision is widely induced by diverse stresses, the role of ribosome collisions in the innate antiviral responses remains unclear.
In this study, we report that viperin restricts viral replication by inhibiting both cellular and viral translation in a manner dependent on its radical SAM activity. Viperin-induced translation inhibition is triggered by colliding ribosomes that subsequently activate the downstream ISR through the eIF2α kinase GCN2. The colliding ribosome sensor ZAK is required to activate the ISR and inhibit translation. We demonstrate that viperin restricts the replication of flaviviruses by limiting viral protein synthesis through this signaling pathway.
Results
Viperin inhibits viral translation
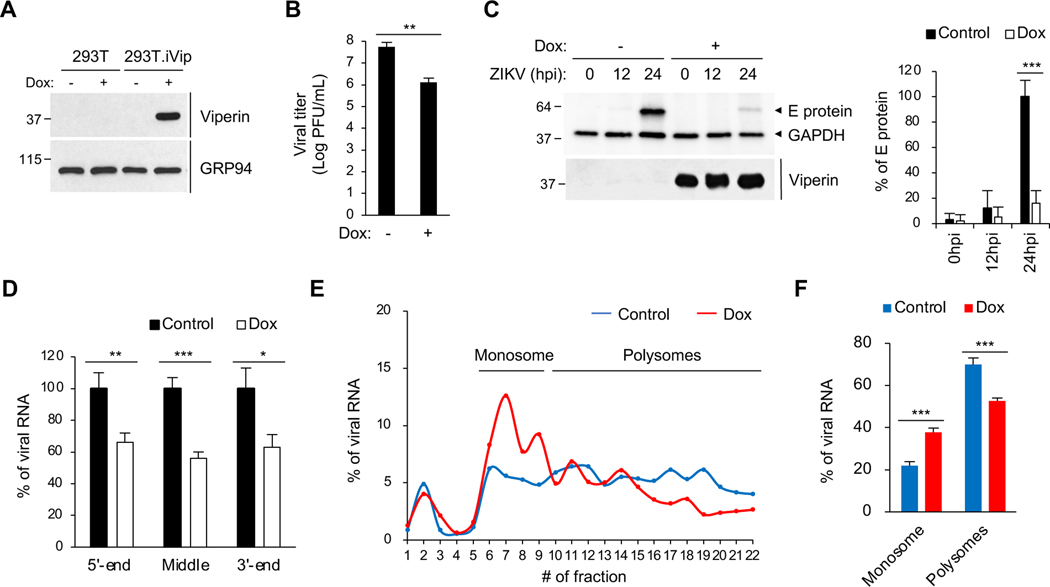
To test the role of viperin in viral replication, we generated a stable 293T cell line expressing doxycycline-inducible viperin (293T.iVip) (Figure 1A). We showed that viperin significantly inhibits ZIKV replication (Figure 1B), consistent with earlier studies (Panayiotou et al., 2018; Van der Hoek et al., 2017; Vanwalscappel et al., 2019; Vanwalscappel et al., 2018), and that it significantly represses the expression of viral protein at 24 hours post-infection (hpi) (Figure 1C, lane 3 and 6). Viperin synthesizes ddhCTP which prematurely terminates RNA transcripts produced by flavivirus RNA-dependent RNA polymerases in vitro (Gizzi et al., 2018). However, it only moderately inhibits viral RNA synthesis in cells infected with ZIKV and DENV-2 (Helbig et al., 2013; Van der Hoek et al., 2017). This could be a result of viperin-mediated inhibition of the synthesis of the RNA-dependent RNA polymerase NS5 of ZIKV (Vanwalscappel et al., 2018). To test whether viperin terminates viral RNA synthesis in ZIKV infected-cells, we monitored viral RNA levels by qRT-PCR using primers targeting the 5’ and 3’ ends or the middle of the viral RNA. Although viperin expression reduced viral RNA transcripts by ~40%, consistent with previous reports (Van der Hoek et al., 2017), all the primers revealed the same level of reduction (Figures 1D and S1A), suggesting that viperin does not cause premature termination of viral RNA transcripts.
Figure 1. Viperin inhibits translation of Zika virus RNA.
(A) Immunoblot analysis of viperin induction in a doxycycline-inducible 293T variant (293T.iVip). Cells were treated with doxycycline (Dox) for 24 h. Cell lysates were analyzed by immunoblotting with anti-viperin and anti-GRP94 antibodies.
(B)Quantification of Zika virus (ZIKV) replication in 293T.iVip cells. Cells were treated with Dox for 24 h and then infected with ZIKV at an MOI of 1 for 24 h, and viral titers were determined by plaque assay. PFU, plaque-forming unit.
(C) Immunoblot analysis of ZIKV envelope (E) protein in 293T.iVip cells. Cells were treated with Dox for 24 h and then infected with ZIKV at an MOI of 1 for 24 h. Cell lysates were analyzed by immunoblotting with anti-flavivirus envelope (E) protein, antiviperin and anti-GAPDH antibodies. Quantification of ZIKV E protein from immunoblot analysis was shown on the right.
(D) qRT-PCR analysis of ZIKV viral RNA in 293T.iVip cells. Cells were treated with Dox and then infected with ZIKV at an MOI of 1 for 24 h. Viral RNA was determined by qRT-PCR targeting the 5’ and 3’ ends or in the middle of the viral RNA and normalized to GAPDH.
(E) Translation analysis of ZIKV RNA in 293T.iVip cells. Cells were treated with Dox for 24 h and then infected with ZIKV at an MOI of 1 for 24 h. Cell lysates were fractionated by sucrose density gradient velocity sedimentation. Viral RNA in sucrose density gradient fractions was determined by qRT-PCR.
(F) Quantification of ZIKV RNA translation in (E). Data are represented as the percentages of ZIKV RNA in monosome and polysome pools.
For (B-D) and (F), data are shown as mean ± SD of three biological repeats (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t test. See also Figures S1 and S2.
The imbalance between the viral RNA and protein levels suggested that viperin could affect translation of viral RNA. To examine this question, we performed polysome profiling coupled with qRT-PCR analysis to directly monitor translation efficiency. The ZIKV genome constitutes a positive-sense, singsle-stranded RNA encoding a polyprotein that is proteolytically processed into individual viral proteins. We measured the distribution of this viral RNA in the polysome profile. Although a small fraction of viral RNA (~5%) remained free (Figure 1E, fraction 1–3, and S1B), most of the viral RNA was associated with ribosomes (Figure 1E, fraction 6–22, and S1B). Importantly, viperin expression caused a drastic shift in viral RNA distribution from actively translating polysomes (Figure 1E, fraction 10–22, and S1B) to the monosome pool (Figure 1E, fraction 6–9, and S1B). Notably, there was a significant increase of viral RNA in the monosome peak and a corresponding decrease in the polysomes (Figure 1F), suggesting that viperin inhibits ZIKV viral RNA translation.
Viperin inhibits host translation in the type I interferon response
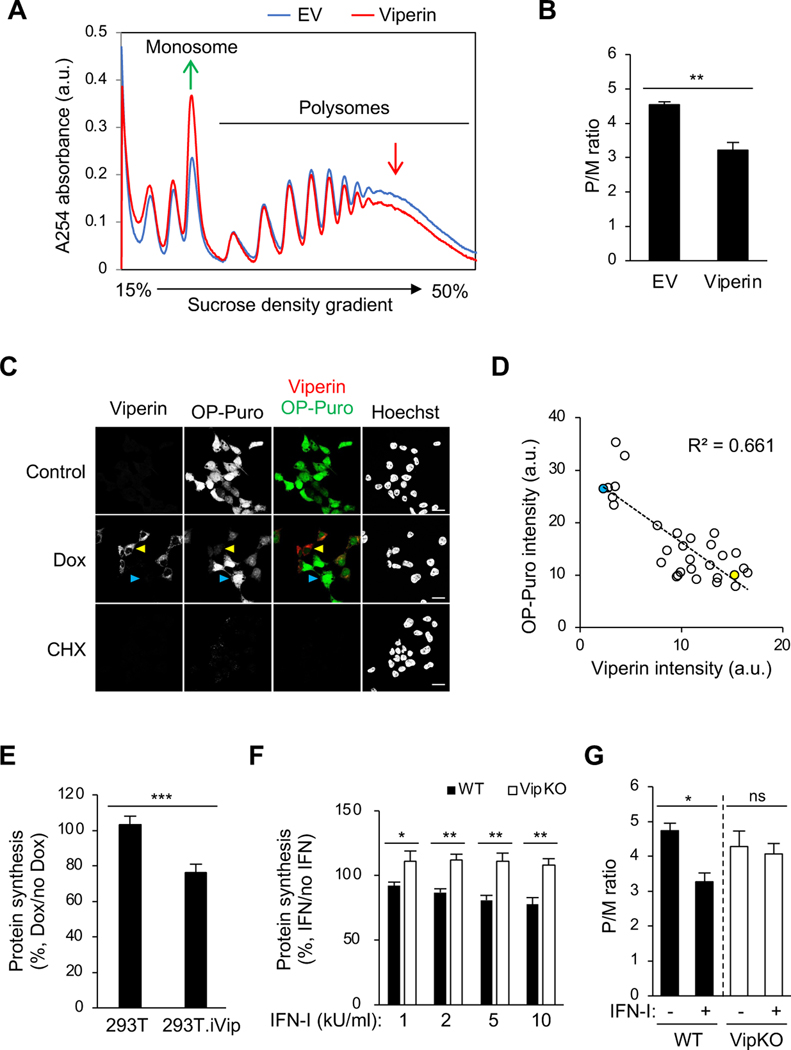
Although the functions of most ISGs remain unclear, several inhibit translation (Schoggins et al., 2011). Given viperin’s broad antiviral activity, we proposed that viperin might function by inhibiting global translation, rather than specific viral RNA translation. To assess this, we again used polysome profiling, which reveals the distribution of the number of ribosomes associated with translating mRNA. We found that viperin induces a global change in the polysome profile, with an increase in monosomes and a decrease in actively translating polysomes (Figure 2A, red line). The decrease in translation was quantified by the ratio of actively translating polysomes to monosomes (P/M ratio), which indicated that viperin significantly inhibits cellular translation (Figure 2B). We confirmed this at the single cell level using O-propargylpuromycin (OP-Puro) labeling (Liu et al., 2012; Nagelreiter et al., 2018), finding a substantial decrease in OP-Puro labeling intensity in doxycycline-treated 293T.iVip cells, (Figure 2C, yellow arrow head). Notably, cells expressing more viperin had reduced protein synthesis compared to those expressing less (Figure 2C; yellow vs cyan arrow heads); viperin expression was inversely correlated with OP-Puro intensity (Figure 2D, R2=0.661). We also showed by flow cytometry that viperin induction inhibits cellular protein synthesis by ~30% (Figure 2E).
Figure 2. Viperin inhibits translation.
(A) Polysome profile analysis of viperin-transfected 293T cells. Cell were transfected with plasmid encoding viperin for 24 h. Cell lysates were cleared by centrifugation, loaded onto a 15–50% sucrose gradient, and subjected to ultracentrifugation. Absorbance was monitored at 254 nm to record the polysome profile. The monosome and polysome pools are indicated. EV, empty vector. a.u., arbitrary unit.
(B) Quantification of polysome profile assay in (A). Data are represented as the polysome-to-monosome (P/M) ratio.
(C) Global protein synthesis analysis by O-propagyl puromycin (OP-Puro) labeling and confocal microscopy in 293T.iVip cells. Cells were treated with Dox, stained by OP-Puro labeling and anti-viperin antibody, and then analyzed by confocal microscopy. High and low viperin expressing cells were indicated by yellow and cyan arrow heads, respectively. Scale bars, 20 μm.
(D) Scatter plot of viperin and OP-Puro fluorescence intensities from individual cells (n=30; R2=0.661). Yellow and cyan dots represent the high and low viperin expressing cells, respectively, as shown in (C). a.u., arbitrary unit.
(E) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in 293T.iVip cells. Cells were treated with Dox, stained by OP-Puro labeling, and analyzed by flow cytometry. Data represent the mean fluorescence intensity ratio between Dox-treated and mock treated cells.
