Abstract
The recapitulation of complex microenvironments that regulate cell behavior during development, disease, and wound healing is key to understanding fundamental biological processes. In vitro, multicellular morphogenesis, organoid maturation, and disease modeling have traditionally been studied either using non-physiological 2D substrates or 3D biological matrices, neither of which replicate the spatiotemporal biochemical and biophysical complexity of biology. Here, we provide a guided overview of recent advances in the programming of synthetic hydrogels that offer precise control over spatiotemporal properties within cellular microenvironments, such as with cell-driven remodeling, bioprinting, or user-defined manipulation of properties (e.g., via light irradiation).
eTOC statement:
In this review, Qazi et al. describe advances in the engineering of hydrogels to explore complex biological processes, such as development, disease, and wound healing. They specifically highlight the spatiotemporal control over hydrogel biochemical and biophysical properties via mechanisms such as cell remodeling or light-mediated changes in hydrogel crosslinking.
1. Overview of cellular microenvironments
Stem and progenitor cell behaviors during development, disease, and wound healing are regulated by spatially and temporally dynamic signals found within their microenvironments (Daley et al., 2008). Microenvironmental features that most strongly influence cell behavior include biophysical properties, such as mechanics, architecture, degradability, and biochemical properties including signaling ligands and bound factors (e.g., growth factors) of the extracellular matrix (ECM) (Madl and Heilshorn, 2018) (Figure 1). The ECM is composed of families of proteins and sugars (e.g., collagens, laminins, glycosaminoglycans, proteoglycans) that not only provide structural stability to tissues and organs but also play an instructive role to guide cell function (Sadtler et al., 2016; Urciuolo et al., 2013). While similar ECM molecules are found throughout the body, the composition and structure vary across individual tissues to guide the specialist functions of the local tissue.
Figure 1:
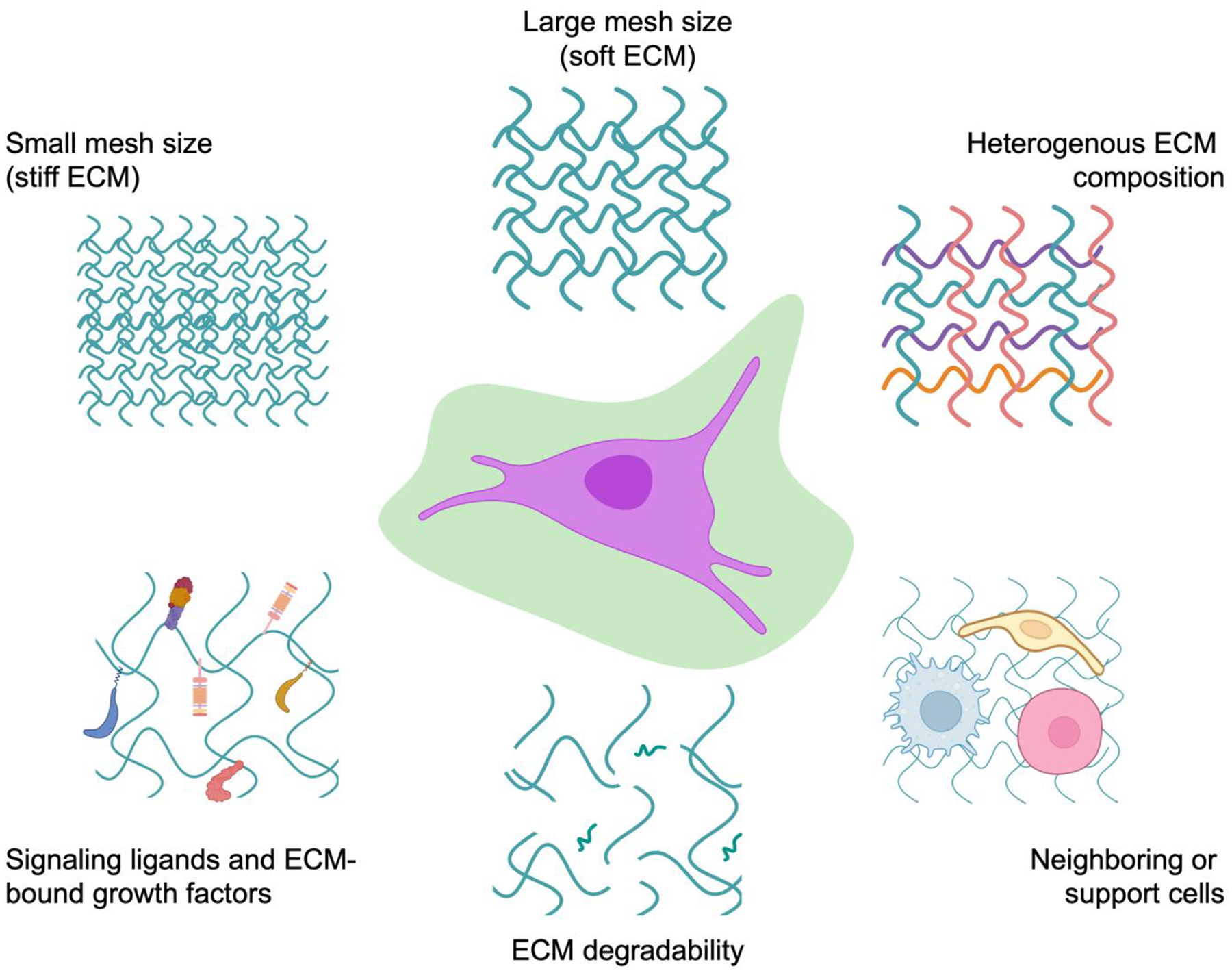
Biophysical and biochemical features of the cellular microenvironment in native ECM. These ECM features include differences in matrix mesh size leading to differences in mechanical properties (e.g., stiffness), chemical composition, bound cell-adhesion sites and signaling ligands such as growth factors, degradability, and neighboring or support cells that can secrete small molecules and soluble cytokines.
Cells interact directly with the ECM via receptors on the cell membrane including integrins, syndecans, and growth factor receptors that connect the ECM to the cellular cytoskeleton and facilitate cell-matrix coupling. These interactions define the activation state, phenotype, and downstream signaling of adult tissue cells as well as stem cells that reside within specialized microenvironments called niches. Cells probe the mechanical properties of their microenvironment via cytoskeletal contractility to apply traction forces at adhesion sites and sense differences in their ability to reorganize and cluster engaged receptors (Roca-Cusachs et al., 2009).
Receptor clustering is dependent on the rigidity (elasticity) and time-dependent malleability (viscoelasticity) of the ECM and differences can affect mechanotransduction, intracellular signaling, and cell fate (Cameron et al., 2011; Huebsch et al., 2010). Traction at the cell-ECM interface also plays an important role in biochemical signaling. Matrix-bound signaling ligands and growth factors (e.g., transforming growth factor beta (TGF-β)) naturally exist in a latent form and are only activated when freed from protective binding proteins via contractile forces exerted by cells (Buscemi et al., 2011). Beyond matrix-bound biochemical cues, recent evidence indicates that the rigidity of the underlying ECM and the cellular contractile state modulates the sensitivity of cells to soluble cues (e.g., growth factors, cytokines) (Crouzier et al., 2011; Shin and Mooney, 2016).
While the properties, composition, and signals presented by the ECM remain relatively stable during homeostasis, they can change dramatically during dynamic processes, as occurs during development, disease, and wound healing (Gattazzo et al., 2014). Cells sense spatial and temporal changes in these biophysical and biochemical properties and respond by altering their behavior, including gene expression, protein synthesis, and downstream cellular functions (Fata et al., 2004). For example, tissue mechanics (400 Pa vs. 60 kPa) influence the embryonic stem cell differentiation into mesodermal lineages in response to Wnt factors (Przybyla et al., 2016), and the ECM that is synthesized and deposited by embryonic cells in the early stages of development plays critical roles in cell survival, migration, differentiation, and tissue morphogenesis (Sakai et al., 2003; Walma and Yamada, 2020). This observation is further evident in loss of function phenotypes in model systems where the alteration or absence of specific ECM components leads to severe abnormalities or even death (Rozario and DeSimone, 2010).
Dynamic spatiotemporal changes in the ECM are necessary for many biological processes. For example, branching morphogenesis in developing organs including the kidney, lung, gut, and mammary gland is regulated by the spatiotemporal degradation and accumulation of various ECM components at leading bud structures (Bonnans et al., 2014; Wiseman et al., 2003). In neuronal development, cell migration, organization, and fate specification are guided by morphogen and biochemical gradients in a spatiotemporal manner (Sagner and Briscoe, 2017). Recent evidence shows that spatial gradients in stiffness (100–300 Pa) exist in vivo and guide embryonic cell and neural crest migration via durotaxis (Shellard and Mayor, 2021). In the context of diseases such as cancer, stiffening of the tumor ECM and anisotropic architectural cues presented by aligned collagen fibers form the basis of malignant cell migration that leads to metastasis (Winkler et al., 2020). Recent advances in sequencing and proteomic technologies have highlighted how diverse signals evolve over time and space in diseased and injured tissues (Klein et al., 2014). In wound healing after injury, the degradation of native ECM triggers cells like fibroblasts to lay down matrix in an environment experiencing temporal shifts in signaling ligands and inflammatory cues secreted by invading immune cells (Moretti et al., 2021). Under optimal conditions, the deposited matrix is transient and provides signaling cues to guide a regenerative response, but under disturbed signaling conditions, excessive deposition results in fibrosis and scar formation.
Studying these biological processes outside of the body has proven challenging, precisely due to some of the complications in recapitulating the complexity of 3D microenvironmental cues that change in both space and time in vitro (Griffith and Swartz, 2006; Ruskowitz and Deforest, 2018). Hydrogels (i.e., water-swollen polymer networks) are evolving to address these challenges, as they can now be programmed to introduce widespread spatiotemporal signals (Brown and Anseth, 2017). In this review, we highlight the current state-of-the-art hydrogel biomaterials that offer spatiotemporal control over vital properties of the microenvironment, as well as guide the reader on the design of such materials for biology. These engineered niches provide a powerful tool to better answer fundamental questions in biology, as well as to lay a groundwork towards building functional tissues.
