Abstract
A hallmark of the innate immune system is its ability to rapidly initiate short-lived or sustained transcriptional programs in a cell- and pathogen-specific manner that is dependent on dynamic chromatin states. Much of the epigenetic landscape is set during cellular differentiation; however, pathogens and other environmental cues also induce changes in chromatin that can either promote tolerance or ‘train’ innate immune cells for amplified secondary responses. We review chromatin processes that enable innate immune cell differentiation and functional transcriptional responses in naive or experienced cells, in concert with signal transduction and cellular metabolic shifts. We discuss how immune chromatin mechanisms are maladapted in disease and novel therapeutic approaches for cellular reprogramming.
Keywords: innate immunity, epigenetics, chromatin, macrophages, inflammation, metabolism, transcription, histone modifications
Introduction
In addition to adapting to tissue-specific cues, innate immune cells are uniquely programmed to appropriately respond to a diverse array of stimuli that range from pathogen infections to non-threatening microbial ligands and metabolites. Certainly, a key tenet of a successful first line immune defense to pathogens is the induction of a signal-specific, cell lineage-specific, and kinetically precise gene expression program. The products of such synchronized gene expression programs occur following pattern recognition receptor (PRR) recognition of conserved features of microorganisms that are absent in the host (pathogen-associated molecular patterns or PAMPs) and host-derived molecules only exposed under conditions of excessive cell death and tissue damage (damage-associated molecular patterns or DAMPs). Subsequent signal transduction and cellular metabolic shifts in neutrophils, monocytes, macrophages, natural killer cells, basophils, dendritic cells or epithelial cells leads to the activation of transcription factors that bind to inducible genes -- cytokines, transcription factors, effector proteins, and metabolic regulators - that enable pathogen clearance, aid adaptive immunity, clear cellular debris, and restore damaged tissues. The different PRRs, signaling pathways, and transcription factors involved in cell differentiation and the transmission of information from the cell surface to the nucleus during an innate immune response continue to be explored. How chromatin mechanisms dictate context-appropriate transcription of precise genes from the larger chromatin landscape is an active area of investigation. These mechanisms are not only central to rapid initial responses to first exposure of a pathogen, but chromatin-associated factors also prime innate immune cells for subsequent re-infections and may be disrupted for anti-inflammatory therapies. Moreover, the contribution of tonic or homeostatic PRR signaling in either priming or repressing chromatin remains poorly understood.
In this review, we discuss the epigenetic regulators that instruct innate immune cell state and functional responses to environmental cues, highlighting overlap between these steps of cellular regulation. We describe the differential chromatin regulation of poised LPS primary response genes versus delayed secondary response genes and the signaling cascades that initiate chromatin changes to enable pro-inflammatory gene transcription. Furthermore, recent work has uncovered how pathogen recognition can stably alter chromatin for tolerized or primed responses to subsequent exposures. Finally, we discuss mutations in human epigenetic factors that lead to inflammatory diseases and the advancement of therapeutic strategies that target epigenetic factors. The advent of new chromatin technologies requiring fewer cells, or single cells, will enable further growth of this field within immunology.
Core components of epigenetic regulation
Gene expression requires the binding of transcription factors to promoters and enhancers, resulting in the recruitment of the transcription apparatus that includes RNA polymerase II (RNA Pol II) and permits transcription initiation, elongation, and termination. However, transcriptional machinery first needs access to genes as DNA is condensed into chromatin and epigenetic mechanisms must permit accessibility of underlying DNA. Broadly, the term epigenetics describes the regulatory mechanisms “outside of or above” the cell’s preconceived DNA code that regulate transcription. Four major mechanisms are considered to be involved in the epigenetic regulation of gene expression patterns: covalent modification of DNA; covalent modification of core histones or histone variants; non-protein-coding RNAs (microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) excellently reviewed here [1,2]; and chromatin remodeling machinery.
Modifications of histones
In eukaryotic cells, 2 meters of DNA is tightly packaged in the nucleus. Specifically, 147bp of nucleotides wrap around an octamer of histones (2 copies each of histone H2A, H2B, H3, and H4) to form a nucleosome, and these nucleosomal units repeat throughout the genome and form increasingly higher-order structures that form chromosomes. Importantly, histone tails have an unstructured N-terminus that protrude from the nucleosomes and are subject to covalent modifications. Epigenetic ‘writers’ catalyze post-translational modifications (e.g. histone methyltransferases, histone acetyltransferases (HATs), kinases), while ‘erasers’ remove these dynamic modifications (e.g. demethylases, histone deacetylases (HDACs)). Regulatory information stored in modified histones is functionally translated by ‘readers’, that dock to defined modified histones via distinct protein domains (e.g. bromodomain, PHD, YEATS). Readers recruit other epigenetic or transcriptional machinery to specific loci thereby serving as the chromatin’s adaptor molecules. These chromatin readers may have similar structural features to adaptor molecules that transduce PRR signaling upstream of transcription [3]. Overall, the combinatorial ‘histone code’, first posited more than twenty years ago [4], is a central orchestrator of gene expression in innate immune cells. For example, myeloid cell identity depends on a combination of histone modifications that result in lineage-inappropriate gene silencing and a separate combination that poises chromatin at inflammatory genes for rapid and robust induction in response to microbial recognition.
DNA and RNA methylation
Methylation of the 5′-carbon of the pyrimidine ring at cytosine nucleotides (5-mC), the most widely studied type of DNA methylation, is catalyzed and maintained by the DNA methyltransferase (DNMT) family members. Ten-eleven translocation (TET) cytosine dioxygenase family members mediate oxidation of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which are critical for active DNA demethylation. In general terms, CpG DNA methylation (DNAme) at promoters inhibits binding of so-called methyl-sensitive transcription factors and recruits repressive DNAme readers (e.g., MeCP2, SETDB1), thus restricting transcription. In contrast, processive oxidation products of DNAme are generally associated with active chromatin and result in loss of DNAme, either passively, through cell division, or actively, via base excision repair. Modifications of RNA have also been shown to regulate gene expression. N6-methyladenosine modification (m6A) on eukaryotic RNA is a common modification which regulates RNA transcript splicing, processing, translation, and decay. In regards to innate immunity, genetic deficiency in METLL3, the methyltransferase catalyzing m6A modification, is associated with reduced NF-κB activity [5] and increased IFN production and viral clearance [6,7].
Chromatin remodeling
Chromatin packaging and topology are important determinants of gene expression. Chromatin undergoes active reorganization of its architecture to permit access of cis-regulatory elements to transcriptional regulators. This is performed by chromatin remodelers, which are ATP dependent translocases that participate in nucleosome sliding, conformational change of the nucleosome, or histone variant exchange. Mammalian chromatin remodelers fall into four families: SWI/SNF (switch/sucrose non-fermenting, ISWI (imitation switch), IN080 (inositol requiring 80), and CHD (chromodomain helicase domain containing). They associate with large protein complexes and have been shown to both promote or prevent transcription. As discussed later, these proteins have an important role in innate immunity by regulating secondary response genes in macrophages following bacterial and viral challenges.
Epigenomic techniques for immunologists
The advent of high-throughput sequencing technologies and novel biochemical methods that survey genomic regulatory regions, chromatin occupancy, and accessibility has significantly advanced our understanding of chromatin biology in cell development and function (Table 1). Many early techniques, such as chromatin immunoprecipitation (ChIP) and DNase I hypersensitivity assays, to assess chromatin occupancy and accessibility required large amounts of material, had poor signal-to-noise ratios and hence often prohibited use by immunologists examining relatively rare but pure primary immune cells. Recently there has been an influx of technologically improved assays (Table 1), including CUT&Run [8], CUT&Tag [9], ATAC-seq [10] and single cell versions of these, that has enabled many immunologists to embrace epigenetic analyses on limited cells. Moreover, protocols such as MINT-ChIP [11] and MulTI-Tag [12] have enabled multiplexing targets in the same cells. Also, epigenome profiling can now be integrated with bulk- and single cell-transcriptomic data using 10x Genomics Single Cell Multiome, simultaneous high-throughput ATAC and RNA expression with sequencing (SHARE-seq) [13,14], or Paired-seq [15]. In addition, chromatin accessibility or RNA measurements coupled with Cytometry by time of flight (CyTOF) as well as spatial genomic and epigenomic techniques will transform our understanding of chromatin regulation in immune cells.
Table 1.