(F) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in immortalized mouse bone marrow-derived macrophage (iBMDM) cell lines from wildtype (WT) and viperin knockout mice (VipKO). Cells were treated with type I interferon (IFN-I) for 18 h, stained by OP-Puro labeling and analyzed by flow cytometry. Data represent the mean fluorescence intensity ratio between IFN-I-treated and mock treated cells. kU, kilounit.
(G) Quantification of polysome profile assay of the IFN-I responses in iBMDM and iBMDM.VipKO cells. Cells were treated with IFN-I for 8 h and analyzed by sucrose density gradient velocity sedimentation. Data are represented as the polysome-to-monosome (P/M) ratio.
For (B) and (E-G), data are shown as mean ± SD of three biological repeats (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t test. ns, not significant.
Viperin is primarily expressed in IFN-stimulated or virally infected cells (Ghosh and Marsh, 2020). To evaluate the function of endogenous viperin during the IFN-I response, we generated immortalized mouse bone marrow-derived macrophage (iBMDM) cell lines from wildtype (WT) and viperin knockout mice (VipKO) and confirmed by immunoblotting that IFN-I only induced viperin in the WT cells (Figure S1C). Using OP-Puro labeling, we found that IFN-I stimulation reduced protein synthesis in WT iBMDM in a dose-dependent manner (Figure 2F, filled bars), while no reduction was seen in iBMDM.VipKO cells (Figure 2F, open bars). We confirmed this by polysome profiling, observing a significant decrease in the P/M ratio only in WT iBMDM (Figure 2G, filled bars), an effect comparable to that of viperin expression in 293T cells (Figure 2B). The decrease results from an increase in monosomes in WT iBMDM (Figure S1D). However, IFN-I stimulation did not affect the polysome profile and P/M ratio in iBMDM.VipKO cells (Figures 2G, open bars, and S1E). Thus, although IFN-I induces hundreds of ISGs, viperin appears to be the critical mediator of translation inhibition during the IFN-I response.
A previous study reported that viperin inhibits RNA synthesis by bacteriophage T7 polymerase (Dukhovny et al., 2018), and premature termination by the viperin product ddhCTP inhibits viral RNA-dependent RNA polymerase activity (Gizzi et al., 2018). Although no evidence supports a role for viperin in cellular RNA synthesis, these observations raise the possibility that viperin might inhibit global translation by limiting transcription. We tested this hypothesis using metabolic labeling with 5-ethynyluridine (5-EU), which measures global RNA synthesis (Jao and Salic, 2008). We found that induction of viperin expression in 293T.iVip did not change the intensity of 5-EU labeling, suggesting no significant effect on transcription (Figure S2A). We also measured RNA reads by next-generation sequencing (RNA-Seq) and found no significant difference in the expression in individual mRNAs between control and viperin-expressing cells (Figure S2B). These results indicate that inhibition of global translation by viperin is not caused by a reduction in transcription.
Radical SAM activity is required for translation inhibition
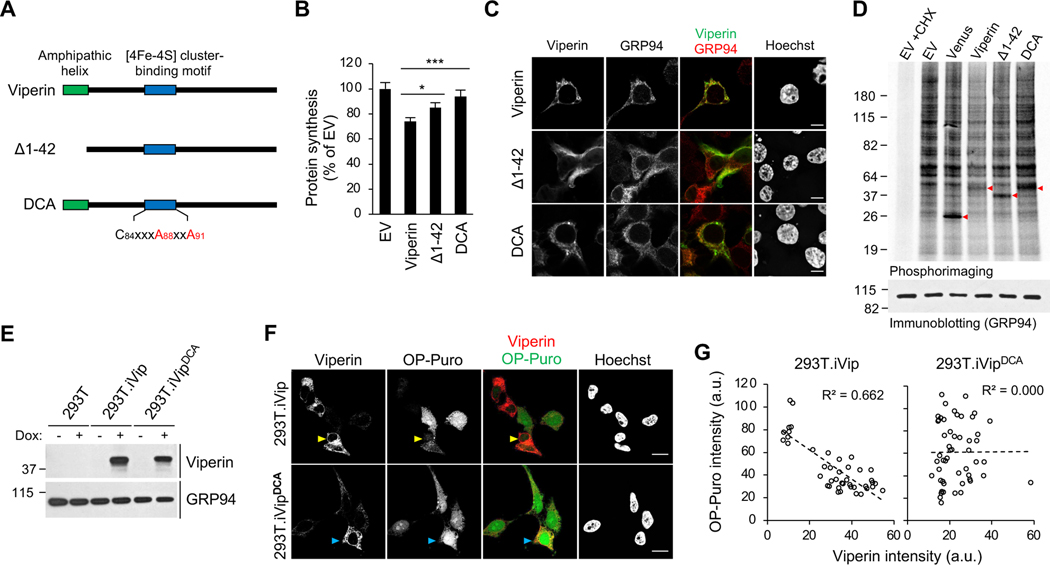
We used mutational analysis to define regions of viperin responsible for translation inhibition. Given that its radical SAM activity is required to restrict the replication of several viruses, including ZIKV, we asked if it is required for translation inhibition. Viperin contains the typical required [4Fe–4S] cluster coordinated by three cysteine residues (Fenwick et al., 2017) and we showed that mutating two of them to alanine (DCA, Figure 3A) resulted in loss of translation inhibition, measured by OP-Puro labeling (Figure 3B). Next we asked whether the localization of viperin on the cytosolic face of the ER via its N-terminal amphipathic helix (Hinson and Cresswell, 2009) is required. Using OP-Puro we found that an ER-localization defective variant of viperin, Δ1–42 (Figures 3A and 3C) (Hinson and Cresswell, 2009), only partially suppressed translation compared to WT viperin (Figure 3B), suggesting that ER localization is important but not necessary. These results were confirmed by monitoring newly synthesized proteins using metabolic labeling with [35S]-methionine/cysteine (Figure 3D). Consistently, we found that the DCA mutant lost the translation inhibition activity (Figure 3D, lane 4 vs 6). Notably, although viperin markedly inhibits metabolic labeling of newly synthesized proteins, the labeled protein profile is virtually unaltered (Figure 3D, lane 2 vs 4), consistent with global inhibition in translation.
Figure 3. Mapping the translation inhibition activity of viperin by mutagenesis.
(A) Schematic illustration of viperin mutants. Viperin, amphipathic helix deletion mutant (Δ1–42), and radical SAM inactive double cysteine-to-alanine mutant (DCA) are shown. The [4Fe-4S] cluster-binding motif (C84xxxC88xxC91) with the mutation sites in red is shown.
(B) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in 293T cells. Cells were transfected with WT and viperin mutants for 24 h, stained by OP-Puro labeling and analyzed by flow cytometry analysis. Data represent the mean fluorescence intensity ratio between cells transfected with the indicated protein and empty vector (EV). Data are shown as mean ± SD of three biological repeats (n = 3). *p < 0.05, ***p < 0.001 by unpaired Student’s t test.
(C) The cellular localization of WT viperin, DCA and Δ1–42 mutants in 293T cells. Cells were transfected with WT and viperin mutants for 24 h, stained by anti-viperin and anti-GRP94 antibodies, and analyzed by confocal immunofluorescence microscopy. GRP94 serves as an endoplasmic reticulum marker. Scale bars, 10 μm.
(D) Metabolic protein labeling in 293T cells transfected with WT and viperin mutants. Cells were pulse labeled with [35S]-methionine/cysteine and analyzed by SDS-PAGE and phosphoimaging. Red arrow heads indicate transfected proteins. EV and Venus fluorescent protein transfections serve as negative controls. Translation inhibitor cycloheximide (CHX) serves as background.
(E) Immunoblot analysis of WT and DCA viperin induction in Dox-inducible 293T variants (293T.iVip and 293T.iVipDCA). Cells were treated with Dox for 24 h. Cell lysates were analyzed by immunoblotting with anti-viperin, anti-GRP94 antibodies.
(F) 293T.iVip and 293T.iVipDCA cells were treated with Dox for 24 h, stained by OP-Puro labeling and anti-viperin antibody and analyzed by confocal microscopy. High viperin expressing cells are indicated by yellow and cyan arrow heads for WT and DCA, respectively. Scale bars, 20 μm.
(G) Scatter plots of viperin and OP-Puro fluorescence intensities from individual cells expressing viperin (n=42; R2=0.662) and DCA mutant (n=51; R2=0.000) shown in (F). a.u., arbitrary unit.
To further investigate the role of the radical SAM activity we generated a doxycycline-inducible 293T cell derivative expressing the DCA mutant (293T.iVipDCA) (Figure 3E, lane 5–6). Consistent with the transient overexpression data (Figure 3B), induction of WT viperin caused translation inhibition (Figure 3F, yellow arrows) whereas induction of the DCA mutant did not (Figure 3F, cyan arrows). We also found that the inverse correlation between cellular viperin expression and translation observed in 293T.iVip cells was abolished in 293T.iVipDCA cells (Figure 3G). Together, these results support a model in which the enzymatic activity of viperin triggers translation inhibition.
The viperin product ddhCTP inhibits translation
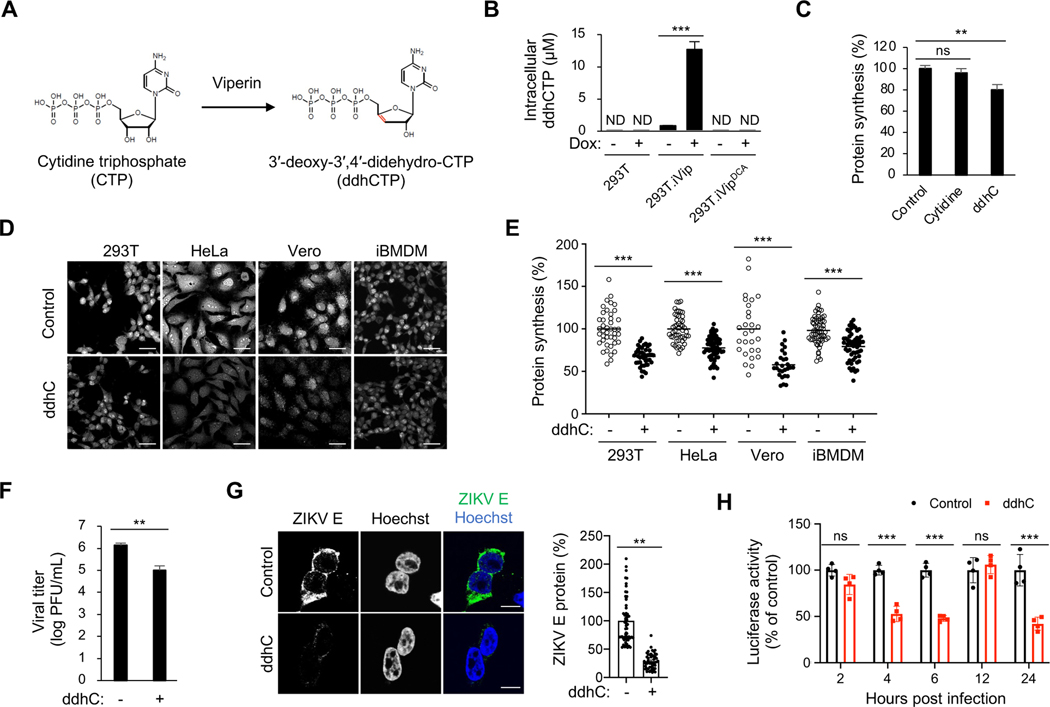
The radical SAM enzymatic activity of viperin is responsible for the synthesis of ddhCTP (Figure 4A) (Gizzi et al., 2018). Consistently, we found that WT viperin, but not the enzymatic inactive DCA mutant, induces ddhCTP synthesis in inducible 293T cells (Figure 4B). To determine whether ddhCTP directly inhibits translation, we treated cells with synthetic ddhC nucleoside, which crosses the plasma membrane and is converted to ddhCTP in the cytosol (Gizzi et al., 2018). Using OP-Puro labeling, we found that ddhC treatment induces a significant decrease in translation (Figure 4C), which phenocopies the consequences of viperin expression. Given that viperin is a highly conserved protein and can be induced in a variety of cell types (Seo et al., 2011a), we performed ddhC treatment and OP-Puro labeling analysis in various cells. We observed that ddhC treatment inhibited translation in human cell lines (293T and HeLa cells), monkey fibroblast cells (Vero cells), and mouse iBMDM (Figures 4D and 4E). We next evaluated the implications of translational inhibition by ddhCTP for viperin’s antiviral activity. We found that synthetic ddhC significantly inhibits ZIKV replication (Figure 4F), consistent with previous reports (Gizzi et al., 2018). Notably, viral protein synthesis was remarkably reduced in cells treated with ddhC (Figure 4G). Using a luciferase reporter ZIKV (Baker et al., 2020), we found that ddhC significantly inhibits ZIKV protein synthesis during initial translation (4 and 6 hpi) and later stages of viral replication (24 hpi) (Figure 4H). Overall, our results suggest that viperin inhibits cellular and viral RNA translation via its enzymatic product, ddhCTP.
