Abstract
The profitability of next-generation biorefineries is acutely contingent on the discovery and utilization of biocatalysts that can valorize lignin. To this end, the metabolic catalogues of diverse microbiota have been mined previously using functional metagenomics in order to identify biocatalysts that can selectively degrade lignin into monoaromatic compounds. Herein, we have further improved the valorization factor of biorefining by deploying functional metagenomics toward the identification of a novel transaminase that can selectively functionalize lignin-derived monoaromatics to produce value-added feedstocks for pharmaceutical synthesis. We implemented a high-throughput colorimetric assay using o-xylylenediamine as the amino donor and successfully identified a transaminase that utilizes the canonical cofactor, pyridoxal 5′-phosphate, to aminate as many as 14 monoaromatic aldehydes and ketones. We subsequently identified the optimal conditions for enzyme activity towards the most favoured amino acceptor, benzaldehyde, including temperature, pH and choice of co-solvent. We also evaluated the specificity of the enzyme towards a variety of amino donors, as well as the optimal concentration of the most favoured amino donor. Significantly, the novel enzyme is markedly smaller than typical transaminases, and it is stably expressed in E. coli without any modifications to its amino acid sequence. Finally, we developed and implemented a computational methodology to assess the activity of the novel transaminase. The methodology is generalizable for assessing any transaminase and facilitates in silico screening of enzyme–substrate combinations in order to develop efficient biocatalytic routes to value-added amines. The computational pipeline is an ideal complement to metagenomics and opens new possibilities for biocatalyst discovery.
We have deployed functional metagenomics toward the identification of a novel transaminase that can selectively functionalize lignin-derived monoaromatics to produce value-added feedstocks for pharmaceutical synthesis.
1. Introduction
Biorefining is loosely defined as the separation, isolation and conversion of cellulose, hemicellulose and lignin from lignocellulosic biomass into fuels, chemicals, materials and energy.1 A number of biocatalytic2 and thermocatalytic3 processes have been developed for biorefining. However, regardless of the mode of conversion employed, biomass comprising a higher content of cellulose and hemicellulose is generally preferred as a feedstock compared to biomass with a higher proportion of lignin.4 The degree to which a resource can be valorized (which we define as the valorization factor) is a function of the ratio of the value of all products that can be manufactured from the resource to the value of the resource. Typically, the greater the number of useful products that can be manufactured from a resource, the greater is its valorization factor; and lignin's poor valorizability can be attributed to its recalcitrance to most biocatalytic or thermocatalytic transformations.5,6 The inability to efficiently valorize lignin represents a lost opportunity for biorefining, especially since lignin has a higher carbon and lower oxygen content compared to both, cellulose and hemicellulose.7 In particular, lignin is arguably one of the most attractive feedstocks for the manufacture of aromatic specialty and fine chemicals that are, among other applications, widely used in pharmaceutical synthesis. Although these chemicals are presently derived from petroleum, they can be sourced from lignin at higher atom economies. The development of conversion platforms that can efficiently valorize lignin is highly desired, particularly biocatalytic processes that are greener, emit lesser CO2 and are more energy efficient than thermocatalytic alternatives.8–13 To this end, functional metagenomics is poised to be a major driver of the discovery of superior biocatalysts for the valorization of lignin. Functional metagenomics provides reliable and easy access to the entire catalogue of microbial biocatalytic transformations without the need to culture any microorganisms in the laboratory.14,15
We had previously reported the identification of 147 novel, whole-cell biocatalysts that selectively degrade lignin using functional metagenomics.16 We employed a whole-cell, GFP-based biosensor to screen 42 520 fosmid clones comprising the environmental DNA of archaea and bacteria from coal beds. Each fosmid ranged between 30- to 50-kilo base pairs in length, and the clones selectively degrade lignin into vanillin and syringaldehyde. Herein, we further improve the valorization factor of lignin use by re-deploying functional metagenomics to discover a novel transaminase that catalyzes the asymmetric synthesis of chiral amines from the monoaromatic degradation products of lignin. It is estimated that roughly 40% of all new chemical entities (NCEs) approved by the US Food & Drug Administration (FDA) contain chiral amines building blocks17 that are the products of enzymes such as imine reductase,18 monoamine oxidase,19 ammonia lyases,20 amine dehydrogenase,21 and transaminases.22,23 Of these, transaminases (TAs) are especially promising for biocatalytic aminations since they can convert monoaromatic aldehydes and ketones into their corresponding amines at high yields and selectivities under mild, aqueous reaction conditions.24 In fact, several chemo-enzymatic schemes employing TAs for the syntheses of enantiopure (R)- or (S)-pharmaceutical intermediates have been reported in recent years.25–30 Amination by a canonical TA commences with the binding of the cofactor pyridoxal 5′-phosphate (PLP) to a lysine residue within the enzyme's active site, which generates an internal (or enzyme-bound) aldimine.31,32 The amino donor binds to the internal aldimine thereafter and molecular rearrangements ensue, which sequentially generate an external (or dissociated) aldimine, a quinonoid, a ketamine, and, finally, pyridoxamine phosphate (PMP) and the oxo product. The latter is released from the active site and is replaced with the amino acceptor, which then reacts with PMP to produce the desired amino product and re-generate PLP.
