Abstract
Human papilloma virus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) is a common cancer worldwide with an unmet need for more effective, less toxic treatments. Currently, both the disease and the treatment of HNSCC cause significant mortality and morbidity. Targeted therapies hold new promise for HPV-negative patients whose tumors harbor oncogenic HRAS mutations. Recent promising clinical results have renewed interest in the development of farnesyltransferase inhibitors (FTIs) as a therapeutic strategy for HRAS-mutant cancers. With the advent of clinical evaluation of the FTI tipifarnib for the treatment of HRAS-mutant HNSCC, we investigated the activity of tipifarnib and inhibitors of HRAS effector signaling in HRAS-mutant HNSCC cell lines. First, we validated that HRAS is a cancer driver in HRAS-mutant HNSCC lines. Second, we showed that treatment with the FTI tipifarnib largely phenocopied HRAS silencing, supporting HRAS as a key target of FTI anti-tumor activity. Third, we performed reverse phase protein array (RPPA) analyses to profile FTI treatment-induced changes in global signaling, and conducted CRISPR/Cas9 genetic loss of function screens to identify previously unreported genes and pathways that modulate sensitivity to tipifarnib. Fourth, we determined that concurrent inhibition of HRAS effector signaling (ERK, PI3K, mTORC1) increased sensitivity to tipifarnib treatment, in part by overcoming tipifarnib-induced compensatory signaling. We also determined that ERK inhibition could block tipifarnib-induced epithelial-to-mesenchymal transition (EMT), providing a potential basis for the effectiveness of this combination. Our results support future investigations of these and other combination treatments for HRAS mutant HNSCC.
Introduction
The three human RAS genes (HRAS, KRAS, and NRAS) comprise the most frequently mutated oncogene family in human cancer and are key targets for anticancer therapeutics (1). One approach to blocking RAS function is inhibition of membrane association. RAS proteins become activated and engage their downstream effectors at the inner leaflet of the plasma membrane. To achieve this required localization, they undergo a series of post-translational modifications. The addition of a C15 farnesyl isoprenoid lipid to the C-terminal membrane targeting motif of RAS is the first and essential step in this process (2), catalyzed by the enzyme farnesyl transferase (FTase).
Two potent and selective FTase inhibitors (FTIs) that block transfer of farnesyl isoprenoids to RAS, tipifarnib (3) and lonafarnib (4), were advanced to Phase III clinical evaluation for cancer treatment and found to be surprisingly nontoxic to normal tissues (5). Unfortunately, these trials were focused on the cancers with the highest frequencies of RAS mutations: pancreatic, colorectal and non-small cell lung cancers, where the predominant mutated RAS isoform is KRAS. Preclinical studies had demonstrated the ability of KRAS and NRAS, but not HRAS, to undergo alternative prenylation and thereby escape the effects of inhibition by FTIs (6). The dismal outcomes of these clinical trials greatly diminished interest in targeting RAS membrane association as an anti-RAS therapeutic strategy (5). The ability of FTIs to effectively inhibit HRAS membrane association suggested that HRAS-driven cancers would be susceptible to FTI treatment. However, the low frequency of HRAS mutations overall in cancer (3%) has long redirected overall anti-RAS drug discovery efforts to focus on KRAS.
The arrival of the era of precision medicine, where actionable targets can be identified even from low occurrence mutations, has altered this mindset. With FTIs already established as effective HRAS inhibitors, and already shown to be well tolerated, cancers harboring HRAS mutations have become of interest for clinical evaluation of FTIs. One such cancer is head and neck squamous cell carcinoma (HNSCC) (7), a common cancer worldwide (8, 9) that affects critical functional structures such as the tongue, pharynx and other parts of the oral cavity. Because of its anatomical locations and the consequences of existing treatments, HNSCC causes significant mortality and morbidity. Conventional treatment options include surgery, radiation and platinum chemotherapy, with more recent utilization of EGFR inhibitors and immunotherapy including PD-1 and PD-L1 inhibitors (10, 11). Yet these treatments often negatively impact ongoing quality of life, even for cured patients, by significantly compromising function and aesthetics. HNSCC patients whose tumors are HPV negative, including those harboring HRAS mutations (5% of HNSCC) (7)), have a worse prognosis than those with HPV positive cancers. More effective treatments with lower toxicities are desperately needed. A recent return to focusing on inhibiting RAS membrane localization in these tumors prompted initiation of the present study.
HRAS is the most frequently mutated RAS isoform in HNSCC (12). Although it was unknown whether mutant HRAS activity is required for tumor maintenance in this tumor type, the observation that resistance to the EGFR inhibitor cetuximab administered to unselected HNSCC patients was frequently accompanied by the appearance of oncogenic mutations in RAS proteins (13) contributed to the perception that HRAS mutant HNSCC would be a candidate for treatment with FTIs. Evaluating this possibility, a small phase I trial of the clinical candidate FTI tipifarnib was initiated in HRAS mutant HNSCC patients. Preliminary reports indicated partial responses in a subset (55%) of 18 patients (14). However, since there are over 50 other FTase substrates, whether the clinical response to FTI treatment can be attributed to HRAS inhibition remained to be resolved. In this study, we silenced HRAS expression and established that it is indeed required for the growth and survival of HRAS mutant HNSCC cell lines. We also demonstrated that treatment with the FTI tipifarnib at doses that blocked HRAS farnesylation and membrane association partially phenocopied genetic loss of HRAS. Using both candidate approaches and unbiased reverse phase protein array (RPPA) pathway activation mapping, we demonstrated that both HRAS knockdown and FTI treatment could result in compensatory increases in ERK MAPK activity and cause reprogramming of pro-survival signaling pathways. We subsequently applied a CRISPR-Cas9 genetic screen to identify clinically relevant targets of drugs that could enhance sensitivity to tipifarnib. Among the top hits were components of both key RAS effector pathways, the RAF-MEK-ERK MAPK cascade and the PI3K-AKT-mTOR pathway. We found that, although HRAS mutant HNSCC lines were surprisingly insensitive to ERK1/2 inhibition alone, addition of either ERK1/2 or PI3K inhibitors enhanced sensitivity to tipifarnib. Finally, we observed that tipifarnib can upregulate markers of epithelial-to-mesenchymal transition (EMT), which can contribute to therapeutic resistance. That FTI-induced upregulation of EMT could be blocked by ERK inhibitor treatment suggested an additional basis for the efficacy of this combination. Our results indicate that these combinations warrant further investigation for the treatment of HRAS mutant HNSCC.
MATERIALS AND METHODS
Further information can be found in the Supplementary Materials.
HNSCC cell lines and culture:
HN30 (HRAS G12D, RRID:CVCL_5525) was a kind gift from Dr. Silvio Gutkind (University of California at San Diego). KYSE30 (HRAS Q61L, RRID:CVCL_1351) was acquired from the Tissue Culture Facility at the Lineberger Comprehensive Cancer Center, UNC-CH, Chapel Hill. UMSCC4 (HRAS G12V, RRID:CVCL_7751), UMSCC11a (HRAS WT, RRID:CVCL_7715), UMSCC43 (HRAS G12V, RRID:CVCL_7755), and UMSCC63 (HRAS G12D, RRID:CVCL_L130) were acquired from the University of Michigan, Ann Arbor, through a Materials Transfer Agreement. Cells were cultured in DMEM and/or Ham's F12 and penicillin/streptomycin under standard conditions at 37°C. Cell lines were STR-authenticated, regularly monitored for Mycoplasma infection using the MycoAlert Mycoplasma detection kit (Lonza), and used no longer than 3 months after thawing.
