Abstract
Small-molecule FLT3 inhibitors have recently improved clinical outcomes for patients with FLT3-mutant acute myeloid leukemia (AML) after many years of development, but resistance remains an important clinical problem. FF-10101 is the first irreversible, covalent inhibitor of FLT3 which has previously shown activity against FLT3 tyrosine kinase inhibitor resistance-causing FLT3 F691L and D835 mutations. We report that FF-10101 is also active against an expanded panel of clinically identified FLT3 mutations associated with resistance to other FLT3 inhibitors. We also demonstrate that FF-10101 can potentially address resistance mechanisms associated with growth factors present in the bone marrow microenvironment but is vulnerable to mutation at C695, the amino acid required for covalent FLT3 binding. These data suggest that FF-10101 possesses a favorable resistance profile that may contribute to improved single agent efficacy when used in patients with FLT3-mutant AML.
Keywords: AML, FLT3, TKI, Resistance, Covalent Inhibitor
Introduction
Oncogenic activating mutations in the receptor tyrosine kinase Fms-Like Tyrosine Kinase 3 (FLT3) are the most common genetic alterations in acute myeloid leukemia (AML) and confer an increased risk of relapse (1–4). Multiple clinically active FLT3 tyrosine kinase inhibitors (TKIs) have recently demonstrated efficacy as monotherapy or in combination with chemotherapy in patients with FLT3-mutant AML. Two phase 3 clinical trials evaluating potent next-generation FLT3 TKIs quizartinib (AC220) (5) and gilteritinib (ASP2215) (6) demonstrated superior response efficacy and overall survival for patients with relapsed/refractory (R/R) FLT3-mutant AML compared to salvage chemotherapy. These findings led to FDA approval of gilteritinib as the first single-agent FLT3 inhibitor in this patient population.
Despite improved activity of next-generation FLT3 TKIs, the composite complete remission rate (CRc) and median overall survival for patients treated with gilteritinib in phase 3 study was 54% and 9.3 months compared to 22% and 5.6 months for chemotherapy, respectively (6). This limited benefit reflects that some patients exhibit primary resistance and responders inevitably acquire secondary resistance if not bridged to allogeneic stem cell transplant. Type II FLT3 inhibitors such as quizartinib and sorafenib bind the inactive conformation of FLT3, and resistance to these TKIs has most often arisen due to tyrosine kinase domain (TKD) mutations at the “gatekeeper” F691 residue and the D835 residue within the activation loop, the latter of which stabilizes the active conformation of FLT3 (7–9). Type I FLT3 inhibitors like crenolanib and gilteritinib target the ATP binding site of FLT3 in its active conformation and were developed to specifically address resistance due to D835 activation loop mutations. Both crenolanib and gilteritinib have demonstrated activity in patients harboring FLT3 D835 mutations although not all patients with such a mutation respond to therapy (6,10), implying that alternate resistance mechanisms also exist. Moreover, drug resistant FLT3 F691L mutations still arise in patients receiving crenolanib and gilteritinib (11,12), highlighting that this mutation remains a clinically relevant problem.
Inadequate response to FLT3 inhibitors is also associated with transient FLT3 signaling inhibition that results from short drug half-life and significant drug-plasma binding (13,14). Gilteritinib’s increased single-agent clinical activity relative to midostaurin is thought to be in part due to better sustained in vivo FLT3 inhibition (15). Levels of FLT3 ligand (FL), the cognate agonist for the FLT3 receptor, have also been found to increase with successive courses of chemotherapy-induced aplasia in patients with AML (16). Exogenous FL exposure has been shown to blunt FLT3 TKI activity in AML cells through persistent ERK activation (17). Other growth factors within the bone marrow microenvironment, such as fibroblast growth factor 2 (FGF2), have also been implicated in FLT3 TKI resistance through apparent protective ERK signaling (18). Recently, our group and others identified parallel signaling pathway activation via secondary mutations of RAS/MAPK effectors as a major mechanism of clinical resistance to gilteritinib and crenolanib (11,12).
There has been a recent resurgence in enthusiasm for the development of covalent kinase inhibitors, valued for their potential to inhibit their target more potently and durably. Such inhibitors have transformed treatment of multiple cancers and hematologic malignancies. One such example has been the use of ibrutinib, the covalent inhibitor of Bruton Tyrosine Kinase (BTK), in patients with chronic lymphocytic leukemia. Nevertheless, despite its success, ibrutinib remains vulnerable to BTK mutation at C481 that removes the sulfhydryl group of the target cysteine required for ibrutinib covalent attachment (19). An analogous drug-resistant loss of a cysteine linker includes a C797S mutation described in patients with Epidermal Growth Factor Receptor (EGFR)-mutated lung adenocarcinoma treated with osimertinib, a clinically effective covalent EGFR inhibitor (20). Mutations of amino acids adjacent to critical drug-binding cysteine residues that structurally disrupt covalent binding are also a theoretical possibility, but this phenomenon has not yet been clinically described in hematologic malignancies.
FF-10101 is a first-in-class, type I covalent inhibitor of FLT3 with activity hypothetically limited only by FLT3 protein turnover. FF-10101 has previously been shown to inhibit proliferation of 32D cells expressing a FLT3-Internal Tandem Duplication (ITD) mutation with quizartinib-resistant D835Y, Y842C/H, and F691L TKD mutations (21). Given its binding mechanism and established activity profile, FF-10101 has the potential to overcome cell intrinsic and extrinsic factors that limit the efficacy of other FLT3 TKIs. Here, we report the activity of FF-10101 against an expanded panel of clinically relevant FLT3 mutations utilizing in vitro cell viability and biochemical experiments as well as structural modeling. These mutations include those identified in patients or by mutagenesis screening to impart resistance to a panel of clinically relevant type I and type II FLT3 inhibitors. We also assess mutations that potentially impair FF-10101’s irreversible binding to FLT3. Lastly, we compare the efficacy of FF-10101 and gilteritinib in overcoming cell extrinsic and intrinsic activators of MAPK signaling identified to induce resistance to FLT3 TKIs. Profiling of FF-10101 against potential resistance mechanisms is important as FF-10101 is currently undergoing phase 1/2a evaluation in patients with R/R AML (NCT03194685). Preliminary results of this study have been recently reported with an overall response rate of 27% (4/30 patients with AML with or without a FLT3 mutation evaluable for response achieved a form of complete remission and 4/30 exhibited partial remission) (22). Notably, patients enrolled in the study had received a median of 3 prior lines of therapy, and a majority of those entering the study with a FLT3 mutation had previously been treated with a FLT3 inhibitor.
Methods
Inhibitors.
FF-10101 and FLA-9430 were provided by FUJIFILM. Gilteritinib (#S7754), quizartinib (#S1526), crenolanib (#S2730), and midostaurin (#S8064) were purchased from Selleckchem.
Cell lines.
