Abstract
Membraneless organelles, such as germ granules and stress granules, are liquid-like condensates formed by phase transition. Recently, we and others have adopted proximity-based labeling methods to determine the composition of these membraneless compartments. Here, we describe the use of TurboID—an engineered promiscuous biotin ligase—to label and purify proteins localizing to Caenorhabditis elegans germ granules, known as P granules. We provide a detailed protocol for visualization of the subcellular localization of biotinylated proteins from dissected gonads, assessment of TurboID enrichment using streptavidin blots, and enrichment of biotinylated proteins under stringent conditions. Altogether, this protocol provides a workflow to unravel the proteome of C. elegans germ granules. Importantly, the assays described here can be applied to interrogate many membraneless organelles, in a diversity of living multicellular organisms.
Graphical abstract:
Keywords: TurboID, Proximity labeling, C. elegans, P granules, RNA granules, Membraneless organelles
Background
Germ granules are evolutionarily conserved membraneless organelles, assembled by weak, transient, and multivalent interactions between proteins and RNAs ( Voronina et al., 2011 ; Banani et al., 2017 ; Trcek and Lehmann, 2019). Due to the nature of liquid-like condensates, germ granules cannot be easily purified by a conventional fractionation-based method. So far, their composition remains largely elusive. Recent advances in enzyme-catalyzed proximity-based labeling provide a powerful approach to define the constituents of germ granules ( Roux et al., 2012 ; Rhee et al., 2013 ; Branon et al., 2018 ). Proximity labeling uses a promiscuous biotin ligase, such as BioID, APEX, or TurboID ( Qin et al., 2021 ). While all three enzymes have been applied to map protein-protein interactions, TurboID possesses at least three advantages in living organisms such as C. elegans ( Price et al., 2021 ; Sanchez et al., 2021 ; Artan et al., 2021 ), i.e., 1) TurboID is active at 20°C, an optimum temperature for C. elegans cultivation ( Branon et al., 2018 ), 2) APEX requires exogenous reagents, which are toxic and cannot easily penetrate the cuticle of worms ( Rhee et al., 2013 ), whereas TurboID uses ATP and biotin, substrates that are readily available in vivo, and 3) TurboID catalyzes biotinylation with greater efficiency than BioID ( Branon et al., 2018 ).
Using the C. elegans germ granule called P granule as a model system, we utilized TurboID in conjunction with mass spectrometry, to comprehensively define its protein components ( Price et al., 2021 ). Here, we describe in detail the workflow to label, visualize, and purify P granule proteins from a strain expressing TurboID-tagged GLH-1, a Vasa helicase known to reside in P granules ( Gruidl et al., 1996 ; Spike et al., 2008 ; Gustafson and Wessel, 2010). Specifically, we use streptavidin blotting analysis, to assess the activity of TurboID, and immunofluorescence staining of dissected gonads, to examine the subcellular localization of biotinylated proteins. We report a stringent purification method to enrich biotinylated proteins, which can be further identified by mass spectrometry ( Price et al., 2021 ). Altogether, our protocol provides the proximity-labeling workflow for defining components of C. elegans germ granules. It is our hope that the level of detail in this protocol enables future investigators to probe diverse phase-separated membraneless organelles in living plants and animals.