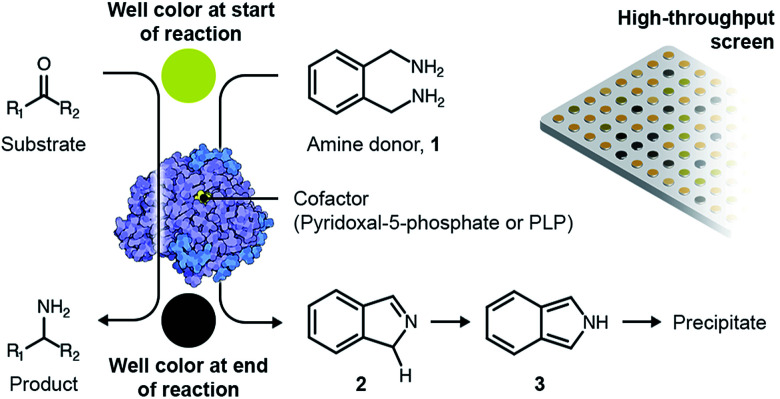
A variety of high-throughput, enzyme-coupled assays have been employed previously to identify TAs.33–37 These assays employ other enzymes in addition to the TA to transform amination intermediates into products that can be detected using rapid analytical procedures such as spectrophotometry. Enzyme-coupled assays are quite sensitive but typically necessitate extensive and expensive optimization before they can be deployed for metagenomic screening.38 As a consequence, we employed a colorimetric assay utilizing o-xylylenediamine (1) as the amino donor to screen novel TAs expressed by an in-house library of E. coli fosmid clones that exhibit activity towards monoaromatic degradation products of lignin.22 Each fosmid once again ranged between 30- to 50-kilo base pairs in length. The reaction between o-xylylenediamine and the internal aldimine within the active site of a likely TA generates 1H-isoindole (2) and PMP. PMP remains in the active site and subsequently reacts with the amino acceptor. However, 1H-isoindole is released into solution, where it tautomerizes to 2H-isoindole (3). The latter then rapidly polymerizes to yield a black precipitate (Scheme 1).
Scheme 1. Overview of the colorimetric assay for high-throughput screening of TAs.
2. Results & discussion
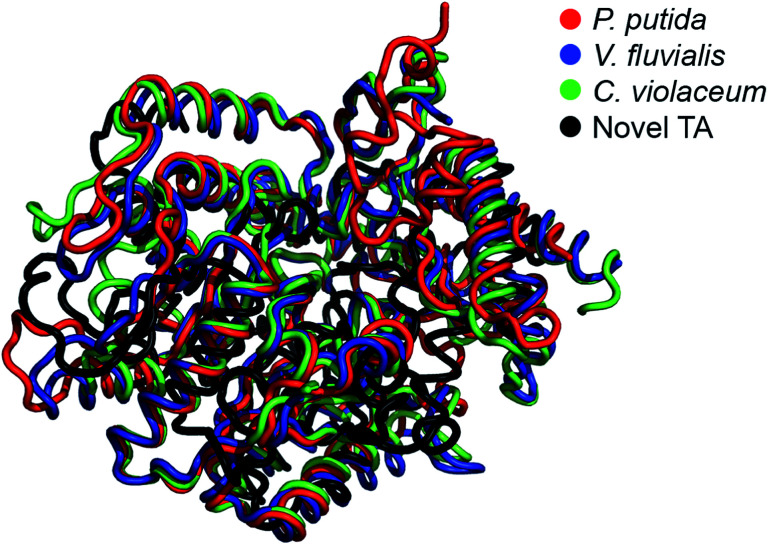
The E. coli fosmid clones were assessed for desired TA activity in 384-well plates. The microcultures were initially propagated in 55 μL LB medium supplemented with 12.5 μg mL−1 of chloramphenicol and 100 μg mL−1l-arabinose. Chloramphenicol is the antibiotic selection marker whereas arabinose ensures that a defined number of copies of the fosmid are maintained within the bacteria. All solvents and chemicals used in this study were purchased from Sigma Aldrich, Alfa Aesar and Santa Cruz Biotechnologies. The microcultures were allowed to grow at 37 °C for 24 hours, after which they were fed 2 mM of PLP, 25 mM of o-xylylenediamine and 5 mM of the amino acceptor, benzaldehyde (4), and then incubated at 30 °C for an additional 48 hours. At the end of the 48 hour incubation, the plates were imaged from above and the RGB values of the pixel corresponding to the centre of each well were extracted using MATLAB. Eight 384-well plates totalling 3072 fosmid clones were screened in this manner and we successfully identified three primary hits. Hit verification subsequently winnowed the number of confirmed TAs to a single candidate, S2A24, which translates to a hit rate of 0.03% (on a clonal basis) or 1 per 122.9-mega base pairs. This hit rate is significantly higher than the median rate that is observed for functional metagenomic screening.39 We later picked the S2A24 clone from the original library and extracted its fosmid using a GeneJET plasmid miniprep kit. Sequencing and annotation of the fosmid confirmed the presence of a novel TA and 37 other unique ORFs. The 371-amino acid sequence of the novel enzyme is roughly 15% shorter than and <30% similar to canonical TAs expressed by Pseudomonas putida,40Vibrio fluvialis41 and Chromobacterium violaceum,35 as well as another TA, pQR1108, that was discovered recently24 (the multiple sequence alignment has been summarized in Fig. S3 in the ESI or ESI package†). Nevertheless, the active site and folds of the novel TA aligns quite well with those of previously discovered candidates (Fig. 1).
Fig. 1. Structural alignment between the model of the novel TA (prepared by homology modeling using I-TASSER) and the TAs expressed by Pseudomonas putida (PDB code: 3A8U), Vibrio fluvialis (PDB code: 3NUI) and Chromobacterium violaceum (PDB code: 4A6R).