Ba/F3 and HS5 cells were a gift from Dr. Neil Shah (University of California, San Francisco). MOLM-14 and MV4–11 cells were a gift from Dr. Scott Kogan (University of California, San Francisco). Stable Ba/F3 cell lines were generated by retroviral spinfection as previously described (7) and all demonstrated cytokine independent growth. Ba/F3 parental cells still cytokine-dependent were continuously cultured in media supplemented with IL3. MOLM-14 (QS) NRAS-mutant and FLT3 TKD mutant cells were generated by culturing MOLM-14 parental cells in media containing escalating doses of quizartinib (0.5 to 20 nmol/L). Quizartinib-resistant cells were subcloned and Sanger sequencing performed. Unless otherwise specified cells lines were cultured in RPMI-1640 with 10% FBS and 1% penicillin/streptomycin/l-glutamine. Cells tested negative for Mycoplasma by the MycoAlert PLUS Mycoplasma Detection Kit (Lonza). Experiments were performed within three months of cell line thawing. Cell line authentication was performed at the University of California, Berkeley, DNA Sequencing Facility using short tandem repeat DNA profiling.
AML patient samples.
De-identified primary AML samples were collected, stored, and studies performed after approval of a protocol from the University of California San Francisco Hematologic Malignancies Tissue Bank by an institutional review board. Informed, written consent according to the Declaration of Helsinki was obtained from patients prior to tissue collection.
Cell-viability and apoptosis experiments.
For cell viability experiments, cells were seeded in 96-well plates and exposed to increasing concentrations of indicated drug for 48 hours. Primary AML samples were rapidly thawed, promptly diluted to a concentration of 1.0 × 106 cells/mL in HS5 conditioned media, and then cultured for 24 hours prior to drug exposure. For doxycycline-inducible NRAS cell line experiments, MOLM-14 and MV4–11 cells transduced with an inducible NRAS-WT or NRAS-mutant vector were stimulated for 24 hours with doxycycline 0.1 μg/mL and then maintained in the same concentration of doxycycline for the duration of an experiment. Cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega) and normalized to untreated control. For cleaved caspase-3 experiments, cells were cultured in complete RPMI or HS5 conditioned media for 24 hours prior to 48-hour drug exposure. CellEvent Caspase-3/7 Green Flow Cytometry Kit (ThermoFisher) was used to measure fraction of apoptotic cells by FACS. To generate conditioned media, 1.0 × 106 HS5 cells were plated in 10 mL of complete RPMI in a 10 cm dish and media harvested after 72 hours, cleared by centrifugation and filtration through a 0.22 µM membrane, and kept at −80°C until use.
Mutagenesis and resistance screen.
Ba/F3 cells harboring randomly mutagenized FLT3-ITD were generated as previously described (7). Ba/F3 cells were selected with 100 or 200 nM FF-10101 in soft agar. Genomic DNA was isolated from resistant Ba/F3 cells and the FLT3 TKD was amplified and sequenced as described (7).
Immunoprecipitation (IP) and immunoblotting assays.
Exponentially growing cells were plated in complete RPMI or HS5 conditioned media when noted and treated with specified drug at indicated concentration. After drug exposure cells were washed in PBS and lysed in buffer (50 mM HEPES, pH 7.4, 10% glycerol, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2) supplemented with protease and phosphatase inhibitors. The lysate was clarified by centrifugation and quantitated by BCA assay (Thermo Scientific). For MOLM-14 IP experiments, FLT3 was immunoprecipitated from 400 ug of total protein using anti-FLT3 antibody (Sino Biological #10445-RP02, RRID:AB_2894912) with samples then resolved on a 10% Bis-Tris gel and transferred to nitrocellulose membranes. Immunoblotting was performed using anti-phosphotyrosine antibody (clone 4G10, EMD Millipore #05–321, RRID:AB_309678) and anti-FLT3 antibody. For remaining western blots, immunoblotting was performed using anti–phospho-FLT3 (Tyr591, Cell Signaling #3461, RRID:AB_331060), anti–phospho-STAT5 (Tyr694, Cell Signaling #9351, RRID:AB_2315225), anti-STAT5 (D2O6Y, Cell Signaling #94205, RRID:AB_2737403), anti–phospho-ERK1/2 (Thr202/Tyr204, Cell Signaling #9101, RRID:AB_331646), anti-ERK1/2 (3A7, Cell Signaling #9107, RRID:AB_10695739), anti-phospho-AKT (Ser473, Cell Signaling #4060, RRID:AB_2315049), anti-AKT (Cell Signaling, #9272, RRID:AB_329827), and anti-FLT3 S-18 antibody (Santa Cruz Biotechnology #sc-480, RRID:AB_2104968).
Structural modeling of FLT3 inhibitors bound to FLT3.
Binding modes between FF-10101 and FLT3 and between quizartinib and FLT3 were investigated with reported co-crystal structures (5X02 and 4RT7) (8,21). Modeled structures of midostaurin and crenolanib bound to FLT3 were constructed with a homology model of active form FLT3 using c-Kit (PDB:1PKG) and co-crystal structures of the drugs reported (4NCT and 6BQP) (23,24).
Data availability.
The data generated in this study are available within the article and its supplementary data files.
Results
FF-10101 displays activity against all mutations associated with resistance to type II FLT3 inhibitors.
For our experiments we utilized Ba/F3 cells transduced to express FLT3-ITD (Ba/F3-ITD) with and without TKD mutations of interest. We first assessed the activity of FF-10101 against a compendium of mutations at the D835 residue that exhibit broad differential sensitivity to type II FLT3 TKIs (25). FF-10101 was previously shown to potently suppress growth of 32D cells harboring isolated FLT3 D835 mutations in the absence of an ITD, including FLT3 D835H/Y/V/E/N (21). We assessed FF-10101 activity against these same mutations in Ba/F3-ITD cells and found FF-10101 still possessed potent anti-proliferative activity against all such cell lines (Fig. 1A). We found similar activity against MOLM-14 FLT-ITD+ human AML cells harboring the same D835H/Y/V/N mutations (Supplementary Fig. 1). Additionally, FF-10101 potently inhibited Ba/F3-ITD cells with FLT3 D835 deletion and four other amino acid substitutions not previously tested against FF-10101 (F/I/A/G, Fig. 1A). Among these mutations, FLT3 D835F/I/del have demonstrated marked in vitro resistance to type II FLT3 inhibitors (25), and D835F/I/A have been clinically identified in patients receiving FLT3 TKI therapy (7,15,26). FLT3 D835del and D835G have separately been catalogued in AML patients in the Sanger Cosmic Database. Altogether, these data reinforce that FF-10101 should be invulnerable to all known type II inhibitor-resistant FLT3 D835 mutations.