Lentivirus shRNA:
shRNA constructs from The RNAi Consortium (TRC) targeting HRAS [TRC 40088, 40091, 10358] were cloned into pLKO.1 (RRID:Addgene_8453). Lentivirus particles were produced in HEK293T cells (RRID:CVCL_0063) using packaging plasmids DNA, pMD2.G (RRID:Addgene_12259) and psPAX2 (RRID:Addgene_12260) from Addgene (15). HNSCC cells were transduced with 0.5 ml of virus-containing supernatant in the presence of polybrene (8 μg/μl) and incubated overnight prior to selection in puromycin (5 μg/μl). Knockdown efficiency was confirmed by western blot.
Proliferation assays:
Proliferation assays were performed as previously described and quantitated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) or alamarBlue (15).
Apoptosis and cell cycle assays:
Cells were subjected to five days of inhibitor treatment or HRAS knockdown. To detect apoptosis, treated cells were analyzed with the Annexin V-FITC staining kit (R&D Systems). For cell cycle analysis, cells were fixed in 70% ethanol overnight and stained with propidium iodide (4 μg/ml) containing RNAse A (100 μg/ml). Flow cytometry was performed using a CytoFLEX S (Beckman Coulter).
Immunoblotting:
Cell lysates were prepared in Laemmli buffer, and equal amounts of protein were subjected to SDS-PAGE, transferred to methanol-activated PVDF membranes, and probed with primary antibodies against the specific targets (largely 1:1,000 dilution), followed by appropriate secondary antibodies (anti-rabbit (NA934V) and anti-mouse (NA931V, RRID:AB_772210) (GE Healthcare) at 1:10,000 dilution). Proteins were detected by chemiluminescence (Amersham ECL Western Blotting Detection Kit, GE Life Sciences) and blots developed using the ChemiDoc MP Imaging System (BioRad, RRID:SCR_019037).
Reverse Phase Protein Array (RPPA) pathway activation mapping:
Samples for RPPA analysis were prepared in 4 biological replicates (16). Briefly, cells were treated with inhibitor or DMSO vehicle control for the indicated times and lysed in T-PER tissue protein extraction reagent (Thermo Fisher). Protein quantity was normalized, lysates were suspended in 2X SDS sample buffer (Bio-Rad) supplemented with β-mercaptoethanol and boiled for 5 minutes, and stored at −80°C until arrayed. Lysates were printed on nitrocellulose coated glass slides in 4 technical replicates and arrays were probed for target proteins. Biotin-labelled secondary antibodies, tyramine-based amplification system, and IRDye 680RD (LI-COR Biosciences) streptavidin fluorescence dye were used to amplify and detect the signal. Slides were scanned and signal intensity calculated using commercially available software (MicroVigene v5.1.0.0, VigeneTech, Inc, RRID:SCR_002820). Technical replicates (4 per sample) were averaged, and the median taken across 4 experimental replicates. Results are reported as the fold-change of up- or down-regulation of drug-treated vs. vehicle-treated samples. Clustered heatmaps were generated using the pheatmap library in R 3.6.2 (RRID:SCR_016418).
CRISPR/Cas9 loss-of-function genetic screens:
A barcoded pooled CRISPR-Cas9 loss-of-function library consisting of 11950 gRNA against 2390 genes has been previously described (17). The pooled plasmid DNA was stock-amplified in Lucigen 10G Elite electrocompetent bacteria and cultured on LB agar plates to achieve a million colonies. After isolation, DNA was packaged in lentiviral particles generated using HEK293T cells. Titered viral particles were delivered to HN30 cells at a low multiplicity of infection (0.3). After puromycin selection, cells were treated with inhibitor or DMSO vehicle control for 2 or 4 weeks. Drug was refreshed every 3-4 days and cells were passaged upon reaching 80% confluency. Cells were maintained at 1000x sequence coverage to prevent artificial selection pressure. Genomic DNA was extracted using Qiagen DNeasy Blood and Tissue kit. Samples were prepared for sequencing by PCR amplification as previously described (18). Sequencing was performed on an Illumina NextSeq 500 with 75bp, single-end reads at a final concentration of 3 pmol DNA. DNA was loaded with a PhiX spike of greater than 20% to enhance signal complexity. The counts of each gRNA were analyzed to identify targets that sensitized or caused resistance to treatment.
Triton X-114 phase separation:
Cells were treated with inhibitor for 24 h, then subjected to phase separation by lysing in pre-condensed Triton X-114 solution (19). Lysates were separated twice into detergent and aqueous phases by incubating at 37°C, then prepared for western blot by addition of 4X SDS buffer and boiling. Immunoblotting was performed to detect endogenous HRAS and the vinculin loading control.
Statistical analysis:
BLISS synergy scores were calculated as previously described (20). All quantitative assays were done at least three times independently and data are represented as the mean +/− SD. A two tailed t-test was used to calculate p-values; p<0.05 was considered significant.
Data Availability Statement:
The data generated in this study are available within the article and its supplementary data files.
Results
HRAS mutant HNSCC cell lines are dependent on HRAS for their proliferation and survival
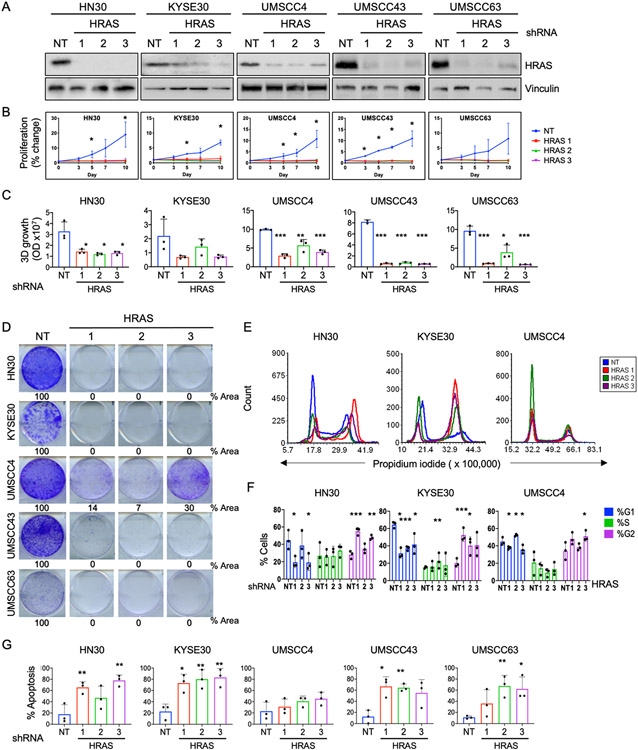
To study the dependence of HNSCC cell lines on mutant HRAS, we used three different lentiviral shRNAs to knock down HRAS expression in a panel of five HRAS mutant HNSCC cell lines. Knockdown was confirmed by immunoblotting (Fig 1A). Anchorage-dependent cell growth over a period of 10 days was evaluated by MTT assay (Fig 1B). HRAS knockdown nearly ablated cell proliferation of all HRAS mutant lines tested. We next evaluated anchorage-independent growth over two weeks. HRAS depletion impaired proliferation in 3D Matrigel of HNSCC cell lines harboring mutant HRAS but not a cell line harboring wildtype (WT) HRAS (Fig 1C and Supplementary Fig S1A, B). HRAS depletion also dramatically impaired 2D clonogenic growth of HRAS mutant cells (Fig 1D).
Figure 1: HRAS mutant HNSCC cell lines are dependent on HRAS for their growth and survival.
A) Western blots showing knockdown of HRAS protein. HRAS mutant HNSCC cell lines were transduced with anti-HRAS or nontargeting (NT) control shRNA and selected in puromycin for 72 h.
