Abstract
Intestinal proteases mediate digestion and immune signaling, while increased gut proteolytic activity disrupts the intestinal barrier and generates visceral hypersensitivity, which in common in irritable bowel syndrome (IBS). However, the mechanisms controlling protease function are unclear. Here we show that members of the gut microbiota suppress intestinal proteolytic activity through production of unconjugated bilirubin. This occurs via microbial β-glucuronidase-mediated conversion of bilirubin conjugates. Metagenomic analysis of fecal samples from patients with post-infection IBS (n=52) revealed an altered gut microbiota composition, in particular a reduction in Alistipes taxa, and high gut proteolytic activity driven by specific host serine proteases compared to controls. Germ-free mice showed 10-fold higher proteolytic activity compared with conventional mice. Colonization with microbiota from high proteolytic activity IBS patients failed to suppress proteolytic activity in germ-free mice, but suppression of proteolytic activity was achieved with colonization using microbiota from healthy donors. High proteolytic activity mice had higher intestinal permeability, a higher relative abundance of Bacteroides and a reduction in Alistipes taxa compared with low proteolytic activity mice. High proteolytic activity IBS patients had lower fecal β-glucuronidase activity and end-products of bilirubin deconjugation. Mice treated with unconjugated bilirubin and β-glucuronidase overexpressing E. coli, which significantly reduced proteolytic activity, while inhibitors of microbial β-glucuronidases increased proteolytic activity. Together, these data define a disease-relevant mechanism of host-microbial interaction that maintains protease homeostasis in the gut.
INTRODUCTION
Proteases constitute approximately 2% of the human genome and serve several important biological functions 1. Infectious, inflammatory, coagulation and neurodegenerative disorders have been attributed in part to aberrant signaling involving the actions of various proteases. The intestinal tract is enriched with a diverse repertoire of proteases of host, microbial and dietary origin. Tight regulation of proteolytic activity (PA) is necessary to prevent tissue injury. In the absence of this regulation, proteases can induce disruption of intestinal barrier function as well as the neuronal sensitization reported in irritable bowel syndrome (IBS) 2–4 and inflammatory bowel disease (IBD) 5,6.
Gastrointestinal infections are one of the most common events inciting the development of IBS, with the subsequent condition termed post-infection IBS (PI-IBS) 7. Infections promote an immune response and perturb the commensal microbiota. While some data show immune activation in PI-IBS patients 8, other work found no evidence for these changes 9. Microbiota data have primarily been descriptive, and certain microbiota changes have been correlated with alterations in pathways associated with epithelial barrier function 10,11. No disease-modifying therapies exist for IBS, with treatment largely limited to symptom control. Attempts to restore microbiota using fecal microbiota transplantation (FMT) have produced mixed results 12,13. Similarly, probiotic therapies have had limited success possibly due to the absence of a well-characterized, targeted mechanism 14.
Pancreatic secretions contain large amounts of digestive proteases and these secretions transit through the intestinal tract as chyme. Older studies have demonstrated significantly higher PA in ileostomy contents compared to feces 15. Additionally, germ free (GF) and microbiota-depleted rodents demonstrate high fecal PA 16. These observations suggest that commensal microbiota directly or indirectly modulate gut PA. Serpins, elafins, siropins and miropins are recognized as inhibitors of specific protease activity 17–19; however, the broader endogenous regulation of PA in the intestinal tract is poorly understood. We hypothesized that microbiota perturbation induced by a gastrointestinal infection produces ineffective control of PA, allowing the development of PI-IBS.
One potential pathway for protease inhibition is through unconjugated bilirubin, a potent inhibitor of proteases 20. Bilirubin is deconjugated in the gut by microbial β-glucuronidase (GUS) enzymes 21, which have been shown to decrease trypsin and chymotrypsin activity in an in vivo colitis model 22. The microbiome and the metabolites it creates via enzymes like GUS suggest it may play a larger role in PA regulation than previously understood. We speculated that the healthy microbiota suppresses PA through gut microbial GUS activity, and that this activity would be impaired in patients with PI-IBS.
RESULTS
Fecal proteolytic activity associates with PI-IBS severity
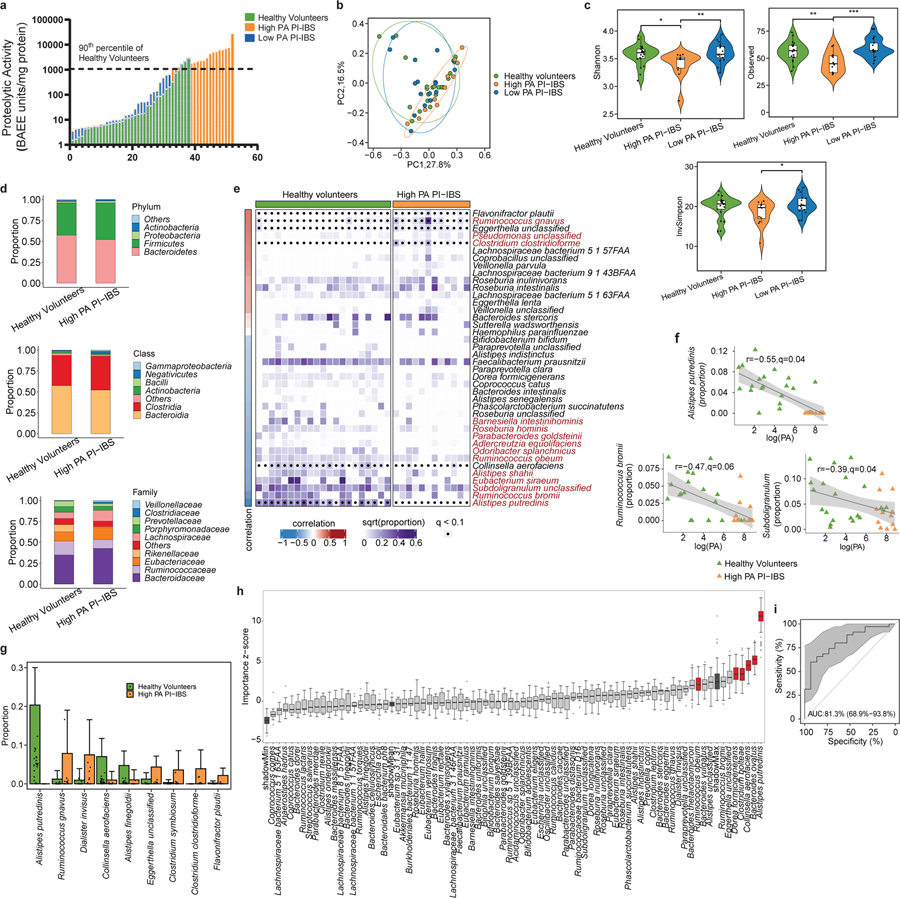
Patients with C. jejuni infection were prospectively followed for potential development of PI-IBS. As shown in our recent epidemiological study, nearly 21% of individuals who were healthy before the infection fulfilled the Rome III criteria for IBS following infection 23. We used a FITC-casein degradation assay to determine PA in fecal supernatants from PI-IBS patients and healthy volunteers 2,24. Thirty seven percent of PI-IBS patients were characterized as high PA (Fig. 1a). The characteristics of healthy volunteers and PI-IBS patients (including both high and low PA) are included in extended data table 1. Bowel movement frequency, stool form and ease of passage scores were all higher in high PA PI-IBS individuals compared to the low PA. We have previously demonstrated that high PA patients have greater symptom severity as well as higher colonic permeability 2.
Figure 1: High proteolytic activity (PA) following C. jejuni infection is characterized by reduced microbiota diversity and taxa loss.
a, PA of PI-IBS and healthy volunteers. High PA in 19/52 PI-IBS patients (90th percentile of healthy volunteers dotted line >1078 BAEE/mg of protein, n=38 healthy, 52 PI-IBS). b, PCoA plot of β-diversity. Microbiota composition in high PA is different from healthy (p=0.05) or low PA PI-IBS (p=0.01) (Bray-Curtis distance, PERMANOVA, with 999 permutations, n= 21 healthy, 12 high PA, 17 low PA PI-IBS). c, Alpha diversity measures between high PA PI-IBS, low PA PI-IBS and healthy volunteers (n=12 high PA PI-IBS, 17 low PA PI-IBS, 21 healthy, linear regressions, *p=0.05,**p=0.01,***p=0.001). d, Higher level taxonomic representation (n=12 high PA PI-IBS, 21 healthy). e, Correlation matrix between fecal PA and taxa. Correlations made taxa with q <0.2 with taxa in red highlighting differential abundance at q<0.1. Spearman correlation coefficient (r) identify taxa correlating with PA. Density reflective of proportions of taxa within healthy or high PA with dots denoting differential at q<0.1. f, Representative scatterplots between logPA and taxa of entire cohort. High PA and healthy volunteers plotted (n=21 healthy, 12 high PA PI-IBS). Grey area: 95% confidence level of linear regression. g, Differentially abundant taxa between healthy and high PA PI-IBS. Alistipes putredinis absent from high PA (n= 21 healthy, 12 high PA, q<0.1). Data are presented as mean + s.d. h, Boruta feature selection algorithm identifying predictive taxa of PA status. Six taxa identified (n=12 high PA, 17 low PA PI-IBS). i, Receiver operating curve of random forests prediction. AUC=0.813, (95% CI 0.69–0.94). Grey area: 95% confidence interval. Boxplots: lower, middle and upper hinges correspond to 25th, 50th and 75th percentiles. Upper and lower whiskers extend to largest and smallest value no further than 1.5 * IQR from respective hinge.
High PA PI-IBS patients exhibit changes in intestinal microbiota
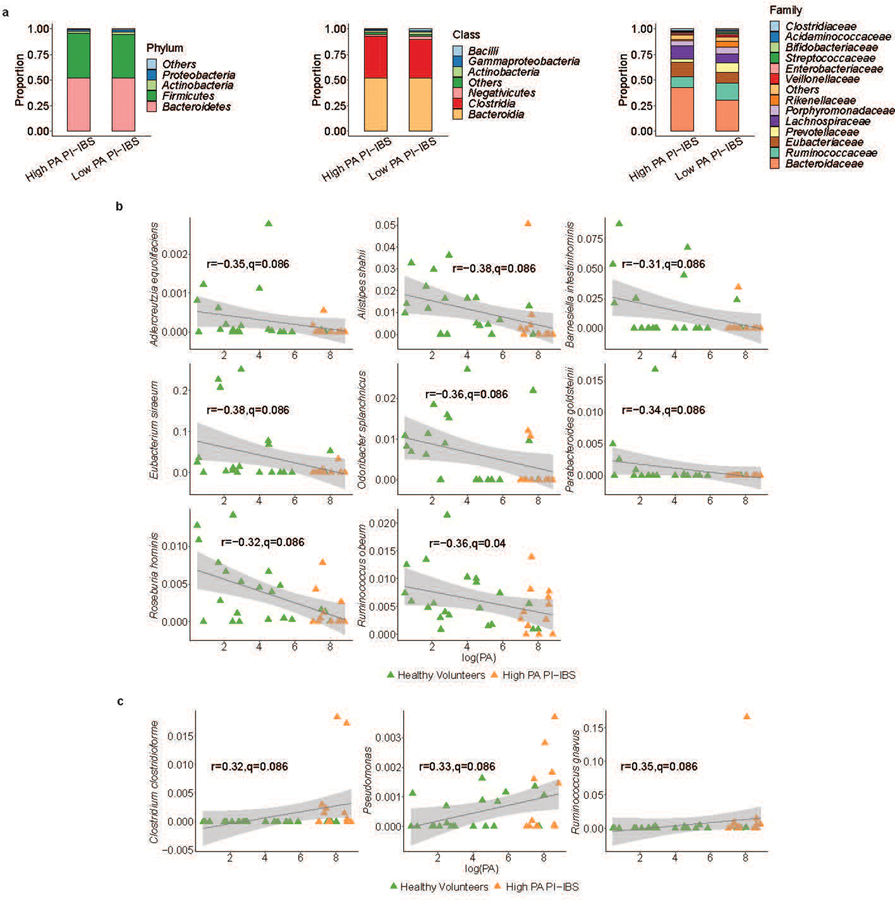
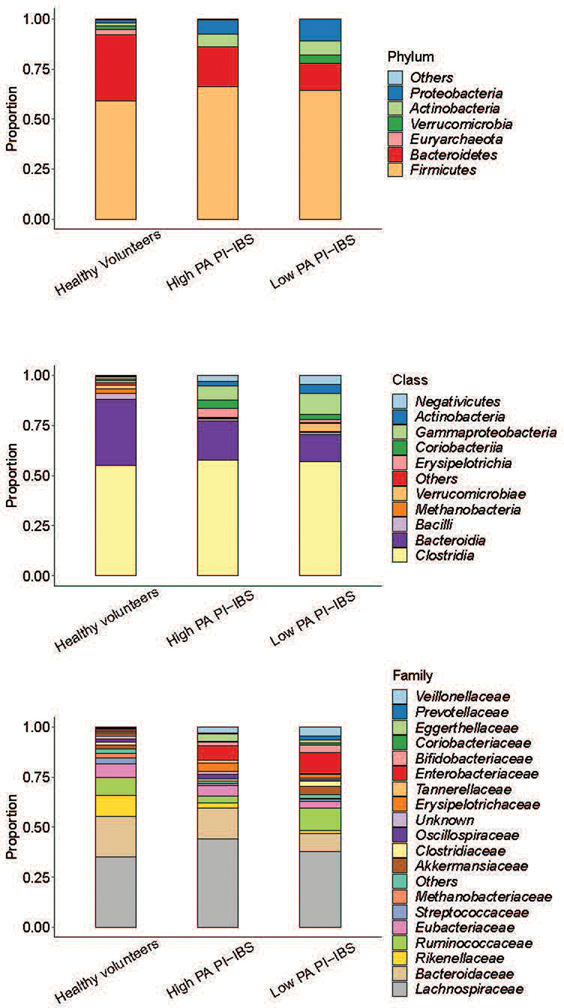
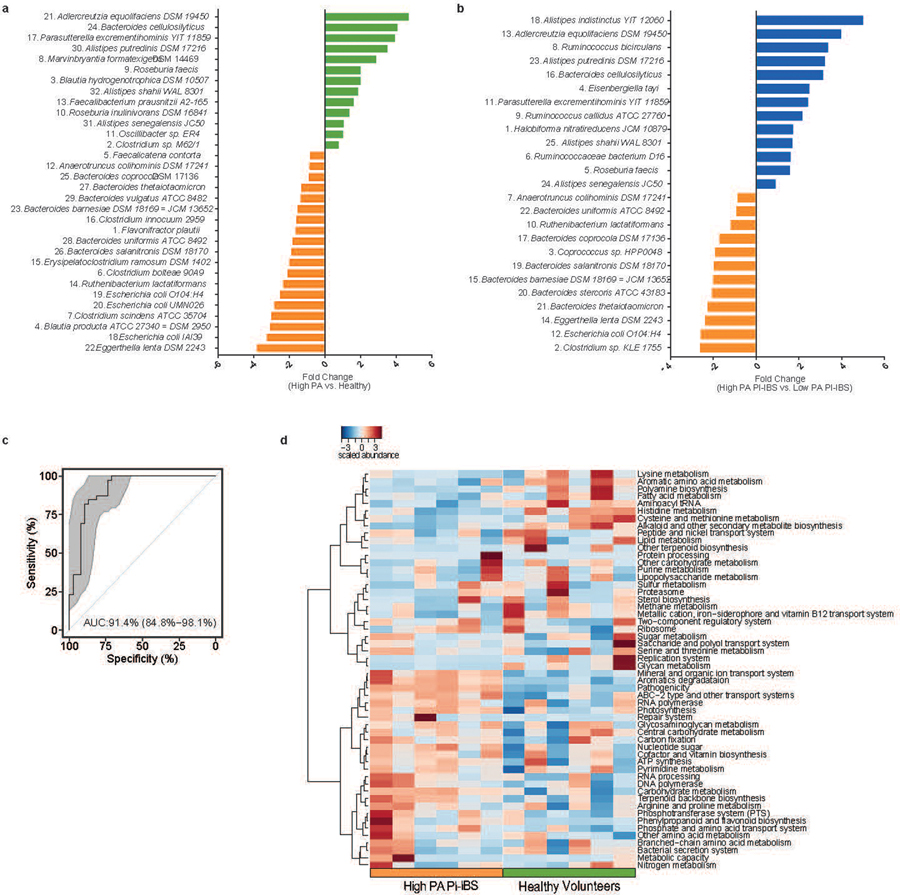
Fecal shotgun metagenomics was used to examine changes in gut microbiota composition. After adjusting for covariates (age, sex, BMI), high PA patients had differences in the beta diversity compared to low PA PI-IBS patients and healthy volunteers, indicating compositional differences in their microbiota (Fig. 1b). Additionally, high PA PI-IBS patients had lower alpha diversity, indicating decreased species richness and evenness compared to low PA PI-IBS and healthy volunteers (Fig. 1c). No major taxonomy shifts were observed at higher phylogenetic levels (phylum and class) with one family different from high PA in both healthy and low PA PI-IBS: Rikenellaceae (Fig. 1d, extended fig. 1a). However, between all three groups, 14 unique bacterial species were found to be differentially abundant (q<0.1, Fig. 1e, colored red). Eleven taxa correlated with low PA status, with the top three species being Alistipes putredinis, Rumincoccus bromii, and Subdoligranulum (Fig 1f; correlation plots for additional differentially abundant taxa with q<0.1 are shown in extended fig. 1, b and c). When only high PA PI-IBS and healthy samples were compared, 9 differentially abundant taxa were identified, of which 3 were in higher abundance in healthy individuals (Fig. 1g). Interestingly, A. putredinis was universally absent in high PI-IBS patients but comprises ~5.0% of the proportional gut taxa in healthy volunteers. A. putredinis was also the only taxa that differentiated the high and low PA PI-IBS groups. A random forest machine learning algorithm with Boruta features identified six bacteria as important predictors of PA status: A. putredinis, Bacteroides ovatus, Collinsella aerofaciens, Clostridium bolteae, Dorea formicigenerans and Ruminococcus obeum (Fig. 1h). The ability of the modeling to predict PA status reported an AUCROC of 0.813 (Fig. 1i).