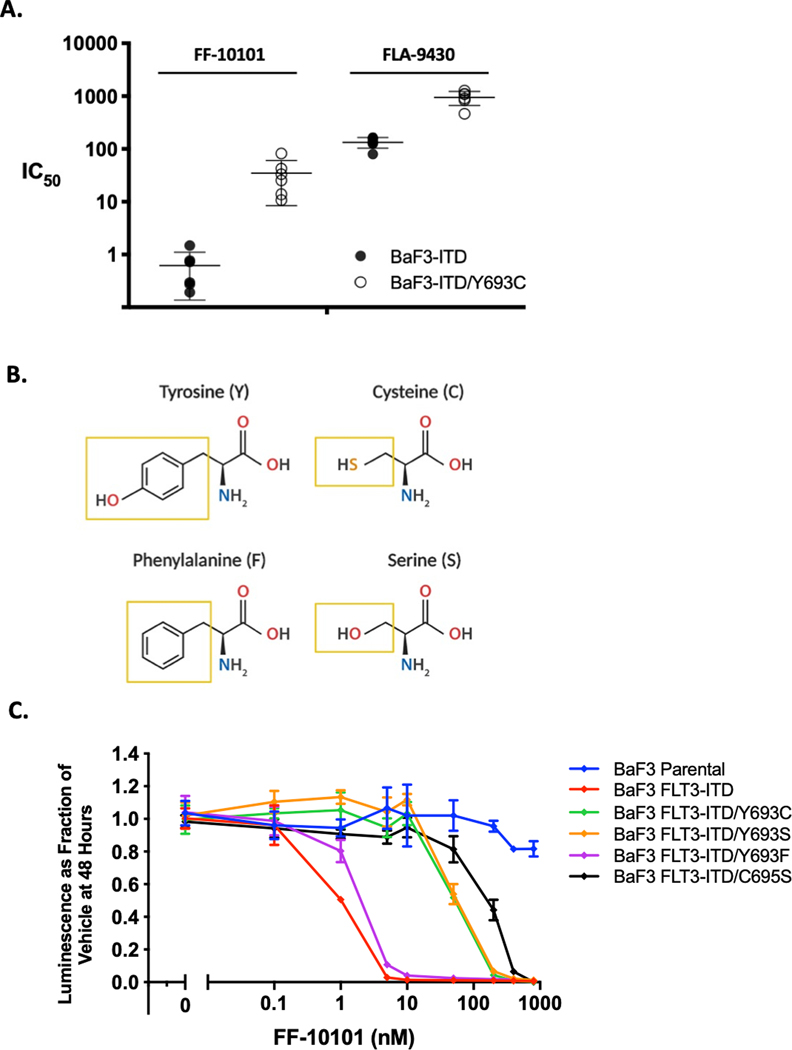
Figure 1. FF-10101 demonstrates favorable in vitro activity against FLT3 KD mutations.
(A) Fold increase in resistance of indicated FLT3-ITD + TKD mutations when compared to FLT3-ITD alone. Mutations with IC50 <10-fold higher than FLT3-ITD defined as Sensitive (blue boxes), those with IC50 10–50-fold higher than FLT3-ITD defined as Moderate Resistance (orange boxes), and those with IC50 >50-fold higher than FLT3-ITD defined as Severe Resistance (red boxes). (B) Assessment of FLT3 autophosphorylation and downstream effectors STAT5, AKT, and ERK by western blot analysis in Ba/F3 cells harboring FLT3-ITD and the indicated TKD mutant isoform in the presence of increasing concentration of FF-10101 for 1 hour. Not all cell lysates were resolved on the same gel and thus total protein for each target are used as loading control and normalization across gels. For all gels, p-ERK and ERK were blotted for simultaneously while membranes with visualized phospho-protein were stripped and re-blotted with total protein antibody for remaining targets. (C) Relative proliferation of all cysteine to serine Ba/F3 mutant cell lines after 48-hour exposure to FF-10101. Error bars represent SD of three technical replicates from a single experiment representative of three independent experiments.
FF-10101 demonstrates favorable in vitro activity against FLT3 TKD mutations causing resistance to type I FLT3 TKIs.
We next evaluated FF-10101 activity against Ba/F3-ITD cells harboring FLT3 F691L and found these to be ∼8-fold more resistant to FF-10101 than cells with FLT3-ITD alone (Fig. 1A), a similar degree of sensitivity previously seen in 32D cells (21). In contrast, Ba/F3-ITD/F691L cells treated with gilteritinib exhibited an ∼14-fold increase in resistance (Fig. 1A). The relative resistance of the F691L mutation compared to FLT3-ITD was approximately the same with crenolanib as with FF-10101 (Fig. 1A), but crenolanib has exhibited less overall potency as a FLT3 inhibitor with higher IC50 values as compared to FF-10101 or gilteritinib (Table 1). Notably, patients treated with gilteritinib and crenolanib have developed F691L mutations at the time of resistance (11,27).We next directly compared the in vitro activity of FF-10101 against other FLT3 TKD mutations predicted to cause resistance to gilteritinib. These mutations were recently identified by our group using a saturation mutagenesis screen and confirmed to confer moderate-to-severe in vitro resistance to gilteritinib (27). These mutations included Y693C or N, G697S, and D698N, all TKD mutations that lie outside the FLT3 activation loop. Notably, a previous mutagenesis screen identified D698N and Y693C as crenolanib-resistant mutations (28), and D698N has also been associated with resistance to the type II FLT3 TKI pexidartinib (8). While G697S has not been previously identified, G697R was isolated in a mutagenesis screen for midostaurin resistance (29). In Ba/F3-ITD cells, G697S, Y693C, and Y693N conferred at least moderate resistance to both gilteritinib and FF-10101 (Fig. 1A). The D698N mutation, however, was sensitive to FF-10101 but caused 17-fold increase in resistance to gilteritinib (relative to FLT3-ITD alone) (Fig. 1A).
Table 1. Mean IC50 of FF-10101 against FLT3 mutant isoforms as compared to other type I and type II FLT3 inhibitors.
Mean reported from at least 3 biologic experimental replicates. IC50 concentrations for specific drugs have been previously reported where indicated.
| FF-10101 | Gilteritinib1 | Crenolanib2,3 | Quizartinib2,3 | Midostaurin | |
|---|---|---|---|---|---|
|
|
|||||
| FLT3-ITD | 1.8 | 1.8 | 11.2 | 0.4 | 5.8 |
| FLT3-ITD/F691L | 14.5 | 25.4 | 55.4 | 151.1 | 6.2 |
| FLT3-ITD/D698N | 5.1 | 31.5 | 85.3 | 0.6 | 65.3 |
| FLT3-ITD/Y693C | 116.1 | 27.9 | 143.0 | 2 2 | 171.7 |
| FLT3-ITD/Y693N | 25.9 | 23.8 | 129.3 | 1.8 | 213.3 |
| FLT3-ITD/G697S | 22.3 | 117.8 | 146.8 | 6.2 | 169.6 |
| FLT3-ITD/K429E | 1.1 | 6.0 | 26.1 | 0.7 | 20.3 |
| FLT3-ITD/N676K | 1.9 | 3.6 | 27.8 | 1.8 | 33.8 |
| FLT3-ITD/D835V | 4.4 | 3.5 | 13.1 | 176.7 | 4.7 |
| FLT3-ITD/D835Y | 11,1 | 1.1 | 13,9 | 56.7 | 6.8 |
| FLT3-ITD/D835H | 8.5 | 1.1 | 12.4 | 14.0 | 3.9 |
| FLT3-ITD/D835F | 13.0 | 1.2 | 16.5 | 457.0 | 5.3 |
| FLT3-ITD/D835I | 7.1 | 0.9 | 17.3 | 222.7 | 5.2 |
| FLT3-ITD/D835del | 6.0 | 4.2 | 184 | 99.3 | 5.3 |
| FLT3-ITD/D835A | 6.5 | 1.3 | 17.5 | 3 2 | 5.0 |
| FLT3-ITD/D835E | 11.2 | 1.4 | 17.4 | 1.8 | 5.8 |
| FLT3-ITD/D835G | 6.3 | 2.3 | 18.2 | 3.0 | 4.6 |
| FLT3-ITD/D835N | 4.7 | 0.9 | 16.8 | 2.1 | 5 8 |
| FLT3-ITD/F691L/D835V | 76.8 | 17.8 | 62.5 | ||
| FLT3-ITD/D698N/D835V | 24.7 | 31.1 | 68.1 | ||
| FLT3-ITD/Y693C/D835V | 399.9 | 34.2 | 156.3 | ||
| FLT3-ITD/C695S | 316.6 | ||||
| FLT3-ITD/C681S | 6.7 | ||||
| FLT3-ITD/C790S | 3.3 | ||||
| FLT3-ITD/C807S | 14.2 | ||||
| FLT3-ITD/C828S | 11.1 | ||||
| FLT3-ITD/C694S | 14.2 | ||||
Tarver TC, et al. Blood Adv. 2020 Feb 11;4(3):514–524.