We subsequently synthesized the gene that encodes a version of the novel TA bearing a N-terminal polyhistidine tag. Its codons were optimized for expression in E. coli and the gene was later cloned into a high-copy pET-15b plasmid under the control of a T7 transcriptional promoter. Additional details about the molecular cloning have been provided in the ESI.† The resulting plasmid is labelled as pET-TA. This plasmid was then transformed into E. coli BL21 (DE3) using the heat shock method, which involves temporarily exposing the bacteria to 42 °C to permeabilize their cell walls in order to facilitate the delivery of the plasmid into the cells.42 The transformed cells were cultured on agar plates supplemented with 100 μg mL−1 of ampicillin, and a single colony was later picked and cultured overnight at 37 °C in 5 mL of LB medium also containing 100 μg mL−1 of ampicillin. This overnight culture was subsequently used as the inoculum for a 10 mL culture in the same ampicillin-supplemented LB medium. The latter culture was propagated at 37 °C until it attained an optical density of 0.6, following which isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium and the incubation temperature was lowered to 25 °C. The concentration of IPTG in the medium was 0.5 mM and the cultures were propagated overnight. The cells were then harvested by centrifugation and lysed, and the TA was purified using Qiagen Ni-NTA spin columns. SDS-PAGE confirmed high expression of the enzyme by cultures of the transformed cells that are induced with IPTG (Fig. S2 in the ESI†).
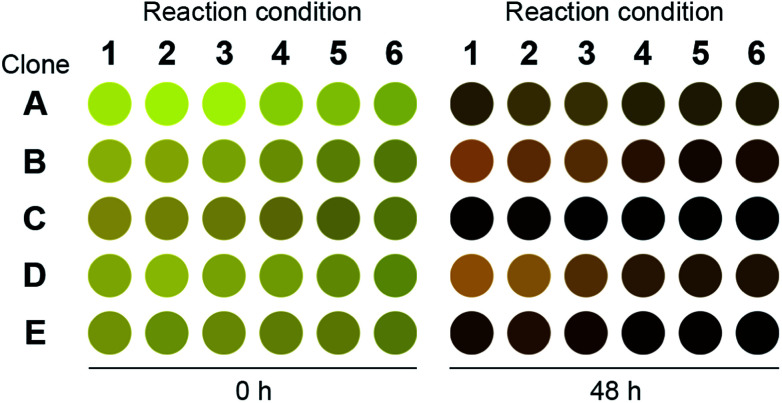
We repeated the o-xylylenediamine assay for 200 μL cultures and lysates of E. coli cells that have been transformed with pET-TA and the S2A24 fosmid. The reactions were conducted in triplicate in a 96-well plate. The culturing and reaction conditions, with the exception of the concentration of o-xylylenediamine, were identical to those of the 384-well plates described earlier. o-Xylylenediamine was supplied at a concentration of either 5 mM or 25 mM. Cultures of E. coli cells transformed with either an empty pET-15b vector or the pET-TA plasmid, but neither of which were supplied with benzaldehyde, were employed as the controls. We observed that the wells harbouring the cell lysates exhibited significantly darker coloration compared to the other wells (Fig. 2). In fact, the lysates of the E. coli cells that had been transformed with pET-TA produced the darkest colour at the end of the 48 hours. We also detected TA activity when the concentration of o-xylylenediamine was reduced from 25 mM to 5 mM. Moreover, the activity is indistinguishable for lysates of the pET-TA cultures and only slightly higher for lysates of the S2A24 clone. However, amination by whole-cell cultures of E. coli transformed with pET-TA and the S2A24 fosmid exhibit marked differences in response to the concentration of o-xylylenediamine, which can be attributed to resistance to transport of the amino donor to the intracellular reaction milieu. Neither control exhibited any TA activity. Although the colour of these reaction mixtures changed, this shift in coloration is attributable to the formation of PMP.22,38 We also verified the results of the colorimetric assay using HPLC. We added 500 μL of methanol to each mixture to quench the reaction. The samples were then centrifuged and the supernatant was injected into a Water Atlantis C18 column (5 μm, 4.6 mm × 250 mm). A gradient of 10 mM ammonium acetate and acetonitrile (ACN) was employed to resolve the species. The gradient commences with 60% ammonium acetate and 40% ACN, transitions to 100% ACN after 10 minutes, and finally reverts back to 60% ammonium acetate and 40% ACN 20 minutes into the run. Under these conditions, benzylamine, α-methylbenzylamine, benzaldehyde and acetophenone elute at 3.6, 4.2, 7.7 and 8.1 minutes, respectively, and the concentrations of benzylamine in the samples align well with the readouts of the colorimetric screen.
Fig. 2. Readout of the 96-well plate o-xylylenediamine assay for the novel TA. Clone A: E. coli transformed with pET-TA but not fed with benzaldehyde; clone B: E. coli transformed with pET-TA; clone C: cell lysate of E. coli transformed with pET-TA; clone D: E. coli transformed with S2A24 fosmid; clone E: cell lysate of E. coli transformed with S2A24 fosmid. With the exception of clone A, which was not provided with any benzaldehyde, all wells are fed with 5 mM benzaldehyde and 2 mM PLP. The volume of each reaction mixture is 200 μL and the plate is maintained at 30 °C and pH 7.5. For reaction conditions 1–3, 5 mM of o-xylylenediamine was supplied to the well. For reaction conditions 4–6, 25 mM of o-xylylenediamine was supplied to the well. The raw image of the plate is included in the ESI.†.