Emerging technologies for immunologists studying chromatin dynamics.
| Technique | Information | Cell Numbers | Reference |
|---|---|---|---|
| Bisulfite-seq | DNA methylation | 100,000 – 500,000 and single cell | [95,96] |
| 5(h)mC-seq | DNA methylation at 5-methylcytosine | 100 – 100,000 | [97] |
| m6A RNA-seq | mRNA methylation | 10 million | [98,99] |
| FAIRE-seq | Chromatin accessibility | 10 million | [100] |
| DNase-seq | Chromatin accessibility | 1 – 10 million | [101] |
| ATAC-seq | Chromatin accessibility | 500 – 50,000 and single cell | [10,102] |
| CUTAC | Chromatin accessibility with the addition of RNA Pol II occupancy | 500 – 50,000 | [103] |
| MNase-seq | Chromatin inaccessibility | 10,000 – 100,000 | [104] |
| GET-seq | Chromatin accessibility and inaccessibility | 5,000 – 20,000 and single cell | [105] |
| SHARE-seq | Chromatin accessibility in concert with RNA transcript levels | single cell | [13,14] |
| ChIP-seq | Chromatin occupancy of specific proteins | 2 – 10 million | [106] |
| CUT&Run | Chromatin occupancy of specific proteins, with reduced background | 500 – 50,000 | [8] |
| CUT&Tag | Chromatin occupancy of specific proteins, with reduced background | 50 – 50,000 and single cell | [9] |
| Spatial-CUT&Tag | Spatial localization of histone modifications in situ | N/A | [107] |
| MINT ChIP | Chromatin occupancy of up to four histone modifications, multiplexed in the same cell population | 50,000 – 100,000 | [11] |
| MulTI-Tag | Chromatin occupancy of multiple histone modifications, multiplexed in the same cell | 500 – 50,000 | [12] |
| Hi-C | Three-dimensional chromatin interactions | 2 – 5 million and single cell | [108,109] |
| ChIA-PET | Three-dimensional chromatin interactions bound by a specific protein | 100 million | [110] |
| IGS | Spatial localization of chromatin | 100 | [111] |
| CyTOF | Protein abundance | 300 | [112] |
Regulation of macrophage identity and plasticity by chromatin mechanisms
Diversity and plasticity are fundamental properties of macrophages, and most work examining epigenetic mechanisms in innate immunity have focused on this cell type. To some extent, the properties of macrophages are imprinted through ontogenetic origin (during embryogenesis from yolk-sac progenitor cells vs. hematopoiesis) but are also heavily dependent on chromatin mechanisms that respond to external or tissue-specific environmental cues. Transcriptomic and epigenetic landscapes of human microglia exposed to an in vitro culture environment signficantly downregulated microglia-specific genes compared to ex vivo microglia, highlighting environment-dependent programming of these macrophages [16]. Similarly, engrafting peritoneal mouse macrophages into the alveolar cavity led to the downregulation of peritoneal-specific gene programs and upregulation of lung macrophage-specific genes [17]. This tissue-specific macrophage identity was shown to be strongly regulated by enhancers [17] and chromatin remodelers, such as BAF/PBAF, to facilitate or prevent transcription factor binding [18,19].
Regulation of silenced facultative heterochromatin, marked by H3K27me3, is also a major orchestrator of macrophage development, polarization, and function. The histone demethylases JMJD3 and UTX specifically facilitate di- and trimethyl H3K27 demethylation and regulate transcription of developmental transcription factors, such as HOX genes [20], and cell fate and patterning proteins, such as Wnt proteins and TGF-β family members [21,22]. JMJD3 is also necessary for expression of IRF4 and the control of macrophage polarization that is required for anti-helminth responses [23]. Similarly, Enhancer of zeste homolog 2 (EZH2), a H3K27 methyltransferase, is required for macrophage cell identity as it mediates Toll-like receptor (TLR)-induced proinflammatory gene expression by directly repressing suppressor of cytokine signaling 3 (SOCS3) expression [24].
Furthermore, work from our group demonstrated a profound role for chromatin reader SP140, associated with inflammatory disease (Table 2), that predominantly occupies heterochromatin marked by H3K27me3 to repress chromatin accessibility and inhibit transcription of lineage-inappropriate genes, including late HOX genes (HOXA7 and HOXA9) [25]. SP140 loss results in severely compromised lineage-defining and microbe-inducible innate transcriptional programs and defective bacterial killing [26]. In addition to histone methylation, histone acetyltransferases and deacetylases serve a role in polarizing macrophages to a pro-inflammatory or alternatively activated state. For example, macrophages that lack HDAC3 become anti-inflammatory [27,28] and polarize towards IL-4 hyperresponsive cells [29]. Inhibition of HDAC3 via butyrate treatment or direct chemical inhibitor alters macrophage metabolism [30], prevents inflammatory responses [31], and enhances anti-microbial responses [30].
Table 2:
Variants of Epigenetic Regulators that associate with susceptibility of Immune Disease.
| Gene | Epigenetic Subclass | Functional Domains | Disease Association |
|---|---|---|---|
| SP100 | Reader | SAND, PHD, Bromodomain | Chronic Obstructive Pulmonary Disease [106] |
| SP110 | Reader | SAND, PHD, Bromodomain | Chronic Lymphocytic Leukemia [107] |
| SP140 | Reader | SAND, PHD, Bromodomain | Crohn’s Disease [108,109], Multiple Sclerosis [110], Chronic Lymphocytic Leukemia [107] |
| BRD1 | Reader | Bromodo main | Rheumatoid Arthritis [111] |
| BRD2 | Readei | Bromodomain | Crohn’s Disease [112], Rheumatoid Arthritis [113] |
| BRWD2/PHIP | Reader | Bromodomain | Sjögren’s Syndrome [114] |
| BRWD3 | Reader | Bromodomain | Pneumonia and COVID19 [115] |
| UHRF1 | Reader | PHD, Tudor, SRA | Type II Diabetes Mellitus [116,117] |
| UHRF2 | Reader | PHD, Tudor, SRA | Asthma [118], Periodontitis [119], Allergy [120], Chronic Sinus Infection [121] |
| CHD1 | Reader | Chromodomain | Type II Diabetes Mellitus [117] |
| CHD4 | Reader | PHD, Chromodomain | Type II Diabetes Mellitus [117] |
| TRIM66 | Reader | PHD, Bromodomain | Type II Diabetes Mellitus [122] |
| L3MBTL3 | Reader | MBT domair | Asthma [123], Multiple Sclerosis [124] |
| DNMT3A | Writer | DNA methyltransferase | Crohn’s Disease [109] |
| DNMT3B | Writer | DNA methyltransferase | Inflammatory Bowel Disease [108,125,126] |
| EHMT2/G9a | Writer | Histone methyltransferase | Type II Diabetes Mellitus [127], Chronic Obstructive Pulmonary Disease [128], Asthma [129], Takayasu Arteritis [130] |
| DOT1L | Writer | Histone methyltransferase | Osteoarthritis [131], Systemic Lupus Erythematosus [132] |
| SUPT3H | Writer | Histone acetyltransferase | Osteoarthritis [128,133] |
| KAT2A | Writer | Histone acetyltransferase | Coronary Artery Disease [134,135], Inflammatory Bowel Disease [136] |
| HDAC4 | Eraser | Histone deacetylase | Chronic Obstructive Pulmonary Disease [128] |
| HDAC7 | Eraser | Histone deacetylase | Inflammatory Bowel Disease [137], Allergy [138], Asthma [139] |
| HDAC9 | Eraser | Histone deacetylase | Osteoarthritis [140] |
| KDM4C | Eraser | Histone demethylase | Systemic Lupus Erythematosus [141], Asthma [142] |
| KDM5A/JARID1A | Eraser | Histone demethylase | Ankylosing Spondylitis [143] |
| TET2 | Eraser | DNA demethylation | Ulcerative Colitis [137], Multiple Sclerosis [110], Chronic Pulmonary Obstructive Disease [144] |
| TET3 | Eraser | DNA demethylation | Systemic Lupus Erythematosus [145] |
| SMARCA2 | Chromatin remodeler | SNF2 ATPase domain | Asthma [146], Hashimoto’s Thyroiditis [147] |
| SMARCA4/BRG1 | Chromatin remodeler | SNF2 ATPase domain | Coronary Artery Disease [148], Multiple Sclerosis [149] |
Kinetics of Macrophage Transcriptional Programs
Much of what we know about chromatin dynamics that regulate transcription after microbial sensing derives from numerous studies on the effect of LPS or Lipid A stimulation and signaling that stems from TLR4 activation. Response to LPS involves the upregulation of genes that are rapidly induced and others whose transcription is delayed [32]. Following LPS stimulation, primary response genes (PRGs) are rapidly induced in the absence of new protein synthesis whereas secondary response genes (SRGs) require new protein synthesis for activation. The promoters of PRGs, such as Tnf, Fos and Nfkbia, are richer in CpG islands and have higher levels of poised RNA Pol II, H3K4me3, H4ac in naive cells compared to SRG promoters [32–37]. These features are characteristic of actively transcribed genes and are associated with higher levels of H3K9/K14 acetylation and H3K4me3 [32,34]. Thus, naive macrophages have poised chromatin landscapes that enable expression of a defined set of PRGs within minutes of cell activation [34,38]. Transcription of these genes do not require chromatin remodeling complexes, such as the SWI/SNF family, or de novo protein synthesis, as their chromatin state is immediately permissive to transcription factor binding and RNA Pol II elongation [32]. SRGs on the other hand, whose transcription peaks around 4 hours post TLR4 stimulation, display low H3K4me3, H4Ac, and no RNA Pol II occupancy in naive macrophages. SRGs, such as Il12b, depend on SWI/SNF remodeling complexes to increase DNA accessibility and transcription [34,38,39]. In addition, PRGs but not SRGs are negatively regulated at baseline by the transcriptional corepressors NCoR/HDAC3 and coREST/HDAC1 to perhaps limit transcription at these poised sites [32].