Figure 4. Viperin’s enzymatic product induces translation inhibition.
(A) Viperin catalyzes the conversion of cytidine triphosphate (CTP) to 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP) (Gizzi et al., 2018).
(B) ddhCTP formation in 293T.iVip and 293T.iVipDCA cells. Cells were treated with Dox for 24 h and analyzed by mass spectrometry. ND, not detected.
(C) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in 293T cells. Cells were treated with cytidine or alcoholic form of ddhCTP (ddhC) for 24 h, stained by OP-Puro labeling and analyzed by flow cytometry. Data represent the mean fluorescence intensity ratio between treated and mock treated cells (Control).
(D) Global protein synthesis analysis in tissue culture cells. Cells were treated with ddhC for 24 h, stained by OP-Puro labeling and analyzed by confocal microscopy. Scale bars, 50 μm.
(E) Quantification of OP-Puro fluorescence intensity from individual cells shown in (D). Data represent the mean fluorescence intensity ratio between ddhC-treated and mock treated cells.
(F) Quantification of ZIKV replication in 293T cells. Cells were treated with ddhC for 24 h and then infected with ZIKV at an MOI of 1 for 24 h, and viral titers were determined by plaque assay. PFU, plaque-forming unit.
(G) Immunofluorescence analysis of the ZIKV E proteins in 293T cells. Cells were treated with ddhC for 24 h and then infected with ZIKV at an MOI of 1 for 24 h, and stained by anti-E protein antibody and analyzed by confocal immunofluorescence microscopy. Scale bars, 10 μm. Cellular viral E protein levels were determined on the right. Cell numbers measured (n) are shown on the top.
(H) 293T cells were treated with ddhC for 24 h and then infected with NanoLuc reporter ZIKV at an MOI of 2. At indicated time post infection, the cells were lysed and measured for luciferase activities.
For (B), (C) and (E-G), data are shown as mean ± SD of three biological repeats (n = 3).
*p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student’s t test. ns, not significant.
Viperin activates the integrated stress response
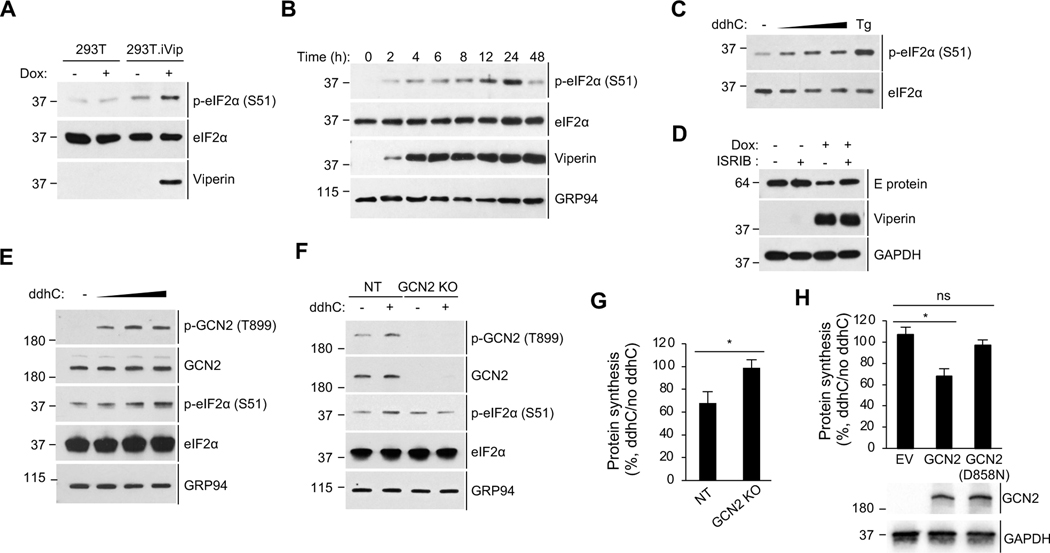
Although viperin inhibits cellular translation the profiles of newly synthesized proteins were unchanged (Figure 3D, lane 2 and 4), suggesting that viperin inhibits translation globally rather than targeting a subset of mRNAs. Given that the IFN-I response down-regulates global translation via the integrated stress response (ISR) and mTORC1 pathway (Ivashkiv and Donlin, 2014), we investigated the role of viperin in activating the molecular components of these pathways in 293T.iVip cells. First, we found that viperin induces eIF2α phosphorylation, a marker of ISR pathway activation (Figure 5A, lane 3-4). Phosphorylation was apparent 3 hours post doxycycline induction, when viperin first became detectable, and peaked between 12–24 hours (Figure 5B). Moreover, we showed that synthetic ddhC nucleoside increased eIF2α phosphorylation in a dose-dependent manner (Figure 5C). In contrast, viperin had no effects on the activation of mTORC1 pathway markers (Figure S3). Given the activation of the ISR by viperin, we asked whether this pathway is involved in the inhibition of viral protein synthesis. Consistently, viperin expression inhibited ZIKV protein synthesis (Figure 5D, lane 1 and 3), but this was abolished by pretreatment of cells with ISRIB (Figure 5D, lane 3–4), a compound that blocks the effects of the ISR activation (Sidrauski et al., 2015a; Sidrauski et al., 2015b). These results suggest that viperin activates the ISR, leading to translation inhibition that serves as an antiviral mechanism.
Figure 5. Viperin activates the integrated stress response (ISR) via the eIF2α kinase GCN2.
(A) Immunoblot analysis of phosphorylation of eIF2α in 293T.iVip cells. Cells were treated with doxycycline (Dox) for 24 h. Cell lysates were analyzed by immunoblotting with anti-phospho-eIF2α (S51), anti-eIF2α and anti-viperin antibodies.
(B) Immunoblot analysis of the induction time course of viperin and the phosphorylation of eIF2α in 293T.iVip cells. Cells were treated with doxycycline (Dox) for the indicated times. Cell lysates were analyzed by immunoblotting with anti-phospho-eIF2α (S51), anti-eIF2, anti-viperin and anti-GRP94 antibodies.
(C) Immunoblot analysis of the phosphorylation of eIF2α in 293T cells treated with 0.5, 1, 2 mM ddhC for 24 h and analyzed by immunoblotting with anti-phospho-eIF2α (S51) and anti-eIF2α antibodies. The ISR activator tapsigargin (Tg) serves as a positive control.
(D) Immunoblot analysis of ZIKV E proteins in 293T.iVip cells. Cells were treated with Dox and an ISR pathway inhibitor ISRIB for 6 h and then infected with ZIKV at an MOI of 1 for 24 h and analyzed by immunoblotting with anti-flavivirus envelope (E) protein, anti-viperin and anti-GAPDH antibodies.
(E) Immunoblot analysis of the phosphorylation of eIF2α and GCN2 in 293T cells. Cells were treated with 0.5, 1, 2 mM ddhC for 24 h and analyzed by immunoblotting with antiphospho-GCN2 (T899), anti-GCN2, anti-phospho-eIF2α (S51), anti-eIF2α and anti-GRP94 antibodies.
(F) Immunoblot analysis of the phosphorylation of eIF2α in GCN2 knockout 293T cells (GCN2 KO) and control cells expressing non-targeting gRNA (NT) treated with ddhC for 24 h. Cell lysates were analyzed by immunoblotting with the indicated antibodies.
(G) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in NT and GCN2 KO cells. Cells were treated with ddhC for 24 h and stained by OP-Puro labeling followed by flow cytometry analysis. Data represent the mean fluorescence intensity ratio between ddhC-treated and mock treated cells.
(H) Global protein synthesis analysis by OP-Puro labeling and flow cytometry analysis in GCN2 KO cells. Cells were transfected with empty vector (EV) or plasmids encoding WT GCN2 or GCN2 kinase mutant [GCN2(D858N)] for 24 h, then treated with ddhC for 24 h, stained by OP-Puro and analyzed by flow cytometry. Data represent the mean fluorescence intensity ratio between ddhC-treated and mock treated cells. The expression of GCN2 proteins is shown below using immunoblot analysis with anti-GCN2 and anti-GAPDH antibodies.
For (G) and (H), data are shown as mean ± SD of three biological repeats (n = 3). *p < 0.05 by unpaired Student’s t test. ns, not significant. See also Figures S3 and S4.
Activation of the ISR inhibits translation in cells subjected to a variety of cellular stresses (Pakos-Zebrucka et al., 2016). Four eIF2α kinases, GCN2, PKR, PERK and HRI, act as sensors for detecting distinct cellular stresses. They are activated by autophosphorylation when the relevant cellular stress is detected (Pakos-Zebrucka et al., 2016). We therefore asked whether synthetic ddhC nucleoside activates each of the kinases by looking at their phosphorylation. We showed that ddhC induces the phosphorylation of GCN2 in a dose-dependent manner but has no effect on the other kinases (Figures 5E and S4). To further evaluate the role of GCN2 we generated GCN2 knockout 293T cells (GCN2 KO) using CRISPR/Cas9. Consistently, we showed that, while ddhC induced eIF2α phosphorylation in control cells (Figure 5F, lane 1–2), it failed to do so in the cells lacking GCN2 (Figure 5F, lane 3–4). Moreover, using OP-Puro labeling, we found that GCN2 is required for the ddhC-induced translation inhibition (Figure 5G). Given that GCN2 kinase activity is required to phosphorylate eIF2α (Inglis et al., 2019), we asked whether this activity mediates translation inhibition by ddhC. GCN2 KO cells complemented with WT GCN2 but not with an inactive GCN2 mutant (D858N), are competent to reduce translation upon ddhC treatment (Figure 5H). These observations support a central role for GCN2 in activation of the ISR by viperin.