Luminal proteolytic activity is driven by host serine proteases
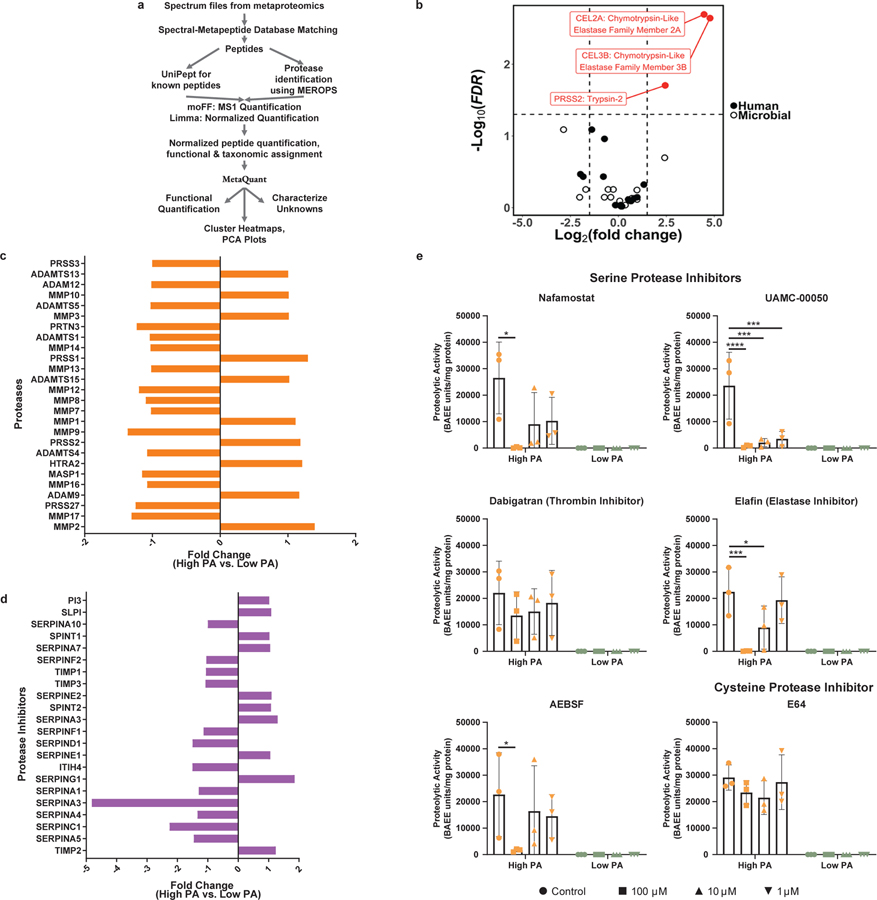
Although high PA has been described previously in IBS, the specific proteases involved have not been characterized. To identify human and microbial peptides and proteins, we developed an untargeted metaproteomics pipeline using Galaxy framework 25–27 (Fig. 2a). Of the peptides and proteins identified, 12% were assigned as proteases. Within high and low PA supernatants, 1413 and 2116 unique peptides were identified (supplementary table 1 for description of proteomics database). 42% peptides in high PA and 36% in low PA fecal samples were of human origin. Only three peptides were differentially abundant, all of which were serine proteases of human origin, and all were higher in abundance in high PA fecal supernatants (Fig. 2b). Chymotrypsin-like pancreatic elastases 2A and 3B both showed 4.5 greater and trypsin-2, a 2.5 greater log2fold abundance in high PA supernatants. Serine proteases have been shown by us and others to disrupt barrier function 2,4. Host colonic mucosa have also been shown to produce serine proteases, including in IBS patients 3,28. To determine mucosal expression, bulk RNA sequencing was performed on rectosigmoid colonic mucosal samples from high and low PA patients. No differences in expression of proteases or protease inhibitors were observed. See supplementary table 2 for complete transcriptomics database. Additionally, proteomics was performed using the SOMAscan© platform.. Expression of several tissue-derived serine proteases was identified but no differences observed between high and low PA biopsies (Fig. 2c). Additionally, protease inhibitor protein abundance was similar (Fig. 2d). Lastly, we used various inhibitors to determine the proteases driving the PA. Only the serine protease (AEBSF, Nafamostat, UAMC-00050) and elastase (Elafin) inhibitors, but not cysteine (E64) and thrombin inhibitor (Dabigatran), suppressed the PA (Fig 2e).
Figure 2: Fecal and tissue proteomics demonstrates serine proteases of human pancreatic origin drive high PA in PI-IBS.
a, Pipeline used for identifying human and microbial fecal proteomic profiles of high PA and low PA feces. b, Metaproteomic analysis of high and low PA fecal samples. Volcano plot highlights feces from high PA volunteers have an increased abundance of host pancreatic proteases (n=7 high PA, 6 low PA, FDR<0.05). Three proteases, chymotrypsin like pancreatic elastase 2A, 3B (FDR=0.002 for each, Standard t-test) and trypsin 2 (FDR=0.02, Standard t-test), all serine family, were identified in greater abundance in high PA samples. c-d, Proteomic analysis of rectosigmoid colonic biopsies using SOMAscan® platform. Comparisons of proteases (c) and protease inhibitors (d). Analysis reveals that production of mucosal derived proteases and inhibitors are comparable regardless of PA phenotype (n=7 high PA, 6 low PA). e, In vitro inhibition of high PA FSN with protease inhibitors. AEBSF (100μM *p=0.042), nafamostat (100μM *p=0.002), UAMC-0050 (100μM ****p=0.0001, 10μM ***p=0.0002, 1μM ***p=0.0004), and elafin (100μM ***p=0.0005, 10μM *p=0.032) significantly inhibited PA in vitro while dabigatran (thrombin inhibitor) and E64 (cysteine protease inhibitor) had no effect. (2-way ANOVA, Tukey’s multiple comparison test, n= 3 high and 3 low PA FSNs per condition). Barplots are presented as mean ± s.d.
High PA associated microbiota fail to suppress luminal PA
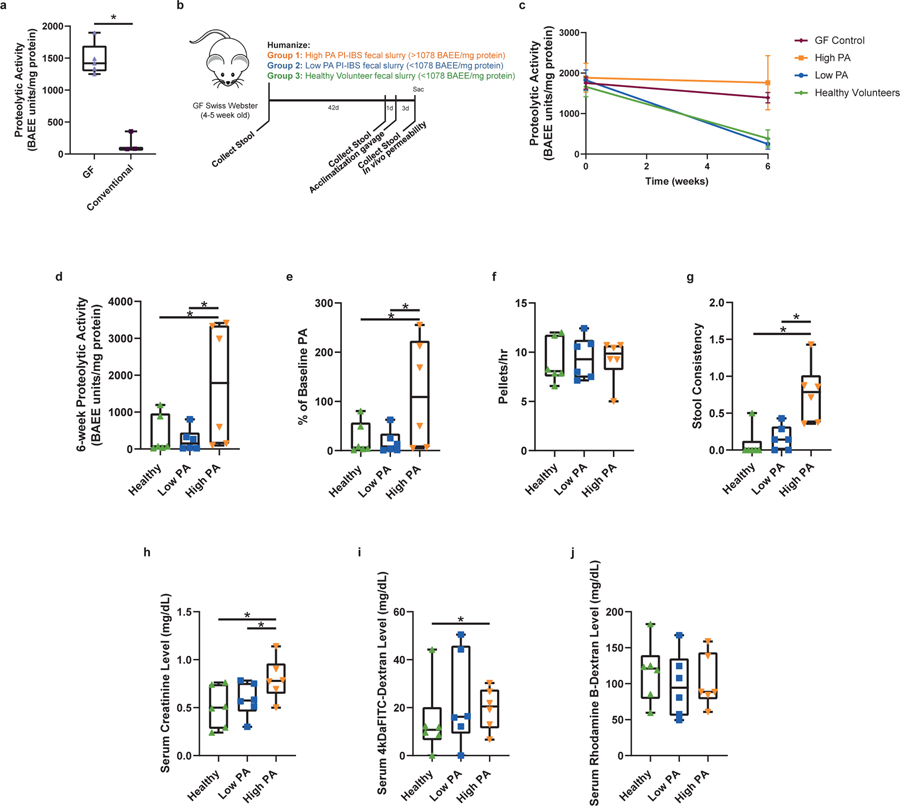
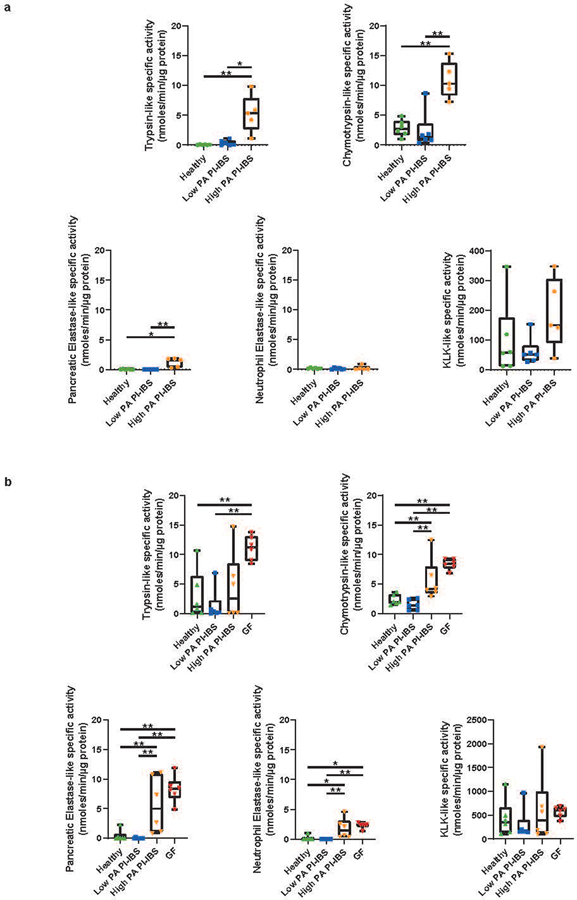
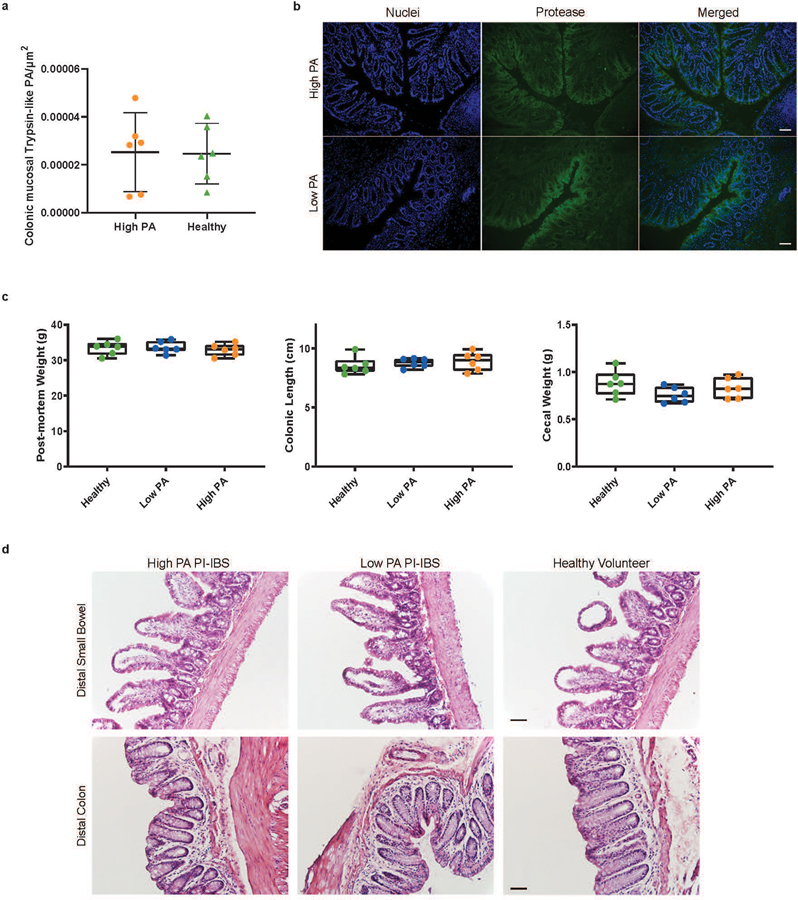
The differences in microbiota, as well as the identification of host proteases driving PA, led us to examine the role of commensal microbiota in regulation of host proteases. To model microbial effects, we used germ free (GF) mice (Swiss-Webster). When compared to specific-pathogen free mice, GF mice had 10-fold higher fecal PA, indicating that the intestinal microbiome plays a role in PA regulation (Fig. 3a). We next humanized GF mice with microbiota from a random set of high PA PI-IBS, low PA PI-IBS patients, and healthy volunteers (Fig. 3b). Mice humanized with healthy or low PA PI-IBS microbiota demonstrated significantly lower PA than mice humanized with high PA (Fig. 3, c and d). Humanization with healthy and low PA PI-IBS microbiota resulted in significant inhibition of PA, whereas high PA humanization showed no change Fig. 3e). In addition to measuring the overall PA using the FITC-casein assay, we assessed enzymatic activity using preferential substrates for a range of serine proteases, and we found high activities for chymotrypsin, pancreatic elastase, and neutrophil elastase in both human donors and humanized mice (extended figure 2b). We also assessed the active PA in GF mice and found activity of trypsin-like, chymotrypsin-like, pancreatic and neutrophilic elastases were similar to high PA humanized mice (extended figure 2b). In situ zymography was used to assess colonic mucosal PA which also showed no differences between high and low PA biopsies (extended figure 3, a and b). Pellet frequency was similar between groups; however, high PA humanized mice had softer pellets when compared to healthy and low PA PI-IBS humanized mice (Fig. 3, f and g). Colonic length, cecal and mouse weight were similar among the three groups (extended figure 3c). Small bowel and colon histopathology were evaluated by an expert pathologist in blinded fashion. No gross differences in cellular morphology or distinction in immune cells (polymorphonuclear leukocytes, macrophages) in either the small intestine or the colon were observed between the different humanization states (extended figure 3d).