Smith CC, et al. Proc Natl Acad Sci U S A. 2014 Apr 8;111(14):5319–24.
Smith CC, et al. Leukemia. 2015 Dec;29(12):2390–2.
Next, we assessed FF-10101 activity against a FLT3 K429E extracellular domain mutation identified in a single patient who developed resistance to crenolanib (11). FF-10101 potently suppressed the proliferation of Ba/F3-ITD/K429E cells (Fig. 1A). FF-10101 was also active against FLT3 N676K, another mutation identified at relapse in an AML patient treated with midostaurin (30).
To better characterize the downstream pathways associated with FF-10101 resistance, we performed western blot analysis of signaling activation in Ba/F3 cells treated with increasing concentrations of FF-10101. Compared to sensitive Ba/F3-ITD cells, drug-resistant Ba/F3-ITD/Y693C cells exhibited persistent FLT3 autophosphorylation and downstream activation of phospho-STAT5, -AKT, and -ERK up to 100 nM of FF-10101. Conversely, FF-10101-sensitive BaF3-ITD cells with N676K, D698N, or D835V mutations showed concentration-dependent inhibition of FLT3 and its downstream signaling (Fig. 1B). To evaluate the possibility that FF-10101 could also inhibit pro-survival pathways downstream of FLT3 in a non-FLT3-dependent manner, we also assessed signaling in cytokine-dependent Ba/F3 parental cells with and without IL3 growth factor stimulation and in the presence or absence of FF-10101. We found that FF-10101 had no apparent influence on IL3-stimulated ERK, AKT, or STAT5 activation (Supplementary Fig. 2), consistent with the expectation that a significant proportion of FF-10101’s inhibitory activity is due to direct FLT3 inhibition.
Compound FLT3 KD mutations cause resistance to FF-10101.
In addition to single point mutations within a TKD, “compound” mutations or two mutations co-occurring in the same TKD are well-described during sequential BCR-ABL-targeted TKI therapy in CML. Such mutations confer additional resistance to ponatinib, the potent third generation BCR-ABL inhibitor (31). While not yet described in AML, analogous compound mutations could emerge with increasing clinical use of FLT3 TKIs with non-overlapping resistance profiles. To assess the efficacy of FF-10101 against compound FLT3 TKD mutations, we tested FF-10101 against Ba/F3-ITD cells containing D835V and three second-site amino acid substitutions that confer resistance to type I FLT3 TKIs. We found that ITD-D698N/D835V and ITD-F691L/D835V induced moderate resistant to FF-10101, while ITD-Y693C/D835V exhibited severe resistance (Table 1).
FF-10101 activity relies upon only a single covalent bond at C695.
Efforts to develop selective, covalent inhibitors have shown that a small molecule may unexpectedly and flexibly target more than one binding cysteine (32), raising the possibility that a drug may rely upon multiple contact cysteine residues for covalent attachment. Previously, the crystal structure of FF-10101 bound to FLT3 demonstrated covalent bond formation at cysteine 695 (21). We sought to confirm C695 as the lone cysteine residue targeted irreversibly by FF-10101 by testing its activity against Ba/F3-ITD cells harboring single serine substitutions at each cysteine residue in the FLT3 TKD. Except for C695S, every C to S substitution in the FLT3 KD demonstrated sensitivity to FF-10101 (Fig. 1C), confirming C695 as the lone site of FF-10101 covalent bonding.
Randomized mutagenesis screening affirms the critical nature of FF-10101 covalent bonding at C695.
We again utilized a well-validated random mutagenesis screening technique in Ba/F3 cells expressing FLT3-ITD to identify additional novel FLT3 TKD mutations that might induce resistance to FF-10101. Based upon a trough plasma concentration of >90 mg/mL achieved in phase 1 clinical study (22), we performed mutagenesis screening using 100 and 200 nM FF-10101 in 300 million cells per drug concentration. We identified FLT3 C695Y in four colonies and FLT3 C695R in another seven colonies out of a total of 12 drug-resistant colonies ultimately isolated. We expect these mutations to be FF-10101-resistant by similar loss of covalent attachment as C695S. FLT3 M664I was also co-identified in two C695-mutant colonies but follow up evaluation in Ba/F3-ITD/M664I cells showed this mutation did not independently induce FF-10101 resistance (Supplementary Fig. 3). An additional drug-resistant colony in which FLT3-ITD was amplified demonstrated no identifiable TKD mutation. These results further confirm that mutation of C695 that renders FF-10101 incapable of covalently binding FLT3 may be a frequent cause of on-target clinical resistance to FF-10101.
Structural modeling of FLT3 inhibitors bound to mutant FLT3 clarify mechanisms of differential TKI sensitivity.
To corroborate our cell viability experiments, we performed structural modeling of FLT3 harboring amino acid substitutions of interest bound to FF-10101 and other clinically developed FLT3 TKIs. We focused attention on the Y693C mutation as it appeared nearly as resistant to FF-10101 as FLT3 C695S, suggesting that cysteine substitution at Y693 compromises FF-10101’s capacity to function as a covalent inhibitor. Utilizing the co-crystal structure of FF-10101 with FLT3, we found that FF-10101 relies upon an aromatic π-π interaction with Y693 to orient the electrophilic acrylamide portion of FF-10101 needed for covalent bond formation near the cysteine residue at 695 (Fig. 2A). This critical π-π interaction is lost with cysteine substitution at residue 693. Crenolanib, midostaurin, and quizartinib also demonstrate π-π interactions with Y693 (Fig. 2A), although quizartinib forms other interactions with FLT3, including a hydrogen bond with the hinge region and hydrophobic interactions in the back-pocket. These additional interactions likely contribute to quizartinib’s retained activity against Y693C. In a previous docking model of gilteritinib with FLT3 (27), it was reported that a phenyl ring within gilteritinib’s structure also formed a π-π interaction with Y693, thus likely also explaining gilteritinib’s modest loss of activity against Y693C and N.
Figure 2. Structural modeling of FLT3 inhibitors bound to resistant mutants.
(A) Mapped interactions focused upon Y693 residue with FF-10101, crenolanib, midostaurin, and quizartinib. Two-way arrows indicate distance (Å) between an aromatic moiety of each respective drug forming a π-π interaction with tyrosine. (B) Hinge region of FLT3 mapped interaction with FF-10101 and midostaurin. (C) FF-10101 and midostaurin mapped against D698. FF-10101 exhibits no interaction while the N-methyl moiety of midostaurin forms hydrogen bond with D698.
When specifically examining the binding mode of FF-10101 in comparison to midostaurin, a mutation at N676 de-stabilized midostaurin’s engagement with the FLT3 hinge region through loss of hydrogen bonding at E692 and K826 (Fig. 2B). FF-10101, on the other hand, exhibited weak or no interaction with the hinge region (Fig. 2B). These findings appeared generally consistent with our cell viability experiments in which BaF3-ITD/N676K cells displayed a relative resistance to midostaurin in terms of increased half maximal inhibitory concentration (Table 1) while remaining sensitive to FF-10101.