Signaling to Chromatin: Histone Phosphorylation
Accumulating evidence demonstrates that signal transduction cascades downstream of PRR activation directly regulate histone or DNA modifications or chromatin-interacting proteins [40] (Figure 1). H3S10ph, H3.3S28ph, and H3.3S31ph are examples of histone residue-specific phosphorylation events that occur downstream of TLR4 activation [41]. After LPS stimulation, MSK1 and MSK2 rapidly phosphorylate H3S28 at promoters and enhancers [41]. In addition, H3S10 located at inducible gene promoters is phosphorylated after NFκB activation, although it is not clear which kinases are responsible for this post-translational event [42–45]. IKKα is recruited to inflammatory genes after NFκB activation [45,46] where it mediates phosphorylation of H3.3S31 within LPS induced gene bodies, such as Tnf, Cxcl2, and Il1a [47]. H3.3S31ph correlates with H3K36me3 density, which is exclusively deposited by the histone methyltransferase SETD2 [47]. The SETD2 catalytic domain binds to H3.3S31ph thereby promoting K36 engagement in the active site [47]. Thus, H3.3S31ph augments SETD2 methyltransferase activity. Other epigenetic enzymes that interact with H3.3S31ph include the H3K27me3 demethylase JMJD3 [48,49] while the PHF1 Polycomb group protein family member (H3K27me3 methyltransferase complex) is ejected by H3.3S31ph [50,51]. How these other enzymes orchestrate gene expression after stimulation in regard to H3.3S31ph modification will need further study. However, Armache et al. reveal that the K36me3 reader and transcriptional corepressor ZMYND11 is already present at a subset of LPS induced genes at baseline and is ejected by dually modified H3.3S31ph/H3.3K36me3 as – providing a mechanism whereby ZMYND11 ejection allows rapid transcription to occur [47]. Thus, many stimulation-induced genes share distinguishing chromatin features: (1) active chromatin states and pre-existing H3.3K36me3, (2) pre-bound ZMYND11 corepressor, (3) stimulation induced H3.3S31ph, and (4) ejection of ZMYND11. Beyond TLR4 stimulation, histone phosphorylation likely features as a transcriptional inducement mechanism downstream of diverse receptors and in different cell types, including adaptive immune cells [40].
Figure 1. Signaling to chromatin mediates inflammatory responses.
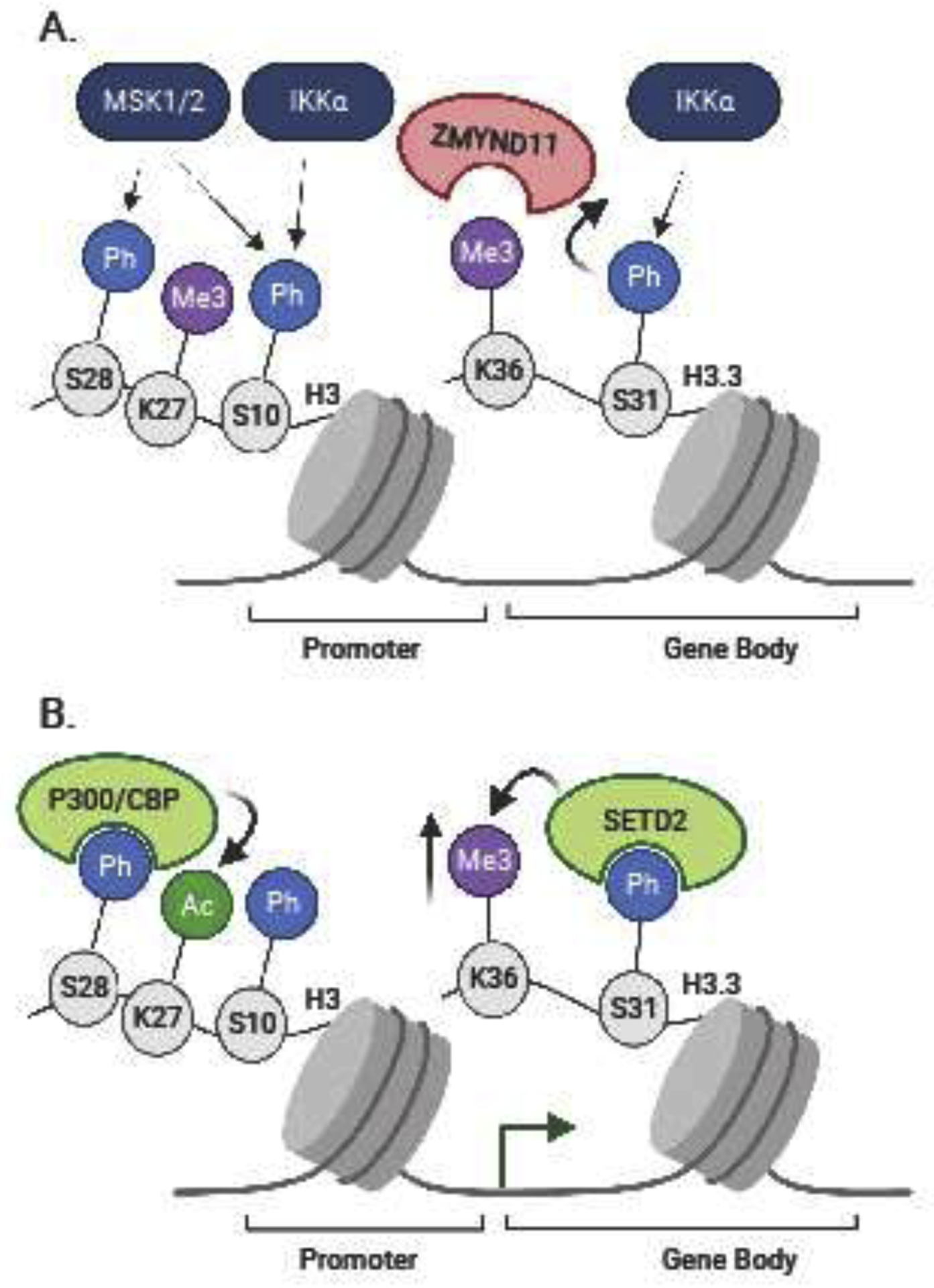
(A) First, kinases, such as MSK1, MSK2, and IKKα, phosphorylate histone modifications, such as H3S28, H3S10, and H3.3S31. Specifically, H3.3S31ph promotes the ejection of ZMYND11 from H3.3K36me3. (B) ZMYND11 ejection allows for increased H3.3K36me3 whereas H3S28ph promotes P300/CBP-mediated acetylation of H3K27 – events that enhance transcription.
Innate Immune Training and Tolerance
Accumulating evidence demonstrates that innate immune cells have adaptive-like features, such as tolerance and training, that depend on alterations to chromatin state. During innate immune training, epigenetic changes or “scaring” persist even after the cell returns to homeostasis following stimulation [52]. This pattern of exposed enhancers and promoters of host-defense genes results in enhanced transcription in response to homologous or heterologous rechallenge. The opposite of trained immunity, “tolerized” innate immune cells are unable to activate gene transcription following restimulation [53,54]. Both innate immune adaptations are rooted in epigenetic reprogramming.
Tolerized genes include pro-inflammatory genes, such as Il6 and Il1b, whereas non-tolerized genes encode antimicrobial effectors, such as Cnlp and Lcn2 [53]. After stimulation, H4Ac, H3K4me3, and Brg1 associated with promoters of LPS inducible genes then dissociate during resolution [33,53,55]. However, upon secondary exposure to LPS, the promoters of tolerized genes do not exhibit a second increase in H4Ac and Brg1 whereas non-tolerized gene promoters display greater and faster H4Ac accumulation than the initial response and maintained H3K4me3 deposition [53]. Inhibition of histone deacetylases or H3K4 demethylases rescues transcription at tolerized genes [53] suggesting that epigenetic enzymes negatively regulate transcription of pro-inflammatory genes and limit pathology associated with inflammation while allowing for pathogen defense. While both tolerized and non-tolerized gene transcription is induced by the same upstream PRR signal transduction, the differential epigenome at these distinct gene sets enables selective and fine-tuned transcription for appropriate immune responses. Thus, this level of epigenetic regulation may be leveraged for selective therapies in the clinic that target inflammation versus antimicrobial responses.
Innate immune training has mostly been observed in monocytes exposed to the fungus C. albicans or its cell wall component β-glucan for a training period, then allowed to return to steady state, and rechallenged (5,7). C. albicans- and β-glucan-induced innate immune training results in a stable increase in H3K4me3 at gene promoters in monocytes and peritoneal macrophages that promotes training [56]. This H3K4me3 deposition is facilitated by lncRNAs [57]. In addition to inflammatory genes, many of the genes with altered promoter H3K4me3 were involved in glycolysis [58] suggesting a metabolic switch accompanies epigenetic reprogramming in innate immune training.
In experimental animal models, the memory of exposure surpasses that of the typical lifespan of innate immune cells. Recent studies have now demonstrated that immune training occurs within the hematopoietic stem cell compartment in bone marrow [59]. Evidence for transmission of trained immunity was also recently demonstrated across generations to murine progeny that survived a sublethal systemic infection with C. albicans [60].