Figure 3: Gnotobiotic mice demonstrate healthy commensal microbiota suppresses host derived PA while high PA PI-IBS microbiota does not.
a, Absence of microbiota characterized by higher PA (Swiss-webster female, 2-tailed Mann-Whitney, n=3 germ free,−5 conventional mice, p=0.04). b, Mouse model of humanization. c, Healthy and low PA microbiota causes host fecal PA drop (n=6 feces/phenotype, dots represent average, presented as mean ± s.d). d, Post-humanization PA is dependent on microbiota. Mice with healthy or low PA PI-IBS microbiota have significantly lower PA compared to high PA humanized mice (383 vs. 246 vs. 1,761 BAEE/mg protein, 2-Way ANOVA Tukey’s, n=6 feces/phenotype, dots represent average, *p=0.03 and *p=0.04 respectively). e, Low PA microbiota suppresses host PA. while mice with high PA microbiota have fecal PA similar to GF mice (% of baseline, healthy 24.5 ± 32.8, low PA 17.8 ± 23.9; high PA 116.3 ± 109.7, One-Way ANOVA-Kruskal-Wallis, n=6 feces/phenotype, dots represent average, *p=0.03). f, Humanized mouse pellet frequency. No difference between humanized groups in pellet production. g, Humanized mouse fecal pellet consistency. High PA humanized mice have significantly looser feces. Scored 0=normal to 4=diarrhea. Two averaged independent observations (One-Way ANOVA-Kruskal-Wallis, n=6 feces/phenotype, dots represent average, *p=0.01 for both). h, Pore pathway permeability. Creatinine levels in high PA humanized mice indicate increased pore pathway permeability (for all permeability pathways, n=6 human feces/phenotype; 3 mice/human feces, dots represent averaged mice; high PA PI-IBS 0.81 ± 0.28 mg/dL; low PA PI-IBS 0.58 ± 0.24; healthy 0.51 ± 0.36, One-Way ANOVA-Kruskal-Wallis, *p=0.05). i, Leak pathway permeability. 4-kDa FITC-dextran levels in high PA humanized mice show increased leak pathway permeability (high PA PI-IBS, 19.1 ± 14.6 mg/dL; low PA PI-IBS 23.9 ± 23.9; healthy 13.7 ± 30.3, One-Way ANOVA-Kruskal-Wallis, *p=0.04). j, Unrestricted pathway permeability. No difference in serum rhodamine levels across humanization states. Boxplots as previously described.
Mice humanized with high PA feces have higher intestinal permeability
Next, in vivo permeability assays were performed to investigate potential differences in permeability pathways between mice with distinct fecal humanized states. Differences in pore (<6Å), leak (<100Å) and unrestricted (no max) permeability pathways were measured by evaluating serum levels of the orally administered tracer molecules creatinine (6 Å), 4kDa-FITC Dextran (28 Å) and 70kDa rhodamine B-Dextran (120 Å), respectively. Compared to healthy and low PA PI-IBS humanized mice, high PA humanized mice had greater serum creatinine and 4-kDa FITC-Dextran (Fig. 3, h and i). Serum levels of rhodamine B-Dextran were similar across groups, indicating that humanization with high PA microbiota did not cause increased permeability through the unrestricted pathway (Fig. 3j).
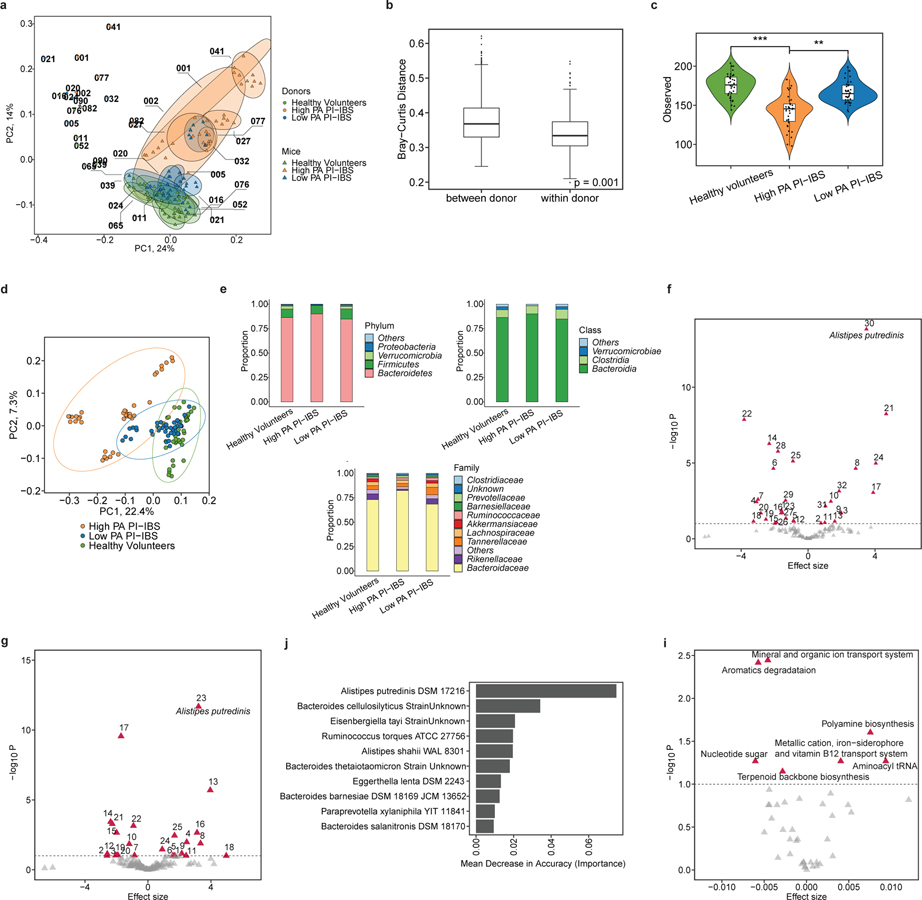
Absence of A. putredinis associated with high PA in mice
Mice humanized with either low PA or high PA microbiota were taxonomically and compositionally more similar within their given cohort than when compared against each other (Fig. 4, a and b). High PA humanized mice had lower alpha diversity than mice humanized with low PA or healthy volunteer microbiota (Fig. 4c). This indicates distinct microbiome composition between high PA PI-IBS and healthy or low PA PI-IBS humanized mice (Fig. 4d). Similar to human samples prior to colonization, no major differences were found at higher taxonomic levels post humanization with the exception of both Rikenellaceae and Bacteroidaceae decreased in high PA mice (Fig. 4e, extended figure 4). Differential abundance analysis found 32 taxa at the species level that were differentially abundant between the healthy volunteer and high PA PI-IBS, while 25 were differential between low PA and high PA PI-IBS humanized groups. A. putredinis was one of the seven taxa found to be in greater abundance in healthy and low PA PI-IBS humanized mice, while several species of Bacteroides were in greater abundance in high PA mice (Fig 4, f and g; for identification of labeled taxa see extended figure 5, a and b). A random forest machine learning algorithm again identified the genus Alistipes as a major predictor of low PA status, with A. putredinis as the most significantly associated species (Fig. 4h). Furthermore, an ROC curve with an AUCROC of 0.914 was generated (extended figure 5c). Nine predicted KEGG pathways were predicted to be significantly different between the healthy and high PA groups (extended figure 5d). The top predicted pathways elevated in high PA humanized mice were “aromatic degradation” and “mineral and organic ion transport”, whereas the “polyamine biosynthesis” pathways were increased in healthy humanized mice (Fig. 4i).
Figure 4: Microbial diversity and composition in humanized mice identify specific microbial taxa predicative of PA status and metabolic pathway differences.
a, Mice humanized with high PA, low PA PI-IBS and healthy volunteer microbiota have differences in microbiota composition (n=6 human feces/phenotype, dots represent average). b, Humanized mice intra-group, not inter-group relatedness. Humanized mice microbiota are compositionally and taxonomically different when compared to each other, but similar within its group (Bray-Curtis, PERMANOVA with 999 permutations, p=0.001). c, Alpha diversity across humanized mice. Healthy volunteer and low PA humanized mice have greater species richness compared to high PA humanized mice (n=6 human feces/phenotype, dots represent average, linear regression on observed species **p=0.01,***p=0.002). d, Beta diversity measures in humanized mice. Microbial composition differs between each humanization state (Bray-Curtis, PERMANOVA with 999 permutations, p=0.01). e, Higher level taxonomic evaluation of engrafted microbiota in humanized mice (n=6 human feces/phenotype, dots represent average). f-g, Volcano plot(s) highlighting strain level differences between high PA and healthy volunteer (f) or high and low PA PI-IBS humanized mice (g). Red icons indicating differences in abundance (q<0.1) with A. putredinis identified (n=6 human feces/phenotype, dots represent average). h, Prediction model of microbiota for PA status by random forests. The top 10 taxa predictive of PA status in mice assessed using mean decrease in accuracy. i, Predicted KEGG pathway differences between high PA and healthy volunteer humanized mice. Differentially abundant KEGG pathways with q<0.1. Effect size denotes average difference in KEGG functional unit between reference and comparison groups. Boxplots as previously described.
Fecal microbiota restoration and A. putredinis supplementation restores protease suppression
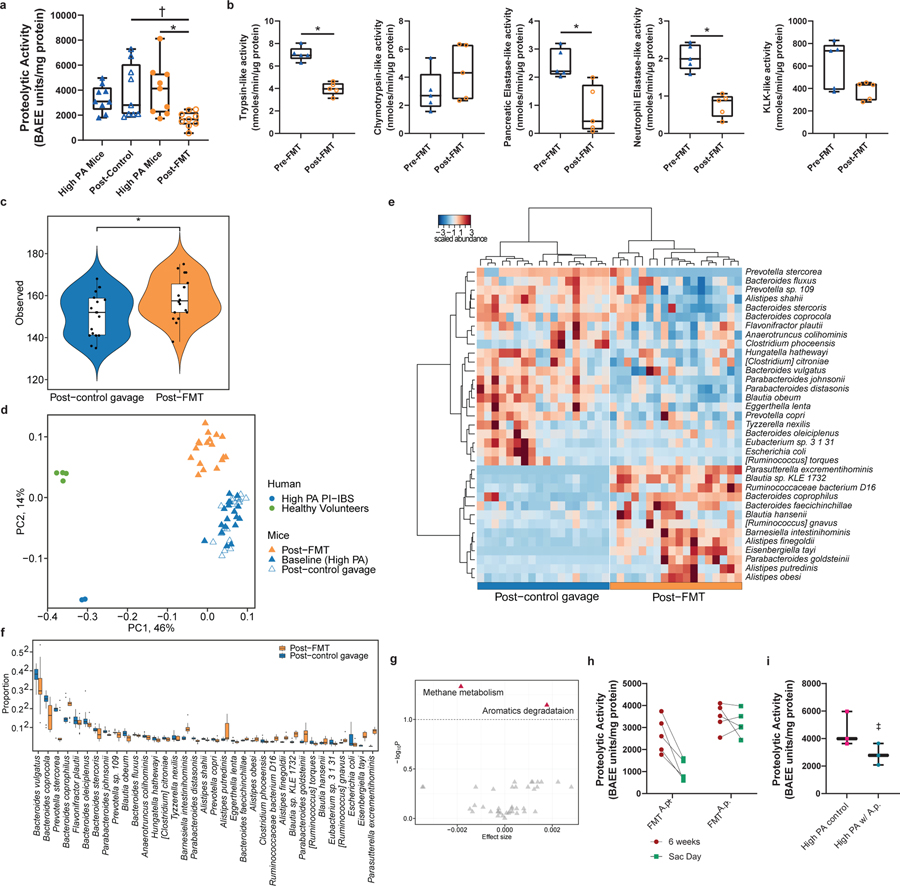
Introducing microbial taxa associated with low PA into the microbiome of high PA individuals may serve as a potential way to lower PA. One strategy for introducing new taxa is through FMT using a healthy low PA donor. High PA PI-IBS humanized mice were administered either a vehicle control, or a fecal slurry made up of a healthy, low PA donor (1.62 BAEE units/mg protein). Fecal PA of FMT treated mice was significantly lower than that of untreated control mice (Fig. 5a) and significant inhibition of PA from baseline in FMT treated but not control mice. On measurement of specific activity, a reduction in the activity of all serine proteases except chymotrypsin and kallikrein was observed in mice given an FMT compared to the control group (Fig. 5b).
Figure 5: Fecal microbiome transfer of low PA microbial communities lowers PA of high PA humanized mice in Alistipes-dependent manner.
a, FMT treatment in high PA mice. FMT with a healthy microbiome decreased PA in high PA humanized mice compared to controls (1676 ± 613.6 vs. 3920± 2178 BAEE units/mg protein, One-Way ANOVA-Kruskal-Wallis †p=0.0235 and 2-sided Wilcoxon-Matched Pairs *p=0.011, 9 mice/group). b, FMT changes proteolytic profile. Trypsin, neutrophilic elastase, and pancreatic elastase activity was lower post-FMT compared to baseline (Paired t-test, Mann-Whitney n=5 mice, trypsin *p=0.003, pancreatic elastase *p=0.04, neutrophil elastase *p=0.006). c, Measurement of alpha diversity. Mice given FMT demonstrate greater microbial diversity compared to control mice (linear regression on observed species, n=9/group, p=0.01). d, Measurement of beta diversity. Bray-Curtis β-diversity ordination demonstrates shifts in microbiota composition following FMT. Mice receiving FMT cluster separately and towards a healthy, low PA microbiome (PERMANOVA, n=9 mice/group, p=0.001). e, Heatmap outlining differentially abundant taxa in mice treated with FMT. Taxa identified differentially abundant at q<0.1 (n=9 mice/group). f, Proportion of differentially abundant bacteria between FMT and control mice. 35 differential bacterial taxa identified at the species level (q<0.1), with 13 in greater abundance after FMT (n=9 mice/group). g, Predicted KEGG pathway differences. Methane metabolism pathway increased, and aromatics degradation decreased in FMT mice (n=9 mice, q<0.1). h, Restoration of low PA phenotype dependent on FMT community. Mice administered a community containing A. putredinis reduced fecal PA while FMT using low PA microbiota lacking A. putredinis was unable to suppress PA. Sac Day is 1-week post-FMT. (2-way ANOVA-Sidak’s, p=0.001, n=5 mice/group). i, High PA feces spiked with A. putredinis attenuates increased PA post-humanization (2-way ANOVA, Sidak’s n=3 mice/group, p=0.05, mean ± s.d). Boxplots as previously described.
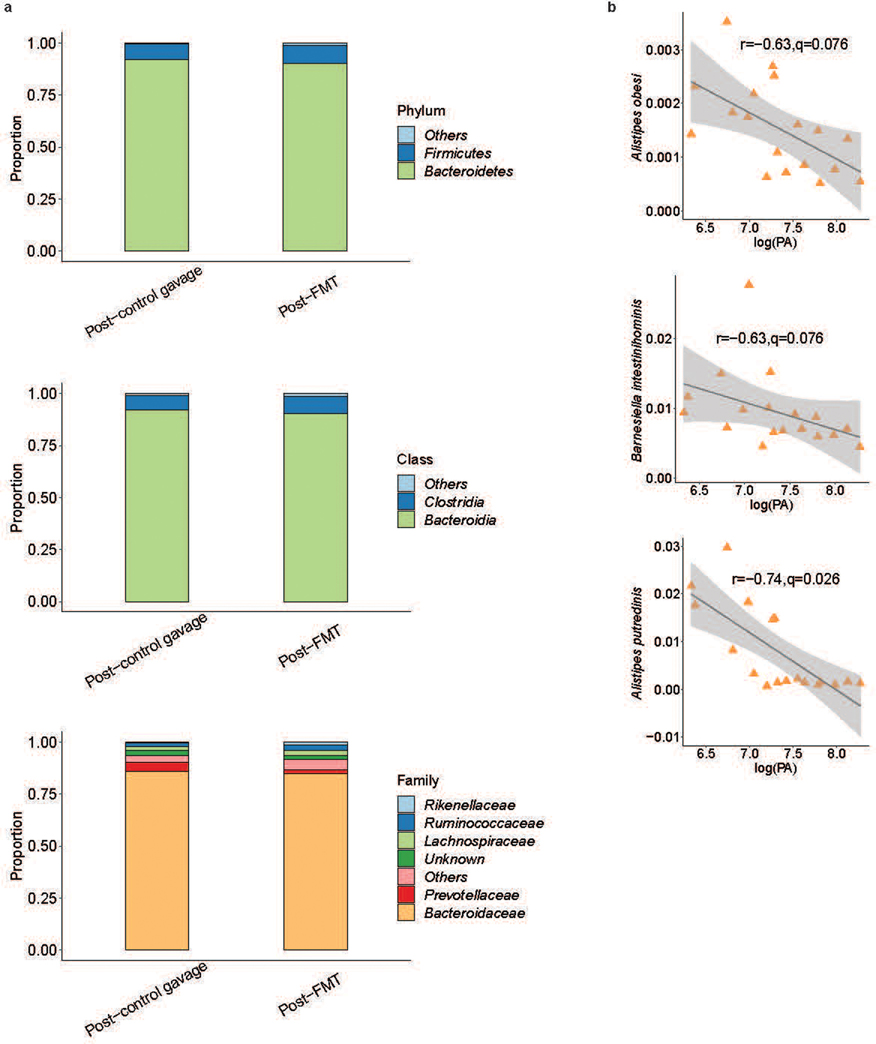
Mice given an FMT had greater alpha diversity (Fig. 5c) as well as beta diversity (Fig. 5d) than vehicle controls indicating greater species diversity and changes in microbiota composition post-FMT. While no differences were observed at phyla and class taxonomic levels, Rikenellaceae, the family of A. putredinis, and 8 additional families were differential post-FMT (extended figure 6a). At the species level there were 35 differentially abundant taxa between the two groups post-FMT (Fig. 5e). Shifts in KEGG pathways post-FMT included increased “methane metabolism” and decreased “aromatics degradation” (Fig. 5f, q<0.1). Interestingly, metabolic by-products of aromatic degradation include amino acid derivatives like phenylethylamine, which are significantly increased in Crohn’s disease 29. Of the differentially abundant taxa, 13 were in greater abundance in the mice receiving FMT. Multiple species of the genus Alistipes were identified (Fig. 5g), which correlated to decreased fecal PA (extended figure 6b).