Materials and Reagents
N2 (Caenorhabditis Genetics Center)
TurboID::glh-1 (The strain is available upon request)
150 mm × 15 mm Petri Dish (Fisher Scientific, catalog number: FB0875714)
22 G syringe needles (Becton Dickinson, catalog number: 305155)
15 mL conical tube (Genesee Scientific, catalog number: 28-103)
Low Binding Microcentrifuge Tubes (Sorenson Bioscience, catalog number: 39640T)
Coverslip 18 × 18 mm (Globe Scientific, catalog number: 1401-10)
OP50-1 Escherichia coli (Caenorhabditis Genetics Center)
Slides for dissection (Globe Scientific, catalog number: 1301)
Slides for microscopy (Fisher Scientific, catalog number: 22-339-408)
Streptavidin Alexa Fluor 488, 2 mg/mL (Invitrogen, catalog number: S11223)
NaCl (VWR, catalog number: BDH9286)
KH2PO4 (Fisher Scientific, catalog number: BP 362-1)
MgSO4 (Fisher Scientific, catalog number: BP213-1)
Lysing Matrix D (MP Biomedicals, catalog number: 6913100)
Tris Base (Fisher Scientific, catalog number: BP152-1)
Sodium Dodecyl Sulfate (Fischer Scientific, catalog number: BP166-100)
Sodium Deoxycholate (Thermo Scientific, catalog number: 89904)
Triton X-100 (Fisher Scientific, catalog number: BP151-500)
cOmplete, Mini EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, catalog number: 11836170001)
PMSF (VWR, catalog number: 97064-898)
Pepstatin A (Sigma-Aldrich, catalog number: P5318)
PierceTM Streptavidin beads (Thermo Scientific, catalog number: 88816)
KCl (VWR, catalog number: BDH9258)
Na2CO3 (Fisher Scientific, catalog number: S263-500)
Urea (Acros Organics, catalog number: 327380010)
Na2HPO4 (Fisher Scientific, catalog number: BP332-1)
KH2PO4 (Fisher Scientific, catalog number: BP362-1)
4× BoltTM LDS Sample Buffer (Fisher Scientific, catalog number: B0007)
SimplyBlue SafeStain (Invitrogen, catalog number: LC6060)
Streptavidin, horseradish peroxidase conjugate (Invitrogen, catalog number: S911)
Antifade mounting medium with DAPI (Vector Laboratories, catalog number: H-1200)
Clarity Western ECL Substrate (Bio-Rad, catalog number: 1705060)
PVDF membrane (Bio-Rad, catalog number: 1620262)
Biotin (Sigma, catalog number: 54639)
DTT (Sigma, catalog number: D0632)
NuPAGE 4–12% Bis-Tris Gel (Invitrogen, catalog number: NP0323BOX)
Paraformaldehyde (Fisher Scientific, catalog number: O4042-500)
Methanol (Fisher Scientific, catalog number: A408-4)
Tween 20 (Fisher Scientific, catalog number: BP337-500)
Tetramisole (Sigma Aldrich, catalog number: L9756-5G)
Nail polish (Electron Microscopy Sciences, catalog number: 72180)
1× M9 (see Recipes)
RIPA buffer (see Recipes)
Solution P (see Recipes)
RIPA buffer with Protease inhibitors (see Recipes)
PBS (see Recipes)
PBST (see Recipes)
Equipment
Open Air Rocker (Fisher Scientific, model: 88861025)
Bead Mill Homogenizer (Fisher Scientific, model: 24)
Large Centrifuge (Beckman Coulter Life Sciences, model: X-30R)
Refrigerated Centrifuge (Eppendorf, model: 5424R)
Rotator (Benchmark Scientific, model: R2024)
Magnetic Separation Stand (Promega, model: Z5343)
Vortex (MSP, model: BV1000)
Mini Gel Tank (Fisher Scientific, catalog number: A25977)
Dissecting microscope (Zeiss, model: Stemi 508)
Inverted Nikon TiE microscope equipped with a CSU-W1 confocal scanner, and a 488 nm and 405 nm lasers (or similar microscope)
Software
ImageJ (https://imagej.nih.gov/ij/)
Procedure
-
Examination of germline biotinylated proteins by fluorescent streptavidin staining
Suspend 30–60 adult worms in M9 buffer, and transfer them to a low binding microcentrifuge tube.
Allow animals to sink to the bottom of the tube, and wash three times with approximately 1 mL of M9 to remove bacteria. Remove M9 between washes by pipetting.
Remove supernatant, add 1 volume (approximately 25 μL) of M9 containing 0.5 mM tetramisole and 0.05% Tween 20, and transfer samples to a dissection slide.
Under a dissecting microscope, dissect worms by decapitating animals near the pharynx: place two 22 G needles on either side of the head, and move the needles in a scissor-like motion ( Day et al., 2020 ).
-
Transfer dissected animals to a low-binding microcentrifuge tube, using a mouth pipette. Add 6 volumes (~150 μL) of 3.3% (w/v) paraformaldehyde in PBST to the sample, and incubate at room temperature for 15 min.
Note: Allow dissected worms to settle to the bottom of the tube by gravity between washes. Centrifugation may damage samples.
Remove fixative by pipetting, wash twice with 1 mL of PBS, and add 1 mL of methanol at -20°C. Incubate samples at -20°C for at least 15 min.
Remove methanol by pipetting, wash samples twice with 1.5 mL of PBST.