Viperin induces ribosome collisions that activate the ISR
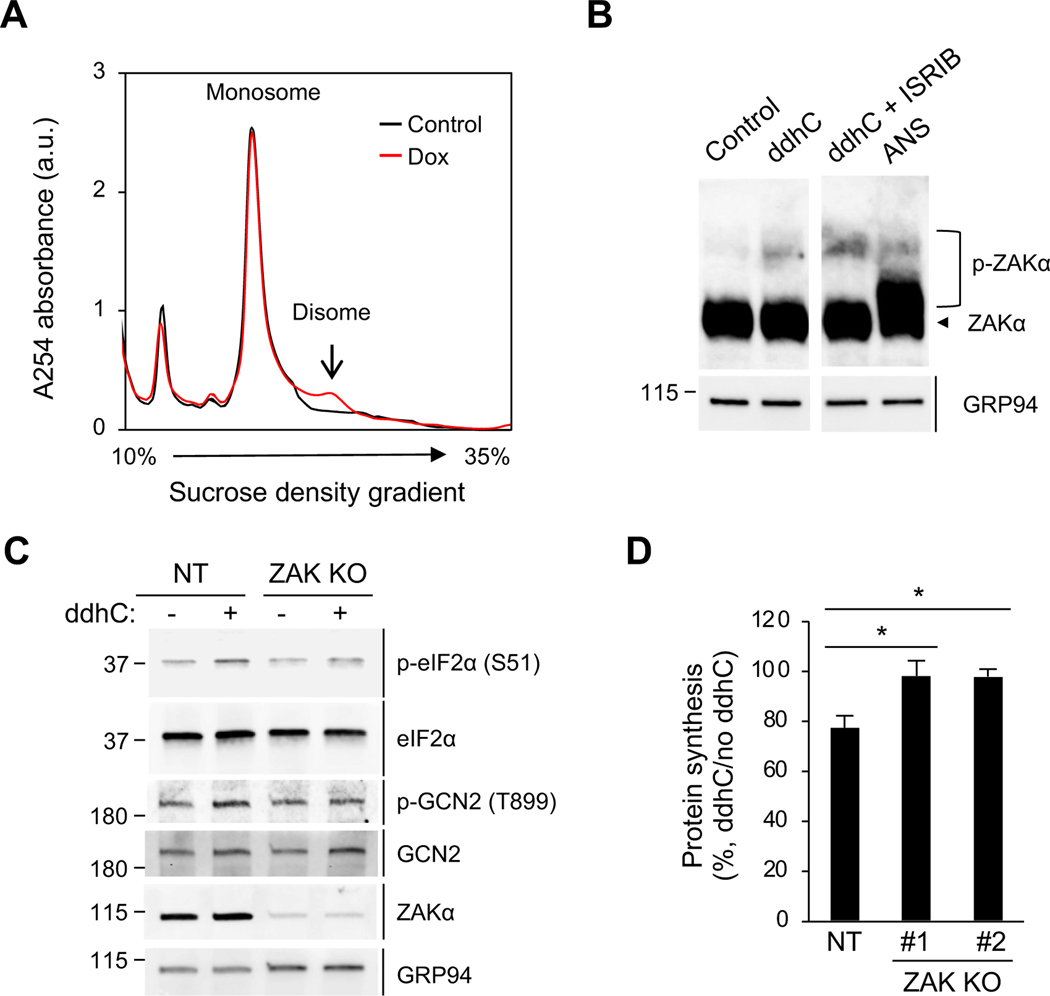
In light of recent studies implicating ribosome collisions in activation of the GCN2 arm of the ISR pathway (Wu et al., 2020; Yan and Zaher, 2021), we asked whether viperin activates GCN2 via induction of ribosome collisions. Ribosome collisions can be directly monitored by the accumulation of RNase-resistant disomes in polysome profiles (Wu et al., 2020; Yan and Zaher, 2021). We found that although the majority of polysomes are digested to monosomes, a fraction of ribosomes resists RNase digestion and sediments as disomes in viperin-expressing cells (Figure 6A, red, and S5A). Consistent with the transient transfection results (Figure 2A), while viperin increases the 80S monosome population the disome population remains unchanged without RNase digestion (Figure S5B). We also found that expression of the DCA mutant fails to induce RNase-resistant disomes (Figure S5C), suggesting that the radical SAM activity and thus ddhCTP are required to trigger ribosome collisions. Recently, Wu et al. identified the MAPKKK ZAK as a sensor for colliding ribosomes. The authors connected ribosome collisions to the GCN2-ISR pathway and showed that the collisions activate the phosphorylation of the α isoform of ZAK (ZAKα). We found that synthetic ddhC nucleotide induces ZAKα phosphorylation (Figure 6B, lane 1–2). Ribosome collisions block translation initiation through the ISR, which reduces the formation of new colliding ribosomes, and blocking the effects of the ISR activation by ISRIB further enhances ribosome collisions and ZAKα phosphorylation (Wu et al., 2020). Consistently, we found here that ZAKα phosphorylation by ddhC was enhanced by ISRIB (Figure 6B, lane 3). To determine whether ZAKα is required for the activation of the ISR by ddhC we generated ZAK KO 293T cell lines using CRISPR/Cas9. We found that ddhC treatment induced the phosphorylation of eIF2α and GCN2 in control cells (Figure 6C, lane 1–2) while no such changes were observed in ZAK KO cells (Figure 6C, lane 3–4). In addition, and consistent with the critical role of ZAK in connecting viperin-induced ribosome collisions and translation inhibition, ddhC failed to inhibit translation in cells lacking ZAK (Figure 6D).
Figure 6. ZAK is required for viperin-induced translation inhibition.
(A) Polysome profile analysis of the formation of RNase-resistant disomes in 293T.iVip cells. Cell were treated with Dox for 24 h. Cell lysates were cleared by centrifugation, digested by RNase, loaded onto a 10–35% sucrose gradient, and subjected to ultracentrifugation. Absorbance was monitored at 254 nm to record the polysome profile. The monosome and disome pools are indicated. a.u., arbitrary unit.
(B) Immunoblot analysis of phosphorylation of ZAKα in 293T cells. Cells were treated with ddhC or co-treated with ISRIB for 24 h and analyzed by phos-tag gel electrophoresis and immunoblotting with anti-ZAK and anti-GRP94 antibodies. Anisomycin (ANS) serves as a positive control. An irrelevant lane was spliced out between lane 2 and 3.
(C) Immunoblot analysis of the phosphorylation of eIF2α and GCN2 in ZAK knockout 293T cells (ZAK KO) and control cells expressing non-targeting gRNA (NT). Cells were treated with ddhC for 24 h. Cell lysates were analyzed by immunoblotting with antiphospho-eIF2α (S51), anti-eIF2α, anti-phospho-GCN2 (T899), anti-GCN2, anti-ZAK and anti-GRP94 antibodies.
(D) NT and two ZAK KO cell lines (#1 and #2) were treated with ddhC for 24 h and stained by OP-Puro labeling followed by flow cytometry. Data are shown as mean ± SD of three biological repeats (n = 3). *p < 0.05 by unpaired Student’s t test. See also Figures S5 and S6.
Viperin restricts viral replication via ribosome collision-dependent translation inhibition
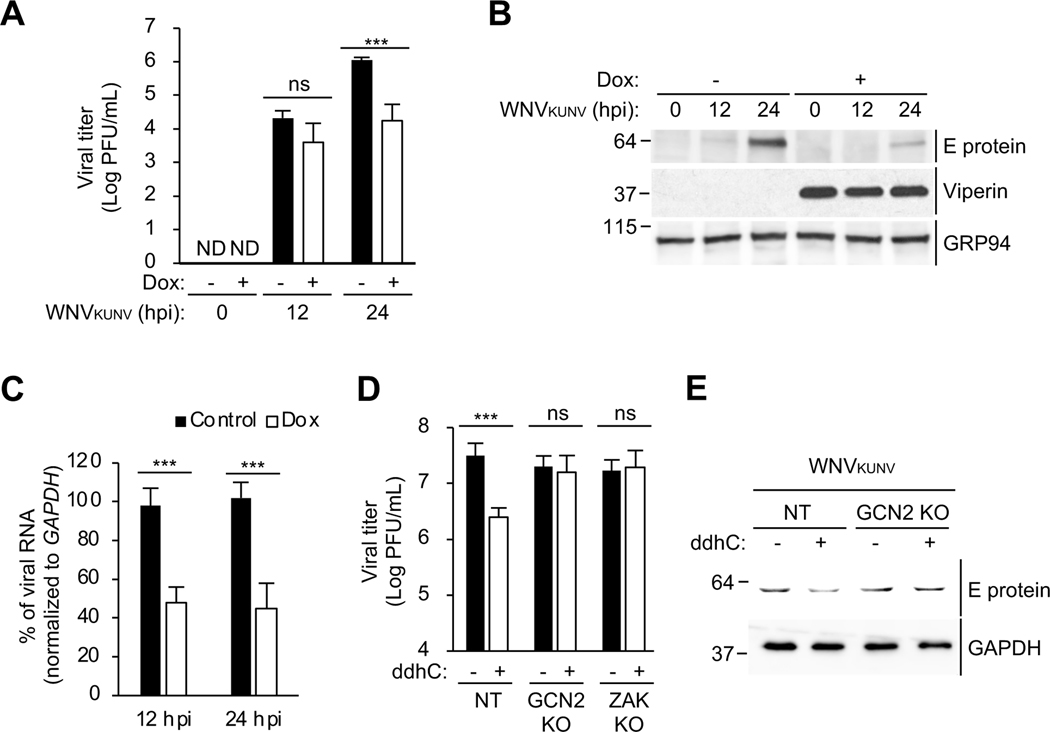
We next evaluated the implications of our mechanistic insights into viperin’s antiviral activity. Viperin was previously shown to restrict the infection of WNV in vivo by an unknown mechanism (Szretter et al., 2011). Consistent with this, viperin induction in 293T.iVip cells significantly reduced the replication of WNV Kunjin strain (WNVKUNV) (Figure 7A). We examined the effects of viperin on WNVKUNV viral protein expression and found ~80% decrease at 24 hpi in viperin-expressing cells (Figure 7B, lane 3 and 6), while viral RNA levels were only moderately attenuated (Figure 7C). We next asked whether GCN2 and ZAK are necessary for inhibiting WNVKUNV replication in response to ddhC treatment. First, we showed that the replication of WNVKUNV is significantly inhibited by ddhC in control cells (Figure 7D), similar to the effect on ZIKV replication (Gizzi et al., 2018). However, ddhC failed to inhibit viral replication in cells lacking GCN2 or ZAK (Figure 7D). Second, ddhC reduced WNVKUNV viral protein expression in control cells (Figure 7E, lane 1–2) but not in GCN2 KO cells (Figure 7E, lane 3–4). Together, these results support a model in which viperin inhibits viral replication by triggering the ISR via ribosome collisions.
Figure 7. GCN2 and ZAK are required for the inhibition of viral protein synthesis and viral replication by viperin.
(A) 293T.iVip cells were treated with Dox and infected with West Nile virus Kunjin strain (WNVKUNV) at an MOI of 1. Viral titers were determined by plaque assay. PFU, plaque-forming unit. hpi, hours post-infection. ND, not detected.
(B)Immunoblot analysis of WNVKUNV E proteins in 293T.iVip cells. 293T.iVip cells were treated with Dox for 24 h, infected with WNVKUNV at an MOI of 1. Cell lysates were analyzed by immunoblotting with anti-flavivirus envelope (E) protein, anti-viperin and anti-GRP94 antibodies. hpi, hours post-infection.
(C) 293T.iVip cells were treated with Dox and then infected with WNVKUNV at an MOI of 1 for 24 h. Cells were harvested for RNA extraction. Viral RNA was determined by qRT-PCR and normalized to GAPDH.
(D-E) NT, GCN2 KO and ZAK KO cells were treated with ddhC for 24 h and infected with WNVKUNV at an MOI of 1 for 24 h. Viral titers were determined by plaque assay (D). PFU, plaque-forming unit. Cell lysates were analyzed by immunoblotting with anti-anti-flavivirus E protein and anti-GAPDH antibodies.
For (A), (C) and (D), data are shown as mean ± SD of three biological repeats (n = 3). ***p < 0.001 by unpaired Student’s t test. ns, not significant.
Discussion
Viperin inhibits replication of several DNA viruses, RNA viruses and retroviruses, and multiple mechanisms have been proposed to explain this. Here we have defined a novel mechanism that is compatible with the ability of viperin to block replication of a wide variety of viruses. We demonstrate that viperin is responsible for the inhibition of protein synthesis observed during the IFN-I response. Mutational analysis showed that this depends on the radical SAM-dependent activity of viperin, which synthesizes the nucleotide triphosphate ddhCTP from the precursor CTP (Gizzi et al., 2018). Importantly, translation inhibition can also be induced in cells treated with exogenous ddhC, which is converted to ddhCTP through the action of well-known intracellular kinases. Our work reveals a mechanism where ddhCTP functions as a key molecular trigger for the ribosome collision sensor ZAK, leading to activation of the integrated stress response (ISR). The pathway is dependent on the eIF2α kinase GCN2 that bridges the ribosome collision signal with global translation inhibition.
Limiting protein synthesis is an effective antiviral mechanism because viral protein synthesis depends on the host translation machinery. However, restricting translation of both host RNA and viral RNA can be considered a ‘scorched earth policy’ when employed to reduce viral replication because of potentially detrimental effects on the host. Viperin moderately attenuates host translation (Figure 2), while significantly reducing viral protein synthesis (Figures 1 and 7). In addition, we found that viperin expression and ddhC treatment led to a small reduction (10% per day) in cell proliferation (Figure S6), whereas the viral replication of ZIKV and WNVKUNV were reduced by approximately 100-fold (Figures 1 and 7), suggesting some selective targeting of viruses. How this might be mediated requires further study. Virus-encoded proteins are essential for almost every step in viral replication. For example, in the case of ZIKV replication, the helicase NS3 and the RNA-dependent RNA polymerase NS5 are required for viral RNA replication (Xu et al., 2019); viral protease NS3 and its cofactor NS2B are critical for ZIKV polyprotein processing and maturation (Lei et al., 2016; Zhang et al., 2016). In addition, viral capsid, envelope and membrane proteins are essential for packaging and producing infectious virus. Thus, inhibiting viral RNA translation not only reduces overall viral protein synthesis but also has major downstream effects on multiple mechanisms that require viral protein activities. Hence, translation attenuation can be amplified in an infected cell by its downstream consequences, resulting in a substantial decrease in viral replication. When the pattern is reproduced in uninfected cells by IFNs released by infected cells, exponential amplification of translation inhibition may effectively protect an infected individual.