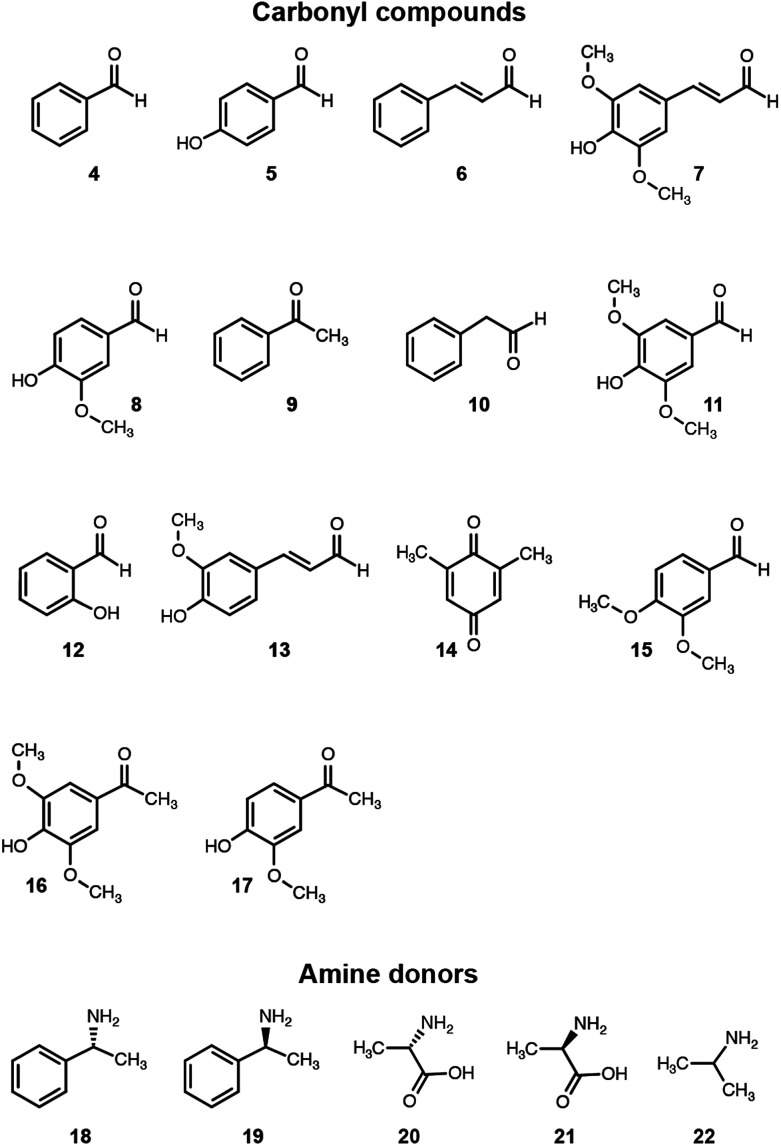
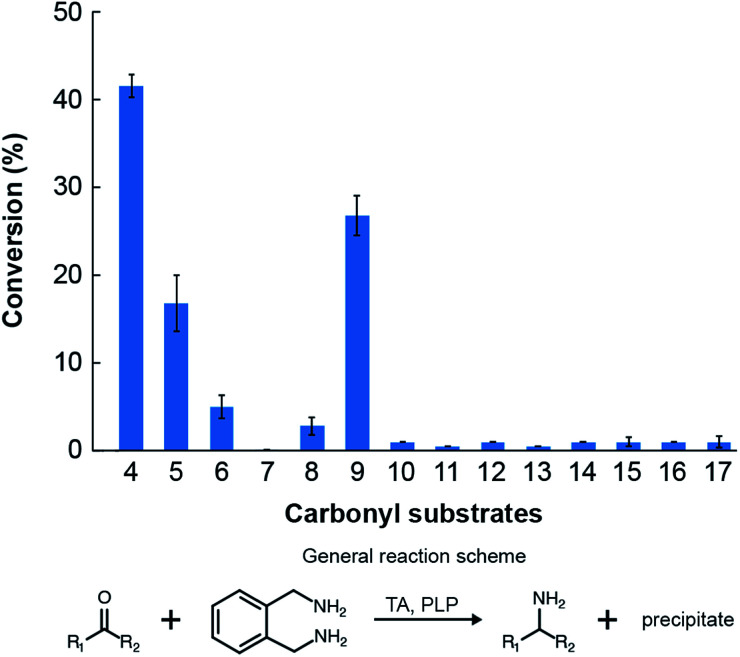
We then assessed the propensity of the novel TA to employ alternative amino donors. Each reaction mixture comprised ∼1 mg mL−1 of crude lysate of cultures of E. coli transformed with the pET-TA plasmid, 2 mM of PLP, 25 mM of the amino donor (Fig. 3) and 5 mM of benzaldehyde (dissolved in DMSO) in 200 μL of a buffered solution of 100 mM potassium phosphate at pH 7.5. As before, the reactions were monitored colorimetrically for 48 hours, after which the mixtures were analyzed for their benzylamine content using HPLC by following the same methodology for sample preparation as described earlier. Each reaction was performed and analyzed in triplicate. We observed that the enzyme has a strong preference for o-xylylenediamine. The conversion of benzaldehyde when o-xylylenediamine is employed as the amino donor is 42%. In contrast, the use of (S)-(−)-1-phenylethylamine (18) results in conversion of 10%, and no benzylamine was formed when (R)-(+)-1-phenylethylamine (19), l-alanine (20), d-alanine (21) and isopropylamine (22) are used as amino donors. We also assessed the potential of the novel TA to aminate other monoaromatic carbonyl compounds that are typically produced by the breakdown of lignin. We used the same reaction conditions as before, but replaced benzaldehyde with other amino acceptors (Fig. 3). After 48 hours had elapsed, the mixtures were vortexed with an equal volume of ethyl acetate and the organic phases were analyzed using GC-MS to determine conversion and confirm 2H-isoindole formation. GC-MS analysis was performed using an Agilent J&W HP-5ms column (25 m length, 0.20 μm film thickness, 0.25 mm inner diameter). Injector and column conditions have been elaborated in the ESI.†
Fig. 3. Amino donors and acceptors evaluated in this study.