While waiting for samples to settle, prepare staining solution in the dark, by making a 1:500 dilution of the streptavidin AF488 stock in PBST.
-
Add 100 μL of staining solution to the samples, and incubate in the dark at 4°C overnight.
Note: All remaining steps should be carried out in the dark and sample exposure to light should be minimized.
Remove as much staining solution as possible by pipetting. Wash three times with 1.5 mL of PBST for 15 min, then once with 1.5 mL of PBS for 5 min.
Remove as much PBS as possible by pipetting, and add 50 μL of antifade mounting medium with DAPI to each tube.
Suspend samples in mounting medium with a glass Pasteur pipette, then transfer ~5 μL of sample to a microscopy slide. Cover sample with a clean coverslip, and seal with nail polish.
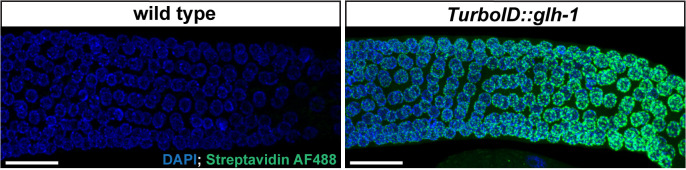
Image samples using confocal fluorescence microscopy (Figure 1).
-
Cultivation of TurboID-tagged C. elegans strains
-
Grow 60,000 synchronized L1 larvae on a 150 mm × 15 mm NGM plate seeded with concentrated OP50-1 Escherichia coli (~2 mL) at 20°C.
Note: Standard cultivation conditions for C. elegans, recipes for Nematode Growth Media (NGM), and production of concentrated bacteria can be found at wormbook.org.
Harvest non-starved gravid worms from plate(s) by pipetting 10 mL of M9 onto the plate, and transferring worm suspension into a 15-mL conical tube.
Spin worms down at 800 × g for 1 min, and replace with fresh M9. Re-spin worms down, replace with fresh M9, then gently invert for 10 min, to remove bacteria.
Wash once more with M9, then ddH2O, and then RIPA.
Resuspend in 2 mL of RIPA buffer with protease inhibitors (4× worm volume).
Quick freeze in liquid nitrogen. Samples can be stored at -80°C before proceeding.
-
-
Homogenization of C. elegans and lysate preparation
Thaw samples, and transfer worm suspension to 2-mL screw cap Lysing Matrix D tubes, which contain ceramic spheres.
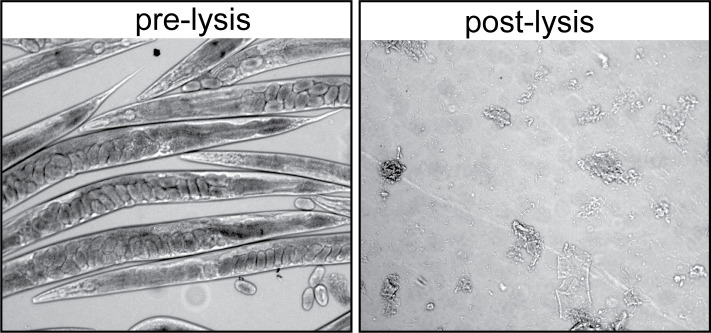
Grind worms 3–4 times (speed =6, time =45 s), cooling samples on ice for 2 min in between, to avoid overheating. Check lysis efficiency under the dissecting microscope (Figure 2).
Quickly spin down in a refrigerated centrifuge. Transfer supernatant to a fresh microcentrifuge tube.
-
Spin twice at 14,000 × g for 8 min. Each time, transfer the supernatant to a fresh tube. Avoid contamination with pellet and lipid interface. Typically, 1500 µL of lysate can be recovered.
Note: Keep samples on ice to prevent protein degradation.
-
Streptavidin affinity pulldown
Save an aliquot of input (20 µL) for the streptavidin blot.
-
Equilibrate streptavidin beads (80 µL per sample) by washing with RIPA buffer with protease inhibitors twice. At each wash, centrifuge the beads at 94 × g for 30 s. Wait 1–2 min to let the beads settle down. Separate beads from wash in a magnetic stand, and remove the supernatant without disturbing the beads.
Note: The amount of streptavidin beads can be adjusted, depending on the abundance of biotinylated proteins.
Mix the protein lysate with the beads.
Incubate with constant rotation (~14 rpm) at 4°C overnight.