Although innate immune training can enhance pathogen responses, maladaptive innate immune training has been proposed to promote chronic immune disease [52]. Furthermore, in addition to peripheral memory, exposure to western diet, exercise, chronic stress, and sleep fragmentation directly alter the chromatin accessibility of the bone marrow progenitor epigenome. Some of these chromatin changes are maintained over time and importantly impact progenitor proliferation, lineage commitment, and functional response to secondary recall challenges [61–63].
Cross-regulation of metabolic and epigenetic pathways in macrophages
As has been thoroughly reviewed previously [64], metabolic and epigenetic pathways are tightly linked (Figure 2). Chromatin regulating proteins rely on available metabolites for their catalytic activity. Thus, when metabolic switches occur following PRR activation or when metabolite availability is altered by the presence of microorganisms, macrophage transcriptional programs adapt or maladapt via epigenetic regulation. For instance, histone acetyltransferases require acetyl coenzyme (acetyl-CoA) for activity, fumarate inhibits the KDM5 family of histone demethylases, and α-ketoglutarate (α-KG) is as cofactor for histone demethylases [65,66]. The tight coupling between metabolism and epigenetic regulation of transcription is also exemplified by recent discoveries of novel epigenetic marks, including histone succinylation [67,68], crotonylation [69], and lactylation [70] that rely on the substrates succinyl-coA, crotonyl-coA and lactate levels, respectively.
Figure 2. Crosstalk between metabolism and innate immune chromatin architecture.
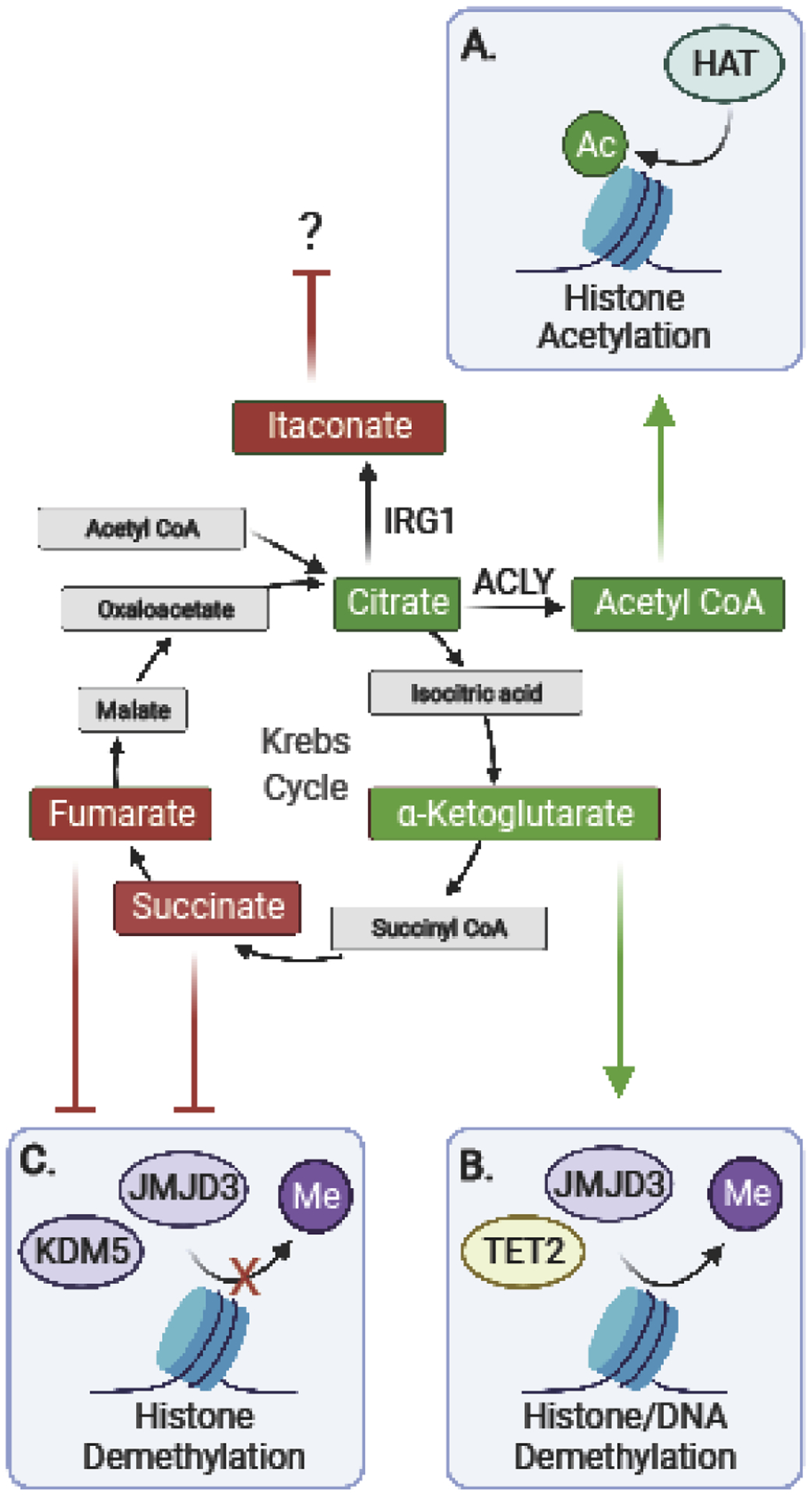
(A) ATP-citrate lyase (ACLY) synthesizes acetyl coenzyme (acetyl-CoA), a coenzyme for histone acetyltransferases (HATs). However, citrate can also be a source for itaconate, an anti-inflammatory metabolite. (B) α -ketoglutarate (α-KG) promotes the function of histone demethylases, such as JMJD3, and DNA demethylases, such as TET2. (C) Fumarate and succinate inhibit histone demethylases, such as JMJD3 and KDM5.
The activity of the tricarboxylic acid (TCA) cycle from which these metabolites are produced is thus a major regulator of chromatin state. Accumulated citrate in LPS activated macrophages is used for production of the potent anti-inflammatory metabolite, itaconate, via the LPS and IFN-inducible enzyme immune-responsive gene 1 protein (IRG1) [71]. Itaconate exerts its anti-inflammatory functions through the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), although how it regulates chromatin modifying enzymes remains to be established.
Succinate acts as a proinflammatory metabolite that directly inhibits histone lysine demethylases (KDM2–7) and the ten-eleven translocation hydroxylases (TET1–3) involved in DNA demethylation. Certainly, in macrophages, an important functional role has been attributed to the αKG/succinate ratio regulating anti-inflammatory versus pro-inflammatory macrophage state. Macrophages polarized with IL4 have increased acetyl-CoA and an increased α-KG:succinate ratio for epigenetic reprogramming via HATS and JMJD3, respectively [72,73]. Conversely, a low α-KG:succinate ratio strengthens the proinflammatory phenotype in LPS activated macrophages. In addition, αKG contributes to endotoxin tolerance after LPS activation [73]. TET2, an LPS inducible DNA demethylase and a target of α-KG was shown to be essential for inflammation resolution [74], and TET2 may be involved in the mechanism by which α-KG promotes LPS tolerance. Thus, pathways involved in α-KG production may be attractive therapeutic targets to reset the epigenome in diseases associated with macrophage malfunction.
Fumarate also has a proinflammatory role in controlling chromatin modifications. Specifically, the accumulation of fumarate in response to pro-inflammatory insults has been shown to be necessary for trained immunity and inflammation by inhibiting KDM5 histone demethylase activity [75]. The inhibition of KDM5 increases the levels of H3K4me3, a marker of active gene transcription at the promoters of Tnf and Il6 cytokines. Notably, fumarate derivatives like dimethyl fumarate (DMF) are currently being used in the clinic to treat autoimmune conditions, including multiple sclerosis (MS) and psoriasis.
In addition to host metabolic pathways, the microbiome serves as the other major source of metabolites in mammals and is emerging as a major influence on host cell epigenetic enzyme activity [76]. Microbiota exclusively metabolize complex carbohydrates derived from dietary fibers in the colonic lumen via fermentative reactions to produce small organic acids, the bulk of which are short chain fatty acids (SCFAs) acetate, propionate, and butyrate – all of which are HDAC inhibitors. Notably, supplementation of SCFAs in germfree mice recapitulated global chromatin states and gene expression patterns observed with complete gut colonization [77]. Other commensal bacteria-derived metabolites, such as inositol-1,4,5-trisphosphate (InsP3), have been shown to stimulate HDAC3 activity in the gut [78]. Recently, butyrate was shown to have a profound impact on macrophage function by promoting antimicrobial transcriptional programs via HDAC3 inhibition [30], but the ability of other microbiota-derived metabolites to dictate innate immune transcription via the epigenome is a fertile area of investigation.