B) Anchorage-dependent 2D proliferation assay following HRAS knockdown. Cells were seeded in triplicate in 12-well plates and viability evaluated by MTT after 10 days. Data represent average +/− SD of three biological replicates. P values are averages of the individual P value of each shHRAS compared to shNT for each time point. *P<0.05
C) Quantification of anchorage-independent 3D colonies formed after HRAS knockdown. Cells were grown in Matrigel for 2 weeks, then stained with alamarBlue. P values as in (B). *P<0.05, **P<0.01, ***P<0.001
D) Representative 2D clonogenic assays showing colony formation after HRAS knockdown. Cells were plated in 6-well plates after lentiviral shRNA transduction and puromycin selection. After 10 days, colonies were stained with crystal violet.
E) Histograms showing cell cycle distribution after HRAS knockdown. After 72 h of shRNA transduction and selection, cells were stained with propidium iodide and subjected to flow cytometry.
F) Quantification of data shown in (E). Data represent average +/− SD of three biological replicates. P values are averages of the individual P value of each shHRAS compared to shNT. *P<0.05, **P<0.01, ***P<0.001
G) Quantification of apoptosis after HRAS knockdown. After 5 days, cells were stained with Annexin V-FITC and propidium iodide and quantified by flow cytometry. Data represent average +/− SD of three biological replicates. P values as in (F).
Next we wanted to determine whether HRAS depletion inhibits growth by causing growth arrest or cell death. We performed cell cycle analysis using flow cytometry. Depletion of HRAS resulted in variable alterations in cell cycle distribution (Fig 1E-F and Supplementary Fig S1C). We then determined, by Annexin-FITC staining, that the HRAS mutant cells were highly susceptible to induction of apoptosis upon HRAS knockdown (Fig 1G and Supplementary Fig S1D), whereas the HRAS WT cell line was not (Supplementary Fig S1D-E). Together, these results indicate that mutant HRAS is important for the proliferation and survival of HRAS mutant HNSCC cells, and, as anticipated, supports mutant HRAS as a therapeutic target in HNSCC.
Treatment of HRAS mutant HNSCC with the FTI tipifarnib inhibits cell growth and induces apoptosis
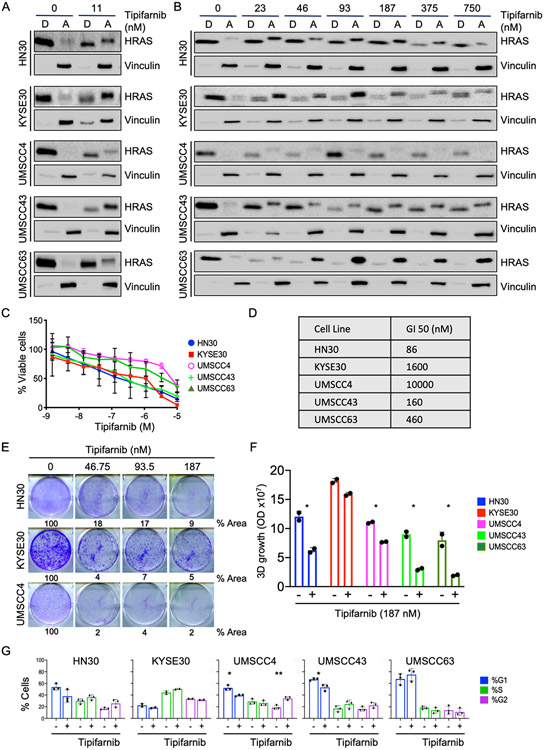
To determine the ability of the FTI tipifarnib to inhibit HRAS farnesylation in HRAS mutant HNSCC, we used Triton-X 114 phase separation to evaluate HRAS partitioning into detergent (farnesylated) versus aqueous (nonfarnesylated) phases. All five HRAS mutant cell lines were sensitive to low nanomolar FTI, displaying both redistribution into the aqueous phase and slower electrophoretic mobility, indicating lack of prenylation, after as little as 24 h of tipifarnib treatment (Fig 2A). Farnesylation is a permanent modification and HRAS has a half-life of 20-24 h (21). Thus, the differing degrees of distribution of endogenous HRAS into the aqueous phase upon FTI treatment likely indicates differential regulation of HRAS transcription and/or translation in each cell line, as the upper band represents only proteins newly synthesized in the presence of FTI. We also observed a variable effect on HRAS protein abundance at higher concentrations of tipifarnib (Fig 2B).
Figure 2: FTI treatment phenocopies HRAS knockdown and inhibits growth of HRAS mutant HNSCC.
A) Western blot showing distribution of HRAS into detergent ("D", membrane) and aqueous ("A", cytosol) phases. Indicated HRAS mutant cells were treated with FTI tipifarnib or DMSO vehicle control for 24 hr and subjected to Triton X-114 phase separation. Nonfarnesylated proteins migrate more slowly (upper bands) than farnesylated proteins (lower bands).
B) Western blot showing variable changes in HRAS abundance upon tipifarnib treatment. Cells treated as in panel (A).
C) AlamarBlue assay quantifying cell viability after tipifarnib treatment for 5 days. Data are average +/− SD of three biological replicates.
D) GI50 of tipifarnib in HRAS mutant HNSCC cell lines in panel (C).
E) Representative 2D clonogenic assay showing quantified growth inhibition (% plate area covered by cells) after treatment with tipifarnib for 10 days.
F) Quantification of 3D colonies formed in Matrigel after treatment with tipifarnib (187 nM). Data are average of two technical replicates. P value: treatment versus DMSO control. *P<0.05
G) Quantification of cell cycle distribution after treatment with tipifarnib. Data are average +/− SD of three biological replicates. P value: treatment versus DMSO control. *P<0.05, **P<0.01
We next evaluated the effect of tipifarnib on cell growth. Cells were treated for 5 days and viability was measured. Like genetic depletion of HRAS, tipifarnib treatment also reduced cell viability, in a dose-dependent manner, albeit with differing sensitivities among the cell lines (Fig 2C and 2D). These lines were also sensitive to tipifarnib-mediated inhibition of 2D clonogenic growth (Fig 2E) and anchorage-independent growth in Matrigel (Fig 2F). Overall, their sensitivity profile in response to tipifarnib was very similar to their response to HRAS depletion. Notably, KYSE30 was the least sensitive to tipifarnib in 3D Matrigel. This cell line has a copy number amplification in EGFR (22) which could explain the lack of sensitivity to FTI treatment in the growth factor-rich environment. As with genetic depletion of HRAS, there was little consistent change in cell cycle distribution upon 72 h of FTI (Fig 2G). Thus, FTI treatment phenocopies HRAS genetic depletion with respect to growth inhibition in HRAS mutant HNSCC cells.
HRAS genetic depletion can cause upregulation of MAPK and AKT signaling pathways
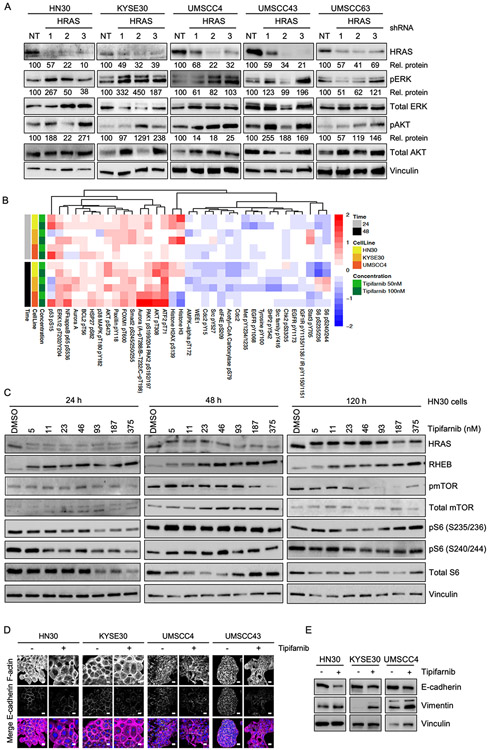
The RAF-MEK-ERK MAP kinase cascade and the PI3K-AKT-mTOR pathways are critical downstream effectors of RAS signaling (1), yet KRAS knockdown in a panel of KRAS mutant pancreatic ductal adenocarcinoma (PDAC) cell lines did not result in consistent loss of either pERK or pAKT (23). We observed that stable depletion of HRAS by lentiviral shRNA resulted in increased pERK and pAKT in a subset of HRAS mutant HNSCC lines (Fig 3A).