2. Traditional platforms to study single cell and multicellular behavior
To study cellular behavior and biological processes involved in development, disease, and wound healing, biologists have traditionally harvested cells from tissue biopsies and cultured them on 2D substrates such as adhesive plastic or ECM-coated glass, or embedded them in commercially available 3D ECM-based matrices like Matrigel. Culture on 2D substrates including tissue culture plastic (TCP) and glass may offer convenience and easy-to-use formats, but often times these culture conditions are not representative of native in vivo conditions due to the flat surface topography, chemically inert surface, and aphysiological stiffness (108-109 Pa) that can be orders of magnitude higher than many soft tissues (102-105 Pa). Coating of 2D surfaces with ECM components like laminin or collagen has been used to provide native biochemical cues to allow attachment and integrin-binding appropriate for cardiomyocytes, neurons, and tissue-resident stem cells (e.g., satellite cells of the skeletal muscle) that reside in well-defined niches inside the body and can spontaneously lose their phenotype when cultured on aphysiological environments ex vivo (Gilbert et al., 2010). However, these methods typically allow coating with only one ECM formulation and do not permit modulation of biophysical properties or spatiotemporal changes in biochemical properties. Maintenance of these cultures can also require complementary addition of soluble cues to the culture medium in lieu of signaling ligands that cells would otherwise naturally encounter in native ECM microenvironments.
Moving to 3D, various natural ECM components have been utilized to fabricate hydrogels for cell culture. Arguably the most commonly used 3D cell culture platform is Matrigel, which is derived from the ECM produced by Engelbreth-Holm-Swarm (EHS) mouse sarcoma cells (Hughes et al., 2010). The major components of Matrigel include laminin, collagen, entactin, and perlecan, whereas many other ECM proteins and peptides have been detected in trace amounts through proteomic profiling studies. Because of its cell-derived origin, Matrigel also contains a variety of growth factors secreted by cells including transforming growth factor (TGF-β), insulin like growth factor (IGF) and fibroblast growth factor (FGF). Together, this mixture of ECM proteins and growth factors contributes to Matrigel’s inherent bioactivity and supports its widespread use in cell biology to study the 3D behavior of single cells or multicellular structures. Due to a composition that consists primarily of basement membrane proteins, Matrigel is particularly well-suited for the ex vivo culture and study of epithelial cells that typically interact with a basement membrane in vivo (Rabata et al., 2020). Similarly, Matrigel may provide bioactive cues that are not naturally found in the microenvironment of other progenitor and tissue-resident cells. Additional drawbacks are associated with the use of Matrigel including a batch to batch variability in its composition that can reduce confidence in the reproducibility of obtained results, limit the ability to modulate biophysical and biochemical properties such as stiffness, signaling ligand density, limit user control over degradability of the 3D environments, and prevent its use for translational applications because of its xenogeneic origin (Aisenbrey and Murphy, 2020). In addition to Matrigel, other naturally derived 3D hydrogels are commonly used, including collagen and fibrin, but both of these materials suffer from similar limitations.
Beyond the culture of cells on 2D surfaces or within 3D natural ECM hydrogels, recent years have seen an exponential rise in more advanced in vitro culture models e.g., organoids (Sato et al., 2009). Organoids are 3D multicellular structures grown in culture from organ-specific stem and progenitor cells that undergo self-assembly through cell-cell and cell-matrix interactions and recapitulate aspects of native tissue or organ architectures and functions in vitro (Marsee et al., 2021). Organoids are powerful tools to model disease, study tissue complexity, and discover potent drugs. Numerous protocols have been published that successfully culture organoids from various tissues including stomach, liver, kidney, lung, and brain (Hofer and Lutolf, 2021). At the most basic level, stem or progenitor cells are cultured in 3D ECM hydrogels comprised of ECM proteins present in native tissues and then exposed to a cocktail of biochemical cues, which can be added serially to match the timing of tissue development or regeneration. By and large, these methods facilitate cellular self-organization through temporal, user-defined manipulation of the composition of soluble factors presented to cells, coupled with the use of ECM-derived culture platforms that can be remodeled by the cells themselves. Due to their reliance on manual intervention and the use of a naturally derived culture substrate that offers limited control over its properties, these culture methods often contribute to heterogeneity in organoid structure, growth, and maturation.
To summarize, while traditional culture methods allow the concentrations of soluble factors present to be easily varied over time to activate or inhibit signaling pathways relevant throughout the growth and differentiation of desired tissues or in disease, most methods offer little control over the spatial availability of such factors. Further, as multicellular constructs grow over time, local gradients of biochemical cues often result, which lead to more complexity in the system that is difficult to control. Thus, there is motivation for more advanced culture systems where various microenvironmental signals can be engineered and tailored to overcome these limitations and redirect overriding stochastic cellular responses.
3. Hydrogels to control cell microenvironments
Hydrogels are water swollen polymer networks that have a long and successful history of use in biomedical applications. Towards cell culture, hydrogels have been designed to mimic specific features of the native extracellular matrix during development, disease, or injury and to study how cells respond to these external cues. This topic has been thoroughly reviewed elsewhere (Madl and Heilshorn, 2018; Nicolas et al., 2020). Further, synthetic hydrogels can be used as alternatives to Matrigel and other naturally derived but poorly controllable ECM-derived proteins (Caliari and Burdick, 2016). Specifically, synthetic hydrogels offer the user a high degree of control over hydrogel biophysical and biochemical properties including mechanics, degradability, architecture, and signaling ligands. This control then allows users to design specific microenvironments for the cell types of interest or those that mimic various aspects of the healthy or diseased native tissue.
Although there are numerous properties that can be engineered into hydrogels, mechanics are of great interest, as mechanical properties (e.g., stiffness, viscoelasticity) can change in tissues due to disease, injury, or aging. Diseases such as cancer cause stiffening of the tumor sites due to the excessive deposition and heterogenous organization of collagen-rich ECM. For example, breast cancer tissues can have elastic moduli that are ~10-fold stiffer than normal mammary tissues (Levental et al., 2009), and adenocarcinoma of the thyroid (44–110 kPa) have been reported to be significantly stiffer than normal thyroid tissues (9–14 kPa) (Lyshchik et al., 2005). Moreover, a diverse variation in mechanical properties (e.g., elastic moduli in the range 1–109 Pa) is observed in healthy tissues and organs in the body ranging from soft bone marrow to stiff bone (Guimarães et al., 2020). Cells sense and respond to mechanical changes through mechanotransduction. Stiffness was originally regarded as a key ECM feature that determined cell phenotype and function, which was supported through seminal studies using variations in hydrogel stiffness (0.1–1 kPa to mimic brain tissue, 8–17 kPa to mimic muscle tissue, and 25–40 kPa to mimic collagenous bone tissue) to alter lineage specification of undifferentiated mesenchymal stromal cells (MSCs) or the maintenance of tissue-specific cells, particularly on substrates mimicking the stiffness of their native tissue microenvironment (Engler et al., 2006; Gilbert et al., 2010). Recognizing that tissues and organs in the body exhibit viscoelastic properties (Chaudhuri et al., 2020), more recent studies have implicated viscoelasticity, temporal changes in material properties, as a key mechanical metric into hydrogel culture environments (Chaudhuri et al., 2016). One way to characterize viscoelasticity is through rheological measurements of the materials’ elastic component (i.e., storage moduli (G’)) and the viscous component (i.e., loss moduli (G”)). Viscoelastic tissues in the body, and the hydrogels designed to mimic their properties, exhibit loss moduli that are ~10–20% of the storage moduli (Chaudhuri et al., 2020). Hydrogel systems have been designed to study cellular mechanobiology and exploit polymers such as polyacrylamide, alginate, hyaluronic acid (HA), and poly(ethylene glycol) (PEG) crosslinked through various chemistries and mechanisms to modulate mechanical properties (Cosgrove et al., 2016; Cruz-Acuña et al., 2017). For example, alginate is a polysaccharide derived from brown algae that undergoes ionic crosslinking in the presence of divalent cations - stiffness can be modulated by changing the concentration of the polymer or the crosslinker ions, whereas viscoelasticity can be modulated by changing the molecular weight of the alginate chains where stress dissipation (relaxation) occurs much quicker in lower molecular weight alginate (Chaudhuri et al., 2015). Although there may be differences in the polymer backbone, functional groups, and crosslinking mechanisms across different systems, there are widespread options that allow precise control over hydrogel mechanics and permit fundamental studies on cell-matrix mechanotransduction.
Cells respond to the mechanics of their microenvironment when they are able to sense these changes via adhesion through surface receptors, such as integrins that bind to corresponding ligands on the ECM and allow cells to generate traction, sense differences in stiffness or viscoelasticity, and retrieve ECM-bound signaling cues like growth factors (Metzger et al., 2016; Wipff and Hinz, 2008). Several ECM components facilitate these cell-matrix interactions in native tissues including laminin and fibronectin, which have cell-binding domains. To replicate these interactions in synthetic hydrogels, peptides mimicking the binding sites found in native ECM proteins have been synthesized and covalently conjugated to polymer backbones (Spicer et al., 2018). These include the arginine-glycine-aspartic acid (RGD) peptide sequence that is derived from fibronectin and the tyrosine-isoleucine-glycine-serine-arginine (YIGSR) peptide sequence derived from laminin. These peptide sequences can be flanked on either side by other amino acids that act as spacers or contain functional groups that permit chemical conjugation to complementary functional groups on the polymer backbone. Studies have shown that cells’ responsiveness to hydrogel properties like viscoelasticity depend critically on the density of these binding peptides (Chaudhuri et al., 2016). While signals like RGD are necessary for cell adhesion, other signaling ligands that can stimulate specific cell functions like differentiation have also been incorporated. These include the covalent conjugation of bone morphogenetic protein-mimicking peptide and N-cadherin mimicking peptide that stimulated 3D encapsulated MSCs to commit to osteogenic and chondrogenic fates, respectively (Bian et al., 2013; Madl et al., 2014).
4. Programmable hydrogels for spatiotemporal control over cells
Despite the extensive work that has been done with hydrogels to engineer cellular microenvironments that mimic the ECM, missing in many of these studies is a focus on spatiotemporal changes in cellular microenvironments. Recent advances in hydrogel design and fabrication technologies have provided unprecedented control over defining spatiotemporally dynamic microenvironments. Although there are many materials, formulations, and techniques to achieve this, we discuss three highly promising approaches and provide recent examples of their application to probe spatiotemporal cell behavior.