Disease Relevance and Therapeutic Opportunities
The rapid rise in the prevalence of chronic immune diseases that cannot be explained by genetics alone lend support to the critical role of environmental factors and epigenetics in these diseases. Analysis of longitudinal twin cohort studies reveal that by the age of 65, 70% of variance in chromatin modifications can be largely attributed to environmental influences [79]. Genome-wide association studies have identified mutations within genetic loci for chromatin readers, writers, and erasers that are significantly associated with inflammatory disease susceptibility (Table 2). However, in addition to mutations that directly affect expression or function of epigenetic enzymes in immune disease, mutations can lie in non-coding epigenetic regulatory regions, such as enhancers. In fact, 60% of autoimmune disease variants map to active immune cell enhancers, but it is unknown how causal variants affect gene transcription as most mutations do not lie in known transcription factor binding motifs [80]. There is also a gap in knowledge in which epigenetic regulators integrate changes from environmental cues and whether genetic variants in these epigenetic regulators are sufficient for disease pathogenesis or require environmental perturbations as a “second hit”. Furthermore, heterogeneity in clinical phenotypes (penetrance and disease expressivity) commonly observed in different patients with the same mutation could be due to contributions from different environmental cues occurring at critical windows of development.
Epigenetic Therapeutics in Inflammatory Disease
Due to the role of chromatin modifying enzymes in dictating precise gene transcription programs in homeostasis and inflammation, targeting this class of proteins raises the possibility to regulate and reduce the magnitude of entire gene expression programs instead of targeting individual inflammatory mediators. Moreover, many epigenetic modulating drugs have current FDA approval for cancer and can potentially be repurposed to treat epigenetic disruptions in the context of autoimmunity or inflammation.
As outlined in the above sections, H3K27me3 prevents promoter accessibility and suppresses the expression of proinflammatory gene programs in macrophages. GSK-J4, an α-ketoglutarate mimic, binds to the catalytic pocket of the H3K27 demethylases JMJD3 and UTX [48]. Thus, GSK-J4 prevents the demethylation of the repressive H3K27me3 and inhibits LPS-induced inflammation [48]. Recently, GSK-J4 prevented abdominal aortic aneurysms and aortic inflammation in mice that stemmed from monocytes and macrophages [81]. Similarly, pharmacological inhibition of EZH2 specifically resolved H3K27me3 at bivalent gene promoters and attenuated cardiac dysfunction in a mouse model of myocardial infarction [82], ameliorated DSS-induced colitis [83], and alleviated lung injury and fibrosis in the LPS-induced acute respiratory distress syndrome model [84].
Class I and Class II pan HDAC inhibitors have exhibited anti-inflammatory effects in vitro and in vivo [31,85–87]. Specific HDAC1 or HDAC3 inhibitors also prevent inflammation in animal models of inflammatory diseases and in peripheral blood mononuclear cells (PBMC) from rheumatoid arthritis patients [85,88]. Despite the reasonable success of HDAC inhibitors as anti-inflammatory agents, the details of how such epigenetic alteration prevents inflammation is unclear. Histone acetylation is exclusively “read” by bromodomains, which have defined lysine acetylation affinities and recruit distinct proteins, such as transcriptional elongation machinery, to the chromatin [32,89,90]. Rather than drugging the enzymes that “write” and “erase” the epigenome, targeting the “readers” of histone modifications may inhibit the function of specific epigenetic modifications without altering the cell’s overall epigenetic landscape determined by “writers” and “erasers.” By designing small molecules that act as histone mimics and bind to reader domains, such as a bromodomain, this class of therapeutics can disrupt histone binding activity. Most work thus far has focused on targeting the bromodomain and extra-terminal (BET) subfamily. I-BET762 (also known as GSK525762A) specifically interacts with BRD2, 3, and 4 and competitively interacts with the acetylated lysine binding pocket of BET bromodomains, acting as a histone mimic [33]. Inhibition of BET proteins prevents the assembly of chromatin activating and transcription elongation complexes at a subset of LPS-inducible promoters [33]. Importantly, I-BET prevented LPS-induced endotoxic shock and bacteria-induced sepsis in mice [33]. Beyond BET family members, other classes of bromodomains are also predicted to have good druggability [91].
Additional chromatin factors such as topoisomerases (TOP) were recently reported to play key roles in infection-induced gene transcription and leveraged for therapeutic benefit. Certainly, TOP inhibition at low and clinically tolerized doses specifically modulated bacteria and virus-inducible inflammatory gene expression programs and demonstrated pre-clinical efficacy in sepsis and COVID-19 [92,93]. Similarly, TOP inhibition can be utilized to repress aberrant gene expression programs upregulated in immune diseases driven by epigenetic reader SP140 loss-of-function [26]. In addition to TOP inhibitors, inhibitors to the positive transcription elongation factor b (P-TEFb) subunit cyclin-dependent kinase 9 (CDK9), also prevent transcription and are being developed as a new line of therapeutics to alleviate inflammation and autoimmunity [94].
Concluding remarks.
Effective innate immunity depends on the fidelity of immune cell differentiation and rapid adaptability of mature cells to the tissue milieu and other environmental factors, such as pathogen invasion. In these contexts, epigenetic mechanisms allow cells to dynamically initiate or terminate transcriptional programs to appropriately respond to changes in the tissue microenvironment, microbial threats, or the resolution of infection. Failure to fine-tune transcriptional programs to prevent exaggerated responses will cause hyper-inflammatory and autoimmune disorders (Figure 3). As our understanding of innate immune ‘training’ and innate immune metabolism grows, it is appealing to speculate that epigenetic mechanisms that integrate these signals may contribute to the persistence of disease-associated phenotypes, even in the absence of the initial trigger. Moreover, there are emerging examples of genetic mutations within chromatin modifying enzymes or chromatin regulatory regions that directly contribute to human immune disorders. Thus, resetting metabolic states to alter epigenetic enzyme function or directly epigenetic enzymes may allow resetting of the disease ‘epigenetic scar’ and restoration of normal transcriptional programs.
Figure 3.
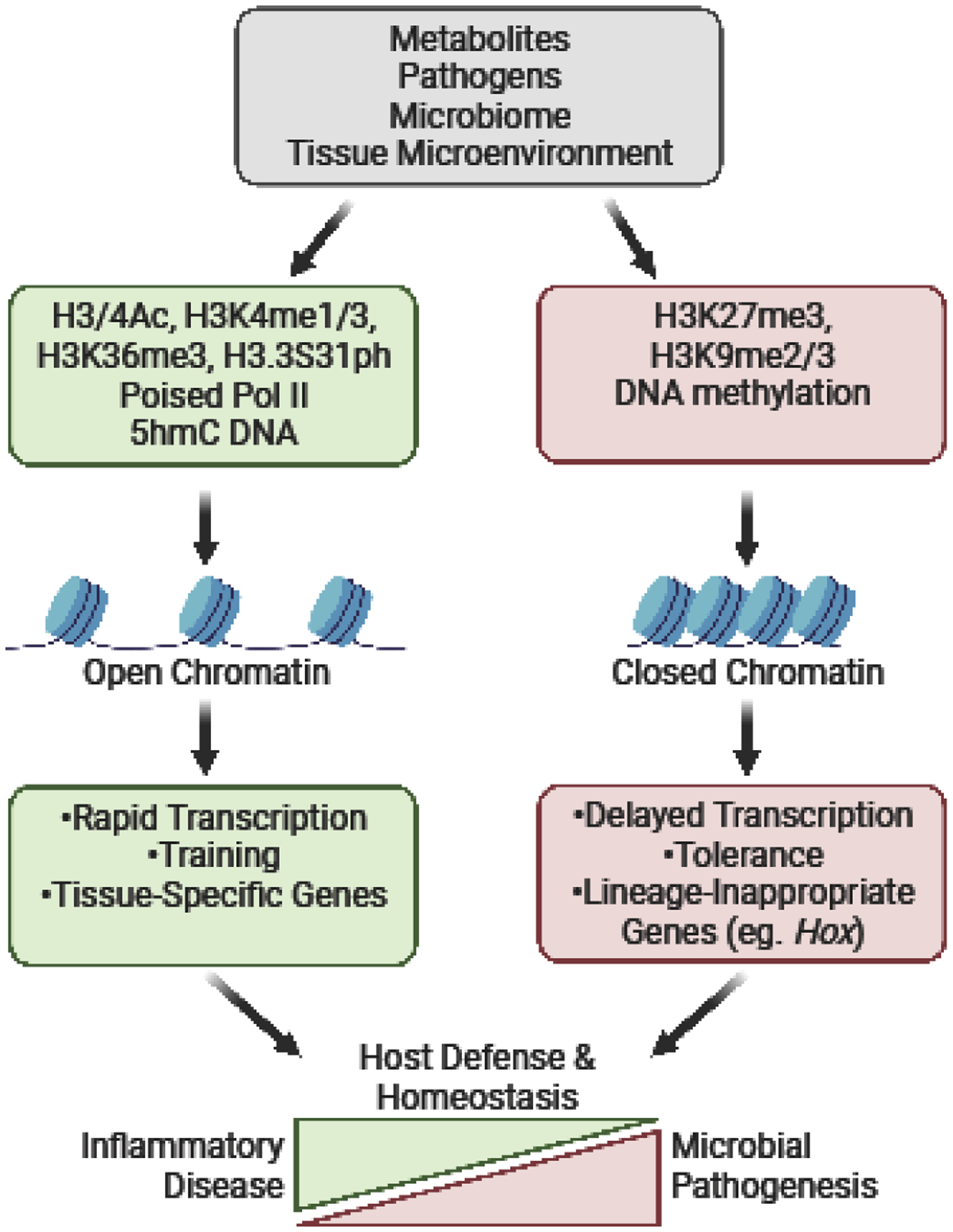
Environmental factors dictate chromatin modifications and accessibility to determine innate immune cell state and function.