In humans and humanized mice, the presence of A. putredinis in the gut microbiota strongly predicted low PA status. We aimed to test if A. putredinis could reverse high PA phenotype by: (1) giving high PA humanized mice an FMT with a community containing A. putredinis (A.p.+) or with a community lacking A. putredinis (A.p.−); and (2) by adding A. putredinis directly to a high PA fecal slurry used to humanize mice. Mice given an FMT with an A.p.+ healthy community had a robust lowering of PA when compared to mice receiving an FMT with an A.p.− healthy donor community (Fig. 5h). The addition of A. putredinis to a high PA fecal slurry also attenuated the marginal increase that was seen with the high PA humanization (Fig. 5i).
GUS-mediated production of unconjugated bilirubin suppresses PA
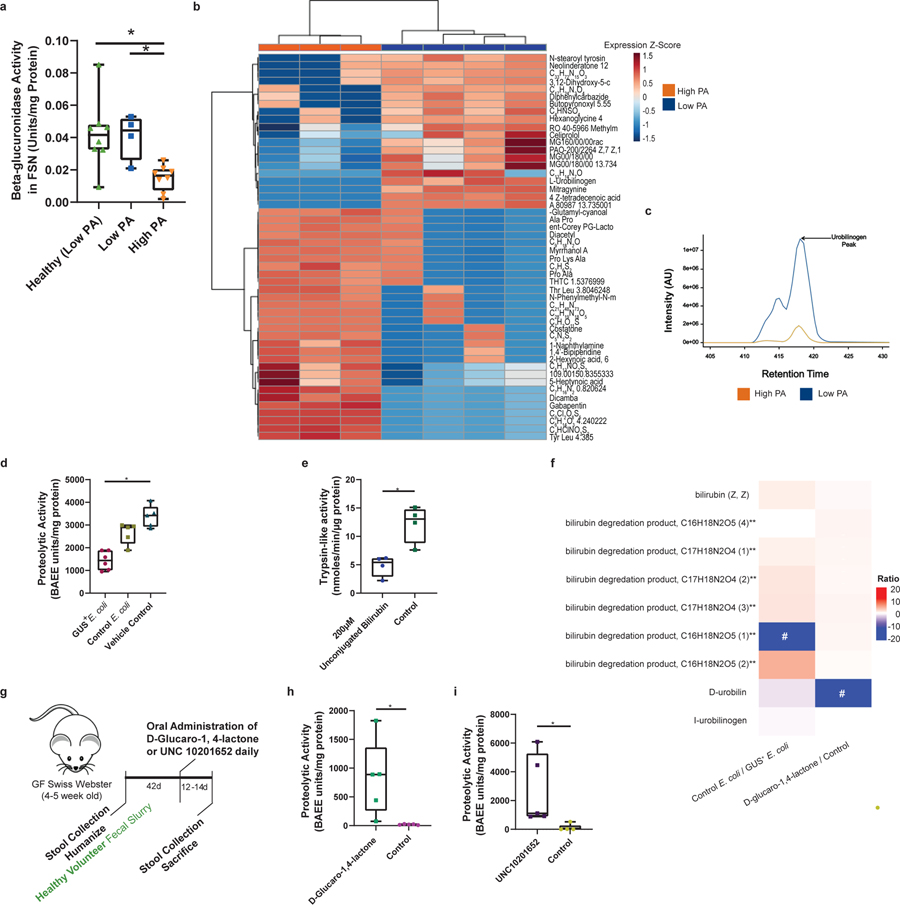
The lack of a known protease inhibitor peptide led us to investigate an alternative mechanism by which luminal proteases could be regulated by the microbiome. Previous literature has correlated the presence of A. putredinis with fecal beta-glucuronidase (GUS) enzyme activity 30, and Alistipes GUS enzymes have been identified in the human and mouse gut microbiomes 21,31. Therefore, we investigated the fecal GUS activity of high PA PI-IBS individuals. High PA PI-IBS patients had significantly lower fecal GUS activity compared to healthy or low PA PI-IBS individuals (Fig. 6a). We then employed an untargeted HPLC-MS/MS metabolomics approach to examine microbial GUS-related metabolites. While a number of differentially abundant metabolites were observed, high PA samples had significantly lower levels of urobilinogen (Fig. 6, b and c, q<0.05). Urobilinogen is generated only when conjugated bilirubin is first deconjugated by gut microbial GUS enzymes.
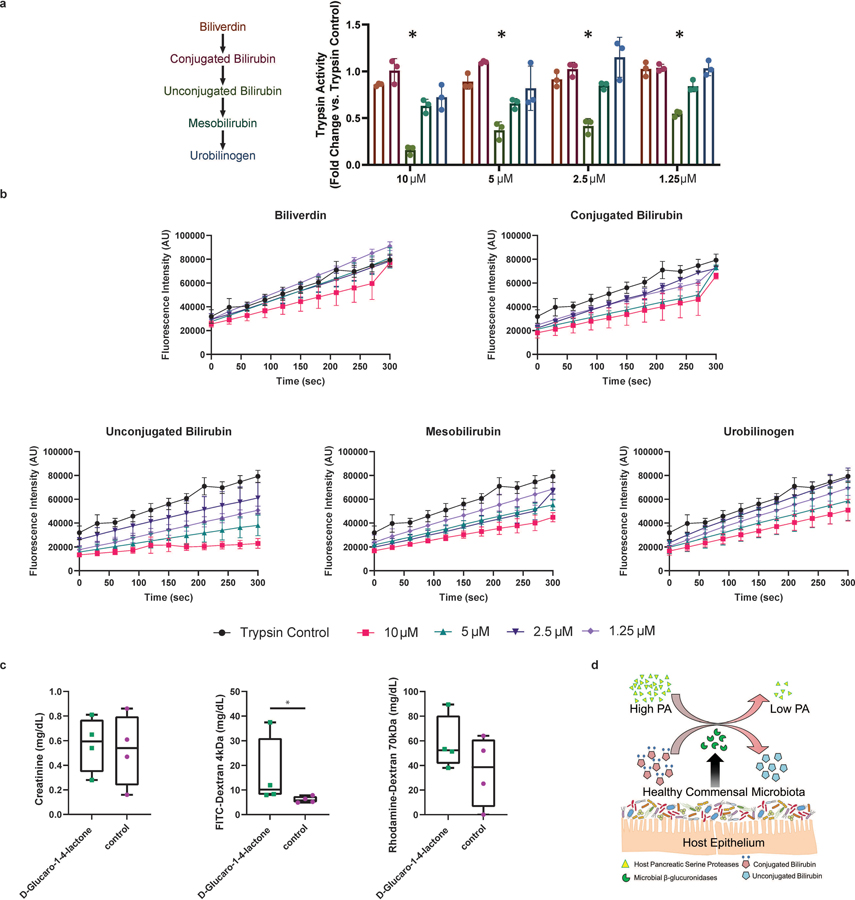
Figure 6: Microbiota mediates PA suppression through microbial β-glucuronidase enzymatic activity and production of unconjugated bilirubin.
a, Beta-glucuronidase (GUS) activity in humans. High PA PI-IBS had lower fecal GUS activity compared to healthy and low PA PI-IBS (One-Way ANOVA-Kruskal-Wallis, *p=0.02 and *p=0.03 respectively, n=4 low PA, 8 high PA and 8 healthy). b, Concentration of urobilinogen in human feces. Untargeted fecal metabolomics identified higher urobilinogen in low PA compared to high PA feces (n=3 high PA and 4 low PA). c, Liquid chromatography tracing of urobilinogen (C18 column, n=3 high PA and 4 low PA PI-IBS). d, GUS suppresses PA in mice. Mice monocolonized with E. coli overexpressing GUS (GUS+) had significantly lower fecal PA compared to controls (1437 ± 411.7 vs. 3371 ± 481.9 BAEE units/mg protein, One-Way ANOVA-Kruskal-Wallis, p=0.004, n=5). e, Unconjugated bilirubin suppresses PA. Mice given unconjugated bilirubin (gavage, 3 days) had less trypsin-like activity compared to controls (4.8 vs 12.2 nmoles/min/μg protein, Mann-Whitney p=0.03, n=4 mice). f, Fecal metabolomics in experimental mice. Increased D-urobilin, #p=0.001 and bilirubin degradation product, C17H20N2O5 #p=0.03, seen in GUS+ and D-Glucaro-1–4-Lactone treated animals respectively (n=4 mice). g, Schematic for GUS inhibitors in humanized mice. h, D-Glucaro-1–4-Lactone increases fecal PA. Mice given D-Glucaro-1–4-Lactone, a non-specific GUS inhibitor, have significantly higher fecal PA than controls (825.3 ± 655.6 vs. 19.3 ± 8.0 BAEE units/mg protein, Mann-Whitney, n=5, p=0.008). i, UNC10201652, a microbial GUS inhibitor, increases fecal PA (2692 ± 2430 vs. 113.8 ± 230.1 BAEE units/mg of protein, Mann-Whitney, n=5, p=0.008). Data presented mean ± s.d. Boxplots as previously described. Human and mouse metabolite intensities were normalized using total ion current for each sample, log-transformed and compared between groups (Student’s t-test). p-values were adjusted using Benjamini-Hochberg method. Metabolites p-value <0.05 and absolute log2 fold change of > 0.5 (0.0 = no change) are mapped.
Increased GUS activity and urobilinogen levels in low PA individuals led us to test whether the administration of GUS could inhibit PA in mice. We monocolonized GF mice (which start with high PA) with either competent control Escherichia coli, or a modified strain of E. coli engineered to constitutively overexpress GUS (GUS+). Mice given the GUS+ E. coli had significantly lower PA than mice receiving a vehicle control gavage (Fig. 6d). Bilirubin mono- and di-glucuronides (conjugated bilirubin) are key endogenous substrates for gut microbial GUS enzymes. Unconjugated bilirubin has been shown to inhibit serine proteases in vitro 20. We tested the in vitro capacity of various metabolites in the bilirubin deconjugation pathway to inhibit purified trypsin. Only unconjugated bilirubin was able to suppress the PA significantly. Mesobilirubin and urobilinogen had some effect, but this was not statistically significant and conjugated bilirubin had no effect (extended figure 7, a and b). Enzyme kinetics and inhibitory concentrations for the tested bilirubin metabolites are provided in extended data table 2. Next, we determined the effects of unconjugated bilirubin on PA in vivo. We administered unconjugated bilirubin (200 μM) orally for 3 days in GF mice. This resulted in a significant reduction in trypsin-like activity when compared to untreated control mice (Fig. 6e). We also found that D-urobilin, an end product of bilirubin deconjugation was increased in GUS+ E. coli treated mice (ratio of control/treated= −12.3, p=0.001) (Fig. 6f). None of the other identified products of bilirubin metabolism pathways were statistically significant. Taken together, these data support the conclusion that both GUS and unconjugated bilirubin play important roles in reducing gut luminal PA.
Inhibition of gut microbial GUS enzymes increases host PA
We next inhibited gut microbial GUS activity in healthy humanized mice to determined effects on the PA (Fig. 6g). Mice receiving D-glucaro-1,4-lactone, a broad-spectrum but modestly potent inhibitor of GUS activity 32 demonstrated significantly higher PA than mice given a control gavage (Fig. 6h). These mice had decreased levels of bilirubin degradation product, C17H20N2O5 (ratio of treated/control=−10, p=0.03 Fig. 6f) and increased permeability for the 4kDa FITC dextran (extended figure 7c). We next employed UNC10201652, a more potent GUS inhibitor that exclusively inhibits gut microbial GUS enzymes 33. Using a similar dosing strategy (Fig. 6g), healthy humanized mice given UNC10201652 demonstrated significantly higher PA compared to control animals (Fig. 6i). Thus, the robust increases in PA produced by GUS inhibitors, and specifically by UNC10201652, indicates that the microbiome is critical for regulation of host PA and microbial GUS activity is clearly important for reducing luminal PA in vivo (extended figure 7d).
DISCUSSION
Imbalance in intestinal proteolytic activity has been demonstrated in variety of conditions, including IBS 2,3,28,34,35, IBD 5,6,36,37 and celiac disease 18,38,39. However, the regulation of PA in the intestinal tract is not well understood. Here we demonstrate that specific proteases of host-origin drive PA and that commensal microbes inhibit proteases through gut microbial β-glucuronidase-mediated production of unconjugated bilirubin. A characteristic feature of high PA phenotype was the absence of commensal A. putredinis, and introduction of A. putredinis containing low PA community into high PA mice or supplementing with A. putredinis alone can suppress the PA. These observations support the conclusions that PI-IBS pathophysiology is driven by loss of critical microbes following intestinal infection, which result in an impaired bilirubin deconjugation and weakened inhibition of specific serine proteases.
Due to the complexity of the luminal environment in the GI tract, it has been difficult to assign the nature or origin of proteases that become elevated in disease states. While high and low PA patients have microbiota differences, fecal metaproteomics did not identify any major differences in the abundance of bacterial proteases. Previous work has in fact shown that human derived proteases make up the bulk of fecal proteases 35. The intestinal epithelium has been identified as a source of luminal proteases, specifically trypsin-3 28. Furthermore, IBD patients have been found to have elevated tissue elastase and thrombin activity 2. Here we found no transcriptomic or proteomic signatures of activated immune pathways, making it likely that intraluminal proteases are not driven by immune cells. Further, mucosal PA was similar between high and low PA colonic biopsies.
Because intestinal infection and subsequent antibiotic use likely depletes microbiota diversity, we speculated that this feature plays an important role in PA regulation. The presence of A. putredinis was the strongest predictor, with high PA patients demonstrating absence of this microbe compared to its 3–5% prevalence in healthy and low PA PI-IBS patients. Alistipes is a genus of Gram-negative, rod-shaped, non-spore forming anaerobic bacteria that currently consists of 13 species 40. Studies have shown the positive impact of Alistipes spp. as oral administration of A. finegoldii decreased colitis in a DSS- mouse model 41 and Alistipes enrichment associated with anti-inflammatory cytokine production in TNBS colitis in NOD2 knockout mice 42. In contrast, one study showed that the development of artificial sweetener-induced ileitis was associated with an increase in Alistipes abundance 43. Further studies are needed to clarify the roles Alistipes spp. play in microbiome dynamics and intestinal health.
Here, we employed GF mice to model the mechanistic influences of the microbiota on host-driven PA. Similar to older observations in GF rats 44, we found GF mice to have 10-fold higher fecal PA compared to specific pathogen free mice. Humanization with healthy commensal microbiota significantly decreased the PA, which was not observed upon humanization with high PA microbiota. Additionally, high PA humanized mice had greater permeability through pore and leak pathways but not unrestricted pathways. This, along with the absence of gross inflammatory changes in the intestine, suggest that the aberrant regulation of PA does not induce frank inflammation, similar to what is noted clinically in IBS patients. One prior study has correlated fecal PA with microbiota composition 45, and increased PA has also associated with increased permeability 24,34,46. Together with these previous observations, the current study demonstrates an essential role of microbiota-based inhibition of the PA, which protects the colonic epithelium from barrier disrupting proteases.