The TA successfully converted 42% of benzaldehyde, 27% of acetophenone (9), 17% of 4-hydroxybenzaldehyde (5) and 7.5% of cinnamaldehyde (6) into their corresponding amines (Fig. 4). In contrast, the conversion of 4-hydroxy-3-methoxybenzaldehyde (8), 3,4-dimethoxybenzaldehyde (15), and 4-hydroxy-3-methoxyacetophenone (17) was less than 5%, and the enzyme exhibited negligible activity towards the other amino acceptors. Incidentally, we also monitored the reaction mixtures colorimetrically. Benzaldehyde and acetophenone yielded the most intense colour, which is consistent with the GC-MS results. However, the colorimetric assay was not as reliable for the coloured substrates (species 6, 11–14 and 16). This is a known limitation of the assay that needs improvement in order to make it universally applicable for use in functional metagenomic screening of TAs that can functionalize any monoaromatic degradation product of lignin.
Fig. 4. Conversions of the amino acceptors considered in this study by the novel TA. Reaction conditions: 5 mM of the individual amino acceptors, 25 mM of o-xylylenediamine and 2 mM PLP in 100 mM potassium phosphate solution buffered at pH 7.5 and maintained at 30 °C.
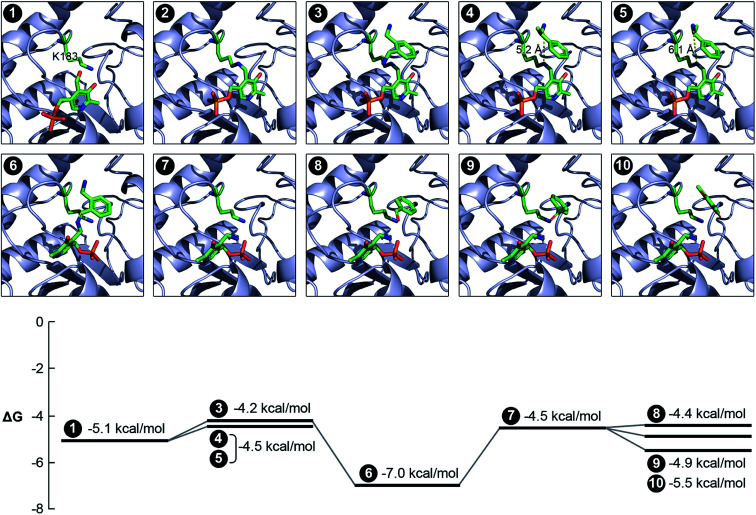
We evaluated the catalytic mechanism of the enzyme using computational modeling. A homology model of the enzyme was constructed using I-TASSER,43 and the structures of all molecules that participate in the catalytic cycle were prepared in Marvin.44 We subsequently docked PLP into the active site of the enzyme using AutoDock Vina.45 The input to the docking calculation comprises an initial set of atomic coordinates for PLP, and the program outputs as many as 9 putative poses of PLP within the active site. A corresponding Gibbs free energy of binding (ΔG) is also computed for each pose. In order to identify the pose that yields the global minimum for ΔG, we selected the most stable pose and re-used its coordinates as the initial value for a subsequent docking calculation. We iterated this methodology until an unchanging pose was outputted. The docked pose of PLP (Fig. 5, Panel 1) aligns exceedingly well with the relative coordinates of PLP in the crystal structures of TAs expressed by Pseudomonas putida (PDB code: 3A8U), Vibrio fluvialis (PDB code: 3NUI) and Chromobacterium violaceum (PDB code: 4A6R), and is in close proximity to the K183 residue of the enzyme. Additionally, the phosphate moiety of PLP is facing outward. This orientation of the phosphate moiety is an important frame of reference for subsequent molecular events. We then used Maestro46 to generate an aldimine bond between the K183 residue and PLP, as well as add charges to individual atoms in the product. The energy of the structure was subsequently minimized in Maestro. The coordinates of all atoms with the exception of the lysine side chain and PLP were fixed in the energy minimization (Fig. 5, Panel 2).
Fig. 5. Computational analysis of the catalytic mechanism of the newly discovered TA. We applied a variety of computational tools to simulate the progression of the reaction within the active site of the enzyme. The Gibbs free energy of binding has also been estimated for each pose. Panel 1: binding of PLP within the active site and the close proximity of the K183 residue; panel 2: reaction between the PLP and the K183 residue to form the internal aldimine; panel 3: docking of o-xylylenediamine within the active site of the aldimine-functionalized TA; panel 4: docking of (S)-(−)-1-phenylethylamine within the active site of the aldimine-functionalized TA; panel 5: docking of (R)-(+)-1-phenylethylamine within the active site of the aldimine-functionalized TA and confirmation of non-reactivity due to the large distance between its nitrogen atom and the nitrogen atom in the aldimine bond; panel 6: formation of the ketamine intermediate on account of the reaction between o-xylylenediamine and PLP; panel 7: formation of PMP in the active site; panel 8: docking of benzaldehyde within the active site of the TA alongside PMP; panel 9: docking of acetophenone within the active site of the TA alongside PMP; panel 10: docking of 2,6-dimethyl-p-benzoquinone within the active site of the TA alongside PMP.