We have shown that viperin inhibits global translation through the GCN2 arm of the ISR. It is well established that viral infections activate the eIF2α kinases PKR, PERK, and GCN2, all of which phosphorylate the initiation factor eIF2α to inhibit global translation. For example, PKR is activated by double-stranded RNA (dsRNA), an indispensable intermediate for the replication of the viral genome for many viruses (Garcia et al., 2006). Viral infection can also induce the unfolded protein response (UPR) which activates PERK (Smith, 2014). GCN2 is directly activated upon the binding of Sindbis virus and HIV-1 genomic RNA (Berlanga et al., 2006; del Pino et al., 2012).
Mutations that inactivate GCN2 increase the susceptibility to infection by DNA viruses, including human adenovirus and mouse cytomegalovirus (Won et al., 2012), and elimination of the GCN2 gene increases susceptibility to infection by RNA viruses (del Pino et al., 2012; Krishnamoorthy et al., 2008). Consistent with these observations, we find that GCN2 plays a critical role in the innate antiviral response, and that the resulting rapid phosphorylation of eIF2α reduces global translation and consequently viral replication (Figures 5 and 7). Moreover, active GCN2 inhibits retroviral infection by phosphorylating retroviral integrases that inhibit the integration of viral genomes (Jaspart et al., 2017). Thus, viperin-induced activation of GCN2 may result in phosphorylation of viral proteins, providing an additional antiviral activity.
Ribosome collisions are widespread on endogenous mRNAs (Guydosh and Green, 2014; Meydan and Guydosh, 2020), and can be resolved by surveillance mechanisms for mRNA [no-go decay (NGD) and nascent peptide ribosome-mediated quality control (RQC)] (Brandman and Hegde, 2016). Ribosome collisions can be induced by cellular stresses, such as UV irradiation or amino acid starvation (Deng et al., 2002; Dong et al., 2000). Failure to resolve ribosome collisions stimulates the autophosphorylation of MAPKKK ZAK, which activates the GCN2 arm of the ISR to inhibit translation initiation and reduce further collisions (Wu et al., 2020). Here we show that viperin induces ribosome collisions and ZAK phosphorylation, leading to the ISR activation and translation inhibition (Figure 6). We also demonstrate that the signaling pathway is required for the inhibition of viral RNA translation and replication (Figure 7). Our results suggest a previously unknown antiviral mechanism operating in which the viperin product ddhCTP triggers ribosome collisions that results in inhibition of viral RNA translation. Persistent ribosome collisions activate the ribotoxic stress response (RSR) by a ZAK-dependent signaling cascade through activation of p38 and c-Jun N-terminal kinase (JNK), which respectively result in cell cycle arrest and apoptosis (Wu et al., 2020). Together with the NGD/RQC and ISR, these multiple layers of regulation suggest that the severity of ribosome collisions determines cell fate (Wu et al., 2020). Notably, although ddhCTP activates the GCN2 arm of the ISR, no RSR activation was observed (Figure S5). Our results suggest that viperin specifically activates the ISR to establish an antiviral state without inducing RSR.
Limitations of the study
Emerging infectious diseases (EIDs) cause a significant burden on public health and global economies. However, antiviral drugs/therapeutics are currently unavailable against most EIDs. Viperin has been shown to inhibit the replication of a broad spectrum of viruses, including several EIDs like ZIKV, WNV, Chikungunya virus, dengue virus, tick-born encephalitis virus, and influenza virus (Ghosh and Marsh, 2020). Here we find that viperin inhibits the viral replication of ZIKV and WNVKUNV by limiting global protein synthesis. Although viruses rely on host translation machinery to produce viral proteins, it is well known that some viruses antagonize IFN-induced translation inhibition. We do not know whether viperin can restrict other EIDs by such antiviral mechanism. Although ddhCTP triggers ribosome collisions, and the downstream ISR pathway, the molecular mechanism by which it initiates this is unclear. It has been shown that ddhCTP can terminate in vitro viral RNA synthesis, which results in early terminated RNA products (Gizzi et al., 2018). ddhCTP may generate defective cellular mRNA which then triggers ribosome collisions. Translation of faulty mRNAs can result in induction of ribosome stalling and collisions (Yan and Zaher, 2019). However, further study is required to determine how ddhCTP induces ribosome collisions. In addition, viperin induces a modest inhibition in cellular translation but greatly reduces viral protein synthesis, suggesting that viperin may employ additional antiviral activities. ddhCTP has been shown to terminate RNA synthesis by several flavivirus RNA-dependent RNA polymerases (Gizzi et al., 2018). Although we failed to directly observe an effect on ZIKV RNA replication by addition of ddhC to cell culture (Figure 1D), the nucleotide may exert two antiviral activities; affecting viral RNA replication and protein synthesis. Moreover, viperin has been shown to modulate cellular nucleotide pools and mitochondrial metabolism (Ebrahimi et al., 2020), which can potentially limit viral RNA and protein syntheses. Clearly, further study is required to determine how these antiviral activities are coordinated to restrict viral replication. Understanding the mechanisms of action of the natural antiviral compound ddhCTP may facilitate the development of broad-spectrum antiviral drugs.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Cresswell (peter.cresswell@yale.edu).
Materials Availability
All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
RNA sequencing data are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) under accession number GEO: GSE142015. Accession number is listed in the key resources table. Original western blot images and microscopy data reported in this paper have been deposited at Mendeley at http://dx.doi.org/10.17632/dyxgt2g7cm.2.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Viperin (MaP.VIP) | Wang et al., 2007 | N/A |
| GRP94 | Enzo Life Sciences | Cat#ADI-SPA-851; RRID: AB_10615790 |
| GAPDH | Proteintech | Cat#60004–1-Ig; RRID: AB_2107436 |
| Phospho-4E-BP1 (Ser65) | Cell Signaling Technology | Cat#9451; RRID: AB_330947 |
| 4E-BP1 | Cell Signaling Technology | Cat#9644; RRID: AB_2097841 |
| Phospho-S6 Ribosomal Protein (Ser235/236) | Cell Signaling Technology | Cat#4857; RRID: AB_2181035 |
| S6 Ribosomal Protein (S6) | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| eIF2α | Cell Signaling Technology | Cat#5324; RRID: AB_10692650 |
| phospho-eIF2α (Ser51) | Cell Signaling Technology | Cat#3597; RRID: AB_390740 |
| phospho-p38 (Thr180/Tyr182) | Proteintech | Cat#9211; RRID: AB_331641 |
| p38 | Proteintech | Cat#9212; RRID: AB_330713 |
| phospho-JNK (Thr183/Tyr185) | Proteintech | Cat#4668; RRID: AB_823588 |
| Flavivirus envelope protein | Millipore | Cat#MAB10216; RRID: AB_827205 |
| GCN2 | Cell Signaling Technology | Cat#3302; RRID: AB_2277617 |
| phospho-GCN2(T899) | Abcam | Cat#ab75836; RRID: AB_1310260 |
| PERK | Cell Signaling Technology | Cat#3192; RRID: AB_2095847 |
| HRI | Proteintech | Cat#20499–1-AP; RRID: AB_10697665 |
| PKR | Cell Signaling Technology | Cat#3072; RRID: AB_2277600 |
| ZAK | Bethyl Laboratories | Cat#A301–993A; RRID: AB_1576612 |
| goat anti-mouse IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11017; RRID: AB_2534084 |
| goat anti-mouse IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21236; RRID: AB_2535805 |
| goat anti-rat IgG, Alexa Fluor 546 | Thermo Fisher Scientific | Cat#A-11081; RRID: AB_2534125 |
| goat anti-rat IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A-11007; RRID: AB_10561522 |
| goat anti-rabbit IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21244; RRID: AB_2535812 |
| goat anti-mouse IgG-HRP | Jackson ImmunoResearch Laboratories | Cat#115–035-146; RRID: AB_2307392 |
| goat anti-rabbit-HRP | Jackson ImmunoResearch Laboratories | Cat#111–005-144; RRID: AB_2337919 |
| goat anti-rat IgG-HRP | Jackson ImmunoResearch Laboratories | Cat#112–005-167; RRID: AB_2338101 |
| Bacterial and virus strains | ||
| NEB 5-α Competent E. coli (High Efficiency) | New England Biolabs | Cat#C2987 |
| Zika virus Cambodia/FSS13025 strain | World Reference Center for Emerging Viruses and Arboviruses at University of Texas Medical Branch at Galveston | NCBI:txid2316109 |
| NanoLuc reporter ZIKV | Baker et al., 2020 | N/A |
| West Nile virus Kunjin strain (WNVKUNV) | Dr. Philip Armstrong | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ddhC | This study | N/A |
| Sucrose | Sigma-Aldrich | Cat#S0389 |
| Dithiothreitol (DTT) | American Bioanalytical | Cat#AB00490 |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | Cat#14190144 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat#11965092 |
| Fetal Bovine Serum | Gemini Bio | Cat#100–106 |
| Gibson Assembly Master Mix | New England Biolabs | Cat#E2611 |
| Q5 Hot Start High-Fidelity 2X Master Mix | New England Biolabs | Cat#M0494 |
| ZymoPURE II Plasmid Midiprep Kit | Zymo Research | Cat#D4201 |
| MEGAscript T7 Transcription Kit | Thermo Fisher Scientific | Cat#AMB13345 |
| ClaI | New England Biolabs | Cat#R0197S |
| EcoRI-HF | New England Biolabs | Cat# R3101S |
| Bovine Serum Albumin (BSA) | American Bioanalytical | Cat#AB01088 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat#15596026 |
| Ethanol 200 proof | Decon Labs | Cat#2716 |
| GlycoBlue | Thermo Fisher Scientific | Cat#AM9515 |
| 4x Laemmli Sample Buffer | Bio-Rad | Cat#1610747 |
| Trans-Blot Turbo™ 5x Transfer Buffer | Bio-Rad | Cat#10026938 |
| Doxycycline | Sigma-Aldrich | Cat#D9891 |
| Universal Type I IFN | PBL Assay Science | Cat#11200–2 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| Glutamax | Thermo Fisher Scientific | Cat#35050061 |
| Saponin | Sigma-Aldrich | Cat#S7900 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| dialyzed fetal bovine serum | Thermo Fisher Scientific | Cat#A3382001 |
| [35S]-methionine/cysteine | PerkinElmer Life Science | Cat#NEG772014MC |
| Tris-HCl | American Bioanalytical | Cat#AB02005 |
| IGEPAL CA-630 | Sigma-Aldrich | Cat#I8896 |
| PhosSTOP | Sigma-Aldrich | Cat#PHOSS-RO |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| RNaseOUT | Thermo Fisher Scientific | Cat#10777019 |
| Rapamycin | Cell Signaling Technology | Cat#9904 |
| 16% paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Goat serum | Thermo Fisher Scientific | Cat#16210064 |
| ProLong Gold Anti-Fade Reagent | Thermo Fisher Scientific | Cat#P36930 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#H3570 |
| O-Propargylpuromycin | Abcam | Cat#ab146664 |