4.1. Temporally dynamic hydrogels for cell-driven remodeling
4.1.1. Overview
Single cells and multicellular structures undergo cycles of breaking down, building, and remodeling their local microenvironments to undergo morphogenetic changes during development and to maintain tissue homeostasis. To support this behavior in vitro, permissive hydrogels have been developed that allow cells to remodel their microenvironment, rather than restricting them within confining static environments (Figure 2). These hydrogel systems are simple platforms with the inherent ability to support specific temporal cell outcomes without user intervention and have provided insight on what properties of the ECM are important for the self-assembly of tissues.
Figure 2:
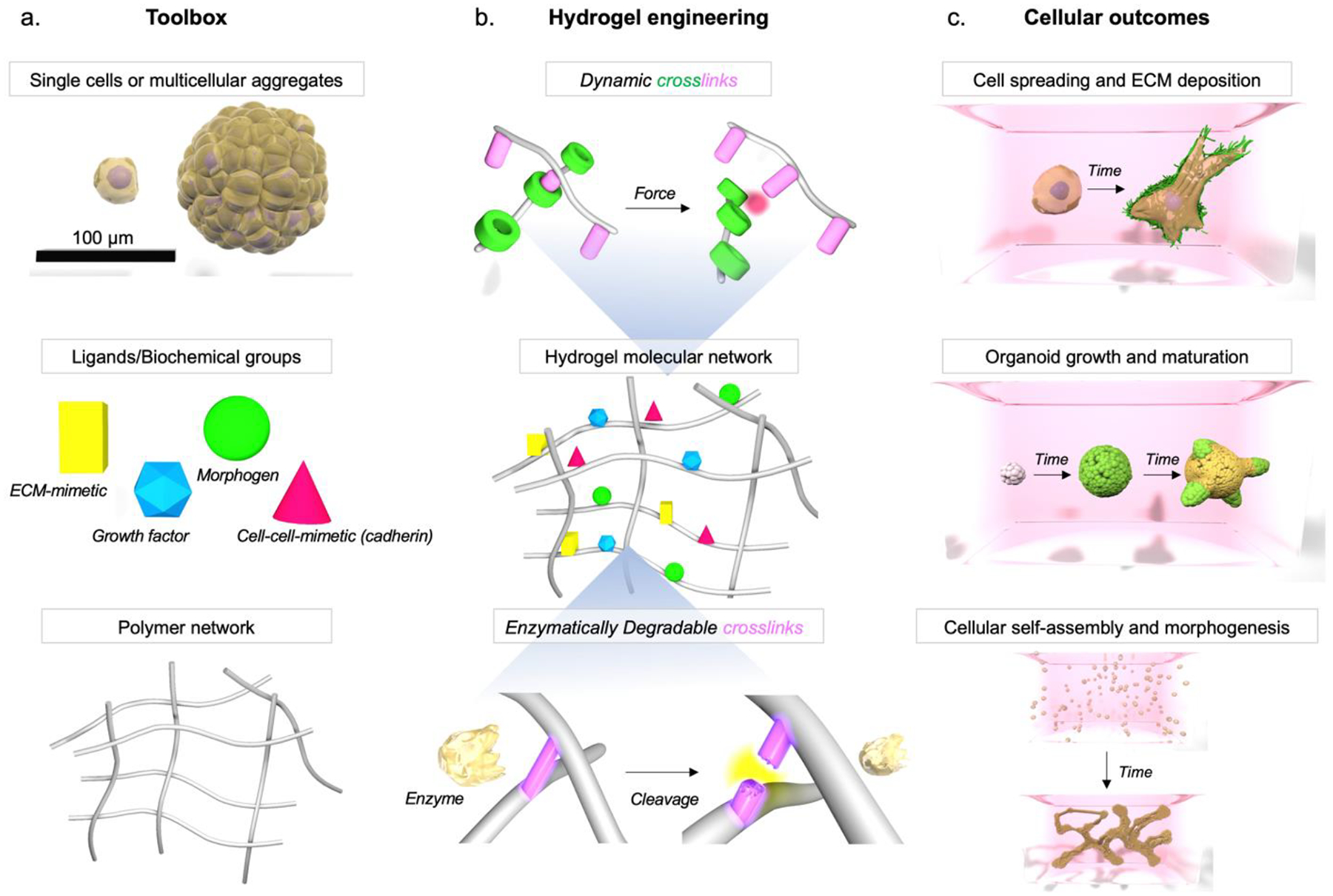
Designing hydrogels for temporal control of cell-driven processes. a. The components of these hydrogel systems include single cells or multicellular aggregates, signaling ligands and other biochemical groups, and a 3D polymer network with chemically reactive functionalities. b. Networks can be engineered to have dynamic crosslinks that break when cells apply forces locally and reform when forces relax (top panel), or have crosslinks that degrade through the action of cell-secreted proteases such as MMPs (bottom panel). c. These engineered hydrogels permit studies of cellular processes for example with single cells that spread and deposit native ECM (top panel), stem cell clusters that mature into tissue-mimetic organoids (middle panel), and homogenously distributed cells that remodel local microenvironments and undergo self-assembly and morphogenesis (bottom panel).
4.1.2. Temporally dynamic hydrogel platforms
One way in which cells remodel the local microenvironment is through the secretion of proteases including matrix metalloproteinases (MMPs) that degrade ECM proteins and allow cells to spread, proliferate, and migrate through dense tissues. This has inspired the design and application of peptide sequences that are susceptible to degradation by MMPs as crosslinkers for 3D hydrogel fabrication (Lutolf et al., 2003). Within these 3D hydrogels, encapsulated cells such as MSCs present MMPs to degrade and restructure their local environment, which allows them to spread, deposit new ECM, generate traction and undergo cell fate specification (Khetan et al., 2013).
Importantly, the level of degradation can be tuned in these materials, providing a programmable platform to control the level of degradation that cells can introduce and a bottom-up approach to control cell fate and organoid maturation (Cruz-Acuña et al., 2019; Trappmann et al., 2017). One drawback with these systems is that because the MMP-degradable peptide is also the crosslinker, it is challenging to decouple stiffness and degradability. Hydrogels crosslinked with these MMP-degradable peptides will eventually lose structural integrity as cells proliferate and increase protease levels. Experiments should thus be planned with realistic timelines (e.g., 1 week of culture or less).
To overcome limitations of protease degradable hydrogels and to provide cells with alternative ways to remodel their environment, synthetic hydrogels have been engineered with dynamic crosslinks – i.e., bonds that break when cells apply local forces and rapidly reform as local stresses relax or when cells migrate away (Yang et al., 2021). Hydrogels based on this system allow the breaking and reforming of dynamic bonds as single cells or multicellular structures grow and exert compressive and tensile forces without elastically pushing cells back to their initial morphology as with stable bonds (Wei et al., 2020). One example of these dynamic hydrogels includes supramolecular guest-host interactions where polymer batches functionalized with different reactive groups (e.g., adamantane: guest, cyclodextrin: host) are dissolved separately and mixed together to achieve instantaneous crosslinking through guest-host interactions. Many dynamic chemical bonds with a range of strengths have been introduced into hydrogels, including hydrogen bonds (Chrisnandy et al., 2021), supramolecular guest-host bonds (Yang et al., 2021), ionic bonds (Chaudhuri et al., 2016), and dynamic covalent bonds (McKinnon et al., 2014a). Each of these bonds has characteristic association/dissociation kinetics that regulate bond breaking and bond reforming. Unfortunately, the reactivity and potential valency of these dynamic bonds means that off-target interactions can occur and reactive functional groups might alter cells in undesirable ways (see considerations box).
CONSIDERATIONS.
| Specification | Details |
|---|---|
| Design specifications for bioprinting of cell-laden bioinks | A key challenge is that properties required for cell survival and tissue deposition are often in conflict with the needs for printability of high fidelity constructs (Cooke and Rosenzweig, 2021). |
| Phototoxicity in light-based manipulation of cell-laden hydrogel properties | In the presence of cells, irradiation can cause cell death through phototoxicity or radical generation. Care must be taken to limit the prolonged exposure to low wavelength light, particularly in the ultraviolet range. (Glass et al., 2018). |
| Ligand and protein bioactivity can be affected by light irradiation | Radical termination can disrupt protein folding, compromising bioactivity of bioactive ligands. The effect of radicals can be mitigated by optimizing the wavelength and intensity of light to reduce the local radical concentration or by including chemical species that terminate or stabilize radicals. |
| Stability of hydrogels with dynamic covalent crosslinks | Dynamic gels will eventually erode over time with media changes and cell remodeling, which could potentially limit the duration of experiments. To overcome this, hydrogels have sometimes involved stabilization with a secondary crosslinking mechanism. |
| Off-target effects of functional groups in dynamic hydrogels | Dynamic chemistries can have off target effects. Aldehyde groups that make up one side of the hydrazone bond can crosslink to a free amine group found in soluble or cell derived proteins to form a weak imine bond. Cyclodextrins used in dynamic guest-host bonds can absorb cholesterol molecules from cell membranes and alter membrane fluidity. Additionally, disruption of ionic bonds in alginate gels could locally change the concentration of calcium, which could impact cell signaling. |
| Imaging cells in 3D hydrogels | Hydrogels can interfere with standard imaging, staining, and immunolabeling methods. Hydrogels may attenuate light, which must be considered when designing experiments. As cells contract fibrous gels, they may densify the material and reduce light penetration to 10s of microns. |
The concentration and type of dynamic bond will control the extent of spreading, allowing users to program how cells remodel their environments. The dynamic nature of these bonds permits stress relaxation and enables the use of these hydrogels as viscoelastic substrates. Dynamic mechanical analysis or rheological characterization are used to determine hydrogel properties, such as stress relaxation times (i.e., the time it takes for stress to relax to half its value for a material held under constant strain) or loss moduli, respectively, where lower relaxation times (seconds to minutes) and higher loss moduli are indicative of higher viscoelasticity, which better mimics that of native tissues (Chrisnandy et al., 2021). One potential challenge with dynamic hydrogels, as with degradable gels, is their lack of long-term stability in culture. This property may cause mechanical properties (e.g., stiffness) to change over time, giving cells a constantly changing environment, which could lead to reduced control over cell outcomes.
Inspired by the naturally fibrous architecture of native ECM macromolecules and proteins, fibrous hydrogel assemblies have also been developed that provide hierarchical structural cues that isotropic hydrogels typically lack. These fiber-based hydrogels may also be designed towards circumventing some of the limitations of dynamic and degradable hydrogels e.g., off target effects of functional groups or disintengration over time, while providing an environment that cells can remodel by pushing and pulling on the fibers (Davidson et al., 2021). These fibrous materials can be fabricated from a range of hydrogel materials. Cell-mediated remodeling can be controlled in these materials by changing the fiber density, which has been exploited to create materials that undergo programmed bending in 3D similar to morphogenic processes in development (Daly et al., 2021a). Although unexplored, fibrous materials have great potential to be used for the culture of multicellular structures like organoids, and for the modular design of fibrous assemblies with varying properties to mimic the diversity of native ECM fibers.