We subsequently docked all the amino donors that were studied previously into the new active site using the same iterative methodology described earlier. Of these, only o-xylylenediamine, (S)-(−)-1-phenylethylamine and (R)-(+)-1-phenylethylamine successfully dock within the active site. Moreover, the distances between the nitrogen atom in the aldimine bond and the nearest nitrogen atom of the amino donor that is a key participant in the sequence of steps that culminates with the formation of PMP and the oxo product are 4.9, 5.2 and 6.1 Å for o-xylylenediamine (Fig. 5, Panel 3), (S)-(−)-1-phenylethylamine (Fig. 5, Panel 4) and (R)-(+)-1-phenylethylamine (Fig. 5, Panel 5), respectively. This result aligns well with the observed conversions of 42% and 10% for o-xylylenediamine and (S)-(−)-1-phenylethylamine, respectively, and confirms the non-reactivity of (R)-(+)-1-phenylethylamine. We then docked the ketamine intermediate of the reaction between PLP and o-xylylenediamine within the active site of the unmodified TA (Fig. 5, Panel 6). The calculation suggests that molecular rearrangements within the intermediate cause it to flip its orientation within the active site in a manner such that its phosphate moiety is now facing inward. This prediction is consistent with the expected behaviour of the molecular in response to steric hindrances within the active site,31,32 as well as the pose of PMP when it has been docked within the active site (Fig. 5, Panel 7). Also, the ΔΔG between the interaction of o-xylylenediamine with the internal aldimine and the interaction of the enzyme with the ketamine intermediate is higher than the corresponding interactions for (S)-(−)-1-phenylethylamine, which substantiates observed differences in the conversions of the two amino donors.
Finally, we docked benzaldehyde (Fig. 5, Panel 8), acetophenone (Fig. 5, Panel 9) and 2,6-dimethyl-p-benzoquinone (14 and Fig. 5, Panel 10) into the active site of the TA with PMP already bound within. Interestingly, the docking calculation does not predict any discernible differences in reactivity of benzaldehyde and acetophenone. In this context, we define reactivity as the distance between the amino moiety of PMP and the carbonyl bond in the amino acceptors. However, the observed conversions of benzaldehyde and acetophenone by the enzyme are 42% and 27%, respectively. Differences in conversion may arise due to an increased resistance to diffusion of acetophenone to the active site compared to benzaldehyde or a higher ΔΔG between the interaction of the enzyme with PMP and benzaldehyde and the interaction of the enzyme with PLP and benzylamine compared to corresponding interactions for acetophenone. The docking calculation also revealed that the distance between the amino moiety of PMP and either carbonyl bond in 2,6-dimethyl-p-benzoquinone is too high for a reaction to occur, which is confirmed by experiments. This suggests that extensive functionalization of the aromatic ring of the amino acceptors introduces significant steric hindrances that greatly impact conversion. Nevertheless, the congruency between experiments and simulation validates the computational analysis that we have implemented to assess the activity of TAs. The computational pipeline is an ideal complement to metagenomics and could be effectively deployed for large-scale in silico screening of enzyme–substrate combinations in order to develop efficient biocatalytic routes for amination of monoaromatic derivatives of lignin for use as pharmaceutical intermediates.
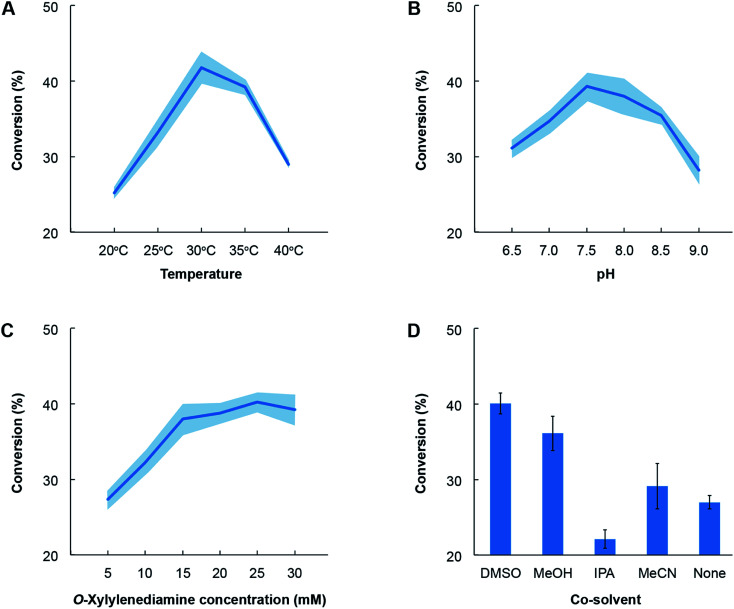
Lastly, we also identified the optimal conditions for activity of the novel TA towards the most favoured amino acceptor, benzaldehyde. o-Xylylenediamine was retained as the amino donor and we quantified the conversion of benzaldehyde to benzylamine as a function of temperature, pH, choice of co-solvent and concentration of the amino donor (Fig. 6). The co-solvent is utilized to dissolve benzaldehyde. All reactions comprised ∼1 mg mL−1 of crude lysate of E. coli transformed with the pET-TA plasmid and 2 mM of PLP in a solution of 100 mM potassium phosphate buffer. Each reaction was undertaken in triplicate for 48 hours and had a final volume of 200 μL; and the benzylamine concentration was quantified using HPLC. In order to assess the influence of temperature on enzymatic activity, we added 5 mM of benzaldehyde dissolved in DMSO and 25 mM of o-xylylenediamine to the reaction mixture and maintained the pH at 7.5. The reactions were then undertaken at five temperatures and we observed that the highest conversion of benzaldehyde was obtained at 30 °C (Fig. 6, Panel A). In order to investigate the influence of pH, we added 25 mM of o-xylylenediamine and 5 mM of benzaldehyde dissolved in DMSO to reaction mixtures maintained at 30 °C and varied the pH between 6.5 and 9. We observed that the conversion peaked at pH 7.5 (Fig. 6, Panel B).