Separate beads from lysate, by placing samples in a magnetic stand. Remove supernatant. Save an aliquot of flow-through (20 µL) for the streptavidin blot.
Wash beads with the following series of buffers to remove nonspecifically bound proteins, each time rotating the samples for 5 min and quickly spinning down at 94 × g, twice with RIPA lysis buffer, once with 1 M KCl, once with 0.1 M Na2CO3, once with 2 M urea in 10 mM Tris-HCl (pH 8), twice again with RIPA lysis buffer, and three times with PBS.
Resuspend beads in 50 µL of PBS. Resuspended beads are ready for on-bead digest and mass spectrometry analysis.
Save an aliquot of resuspended beads (10 µL) for the streptavidin blot.
-
Streptavidin blotting analysis
Elute proteins from resuspended beads using 20 µL of 4× LDS Sample Buffer supplemented with 20 mM DTT and 2 mM biotin. Boil samples at 95°C for 10 min. Vortex briefly and transfer the supernatant (~30 µL) to a fresh tube.
Load 5 µL/1,500 µL (0.33%) input, 5 µL/1,500 µL (0.33%) flow-through, and 3.3 µL/50 µL (6.67%) pulldown samples in a 4–12% Bis-Tris Gel, and run the gel according to the manufacturer's instructions.
Transfer samples to a PVDF membrane at 20V for 45 min.
Incubate the PVDF membrane overnight with Streptavidin-HRP (1:4,000).
-
Develop with Clarity Western ECL substrates (1:1) for 5 min.
Note: The membrane can be stained with Coomassie blue dyes for the loading control.
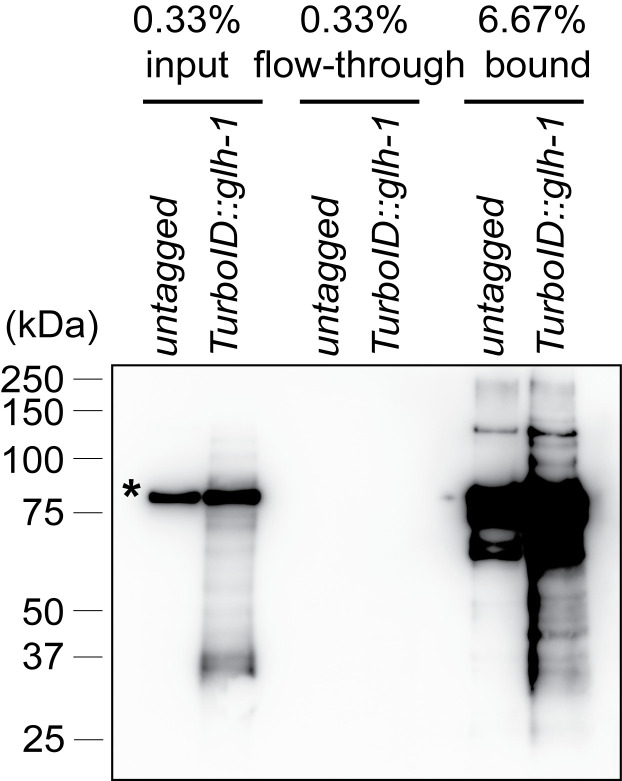
If streptavidin affinity pulldown is successful, biotinylated proteins should be depleted in the flow-through fraction, and enriched in the bound fraction (Figure 3).
Figure 1. Streptavidin-Alexa Fluor 488 staining of gonads dissected from wild-type and TurboID::glh-1 animals.
Representative maximum intensity projection of z-slices spanning the depth of the gonad in the late pachytene region. Robust perinuclear signals are observed in TurboID::glh-1 gonads, but not in wild-type gonads. DAPI stains DNA. Streptavidin-Alexa Fluor 488 stains biotinylated molecules. Scale bar = 20 μm. Images were acquired using a 60× water immersion objective.
Figure 2. Homogenization of C. elegans samples.
Pre-lysis and post-lysis suspension images were acquired using a 10× objective. After successful homogenization, intact worms are no longer visible and samples become turbid.
Figure 3. Streptavidin-horseradish peroxidase blotting.
Biotinylated proteins are present in the input, depleted in the flow-through and enriched in the pulldown fraction prepared from wild-type C. elegans and a strain expressing TurboID::GLH-1. An asterisk marks the endogenously biotinylated mitochondrial protein PCCA-1.