Figure 3: HRAS depletion and tipifarnib treatment can cause compensatory upregulation of ERK MAPK and AKT signaling pathways; induction of morphological changes.
A) Western blot showing increased levels of pERK1/2 and pAKT in some cell lines after HRAS knockdown. Cells were transduced with 3 different anti-HRAS shRNAs or NT control and selected in puromycin for 72 h prior to blotting for phosphorylated and total ERK and AKT. Densitometry was used to quantitate ratios of phosphorylated to total proteins ("Rel. proteins"), normalized to NT control.
B) Heatmap showing the top 35 differentially regulated proteins or phosphoproteins in the indicated cells treated with tipifarnib or DMSO vehicle for 24 or 48 h. Cell lysates were subjected to RPPA analysis. Red: increase; blue: decrease.
C) Western blots of HN30 cells showing signaling downstream of HRAS and RHEB after 24, 48 or 120 h of treatment with tipifarnib at the indicated concentrations.
D) Immunofluorescence images showing expression and distribution of F-actin and E-cadherin in cells treated with tipifarnib (200 nM) or DMSO vehicle. In the merge image, F-actin is shown in magenta, E-cadherin in green and the nucleus in blue. Scale bar = 20 μm.
E) Western blots showing expression of E-cadherin and vimentin after treatment as in (D).
Tipifarnib treatment partially phenocopies HRAS genetic depletion, causing compensatory increases in both ERK MAPK and AKT signaling
During the original development of FTIs more than 20 years ago, many studies were performed to assess their effects on cellular phenotypes such as morphology, proliferation, survival, cell cycle progression and tumorigenicity. With the advent of more modern technologies to address cell signaling in a higher throughput manner, it has now become possible to more thoroughly examine the signaling changes that occur when farnesylation is blocked. In order to understand the signaling changes conferred by tipifarnib treatment in HRAS mutant HNSCC cells, we performed unbiased RPPA pathway activation mapping (16). We treated three cell lines (HN30, KYS30 and UMSCC4) with FTI for 24 or 48 h and prepared lysates for analysis of ~200 phosphorylated and total proteins. RPPA pathway activation mapping revealed an increase in pERK1/2 and pAKT at both time points following FTI treatment (Fig 3B). Tipifarnib-mediated compensatory increases in pERK have also been observed in HRAS mutant thyroid cancer cells (24). Additionally, several other RAS signaling-associated kinases and transcription factors were upregulated, including PAK1, p38 (MAPK14), NF-kB and ATF2. Conversely, we observed decreases in several receptor tyrosine kinases such as EGFR, c-Kit and MET, and targets of the mTOR pathway such as p70 ribosomal protein S6 kinase. We noted a modest increase of inhibitory phosphorylation of the anti-apoptotic protein BCL2, suggesting an early attempt by the cell to counter the stress of shutting down multiple pathways by inducing apoptosis.
Tipifarnib treatment inhibits RHEB farnesylation but not mTOR signaling
The spectrum of biological activities of farnesylated proteins (25-27) has greatly complicated our ability to understand the mechanistic consequences of inhibiting farnesyltransferase. Other farnesylated proteins include RAS-related small GTPases such as RHEB, a key regulator of mTOR signaling. RHEB must be farnesylated in order to promote S6 kinase activation through mTOR, and has been suggested to promote RAS-independent consequences of FTIs (28, 29). Therefore, we investigated the effect of tipifarnib treatment on RHEB farnesylation and mTOR signaling in HNSCC. Tipifarnib induced complete loss of farnesylation as indicated by gel shift (Fig 3C), accompanied by a possible attempt at compensation by upregulating RHEB expression (Fig 3C). However, despite the robust inhibition of RHEB processing at 24 h at the lowest dose of FTI, we did not observe a direct effect on activity of mTOR as measured by its own phosphorylation status. Phosphorylation of mTOR did decrease upon long term treatment at higher concentrations. We also observed a variable decrease in the activity of mTOR substrates S6K and S6 at 24 and 48 h (Fig 3B and 3C); however, this was restored over long term treatment, perhaps due to compensatory activation of ERK and/or AKT. Thus, tipifarnib-mediated loss of RHEB farnesylation is insufficient to prevent activation of mTOR in HRAS mutant HNSCC.
Tipifarnib induces morphological changes and epithelial-to-mesenchymal transition
Some of the earliest recognized effects of FTIs included alterations in actin-mediated cell morphology (30-32). Such alterations can reflect changes in epithelial-to-mesenchymal-transition (EMT) status. However, this had not been investigated in HNSCC. We therefore applied confocal immunofluorescence microscopy to investigate cell morphology and the F-actin cytoskeleton in the HRAS mutant HNSCC cell lines. We observed remarkable heterogeneity in cell morphology and F-actin structures, in both treated and untreated conditions (Fig 3D). For instance, whereas KYSE30 cells are round and have significant cortical actin, UMSCC4 cells are smaller and have a fibroblast-like morphology with F-actin stress fibers. In line with these observations, we also observed heterogeneity in basal EMT status based on immunoblotting for the epithelial marker E-cadherin and the mesenchymal marker vimentin (Supplementary Figs S2A, 2B). KYSE30 cells are characterized by high E-cadherin levels and hardly any vimentin, indicative of a more epithelial-like basal state, in line with the observed cell morphology. In contrast, UMSCC4 cells have significantly higher vimentin than E-cadherin levels, indicating a more mesenchymal-like basal state, also in line with their morphology. Together, both the imaging and the western blot studies indicate that all the HNSCC lines are in a partial EMT state under basal conditions. Imaging studies showed that tipifarnib increased cell size and decreased E-cadherin expression (Fig 3D), which was confirmed by western blot (Fig 3E). Conversely, the mesenchymal marker vimentin was increased upon tipifarnib treatment (Fig 3E), in contrast to early findings in RAS-transformed fibroblast model systems, which reverted to a more flattened epithelial-like morphology upon treatment with FTIs (30, 31). These data indicate that tipifarnib treatment can induce EMT in HRAS mutant HNSCC, regardless of their relative sensitivity to tipifarnib-mediated growth suppression. EMT is associated with resistance and poor treatment outcome in cancers treated with conventional cytotoxic therapies (33, 34). We hypothesize that induction of EMT could similarly cause emergence of resistance in tipifarnib treated HNSCC tumors.