Bottom-up systems such as the ones discussed here mimic features of the native ECM and allow cells to conduct essential cell functions. One of these is the deposition of nascent matrix deposited by cells in the pericellular space consisting of numerous proteins including fibronectin and laminin. Cells begin producing their own nascent ECM within hours of culture in 3D hydrogels and this ECM becomes an additional interface between the cell and the synthetic hydrogel (Loebel et al., 2019, 2020). Metabolic labeling techniques have been employed as an effective approach to visualize much of the cell-secreted ECM, revealing that a pericellular matrix surrounds encapsulated cells that grows over time and that likely reduces signaling between cells and the hydrogel. These aspects must be considered when using any engineered systems for long-term temporal studies.
4.1.3. Representative applications to study cell behavior
While hydrogels consisting of dynamic bonds and fiber-based assemblies are recent advances, protease degradable hydrogels have been around for many years and widely used to probe a variety of biological questions. In single cell systems, matrix degradability has allowed researchers to study how encapsulated mesenchymal stromal cells exert traction forces in covalently crosslinked systems to regulate cell fate (Khetan et al., 2013). In the area of stem cell biology, Madl et al. used a dynamic hydrogel system with varied levels of degradability to demonstrate that temporal cell-mediated matrix remodeling is critical to the maintenance of neural progenitor cell stemness (Madl et al., 2017). The establishment of cell-cell contacts and promotion of beta-catenin signaling that was central to this biological response was only possible in dynamic hydrogels and was not observed in static hydrogels.
Beyond single cells, protease degradable hydrogels have also been used to study multicellular processes for example vascular sprouting. For self-assembly of multicellular structures such as vascularized networks, cells must be able to both invade the material structure and degrade its network (Lutolf et al., 2003). Multicellular sprouting and lumen formation by endothelial cells from engineered parental vessels can be enabled in synthetic hydrogels by incorporating MMP degradable crosslinks and adhesive ligands that allow sprouting cells to locally degrade the matrix through MMP secretion (Liu et al., 2021). MMP-degradable hydrogels have also been used as an interstitial matrix within microporous granular hydrogels with spatially complex interconnected porosity to investigate endothelial cell sprouting and growth from spheroids of endothelial and mesenchymal stromal cells that act as a point source of cells (Qazi et al., 2021). In another system, endothelial and mesenchymal stromal cells distributed homogenously as single cells throughout a 3D MMP-degradable hydrogel locally remodeled their microenvironment and morphed into vessel-like structures (Blache et al., 2016).
Intestinal organoids respond to MMP degradable gels differently based on their maturation status, with non-differentiated expanding organoids losing their stem-like properties in degradable gels, while differentiated organoids maintain higher viability in degradable gels (Cruz-Acuña et al., 2017; Gjorevski et al., 2016). This could be due to the role of compressive stresses during different stages of morphogenesis. As cells break down their local microenvironment, this gives them the ability to expand into new space and also reduce stress built up from growth, which exerts compressive forces on the ECM. Hydrogels with dynamic bonds allow cells greater flexibility in spreading, migration, and local remodeling. Multicellular structures like iPSC-derived organoids (Indana et al., 2021) and differentiated intestinal organoids (Chrisnandy et al., 2021) benefit from dynamic hydrogels that allow for cell expansion and crypt budding, respectively.
4.2. Spatial control and patterning of hydrogels with bioprinting
4.2.1. Overview
Native tissues and organs exhibit a high degree of spatial organization, including the intricate arrangement of cells and hierarchically structured ECM. This spatial organization is compromised with injury and disease, leading to aberrant cell signaling and tissue remodeling. Modeling micrometer scale spatial organization with traditional methods of fabricating bulk cell-laden hydrogels presents numerous and often insurmountable challenges, which are now being overcome with new 3D fabrication techniques such as biofabrication and bioprinting. Bioprinting is a rapidly growing field of tissue engineering that enables excellent control over the three-dimensional spatial deposition of cells, materials, and other signaling factors, and has opened new doors to study biological processes (Figure 3).
Figure 3:
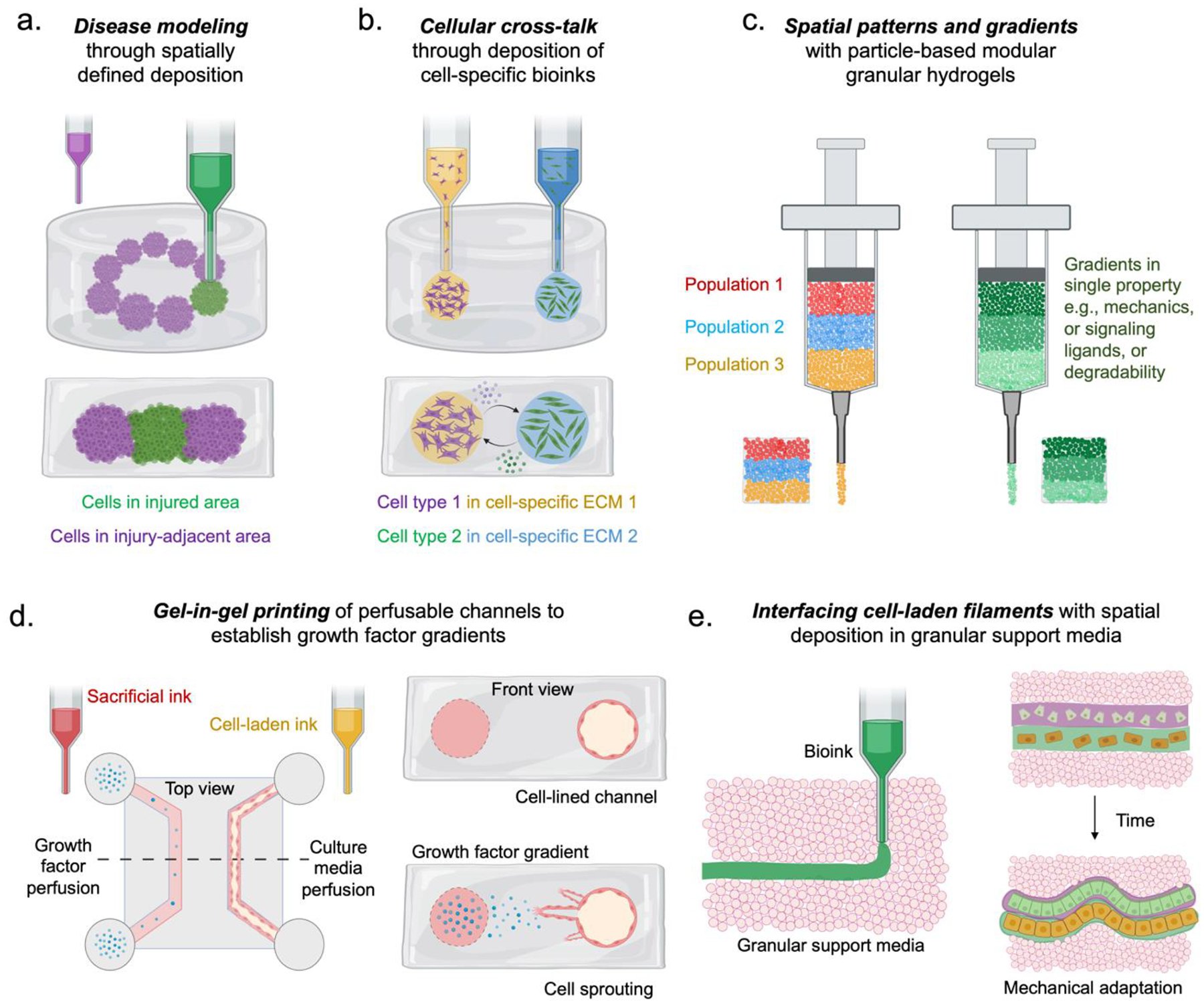
Spatial control of hydrogels with bioprinting and applications to probe cell behavior. Extrusion bioprinting has been used to (a) create disease models with spatially defined regions of abnormally functioning cells, and to (b) study cellular cross-talk by local deposition in cell-specific microenvironments. (c) Other hydrogel formulations such as particle-based modular granular hydrogels have been used to create distinct or gradual gradients in material properties. (d) Suspension bath printing has been used to create perfusable channels with cell attachment to probe spatially regulated growth factor gradients and angiogenic response. (e) Deposition of free-standing filaments is possible with extrusion into a granular support medium that allows time-dependent cell-driven interfacing with adjacent filaments and mechanical adaptation.
4.2.2. Bioprinting hydrogels for spatial control
The most widely reported method in bioprinting is based on the mixing of cells and hydrogel solutions to form a bioink (Moroni et al., 2018). This bioink is then either extruded through a needle into a defined geometry prior to crosslinking (extrusion bioprinting) or a bath of bioink is crosslinked layer-by-layer with the application of specific wavelengths of light (digital light processing (DLP) based bioprinting). Bioinks can be harnessed in several ways, including being extruded to form pre-programmed 3D geometries, used to deposit defined volumes of cell-laden materials into compartmentalized scaffolds, or used to create spatially defined gel-in-gel compartments. Recent advances in light-activated biomaterials have enabled the production of high fidelity constructs with spatial and temporal control of cells through chemical and mechanical stimuli (Morgan et al., 2020; Ouyang et al., 2020). Extrusion bioprinting has been used to create 3D constructs with heterogenous cell-containing environments where spatial organization can be pre-programmed and defined by the user (Kolesky et al., 2014). Many of the hydrogels described already have the potential to be processed using bioprinting techniques.
Tissue injury often leads to disruption of cell viability, behavior, and signaling in spatially localized regions. Models where the interaction between cells in injured and injury-adjacent regions can be studied are challenging to create with traditional material platforms. Bioprinting methods have been developed to address this, namely through the printing of hydrogels into suspension baths. Specifically, shear-thinning and self-healing hydrogels enable recapitulation of this spatial arrangement by acting as suspension baths where cells or materials, including multicellular structures such as spheroids or organoids, can be deposited. Extrusion needles or micromanipulators to deposit spheroids can be inserted or dragged through these hydrogels that undergo transient bond breaking (shear-thinning) and rapid structural recovery (self-healing) (Ayan et al., 2020; Daly et al., 2021b). The deposition of cell-laden bioinks with distinct formulations enables the co-culture of different cell populations in their niche-mimicking microenvironments.