Recipes
-
1× M9
3 g KH2PO4
6 g Na2HPO4
5 g NaCl
1 mL of 1 M MgSO4
Bring up volume to 1 L with ddH2O
Sterilize by autoclaving and store at room temperature.
Note: MgSO4 can be added after autoclaving to prevent precipitation.
-
RIPA buffer
50 mM Tris-HCl (pH 7.5)
150 mM NaCl
0.125% SDS
0.125% sodium deoxycholate
1% Triton X-100 in Millipore water
Filter-sterilize and store at 4°C.
-
Solution P
90 mg PMSF
2 mg Pepstatin A
5 mL of Ethanol
Store at -20°C.
-
RIPA buffer with protease inhibitors
Crush 1 tablet of cOmplete protease inhibitor cocktail and add to 10 mL of RIPA. Vortex briefly until dissolved.
Add 100 μL of solution P.
Use immediately or store at -80°C.
-
PBS
Dissolve the following in 800 mL of ddH2O:
80 g NaCl
2 g KCl
14.4 g Na2HPO4
2.4 g KH2PO4
Adjust pH to 7.4
Bring up volume to 1 L with ddH2O.
Sterilize by autoclaving, and store at room temperature.
-
PBST
Add 25 μL of Tween 20 to 50 mL of PBS.
Store at room temperature.
Acknowledgments
We thank D. Schoenberg for comments and the Neuroscience Imaging Core for instruments (P30NS104177). OP50-1 Escherichia coli was provided by the Caenorhabditis Genetics Center supported by NIH (P40-OD010440). This work was supported by NIH Pathway to Independence Award (R00GM124460) and Maximizing Investigators' Research Award (R35GM142580) to W.T.
This protocol was derived from the original research paper “Proximity labeling identifies LOTUS domain proteins that promote the formation of perinuclear germ granules in C. elegans” ( Price et al., 2021 ).
Competing interests
The authors declare that they have no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Artan M., Barratt S., Flynn S. M., Begum F., Skehel M., Nicolas A. and de Bono M.(2021). Interactome analysis of Caenorhabditis elegans synapses by TurboID-based proximity labeling . J Biol Chem 297(3): 101094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banani S. F., Lee H. O., Hyman A. A. and Rosen M. K.(2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18(5): 285-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branon T. C., Bosch J. A., Sanchez A. D., Udeshi N. D., Svinkina T., Carr S. A., Feldman J. L., Perrimon N. and Ting A. Y.(2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36(9): 880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day N. J., Wang X. and Voronina E.(2020). In Situ Detection of Ribonucleoprotein Complex Assembly in the C. elegans Germline using Proximity Ligation Assay . J Vis Exp (159): 10.3791/60982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gruidl M. E., Smith P. A., Kuznicki K. A., McCrone J. S., Kirchner J., Roussell D. L., Strome S. and Bennett K. L.(1996). Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans . Proc Natl Acad Sci U S A 93(24): 13837-13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gustafson E. A. and Wessel G. M.(2010). Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays 32(7): 626-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Price I. F., Hertz H. L., Pastore B., Wagner J. and Tang W.(2021). Proximity labeling identifies LOTUS domain proteins that promote the formation of perinuclear germ granules in C. elegans . Elife 10: e72276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin W., Cho K. F., Cavanagh P. E. and Ting A. Y.(2021). Deciphering molecular interactions by proximity labeling. Nat Methods 18(2): 133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rhee H. W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., Carr S. A. and Ting A. Y.(2013). Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339(6125): 1328-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roux K. J., Kim D. I., Raida M. and Burke B.(2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196(6): 801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanchez A. D., Branon T. C., Cote L. E., Papagiannakis A., Liang X., Pickett M. A., Shen K., Jacobs-Wagner C., Ting A. Y. and Feldman J. L.(2021). Proximity labeling reveals non-centrosomal microtubule-organizing center components required for microtubule growth and localization. Curr Biol 31(16): 3586-3600 e3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spike C., Meyer N., Racen E., Orsborn A., Kirchner J., Kuznicki K., Yee C., Bennett K. and Strome S.(2008). Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins . Genetics 178(4): 1973-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trcek T. and Lehmann R.(2019). Germ granules in Drosophila . Traffic 20(9): 650-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voronina E., Seydoux G., Sassone-Corsi P. and Nagamori I.(2011). RNA granules in germ cells. Cold Spring Harb Perspect Biol 3(12): a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]