CRISPR/Cas9 loss-of-function genetic screen identifies both expected and novel targets for sensitization of HRAS mutant HNSCC to tipifarnib
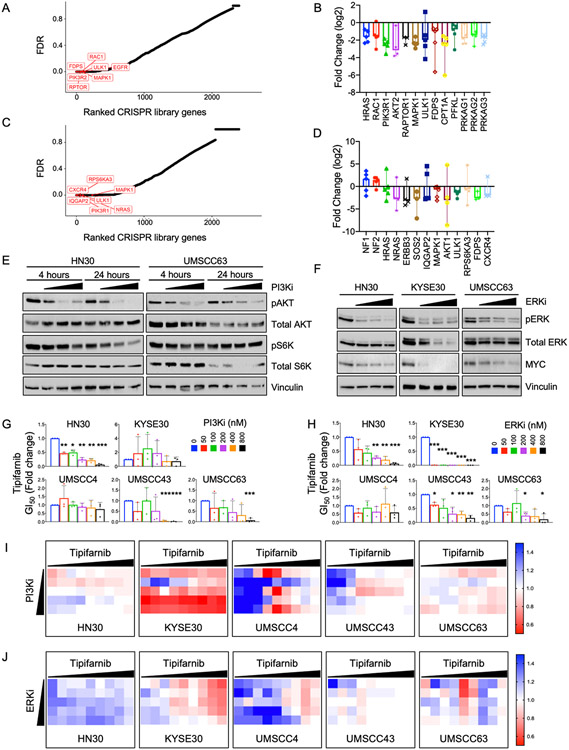
Acquired treatment resistance is a challenge for all targeted cancer therapies. Forecasting the potential mechanisms of resistance and improving sensitization to existing therapies is crucial for successful cancer treatment. We used lentiviral transduction to deliver a barcoded CRISPR/Cas9 druggable genome library, targeting ~2500 genes with 5 guide RNAs each (17), to tipifarnib-treated HRAS mutant HNSCC HN30 cells. Following puromycin selection, cells were treated for two or four weeks at 1000x library coverage with tipifarnib at the GI25, a dose that caused some inhibition of FTase without substantially inhibiting cell growth. Surviving cells were pooled and sequenced to identify enriched or depleted sgRNAs (see counts in Supplementary Tables 1 and 2). The sgRNA counts were entered in HitSelect, a published algorithm (35), to calculate rank and effect size. We compared the top 10% of genes from each time point, using stringent criteria of at least 3 active sgRNAs of the 5 total for each gene. At the two-week time point, the top hit (Fig 4A) was FDPS, encoding farnesyl diphosphate synthase, which catalyzes the production of farnesyl isoprenoids. Loss of FDPS enhanced cell sensitivity to the FTI tipifarnib, consistent with impaired synthesis of the isoprenoids required for farnesylation. Other top hits included regulators of PI3K-AKT-mTOR signaling (RAC1, PIK3R1/2, AKT2, RPTOR) and regulators of metabolism (RPTOR, ULK1, PFKL, CPT1A) (Fig 4A, 4B). IQGAP2, which interacts with RAC1 and RHO GTPases (36) and inhibits EMT (37) was the top hit at the 4-week time point, consistent with the possibility that tipifarnib induction of EMT may limit its efficacy in HRAS mutant HNSCC. Other top hits included positive regulators of RTK-RAS signaling (ERBB3, SOS2, HRAS and NRAS) and protein translation (RPS6KA3) (Fig 4C and 4D). Conversely to the depletion of the above genes, we also observed enrichment of the RASGAP NF1 and tumor suppressor NF2, encoding MERLIN (Fig 4D). These inhibit RAS signaling and their loss has been linked to resistance to tipifarnib in HRAS mutant thyroid cancer models (24).
Figure 4. CRISPR-Cas9 loss-of-function (LOF) screen identifies known and novel targets to sensitize cells to tipifarnib treatment.
A) False discovery rate (FDR) for each gene in the CRISPR library at the 2 week time point. Top hits are highlighted in red.
B) Fold change of gRNA against indicated genes in the library after 2 weeks of treatment. Data represented as median with range.
C) False discovery rate (FDR) for each gene in the CRISPR library at the 4 week time point. Top hits are highlighted in red.
D) Fold change for gRNA against indicated genes in the library after 4 weeks of treatment. Data represented as median with range.
E) Western blot showing target inhibition of PI3K signaling pathway by PI3K-alpha inhibitor alpelisib. Cells were treated for 4 or 24 h at 100, 200 and 400 nM.
F) Western blot showing target inhibition of ERK MAPK pathway by ERK1/2 inhibitor SCH772984. Cells were treated for 48 h at 100, 200 and 400 nM.
G) Fold change in tipifarnib GI50 when combined with indicated concentrations of PI3K-alpha inhibitor alpelisib. P values: combination versus tipifarnib alone. *P<0.05, **P<0.01, ***P<0.001
H) Fold change in tipifarnib GI50 when combined with indicated concentrations of ERK1/2 inhibitor SCH772984. P values: combination versus tipifarnib alone. *P<0.05, **P<0.01, ***P<0.001
I) Bliss scores quantifying synergy between tipifarnib (1.5 nM - 10 μM) and PI3K-alpha inhibitor (50 - 800 nM).
J) Bliss scores quantifying synergy between tipifarnib (1.5 nM - 10 μM) and ERK1/2 inhibitor (50 - 800 nM).
At the 4-week time point, another top hit was CXCR4, a GPCR receptor for the CXCL12 ligand, also known as SDF-1, that drives metastasis homing. CXCL12/CXCR4 signaling exerts its effects through a plethora of RAS- and RHO-mediated pathways (38). Overexpression of either component has been identified as a potential biomarker for tipifarnib activity in blood cancers (39). The serine/threonine kinase ULK1, which initiates autophagy, was a top sensitizer to tipifarnib in the CRISPR screens at both time points. We also performed KEGG pathway enrichment analysis for the top 10% of hits from each dataset (Supplementary Fig S3A). Prominent among these were genes regulating metabolic pathways, indicating that metabolic reprogramming is an important component of the response to tipifarnib. We and others have demonstrated that MAPK inhibition sensitizes KRAS mutant PDAC to autophagy inhibition (40, 41) and here we observed that tipifarnib can upregulate compensatory MAPK signaling in HRAS mutant HNSCC. To begin to validate these interesting hits, we performed preliminary experiments by treating HRAS mutant HNSCC cells with a combination of tipifarnib and either the autophagy inhibitor chloroquine or the CXCR4 inhibitor WZ811. In a subset of cell lines these combinations were synergistic, as indicated by Bliss scores less than one (Supplementary Figs S3B, 3C). Interestingly, T cell receptor signaling was also prominent, supporting the idea that FTIs could be combined with immunotherapy for improved clinical benefit in HRAS mutant HNSCC. These screens identify potential combinations that warrant further investigation in future studies.
Combination of the FTI tipifarnib with ERKi, PI3Ki or mTORi causes apoptosis in HRAS mutant HNSCC
Both RPPA (Fig 3B) and KEGG pathway analysis (Supplementary Fig S3A) also confirmed that many hits in our screen regulate the ERK MAPK and PI3K-AKT-mTOR pathways. We first selected the ERK1/2-selective inhibitor SCH772984 and the PI3K alpha-selective inhibitor alpelisib for further investigation. Cells were treated for 5 days with the combinations of tipifarnib with a dose response of each inhibitor at doses that partially or completely inhibited their respective targets (Fig 4E and 4F). Cell viability was quantified by alamarBlue, and the GI50 (Fig 4G and 4H) and Bliss synergy scores were calculated (Fig 4I and 4J). Both inhibitors sensitized cells to tipifarnib and enhanced the growth inhibition induced by either single agent alone (Fig 4G and 4H, Supplementary Fig S3D), with GI50s that were many fold lower than the doses required to inhibit signaling.