In concordance with FF-10101’s observed activity against the D698N mutation, we found that FF-10101 did not interact with D698. In contrast, midostaurin engaged D698 by a hydrogen bond (Fig. 2C). In prior modeling, gilteritinib docking also required an aspartic acid at the FLT3 698 position to maintain a hydrogen bonding interaction network with N701 as loss of this network allowed N701 to sterically clash with gilteritinib in a manner that disfavored drug binding (27). Crenolanib also interacts with D698 by formation of a salt bridge (28). These findings likely explain FF-10101’s greater relative activity against D698N in comparison to other type I FLT3 TKIs.
Finally, we analyzed the binding mode of FF-10101 with FLT3 G697S given the severe gilteritinib resistance seen with this mutation. Prior gilteritinib docking implicated loss of a CH-π interaction with G697 and steric clash of the G697S sidechain with gilteritinib as likely contributors to severe resistance (27). Interestingly, our modeling did not predict any unfavorable interaction between FF-10101 and G697 or G697S (Supplementary Fig. 4A-B). A molecular dynamics simulation using midostaurin previously suggested that a G697R mutation may shield access of midostaurin to the type I inhibitor binding pocket via disrupted orientation of the P-loop (33). Such a dynamic structural mechanism could also explain the moderate resistance of G697S to FF-10101 in the absence of direct interaction of the residue with FF-10101.
An aromatic π-π interaction between FF-10101 and Y693 regulates FF-10101 covalent binding at C695.
Using the information implied by our structural modeling, we next employed a combined pharmacologic and mutagenesis approach to confirm the mechanism by which Y693C interferes with FF-10101 covalent binding. First, we utilized FLA-9430, a compound identical to FF-10101 except for absence of the acrylamide double bond necessary for covalent bond formation with FLT3, thus rendering FLA-9430 a reversible inhibitor (21) (Supplementary Fig. 4C). Using cell viability experiments in Ba/F3-ITD cells, we found that Y693C exhibited an ∼100-fold increase in relative resistance to FF-10101 as compared to FLT3-ITD alone, while FLA-9430 exhibited only a ∼10-fold increase, strongly suggesting that Y693C impedes FF-10101’s covalent bond to FLT3 (Fig. 3A). To further establish the importance of the aromatic sidechain of tyrosine at residue 693, we generated Ba/F3-ITD cells with either a phenylalanine or serine substitution at Y693 (Fig. 3B). We reasoned that a substitute phenylalanine sidechain that recapitulates the native aromatic interaction with Y693 would remain sensitive to FF-10101 while a non-aromatic serine substitution would behave similarly to Y693C. Indeed, we found that Ba/F3-ITD/Y693F cells retained sensitivity to FF-10101 while cells with Y693S were as resistant to FF-10101 as those with Y693C (Fig. 3C). These findings suggest any non-aromatic amino acid substitution at residue 693 would be particularly resistant to FF-10101 and possibly drive clinical relapse in patients.
Figure 3. FF-10101 activity relies upon a single covalent bond at C695 that may be influenced by an adjacent TKD mutation at Y693.
(A) Scatter dot plot showing relative IC50 of FF-10101 or FLA-9430 against Ba/F3-ITD and Ba/F3-ITD/Y693C mutant cell lines from proliferation experiments. Each IC50 measured in six separate experiments with bars indicating mean and SD. (B) Chemical structures of amino acids evaluated at residue 693 with respective sidechains highlighted (yellow squares). (C) Relative proliferation after 48-hour exposure of indicated mutant Y693 cell lines to serial concentrations of FF-10101 in a single experiment with three technical replicates that is representative of three independent experiments. C695S dose-response curve used as relative indicator for loss of covalent drug binding.
FF-10101 may overcome cell extrinsic FLT3 TKI resistance.
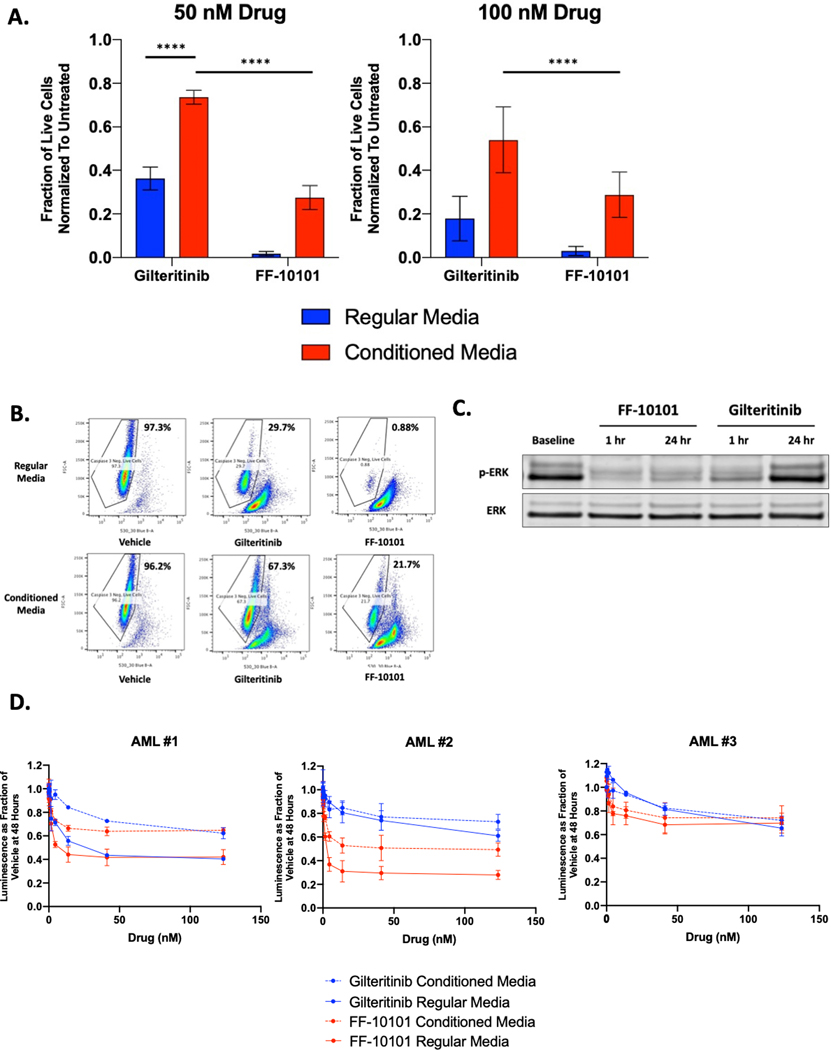
Early use of FLT3 inhibitors in AML patients demonstrated clearance of circulating blasts in the peripheral blood with minimal blast reduction in the bone marrow (34). Cytokine and bone marrow stroma-stimulated ERK activation has since been identified as a critical pro-survival signal for FLT3-ITD+ blasts during FLT3 TKI treatment (17,18). We compared the capacity of FF-10101 or gilteritinib to induce apoptosis in FLT3-ITD+ human AML cells cultured in the presence of conditioned media harvested from an HS5 immortalized human bone marrow stromal cell line that secretes FGF2 and other supportive growth factors that cause resistance to FLT3 TKIs (18,35). We found that FF-10101 induced significantly more apoptosis of MOLM-14 cells in HS5 conditioned media compared to gilteritinib at 50 and 100 nM doses of each drug (Fig. 4A-B). These results appeared to be in part cell line and dose-dependent as in MV4–11 cells, another FLT3-ITD+ human AML cell line, FF-10101 also exhibited greater cytotoxicity at a 50 nM drug dose but similar cytotoxicity as gilteritinib at 100 nM (Supplementary Fig. 5A).