Highlights.
Chromatin dynamics regulate pattern, timing and magnitude of gene expression
Signal transduction downstream of TLR4 regulate histones for inducible gene transcription
Metabolic shifts and epigenetics of innate immune cells are intimately linked
Chromatin dynamics enable memory of microbial exposure for tolerance or trained immunity
Epigenetic therapies demonstrate promise for inflammatory disease
Acknowledgements
Figures were created using BioRender. This study was supported by NIH R01DK119996 (K.L.J.), Canadian Institutes of Health Research (CIHR) postdoctoral fellowship (H.A.), NIH F31DK127518 (I.F.), and K.L.J is a John Lawrence MGH Research Scholar, 2020-2025.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA: Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol 2014, 14:361–376. [DOI] [PubMed] [Google Scholar]
- 2.Robinson EK, Covarrubias S, Carpenter S: The how and why of lncRNA function: an innate immune perspective. Biochim Biophys Acta (BBA) Gene Regul Mech 2020, 1863:194419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huoh YS, Hur S: Death domain fold proteins in immune signaling and transcriptional regulation. FEBS J 2021. 10.1111/febs.15901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD: Translating the histone code. Science 2001, 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Li B, Arumugam S, Lu Q, Mankash SM, Li J, Sun B, Li J, Flavell RA, Li H-B et al. : m6A mRNA methylation-directed myeloid cell activation controls progression of NAFLD and obesity. Cell Rep 2021, 37:109968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N, Le-Trilling VTK et al. : m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 2019, 20:173–182. [DOI] [PubMed] [Google Scholar]
- 7.Qiu W, Zhang Q, Zhang R, Lu Y, Wang X, Tian H, Yang Y, Gu Z, Gao Y, Yang X et al. : N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat Commun 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skene PJ, Henikoff S: An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S: CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ: ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 2015, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Galen P, Viny AD, Ram O, Ryan RJH, Cotton MJ, Donohue L, Sievers C, Drier Y, Liau BB, Gillespie SM et al. : A multiplexed system for quantitative comparisons of chromatin landscapes. Mol Cell 2016, 61:170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meers MP, Janssens DH, Henikoff S: Multifactorial Chromatin Regulatory Landscapes at Single Cell Resolution. bioRxiv; 2021. 10.1101/2021.07.08.451691. [DOI] [Google Scholar]
- 13.Chung H, Parkhurst CN, Magee EM, Phillips D, Habibi E, Chen F, Yeung BZ, Waldman J, Artis D, Regev A: Joint single-cell measurements of nuclear proteins and RNA in vivo. Nat Methods 2021, 18:1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Zhang B, Lafave LM, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T et al. : Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 2020, 183:1103–1116.e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R, Lucero J, Behrens MM, Hu M, Ren B: An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat Struct Mol Biol 2019, 26:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP et al. : An environment-dependent transcriptional network specifies human microglia identity. Science 2017, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I: Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014, 159:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAndrew MJ, Gjidoda A, Tagore M, Miksanek T, Floer M: Chromatin remodeler recruitment during macrophage differentiation facilitates transcription factor binding to enhancers in mature cells. J Biol Chem 2016, 291:18058–18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang K, Park SH, Chen J, Qiao Y, Giannopoulou E, Berg K, Hanidu A, Li J, Nabozny G, Kang K et al. : Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 2017, 47:235–250.e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G: The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130:1083–1094. [DOI] [PubMed] [Google Scholar]
- 21.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK et al. : Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441:349–353. [DOI] [PubMed] [Google Scholar]
- 22.Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B: Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc Natl Acad Sci U S A 2006, 103:6994–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T et al. : The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010, 11:936–944. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Wang Y, Yuan J, Li N, Pei S, Xu J, Luo X, Mao C, Liu J, Yu T et al. : Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J Exp Med 2018, 215:1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.•.Mehta S, Cronkite DA, Basavappa M, Saunders TL, Adiliaghdam F, Amatullah H, Morrison SA, Pagan JD, Anthony RM, Tonnerre P et al. : Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Sci Immunol 2017, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]; Work from our group has uncovered the importance of the immune cell chromatin reader, SP140, in maintaining macrophage identity. Here we demonstrate the epigenetic mechanism by which SP140 represses chromatin. Since SP140 loss is associated with multiple immune diseases, this work is important for understanding how SP140 prevents chronic inflammation.
- 26.Amatullah H, Digumarthi S, Fraschilla I, Adiliaghdam F, Bonilla G, Wong LP, Sadreyev RI, Jeffrey KL: Identification of topoisomerase as a precision-medicine target in chromatin reader SP140-driven Crohn’s disease. 2022. 10.1101/2021.09.20.461083. 82. [DOI]
- 27.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G: Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A 2012, 109:E2865–E2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leus NG, van der Wouden PE, van den Bosch T, Hooghiemstra WTR, Ourailidou ME, Kistemaker LE, Bischoff R, Gosens R, Haisma HJ, Dekker FJ: HDAC 3-selective inhibitor RGFP966 demonstrates anti-inflammatory properties in RAW 264.7 macrophages and mouse precision-cut lung slices by attenuating NF-kappaB p65 transcriptional activity. Biochem Pharmacol 2016, 108:58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D et al. : Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev 2011, 25:2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.••.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E et al. : The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 2019, 50:432–445 e437 [DOI] [PMC free article] [PubMed] [Google Scholar]; Work from this group has pioneered the examination of signal transduction events after microbial recognition that can initiate epigenetic changes to enable rapidly inducible gene transcription in innate immune cells. This study, specifically, determines the function of the H3.3S31 modification that is increased after LPS stimulation.
- 31.Chang PV, Hao L, Offermanns S, Medzhitov R: The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014, 111:2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hargreaves DC, Horng T, Medzhitov R: Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009, 138:129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H et al. : Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST: A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 2009, 138:114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smale ST: Selective transcription in response to an inflammatory stimulus. Cell 2010, 140:833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natoli G, Ghisletti S, Barozzi I: The genomic landscapes of inflammation. Genes Dev 2011, 25:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatt DM, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST: Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 2012, 150:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST: Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev 2006, 20:282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinmann AS, Plevy SE, Smale ST: Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 1999, 11:665–675. [DOI] [PubMed] [Google Scholar]
- 40.Martínez De Paz A, Josefowicz SZ: Signaling-to-chromatin pathways in the immune system. Immunol Rev 2021, 300:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josefowicz SZ, Shimada M, Armache A, Li CH, Miller RM, Lin S, Yang A, Dill BD, Molina H, Park HS et al. : Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol Cell 2016, 64:347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI: Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001, 292:110–113. [DOI] [PubMed] [Google Scholar]
- 43.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC: The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J 1999, 18:4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS: A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 2003, 423:659–663. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto Y, Verma UN, Prajapati S, Kwak Y-T, Gaynor RB: Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 2003, 423:655–659. [DOI] [PubMed] [Google Scholar]
- 46.Thorne JL, Ouboussad L, Lefevre PF: Heterochromatin protein 1 gamma and IκB kinase alpha interdependence during tumour necrosis factor gene transcription elongation in activated macrophages. Nucleic Acids Res 2012, 40:7676–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.••.Armache A, Yang S, Martinez de Paz A, Robbins LE, Durmaz C, Cheong JQ, Ravishankar A, Daman AW, Ahimovic DJ, Klevorn T et al. : Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 2020, 583:852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]; Work from this group has pioneered the examination of signal transduction events after microbial recognition that can initiate epigenetic changes to enable rapidly inducible gene transcription in innate immune cells. This study, specifically, determines the function of the H3.3S31 modification that is increased after LPS stimulation.
- 48.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H et al. : A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012, 488:404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S et al. : Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 2009, 28:3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D: Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 2008, 28:2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews FH, Gatchalian J, Krajewski K, Strahl BD, Kutateladze TG: Regulation of methyllysine readers through phosphorylation. ACS Chem Biol 2016, 11:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Netea MG, Dominguez-Andres J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, Joosten LAB, van der Meer JWM, Mhlanga MM, Mulder WJM et al. : Defining trained immunity and its role in health and disease. Nat Rev Immunol 2020, 20:375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster SL, Hargreaves DC, Medzhitov R: Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 2007, 447:972–978. [DOI] [PubMed] [Google Scholar]
- 54.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA et al. : Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014, 345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G: Latent enhancers activated by stimulation in differentiated cells. Cell 2013, 152:157–171. [DOI] [PubMed] [Google Scholar]
- 56.Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C et al. : Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 2012, 12:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fanucchi S, Fok ET, Dalla E, Shibayama Y, Borner K, Chang EY, Stoychev S, Imakaev M, Grimm D, Wang KC et al. : Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat Genet 2019, 51:138–150. [DOI] [PubMed] [Google Scholar]
- 58.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A et al. : mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345:1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.•.Khan N, Downey J, Sanz J, Kaufmann E, Blankenhaus B, Pacis A, Pernet E, Ahmed E, Cardoso S, Nijnik A et al. : M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell 2020, 183:752–770 e722 [DOI] [PMC free article] [PubMed] [Google Scholar]; It was previously shown that Bacille Calmette-Guéren (BCG) or b-glucan reprograms HSCs to protect against Mycobacterium tuberculosis (Mtb). However, this study provides evidence that Mtb infection reprograms HSCs to suppress myelopoiesis and impair control of chronic Mtb infection.