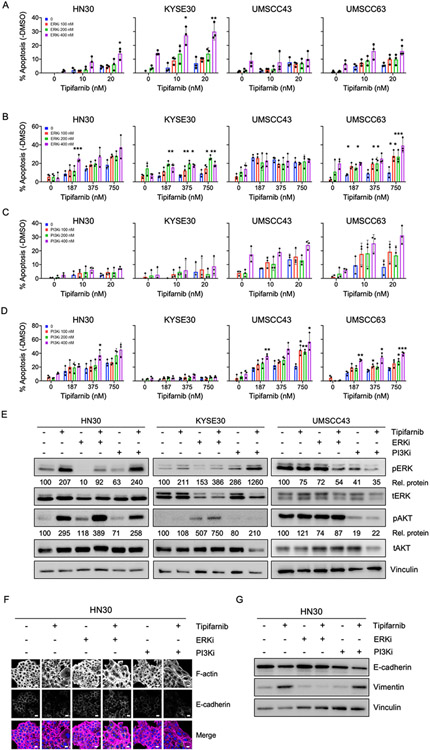
To determine whether these combinations were simply cytostatic or could also induce cell death, we treated cells with tipifarnib in combination with a range of concentrations of PI3K-alpha inhibitor or ERK inhibitor that caused complete to partial inhibition of their respective targets (Figs 4E-F). Treatment with single agents caused a modest amount of apoptosis which trended higher upon combination treatment (Fig 5A-5D). Some lines were sensitive to both combinations, while others were sensitive to only one. For example, HN30 and UMSCC63 were sensitive to FTI + ERKi and FTI + PI3Ki. KYSE30 was sensitive to FTI + ERKi while UMSCC43 was sensitive only to FTI + PI3Ki, and only at higher doses. Sensitivity to the combinations generally correlated to sensitivity to the individual agents. Bliss scores indicated synergy to both or either of the inhibitors (Supplementary Fig S4A-D).
Figure 5: Combination of FTI tipifarnib with ERKi or PI3Ki enhances apoptosis and induces EMT in HRAS mutant HNSCC.
A-D) Quantification of apoptosis in cells treated with low (10-20 nM, panels A,C) or high doses (187-750 nM, panels B,D) of tipifarnib and 100-400 nM of ERKi (SCH772984) or PI3Ki (alpelisib) for five days. Annexin-FITC positive cells were quantified and negative control DMSO values were subtracted. Data represent average +/− SD of three independent replicates. P value: combination versus tipifarnib alone. *P<0.05, **P<0.01, ***P<0.001
E) Western blots showing target inhibition of ERK1/2 and AKT. Cells were treated for 24 h with tipifarnib (200 nM) and ERKi (400 nM) or PI3Ki (400 nM) in cell lines displaying differential sensitivity profiles to the combinations. Densitometry was used to quantitate ratios of phosphorylated to total proteins ("Rel. proteins"), normalized to DMSO control.
F) Immunofluorescence images showing expression and distribution of F-actin and E-cadherin in response to the combinations or drugs alone, treated as in (E). In the merge image, F-actin is shown in magenta, E-cadherin in green and the nucleus in blue. Scale bar = 20 μm.
G) Western blots showing expression of E-cadherin and vimentin in response to the combinations or drugs alone, treated as in (E).
In agreement with the genetic sensitizer screen, RPPA analysis also showed that FTI treatment causes upregulation of MAPK/ERK and PI3K/AKT pathway signatures (Supplementary Fig S4E and 4F). We sought to analyze the signaling changes in these pathways upon combination treatments in three different cell lines with differential sensitivity to these inhibitors. We observed a compensatory increase in phosphorylation of AKT and ERK1/2 in cell lines insensitive to the combinations of FTI/PI3Ki (KYSE30) and FTI/ERKi (UMSCC43), respectively (Fig 5E), possibly explaining their insensitivity to these particular combinations.
To evaluate whether combining tipifarnib with ERKi or PI3Ki would rescue the EMT induced by tipifarnib alone, we analyzed the expression of E-cadherin and vimentin by immunofluorescence imaging and western blotting. Tipifarnib alone, PI3K inhibitor alone or the combination decreased E-cadherin and increased vimentin, indicating an EMT (Fig 5F and 5G, and Supplementary Figs S5A-D). In contrast, ERK inhibitor alone or the combination maintained E-cadherin levels and prevented the increase in vimentin expression induced by tipifarnib alone. This indicates the reverse of EMT (i.e., MET). These results show that the combination of FTI with ERK inhibition may be superior to single agent and could prevent or ameliorate emergence of resistance due to EMT. To extend this result, in addition to the ERK1/2-selective inhibitor SCH772984, we also determined that tipifarnib synergistically enhanced apoptosis when combined with BVD-523/ulixertinib, a clinical candidate ERK inhibitor (Supplementary Figs S6A-C).
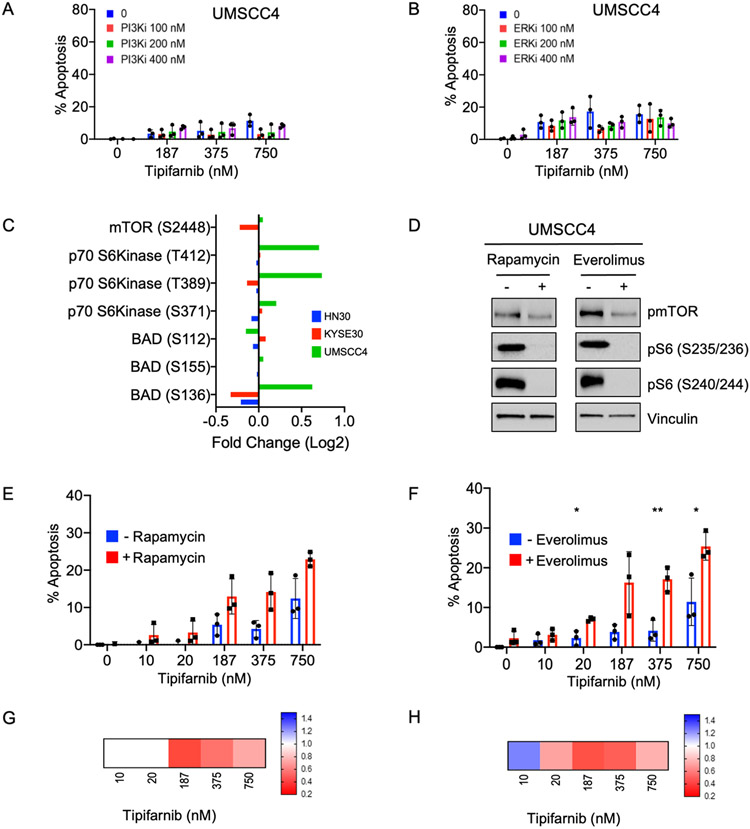
Finally, we sought to identify a combination that could overcome the resistance of UMSCC4 to tipifarnib with or without PI3Ki or ERKi (Figs 6A and 6B). RPPA analysis showed that, uniquely in UMSCC4 cells, FTI increased activation of p70 S6K and phosphorylation of its substrate BAD at the inhibitory site S136 (Fig 6C), which decreases pro-apoptotic BAD activity and promotes cell survival. As p70 S6K is a direct substrate of mTOR, we speculated that blocking S6 activation using the mTOR inhibitor rapamycin could overcome this survival-promoting consequence of FTI treatment in UMSCC4 cells. Indeed, we found that rapamycin or another clinical analog, everolimus, effectively shut down the activity of mTOR and p70 S6K (Fig 6D), and the combination of tipifarnib with either agent increased apoptosis compared to single agent alone (Figs 6E, 6F). The synergy of these combinations was shown by Bliss scores of less than one (Figs 6G, 6H).
Figure 6: Inhibition of mTOR/S6 activity is critical to cause cell death in response to tipifarnib treatment of refractory cells.
A-B) Quantification of apoptosis in tipifarnib-refractory UMSCC4 cells. Cells were treated for 5 days with the indicated doses of tipifarnib and (A) the PI3Kalpha-selective inhibitor alpelisib or (B) the ERK1/2-selective inhibitor, SCH772984. Annexin-FITC positive cells were quantified and normalized to control. Data represent the average of three independent replicates; values above zero are shown.