Fig. 6. Optimization of reaction conditions for cell lysates for E. coli transformed with pET-TA. All reactions comprised ∼1 mg mL−1 of crude lysate of E. coli transformed with the pET-TA plasmid and 2 mM of PLP in a solution of 100 mM potassium phosphate. Each reaction was undertaken in triplicate for 48 hours and had a final volume of 200 μL; and the benzylamine concentration was quantified using HPLC. The common reaction mixture was suitably modified to deduce the effect of temperature (panel A), pH (panel B), o-xylylenediamine (panel C) and choice of co-solvent (panel D). The activity of the enzyme exhibits canonical behaviour in response to changes in temperature, pH and concentration of the substrate. Each experiment was performed in triplicate and the shaded region denotes the standard error in the measurements.
Likewise, the impact of the concentration of o-xylylenediamine on conversion was measured by varying its levels in reaction mixtures comprising 5 mM of benzaldehyde in DMSO and maintained at pH 7.5 and 30 °C. We observed that conversion of benzaldehyde exhibits a hyperbolic response to the concentration of o-xylylenediamine and plateaus near 25 mM (Fig. 6, Panel C). The activity of the enzyme exhibits canonical behaviour in response to changes in temperature, pH and concentration of the substrate. Finally, since lignin-derived monoaromatic compounds are poorly soluble in aqueous solutions, we also evaluated several co-solvents for their ability to enhance conversion by improving solubility of benzaldehyde in the reaction mixture. We prepared 10% (volumetric basis) solutions of benzaldehyde in DMSO, acetonitrile (MeCN), isopropyl alcohol (IPA) and methanol (MeOH), and added them to the reaction mixtures to attain a final benzaldehyde concentration of 5 mM. We also directly added benzaldehyde to the reaction mixture without any co-solvent. To our surprise, we observed that the reaction mixture without any co-solvents attained a higher conversion than the mixture that was supplied with benzaldehyde dissolved in IPA (Fig. 6, Panel D). The use of DMSO yielded the highest conversion, followed by MeOH and MeCN.
3. Conclusions
In summary, we employed a high-throughput colorimetric assay using o-xylylenediamine as the amino donor and PLP to identify a novel TA from a subset of our in-house metagenomic library. This is the first ever study to report the successful use of functional metagenomics toward the discovery of a TA. The enzyme successfully transforms several monoaromatic aldehydes and ketones to their corresponding amines in vivo, although the highest conversions were observed when it is used as a constituent of the cell lysate. We then evaluated the specificity of the enzyme towards a variety of amino donors, as well as optimal concentration of the most favoured amino donor, and also identified the optimal conditions for enzyme activity towards the most favoured amino acceptor, benzaldehyde, including temperature, pH and choice of co-solvent. Finally, we developed and implemented a computational methodology to assess its activity. The methodology is generalizable to other enzymes in its class, which facilitates in silico screening of enzyme–substrate combinations and the development of novel biocatalytic routes for amination of monoaromatic derivatives of lignin for use as pharmaceutical intermediates. Significantly, since the enzyme is active towards products of the degradation of lignin that is catalyzed by other enzymes that we have discovered previously, it can be readily deployed in biorefining operations for one-pot valorization of lignin.
Conflicts of interest
The authors declare that they do not have any conflicts of interest.
Supplementary Material
Acknowledgments
The authors would like to thank Joe Ho and Kateryna Ievdokymenko for their assistance with the experimentation. We thank the Natural Sciences & Engineering Research Council (NSERC), Canadian Foundation for Innovation and Mitacs for their generous funding.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02764a
Notes and references
- Luo L. van der Voet E. Huppes G. Bioresour. Technol. 2010;101:5023–5032. doi: 10.1016/j.biortech.2009.12.109. [DOI] [PubMed] [Google Scholar]
- Heux S. Meynial-Salles I. O'Donohue M. Dumon C. Biotechnol. Adv. 2015;33:1653–1670. doi: 10.1016/j.biotechadv.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Wettstein S. Alonso D. Gürbüz E. Dumesic J. Curr. Opin. Chem. Eng. 2012;1:218–224. doi: 10.1016/j.coche.2012.04.002. [DOI] [Google Scholar]
- Parajuli R. Dalgaard T. Jørgensen U. Adamsen A. Knudsen M. Birkved M. Gylling M. Schjørring J. Renewable Sustainable Energy Rev. 2015;43:244–263. doi: 10.1016/j.rser.2014.11.041. [DOI] [Google Scholar]