While microbiota restoration has been of interest for treating IBS, mixed results have been observed with probiotics 14 and FMT trials have largely been unsuccessful 12,47–49 with a few notable exceptions 13,50. In our mouse model, introduction of a healthy low PA community into high PA humanized mice via FMT caused a drop in PA and changes in both alpha and beta diversity. Interestingly, we also found that reduced PA was not a universal phenomenon after FMT with healthy donor communities, akin to the variabilities observed in clinical FMT studies. The presence of low PA predictors like A. putredinis was critical in lowering PA, as FMT from donors lacking these taxa did not result in suppression of PA. Recent studies have suggested that FMT engraftment depends on both the donor and recipient microbiome pre-FMT 51, and that successful treatment may be dependent on the presence of specific bacterial taxa 52. Additional studies are necessary to unravel the essential features of engraftment dynamics, as well as the key microbial taxa that associate with PA suppression and correction of barrier dysfunction.
Inhibition of host proteases could arise from microbial protease inhibitors, microbial influences on host protease inhibitors, and through changes in the luminal milleu 19,39,53. In a prior study, the presence of Alistipes was associated with bacterial β-glucuronidase (GUS) activity 30, which is critical for the deconjugation of bilirubin 30,54. Alistipes β-glucuronidases have also been established to be present in both the human and mouse gut microbiomes 21,31. Compared to the low PI-IBS individuals, high PA patients had both lower fecal levels of GUS activity and lower fecal levels of urobilinogen, an end product of deconjugated bilirubin metabolism 55. We demonstrate that unconjugated bilirubin, but not some of the other products in bilirubin deconjugation pathway, significantly decreased trypsin activity in vitro and in vivo 20. Furthermore, we show that E. coli with constitutive GUS expression suppresses PA in vivo and inhibition of GUS converts a healthy, low PA humanized mice into high PA.. The results we obtain identify a microbiome-dependent mechanism for host PA regulation and also provide an exciting potential therapeutic target for the treatment of the high PA associated with gut diseases.
Although specific taxa like A. putredinis predict low PA, further studies are required to determine if their presence is essential for suppression of PA. GUS enzymes are commonly expressed by bacteria 21,31 and there is significant variability in the efficiency of different gut microbial GUS enzyme orthologs in the processing substrates like estrogen 56–58. As bilirubin processing by microbial GUS enzymes has not been studied in detail, further studies will need to determine if certain taxa are more efficient at inhibition of PA via this deconjugation activity. It is also plausible that additional named or unnamed metabolites in bilirubin deconjugation pathway or metabolites other than those in bilirubin deconjugation pathway also play a role in inhibition of serine proteases.
The tissue and fecal proteomics assays are limited by sensitivity and may have missed low abundance proteases or protease inhibitors. Determination of specific proteases driving the PA is limited by the poor specificity of the tracers as well as inhibitors. However, the complementary experiments using proteomics, substrate assays, inhibitors, all point towards trypsin and elastases as drivers of PA in these patients. Although humanized models have limitations in terms of immune development and the potential for incomplete engraftment of the donor microbiome 59,60, our model was found to be consistent with others 61 and was able to recapitulate the PA phenotypes, proteolytic profiles, and the physiological changes in permeability observed in humans. The complimentary human and mice findings presented here establish that the intestinal microbiome is critical for inhibiting host-derived proteases and maintaining barrier function through their GUS-mediated production of unconjugated bilirubin. They suggest that microbiota-based therapies may potentially target protease imbalances and the resulting phenotypes of impaired barrier function and visceral hypersensitivity in IBS and other gastrointestinal disorders.
METHODS
Study Design
The goal of this study was to determine the source and regulation of proteases within the gastrointestinal tract of individuals with high fecal proteolytic activity (PA). Within 7 to 10 days of notification of Campylobacter cases, Minnesota Department of Health conducts interviews with patients or guardians who are culture positive. PI-IBS development was initially assessed by sending surveys to patients aged 18–80 years from 6 to 9 months following a positive culture for Campylobacter species, recruiting individuals from November 1, 2011, to September 30, 2019. This was done by mail, or by telephone interviews for non-responders. Participants were then asked to complete ta Rome III IBS questionnaire. The identified patients were then invited for recruitment at Mayo Clinic where the study physician confirmed IBS and clarified all inclusion and exclusion criteria have been met and participants understand the instructions for collection of stool samples and metadata (bowel diary, psychological and other questionnaires). Healthy volunteers were recruited through advertisements. They completed a screening questionnaire to confirm absence of any ongoing GI symptoms or prior GI diagnoses. Both male and female participants were recruited for the study.
A total of 52 PI-IBS patients defined by Rome III criteria and 38 healthy volunteers were recruited in an observational study. The Rome III criteria were used to define IBS and its subtypes based on the presence of abdominal pain or discomfort at least 2–3 times a month and alterations in bowel function (stool consistency or frequency) for ≥6 months to demonstrate chronicity of symptoms62. Those with a history of abdominal surgery (except hernia, C-section, hysterectomy, appendectomy or cholecystectomy), inflammatory bowel disease, microscopic colitis, or celiac disease were excluded. Additionally, recruited volunteers were not pregnant at the time of the study. Use of tobacco or alcohol for the duration of the study was prohibited. Following medications were prohibited 7 days prior to study participation: those affecting gastrointestinal transit, serotonergic agents, anti-cholinergic agents, antimuscarinics, narcotics, peppermint oil, antibiotics or new probiotics. Ingestion of artificial sweeteners such as Splenda™ (sucralose), Nutrasweet ™ (aspartame), lactulose or mannitol was prohibited for 2 days before the start and during the study. All subjects taking part in the study were asked to complete the Hospital Anxiety and Depression Scale (HADS) and a 7-day bowel diary. All participants completed the Hospital anxiety and depression scale (HADS). PI-IBS patients also completed the Symptom Checklist-90 (SCL-90), IBS Symptom severity scale (IBS-SSS), IBS-quality of life (IBS-QoL) questionnaire as well as the Long Bowel Disease questionnaire (BDQ). Validated IBS-Symptom Severity Scale (IBS-SSS)63 and IBS-Quality of Life (IBS-QoL) scales were used64. Established Bristol Stool scale was used to determine stool form65. For assessment of psychological symptoms, validated Hospital Anxiety and Depression scale were used66. Mayo Clinic Institutional Review Board approved human studies and all participants provided a written informed consent (IRB protocol: 12-006529; ClinicalTrials.gov identifier: NCT03266068). Fecal PA was evaluated in these individuals. Feces from the cohort were used to humanize germ free Swiss Webster mice under the Mayo Clinic Institutional Animal Care and Use Committee approved protocol (Protocol #A00003420-18-R20) after which relationships between microbiota, proteases and the mechanisms of inhibition of these proteases was evaluated.
Generation of fecal supernatant
Fecal supernatants (FSNs) were made fresh prior to each experiment. Feces from patients (0.1g) or mice (1 pellet) was added to 0.8mL of phosphate buffered saline (PBS) and subsequently homogenized with a pellet pestle for 5–10 seconds (Sigma-Aldrich, St. Louis, MO, USA). Homogenates were spun twice at 5,000 g for 10 min at 4°C and then added to a 0.22 μm Spin-X tube filter (Corning Life Sciences, Durham, NC, USA). Samples were filtered at 4°C, 10,000 g for 5 min and FSN was stored on ice until use.
Measurement of overall proteolytic activity
Pierce Fluorescent Protease Assay Kit (Thermo Scientific, St. Peters, MO, USA) was used to measure fecal PA of prepared FSNs in triplicate. A 5 μL sample of FSN was added to 195 μL of fluorescein isothiocyanate (FITC)-labeled casein in a 96-well black bottom plate. Samples incubated with substrate for 10 min at room temperature and subsequently read using a Synergy Mx Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA). Fluorescent intensity as a result of proteolytic digestion of FITC-casein was determined with a standard curve and then normalized to protein content in the FSN measured using the Bradford assay (Bio-Rad, Hercules, CA, USA). Fecal samples were categorized as high PA and low PA based on the cut-off for healthy volunteers. 90th percentile of healthy volunteers was used as the cutoff (>1078 BAEE/mg of protein). PI-IBS individuals were identified as either high or low PA came from an analysis of the distribution of PA in healthy volunteers which followed a log-normal distribution. Within the healthy volunteer cohort, there is a clear bimodality in the distribution, indicating the presence of two populations. We used a 90% threshold (>1078 BAEE/mg of protein) to define the right-sided distribution of the overall population as this subgroup within the healthy cohort was clearly different from the rest in their fecal PA. As a consequence of this, the cutoff of 90% was used and provides the biological and statistical rationale for categorizing PI-IBS subjects into either high and low PA groups.
Measurement of activity of specific proteases
Trypsin-like, chymotrypsin-like, neutrophil elastase, pancreatic elastase and kallikrein proteolytic activities were measured using specific substrates N-p-Tosyl-Gly-Pro-Arg 7-amido-methylcoumarin HCl (AMC), Suc-Ala-Ala-Pro-Phe-AMC, Suc-Ala-Ala-Pro-Val-AMC, Suc-Ala-Ala-Ala-AMC and Pro-Phe-Arg-AMC respectively. FSNs (5 μL) were mixed with the specific substrate (100μM) in a buffer of Tris-HCl (50mM, pH 8.0) with CaCl2 (10mM). Substrate solution was automatically dispensed into a black bottom microtiter plate for a total reaction volume of 200 μL. Cleaved substrate was evaluated by change in fluorescence using a BioTek Synergy™ Mx Microplate Reader (Excitation/Emission = 380/460), taking measurements kinetically at 37°C for 15 minutes. Enzymatic activity was calculated for each enzyme (nmoles/min) using a standard curve generated for free AMC which was then converted to specific activity (nmoles/min/μg) by using the protein concentration obtained using the Bradford method.
In vitro inhibition of high PA FSNs using protease inhibitors
AEBSF (Sigma Aldrich, A8456), nafamostat mesolate (Sigma Aldrich, N0289), elafin (Sigma Aldrich, E7280), dabigatran (Sigma Aldrich, SML2370), E-64 (Sigma Aldrich, E3132) and UAMC-0050 inhibitors were used in vitro to profile the proteases present in high PA and low PA FSNs. A 5μL sample of FSNs, prepared as previously described, was added to a 96-well black bottom plate containing 20μL of the tested inhibitor. Each inhibitor was tested at a final concentration of 100, 10 and 1μM in the wells. FITC-labeled casein (175μL) was then immediately added to the wells and allowed to incubate for 10 min at room temperature. Fluorescent intensity as a result of proteolytic digestion of FITC-casein was read. A standard curve was used to determine proteolytic activity which was then normalized to protein content in the FSN which had been previously measured Bradford assay, similar to the aforementioned measurement of fecal proteolytic activity.
In situ zymography
High and low PA distal colonic tissue was assessed for potential tissue contributions to PA. Biopsy sections (8μm) that were freshly embedded in OCT, sliced and allowed to air dry on microscope slides. These sections were then washed with PBS containing 2% Tween-20. The sections were then stained with a green-fluorescent SYTOX Green Nuclear Stain (ThermoFisher, S7020) for 15 minutes in a humid chamber at room temperature. The sections were then covered with a low melt agar overlay (0.3%) containing N-p-Tosyl Gly-Pro-Arg 7-amido-4-methylcoumarin hydrochloride (100μg/mL) prepared in 50mM Tris-HCl pH 8.0 containing 10mM CaCl2. The substrate agar overlay allows for the visualization of localized tissue specific proteolytic activity. A single slide for each sample was then incubated at 4°C overnight and served as a background control for that sample while a paired slide incubated at 37°C overnight and served as the test slide. Fluorescence intensity was measured under the same settings for both the 4°C and 37°C incubated slides with images obtained using an epifluorescence microscope. Area corrected image intensities were collected using ImageJ Java 1.8.0_172 software and reported as the difference between the testing slide and the background control. Representative images were pseudocolored blue for nuclei and green for tissue enzymatic activity.
Shotgun metagenomics of human samples
Sequencing
DNA is purified from human stool samples using MO BIO Laboratories PowerSoil DNA Isolation Kit, MP Biomedicals FastPrep-24 5G Homogenizer and an automated high throughput nucleic acid isolation system (Perkin Elmer Chemagic MSM I) based on proprietary M-PVA bead technology. The concentration measured using Qubit dsDNA HS Kit (PN Q32854 Thermo Fisher Scientific Inc., Waltham, MA). Libraries were sequenced on Illumina HiSeq 4000, PE150. Sequence reads post-processing and taxonomy calling. Raw reads were processed using BBDuk v38.73 67 to remove adapters and perform read quality filtering.
Bioinformatics
Processed reads were analyzed using the SHOGUN v1.0.8 taxonomy profiler 68, using the BURST aligner option 69. Profiling of the functional content was performed using SHOGUN v1.0.8 to obtain KEGG orthology entries, modules and pathways, as well as HUMAnN2 v2.8.1 70 using its UniRef90 database to obtain UniRef90 71 gene family abundance. Functional profiling against targeted databases was also performed using HUMAnN2, using the MEROPS peptidase database release 12.0 27,70.
Measurement of β-glucuronidase (GUS) activity
GUS enzymatic activity was measured using the substrate 4-Nitrophenyl-β-D-glucuronide (10mM, pH 7.0) 72. FSNs (50 μL) were dispensed in duplicate into a 96-well clear bottom plate. A plate dispenser was used to automatically add 50 μL of substrate to each well. The reaction was moved immediately to a BioTek Synergy™ Mx Microplate Reader, pre-warmed to 370C. Plates were read at 405nm every 5 min for one hour. Concentrations of GUS (U/mg of protein) were determined based off a standard curve of pure GUS enzyme and standardized to the total amount of protein as determined by the Bradford assay.
Fecal metaproteomics
In-gel digestion
Gel bands were washed using 1:100mM ammonium bicarbonate:acetonitrile and subsequently dehydrated using acetonitrile. Proteins were reduced and alkylated using 10 mM dithiothreitol in 50mM ammonium bicarbonate and 55 mM iodoacetamide in 50mM ammonium bicarbonate in dark, respectively. The gel pieces were washed with 1:1 acetonitrile:100mM ammonium bicarbonate and dehydrated with acetonitrile. Trypsin (5ng/μl in 50mM ammonium bicarbonate and 5mM calcium chloride) was added for in-gel digestion overnight at 37 °C. Peptides were extracted sequentially using 50% acetonitrile 0.3% formic acid and 80% acetonitrile 0.3% formic acid. Peptides were desalted using the MCX-type STAGE tips (3M Empore SDB-XC filters) and dried in vacuo for storage at −80°C.
Liquid chromatography
An Eksigent 425 system was coupled to an AB Sciex TripleTOF 5600+ mass spectrometer. The liquid chromatography system uses a trap column (ChromXP 5 μm ChromXP C18CL 120 Å 10×0.3mm) and an analytical column (Eksigent 3 μm ChromXP C18CL 120 Å 150×0.3mm). The analytical column temperature was maintained at 35 °C. LCMS grade solvents were purchased from Honeywell and composed of buffer A (0.1% formic acid in water (v/v)) and buffer B (0.1% formic acid in acetonitrile (v/v)). Peptides were reconstituted in 10μl 2% solvent B and 8μl was loaded at 10 μl/min for 3 minutes onto the trap column. Analytical separation was established by maintaining 3% B during loading. After a valve switch event, peptides were resolved using the analytical column at a flow rate of 5 μl/min with a linear gradient of 3–25% HPLC buffer B for 38 minutes, followed by a 5 min linear gradient to 32% B. Following the peptide elution window, in 2 min the gradient was increased to 80% B for 3 min. Initial chromatographic conditions were restored in 1 min and maintained for 8 min.
Mass spectrometry
Peptides were ionized using the SCIEX DuoSpray ion source with a 50 μm ESI electrode. Data was acquired using an ion spray voltage floating of 5.5 kV, curtain gas of 35 PSI, GS1 of 19 PSI, GS2 of 15 PSI and an interface heater temperature of 75 °C. The Information Dependent Acquisition (IDA) mode was used to scan ions with charge states +2-+5 with intensity greater than 150 units. Exclusion time was 15 seconds and tolerance was set to 100 ppm. Survey scans (MS1) were acquired in 250 ms over a mass range 400–1250 m/z. Following the survey scan, twenty MS/MS spectra were acquired over a mass range 100–1500 m/z using a collision energy setting of 35 spreading 5 eV.