C) Fold change in mTOR and mTOR-regulated signaling showing increases in activity only in tipifarnib-refractory cells. Indicated cell lines were treated with tipifarnib and subjected to RPPA analysis as in Fig 3B.
D) Western blot showing target inhibition of mTOR signaling. UMSCC4 cells were treated with mTORCi rapamycin (15 nM) or everolimus (25 nM) for 24 h.
E-F) Apoptosis assays showing increased efficacy of tipifarnib in refractory cells in the presence of mTORCi. UMSCC4 cells were treated with tipifarnib alone or in combination with rapamycin or everolimus. Data represent the average +− SD of three independent replicates. *P<0.05, **P<0.01, ***P<0.001
G-H) Bliss scores quantifying synergy between tipifarnib (10-750 nM) and mTOR inhibitors rapamycin (15 nM) and everolimus (25 nM).
Discussion
Based on promising results from an initial phase I clinical trial in HNSCC, the farnesyl transferase inhibitor tipifarnib is currently in directed phase II clinical trials for the treatment of recurrent or metastatic HRAS mutant HNSCC (NCT02383927, NCT03719690) and was recently given fast track designation by the FDA for review. Although HRAS mutation is currently the key selection criterion for trial enrollment, the role of mutant HRAS as an independent driver of HNSCC growth was unknown. Previous reports had focused on its possible role in the context of HNSCC resistance to EGFR inhibitors erlotinib and cetuximab (42, 43). Our direct demonstration that mutant HRAS protein is required for the growth and survival of HNSCC firmly credentials it as a therapeutic target in this disease. In agreement, a study published while this manuscript was in preparation showed that only HRAS mutant, but not HRAS WT, HNSCC cell lines and PDX models were susceptible to growth inhibition in vitro and in vivo by tipifarnib (44). However, although we have shown here that tipifarnib is fully capable of blocking HRAS farnesylation, membrane association, and signaling in HNSCC, it is not clear that inhibition of HRAS alone is sufficient to explain tipifarnib efficacy. Neither HRAS mutation nor dependence is fully predictive of tipifarnib response. This should not be surprising, considering that FTIs are not anti-RAS drugs, but rather are highly selective inhibitors of FTase, which has numerous farnesylated substrates (26, 27).
Our panel of HRAS mutant HNSCC cell lines represented the anatomic and histological diversity of the head and neck region. We observed remarkable heterogeneity in basal morphology as well as response to tipifarnib treatment. Through unbiased pathway activation mapping and genetic screens, we found that the signaling changes induced by tipifarnib included compensatory upregulation of MAPK and PI3K-AKT, the two most prominent RAS effector pathways. Upregulation of these pathways has recently also been identified in tipifarnib-treated thyroid cancer models (24) but not bladder carcinoma (45) or HNSCC (44). We suspect that differences in experimental conditions explain the latter. Our work indicates that combining tipifarnib with inhibitors of these pathways is a logical next step. Patients are typically treated with the maximum tolerated dose of targeted therapies. Our data suggests that partial inhibition of two targets simultaneously could be therapeutically advantageous and may help in reducing drug-associated toxicities.
Likewise, our genetic loss of function screen also indicated that inhibitors of the MAPK and PI3K-AKT pathways should sensitize these tumors to FTIs. We validated these combinations and established that sensitivity to a particular combination correlated with lack of compensation by the alternative pathway. Previous reports from RAS-transformed fibroblasts and thyroid cancers support greater efficacy of the combination of FTI with MEK or PI3K inhibitor as more effective than FTI alone (24, 46). We observed that tipifarnib induced an ERK-mediated signaling signature, which led us to co-target the terminal node of the MAPK signaling pathway. HRAS promotes EMT in an ERK-dependent manner (47). EMT in turn promotes chemoresistance (33, 34). Based on our finding that tipifarnib induces EMT in HRAS mutant HNSCC despite inhibiting HRAS itself, and that combining tipifarnib with ERK inhibition but not PI3K inhibition prevents this induction, we propose that future trials preferentially assess this combination specifically in HRAS mutant HNSCC.
It will also be interesting to further validate other hits from our genetic sensitizer screen. Some, such as CXCR4 or ULK1, already have inhibitors under development or in the pipeline for clinical trials and are potentially actionable; others, such as IQGAP2, are more useful to further explore mechanisms of FTI sensitivity and resistance. Additionally, it will be of great interest to further examine our preliminary data indicating that the mTOR inhibitors rapamycin and everolimus can overcome tipifarnib-driven inhibition of the pro-apoptotic protein BAD, thereby promoting apoptosis. Clinical trials have demonstrated that everolimus lacks activity as monotherapy or in combination with cisplatin and cetuximab in unselected HNSCC patients. However, a recent abstract presentation on a small phase II trial (NCT01111058) reported that HNSCC patients with TP53 mutations fared better on everolimus than those without such mutations (48). HRAS mutations and TP53 mutations are typically mutually exclusive in HNSCC (7). It will be interesting to determine whether the HRAS mutant cohort benefits from everolimus in combination with tipifarnib. In any case, recent encouraging reports showing durable responses in the small series of HRAS mutant HNSCC patients treated with tipifarnib (49, 50) suggest that any new findings relevant to enhancing tipifarnib sensitivity and overcoming resistance will be welcomed for further clinical improvement of this difficult disease.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) to ADC, CJD and EFP (P01CA203657, R01CA42978 and R35CA232113). SJ was supported by a fellowship from the Royster Society of Fellows. CMG and GAH were supported by fellowships from the NIH (F32CA221005; F32CA200313 and T32CA009156, respectively). AMW was supported by a fellowship from the American Cancer Society (PF-18-061). JND was supported by fellowships from the Slomo and Cindy Silvian Foundation and the NIH (F30CA243253 and T32 CA071341).
Abbreviations:
- EMT
epithelial-mesenchymal transition
- ERKi
ERK inhibitor
- FTI
farnesyltransferase inhibitor
- HNSCC
head and neck squamous cell carcinoma
Footnotes
Conflict of interest statement:
M. Pierobon and EF Petricoin III are inventors on US Government and University assigned patents and patent applications. As inventors, they are entitled to receive royalties as provided by US Law and George Mason University policy. They receive royalties from TheraLink Technologies and are consultants to and/or shareholders of Avant Diagnostics, Inc. EF Petricoin III is also consultant and shareholder of Perthera, Inc. CJ Der is a consultant/advisory board member for Mirati Therapeutics, Deciphera 950 Pharmaceuticals and Anchiano Therapeutics. CJ Der has received research funding support from Mirati Therapeutics and Deciphera Pharmaceuticals. CJ Der has consulted for Ribometrix, Jazz Therapeutics, SVB Leerink, Axon Advisors LLC, Third Bridge, Sanofi, SmartAnalyst, Turning Point Therapeutics and Eli Lilly. AD Cox has consulted for Eli Lilly and Mirati Therapeutics. The remaining authors declare no conflicts of interest.