Figure 4. As a covalent FLT3 inhibitor, FF-10101 may overcome off-target mechanisms that impede the activity of reversible FLT3 inhibitors.
(A) Apoptosis measured in MOLM-14 cells following 48-hour treatment with gilteritinib or FF-10101 at indicated concentrations in regular complete RPMI media or HS5 conditioned media. Live cells negative for caspase-3 were normalized to untreated control cells. Results represent aggregate data from three independent experiments each with two technical replicates. Error bars represent SD and statistical analysis performed using paired t-test. ****, P<0.0001. (B) Representative FACS plot to demonstrate gating of cell populations negative for CellEvent (caspase-3 stain) in 50 nM drug experiment summarized in (A). (C) Assessment of ERK phosphorylation by western blot in MOLM-14 cells treated with FF-10101 (50 nM) or gilteritinib (50 nM) for 1 and 24-hours in HS5 conditioned media. (D) 48-hour dose-response evaluation of FF-10101 and gilteritinib against three primary FLT3-mutant AML samples using CellTiterGlo. Cells were continuously cultured in either regular (solid line) or conditioned media (dotted line) during drug exposure. Each graph in panel (D) is representative of a single experiment with three technical replicates performed in two independent experiments for each primary sample.
To explain these findings, we hypothesized that FF-10101 could suppress adaptive ERK reactivation in response to FLT3 inhibition, a phenomenon that has been previously observed and is augmented by conditioned media (36). In MOLM-14 cells, we found that FF-10101 exhibited superior inhibition of FLT3 phosphorylation by FF-10101 after 1 and 24 hours of 50 nM drug exposure as compared to gilteritinib (Supplementary Fig. 2B). While these changes were subtle, FF-10101 did strikingly suppress rebound ERK re-activation after 24 hours of drug exposure while gilteritinib did not (Fig. 4C).
We next assessed whether FF-10101 could exert similar activity in three primary FLT3-mutant AML samples also supported by HS5 conditioned media. The clinical characteristics of these AML patients are summarized in Supplementary Table 1. Of these three samples, two (AML #1 and #3) were obtained from patients with newly diagnosed FLT3-ITD+ AML while the third (AML #2) came from a patient with new AML and a FLT3 D835V mutation. When compared directly with gilteritinib in either regular or protective HS5 conditioned media, we found that FF-10101 again appeared to induce greater cytotoxicity of AML cells at drug doses at or below 50 nM in all three samples (Fig. 4D). At doses of 100 nM (the approximate plasma concentration of gilteritinib at steady state in patients receiving 120 mg/day), the activity of FF-10101 and gilteritinib appeared similar in the two FLT3-ITD+ samples while FF-10101 induced greater cytotoxicity in AML #2. These findings suggest that FF-10101 may have greater potential to overcome bone marrow microenvironment-mediated TKI resistance than gilteritinib and consequently be more active for a subset of patients with FLT3-mutant AML, especially at sub maximal plasma concentrations.
Cell intrinsic NRAS co-mutations that cause resistance to most FLT3 TKIs are likely to also cause resistance to FF-10101.
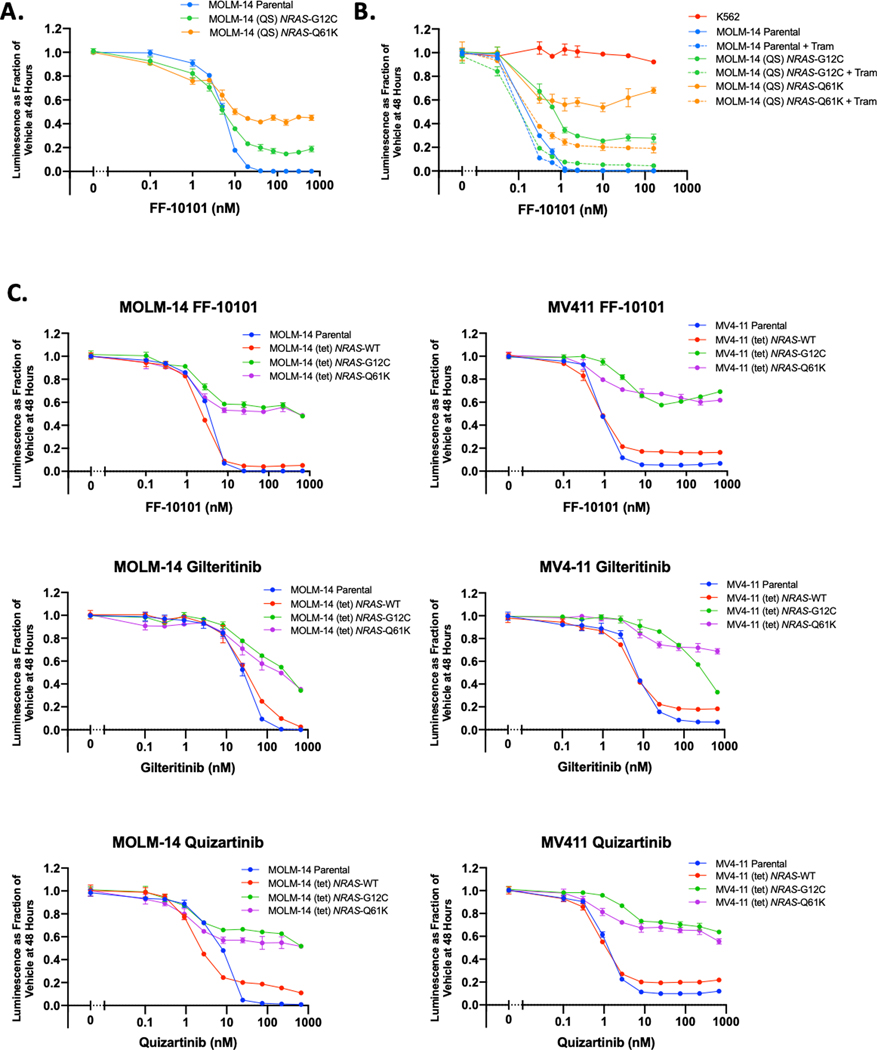
Finally, we evaluated FF-10101 against MOLM-14 cells harboring secondary NRAS-G12C and Q61K mutations following long-term selection with quizartinib (herein MOLM-14 (QS) NRAS-G12C or Q61K cells). Secondary NRAS mutations have been associated with clinical gilteritinib resistance, and MOLM (QS) NRAS-mutant cells have previously demonstrated in vitro resistance to gilteritinib (12). We found that while FF-10101 modestly suppressed proliferation, MOLM-14 (QS) NRAS-mutant cells exhibited relative resistance to FF-10101 (Fig. 5A). Consistent with previous reports with gilteritinib (12), addition of a MEK inhibitor (trametinib) at low dose abrogated a significant degree of this resistance (Fig. 5B). We affirmed that mutant NRAS alone is sufficient to induce this resistance by using MOLM-14 and MV4–11 cells transduced with a doxycycline-inducible expression vector containing NRAS-WT, NRAS-G12C, or NRAS-Q61K (MOLM-14 (tet) and MV4–11 (tet) cells). We previously demonstrated that 24-hour induction of mutant NRAS isoform expression in MOLM-14 (tet) and MV4–11 (tet) cells activates expected downstream pro-survival signaling via ERK and AKT (12). Induction of NRAS-G12C or NRAS-Q61K generated significant resistance to FF-10101, gilteritinib, and quizartinib in both MOLM-14 (tet) and MV4–11 (tet) cells (Fig. 5C), indicating that patients treated with FF-10101 are likely to remain vulnerable to relapse driven by oncogenic MAPK pathway activation.