Colonic tissue proteomics
Biopsies were collected from the sigmoid colon and completed by a single endoscopist (MG). A flexible sigmoidoscopy was done following a tap water enema, with biopsies using a large capacity biopsy forceps (no pin) 25cm from the anal verge and immediately snap frozen in liquid nitrogen and stored at −80°C until use. These were cryopulverized using a freezer mill. 200uL of extraction buffer (T-PER tissue protein extraction agent, Thermo Scientific), plus 2 μL of Halt Protease Inhibitor Cocktail (Thermo Scientific) was added to each tube. Samples were homogenized on ice using an Omni PowerGen Homogenizer with Omni tips for soft tissues (Thermo Scientific) for 30 seconds until no fragments were visible. Samples were centrifuged at 4°C, 14,000g, for 10 minutes. Supernatant were collected and filtered using a 0.2μm Spin-X tube, centrifuged at 4°C, 5min at max speed. Samples were shipped on ice to Washington University St. Louis to be run on the SOMAscan® 1.3 K cell and tissue platform.
Colonic tissue transcriptomics
Sigmoid colon biopsies were collected from volunteers, placed in RNA later (Fisher) had and immediately snap frozen in liquid nitrogen. Total RNA was extracted by using an miRNeasy Kit from Qiagen followin manufacturer instructions and measured with a RiboGreen RNA quantification kit (Invitrogen). RNA integrity and profile were analyzed using an Agilent BioAnalyzer/TapeStation, followed by library preparation with Illumina’s TruSeq Stranded mRNA Library Prep Kit without polyA selection. Final libraries were measured using Picogreen and quality assessed using the Agilent Tapestation. TruSeq stranded mRNA libraries with unique dual-indexed (UDI) were generated (Illumina). All libraries were pooled and sequenced on a NovaSeq 2×150-bp Charter-Service RNA-Seq run, generating ≥20 Million reads for each library with mean quality scores for all libraries ≥Q30 on a 150 PE flow cell lane. RNA-seq fastq files for samples were obtained via Illumina sequencing platform with QC performed on the FASTQ files using RSeQC software version used was v3.0.1 (http://rseqc.sourceforge.net/). Data analysis was done using Mayo Analysis Pipeline for RNA Sequencing (MAPRSeq v3.1.3) with paired end reads were aligned using STAR-2.6.1c and alignment to the human genome reference hg38.
Fecal metabolomics
HPLC-MS
Fecal samples were deproteinized with six times volume of cold acetonitrile:methanol (1:1 ratio), kept on ice with intermittent vortexing for 30 minutes at 4C, then centrifuged at 18000xg. 13C6-phenylalanine (3μl at 250ng/μl) was added as internal standard to each sample prior to deproteinization. The supernatants were divided into 2 aliquots and dried down for analysis on a Quadrupole Time-of-Flight Mass Spectrometer (Agilent Technologies 6550 Q-TOF) coupled with an Ultra High Pressure Liquid Chromatograph (1290 Infinity UHPLC Agilent Technologies). Profiling data were acquired under both positive and negative electrospray ionization conditions over a mass range of 100 – 1200 m/z at a resolution of 10,000–35,000 (separate runs). Metabolite separation was achieved using two columns of differing polarity, a hydrophilic interaction column (HILIC, ethylene-bridged hybrid 2.1 × 150 mm, 1.7 mm; Waters) and a reversed-phase C18 column (high-strength silica 2.1 × 150 mm, 1.8 mm; Waters). For each column, the run time is 20 min using a flow rate of 400 ul/min. A total of four runs per sample was performed to give maximum coverage of metabolites. Samples were injected in duplicate or triplicate, and a quality control sample, made up of a subset of samples from the study was injected several times during a run. All raw data files obtained were converted to compound exchange file format using Masshunter DA reprocessor software (Agilent). Mass Profiler Professional (Agilent) was used for data alignment and to convert each metabolite feature (m/z × intensity × time) into a matrix of detected peaks for compound identification.
Bioinformatics
An unsupervised principal component analysis, ANOVA, 3D plot and heat map and a Partial Least Square discrimination analysis (PLS-DA) comparison between groups were obtained for analysis. This gives a list of accurate mass molecular weights of differentially expressed components that was run against the Metlin database to give putative identification (IDs). The list of components would have a putative ID or a mass (m/z) value depending on whether match was found. Components that were assigned putative IDs were further examined by comparison to a purchased reference standard of the proposed compound. Mass accuracy of the Q-TOF method was <5ppm with retention time precision better than 0.2%. A 1.2x fold change can be detected with a precision of 4%.
Raw LC-MS data files are processed using a custom in house constructed pipeline. Data files were first segregated by the analytical mode: negative C18, positive C18, negative HILIC, or positive HILIC. Each segregated file set was processed independently using the pipeline. This pipeline started by loading data files of a segregated set into Agilent ProFinder software. ProFinder was configured to perform retention time alignment of all data files, find peaks, match their mass and retention time (wherever possible) against METLIN database for compound identification, and produce a matrix of compounds and their intensities in respective samples. The compound-intensity matrix was loaded into an in house developed pipeline for differential expression analysis. This process started by filtering low intensity metabolites by removing compounds that do not have an intensity of at least 5000 in at least 75% of the samples. Next, MetaboanalystR functions were utilized for data normalization, differential expression analysis and visualization. To account for intra- and inter-batch effects, metabolite data in each sample were normalized using total ion current detected in the respective sample. Normalized data was log2 transformed. Transformed intensities of each metabolite observed across all samples were centered and scaled via mean centering. Processed metabolite data were analyzed by multivariate approach such as Principal Component Analysis (PCA) to reveal data heterogeneity, groupings, outliers and trends. Hierarchical clustering analysis was performed to assess clustering of biological replicates, technical replicates, and metabolite clusters that correlate with different subsets of clinical variables. Transformed intensities of each metabolite were compared between high and low PA groups using Students’ unpaired t-test. Resulting p-values were adjusted for multiple testing using Benjamini-Hochberg method 73(FDR). Metabolites with an FDR-adjusted p-values or q-values <= 0.05 and an absolute log2[fold change] >= 1.5 (where a value of 0.0 signifies no change) between two groups were considered as statistically significantly differentially expressed and saved for further analyses.
For metabolomic analysis in mice, fecal samples were collected and sent to Metabolon Inc. where global metabolic profiling was completed with using the Metabolon platform (Metabolon Inc., NC, USA).
Humanization of germ-free mice
Animal experiments were approved by the Mayo Clinic Institutional Animal Care and Use Committee (Protocol #A00003420-18-R20). Swiss Webster, germ-free female mice were housed in flexible film isolators (CBC, WI, USA) and given free access to both autoclave food and water. 4–6 week old mice were given an oral gavage of patient fecal slurry (200 μL human fecal suspension) prepared 1:2 with pre-reduced PBS in an anaerobic growth chamber, kept at 37°C, and supplied with 15% CO2, 5% H2 and 80% N2 (Coy Laboratory Products, Grass Lake, MI). Upon humanization mice were housed in individually ventilated Maxiseal cages (Arrowmight, Hereford, England, 3–5 mice/cage) for 6 weeks to allow for microbiome engraftment. Mouse pellets were collected at baseline (pre-gavage), 6-weeks post, and during various treatments. Animals were treated in accordance with the Mayo Clinic Institutional Animal Care and Use Committee under the approved protocol #A00003420-18-R20. Swiss Webster, germ-free female mice were initially housed in flexible film isolators with free access to both autoclaved food and water. After humanization in randomized order, mice were housed in individually ventilated, sterilized Maxiseal cages with, autoclaved food and water, sterile woodchip bedding and an enrichment bedding of crink-l’nest. Mice numbers never exceed 5 mice/cage, and on average were housed 3 mice to a cage. Mice received bedding and cage changes every 2-weeks, with food and water replaced as needed. Mice were kept on a 12h on/12h off light cycle and handled only during the daytime to avoid disruptions in circadian rhythm. Mice were kept in a temperature and humidity-controlled room with temperature ranging from 64 to 76 °F and a relative humidity between 30% and 40%. The operator was not blinded to the assigned humanization group.
Scoring of fecal pellet frequency and consistency
Humanized mice were transferred to sterile cages with a wire rack bottom. Mice were left alone for one hour after which the total number of pellets were counted in each cage. Humanized mice representative of each human microbiome was then averaged together to give the number of pellets produced per hour for the respective humanized state. Pellets collected from humanized mice were scored 0–4 based on a consistency scale previously reported 74. Scoring of fecal pellet consistency was defined as: 0 = normal/formed, 1 = loose, but formed pellet, 2 = no form, lose with some diarrhea, 3 = diarrhea and 4 = severe, watery diarrhea. Two independent measurements from two separate researchers were recorded from which the average pellet consistency was calculated for each mouse. Averages of each human microbiome were then reported as an overall average for each humanized state (average of averages).
Shotgun metagenomics in humanized mice
Sequencing
Upon submission, the DNA samples are quantified using a fluorimetric PicoGreen assay. The purity of the samples are also assessed via a Nanodrop. For a sample to pass QC, it needs to quantify greater than 0.2 ng/ul. If the samples pass QC they enter the TruSeq NexteraXT DNA library preparation queue. gDNA samples are converted to Illumina sequencing libraries using Illumina’s NexteraXT DNA Sample Preparation Kit (Cat. # FC-131-1096). The NexteraXT protocol was followed except that all steps were performed on a quarter reaction scale. In summary, 0.25 ng of gDNA is simultaneously fragmented and tagged with a unique adapter sequence. This “tagmentation” step is mediated by a transposase. The tagmented DNA is simultaneously indexed and amplified 12 PCR cycles. Final library size distribution is validated using capillary electrophoresis and quantified using fluorimetry (PicoGreen). Pooled libraries are denatured and diluted to the appropriate clustering concentration. The libraries are then loaded onto the NovaSeq paired end flow cell and clustering occurs on board the instrument. Once clustering is complete, the sequencing reaction immediately begins using Illumina’s 2-color SBS chemistry. Upon completion of read 1, 2 separate 8 or 10 base pair index reads are performed. Finally, the clustered library fragments are re-synthesized in the reverse direction thus producing the template for paired end read 2.
Bioinformatics
Base call (.bcl) files for each cycle of sequencing are generated by Illumina Real Time Analysis (RTA) software. The base call files and run folders are streamed to servers maintained at the Minnesota Supercomputing Institute. Primary analysis and de-multiplexing are performed using Illumina’s bcl2fastq v2.20. The end result of the bcl2fastq workflow is de-multiplexed FASTQ files that are released to client accounts for subsequent analysis by the mapping software and aligner of their own choosing. The microbiome data were summarized into α-diversity, β-diversity and taxa abundances. α-diversity reflects species richness and evenness within bacterial populations. Three alpha diversity indices (within sample diversity) were calculated on the rarefied data: observed feature number, Shannon diversity index and Inverse Simpson index. A linear model (or linear mixed effects model with subject-level random intercept) was used for testing the association with α-diversity. β-diversity reflects the shared diversity between bacterial populations in terms of ecological distance. Bray-Curtis distance, a β-diversity measure, was calculated on the rarefied data. PERMANOVA was used for testing the association with β-diversity (“adonis” function in package “vegan” 75 v2.5.6, 999 permutations). Permutation test was used to test the difference between within-donor distance and between-donor distance (999 permutations). To visualize the relationships among samples, principal coordinate analysis (PCoA) with Bray-Curtis distance method was applied. Taxa associations were performed at the phylum, class, order, family, genus and species and/or strain level. Functional associations were performed at the KEGG pathway level 3. Taxa/pathways with prevalence less than 10% or with the maximum proportion less 0.2% were excluded from testing to reduce the number of the tests. For human data, differential taxa abundance analysis was performed on the relative abundance data at each taxonomic rank using a permutation test (999 permutations). The permutation test was based on F-statistic of a linear model (square-root transformed taxa normalized abundance as the response variable) as the test statistics. For mice data, differential taxa abundance analysis was analyzed by fitting a generalized mixed effects model (“glmmPQL” function in package “MASS”76 v7.3.53) using Penalized Quasi-Likelihood (family = “quasipossion”) to account for overdispersion and correlations between samples from the same donor. log transformed GMPR size factor 77 was included as the offset to address sequence depth variation. To identify differentially abundant KEGG pathways associated with variable of interest, we fit a linear mixed effects model to the arcsine square root transformed relative abundance data of KEGG functions (level3). To test the association between taxa and log transformed PA value, spearman correlation analysis was used. For all taxa association analysis, false discovery rate (FDR) control (Benjamini-Hochberg procedure 78) was used for multiple testing correction (“p.adjust” function in package “stats” v3.6.1). FDR-adjusted p-values or q-values < 0.10 were considered significant.
Random forests 79 was used for predictive modeling based on the microbiota profile (genus-level relative abundance data) using default parameters of the R implementation of the algorithm (R package “randomForest”). Leave-one-out cross-validation was used to assess the classification accuracy in human data, and leave-one-donor-out was applied in mice data to account for potential dependency of the microbiome derived from the same donor. Receiver operating characteristic (ROC) curve 80 (“roc” function in package “pROC” v1.16.2) was used for assessing the prediction performance. The Boruta algorithm73 was used to select the taxa that have significant predictive power (R package “Boruta” v7.0.0). All the statistical analyses were performed in R-3.6.1 (R Development Core Teams).
In vivo permeability
Mice were removed from standard housing and fasted but allowed access to water for 2 hours prior to experimentation. Mice were then given a gavage solution (200 μL) containing 3 tracer molecules: creatinine (100mg/mL), 4kDa FITC-Dextran (60mg/mL) and 70kDa rhodamine B-dextran (Sigma). Five hours after gavage, blood was collected via intra-cardiac puncture and serum was separated using a 1.1 mL Z-gel micro tube (Sarstedt, NC, USA) by centrifugation at 10,000 g for 10 min at 4°C. Creatinine was measured using a COBAS c311 (Roche diagnostics) by the Mayo Clinic Immunochemical Core. Serum FITC and rhodamine B-dextran was evaluated using a Synergy Mx (BioTek) plate reader at excitation(s) of 485 and 553nm and emission wavelengths of 528 and 627nm respectively.
Tissue histology
Small intestinal and colonic tissue was fixed in 4% paraformaldehyde, embedded in OCT, sectioned (10 μm thickness) using a cryostat (Leica Biosystems, Lincolnshire, IL, USA) and mounted on slides. A standard hematoxylin and eosin staining protocol was followed. Slides were then graded in blinded fashion by a pathologist to assess gross differences in morphology and immune or other cellular infiltration.
Monocolonization of germ-free mice with β-glucuronidase (GUS) expressing E. coli
E. coli containing plasmid pOPS0750, a gift from Philip Poole (Addgene plasmid # 115511; http://n2t.net/addgene:115511; RRID:Addgene_115511) was grown in Luria Bertani (LB) broth + Kanamycin at 20 μg/mL overnight. Plasmid DNA was extracted using a Qiaprep Spin Miniprep Kit (Qiagen) according to manufacturer’s instructions and DNA concentration measured using a Qubit dsDNA kit (ThermoFisher). ArcticExpress (DE3) Competent E. coli (Agilent) were transformed (50 ng of plasmid) per the instructions provided. Transformed bacteria were plated on LB agar plates with Kanamycin (20 μg/mL) and Tetracycline (20 μg/mL) and incubated at 37°C overnight. Individual colonies were streaked for isolation with single colonies picked and then grown in LB broth. Overnight cultures were spun for 5 min at max speed and resuspended to an OD600 of 1.0 (10^9 cells/mL) in sterile PBS. 200 μL of transformed bacteria, or DE3 competent E. coli were used to monocolonize 10-week old female germ-free Swiss Webster Mice in a random order. Confirmation of monocolonization by either control or GUS+ E. coli was done by collecting pellets at day 7, and inoculating pellets in selective media at 37°C, examining for growth overnight. Pellets from culture positive, monocolonized animals were then used to assess fecal PA as described earlier in the methods.
Unconjugated bilirubin treatment in germ free mice
Unconjugated bilirubin (Sigma-Aldrich, B4126) was prepared (200 μM) in dimethyl sulfoxide (DMSO, 0.4%) immediately before use on ice in the dark. Mice were either given 200 μL once/day of unconjugated bilirubin (treated) or DMSO as a control (vehicle) over the course of 3 days. After day 3 of treatment, pellets were collected and trypsin-like activity in pellets was measured.