Tissues often have gradient structures, and in diseased tissues the mechanical and chemical environments can be drastically different to neighboring healthy tissues. These gradient features can be replicated in bioprinted constructs by mixing materials of different stiffness or chemistries; however, the challenge is to understand the size scale over which the gradient acts. To address this, microfluidic printheads have been used to combine multiple materials at controlled ratios on their way to the extruding orifice, giving rise to a dynamically-controllable extrusion system (Pedron et al., 2015; Xin et al., 2020). Similarly, with changing light exposure, the stiffness or ligand density of photo-tunable constructs can be varied in the range of hundreds of micrometers throughout their structures (Grigoryan et al., 2021; Vega et al., 2018; Wang et al., 2022). Modular systems like granular hydrogels also permit the creation of spatial gradients. Granular hydrogels are assembled through the packing of hydrogel microparticles and are extrudable through syringes and printer nozzles to allow the creation of macroscale centimeter-scale constructs with micrometer-scale feature resolutions. Particles with distinct properties can be fabricated in separate batches and used as individual inks in tandem to create spatial patterns, including with the loading of multiple particle types into a single syringe for extrusion (Darling et al., 2018). These constructs could mimic naturally encountered gradients in mechanical properties, structural cues, or biochemical ligands and could be a useful platform to screen cell responses to a variety of cues (Xin et al., 2020).
4.2.3. Representative applications of hydrogels to study cell behavior
Traditionally, disease models and testing of drug candidates have relied on 2D culture environments or in vivo models. In contrast, the development of programmable biomaterials have enabled the use of 3D culture microenvironments that afford more physiologically relevant systems that are likely to provide a higher predictive value for clinical scenarios (Langhans, 2018). For example, mini-brains have been 3D printed with spatially distinct regions populated with macrophages and glioblastoma cells to model and probe intercellular interactions and to enable the testing of potential drug candidates (Heinrich et al., 2019). With spatially controlled environments, patient-derived cells can be introduced into the models to determine their responses to different therapeutics and different doses. These approaches are of particular interest in cancer research. In the native tumor microenvironment there are interactions between cancer cells, cancer-associated fibroblasts (CAFs) and endothelial cells. Traditional cancer screening models only considered single cell populations but with the introduction of 3D co-cultures, CAFs can be seeded alongside cancer cells or in spatially distinct co-cultures. In a colorectal cancer model, cancer organoids were seeded in hyaluronic acid/gelatin hydrogels and cultured for 48 hours. With the addition of CAFs to the surface of the hydrogel, organoid growth was hugely accelerated in a much more physiologically relevant manner. Further, with CAFs seeded on the surface of 3D scaffolds, they could be easily removed with trypsinisation for the accurate assessment of cancer cell survival against standard-of-care therapeutics (Luo et al., 2021). With defined spatial environments, the migration of tumour cells (Mohseni Garakani et al., 2021) and the chemoattractive formation of vascular networks towards a tumour compartment can also be assessed (Molley et al., 2020). Patient-derived cells can then be introduced into these models to begin screening of therapeutics for personalized medicine.
Any in vivo tissue environment consists of heterogenous populations of cells that secrete distinct soluble signals and deposit cell-specific ECM that together contribute to the complexity of tissue microenvironments. Studying the role of cellular diversity becomes especially important in the context of diseases and regenerative processes where support cells have been known to exacerbate disease such as fibroblasts in cancer (Labernadie et al., 2017), or support regeneration for example macrophages in wound healing (Sadtler et al., 2016). In cartilage tissue engineering, co-culture of differentiated chondrocytes and mesenchymal stromal cells has been shown to accelerate tissue matrix formation in chitosan based hydrogels (Scalzone et al., 2019). Hydrogels have been used in various contexts to develop heterogenous co-culture platforms to study cellular interactions and implications of ECM properties on cellular outcomes.
To study cell response to gradients in soluble cues, suspension bath printing can be used to deposit sacrificial inks within 3D support hydrogels which can later be perfused with media to create hollow channels. Soluble factors can be introduced into reservoirs connected to one of the hollow channels and cells can be introduced into an adjacent channel. This setup has been used to study how soluble factor gradients stimulate endothelial sprouting from parent engineered vessels lined with endothelial cells (Song et al., 2018; Szklanny et al., 2021). Support hydrogels typically need to be stabilized with secondary covalent crosslinking to permit long-term studies, but other alternative support media have also been reported for bioprinting applications. For example, granular support bath consisting of packed microgels is an emerging platform to deposit free standing mixtures of cells suspended in natural or synthetic ECM (Bhattacharjee et al., 2015; Lee et al., 2019). Microgels can be made from a variety of materials including alginate and agarose through emulsification or fragmentation, and they behave as solid materials when packed together. Importantly, these materials are shear-thinning and self-healing such that printing nozzles can be inserted and dragged through the granular media to deposit cells or materials in precise 3D locations (Hinton et al., 2015; Jeon et al., 2020). In this platform, cells remodel the deposited material and undergo mechanical adaptation in the form of buckling, bending, or contraction, enabling studies on the mechanical forces involved during tissue maturation (Morley et al., 2019).
4.3. Spatiotemporal control of hydrogels with light
4.3.1. Overview
As opposed to hydrogel systems that permit cell-driven remodeling and bioprinting, other hydrogel systems have been engineered that allow on-demand user-defined spatial and temporal manipulation of hydrogel properties using external cues. In vivo, the spatial and temporal presentation of growth factors and ECM ligands is tightly regulated within specific tissue niches, and dysregulation of these cues leads to disease or tissue dysfunction. Additionally, mechanical changes occur across a range of diseases (e.g., fibrosis). Although various cues have been used including light, ultrasound, magnetism, and mechanical vibrations, here we focus exclusively on using light as a tool to modify cellular microenvironments (Figure 4). Light can be used to stimulate photosensitive chemical species incorporated within hydrogels, leading to reactions that either break existing bonds or form new ones. Cytocompatible and bioorthogonal light-responsive reactions have been developed that allow hydrogel degradation or softening through bond breaking or hydrogel stiffening and signaling ligand immobilization through bond forming within cell-laden hydrogels with spatiotemporal control. These exciting developments give the experimentalist exquisite control over where and when cellular microenvironments are modified to probe or perturb complex cell behaviors.
Fig. 4:
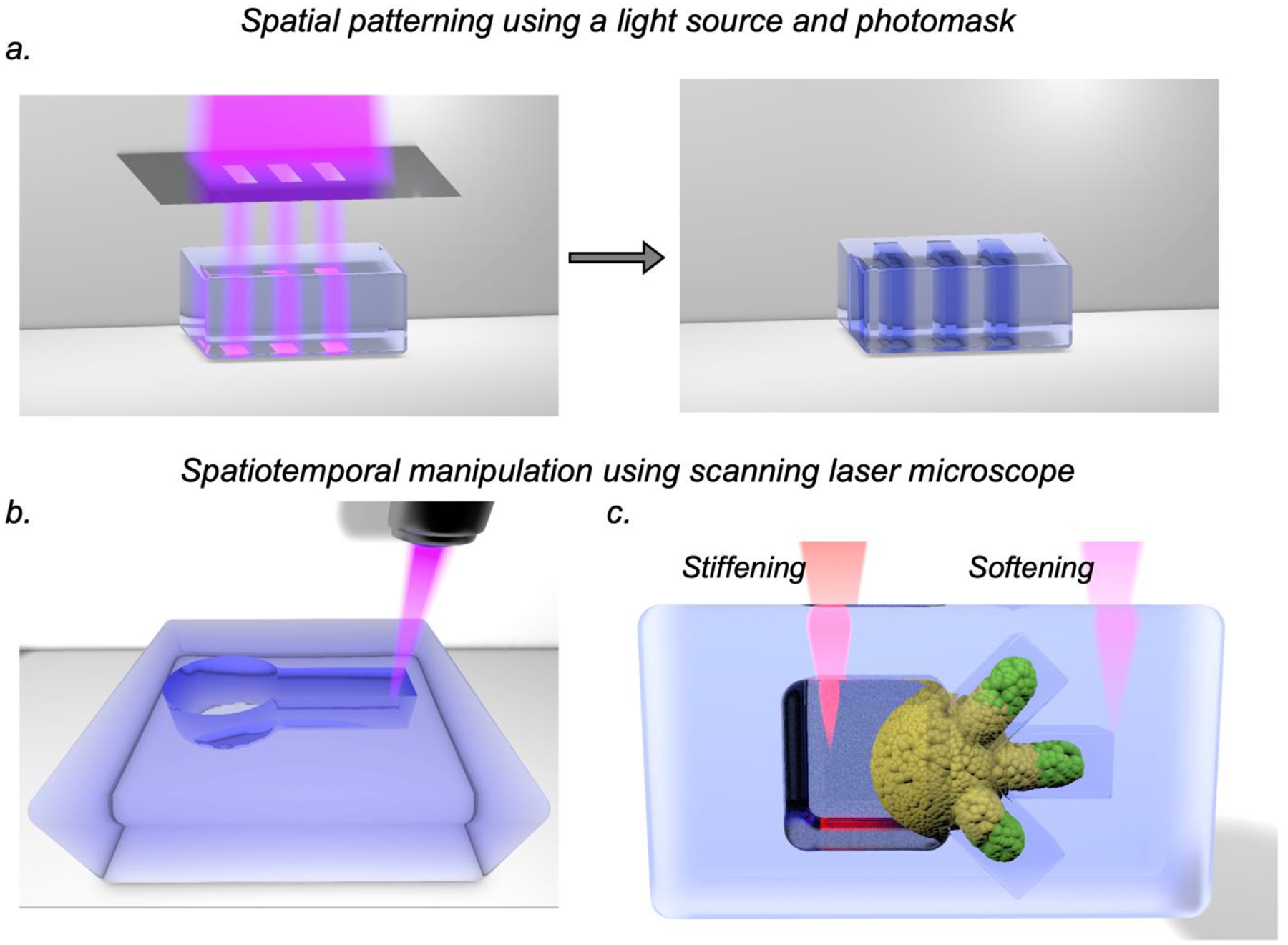
Spatiotemporal control of hydrogel properties with light. a. Spatial patterning of hydrogels can be achieved by directing light through a pre-designed photomask into a hydrogel where spatial regions exposed to light undergo changes in mechanics or signaling ligand composition. Precise patterning of hydrogels with sub-micron resolution in the x,y,z planes can be achieved with a laser scanning microscope. This technique allows (b) local photo-degradation to create channels or other microscale features, or (c) peri-cellular alteration in mechanics through photo-stiffening or photo-softening to control and perturb cellular processes.