Figure 5. Secondary activating NRAS mutations are not overcome by FF-10101.
(A) Relative proliferation of MOLM-14 parental cells and MOLM-14 (QS) NRAS-G12C or NRAS-Q61K cells following 48-hour exposure to FF-10101 at serial concentration. (B) Relative proliferation MOLM-14 parental or MOLM-14 (QS) NRAS-mutant cells following 48-hour exposure to serial concentrations of FF-10101 in combination with a fixed dose of trametinib 5 nM. (C) 48-hour dose-response evaluation of FF-10101 (top row), gilteritinib (middle row), and quizartinib (bottom row) in MOLM-14 (tet) and MV4–11 (tet) cells. Expression of NRAS-WT or NRAS mutants G12C and Q61K was induced with doxycycline for 24 hours prior to beginning drug exposure. All panels are single experiments performed with three technical replicates and representative of three independent experiments.
Discussion
The clinical utility of FLT3 inhibition in FLT3-mutant AML has been increasingly realized with the approvals of midostaurin in combination with chemotherapy for newly diagnosed patients (37) and gilteritinib as monotherapy for patients with R/R disease. Rigorous preclinical evaluation of several FLT3 TKIs, correlative studies from clinical trials, and serial FLT3 sequencing in AML patients treated with FLT3 TKIs has generated significant insight into the requisite traits for a clinically effective FLT3 inhibitor, most importantly the capability of an inhibitor to overcome FLT3 and non-FLT3 mechanisms of resistance.
Here, we present evidence that the first covalent FLT3 inhibitor FF-10101 is highly active against the majority of TKD mutations that render patients resistant to other FLT3 TKIs in clinical use. Importantly, FF-10101 exerts greater in vitro potency than gilteritinib against the gatekeeper F691L mutation. It should be noted that gilteritinib exhibited a similar preclinical profile and may effectively suppress an F691L clone when administered at doses higher than the FDA-approved dose of 120 mg (27). Nevertheless, such a therapeutic strategy has not been evaluated clinically. In the absence of such experience, these data suggest that FF-10101 could serve as an effective second line TKI for FLT3-mutant AML patients who relapse with known resistance-conferring FLT3 TKD mutations.
FF-10101 is also generally active against several FLT3 mutations identified in mutagenesis screening evaluating resistance to other FLT3 inhibitors although loses significant activity when covalent binding is compromised. We confirm that this impairment from mutation at Y693 is due to loss of a π-π interaction required to position FF-10101 for covalent bond formation at C695. Mutations at C695 or Y693 have not been isolated yet in a patient treated with a FLT3 TKI, but our analysis suggests that both mutations should be carefully monitored for in patients receiving FF-10101. Altogether, our studies reinforce growing evidence that unique on-target resistance profiles should be considered when selecting the most likely effective FLT3 inhibitor for individual patients.
Finally, we demonstrate that FF-10101 often exerts greater cytotoxicity than gilteritinib at similar dose levels (<100 nM) in AML cell lines and primary AML blasts cultured in a conditioned media model of the tumor microenvironment, possibly due to suppression of ERK re-activation following extended drug exposure. These results could be explained in multiple ways. First, FF-10101 possesses a differential inhibitory profile and targets non-FLT3 kinases that also stimulate ERK via parallel pathways (i.e. KIT, IRAK4) (21). Alternatively, the degree of ERK reactivation in AML cells is still modulated by FLT TKIs in a dose dependent manner (36), suggesting that ERK reactivation is partially driven by FLT3 signaling. AML cells also can express FL to stimulate autocrine FLT3 signaling (38). These observations support the possibility that superior FLT3 receptor occupancy owing to FF-10101’s covalent binding may play a role in increased suppression of ERK reactivation when compared to gilteritinib. Lastly, other covalent inhibitors have been shown to stabilize and prolong their target kinase’s half-life while silencing downstream pathways several hours after drug washout, suggesting that covalent modification may lead to slower kinase turnover and reduced native kinase synthesis (39). It is possible that FF-10101 could influence FLT3 receptor expression in a similar manner.
Nevertheless, secondary RAS/MAPK activating mutations have already arisen as a primary off-target resistance mechanism to gilteritinib and appear to confer resistance to FF-10101. Parallel or partially redundant signaling pathway activation may become a relevant mediator of clinical resistance to FF-10101 as on-target FLT3 inhibition is maximized, although evidence for this remains to be seen in clinical studies.
Careful structural and functional characterization of investigational FLT3 inhibitors such as FF-10101 as reported in this study are critical to anticipate mechanisms of resistance and identify optimal clinical uses. FLT3 TKI-naïve and TKI-exposed patients harbor genetically heterogeneous populations of FLT3-ITD, TKD, and co-mutant subclones that determine clinical response to TKIs with unique resistance profiles (40). Inter- and intra-patient clonal heterogeneity will likely be increasingly recognized with more widespread use of clinical NGS, high throughput single-cell genotyping, and other molecular analysis of AML tumors at multiple therapeutic timepoints. With careful resistance profiling of any FLT3 TKI, the nature of drug-resistant subclones that could gain competitive fitness under selective drug pressure may be reasonably inferred and this information used to optimally sequence TKI therapy for an individual patient. In the future, selection of FLT3-mutant AML therapy may then be guided by mutational analysis and side effect profiling akin to how BCR-ABL inhibitors are selected in BCR-ABL-driven leukemias.
Notably, in recent reports of the phase 1/2a evaluation of FF-10101 in patients with R/R AML, four patients who experienced a partial remission had previously been treated with a FLT3 TKI and one patient who achieved a complete remission had already exhibited disease progression on gilteritinib (22). These results and our findings show that FF-10101 has potential to provide clinical benefit to AML patients that acquire certain FLT3 TKD resistance mutations or following treatment with other FLT3 inhibitors.
Supplementary Material
Acknowledgments
Financial Support:
C.C.S is a Damon Runyon-Richard Lumsden Foundation Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-99–18). This work was also supported by a Research Scholar Grant from the American Cancer Society (132032-RSG-18–063-01-TBG).
Disclosure of Conflicts of Interest:
C.C.S. has received research support from Astellas Pharma, Revolution Medicines, AbbVie and FUJIFILM Corporation and has served as an advisory board member for Astellas Pharma, Genentech and Daiichi Sankyo. D.T. and M.A. are employees of FUJIFILM. A.C.L. has received research support from Astellas Pharma, Autolus Therapeutics, Jazz Pharmaceuticals, Kadmon Corporation, Kite Pharma, and Pharmacyclics and has served as a consultant to AbbVie, Amgen, and Pfizer. The remaining authors declare no competing financial interests.