In vitro trypsin inhibition by unconjugated bilirubin
Metabolites within the bilirubin pathway were tested for the ability to inhibit trypsin activity in vitro. Biliverdin (Sigma Aldrich, 30891), mesobilirubin (Frontier Specialty Chemical, M588-9), urobilinogen (Mybiosource, MBS173206, unconjugated bilirubin (Sigma Aldrich, B4126) and conjugated bilirubin (Frontier Scientific, B850) were reconstituted to 1mg/mL. These stock solutions were then used to prepare a series of dilutions. 69μL of buffer (0.046M Tris-HCl, 0.0115M CaCl2, pH 8.1) was added to a black bottom plate along with 3μL of trypsin (10ng/μL) and 3μL of either unconjugated or conjugated bilirubin. A 100μM substrate solution of N-p-Tosyl-Gly-Pro-Arg 7-amido-methylcoumarin HCl was prepared in buffer as previously described and kept on ice. 75μL of substrate was automatically dispensed into each well to give a final reaction volume of 150μL. Reactions were briefly shaken and then transferred to a Synergy Mx Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA) and read kinetically every 30 seconds for a total of 5 min at 37°C. ΔRFU/min were calculated in the linear range and trypsin activity was expressed as a ratio of the fluorescence measured in the control trypsin well to the fluorescence measured in the presence of the potential metabolite inhibitor.
Fecal Microbiota Transfer (FMT)
To test the efficacy of an FMT in lowering fecal PA, 6-weeks post humanization, high PA humanized mice were given an oral gavage of healthy volunteer fecal slurry (200 μL, 1:2 with pre-reduced PBS under anaerobic conditions). Pellets were collected at day 3 and 1-week post FMT and assessed for fecal PA and changes in the microbiome.
Mice humanization with or without A. putredinis spiked feces
High PA PI-IBS patient fecal slurries were prepared as described with the following modifications. Mice were randomly divided into two groups, one receiving high PA fecal slurry and the other given high PA fecal slurry with A. putredinis (ATCC® 29800™) added. To generate the A. putredinis positive high PA fecal slurry, 1 mL of an overnight culture of A. putredinis OD 1 (10^9 cells/mL) was mixed with approximately 500 mg of high PA feces under anaerobic conditions. 4–6-week-old Swiss Webster mice were given an oral gavage (200 μL) of either high PA fecal slurry (high PA control) or high PA fecal slurry “spiked” with A. putredinis. Mice were housed for 6 weeks after which pellets collected from baseline and 6-weeks post gavage were evaluated for changes in fecal PA. The operator was not blinded to the assigned humanization group.
GUS inhibition in humanized mice
The role of GUS enzymes in regulating luminal PA was tested in healthy, low PA humanized mice using two inhibitors of GUS: broad spectrum inhibitor D-Glucaro-1,4-lactone and the loop 1 microbial specific GUS inhibitor UNC10201652. After 6-weeks post humanization with healthy humanized mice we administered either D-Glucaro-1,4-lactone (200 mg/kg), or UNC10201652 ((9(5-Morpholin-4-yl-8-piperazin-1-yl-1,2,3,4-tetrahydro-7-thia-6,9,10,11-tetraaza-benzo[c]fluorene) obtained from Dr. Matthew Redinbo, UNC Chapel Hill) at 1 mg/kg via oral gavage (200 μL) daily for 12 and 14 days respectively. Control mice for both treatments were run in parallel. Pellets were collected 3 hr post treatment as well on days 12 and 14 before sacrifice. Fecal PA was assessed as previously described and compared between the inhibitor treated and vehicle control mice. The operator was not blinded to the assigned humanization group.
Statistics and Reproducibility
All continuous variables are shown as means ± SD, and as frequencies and percentages for categorical variables. Non-Gaussian distributions were assumed, and statistical tests of using either 2-way Mann-Whitney U test or 2-way Wilcoxon signed-ranks were done to analyze data sets. When more than 2 groups were compared, non-parametric one-way analysis of variance (ANOVA) test (Kruskal-Wallis, Dunn’s Multiple Comparison) and 2-way ANOVA for paired comparisons with post hoc comparisons (Sidak’s, Tukey) were used. All experiments were conducted in triplicate unless otherwise noted and provided as a mean ± SD with results considered statistically significant at a p<0.05.
Extended Data
Extended Data Fig. 1. Taxa differentially abundant between low and high PA status and correlation with fecal PA.
a, The microbiota of high and low PA PI-IBS patients (n=12 high PA PI-IBS, 17 low PA PI-IBS) were compared at the phyla, class, and family levels. At both phyla and class levels of taxonomy, no significant differences were observed between the two groups of patients. However, one taxon, Rikenellaceae, was differential at the family level (FDR<0.05). Rikenellaceae, the family of A. putredinis and was in higher abundance in low PA PI-IBS subjects with an average abundance of 5.3%, compared to 1.1% in high PA PI-IBS individuals. b, Correlation scatterplots for differentially abundant taxa negatively correlating with PA. In addition to the top 3, an additional 8 differentially abundant (11/14) taxa correlated with low fecal PA. They exhibited weak negative correlation (0.31 ≤ |r| ≤ 0.38). High PA (orange) and healthy volunteers (green) are plotted. Correlation coefficients (r) and q-values are generated from comparisons within the entire cohort to assess the relationship between PA and taxa abundance (n=21 HV, 12 high PA PI-IBS, 17 low PA PI-IBS). Grey area shows 95% confidence level for linear smooths. c, Correlation scatterplots for differentially abundant taxa that positively correlate with PA. Of the 14 differential taxa, only 3 positively correlated with high fecal PA. Clostridium clostridioforme, Pseudomonas and Ruminococcus gnavus all had a weak, positive correlation (0.32 ≤ |r| ≤ 0.35). Plots were generated as stated above. Grey area shows 95% confidence level for linear smooths.
Extended Data Fig. 2. Specific serine protease activity assays in both high PA human and high PA humanized mouse fecal supernatants.
a, Specific serine protease activity of human fecal samples. Using an enzyme preferential substrate assay, fecal trypsin (*p=0.049, **p=0.004), chymotrypsin (**p=0.004) and pancreatic elastase (*p=0.03, **p=0.007) activities were increased in fecal supernatants generated from high PA individuals compared to both low PA PI-IBS and healthy volunteers (One-way ANOVA, multiple comparisons Kruskal-Wallis, n=6 FSNs/group). b, Specific activity of serine proteases in humanized mice and germ-free mice. Mice humanized with high PA microbiota had increased chymotrypsin (*p=0.049, **p=0.004), pancreatic elastase (healthy **p=0.008, low PA PI-IBS **p=0.002) and neutrophil elastase activity (*p=0.01, **p=0.002) in fecal supernatants compared to either healthy volunteer or low PA PI-IBS humanized mice. Decreased trypsin (healthy **p=0.008, low PA PI-IBS **p=0.002), chymotrypsin (**p=0.002), pancreatic elastase (**p=0.002) and neutrophil elastase (*p=0.02, **p=0.004) activity were seen in healthy and low PA PI-IBS humanized mice compared to the germ-free (GF) state. (One-way ANOVA, multiple comparisons Kruskal-Wallis, n=3 mice tested/humanization, 6 humanizations/phenotype, n=6 germ-free mice). Boxplots: lower, middle and upper hinges correspond to 25th, 50th and 75th percentiles. Upper and lower whiskers extend to the largest and smallest value no further than 1.5 * IQR from the respective hinge.
Extended Data Fig. 3. Tissue protease activity, gross morphology or histopathology of the high PA, low PA and healthy volunteer humanized mice.
a, In situ zymography for trypsin-like activity in mouse colonic tissue. No differences were observed between high PA and healthy humanized mice (n=6 mice/group, data presented as mean ±SD). b, Representative in situ zymography image of high PA and healthy mouse tissue. SYTOX Green Nuclear Stain (ThermoFisher, S7020) pseudocolored blue, N-p-Tosyl_Gly-Pro-Arg 7-amido-4-methylcoumarin hydrochloride cleavage pseudocolored green (Scale bar 50μm), c, Mouse weight, cecal weight and colonic length of humanized mice. Post-mortem weight was collected on humanized mice after which the gastrointestinal tract was removed and cecal weight was recorded. Colonic length measurements were done from proximal cecum to the distal rectum (n=6 human feces/phenotype, dots represent average). Boxplots: lower, middle and upper hinges correspond to 25th, 50th and 75th percentiles. Upper and lower whiskers extend to the largest and smallest value no further than 1.5 * IQR from the respective hinge. d, Histological examination of the gastrointestinal tract of humanized mice. Distal small bowel and distal colon tissue sections were evaluated by a pathologist (RG) in blinded manner. No observed differences in inflammation and presence of immune cells between humanized mice (n= 6 mice scored/phenotype, Scale bar 50μm).
Extended Data Fig. 4. Higher level taxonomic evaluation of human fecal samples used for humanization.
Comparisons were made between the microbiota of healthy, low PA and high PA PI-IBS fecal slurries used for mouse humanization (n=6 human feces/phenotype) which showed no significant differences at phylum, class and family levels.
Extended Data Fig. 5. Differentially abundant taxa and KEGG pathways between healthy, low PA and high PA PI-IBS humanized mice.
a, Comparison between healthy and high PA PI-IBS humanized mice engrafted microbiota. 32 differentially abundant taxa were identified between high PA with healthy humanized mice. Of the identified taxa, 13 were in greater fold abundance in healthy humanized mice compared to high PA PI-IBS mice. Colors denote greater abundance in the respective humanized group (green: healthy, orange: high PA PI-IBS, n=6 feces/phenotype). Numbers labeling the taxa correspond to the labels presented in the main manuscript, Fig. 4f. b, Differences in observed taxa between engrafted microbiota of low PA and high PA PI-IBS humanized mice. Microbiome analysis showed 25 differential taxa between low PA and high PA PI-IBS humanized groups, with 13 in greater abundance in low PA, and 12 in high PA. Colors denote greater abundance in the respective humanized group (blue: low PA PI-IBS, orange: high PA PI-IBS n=6 feces/phenotype). Numbers adjacent to taxa correspond to labels provided in main manuscript Figure 4g. c, Receiver operating curve assessing random forest ability to predict PA status based on taxa in humanized mice. The ability of random forest modelling algorithm to predict PA status based on selected taxa was assessed in humanized mice with an area under curve (AUC) of 0.914, (95% CI 0.848–0.981). Grey area shows confidence shape. d, Heatmap of predicted KEGG pathway differences between high PA and healthy humanized mice (n=6 feces/phenotype).
Extended Data Fig. 6. Fecal microbiome transfer in high PA humanized mice results in a compositional changes.
a, Microbiome profiles of high PA humanized mice receiving either a control or an FMT with healthy microbiota (n=9 mice/group) were compared at the phyla, class, and family levels. At both phyla and class levels of taxonomy, no significant differences were observed between the two groups of mice. However, at the family level, 9 differential taxa were observed. With increased abundance of Prevotellaceae, Eubacteriaceae, Enterobacteriaceae, Bacteroidaceae, and Clostridiaceae in controls while Odoribacteraceae, Rikenellaceae, Barnesiellaceae and Sutterellaceae were more abundant in mice receiving FMT (FDR<0.1).b, Correlation scatterplots for differentially abundant taxa that negatively correlate with PA post-FMT treatment. After FMT, three bacterial species were found to negatively correlate with fecal PA, all at a q< 0.1. Taxa identified were A. putredinis, Barnesiella intestinihominis, and A. obesi. All of these taxa had strong negative correlations with PA status in mice post-FMT (0.6 ≤ |r| ≤ 0.79). Correlation coefficients (r) and q-values are generated from comparisons within FMT and control animals to assess the relationship between PA and differentially abundant taxa (n=9 mice/group). Grey area shows 95% confidence level for linear smooths.
Extended Data Fig. 7. In vitro trypsin activity is suppressed by unconjugated bilirubin, and inhibition of GUS enzymes results in increased intestinal permeability.
a, Trypsin activity in the presence of metabolites within the bilirubin deconjugation pathway. Compared to the other metabolites used for experimentation, unconjugated bilirubin was the only metabolite that significantly inhibited trypsin activity across all concentrations examined. Data presented as Δfluorescence/time, normalized to a trypsin-only control (2-Way ANOVA, Tukey’s multiple comparison test, n=3 *p=0.001, data presented as mean ±SD). b, Time course inhibition of trypsin activity in the presence of bilirubin metabolites (n=3 biologically independent replicates, data presented as mean ±SD). c, Measurement of intestinal permeability in D-Glucaro-1,4-lactone treated humanized mice. Serum 4-kDa FITC-dextran levels were greater in healthy humanized mice treated with D-Glucaro-1,4-lactone indicating inhibition of GUS enzymes causes an increase in leak pathway permeability (2-sided Mann-Whitney, n=4/group *p=0.03). Boxplots: lower, middle and upper hinges correspond to 25th, 50th and 75th percentiles. Upper and lower whiskers extend to the largest and smallest value no further than 1.5 * IQR from the respective hinge. d, Proposed mechanism of microbial based inhibition of host proteases via the production of GUS enzymes.
Table 1.
Demographic and clinical characteristics of study volunteers
| Healthy (n=38) | High PA PI-IBS (n=19) | Low PA PI-IBS (n=33) | ANOVA | |
|
| ||||
| Gender (Female, Male) | 24, 14 | 11, 8 | 19, 14 | |
| Age (Mean, SD) | 43, 14 | 43, 15 | 48, 18 | 0.36 |
| BMI (Mean, SD) | 28.16, 5.10 | 31.6, 8.45 | 29.8, 6.47 | 0.35 |
| Race (% Caucasian, total n) | 93.5, 31 | 100, 16 | 100, 29 | |
| HADS - Anxiety (Mean, SD, n) | 2.65, 2.85, 31 | 5.38, 3.91, 16 | 5.24, 2.66, 29 | <0.001 |
| HADS - Depression (Mean, SD, n) | 0.77, 1.2, 31 | 2.75, 2.98, 16 | 2.83, 2.56, 29 | <0.0001 |
| BM/Day (Mean, SD, n) | 1.36, 0.54, 37 | 2.80, 1.16, 19 | 1.77, 0.77, 31 | <0.0001 |
| Bristol Stool Form (Mean, SD, n) | 4.07, 0.67, 37 | 5.07, 0.55, 19 | 4.27, 1.11, 31 | <0.0001 |
| Ease of Passage (Mean, SD, n) | 4.00, 0.14, 37 | 4.63, 0.56, 19 | 4.19, 0.46, 31 | <0.0001 |
|
| ||||
| High PA PI-IBS | Low PA PI-IBS | p-value (t-test) | ||
|
| ||||
| IBS Subtype (C, D, M) | 0, 11, 8 | 3, 19, 11 | ||
| IBS SSS (Mean, SD) | 274.4, 118.6 | 203.3, 111.2 | 0.08 | |
| IBS QOL (Mean, SD) | 28.78, 18.22 | 21.58, 17.80 | 0.13 | |
Statistical analysis: Non-parametric One-way ANOVA with Dunn’s multiple comparison post-hoc and Student’s t-test. Standard deviation (SD), Body mass index (BMI, kg/m2), Hospital anxiety and depression score (HADS; 0–7 normal, 8–10 borderline abnormal, 11–21 abnormal), Bowel movement (BM), Bristol stool form (1:constipation-7:diarrhea), Ease of passage scale (1–7), IBS subtype (C: Constipation, D: Diarrhea, M: Mixed) Symptom severity score (SSS; 75–175 mild 175–300 moderate, >300 severe), Quality of life (QOL; 34–170).
Table 2.
Enzymatic kinetics of trypsin inhibition by metabolites in bilirubin deconjugation pathway
| Inhibitor Name | Structure | Kobs (s−1) | k2/Ki (M−1s−1) | IC50 (μM) |
|---|---|---|---|---|
|
| ||||
| Biliverdin |
|
0.001822 | 181.8182 | 12.68 |
| Conjugated Bilirubin |
|
0.004937 | 500 | 4.679 |
| Unconjugated Bilirubin |
|
0.006642 | 666.6667 | 3.478 |
| Mesobilirubin |
|
0.005981 | 588.2353 | 3.862 |
| Urobilinogen |
|
0.004785 | 476.1905 | 4.828 |
Inhibition constants were measured in a 0.046M Tris-HCl, 0.0115M CaCl2, held at a pH 8.1 and 37°C. 10ug/mL of bovine pancreatic trypsin was used with the substrate N-p-Tosyl-Gly-Pro-Arg 7-amido-methylcoumarin HCl stock at 100μM. Each well has a final volume of 150μL: 3μL trypsin, 3μL of inhibitor, 69μL of buffer, 75μL of buffered substrate. The second-order inhibition rate (k2/Ki (M−1s−1)) was generated by carrying out a series of experiments at different concentrations of the potential inhibitory metabolite(s). Non-linear regression modelling was done to determine the IC50 values for the metabolites within the bilirubin deconjugation pathway (n=3 replicates/metabolite).