| L-Ascorbic acid | Sigma-Aldrich | Cat#A4544 |
| Copper sulfate | Sigma-Aldrich | Cat#C1297 |
| Azide-fluor 488 | Sigma-Aldrich | Cat#760765 |
| Cycloheximide | Sigma-Aldrich | Cat#C4859 |
| Potassium acetate | Sigma-Aldrich | Cat#P5708 |
| Magnesium chloride | Sigma-Aldrich | Cat#M8266 |
| RNase A | Sigma-Aldrich | Cat#R6513 |
| Sodium chloride | American Bioanalytical | Cat#AB01915 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat#EDS |
| Acetonitrile | Sigma-Aldrich | Cat#34851 |
| Methanol | Sigma-Aldrich | Cat#34860 |
| Water | American Bioanalytical | Cat#AB02128 |
| Formic acid | Thermo Fisher Scientific | Cat#A117–50 |
| DMEM, high glucose, no glutamine, no methionine, no cystine | Thermo Fisher Scientific | Cat#21013024 |
| Thapsigargin | Sigma-Aldrich | Cat#T9033 |
| Sodium arsenite | Sigma-Aldrich | Cat#S7400 |
| Polyinosinic:polycytidylic acid [poly(I:C)] | Sigma-Aldrich | Cat#P9582 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668500 |
| OptiMEM | Thermo Fisher Scientific | Cat#11058021 |
| Integrated stress response inhibitor (ISRIB) | Sigma-Aldrich | Cat#SML0843 |
| 5-Ethynyluridine | Abcam | Cat#ab146642 |
| Tween-20 | Sigma-Aldrich | Cat#P7949 |
| Triton X-100 | American Bioanalytical | Cat#AB02025 |
| Critical commercial assays | ||
| Luna Universal One-Step RT-qPCR Kit | New England Biolabs | Cat#E3005 |
| Bradford protein assay | Bio-Rad | Cat#5000001 |
| Power SYBR Green (Thermo Fisher Scientific) | Thermo Fisher Scientific | Cat#4367659 |
| CellTrace CFSE Cell Proliferation Kit (Invitrogen) | Thermo Fisher Scientific | Cat#C34570 |
| Annexin V-FITC Apoptosis Detection Kit | Thermo Fisher Scientific | Cat#BMS500FI |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat#18064014 |
| NEBNext rRNA Depletion Kit (NEB) | New England Biolabs | Cat#E6310 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020594 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE142015 |
| Raw Western blot and microscopy images | This paper; Mendeley Data | DOI:10.17632/dyxgt2g7cm.2 |
| Experimental models: Cell lines | ||
| Human: HEK293T | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| African green monkey: Vero | ATCC | Cat#CCL-81; RRID: CVCL_0059 |
| Human: HeLa | ATCC | Cat#CCL-2; RRID: CVCL_0030 |
| Hamster: BHK-21 | ATCC | Cat#CCL-10; RRID: CVCL_1915 |
| Mouse: iBMDM | This study | N/A |
| Mouse: iBMDM.VipKO | This study | N/A |
| Human: 293T.iVip | This study | N/A |
| Human: HEK293T-Cas9 | Dr. Jorge Galán Chang et al., 2016 |
N/A |
| Human: 293T.GCN2KO | This study | N/A |
| Human: 293T.ZAKKO | This study | N/A |
| Experimental models: Organisms | ||
| Mouse: B6 (C57BL/6J) | Jackson Laboratory | Cat#000664; RRID: IMSR_JAX:000664 |
| Mouse: Viperin KO (B6.129-Rsad2tm1Kchc/CresJ) | Seo et al., 2011 | RRID:IMSR_JAX:032321 |
| Oligonucleotides | ||
| All standard primers for qRT-PCR | This study | Table S1 |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher Scientific | Cat#V79020 |
| pcDNA3.1-Venus | Hee and Cresswell, 2017 | N/A |
| pcDNA3.1-viperin | Seo et al., 2011 | N/A |
| pcDNA3.1-viperin (d1–42) | Seo et al., 2011 | N/A |
| pcDNA3.1-viperin (DCA) | Seo et al., 2011 | N/A |
| pcDNA3.1-GCN2 | This study | N/A |
| pcDNA3.1-GCN2(D858N) | This study | N/A |
| pCMV-VSV-G | Addgene | Cat#8454 |
| psPAX2 | Addgene | Cat#12260 |
| non-targeting control gRNA | Addgene | Cat#80263 |
| GCN2 gRNA 1 | Addgene | Cat#75875 |
| GCN2 gRNA 2 | Addgene | Cat#75876 |
| ZAK gRNA 1 | Addgene | Cat#75596 |
| ZAK gRNA 2 | Addgene | Cat#75597 |
| pTRIPZ-EV | This study | N/A |
| pTRIPZ-viperin | This study | N/A |
| pTRIPZ-viperin (DCA) | This study | N/A |
| pFLZIKV | Shan et al., 2016 | N/A |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | RRID:SCR_003070 |
| STAR | Dobin et al., 2013 | RRID:SCR_004463 |
| Bedtools Multicov | Quinlan and Hall, 2010 | RRID:SCR_006646 |
| FlowJo | FlowJo LLC | RRID:SCR_008520 |
| Other | ||
| Mini-PROTEAN TGX Stain-Free™ Protein Gels | Bio-Rad | Cat#4568084 |
| SuperSep Phos-tag gel | Wako Chemicals | Cat#198–17981 |
| Immobilon-P PVDF membrane | Sigma-Aldrich | Cat#IPVH00010 |
Experimental Model and Subject Details
Cells
HEK293T, HeLa and Vero cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gemini Bio). To generate doxycycline-inducible cell lines, a pTRIPZ lentivirus was used to transduce HEK293T cells. Cells were selected with puromycin (Thermo Fisher Scientific) at 5 μg/mL to obtain stable cell lines. Cells were induced at 0.5 μg/mL doxycycline (Dox) for 24 h unless stated otherwise. To generate GCN2 and ZAK knockout cell lines, HEK293T-Cas9 cells were transduced with a lentivirus encoding the guide RNA of interest. Cells were selected with puromycin (Thermo Fisher Scientific) at 5 μg/mL to obtain stable cell lines. HEK293T-Cas9 was obtained from Dr. Jorge E. Galán at Yale University (Chang et al., 2016). iBMDM and iBMDM.VipKO were generated and cultured as previously described (De Nardo et al., 2018). In brief, mouse bone marrow cells were isolated from B6 or viperin KO mice. Cells were cultured in culture medium (DMEM, 5% FBS, 1% Pen/Strep) with 30% L929 cell conditioned medium containing macrophage colony stimulating factor (MCSF). After 6 days, cells were infected with Cre-J2 virus for 24 h and then cultured in culture medium with 20% L929 cell conditioned medium for 24 h. The infection was repeated once the following day. After 7 days, cells were cultured in culture medium containing decreasing L929 cell conditioned medium until cells were able to proliferate in the absence of L929 cell conditioned medium. For inducing the expression of ISGs, cells were treated with 10,000 U/mL of Universal Type I interferon (PBL) unless stated otherwise.
Viruses and viral infections
Zika virus (ZIKV) Cambodia strain was obtained from Dr. Erol Fikrig at Yale University. West Nile virus Kunjin strain (WNVKUNV) was obtained from Dr. Philip M. Armstrong at The Connecticut Agricultural Experiment Station. High titer stocks of ZIKV Cambodia strain were obtained by passage in Vero cells. High titer stocks of WNVKUNV were obtained by passage in BHK-21 cells. Monolayers of cells were initially adsorbed with either WNVKUNV or ZIKV at the indicated multiplicity of infection (MOI) in 2% FBS/DMEM for 1h at 37°C. Unbound virus was removed, and cells were maintained in 10% FBS/DMEM at 37°C. Viral replication was measured by plaque assay on Vero cells for ZIKV or BHK-21 cells for WNVKUNV. For NanoLuc reporter ZIKV infection (Baker et al., 2020), cells were infected at an MOI of 2. At indicated time post infection, the cells were lysed and measured for luciferase activities (Puig-Basagoiti et al., 2005).
Method Details
Antibodies
Primary antibodies used were anti-viperin (MaP.VIP) (Wang et al., 2007), anti-GRP94 (Enzo), anti-GAPDH (Proteintech), anti-phospho-4EBP1 (Cell Signaling), anti-4EBP1 (Cell Signaling), anti-PRS6 (Cell Signaling), anti-phospho-RPS6 (Cell Signaling; 2217), anti-phospho-eIF2α (S51) (Cell Signaling), anti-eIF2α (Cell Signaling), anti-Flaviviral E protein (Millipore), anti-GCN2 (Cell Signaling), anti-phospho-GCN2 (T899) (Abcam), anti-PERK (Cell Signaling), anti-HRI (Proteintech), anti-PKR (Cell Signaling), anti-ZAK (Bethyl). All secondary antibodies used for immunofluorescence imaging (goat antimouse IgG coupled to Alexa Fluor 488, Alexa Fluor 647, goat anti-rat IgG coupled to Alexa Fluor 546, Alexa Fluor 594 and goat anti-rabbit IgG coupled to Alexa Fluor 647) were purchased from Invitrogen, and those for immunoblotting (goat anti-mouse IgG and anti-rabbit and anti-rat IgG coupled to horseradish peroxidase and alkaline phosphatase) were purchased from Jackson ImmunoResearch.
Plasmid construction
pcDNA3.1-viperin and viperin mutants have been previously described (Seo and Cresswell, 2013; Seo et al., 2011b).
The coding sequence of human GCN2 (GenBank: BC146319.1) was purchased from Dharmacon and subcloned into pcDNA3.1 vector. GCN2(D858N) was generated by Gibson Assembly Kit (NEB). For CRISPR-Cas9 knockout, lentiviruses were generated with pCMV-VSV-G (Addgene, 8454), psPAX2 (Addgene, 12260) and guide RNA encoding plasmids, GCN2 gRNAs (75875 and 75876, Addgene), ZAK gRNAs (Addgene, 75596 and 75597) and non-targeting control gRNA (Addgene, 80263). All primers used for molecular cloning and qRT-PCR were synthesized by the Keck facility at Yale University. Plasmids were purified using Qiagen and Zymo Research kits.
Metabolic labeling
HEK293T cells were transfected with pcDNA3.1 plasmids encoding indicated proteins for 24 h. Cells were starved in cysteine- and methionine-free DMEM supplemented with 5% dialyzed fetal bovine serum (Invitrogen), 1% Glutamax and 1% penicillin/streptomycin for 1h. Cells were metabolically labeled with [35S]-methionine/cysteine (PerkinElmer Life Science) for 15min, lysed for SDS-PAGE and analyzed by phosphoimaging and immunoblotting. Translation was estimated by areas under the curve using ImageJ and normalized to empty vector control.
RNA-Seq
Total RNA was purified by Trizol (Thermo Fisher Scientific) according to the manufacturer’s instructions, and rRNAs were depleted by NEBNext rRNA Depletion Kit (NEB). RNA-seq libraries were constructed following Illumina Tru-seq stranded mRNA protocol. The libraries were sequenced on HiSeq 2500 with 76 bp single-end run. Sequencing reads were aligned to the hg38 genome by STAR and counted using BEDTools multicov (Dobin et al., 2013; Quinlan and Hall, 2010).
ddhCTP detection and synthetic ddhC nucleoside treatments
Cell pellets were dissolved in 500 μL of LCMS extraction buffer (40:40:20 ACN/MeOH/H2O, 0.1 M formic acid) containing heavy label standards for 13C10 15N5-ATP, 13C9 15N2-UTP, 13C9 15N3-CTP, and 13C9 15N3-ddhCTP. The mixture was then dried in a rotary lyophilizer, resuspended in water, and sonicated. The extracts were then centrifuged at 15,000 x g for 10min before being analyzed by LCMS as previously described (Gizzi et al., 2018). ddhC was synthesized as previously described (Gizzi et al., 2018). For synthetic ddhC nucleoside treatments, cells were treated with ddhC in 10% FBS/DMEM at a concentration of 1 mM for 24 h unless stated otherwise.
O-propargyl puromycin (OP-Puro) labeling
Cells were incubated with 50 μM of OP-Puro in 10% FBS/DMEM for 1h. Cells were harvested and washed with ice-cold PBS. Cells were fixed with 1% paraformaldehyde in PBS for 15 min on ice, washed in PBS, then permeabilized in permeabilization buffer (3% FBS and 0.1% saponin in PBS) for 5 min at room temperature. The Click chemistry reaction was performed by incubating cells with Click solution (1 mM CuSO4, 5 μM azide-fluor 488, 150 mM Tris, pH8.5 and 100 mM ascorbic acid adding before use) for 30 min at room temperature. The cells were washed three times in permeabilization buffer and resuspended in PBS for flow cytometry analysis or confocal imaging.