References
- 1.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 2011;13:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.End DW, Smets G, Todd AV, Applegate TL, Fuery CJ, Angibaud P, et al. Characterization of the antitumor effects of the selective farnesyl protein transferase inhibitor R115777 in vivo and in vitro. Cancer Res 2001;61:131–7 [PubMed] [Google Scholar]
- 4.Liu M, Bryant MS, Chen J, Lee S, Yaremko B, Lipari P, et al. Antitumor activity of SCH 66336, an orally bioavailable tricyclic inhibitor of farnesyl protein transferase, in human tumor xenograft models and wap-ras transgenic mice. Cancer Res 1998;58:4947–56 [PubMed] [Google Scholar]
- 5.Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res 2015;21:1819–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 1997;272:14459–64 [DOI] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424 [DOI] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33 [DOI] [PubMed] [Google Scholar]
- 10.Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G Jr., et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915–28 [DOI] [PubMed] [Google Scholar]
- 11.Kao HF, Lou PJ. Immune checkpoint inhibitors for head and neck squamous cell carcinoma: Current landscape and future directions. Head Neck 2019;41 Suppl 1:4–18 [DOI] [PubMed] [Google Scholar]
- 12.Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res 2020;80:2969–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braig F, Voigtlaender M, Schieferdecker A, Busch CJ, Laban S, Grob T, et al. Liquid biopsy monitoring uncovers acquired RAS-mediated resistance to cetuximab in a substantial proportion of patients with head and neck squamous cell carcinoma. Oncotarget 2016;7:42988–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho A, Chau N, Bauman J, Bible K, Chintakuntlawar A, Cabanillas ME, et al. Preliminary results from a phase II trial of tipifarnib in squamous cell carcinomas (SCCs) with HRAS mutations. Ann Oncol 2018;29:146O [Google Scholar]
- 15.Zhou B, Ritt DA, Morrison DK, Der CJ, Cox AD. Protein Kinase CK2α Maintains Extracellular Signal-regulated Kinase (ERK) Activity in a CK2α Kinase-independent Manner to Promote Resistance to Inhibitors of RAF and MEK but Not ERK in BRAF Mutant Melanoma. J Biol Chem 2016;291:17804–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF 3rd, Pierobon M. Reverse Phase Protein Microarrays. Methods Mol Biol 2017;1606:149–69 [DOI] [PubMed] [Google Scholar]
- 17.Ozkan-Dagliyan I, Diehl JN, George SD, Schaefer A, Papke B, Klotz-Noack K, et al. Low-Dose Vertical Inhibition of the RAF-MEK-ERK Cascade Causes Apoptotic Death of KRAS Mutant Cancers. Cell Rep 2020;31:107764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taguchi Y, Schätzl HM. Small-scale Triton X-114 Extraction of Hydrophobic Proteins. Bio Protoc 2014;4:e1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 2015;3:e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulsh LS, Shih TY. Metabolic turnover of human c-rasH p21 protein of EJ bladder carcinoma and its normal cellular and viral homologs. Mol Cell Biol 1984;4:1647–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang Y, Zhang TT, et al. Consistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridization. Clin Cancer Res 2013;19:5867–78 [DOI] [PubMed] [Google Scholar]
- 23.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 2016;29:75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Untch BR, Dos Anjos V, Garcia-Rendueles MER, Knauf JA, Krishnamoorthy GP, Saqcena M, et al. Tipifarnib Inhibits HRAS-Driven Dedifferentiated Thyroid Cancers. Cancer Res 2018;78:4642–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer 2011;11:775–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol 2004;343:417–33 [DOI] [PubMed] [Google Scholar]
- 27.Zverina EA, Lamphear CL, Wright EN, Fierke CA. Recent advances in protein prenyltransferases: substrate identification, regulation, and disease interventions. Curr Opin Chem Biol 2012;16:544–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basso AD, Mirza A, Liu G, Long BJ, Bishop WR, Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem 2005;280:31101–8 [DOI] [PubMed] [Google Scholar]
- 29.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem 2003;278:32493–6 [DOI] [PubMed] [Google Scholar]
- 30.Cox AD, Garcia AM, Westwick JK, Kowalczyk JJ, Lewis MD, Brenner DA, et al. The CAAX peptidomimetic compound B581 specifically blocks farnesylated, but not geranylgeranylated or myristylated, oncogenic ras signaling and transformation. J Biol Chem 1994;269:19203–6 [PubMed] [Google Scholar]
- 31.Prendergast GC, Davide JP, deSolms SJ, Giuliani EA, Graham SL, Gibbs JB, et al. Farnesyltransferase inhibition causes morphological reversion of ras-transformed cells by a complex mechanism that involves regulation of the actin cytoskeleton. Mol Cell Biol 1994;14:4193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki N, Del Villar K, Tamanoi F. Farnesyltransferase inhibitors induce dramatic morphological changes of KNRK cells that are blocked by microtubule interfering agents. Proc Natl Acad Sci U S A 1998;95:10499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Morais EF, Rolim LSA, de Melo Fernandes Almeida DR, de Farias Morais HG, de Souza LB, de Almeida Freitas R. Biological role of epithelial-mesenchymal-transition-inducing transcription factors in head and neck squamous cell carcinoma: A systematic review. Arch Oral Biol 2020;119:104904. [DOI] [PubMed] [Google Scholar]
- 34.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz AA, Qin H, Ramalho-Santos M, Song JS. HiTSelect: a comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res 2015;43:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, et al. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol 1996;16:4869–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L, Shao Y, Ren L, Liu X, Li Y, Xu J, et al. IQGAP2 Inhibits Migration and Invasion of Gastric Cancer Cells via Elevating SHIP2 Phosphatase Activity. Int J Mol Sci 2020;21:1968–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res 2014;124:31–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gualberto A, Scholz C, Janes MR, Kessler L, Raza A. The CXCL12/CXCR4 Pathway As a Potential Target of Tipifarnib in Acute Myeloid Leukemia and Myelodysplastic Syndromes. Blood 2017;130:3957 [Google Scholar]
- 40.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 2019;25:628–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 2019;25:620–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hah JH, Zhao M, Pickering CR, Frederick MJ, Andrews GA, Jasser SA, et al. HRAS mutations and resistance to the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in head and neck squamous cell carcinoma cells. Head Neck 2014;36:1547–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rampias T, Giagini A, Siolos S, Matsuzaki H, Sasaki C, Scorilas A, et al. RAS/PI3K crosstalk and cetuximab resistance in head and neck squamous cell carcinoma. Clin Cancer Res 2014;20:2933–46 [DOI] [PubMed] [Google Scholar]
- 44.Gilardi M, Wang Z, Proietto M, Chillà A, Calleja-Valera JL, Goto Y, et al. Tipifarnib as a Precision Therapy for HRAS-Mutant Head and Neck Squamous Cell Carcinomas. Mol Cancer Ther 2020;19:1784–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HW, Sa JK, Gualberto A, Scholz C, Sung HH, Jeong BC, et al. A Phase II Trial of Tipifarnib for Patients with Previously Treated, Metastatic Urothelial Carcinoma Harboring HRAS Mutations. Clin Cancer Res 2020;26:5113–19 [DOI] [PubMed] [Google Scholar]
- 46.Du W, Liu A, Prendergast GC. Activation of the PI3'K-AKT pathway masks the proapoptotic effects of farnesyltransferase inhibitors. Cancer Res 1999;59:4208–12 [PubMed] [Google Scholar]
- 47.Li Q, Mattingly RR. Restoration of E-cadherin cell-cell junctions requires both expression of E-cadherin and suppression of ERK MAP kinase activation in Ras-transformed breast epithelial cells. Neoplasia 2008;10:1444–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nathan CAO, Hayes DN, Harismendy O, Flores J, Moore-Medlin T, Gutkind JS, et al. Multi-Institutional Randomized Double-Blind Phase II Trial of Everolimus vs. Placebo as Adjuvant Therapy in Patients with Locally Advanced Squamous Cell Cancer of the Head and Neck (SCCHN). Int J Radiat Oncol Biol Phys 2020;106:1116 [Google Scholar]
- 49.Caruso C Tipifarnib targets HRAS-mutant cancers. Cancer Discov 2019;9:1637–3834437193 [Google Scholar]
- 50.Ho A, Brana I, Haddad R, Bauman J, Bible K, Faugeras L, et al. Preliminary results from a phase 2 trial of tipifarnib in squamous cell carcinomas (SCCs) with HRAS mutations. Mol Cancer Ther 2019;18:Abstract nr PR08 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files.