4.3.2. Light-responsive hydrogels
Chemical reactions that allow the formation of new bonds within a hydrogel have been leveraged to modulate mechanical properties or chemical composition (Deforest et al., 2009; Tam et al., 2017). These reactions may consume functional groups present on the polymer backbone that remain after initial hydrogel formation, and the extent of additional reaction determines the degree of stiffening or the concentration of ligands that can be introduced in the secondary modification. Several technical considerations are involved when using photochemical reactions to alter mechanical properties or the presentation of biochemical cues, and the reader is guided to the considerations box in this paper for further details.
As one example, hydrogel stiffening is facilitated by the photoinduced formation of crosslinks when light is directed to regions of the cell-laden hydrogel that have been infiltrated with additional photoinitiator and reactive crosslinker molecules (Zheng et al., 2017). The degree of stiffening can be dependent on light intensity, duration of light exposure, and availability of reactive functional groups. For spatially defined studies, photomasks with predefined patterns can be used to modulate macroscale regions within the hydrogel or alternatively laser microscopes can be used to modulate microscale regions in the cellular microenvironment typically with a sub-micron resolution in the xyz planes (Figure 4a,b). These hydrogel stiffening strategies have been used extensively to replicate disease or injury-associated tissue stiffening to study cellular phenotype, mechanotransduction, migration, and differentiation.
Immobilization of signaling ligands onto a pre-crosslinked cell-laden hydrogel also utilizes addition reactions (Fisher et al., 2018). Photoaddition reactions have been used to replicate these spatiotemporally dynamic signals. For this to occur, ligands must contain a photo-reactive group that permits immobilization to the polymer network on exposure to light. The conjugation of cell-adhesive RGD ligand is perhaps the most widely used example of this method. Vega et al. used a sliding photomask to create spatial gradients of hydrogel bound RGD through a light-initiated thiol-ene reaction between norbornene functional groups on the polymer backbone and thiol-containing peptides (Vega et al., 2018). Other chemistries (e.g., acrylate functional groups) and techniques (e.g., two photon patterning) can also be used to spatially control ligand presentation in three dimensions (Lee et al., 2008). Another strategy to spatiotemporally expose cells to signaling ligands is through selective light-induced removal of a caging or protective chemical group from a bound ligand. Caging groups prevent cells from sensing the signaling ligand, but their photocleavage and release at a user-defined time point and in spatially determined regions can reveal the signaling ligand to the cells. Spatiotemporal removal of caging groups to expose bound RGD has been shown to modulate in vivo cell adhesion, inflammation, and vascularization (Lee et al., 2015).
Spatiotemporal hydrogel softening can be achieved through photocleavage reactions which break crosslinking bonds on exposure to light. These subtractive reactions reduce hydrogel crosslinking density in regions exposed to light and can be used to interrogate cellular mechanotransduction before and after a user-directed change, or to direct migration or tissue organization in eroded or softened regions of the cellular microenvironment (Arakawa et al., 2017). Molecules containing ortho-nitrobenzyl and coumarin functionalities undergo photoinduced cleavage and can be used to decrease the mechanical stiffness of the hydrogel (Levalley et al., 2020). These subtractive reactions can similarly be used to remove polymer-bound signaling ligands; for this, a photocleavable group can be incorporated as a link between a biochemical ligand and the polymer backbone, such that cleavage of this group on exposure to light results in the release of the ligand in regions and at times defined by the user. Dynamic removal of photocleavable RGD has been used to abrogate cell-matrix interactions and promote chondrogenic differentiation of hMSCs (Kloxin et al., 2009).
4.3.3. Representative applications to study cell behavior
Light-responsive hydrogels have been applied to study mechanistic aspects of tissue or organ fibrosis, which can occur in response to acute injury or chronic disease and is characterized by excessive deposition of ECM (e.g., collagen by activated fibroblasts). Excessive ECM deposition leads to stiffening of the microenvironment and could lead to tissue failure, as observed in fibrotic heart valves that fail to regulate blood flow (Kloxin et al., 2010). Fibroblasts are sensitive to changes in the mechanics of their microenvironments and the tissue stiffening that occurs after muscle injury or liver fibrosis triggers the differentiation of resident fibroblasts into activated myofibroblasts that are responsible for depositing excessive ECM. Spatiotemporally dynamic hydrogels have been used to study this transition between fibroblast states. For example, Caliari et al. used a dual crosslinking hydrogel that undergoes user-defined gradual stiffening to probe the transition of liver-derived hepatic stellate cells into activated myofibroblasts (Caliari et al., 2016). Using this system, hydrogel stiffness was varied from ~2 kPa up to ~35 kPa, providing a broad physiologically relevant range to probe differences in cell behavior. More recently, photo-stiffening hydrogels (~2 kPa to ~40 kPa) were used to replicate the increase in tissue stiffness following muscle injury (~5 kPa to ~22 kPa) and study the biological impact of such dynamic stiffening on muscle stem cells, including the promotion of migration and proliferation towards facilitating regeneration of the injured muscle (Silver et al., 2021).
To study the effect of mechanical softening on the reversibility of myofibroblast differentiation, Kloxin and colleagues used photocleavable crosslinkers to generate stiffness gradients within a single hydrogel to show that photodegradation reduces the number of activated fibroblasts (Kloxin et al., 2010). By varying the time of exposure to light in photodegradable hydrogels, the authors dynamically altered mechanical properties from ~32 kPa (minimal exposure to light) to ~5 kPa (maximum exposure to light). Additionally, eroding specific regions of the hydrogel in 3D space can be used to direct the spatial migration or expansion of single or multicellular structures into the space voided by photodegradation (Arakawa et al., 2017). This has been used to direct the extension of motor neurons (McKinnon et al., 2014b) and intestinal organoid crypts (Gjorevski et al., 2022).
Viscoelastic hydrogels have been used extensively to study the cellular response to environments with varying viscoelastic character (Chaudhuri et al., 2020). However, photoinduced viscoelastic materials are required to study changes in spatial and temporal viscoelastic behavior. To temporally modulate viscoelastic behavior, a photoaddition reaction was used to quench the reactive groups involved in viscoelastic crosslink exchange, effectively turning off the viscoelastic behavior of the material. This method was used to decouple the effects of viscoelasticity and elastic modulus on fibroblast mechanotransduction (Carberry et al., 2020). Conversely, viscoelasticity could be “turned off” by using light to generate network tethered radicals that could participate in reversible addition-fragmentation reactions with network tethered allyl sulfide groups, which effectively form and break crosslinks continuously. This photoinduced viscoelastic behavior was used to spatially probe the retraction of individual protrusions of hMSCs (Marozas et al., 2019).
Organoids have emerged as advanced models to study and perturb developmental processes in vitro. Light-mediated alterations to the local mechanics or biochemical cue availability are uniquely well-suited to study organoids, as they provide the spatiotemporal resolution necessary to alter single or population-wide cellular behavior, both of which may be of interest to organoid biologists. Two recent papers have designed systems to locally alter matrix mechanics, either by softening or stiffening, to induce changes on a smaller population of cells, and thus guide symmetry breaking and budding or branching events. Specifically, local matrix softening using PEG-based hydrogels (where elastic modulus reduced to less than half its original value) using photocleavable nitrobenzyl crosslinkers facilitated deterministic crypt formation with optimized light doses to photopattern softened regions matching intestinal crypt dimensions (Gjorevski et al., 2022). Using the opposite approach, live 4D bioprinting was used to photo-stiffen specific regions around organoids or organotypic cultures. This method added photocrosslinkable coumarin based hydrogel precursors to already formed ECM-based hydrogels, either Matrigel or collagen, then used multiphoton light to crosslink new hydrogels within the existing hydrogel, in essence stiffening the environment in specific regions (from ~2 to 22 kPa). This method was used to confine or pattern intestinal organoid growth and crypt formation, as well as to stiffen regions and direct lung epithelial branching (Urciuolo et al., 2020). Finally, while most of the work in this arena is with intestinal organoids, it is likely that these types of local changes to matrix mechanics can be more broadly applied to control similar events in other branching and budding organoids, such as mammary or salivary tissues, based on studies more fundamentally describing how such events are regulated (Wang et al., 2021).
5. Concluding Remarks and Future Outlook
Cellular behavior involved in complex biological processes are dynamic and are regulated by biophysical and biochemical signals that vary in both space and time. Traditional methods of cell culture involving 2D substrates and 3D naturally-derived matrices like Matrigel neither mimic this complexity nor provide the experimentalist with control over local properties. In this review, we have provided the reader a guided overview of synthetic hydrogels and technologies that allow control over the spatiotemporal properties of cellular microenvironments. In mimicking the dynamic biophysical and biochemical properties of the native ECM, these hydrogel platforms enable novel insights into disease modeling, inter-cellular communication, development, tissue engineering, and mechanobiology.
Biomaterials scientists are continually developing new and better materials that enhance functionality, permit control over multiple signals, and provide improved spatiotemporal control. These are either materials that permit cell-driven remodeling, or that allow spatial control and patterning, and those that can be manipulated by the user with external cues such as light. These materials have been adapted for use not only to study single cell biology, but increasingly to probe the function of multicellular structures that grow or mature over the culture period including multicellular spheroids or stem cell-derived organoids. The outcome, much to the benefit of basic biologists, is that we are now capable of exploring how spatiotemporally dynamic signals like gradients of soluble cues, matrix-bound biochemical ligands, changes in mechanics, and paracrine signals secreted by neighboring cells impact biological processes.
With the availability and widespread adoption of these hydrogel platforms for various studies, future work must benchmark in vitro cellular response to varying spatiotemporal signals against in vivo processes to get a better understanding of how faithfully these in vitro systems can recapitulate or predict outcomes. At present, dynamic hydrogels largely focus on replicating a singular aspect from among a complex array of ECM properties – e.g., mechanics or gradients in soluble cues or exposure to a signaling ligand. While this reductionist approach has helped in elucidating signaling mechanisms and cell function, future work will likely begin to build in increasing complexity where multiple spatiotemporal properties can be controlled combinatorially (Rosales and Anseth, 2016). When this happens, it will be important to have complementary approaches to characterize the resulting cell behavior.