Polysome profiling
Cells grown to 80%–90% confluence were treated with 50 μg/mL cycloheximide (CHX) at 37°C for 10 min, washed with ice-cold PBS, incubated with 50 μg/mL CHX/PBS at 4°C for 20 min, and lysed by adding polysome lysis buffer (200 mM KOAc, 50 mM Tris, pH 7.4, 10 mM MgCl2, 1 mM DTT, 0.5% IGEPAL CA-630, RNaseOUT, and protease inhibitor cocktail (Sigma)) on ice for 15 min. Cell lysates were centrifuged at 14,000 rpm for 15 min to remove cell debris. Polyribosomes were resolved on 15%–50% linear sucrose gradients and fractionated as described previously (Hsu et al., 2018). RNase-digested profiles were performed as described previously with minor modifications (Wu et al., 2020). In brief, cells grown to 80%–90% confluence were washed with ice-cold PBS and lysed by adding RNase-polysome lysis buffer (200 mM KOAc, 50 mM Tris, pH 8.0, 10 mM MgCl2, 1 mM DTT, 0.5% IGEPAL CA-630, and protease inhibitor cocktail (Sigma)) on ice for 15 min. Cell lysates were centrifuged at 14,000 rpm for 15 min to remove cell debris. Clear lysates containing ~5 OD260 units of total RNA were treated with RNase A (Sigma-Aldrich, R6513) at 150 μg/mL for 30 min at RT with agitation. Digested lysates were resolved on 10%–35% linear sucrose gradients (200 mM KOAc, 50 mM Tris, pH 8.0, 10 mM MgCl2) using Beckmann Coulter SW41 Ti rotors at 40,000 rpm for 4°C for 2 hr. For RNA detection, RNA was Trizol isolated according to the manufacturer’s instruction, quantified by RT-PCR (Luna Universal One-Step RT-qPCR Kit; New England Biolabs) and analyzed by Mx3000P (Stratagene).
Immunoblotting
Cells were harvested, washed with ice-cold PBS, lysed on ice for 10 min in NP40 lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1% IGEPAL CA-630, PhosSTOP (Roche), and protease inhibitor cocktail (Sigma)) and centrifuged at 4°C. Protein concentration was measured by Bradford protein assay (BioRad) as described in the manufacturer’s instructions. Cell lysates were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore). For phospho-protein analysis, phos-tag gel (Wako) was used. Anti-ZAK immunoblots were performed as described previously (Wu et al., 2020).
Confocal immunofluorescence microscopy
Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature and blocked with 10% normal goat serum in PBST (0.2% Tween-20 in PBS). Primary and secondary stainings were performed in 2.5% normal goat serum/PBST. Primary antibodies used were the same as used for immunoblotting. Secondary antibodies were the Alexa Fluor series from Thermo Fisher Scientific. Slides were mounted with ProLong Gold Anti-Fade Reagent (Thermo Fisher Scientific), imaged by Leica SP8 model confocal microscope and analyzed using ImageJ.
In vitro transcription
ZIKV RNA was generated as described previously (Shan et al., 2016). In brief, ZIKV pFLZIKV cDNA was digested by restriction enzymes to generate linear DNAs for in vitro transcription. DNA was converted to RNA using MEGAscript T7 Transcription Kit (Thermo Fisher Scientific). RNA was purified using TRIzol extraction. ClaI and EcoRI were used to generate the DNA templates for the full-length and truncated ZIKV RNAs respectively.
Quantitative RT-PCR
RNA was extracted from cells using Trizol according to the manufacturer’s instructions. For two-step qRT-PCR, RNA was converted to cDNA using SuperScript II Reverse Transcriptase (Invitrogen) and quantified using Power SYBR Green (Thermo Fisher Scientific). For one-step qRT-PCR, RNA was quantified using Luna Universal One-Step RT-qPCR Kit (NEB). Analyses were performed using an Mx3000P (Stratagene). The primers are listed in Table S1.
Flow cytometry
Cells were harvested, washed with PBS, and collected using an Accuri C6 CSampler (BD Biosciences). Analysis was performed using FlowJo software.
Quantification and Statistical Analysis
If not stated otherwise, statistical significance was tested by unpaired two-tailed Student’s t test. Only p values less than 0.05 were considered statistically significant [not significant (ns), p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). Confocal immunofluorescence and immunoblot analyses were quantified using ImageJ.
Supplementary Material
Highlights.
Viperin regulates cellular translation to restrict viral translation and replication
Viperin activates the GCN2-arm of the integrated stress response
Viperin induces ribosome collision-mediated translation inhibition
Viperin’s product ddhCTP induces ribosome collision
Acknowledgments
We thank Dr. Yue Li for assistance in designing viperin mutants. The Cambodia Zika strain was a gift from Dr. Erol Fikrig at Yale University. Kunjin virus was a gift from Philip M. Armstrong at The Connecticut Agricultural Experiment Station. We thank Dr. Brett Lindenbach for the Brandel density gradient fractionation system. J.H. was supported by the Cancer Research Institute Irvington Postdoctoral Fellowship, J.B.P. is supported by 5 T32 HL 7974–19 and M.L.R. is supported by Grant Number: KL2 TR001862. J.L. was supported by postdoctoral fellowships from the International Human Frontier Science Program Organization (LT000037/2018-L) and Jane Coffin Childs Memorial Fund. This work was supported by NIH grants RO1 AI059167 (P.C.), P01 GM118303 (S.C.A.), R21-AI133329 (T.L.G. and S.C.A.) and the Price Family Foundation (S.C.A.). L.D.H., J.M.W and G.B.E. thank the Ministry of Business Innovation & Employment for support of this work (Endeavour Fund, Contract UOOX1904 (N.Z.)). We acknowledge the Albert Einstein Anaerobic Structural and Functional Genomics Resource (http://www.nysgxrc.org/psi3/anaerobic.html) and the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. The Graphical Abstract is created with BioRender.com.
Footnotes
Declaration of Interests
T.L.G. and S.C.A. have filed intellectual property around the use of ddhC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker C, Xie X, Zou J, Muruato A, Fink K, and Shi PY (2020). Using recombination-dependent lethal mutations to stabilize reporter flaviviruses for rapid serodiagnosis and drug discovery. EBioMedicine 57, 102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, and Barber GN (2000). Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13, 129–141. [DOI] [PubMed] [Google Scholar]
- Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, Carrasco L, and de Haro C. (2006). Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J 25, 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, and Hegde RS (2016). Ribosome-associated protein quality control. Nat Struct Mol Biol 23, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton-Smith C, and Elliott RM (2012). Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J Virol 86, 11548–11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SJ, Song J, and Galan JE (2016). Receptor-Mediated Sorting of Typhoid Toxin during Its Export from Salmonella Typhi-Infected Cells. Cell Host Microbe 20, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC. and Cresswell P. (2001). Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A 98, 15125–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nardo D, Kalvakolanu DV, and Latz E. (2018). Immortalization of Murine Bone Marrow-Derived Macrophages. Methods Mol Biol 1784, 35–49. [DOI] [PubMed] [Google Scholar]
- del Pino J, Jimenez JL, Ventoso I, Castello A, Munoz-Fernandez MA, de Haro C, and Berlanga JJ (2012). GCN2 has inhibitory effect on human immunodeficiency virus-1 protein synthesis and is cleaved upon viral infection. PLoS One 7, e47272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, and Sonenberg N. (2002). Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol 12, 1279–1286. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, and Hinnebusch AG (2000). Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6, 269–279. [DOI] [PubMed] [Google Scholar]
- Dukhovny A, Shlomai A, and Sklan EH (2018). The antiviral protein Viperin suppresses T7 promoter dependent RNA synthesis-possible implications for its antiviral activity. Sci Rep 8, 8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi KH, Howie D, Rowbotham JS, McCullagh J, Armstrong FA, and James WS (2020). Viperin, through its radical-SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett 594, 1624–1630. [DOI] [PubMed] [Google Scholar]
- Fenwick MK, Li Y, Cresswell P, Modis Y, and Ealick SE (2017). Structural studies of viperin, an antiviral radical SAM enzyme. Proc Natl Acad Sci U S A 114, 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, Chin KC, Aw P, George J, Kuznetsov VA, Schreiber M, et al. (2007). Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis 1, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, and Esteban M. (2006). Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70, 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, and Marsh ENG (2020). Viperin: An ancient radical SAM enzyme finds its place in modern cellular metabolism and innate immunity. J Biol Chem 295, 11513–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, et al. (2018). A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, and Green R. (2014). Dom34 rescues ribosomes in 3’ untranslated regions. Cell 156, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig KJ. Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, and Beard MR (2013). Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the Cterminal end of viperin. PLoS Negl Trop Dis 7, e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson ER, and Cresswell P. (2009). The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem 284, 4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JC, Reid DW, Hoffman AM, Sarkar D, and Nicchitta CV (2018). Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 24, 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis AJ, Masson GR, Shao S, Perisic O, McLaughlin SH, Hegde RS, and Williams RL (2019). Activation of GCN2 by the ribosomal P-stalk. Proc Natl Acad Sci U S A 116, 4946–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Chuang JH, and Ackerman SL (2016). Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, and Donlin LT (2014). Regulation of type I interferon responses. Nat Rev Immunol 14, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, and Salic A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspart A, Calmels C, Cosnefroy O, Bellecave P, Pinson P, Claverol S, GuyonnetDuperat V, Dartigues B, Benleulmi MS, Mauro E, et al. (2017). GCN2 phosphorylates HIV-1 integrase and decreases HIV-1 replication by limiting viral integration. Sci Rep 7, 2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, and Guo JT (2008). Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol 82, 1665–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, et al. (2010). Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol 84, 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Santos Rocha C, Hirao LA, Mendes EA, Tang Y, Thompson GR 3rd, Wong JK, and Dandekar S. (2017). HIV Exploits Antiviral Host Innate GCN2-ATF4 Signaling for Establishing Viral Replication Early in Infection. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy J, Mounir Z, Raven JF, and Koromilas AE (2008). The eIF2alpha kinases inhibit vesicular stomatitis virus replication independently of eIF2alpha phosphorylation. Cell Cycle 7, 2346–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Hansen G, Nitsche C, Klein CD, Zhang L, and Hilgenfeld R. (2016). Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 353, 503–505. [DOI] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, and Salic A. (2012). Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A 109, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, and Guydosh NR (2020). Disome and Trisome Profiling Reveal Genomewide Targets of Ribosome Quality Control. Mol Cell 79, 588–602 e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelreiter F, Coats MT, Klanert G, Gludovacz E, Borth N, Grillari J, and Schosserer M. (2018). OPP Labeling Enables Total Protein Synthesis Quantification in CHO Production Cell Lines at the Single-Cell Level. Biotechnol J 13, e1700492. [DOI] [PubMed] [Google Scholar]
- Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, Wilkinson J, Bye CR, Wright TK, Rambukwelle D, et al. (2012). HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120, 778–788. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM (2016). The integrated stress response. EMBO Rep 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, Upadhyay AS, and Overby AK (2018). Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Basagoiti F, Deas TS, Ren P, Tilgner M, Ferguson DM, and Shi PY (2005). High-throughput assays using a luciferase-expressing replicon, virus-like particles, and full-length virus for West Nile virus drug discovery. Antimicrob Agents Chemother 49, 4980–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, and Rice CM (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JY, and Cresswell P. (2013). Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog 9, e1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JY, Yaneva R, and Cresswell P. (2011a). Viperin: a multifunctional, interferoninducible protein that regulates virus replication. Cell Host Microbe 10, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JY, Yaneva R, Hinson ER, and Cresswell P. (2011b). Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332, 1093–1097. [DOI] [PubMed] [Google Scholar]
- Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, et al. (2016). An Infectious cDNA Clone of Zika Virus to Study Viral Virulence, Mosquito Transmission, and Antiviral Inhibitors. Cell Host Microbe 19, 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, and Walter P. (2015a). The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, Sokabe M, Mendez AS, Newton BW, Tang EL, et al. (2015b). Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife 4, e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA (2014). A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol 5, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, and Diamond MS (2011). The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol 85, 11557–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay AS, Vonderstein K, Pichlmair A, Stehling O, Bennett KL, Dobler G, Guo JT, Superti-Furga G, Lill R, Overby AK, et al. (2014). Viperin is an ironsulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell Microbiol 16, 834–848. [DOI] [PubMed] [Google Scholar]
- Van der Hoek KH. Eyre NS, Shue B, Khantisitthiporn O, Glab-Ampi K, Carr JM, Gartner MJ, Jolly LA, Thomas PQ, Adikusuma F, et al. (2017). Viperin is an important host restriction factor in control of Zika virus infection. Sci Rep 7, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwalscappel B, Gadea G, and Despres P. (2019). A Viperin Mutant Bearing the K358R Substitution Lost its Anti-ZIKA Virus Activity. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwalscappel B, Tada T, and Landau NR (2018). Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 522, 199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hinson ER, and Cresswell P. (2007). The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96-105. [DOI] [PubMed] [Google Scholar]
- Won S, Eidenschenk C, Arnold CN, Siggs OM, Sun L, Brandl K, Mullen TM, Nemerow GR, Moresco EM, and Beutler B. (2012). Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol 86, 1802–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, and Green R. (2020). Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 182, 404–416 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Ci Y, Wang L, Yang Y, Zhang L, Xu C, Qin C, and Shi L. (2019). Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res 47, 8693–8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL, and Zaher HS (2019). How do cells cope with RNA damage and its consequences? J Biol Chem 294, 15158–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL, and Zaher HS (2021). Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol Cell 81, 614–628 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C, and Luo D. (2016). Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 354, 15971600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) under accession number GEO: GSE142015. Accession number is listed in the key resources table. Original western blot images and microscopy data reported in this paper have been deposited at Mendeley at http://dx.doi.org/10.17632/dyxgt2g7cm.2.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Viperin (MaP.VIP) | Wang et al., 2007 | N/A |
| GRP94 | Enzo Life Sciences | Cat#ADI-SPA-851; RRID: AB_10615790 |
| GAPDH | Proteintech | Cat#60004–1-Ig; RRID: AB_2107436 |
| Phospho-4E-BP1 (Ser65) | Cell Signaling Technology | Cat#9451; RRID: AB_330947 |
| 4E-BP1 | Cell Signaling Technology | Cat#9644; RRID: AB_2097841 |
| Phospho-S6 Ribosomal Protein (Ser235/236) | Cell Signaling Technology | Cat#4857; RRID: AB_2181035 |
| S6 Ribosomal Protein (S6) | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| eIF2α | Cell Signaling Technology | Cat#5324; RRID: AB_10692650 |
| phospho-eIF2α (Ser51) | Cell Signaling Technology | Cat#3597; RRID: AB_390740 |
| phospho-p38 (Thr180/Tyr182) | Proteintech | Cat#9211; RRID: AB_331641 |
| p38 | Proteintech | Cat#9212; RRID: AB_330713 |
| phospho-JNK (Thr183/Tyr185) | Proteintech | Cat#4668; RRID: AB_823588 |
| Flavivirus envelope protein | Millipore | Cat#MAB10216; RRID: AB_827205 |
| GCN2 | Cell Signaling Technology | Cat#3302; RRID: AB_2277617 |
| phospho-GCN2(T899) | Abcam | Cat#ab75836; RRID: AB_1310260 |
| PERK | Cell Signaling Technology | Cat#3192; RRID: AB_2095847 |
| HRI | Proteintech | Cat#20499–1-AP; RRID: AB_10697665 |
| PKR | Cell Signaling Technology | Cat#3072; RRID: AB_2277600 |
| ZAK | Bethyl Laboratories | Cat#A301–993A; RRID: AB_1576612 |
| goat anti-mouse IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11017; RRID: AB_2534084 |
| goat anti-mouse IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21236; RRID: AB_2535805 |
| goat anti-rat IgG, Alexa Fluor 546 | Thermo Fisher Scientific | Cat#A-11081; RRID: AB_2534125 |
| goat anti-rat IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A-11007; RRID: AB_10561522 |
| goat anti-rabbit IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21244; RRID: AB_2535812 |
| goat anti-mouse IgG-HRP | Jackson ImmunoResearch Laboratories | Cat#115–035-146; RRID: AB_2307392 |
| goat anti-rabbit-HRP | Jackson ImmunoResearch Laboratories | Cat#111–005-144; RRID: AB_2337919 |
| goat anti-rat IgG-HRP | Jackson ImmunoResearch Laboratories | Cat#112–005-167; RRID: AB_2338101 |
| Bacterial and virus strains | ||
| NEB 5-α Competent E. coli (High Efficiency) | New England Biolabs | Cat#C2987 |
| Zika virus Cambodia/FSS13025 strain | World Reference Center for Emerging Viruses and Arboviruses at University of Texas Medical Branch at Galveston | NCBI:txid2316109 |
| NanoLuc reporter ZIKV | Baker et al., 2020 | N/A |
| West Nile virus Kunjin strain (WNVKUNV) | Dr. Philip Armstrong | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ddhC | This study | N/A |
| Sucrose | Sigma-Aldrich | Cat#S0389 |
| Dithiothreitol (DTT) | American Bioanalytical | Cat#AB00490 |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | Cat#14190144 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat#11965092 |
| Fetal Bovine Serum | Gemini Bio | Cat#100–106 |
| Gibson Assembly Master Mix | New England Biolabs | Cat#E2611 |
| Q5 Hot Start High-Fidelity 2X Master Mix | New England Biolabs | Cat#M0494 |
| ZymoPURE II Plasmid Midiprep Kit | Zymo Research | Cat#D4201 |
| MEGAscript T7 Transcription Kit | Thermo Fisher Scientific | Cat#AMB13345 |
| ClaI | New England Biolabs | Cat#R0197S |
| EcoRI-HF | New England Biolabs | Cat# R3101S |
| Bovine Serum Albumin (BSA) | American Bioanalytical | Cat#AB01088 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat#15596026 |
| Ethanol 200 proof | Decon Labs | Cat#2716 |
| GlycoBlue | Thermo Fisher Scientific | Cat#AM9515 |
| 4x Laemmli Sample Buffer | Bio-Rad | Cat#1610747 |
| Trans-Blot Turbo™ 5x Transfer Buffer | Bio-Rad | Cat#10026938 |
| Doxycycline | Sigma-Aldrich | Cat#D9891 |
| Universal Type I IFN | PBL Assay Science | Cat#11200–2 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| Glutamax | Thermo Fisher Scientific | Cat#35050061 |
| Saponin | Sigma-Aldrich | Cat#S7900 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| dialyzed fetal bovine serum | Thermo Fisher Scientific | Cat#A3382001 |
| [35S]-methionine/cysteine | PerkinElmer Life Science | Cat#NEG772014MC |
| Tris-HCl | American Bioanalytical | Cat#AB02005 |
| IGEPAL CA-630 | Sigma-Aldrich | Cat#I8896 |
| PhosSTOP | Sigma-Aldrich | Cat#PHOSS-RO |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| RNaseOUT | Thermo Fisher Scientific | Cat#10777019 |
| Rapamycin | Cell Signaling Technology | Cat#9904 |
| 16% paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Goat serum | Thermo Fisher Scientific | Cat#16210064 |
| ProLong Gold Anti-Fade Reagent | Thermo Fisher Scientific | Cat#P36930 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#H3570 |
| O-Propargylpuromycin | Abcam | Cat#ab146664 |
| L-Ascorbic acid | Sigma-Aldrich | Cat#A4544 |
| Copper sulfate | Sigma-Aldrich | Cat#C1297 |
| Azide-fluor 488 | Sigma-Aldrich | Cat#760765 |
| Cycloheximide | Sigma-Aldrich | Cat#C4859 |
| Potassium acetate | Sigma-Aldrich | Cat#P5708 |
| Magnesium chloride | Sigma-Aldrich | Cat#M8266 |
| RNase A | Sigma-Aldrich | Cat#R6513 |
| Sodium chloride | American Bioanalytical | Cat#AB01915 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat#EDS |
| Acetonitrile | Sigma-Aldrich | Cat#34851 |
| Methanol | Sigma-Aldrich | Cat#34860 |
| Water | American Bioanalytical | Cat#AB02128 |
| Formic acid | Thermo Fisher Scientific | Cat#A117–50 |
| DMEM, high glucose, no glutamine, no methionine, no cystine | Thermo Fisher Scientific | Cat#21013024 |
| Thapsigargin | Sigma-Aldrich | Cat#T9033 |
| Sodium arsenite | Sigma-Aldrich | Cat#S7400 |
| Polyinosinic:polycytidylic acid [poly(I:C)] | Sigma-Aldrich | Cat#P9582 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668500 |
| OptiMEM | Thermo Fisher Scientific | Cat#11058021 |
| Integrated stress response inhibitor (ISRIB) | Sigma-Aldrich | Cat#SML0843 |
| 5-Ethynyluridine | Abcam | Cat#ab146642 |
| Tween-20 | Sigma-Aldrich | Cat#P7949 |
| Triton X-100 | American Bioanalytical | Cat#AB02025 |
| Critical commercial assays | ||
| Luna Universal One-Step RT-qPCR Kit | New England Biolabs | Cat#E3005 |
| Bradford protein assay | Bio-Rad | Cat#5000001 |
| Power SYBR Green (Thermo Fisher Scientific) | Thermo Fisher Scientific | Cat#4367659 |
| CellTrace CFSE Cell Proliferation Kit (Invitrogen) | Thermo Fisher Scientific | Cat#C34570 |
| Annexin V-FITC Apoptosis Detection Kit | Thermo Fisher Scientific | Cat#BMS500FI |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat#18064014 |
| NEBNext rRNA Depletion Kit (NEB) | New England Biolabs | Cat#E6310 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020594 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE142015 |
| Raw Western blot and microscopy images | This paper; Mendeley Data | DOI:10.17632/dyxgt2g7cm.2 |
| Experimental models: Cell lines | ||
| Human: HEK293T | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| African green monkey: Vero | ATCC | Cat#CCL-81; RRID: CVCL_0059 |
| Human: HeLa | ATCC | Cat#CCL-2; RRID: CVCL_0030 |
| Hamster: BHK-21 | ATCC | Cat#CCL-10; RRID: CVCL_1915 |
| Mouse: iBMDM | This study | N/A |
| Mouse: iBMDM.VipKO | This study | N/A |
| Human: 293T.iVip | This study | N/A |
| Human: HEK293T-Cas9 | Dr. Jorge Galán Chang et al., 2016 |
N/A |
| Human: 293T.GCN2KO | This study | N/A |
| Human: 293T.ZAKKO | This study | N/A |
| Experimental models: Organisms | ||
| Mouse: B6 (C57BL/6J) | Jackson Laboratory | Cat#000664; RRID: IMSR_JAX:000664 |
| Mouse: Viperin KO (B6.129-Rsad2tm1Kchc/CresJ) | Seo et al., 2011 | RRID:IMSR_JAX:032321 |
| Oligonucleotides | ||
| All standard primers for qRT-PCR | This study | Table S1 |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher Scientific | Cat#V79020 |
| pcDNA3.1-Venus | Hee and Cresswell, 2017 | N/A |
| pcDNA3.1-viperin | Seo et al., 2011 | N/A |
| pcDNA3.1-viperin (d1–42) | Seo et al., 2011 | N/A |
| pcDNA3.1-viperin (DCA) | Seo et al., 2011 | N/A |
| pcDNA3.1-GCN2 | This study | N/A |
| pcDNA3.1-GCN2(D858N) | This study | N/A |
| pCMV-VSV-G | Addgene | Cat#8454 |
| psPAX2 | Addgene | Cat#12260 |
| non-targeting control gRNA | Addgene | Cat#80263 |
| GCN2 gRNA 1 | Addgene | Cat#75875 |
| GCN2 gRNA 2 | Addgene | Cat#75876 |
| ZAK gRNA 1 | Addgene | Cat#75596 |
| ZAK gRNA 2 | Addgene | Cat#75597 |
| pTRIPZ-EV | This study | N/A |
| pTRIPZ-viperin | This study | N/A |
| pTRIPZ-viperin (DCA) | This study | N/A |
| pFLZIKV | Shan et al., 2016 | N/A |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | RRID:SCR_003070 |
| STAR | Dobin et al., 2013 | RRID:SCR_004463 |
| Bedtools Multicov | Quinlan and Hall, 2010 | RRID:SCR_006646 |
| FlowJo | FlowJo LLC | RRID:SCR_008520 |
| Other | ||
| Mini-PROTEAN TGX Stain-Free™ Protein Gels | Bio-Rad | Cat#4568084 |
| SuperSep Phos-tag gel | Wako Chemicals | Cat#198–17981 |
| Immobilon-P PVDF membrane | Sigma-Aldrich | Cat#IPVH00010 |