- 60.•.Katzmarski N, Dominguez-Andres J, Cirovic B, Renieris G, Ciarlo E, Le Roy D, Lepikhov K, Kattler K, Gasparoni G, Handler K et al. : Transmission of trained immunity and heterologous resistance to infections across generations. Nat Immunol 2021, 22:1382–1390 [DOI] [PubMed] [Google Scholar]; This recent paper provides evidence that parents who can resolve infections can pass on enhanced protection to their offspring over multiple generations. For example, Candida albicans infection in parental mice epigenetically reprograms myeloid progenitors for enhanced transcription of inflammatory genes beneficial during E. coli infection.
- 61.Christ A, Gunther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, Scholz CJ, Oosting M, Haendler K, Bassler K et al. : Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 2018, 172:162–175 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.•.Frodermann V, Rohde D, Courties G, Severe N, Schloss MJ, Amatullah H, McAlpine CS, Cremer S, Hoyer FF, Ji F et al. : Exercise reduces inflammatory cell production and cardiovascular inflammation via instruction of hematopoietic progenitor cells. Nat Med 2019, 25:1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that exercise reduces chromatin accessibility in hematopoietic progenitor cells to prevent transcription of proliferation and lineage defining genes and is associated with improved survival during sepsis. On the contrary, a sedentary lifestyle provides an increased supply of leukocytes during atherosclerosis that may exacerbate disease.
- 63.Barrett TJ, Corr EM, Van Solingen C, Schlamp F, Brown EJ, Koelwyn GJ, Lee AH, Shanley LC, Spruill TM, Bozal F et al. : Chronic stress primes innate immune responses in mice and humans. Cell Rep 2021, 36:109595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Etchegaray JP, Mostoslavsky R: Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell 2016, 62:695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chisolm DA, Weinmann AS: Connections between metabolism and epigenetics in programming cellular differentiation. Annu Rev Immunol 2018, 36:221–246. [DOI] [PubMed] [Google Scholar]
- 66.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y: Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439:811–816. [DOI] [PubMed] [Google Scholar]
- 67.Yokoyama A, Katsura S, Sugawara A: Biochemical analysis of histone succinylation. Biochem Res Int 2017, 2017:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, Tan L, Yang P, Lee JH, Li XJ et al. : KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N et al. : Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M et al. : Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E et al. : Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben-Sahra I, Byles V, Polynne-Stapornkul T et al. : AktmTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. eLife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.•.Liu P-S, Wang H, Li X, Chao T, Teav T, Christen S, Di Conza G, Cheng W-C, Chou C-H, Vavakova M et al. : α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017, 18:985–994 [DOI] [PubMed] [Google Scholar]; Here, Liu et al. elucidate how α-ketoglutarate (α-KG) accumulation allows JMJD3 to promote the epigenetic reprogramming necessary for IL-4 dependent alternative activation of macrophages. Furthermore, α-KG suppresses NF-κB activity and promotes LPS tolerance in a JMJD3-independent mechanism. This work builds off previous studies on the requirement of α-KG for JMJD3 activity and anti-helminth immunity by defining the metabolic requirements for macrophage polarization.
- 74.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X et al. : Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015, 525:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.•.Arts RJ, Novakovic B, Ter Horst R, Carvalho A, Bekkering S, Lachmandas E, Rodrigues F, Silvestre R, Cheng SC, Wang SY et al. : Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab 2016, 24:807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identifies the metabolic pathways necessary for induction of trained immunity in monocytes. For example, they show fumarate inhibits KDM5 histone demethylases to program monocytes for enhanced future response, and fumarate exposure alone can induce trained immunity.
- 76.Amatullah H, Jeffrey KL: Epigenome-metabolome-microbiome axis in health and IBD. Curr Opin Microbiol 2020, 56:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM: Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 2016, 64:982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.••.Wu SE, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, Karns R, Denson LA, Haslam DB, Alenghat T: Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature 2020, 586:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]; Not only can commensal bacteria-derived butyrate inhibit HDAC3 activity, but this study now shows that commensal bacteria, such as E. coli, can produce inositol-1,4,5-triphosphate (InsP3) that counters the effect of butyrate. This is because InsP3 enhances HDAC3 activity in the intestine to promote recovery after intestinal damage.
- 79.Cheung P, Vallania F, Warsinske HC, Donato M, Schaffert S, Chang SE, Dvorak M, Dekker CL, Davis MM, Utz PJ et al. : Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 2018, 173:1385–1397.e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA et al. : Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis FM, Tsoi LC, Melvin WJ, denDekker A, Wasikowski R, Joshi AD, Wolf S, Obi AT, Billi AC, Xing X et al. : Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms. J Exp Med 2021, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Julie R, Déborah G, Sylvanie R, Virginie T, Anaïs D, Alphonse C, Jean-Paul H, Zina B, Claire V, Delphine B, Dominique G, Marjorie B, Vincent R, Eric D, Ebba B: Fraineau Sylvain Ezh2 as an epigenetic checkpoint regulator during monocyte differentiation: a potential target to improve cardiac repair after myocardial infarction. 2022. 10.1101/2021.02.17.428828. [DOI]
- 83.Zhou J, Huang S, Wang Z, Huang J, Xu L, Tang X, Wan YY, Li Q-J, Symonds ALJ, Long H et al. : Targeting EZH2 histone methyltransferase activity alleviates experimental intestinal inflammation. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bao X, Liu X, Liu N, Zhuang S, Yang Q, Ren H, Zhao D, Bai J, Zhou X, Tang L: Inhibition of EZH2 prevents acute respiratory distress syndrome (ARDS)-associated pulmonary fibrosis by regulating the macrophage polarization phenotype. Respir Res 2021, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Segain JP, Raingeard de la Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottiere HM, Galmiche JP: Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 2000, 47:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luhrs H, Gerke T, Muller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T: Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol 2002, 37:458–466. [DOI] [PubMed] [Google Scholar]
- 87.Vojinovic J, Damjanov N: HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol Med 2011, 17:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein K, Gay S: Epigenetics in rheumatoid arthritis. Curr Opin Rheumatol 2015, 27:76–82. [DOI] [PubMed] [Google Scholar]
- 89.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T et al. : Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K: The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005, 19:523–534. [DOI] [PubMed] [Google Scholar]
- 91.Filippakopoulos P, Knapp S: Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014, 13:337–356. [DOI] [PubMed] [Google Scholar]
- 92.Ho JSY, Mok BW-Y, Campisi L, Jordan T, Yildiz S, Parameswaran S, Wayman JA, Gaudreault NN, Meekins DA, Indran SV et al. : TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell 2021, 184:2618–2632.e2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rialdi A, Campisi L, Zhao N, Lagda AC, Pietzsch C, Ho JSY, Martinez-Gil L, Fenouil R, Chen X, Edwards M et al. : Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 2016, 352: aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujinaga K: P-TEFb as a promising therapeutic target. Molecules 2020, 25:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feil R, Charlton J, Bird AP, Walter J, Reik W: Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res 1994, 22:695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G: Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 2014, 11:817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S et al. : Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 2011, 42:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M et al. : Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485:201–206. [DOI] [PubMed] [Google Scholar]
- 99.Meyer KD, Saletore Y, Zumbo P, Elemento O, Christopher Samie: Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell 2012, 149:1635. –1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD: FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 2007, 17:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE: High-resolution mapping and characterization of open chromatin across the genome. Cell 2008, 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ: Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Henikoff S, Henikoff JG, Kaya-Okur HS, Ahmad K: Efficient chromatin accessibility mapping in situ by nucleosometethered tagmentation. eLife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pajoro A, Muiño JM, Angenent GC, Kaufmann K: Profiling Nucleosome Occupancy by MNase-seq: Experimental Protocol and Computational Analysis. Methods Mol Biol 2018, 1675:167–181. [DOI] [PubMed] [Google Scholar]
- 105.Tedesco M, Giannese F, Lazarević D, Giansanti V, Rosano D, Monzani S, Catalano I, Grassi E, Zanella ER, Botrugno OA et al. : Chromatin velocity reveals epigenetic dynamics by single-cell profiling of heterochromatin and euchromatin. Nat Biotechnol 2021. 10.1038/s41587-021-01031-1. [DOI] [PubMed] [Google Scholar]
- 106.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP et al. : Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng Y, Bartosovic M, Kukanja P, Zhang D, Liu Y, Su G, Enninful A, Bai Z, Castelo-Branco G, Fan R: Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 2022, 375:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO et al. : Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P: Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH et al. : An oestrogen-receptor-α-bound human chromatin interactome. Nature 2009, 462:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Payne AC, Chiang ZD, Reginato PL, Mangiameli SM, Murray EM, Yao C-C, Markoulaki S, Earl AS, Labade AS, Jaenisch R et al. : In situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science 2021, 371: eaay3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD: Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 2009, 81:6813–6822. [DOI] [PubMed] [Google Scholar]