It was our intention with this review to narrow the gap between biologists and these advanced engineering tools to create biomimetic 3D matrices. Many biologists may prefer the simplicity of 2D substrates or naturally-derived Matrigel, but we anticipate that they will soon recognize how powerful dynamic hydrogel-based platforms can be in providing reliable and reproducible data while also giving the user complete control of spatiotemporal properties. Collaborations with biomaterials scientists and bioengineers will not only accelerate the adoption of these platforms but will also provide necessary feedback to continue improving hydrogel design.
Acknowledgements
This work was supported by the National Science Foundation through the Center for Engineering MechanoBiology STC (CMMI: 15-48571 to J.A.B.), the UPenn MRSEC program (DMR-1720530 to J.A.B.), and grant RECODE 2033723 (to K.S.A.), as well as through the National Institutes of Health (R01AR077362, R01AR056624 to J.A.B., and R01DE016523, R01DE120921 to K.S.A.) and the German Science Foundation (QA 58/1-1 to T.H.Q.). Schematics were created with Biorender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Aisenbrey EA, and Murphy WL (2020). Synthetic alternatives to Matrigel. Nat. Rev. Mater 5, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa CK, Badeau BA, Zheng Y, and DeForest CA (2017). Multicellular Vascularized Engineered Tissues through User-Programmable Biomaterial Photodegradation. Adv. Mater 29, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayan B, Heo DN, Zhang Z, Dey M, Povilianskas A, Drapaca C, and Ozbolat IT (2020). Aspiration-assisted bioprinting for precise positioning of biologics. Sci. Adv 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, and Angelini TE (2015). Writing in the granular gel medium. Sci. Adv 1, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Guvendiren M, Mauck RL, and Burdick JA (2013). Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. Proc. Natl. Acad. Sci 110, 10117–10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blache U, Metzger S, Vallmajo-Martin Q, Martin I, Djonov V, and Ehrbar M (2016). Dual Role of Mesenchymal Stem Cells Allows for Microvascularized Bone Tissue-Like Environments in PEG Hydrogels. Adv. Healthc. Mater 5, 489–498. [DOI] [PubMed] [Google Scholar]
- Bonnans C, Chou J, and Werb Z (2014). Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TE, and Anseth KS (2017). Spatiotemporal hydrogel biomaterials for regenerative medicine. Chem. Soc. Rev 46, 6532–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi L, Ramonet D, Klingberg F, Formey A, Smith-Clerc J, Meister JJ, and Hinz B (2011). The single-molecule mechanics of the latent TGF-β1 complex. Curr. Biol 21, 2046–2054. [DOI] [PubMed] [Google Scholar]
- Caliari SR, and Burdick JA (2016). A practical guide to hydrogels for cell culture. Nat. Methods 13, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, and Burdick JA (2016). Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci. Rep 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AR, Frith JE, and Cooper-White JJ (2011). The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials 32, 5979–5993. [DOI] [PubMed] [Google Scholar]
- Carberry BJ, Rao VV, and Anseth KS (2020). Phototunable Viscoelasticity in Hydrogels Through Thioester Exchange. Ann. Biomed. Eng 48, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, and Mooney DJ (2015). Substrate stress relaxation regulates cell spreading. Nat. Commun 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, et al. (2016). Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater 15, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ, and Shenoy VB (2020). Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrisnandy A, Blondel D, Rezakhani S, Broguiere N, and Lutolf MP (2021). Synthetic dynamic hydrogels promote degradation-independent in vitro organogenesis. Nat. Mater 2021 1–9. [DOI] [PubMed] [Google Scholar]
- Cooke ME, and Rosenzweig DH (2021). The rheology of direct and suspended extrusion bioprinting. APL Bioeng. 5, 011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove BD, Mui KL, Driscoll TP, Caliari SR, Mehta KD, Assoian RK, Burdick JA, and Mauck RL (2016). N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater 15, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzier T, Fourel L, Boudou T, Albigès-Rizo C, and Picart C (2011). Presentation of BMP-2 from a soft biopolymeric film unveils its activity on cell adhesion and migration. Adv. Mater 23, 111–118. [DOI] [PubMed] [Google Scholar]
- Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, García-Hernández V, Miller AJ, Spence JR, Nusrat A, et al. (2017). Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol 19, 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Acuña R, Mulero-Russe A, Clark AY, Zent R, and García AJ (2019). Identification of matrix physicochemical properties required for renal epithelial cell tubulogenesis by using synthetic hydrogels. J. Cell Sci 132, jcs226639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Peters SB, and Larsen M (2008). Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci 121, 255–264. [DOI] [PubMed] [Google Scholar]
- Daly AC, Prendergast ME, Hughes AJ, and Burdick JA (2021a). Bioprinting for the Biologist. Cell 184, 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AC, Davidson MD, and Burdick JA (2021b). 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling NJ, Sideris E, Hamada N, Carmichael ST, and Segura T (2018). Injectable and Spatially Patterned Microporous Annealed Particle (MAP) Hydrogels for Tissue Repair Applications. Adv. Sci 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MD, Prendergast ME, Ban E, Xu KL, Mickel G, Mensah P, Dhand A, Janmey PA, Shenoy VB, and Burdick JA (2021). Programmable and contractile materials through cell encapsulation in fibrous hydrogel assemblies. Sci. Adv 7, 8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforest CA, Polizzotti BD, and Anseth KS (2009). Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater 8, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, and Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689. [DOI] [PubMed] [Google Scholar]
- Fata JE, Werb Z, and Bissell MJ (2004). Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SA, Tam RY, Fokina A, Mahmoodi MM, Distefano MD, and Shoichet MS (2018). Photo-immobilized EGF chemical gradients differentially impact breast cancer cell invasion and drug response in defined 3D hydrogels. Biomaterials 178, 751–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattazzo F, Urciuolo A, and Bonaldo P (2014). Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta - Gen. Subj 1840, 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, and Blau HM (2010). Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, and Lutolf MP (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Nikolaev M, Brown TE, Mitrofanova O, Brandenberg N, DelRio FW, Yavitt FM, Liberali P, Anseth KS, and Lutolf MP (2022). Tissue geometry drives deterministic organoid patterning. Science 375, eaaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass S, Trinklein B, Abel B, and Schulze A (2018). TiO2 as photosensitizer and photoinitiator for synthesis of photoactive TiO2-PEGDA hydrogel without organic photoinitiator. Front. Chem 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LG, and Swartz MA (2006). Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol 7, 211–224. [DOI] [PubMed] [Google Scholar]
- Grigoryan B, Sazer DW, Avila A, Albritton JL, Padhye A, Ta AH, Greenfield PT, Gibbons DL, and Miller JS (2021). Development, characterization, and applications of multi-material stereolithography bioprinting. Sci. Rep 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães CF, Gasperini L, Marques AP, and Reis RL (2020). The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater 5, 351–370. [Google Scholar]
- Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, and Prakash J (2019). 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Adv. Mater 31, 1–9. [DOI] [PubMed] [Google Scholar]
- Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, and Feinberg AW (2015). Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer M, and Lutolf MP (2021). Engineering organoids. Nat. Rev. Mater 6, 402–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali O. a, Bencherif SA, Rivera-Feliciano J, and Mooney DJ (2010). Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater 9, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Postovit LM, and Lajoie GA (2010). Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890. [DOI] [PubMed] [Google Scholar]
- Indana D, Agarwal P, Bhutani N, Chaudhuri O, Indana D, Chaudhuri O, Agarwal P, and Bhutani N (2021). Viscoelasticity and Adhesion Signaling in Biomaterials Control Human Pluripotent Stem Cell Morphogenesis in 3D Culture. Adv. Mater 33, 2101966. [DOI] [PubMed] [Google Scholar]
- Jeon S, Heo JH, Kim MK, Jeong W, and Kang HW (2020). High-Precision 3D Bio-Dot Printing to Improve Paracrine Interaction between Multiple Types of Cell Spheroids. Adv. Funct. Mater 30, 2005324. [Google Scholar]
- Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, and Burdick JA (2013). Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater 12, 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein O, Strohschein K, Nebrich G, Oetjen J, Trede D, Thiele H, Alexandrov T, Giavalisco P, Duda GN, von Roth P, et al. (2014). MALDI imaging mass spectrometry: Discrimination of pathophysiological regions in traumatized skeletal muscle by characteristic peptide signatures. Proteomics 14, 2249–2260. [DOI] [PubMed] [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, and Anseth KS (2009). Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin AM, Benton JA, and Anseth KS (2010). In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials 31, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, and Lewis JA (2014). 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater 26, 3124–3130. [DOI] [PubMed] [Google Scholar]
- Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, González-Tarragó V, Elosegui-Artola A, Albertazzi L, et al. (2017). A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol 19, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans SA (2018). Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, and Feinberg AW (2019). 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487. [DOI] [PubMed] [Google Scholar]
- Lee SH, Moon JJ, and West JL (2008). Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials 29, 2962–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TT, García JR, Paez JI, Singh A, Phelps EA, Weis S, Shafiq Z, Shekaran A, Del Campo A, and García AJ (2015). Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater 14, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levalley PJ, Neelarapu R, Sutherland BP, Dasgupta S, Kloxin CJ, and Kloxin AM (2020). Photolabile Linkers: Exploiting Labile Bond Chemistry to Control Mode and Rate of Hydrogel Degradation and Protein Release. J. Am. Chem. Soc 142, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. (2009). Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 139, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Long H, Zeuschner D, Räder AFB, Polacheck WJ, Kessler H, Sorokin L, and Trappmann B (2021). Synthetic extracellular matrices with tailored adhesiveness and degradability support lumen formation during angiogenic sprouting. Nat. Commun 2021 12, 3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel C, Mauck RL, and Burdick JA (2019). Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel C, Kwon MY, Wang C, Han L, Mauck RL, and Burdick JA (2020). Metabolic Labeling to Probe the Spatiotemporal Accumulation of Matrix at the Chondrocyte–Hydrogel Interface. Adv. Funct. Mater 30, 1909802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fong ELS, Zhu C, Lin QXX, Xiong M, Li A, Li T, Benoukraf T, Yu H, and Liu S (2021). Hydrogel-based colorectal cancer organoid co-culture models. Acta Biomater. 132, 461–472. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, and Hubbell JA (2003). Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics. Proc. Natl. Acad. Sci 100, 5413–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyshchik A, Higashi T, Asato R, Tanaka S, Ito J, Hiraoka M, Brill AB, Saga T, and Togashi AK (2005). Elastic Moduli Of Thyroid Tissues Under Compression. 110, 101–110. [DOI] [PubMed] [Google Scholar]