References
- 1.Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002;99:4326–35 [DOI] [PubMed] [Google Scholar]
- 3.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 2001;98:1752–9 [DOI] [PubMed] [Google Scholar]
- 4.Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood 2002;100:4372–80 [DOI] [PubMed] [Google Scholar]
- 5.Cortes JE, Khaled S, Martinelli G, Perl AE, Ganguly S, Russell N, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2019;20:984–97 [DOI] [PubMed] [Google Scholar]
- 6.Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. N Engl J Med 2019;381:1728–40 [DOI] [PubMed] [Google Scholar]
- 7.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012;485:260–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith CC, Zhang C, Lin KC, Lasater EA, Zhang Y, Massi E, et al. Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 “Gatekeeper” F691L Mutation with PLX3397. Cancer Discov 2015;5:668–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Man CH, Fung TK, Ho C, Han HH, Chow HC, Ma AC, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood 2012;119:5133–43 [DOI] [PubMed] [Google Scholar]
- 10.Cortes JE, Kantarjian HM, Kadia TM, Borthakur G, Konopleva M, Garcia-Manero G, et al. Crenolanib besylate, a type I pan-FLT3 inhibitor, to demonstrate clinical activity in multiply relapsed FLT3-ITD and D835 AML. Journal of Clinical Oncology 2016;34:7008- [Google Scholar]
- 11.Zhang H, Savage S, Schultz AR, Bottomly D, White L, Segerdell E, et al. Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat Commun 2019;10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov 2019;9:1050–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levis M BP, Smith BD, Stine A, Pham R, Stone R, Deangelo D, Galinsky I, Giles F, Estey E, Kantarjian H, Cohen P, Wang Y, Roesel J, Karp JE, Small D. Plasma inhibitory activity (PIA): a pharmacodynamic assay reveals insights into the basis for cytotoxic response to FLT3 inhibitors. Blood 2006;108:3477–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratz KW CJ, Roboz GJ, Rao N, Arowojolu O, Stine A, Shiotsu Y, Shudo A, Akinaga S, Small D, Karp JE, Levis M. A pharmacodynamic study of the FLT3 inhibitor KW-2449 yields insight into the basis for clinical response. Blood 2009;113:3938–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LY, Hernandez D, Rajkhowa T, Smith SC, Raman JR, Nguyen B, et al. Preclinical studies of gilteritinib, a next-generation FLT3 inhibitor. Blood 2017;129:257–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, Yang X, Knapper S, White P, Smith BD, Galkin S, et al. FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood 2011;117:3286–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Sexauer A, Levis M. Bone marrow stroma-mediated resistance to FLT3 inhibitors in FLT3-ITD AML is mediated by persistent activation of extracellular regulated kinase. Br J Haematol 2014;164:61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traer E, Martinez J, Javidi-Sharifi N, Agarwal A, Dunlap J, English I, et al. FGF2 from Marrow Microenvironment Promotes Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia. Cancer Res 2016;76:6471–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagal-Shamanna R, Jain P, Patel KP, Routbort M, Bueso-Ramos C, Alhalouli T, et al. Targeted multigene deep sequencing of Bruton tyrosine kinase inhibitor-resistant chronic lymphocytic leukemia with disease progression and Richter transformation. Cancer 2019;125:559–74 [DOI] [PubMed] [Google Scholar]
- 20.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaura T NT, Uda K, Ogura H, Shin W, Kurokawa N, Saito K, Fujikawa N, Date T, Takasaki M, Terada D, Hirai A, Akashi A, Chen F, Adachi Y, Ishikawa Y, Hayakawa F, Hagiwara S, Naoe T, Kiyoi H. A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells with FLT3 mutations. Blood 2018;131:426–38 [DOI] [PubMed] [Google Scholar]
- 22.Levis MJ, Smith CC, Perl AE, Schiller GJ, Fathi AT, Roboz GJ, et al. Phase 1 first-in-human study of irreversible FLT3 inhibitor FF-10101–01 in relapsed or refractory acute myeloid leukemia. Journal of Clinical Oncology 2021;39:7008- [Google Scholar]
- 23.Alexeeva M, Aberg E, Engh RA, Rothweiler U. The structure of a dual-specificity tyrosine phosphorylation-regulated kinase 1A-PKC412 complex reveals disulfide-bridge formation with the anomalous catalytic loop HRD(HCD) cysteine. Acta Crystallogr D Biol Crystallogr 2015;71:1207–15 [DOI] [PubMed] [Google Scholar]
- 24.Counago RM, de Souza GP, dos Reis CV, Ramos PZ, Drewry D, Massirer KB, Arruda P, Edwards AM, Elkins JM, Structural Genomics Consortium (SGC). Crystal Structure of the Human CAMKK2B in complex with Crenolanib 2017
- 25.Smith CC, Lin K, Stecula A, Sali A, Shah NP. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia 2015;29:2390–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen K, Travins J, Wang F, David MD, Artin E, Straley K, et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov 2017;7:478–93 [DOI] [PubMed] [Google Scholar]
- 27.Tarver TC, Hill JE, Rahmat L, Perl AE, Bahceci E, Mori K, et al. Gilteritinib is a clinically active FLT3 inhibitor with broad activity against FLT3 kinase domain mutations. Blood Adv 2020;4:514–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CC LE, Lin KC, Wang Q, McCreery MQ, Stewart WK, Damon LE, Perl AE, Jeschke GR, Sugita M, Carroll M, Kogan SC, Kuriyan J, Shah NP. Crenolanib is a selective type I pan-FLT3 inhibitor. Proc Natl Acad Sci U S A 2014;111:5319–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cools J, Mentens N, Furet P, Fabbro D, Clark JJ, Griffin JD, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res 2004;64:6385–9 [DOI] [PubMed] [Google Scholar]
- 30.Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood 2006;107:293–300 [DOI] [PubMed] [Google Scholar]
- 31.Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, et al. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell 2014;26:428–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A 2014;111:E4869–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CC, Chuang YC, Liu YL, Yang CN. A molecular dynamics simulation study for variant drug responses due to FMS-like tyrosine kinase 3 G697R mutation. Rsc Advances 2017;7:29871–81 [Google Scholar]
- 34.Stone RM, DeAngelo DJ, Klimek V, Galinsky I, Estey E, Nimer SD, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood 2005;105:54–60 [DOI] [PubMed] [Google Scholar]
- 35.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood 1995;85:997–1005 [PubMed] [Google Scholar]
- 36.Bruner JK, Ma HS, Li L, Qin ACR, Rudek MA, Jones RJ, et al. Adaptation to TKI Treatment Reactivates ERK Signaling in Tyrosine Kinase-Driven Leukemias and Other Malignancies. Cancer Res 2017;77:5554–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med 2017;377:454–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng R, Levis M, Piloto O, Brown P, Baldwin BR, Gorin NC, et al. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood 2004;103:267–74 [DOI] [PubMed] [Google Scholar]
- 39.Strelow JM. A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov 2017;22:3–20 [DOI] [PubMed] [Google Scholar]
- 40.Smith CC, Paguirigan A, Jeschke GR, Lin KC, Massi E, Tarver T, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood 2017;130:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files.