Supplementary Material
Acknowledgements:
Authors acknowledge Ms. Lori Anderson and Ms. Kristy Zodrow for administrative support. We thank the Center for Mass Spectrometry and Proteomics at the University of Minnesota for resources used to generate and analyze fecal metaproteomics data.
DK 103911 through the National Institute of Health to MG.
DK 120745 through the National Institute of Health to MG.
DK 127998 through the National Institute of Health to MG
Support was provided through Mayo Clinic Research Pipeline K2R Program Award to MG. Supported by Pilot & Feasibility Award from Mayo Clinic Cell Signaling in Gastroenterology via P30DK084567 through the National Institute of Health to MG
These grants provided funds for data collection, analysis and personnel support.
Footnotes
Competing interests: Dr. Grover has disclosure to Mayo Clinic ventures on microbes described in this manuscript. None to disclose for other co-authors.
Code availability
Proteomics codes can be found on:
https://github.com/galaxyproteomics/metaquantome
https://github.com/galaxyproteomics/tools-galaxyp/tree/master/tools/unipept
https://bioconductor.org/packages/release/bioc/html/PECA.html.
Microbiome codes are available at:
Data, and materials availability:
Raw data for human and mouse metagenomics are publicly available via sequence read archive (SRA) under the BioProject accession IDs PRJNA705217 and PRJNA705695 respectively. Human RNAseq data is deposited in the Gene Expression Omnibus (GEO) under accession number GSE168759. Fecal metaproteomic data is available at PRIDE PXD025127 and descriptors provided in Supplementary file 1. Human colonic proteomics dataset is available as figure 2 source data. Human and mice metabolomic raw reads are available at Metabolomics Workbench via ST002094 and ST002090 respectively (https://www.metabolomicsworkbench.org/).
References
- 1.Turk B Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov 5, 785–799, doi: 10.1038/nrd2092 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Edogawa S et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut 69, 62–73, doi: 10.1136/gutjnl-2018-317416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cenac N et al. Role for protease activity in visceral pain in irritable bowel syndrome. J Clin Invest 117, 636–647, doi: 10.1172/JCI29255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annahazi A et al. Luminal cysteine-proteases degrade colonic tight junction structure and are responsible for abdominal pain in constipation-predominant IBS. Am J Gastroenterol 108, 1322–1331, doi: 10.1038/ajg.2013.152 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Denadai-Souza A et al. Functional proteomic profiling of secreted serine proteases in health and inflammatory bowel disease. Sci Rep 8, 7834, doi: 10.1038/s41598-018-26282-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galipeau HJ et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology, doi: 10.1053/j.gastro.2020.12.004 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Klem F et al. Prevalence, Risk Factors, and Outcomes of Irritable Bowel Syndrome After Infectious Enteritis: A Systematic Review and Meta-analysis. Gastroenterology 152, 1042–1054 e1041, doi: 10.1053/j.gastro.2016.12.039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbara G et al. Rome Foundation working team report on post-infection irritable bowel syndrome. Gastroenterology 156, 46–58 e47, doi: 10.1053/j.gastro.2018.07.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wouters MM et al. Psychological comorbidity increases the risk for postinfectious IBS partly by enhanced susceptibility to develop infectious gastroenteritis. Gut 65, 1279–1288, doi: 10.1136/gutjnl-2015-309460 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Sundin J et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther 41, 342–351, doi: 10.1111/apt.13055 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Jalanka-Tuovinen J et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 63, 1737–1745, doi: 10.1136/gutjnl-2013-305994 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Xu D et al. Efficacy of fecal microbiota transplantation in irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol 114, 1043–1050, doi: 10.14309/ajg.0000000000000198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Salhy M, Hatlebakk JG, Gilja OH, Brathen Kristoffersen A & Hausken T Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut 69, 859–867, doi: 10.1136/gutjnl-2019-319630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford AC, Harris LA, Lacy BE, Quigley EMM & Moayyedi P Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharmacol Ther 48, 1044–1060, doi: 10.1111/apt.15001 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Bohe M, Borgstrom A, Genell S & Ohlsson K Determination of immunoreactive trypsin, pancreatic elastase and chymotrypsin in extracts of human feces and ileostomy drainage. Digestion 27, 8–15, doi: 10.1159/000198913 (1983). [DOI] [PubMed] [Google Scholar]
- 16.Genell S & Gustafsson BE Impaired enteric degradation of pancreatic endopeptidases in antibiotic-treated rats. Scand J Gastroenterol 12, 801–809, doi: 10.3109/00365527709181723 (1977). [DOI] [PubMed] [Google Scholar]
- 17.Vergnolle N Protease inhibition as new therapeutic strategy for GI diseases. Gut 65, 1215–1224, doi: 10.1136/gutjnl-2015-309147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galipeau HJ et al. Novel role of the serine protease inhibitor elafin in gluten-related disorders. Am J Gastroenterol 109, 748–756, doi: 10.1038/ajg.2014.48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motta JP et al. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci Transl Med 4, 158ra144, doi: 10.1126/scitranslmed.3004212 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Qin X Inactivation of digestive proteases by deconjugated bilirubin: the possible evolutionary driving force for bilirubin or biliverdin predominance in animals. Gut 56, 1641–1642, doi: 10.1136/gut.2007.132076 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollet RM et al. An atlas of beta-glucuronidases in the human intestinal microbiome. Structure 25, 967–977 e965, doi: 10.1016/j.str.2017.05.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y et al. Bacterial beta-glucuronidase alleviates dextran sulfate sodium-induced colitis in mice: A possible crucial new diagnostic and therapeutic target for inflammatory bowel disease. Biochem Biophys Res Commun 513, 426–433, doi: 10.1016/j.bbrc.2019.03.196 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Berumen A et al. Characteristics and Risk Factors of Post-Infection Irritable Bowel Syndrome After Campylobacter Enteritis. Clin Gastroenterol Hepatol, doi: 10.1016/j.cgh.2020.07.033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon H et al. Increased pancreatic protease activity in response to antibiotics impairs gut barrier and triggers colitis. Cell Mol Gastroenterol Hepatol 6, 370–388 e373, doi: 10.1016/j.jcmgh.2018.05.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank C et al. Disseminating metaproteomic informatics capabilities and knowledge using the Galaxy-P framework. Proteomes 6, doi: 10.3390/proteomes6010007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Easterly CW et al. metaQuantome: An Integrated, Quantitative Metaproteomics Approach Reveals Connections Between Taxonomy and Protein Function in Complex Microbiomes. Mol Cell Proteomics 18, S82–S91, doi: 10.1074/mcp.RA118.001240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawlings ND et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46, D624–D632, doi: 10.1093/nar/gkx1134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolland-Fourcade C et al. Epithelial expression and function of trypsin-3 in irritable bowel syndrome. Gut 66, 1767–1778, doi: 10.1136/gutjnl-2016-312094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs JP et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol 2, 750–766, doi: 10.1016/j.jcmgh.2016.06.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores R et al. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PLoS One 7, e39745, doi: 10.1371/journal.pone.0039745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creekmore BC et al. Mouse Gut Microbiome-Encoded beta-Glucuronidases Identified Using Metagenome Analysis Guided by Protein Structure. mSystems 4, doi: 10.1128/mSystems.00452-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macfadyen A & Ho KJ D-glucaro-1,4-lactone: its excretion in the bile and urine and effect on the biliary secretion of beta-glucuronidase after oral administration in rats. Hepatology 9, 552–556, doi: 10.1002/hep.1840090408 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Pellock SJ et al. Gut microbial beta-glucuronidase inhibition via catalytic cycle interception. ACS Cent Sci 4, 868–879, doi: 10.1021/acscentsci.8b00239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gecse K et al. Increased faecal serine protease activity in diarrhoeic IBS patients: a colonic lumenal factor impairing colonic permeability and sensitivity. Gut 57, 591–599, doi: 10.1136/gut.2007.140210 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Tooth D et al. Characterisation of faecal protease activity in irritable bowel syndrome with diarrhoea: origin and effect of gut transit. Gut 63, 753–760, doi: 10.1136/gutjnl-2012-304042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll IM & Maharshak N Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J Gastroenterol 19, 7531–7543, doi: 10.3748/wjg.v19.i43.7531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jablaoui A et al. Fecal serine protease profiling in inflammatory bowel diseases. Front Cell Infect Microbiol 10, 21, doi: 10.3389/fcimb.2020.00021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caminero A et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat Commun 10, 1198, doi: 10.1038/s41467-019-09037-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarville JL et al. A commensal Bifidobacterium longum strain prevents gluten-related immunopathology in mice through expression of a serine protease inhibitor. Appl Environ Microbiol 83, doi: 10.1128/AEM.01323-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker BJ, Wearsch PA, Veloo ACM & Rodriguez-Palacios A The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol 11, 906, doi: 10.3389/fimmu.2020.00906 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dziarski R, Park SY, Kashyap DR, Dowd SE & Gupta D Pglyrp-regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One 11, e0146162, doi: 10.1371/journal.pone.0146162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butera A et al. Nod2 deficiency in mice is associated with microbiota variation favouring the expansion of mucosal CD4+ LAP+ regulatory cells. Sci Rep 8, 14241, doi: 10.1038/s41598-018-32583-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Palacios A et al. The artificial sweetener Splenda promotes gut proteobacteria, dysbiosis, and myeloperoxidase reactivity in Crohn’s disease-like ileitis. Inflamm Bowel Dis 24, 1005–1020, doi: 10.1093/ibd/izy060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genell S, Gustafsson BE & Ohlsson K Quantitation of active pancreatic endopeptidases in the intestinal contents of germfree and conventional rats. Scand J Gastroenterol 11, 757–762 (1976). [PubMed] [Google Scholar]
- 45.Carroll IM et al. Fecal protease activity is associated with compositional alterations in the intestinal microbiota. PLoS One 8, e78017, doi: 10.1371/journal.pone.0078017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roka R et al. Colonic luminal proteases activate colonocyte proteinase-activated receptor-2 and regulate paracellular permeability in mice. Neurogastroenterol Motil 19, 57–65, doi: 10.1111/j.1365-2982.2006.00851.x (2007). [DOI] [PubMed] [Google Scholar]
- 47.Aroniadis OC et al. Faecal microbiota transplantation for diarrhoea-predominant irritable bowel syndrome: a double-blind, randomised, placebo-controlled trial. Lancet Gastroenterol Hepatol 4, 675–685, doi: 10.1016/S2468-1253(19)30198-0 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Halkjaer SI et al. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut 67, 2107–2115, doi: 10.1136/gutjnl-2018-316434 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Holster S et al. The effect of allogenic versus autologous fecal microbiota transfer on symptoms, visceral perception and fecal and mucosal microbiota in irritable bowel syndrome: a randomized controlled study. Clin Transl Gastroenterol 10, e00034, doi: 10.14309/ctg.0000000000000034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goll R et al. Effects of fecal microbiota transplantation in subjects with irritable bowel syndrome are mirrored by changes in gut microbiome. Gut Microbes 12, 1794263, doi: 10.1080/19490976.2020.1794263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smillie CS et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240 e225, doi: 10.1016/j.chom.2018.01.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuno S et al. Bifidobacterium-rich fecal donor may be a positive predictor for successful fecal microbiota transplantation in patients with irritable bowel syndrome. Digestion 96, 29–38, doi: 10.1159/000471919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mkaouar H et al. Siropins, novel serine protease inhibitors from gut microbiota acting on human proteases involved in inflammatory bowel diseases. Microb Cell Fact 15, 201, doi: 10.1186/s12934-016-0596-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Awolade P et al. Therapeutic significance of beta-glucuronidase activity and its inhibitors: A review. Eur J Med Chem 187, 111921, doi: 10.1016/j.ejmech.2019.111921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamoud AR, Weaver L, Stec DE & Hinds TD Jr. Bilirubin in the liver-gut signaling axis. Trends Endocrinol Metab 29, 140–150, doi: 10.1016/j.tem.2018.01.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ervin SM et al. Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens. J Biol Chem 294, 18586–18599, doi: 10.1074/jbc.RA119.010950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatt AP et al. Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy. Proc Natl Acad Sci U S A 117, 7374–7381, doi: 10.1073/pnas.1918095117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace BD et al. Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol 22, 1238–1249, doi: 10.1016/j.chembiol.2015.08.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koren O et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480, doi: 10.1016/j.cell.2012.07.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arrieta MC et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7, 307ra152, doi: 10.1126/scitranslmed.aab2271 (2015). [DOI] [PubMed] [Google Scholar]
- 61.De Palma G et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med 9, doi: 10.1126/scitranslmed.aaf6397 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Longstreth GF et al. Functional bowel disorders. Gastroenterology 130, 1480–1491, doi: 10.1053/j.gastro.2005.11.061 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Francis CY, Morris J & Whorwell PJ The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Ther 11, 395–402, doi: 10.1046/j.1365-2036.1997.142318000.x (1997). [DOI] [PubMed] [Google Scholar]
- 64.Drossman DA et al. Further validation of the IBS-QOL: a disease-specific quality-of-life questionnaire. Am J Gastroenterol 95, 999–1007, doi: 10.1111/j.1572-0241.2000.01941.x (2000). [DOI] [PubMed] [Google Scholar]
- 65.Blake MR, Raker JM & Whelan K Validity and reliability of the Bristol Stool Form Scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 44, 693–703, doi: 10.1111/apt.13746 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Zigmond AS & Snaith RP The hospital anxiety and depression scale. Acta Psychiatr Scand 67, 361–370, doi: 10.1111/j.1600-0447.1983.tb09716.x (1983). [DOI] [PubMed] [Google Scholar]
- 67.BBMap v. 38.73 (https://sourceforge.net/projects/bbmap/, 2019).
- 68.Hillmann B et al. SHOGUN: a modular, accurate and scalable framework for microbiome quantification. Bioinformatics 36, 4088–4090, doi: 10.1093/bioinformatics/btaa277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Ghalith G & Knights D BURST enables mathematically optimal short-read alignment for big data. bioRxiv, 2020.2009.2008.287128, doi: 10.1101/2020.09.08.287128 (2020). [DOI] [Google Scholar]
- 70.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15, 962–968, doi: 10.1038/s41592-018-0176-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzek BE, Huang H, McGarvey P, Mazumder R & Wu CH UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288, doi: 10.1093/bioinformatics/btm098 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Flores R, Shi J, Gail MH, Ravel J & Goedert JJ Assessment of the human faecal microbiota: I. Measurement and reproducibility of selected enzymatic activities. Eur J Clin Invest 42, 848–854, doi: 10.1111/j.1365-2362.2012.02660.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kursa MB & Rudnicki WR Feature Selection with the Boruta Package. J Stat Softw 36, 1–13, doi:DOI 10.18637/jss.v036.i11 (2010). [DOI] [Google Scholar]
- 74.Fitzpatrick LR et al. Bacillus coagulans GBI-30, 6086 limits the recurrence of Clostridium difficile-Induced colitis following vancomycin withdrawal in mice. Gut Pathogens 4, 13, doi: 10.1186/1757-4749-4-13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dixon P VEGAN, a package of R functions for community ecology. J Veg Sci 14, 927–930 (2003). [Google Scholar]
- 76.Ripley B, Venables B, Bates DM, Hornik K, Gebhardt A, Firth D Package “MASS” Cran R 538 (2013). [Google Scholar]
- 77.Chen L et al. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. Peerj 6, doi:ARTN e460010.7717/peerj.4600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benjamini Y & Hochberg Y Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B 57, 289–300, doi:DOI 10.1111/j.2517-6161.1995.tb02031.x (1995). [DOI] [Google Scholar]
- 79.Breiman L Random forests. Mach Learn 45, 5–32, doi:Doi 10.1023/A:1010933404324 (2001). [DOI] [Google Scholar]
- 80.Robin X et al. pROC: an open-source package for R and S plus to analyze and compare ROC curves. Bmc Bioinformatics 12, doi:Artn 7710.1186/1471-2105-12-77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for human and mouse metagenomics are publicly available via sequence read archive (SRA) under the BioProject accession IDs PRJNA705217 and PRJNA705695 respectively. Human RNAseq data is deposited in the Gene Expression Omnibus (GEO) under accession number GSE168759. Fecal metaproteomic data is available at PRIDE PXD025127 and descriptors provided in Supplementary file 1. Human colonic proteomics dataset is available as figure 2 source data. Human and mice metabolomic raw reads are available at Metabolomics Workbench via ST002094 and ST002090 respectively (https://www.metabolomicsworkbench.org/).