- 113.Lutz SM, Cho MH, Young K, Hersh CP, Castaldi PJ, McDonald ML, Regan E, Mattheisen M, Demeo DL, Parker M et al. : A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genetics 2015. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Law PJ, Berndt SI, Speedy HE, Camp NJ, Sava GP, Skibola CF, Holroyd A, Joseph V, Sunter NJ, Nieters A et al. : Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun 2017, 8:14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA et al. : Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R et al. : Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genetics 2010, 42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.International Multiple Sclerosis Genetics C, Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D et al. : Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013, 45:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Knevel R, Klein K, Somers K, Ospelt C, Houwing-Duistermaat JJ, van Nies JA, de Rooy DP, de Bock L, Kurreeman FA, Schonkeren J et al. : Identification of a genetic variant for joint damage progression in autoantibody-positive rheumatoid arthritis. Ann Rheum Dis 2014, 73:2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ostrowski J, Paziewska A, Lazowska I, Ambrozkiewicz F, Goryca K, Kulecka M, Rawa T, Karczmarski J, Dabrowska M, Zeber-Lubecka N et al. : Genetic architecture differences between pediatric and adult-onset inflammatory bowel diseases in the Polish population. Sci Rep 2016, 6:39831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mahdi H, Fisher BA, Källberg H, Plant D, Malmström V, Rönnelid J, Charles P, Ding B, Alfredsson L, Padyukov L et al. : Specific interaction between genotype, smoking and autoimmunity to citrullinated ä-enolase in the etiology of rheumatoid arthritis. Nat Genetics 2009, 41:1319–1324. [DOI] [PubMed] [Google Scholar]
- 121.Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, Grundahl KM, Kelly JA, Dozmorov MG, Miceli-Richard C, Bowman S et al. : Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren’s syndrome. Nat Genet 2013, 45:1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Sonmez T, Coker D, Symons A, Esparza-Gordillo J, Aslibekyan S, Auton A: Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genetics 2021, 53:801–808. [DOI] [PubMed] [Google Scholar]
- 123.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N et al. : Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genetics 2018, 50:1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vujkovic M, Keaton JM, Lynch JA, Miller DR, Zhou J, Tcheandjieu C, Huffman JE, Assimes TL, Lorenz K, Zhu X et al. : Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1. 4 million participants in a multi-ancestry meta-analysis. Nat Genet 2020, 52:680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu Z, Zhu X, Liu CL, Shi H, Shen S, Yang Y, Hasegawa K, Camargo CA Jr, Liang L: Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J 2019:54. [DOI] [PubMed] [Google Scholar]
- 126.Divaris K, Monda KL, North KE, Olshan AF, Lange EM, Moss K, Barros SP, Beck JD, Offenbacher S: Genome-wide association study of periodontal pathogen colonization. J Dent Res 2012, 91:21S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, Christiani DC, Liang L: A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet 2018, 50:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, Tung JY, Hinds DA: Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun 2017, 8:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M et al. : A cross-population atlas of genetic associations for 220 human phenotypes. Nature Genetics 2021, 53:1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ierodiakonou D, Coull BA, Zanobetti A, Postma DS, Boezen HM, Vonk JM, McKone EF, Schildcrout JS, Koppelman GH, Croteau-Chonka DC et al. : Pathway analysis of a genome-wide gene by air pollution interaction study in asthmatic children. Journal of Exposure Science & Environmental Epidemiology 2019, 29:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andlauer TF, Buck D, Antony G, Bayas A, Bechmann L, Berthele A, Chan A, Gasperi C, Gold R, Graetz C et al. : Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci Adv 2016, 2:e1501678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji SG et al. : Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017, 49:256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hubenthal M et al. : Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016, 48:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bonàs-Guarch S, Guindo-Martínez M, Miguel-Escalada I, Grarup N, Sebastian D, Rodriguez-Fos E, Sánchez F, Planas-Fèlix M, Cortes-Sánchez P, González S et al. : Re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes. Nature Communications 2018:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim W, Prokopenko D, Sakornsakolpat P, Hobbs BD, Lutz SM, Hokanson JE, Wain LV, Melbourne CA, Shrine N, Tobin MD et al. : Genome-Wide Gene-by-Smoking Interaction Study of Chronic Obstructive Pulmonary Disease. Am J Epidemiol 2021, 190:875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu Z, Guo Y, Shi H, Liu C-L, Panganiban RA, Chung W, O’Connor LJ, Himes BE, Gazal S, Hasegawa K et al. : Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. Journal of Allergy and Clinical Immunology 2020, 145:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Renauer PA, Saruhan-Direskeneli G, Coit P, Adler A, Aksu K, Keser G, Alibaz-Oner F, Aydin SZ, Kamali S, Inanc M et al. : Identification of Susceptibility Loci inIL6,RPS9/LILRB3, and an Intergenic Locus on Chromosome 21q22 in Takayasu Arteritis in a Genome-Wide Association Study. Arthritis & Rheumatology 2015, 67:1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FMF, Doherty SA, Hart DJ, Hofman A, Luyten FP, Maciewicz RA, Mangino M et al. : Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proceedings of the National Academy of Sciences 2012, 109:8218–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Song Q, Lei Y, Shao L, Li W, Kong Q, Lin Z, Qin X, Wei W, Hou F, Li J et al. : Genome-wide association study on Northern Chinese identifies KLF2, DOT1L and STAB2 associated with systemic lupus erythematosus. Rheumatology (Oxford) 2021, 60:4407–4417. [DOI] [PubMed] [Google Scholar]
- 140.Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. The Lancet 2012, 380:815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van der Harst P, Verweij N: Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res 2018, 122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koyama S, Ito K, Terao C, Akiyama M, Horikoshi M, Momozawa Y, Matsunaga H, Ieki H, Ozaki K, Onouchi Y et al. : Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet 2020, 52:1169–1177. [DOI] [PubMed] [Google Scholar]
- 143.Huang C, Haritunians T, Okou DT, Cutler DJ, Zwick ME, Taylor KD, Datta LW, Maranville JC, Liu Z, Ellis S et al. : Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans. Gastroenterology 2015, 149:1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T et al. : Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015, 47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, Helmer Q, Tillander A, Ullemar V, Van Dongen J et al. : Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nature Genetics 2017, 49:1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han Y, Jia Q, Jahani PS, Hurrell BP, Pan C, Huang P, Gukasyan J, Woodward NC, Eskin E, Gilliland FD et al. : Genome-wide analysis highlights contribution of immune system pathways to the genetic architecture of asthma. Nature Communications 2020:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Styrkarsdottir U, Lund SH, Thorleifsson G, Zink F, Stefansson OA, Sigurdsson JK, Juliusson K, Bjarnadottir K, Sigurbjornsdottir S, Jonsson S et al. : Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet 2018, 50:1681–1687. [DOI] [PubMed] [Google Scholar]
- 148.Alarcon-Riquelme ME, Ziegler JT, Molineros J, Howard TD, Moreno-Estrada A, Sanchez-Rodriguez E, Ainsworth HC, Ortiz-Tello P, Comeau ME, Rasmussen A et al. : Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture. Arthritis Rheumatol 2016, 68:932–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yucesoy B, Kaufman KM, Lummus ZL, Weirauch MT, Zhang G, Cartier A, Boulet L-P, Sastre J, Quirce S, Tarlo SM et al. : Genome-Wide Association Study Identifies Novel Loci Associated With Diisocyanate-Induced Occupational Asthma. Toxicological Sciences 2015, 146:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Wordsworth BP: The histone demethylase JARID1A is associated with susceptibility to ankylosing spondylitis. Genes & Immunity 2011, 12:395–398. [DOI] [PubMed] [Google Scholar]
- 151.Moll M, Jackson VE, Yu B, Grove ML, London SJ, Gharib SA, Bartz TM, Sitlani CM, Dupuis J, O’Connor GT et al. : A systematic analysis of protein-altering exonic variants in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol 2021, 321:L130–L143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yin X, Kim K, Suetsugu H, Bang S-Y, Wen L, Koido M, Ha E, Liu L, Sakamoto Y, Jo S et al. : Meta-analysis of 208370 East Asians identifies 113 susceptibility loci for systemic lupus erythematosus. Annals of the Rheumatic Diseases 2021, 80:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Almoguera B, Vazquez L, Mentch F, Connolly J, Pacheco JA, Sundaresan AS, Peissig PL, Linneman JG, McCarty CA, Crosslin D et al. : Identification of Four Novel Loci in Asthma in European American and African American Populations. American Journal of Respiratory and Critical Care Medicine 2017, 195:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brčić L, Barić A, Benzon B, Brekalo M, Gračan S, Kaličanin D, Škrabić V, Zemunik T, Barbalić M, Novak I et al. : AATF and SMARCA2 are associated with thyroid volume in Hashimotočs thyroiditis patients. Scientific Reports 2020:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155..Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC et al. : A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015, 47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.International Multiple Sclerosis Genetics, C: Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]