- Madl CM, and Heilshorn SC (2018). Engineering Hydrogel Microenvironments to Recapitulate the Stem Cell Niche. Annu. Rev. Biomed. Eng 20, 21–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl CM, Mehta M, Duda GN, Heilshorn SC, and Mooney DJ (2014). Presentation of BMP-2 mimicking peptides in 3D hydrogels directs cell fate commitment in osteoblasts and mesenchymal stem cells. Biomacromolecules 15, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl CM, Lesavage BL, Dewi RE, Dinh CB, Stowers RS, Khariton M, Lampe KJ, Nguyen D, Chaudhuri O, Enejder A, et al. (2017). Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater 16, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozas IA, Cooper-White JJ, and Anseth KS (2019). Photo-induced viscoelasticity in cytocompatible hydrogel substrates. New J. Phys 21, 045004. [Google Scholar]
- Marsee A, Roos FJM, Verstegen MMA, Roos F, Verstegen M, Clevers H, Vallier L, Takebe T, Huch M, Peng WC, et al. (2021). Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 28, 816–832. [DOI] [PubMed] [Google Scholar]
- McKinnon DD, Domaille DW, Cha JN, and Anseth KS (2014a). Biophysically Defined and Cytocompatible Covalently Adaptable Networks as Viscoelastic 3D Cell Culture Systems. Adv. Mater 26, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon DD, Brown TE, Kyburz KA, Kiyotake E, and Anseth KS (2014b). Design and characterization of a synthetically accessible, photodegradable hydrogel for user-directed formation of neural networks. Biomacromolecules 15, 2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Blache U, Lienemann PS, Karlsson M, Weber FE, Weber W, and Ehrbar M (2016). Cell-Mediated Proteolytic Release of Growth Factors from Poly(Ethylene Glycol) Matrices. Macromol. Biosci 16, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Mohseni Garakani M, Ahangar P, Watson S, Nisol B, Wertheimer MR, Rosenzweig DH, and Ajji A (2021). A novel 3D co-culture platform for integrating tissue interfaces for tumor growth, migration and therapeutic sensitivity: “PP-3D-S.” Mater. Sci. Eng. C 112566. [DOI] [PubMed] [Google Scholar]
- Molley TG, Wang X, Hung T. tyng, Jayathilaka PB, Yang JL, and Kilian KA (2020). Geometrically Structured Microtumors in 3D Hydrogel Matrices. Adv. Biosyst 4, 1–8. [DOI] [PubMed] [Google Scholar]
- Moretti L, Stalfort J, Barker TH, and Abebayehu D (2021). The interplay of fibroblasts, the extracellular matrix, and inflammation in scar formation. J. Biol. Chem 298, 101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan FLC, Moroni L, and Baker MB (2020). Dynamic Bioinks to Advance Bioprinting. Adv. Healthc. Mater 9, 1901798. [DOI] [PubMed] [Google Scholar]
- Morley CD, Ellison ST, Bhattacharjee T, O’Bryan CS, Zhang Y, Smith KF, Kabb CP, Sebastian M, Moore GL, Schulze KD, et al. (2019). Quantitative characterization of 3D bioprinted structural elements under cell generated forces. Nat. Commun 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni L, Boland T, Burdick JA, De Maria C, Derby B, Forgacs G, Groll J, Li Q, Malda J, Mironov VA, et al. (2018). Biofabrication: A Guide to Technology and Terminology. Trends Biotechnol. 36, 384–402. [DOI] [PubMed] [Google Scholar]
- Nicolas J, Magli S, Rabbachin L, Sampaolesi S, Nicotra F, and Russo L (2020). 3D Extracellular Matrix Mimics: Fundamental Concepts and Role of Materials Chemistry to Influence Stem Cell Fate. Biomacromolecules 21, 1968–1994. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Armstrong JPK, Lin Y, Wojciechowski JP, Lee-Reeves C, Hachin D, Zhou K, Burdick JA, and Stevens MM (2020). Expanding and Optimizing 3D Bioprinting Capabilities using Complementary Network Bioinks. Sci. Adv 6, eabc5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedron S, Becka E, and Harley BA (2015). Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment. Adv. Mater 27, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Przybyla L, Lakins JN, and Weaver VM (2016). Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation. Cell Stem Cell 19, 462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi TH, Wu J, Muir VG, Weintraub S, Gullbrand SE, Lee D, Issadore D, and Burdick JA (2021). Anisotropic Rod-Shaped Particles Influence Injectable Granular Hydrogel Properties and Cell Invasion. Adv. Mater 2109194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabata A, Fedr R, Soucek K, Hampl A, and Koledova Z (2020). 3D Cell Culture Models Demonstrate a Role for FGF and WNT Signaling in Regulation of Lung Epithelial Cell Fate and Morphogenesis. Front. Cell Dev. Biol 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Gauthier NC, Del Rio A, and Sheetz MP (2009). Clustering of α5β1 integrins determines adhesion strength whereas αvβ3 and talin enable mechanotransduction. Proc. Natl. Acad. Sci. U. S. A 106, 16245–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales AM, and Anseth KS (2016). The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater 1, 15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozario T, and DeSimone DW (2010). The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol 341, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskowitz ER, and Deforest CA (2018). Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater 3, 17087. [Google Scholar]
- Sadtler K, Powell JD, Wolf MT, Elisseeff JH, Estrellas K, Pardoll DM, Wagner KR, Tam AJ, Patel CH, Allen BW, et al. (2016). Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 352, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, and Briscoe J (2017). Morphogen interpretation: concentration, time, competence, and signaling dynamics. Wiley Interdiscip. Rev. Dev. Biol 6, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Larsen M, and Yamada KM (2003). Fibronectin requirement in branching morphogenesis. Nature 423, 876–881. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Scalzone A, Ferreira AM, Tonda-Turo C, Ciardelli G, Dalgarno K, and Gentile P (2019). The interplay between chondrocyte spheroids and mesenchymal stem cells boosts cartilage regeneration within a 3D natural-based hydrogel. Sci. Rep 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellard A, and Mayor R (2021). Collective durotaxis along a self-generated stiffness gradient in vivo. Nature 600, 690–694. [DOI] [PubMed] [Google Scholar]
- Shin JW, and Mooney DJ (2016). Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias. Proc. Natl. Acad. Sci. U. S. A 113, 12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JS, Günay KA, Cutler AA, Vogler TO, Brown TE, Pawlikowski BT, Bednarski OJ, Bannister KL, Rogowski CJ, McKay AG, et al. (2021). Injury-mediated stiffening persistently activates muscle stem cells through YAP and TAZ mechanotransduction. Sci. Adv 7, eabe4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KH, Highley CB, Rouff A, and Burdick JA (2018). Complex 3D-Printed Microchannels within Cell-Degradable Hydrogels. Adv. Funct. Mater 28, 1801331. [Google Scholar]
- Spicer CD, Pashuck ET, and Stevens MM (2018). Achieving Controlled Biomolecule-Biomaterial Conjugation. Chem. Rev 118, 7702–7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklanny AA, Machour M, Redenski I, Chochola V, Goldfracht I, Kaplan B, Epshtein M, Simaan Yameen H, Merdler U, Feinberg A, et al. (2021). 3D Bioprinting of Engineered Tissue Flaps with Hierarchical Vessel Networks (VesselNet) for Direct Host-To-Implant Perfusion. Adv. Mater 33, 2102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam RY, Smith LJ, and Shoichet MS (2017). Engineering Cellular Microenvironments with Photo- and Enzymatically Responsive Hydrogels: Toward Biomimetic 3D Cell Culture Models. Acc. Chem. Res 50, 703–713. [DOI] [PubMed] [Google Scholar]
- Trappmann B, Baker BM, Polacheck WJ, Choi CK, Burdick JA, and Chen CS (2017). Matrix degradability controls multicellularity of 3D cell migration. Nat. Commun 8, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, et al. (2013). Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun 4, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urciuolo A, Poli I, Brandolino L, Raffa P, Scattolini V, Laterza C, Giobbe GG, Zambaiti E, Selmin G, Magnussen M, et al. (2020). Intravital three-dimensional bioprinting. Nat. Biomed. Eng 4, 901–915. [DOI] [PubMed] [Google Scholar]
- Vega SL, Kwon MY, Song KH, Wang C, Mauck RL, Han L, and Burdick JA (2018). Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nat. Commun 9, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walma DAC, and Yamada KM (2020). The extracellular matrix in development. Development 147, dev175596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li W, Mille LS, Ching T, Luo Z, Tang G, Garciamendez CE, Lesha A, Hashimoto M, and Zhang YS (2022). Digital Light Processing Based Bioprinting with Composable Gradients. Adv. Mater 34, 2107038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Matsumoto K, Lish SR, Cartagena-Rivera AX, and Yamada KM (2021). Budding epithelial morphogenesis driven by cell-matrix versus cell-cell adhesion. Cell 184, 3702–3716.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Schnellmann R, Pruitt HC, and Gerecht S (2020). Hydrogel Network Dynamics Regulate Vascular Morphogenesis. Cell Stem Cell 27, 798–812.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, and Werb Z (2020). Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun 11, 5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff PJ, and Hinz B (2008). Integrins and the activation of latent transforming growth factor β1 - An intimate relationship. Eur. J. Cell Biol 87, 601–615. [DOI] [PubMed] [Google Scholar]
- Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, and Werb Z (2003). Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J. Cell Biol 162, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin S, Dai J, Gregory CA, Han A, and Alge DL (2020). Creating Physicochemical Gradients in Modular Microporous Annealed Particle Hydrogels via a Microfluidic Method. Adv. Funct. Mater 30, 1907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wei K, Loebel C, Zhang K, Feng Q, Li R, Wong SHD, Xu X, Lau C, Chen X, et al. (2021). Enhanced mechanosensing of cells in synthetic 3D matrix with controlled biophysical dynamics. Nat. Commun 2021 121 12, 3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Hu J, Wang H, Huang J, Yu Y, Zhang Q, and Cheng Y (2017). Dynamic Softening or Stiffening a Supramolecular Hydrogel by Ultraviolet or Near-Infrared Light. ACS Appl. Mater. Interfaces 9, 24511–24517. [DOI] [PubMed] [Google Scholar]