- Yamaguchi A. Mimura N. Shirai M. Sato O. Sci. Rep. 2017;7:46172. doi: 10.1038/srep46172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas A. Beckham G. Biddy M. Chandra R. Chen F. Davis M. Davison B. Dixon R. Gilna P. Keller M. Langan P. Naskar A. Saddler J. Tschaplinski T. Tuskan G. Wyman C. Science. 2014;344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Rinaldi R. Jastrzebski R. Clough M. Ralph J. Kennema M. Bruijnincx P. Weckhuysen M. Angew. Chem., Int. Ed. 2016;55:8164–8215. doi: 10.1002/anie.201510351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. Luque R. Matharu A. Annu. Rev. Chem. Biomol. Eng. 2012;3:183–207. doi: 10.1146/annurev-chembioeng-062011-081014. [DOI] [PubMed] [Google Scholar]
- Truppo M. D. ACS Med. Chem. Lett. 2017;8:476–480. doi: 10.1021/acsmedchemlett.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. Zengler K. Nat. Rev. Microbiol. 2004;2:141–150. doi: 10.1038/nrmicro819. [DOI] [PubMed] [Google Scholar]
- Whitman W. B. Coleman D. C. Wiebe W. J. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz P. Eck J. Nature. 2005;3:510–516. doi: 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- Yadav V. G. Stephanopoulos G. ChemSusChem. 2014;7:1847–1853. doi: 10.1002/cssc.201301219. [DOI] [PubMed] [Google Scholar]
- Taupp M. Mewis K. Hallam S. J. Curr. Opin. Biotechnol. 2011;22:465–472. doi: 10.1016/j.copbio.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Armstrong Z. Mewis K. Strachan C. Hallam S. J. Curr. Opin. Chem. Biol. 2015;29:18–25. doi: 10.1016/j.cbpa.2015.06.032. [DOI] [PubMed] [Google Scholar]
- Ho J. C. H. Pawar S. V. Hallam S. J. Yadav V. G. ACS Synth. Biol. 2018;7:392–398. doi: 10.1021/acssynbio.7b00412. [DOI] [PubMed] [Google Scholar]
- Aleku G. A. France S. P. Man H. Mangas-Sanchez J. Montgomery S. L. Sharma M. Leipold F. Hussain S. Grogan G. Turner N. J. Nat. Chem. 2017;9:961–969. doi: 10.1038/nchem.2782. [DOI] [PubMed] [Google Scholar]
- Leipold F. Hussain S. Ghislieri D. Turner N. J. ChemCatChem. 2013;5:3505–3508. doi: 10.1002/cctc.201300539. [DOI] [Google Scholar]
- Ghislieri D. Green A. P. Pontini M. Willies S. C. Rowles I. Frank A. Grogan G. Turner N. J. J. Am. Chem. Soc. 2013;135:10863–10869. doi: 10.1021/ja4051235. [DOI] [PubMed] [Google Scholar]
- Heberling M. M. Wu B. Bartsch S. Janssen D. B. Curr. Opin. Chem. Biol. 2013;17:250–260. doi: 10.1016/j.cbpa.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Abrahamson M. J. Vázquez-Figueroa E. Woodall N. B. Moore J. C. Bommarius A. S. Angew. Chem., Int. Ed. 2012;51:3969–3972. doi: 10.1002/anie.201107813. [DOI] [PubMed] [Google Scholar]
- Green A. P. Turner N. J. O'Reilly E. Angew. Chem., Int. Ed. 2014;53:10714–10717. doi: 10.1002/anie.201406571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. Berglund P. Green Chem. 2017;19:333–360. doi: 10.1039/C6GC02328B. [DOI] [Google Scholar]
- Baud D. Jeffries J. W. E. Moody T. S. Ward J. M. Hailes H. C. Green Chem. 2017;19:1134–1143. doi: 10.1039/C6GC02769E. [DOI] [Google Scholar]
- Saville C. K. Janey J. M. Jarvis W. R. Colbeck J. C. Krebber A. Fleitz F. J. Brands J. Science. 2010;329:305–310. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- Sehl T. Hailes H. C. Ward J. M. Wardenga R. Von Lieres E. Offermann H. Westphal R. Pohl M. Rother D. Angew. Chem., Int. Ed. 2013;52:6772–6775. doi: 10.1002/anie.201300718. [DOI] [PubMed] [Google Scholar]
- Frodsham L. Golden M. Hard S. Kenworthy M. N. Klauber D. J. Leslie K. Macleod C. Meadows R. E. Mulholland K. R. Reilly J. Squire C. Tomasi S. Watt D. Wells A. S. Org. Process Res. Dev. 2013;17:1123–1130. doi: 10.1021/op400133d. [DOI] [Google Scholar]
- Richter N. Simon R. C. Kroutil W. Ward J. M. Hailes H. C. Chem. Commun. 2014;50:6098–6100. doi: 10.1039/C3CC49080G. [DOI] [PubMed] [Google Scholar]
- Lichman B. R. Lamming E. D. Pesnot T. Smith J. M. Hailes H. C. Ward J. M. Green Chem. 2015;17:852–855. doi: 10.1039/C4GC02325K. [DOI] [Google Scholar]
- Richter N. Simon R. C. Lechner H. Kroutil W. Ward J. M. Hailes H. C. Org. Biomol. Chem. 2015;13:8843–8851. doi: 10.1039/C5OB01204J. [DOI] [PubMed] [Google Scholar]
- Koszelewski D. Tauber K. Faber K. Kroutil W. Trends Biotechnol. 2010;28:324–332. doi: 10.1016/j.tibtech.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Mathew S. Yun H. ACS Catal. 2012;2:993–1001. doi: 10.1021/cs300116n. [DOI] [Google Scholar]
- Truppo M. D. Rozzell J. D. Moore J. C. Turner N. J. Org. Biomol. Chem. 2009;7:395–398. doi: 10.1039/B817730A. [DOI] [PubMed] [Google Scholar]
- Schätzle S. Höhne M. Redestad E. Robins K. Bornscheuer U. T. Anal. Chem. 2009;81:8244–8248. doi: 10.1021/ac901640q. [DOI] [PubMed] [Google Scholar]
- Kaulmann U. Smithies K. Smith M. E. B. Hailes H. C. Ward J. M. Enzyme Microb. Technol. 2007;41:628–637. doi: 10.1016/j.enzmictec.2007.05.011. [DOI] [Google Scholar]
- Hopwood J. Truppo M. D. Turner N. J. Lloyd R. C. Chem. Commun. 2011;47:773–775. doi: 10.1039/C0CC02919J. [DOI] [PubMed] [Google Scholar]
- Hwang B.-Y. Kim B.-G. Enzyme Microb. Technol. 2004;34:429–436. doi: 10.1016/j.enzmictec.2003.11.019. [DOI] [Google Scholar]
- Baud D. Ladkau N. Moody T. S. Ward J. M. Hailes H. C. Chem. Commun. 2015;51:17225–17228. doi: 10.1039/C5CC06817G. [DOI] [PubMed] [Google Scholar]
- Culligan E. Sleator R. Marchesi J. Hill C. Virulence. 2014;5:399–412. doi: 10.4161/viru.27208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel E. U. Ramos J. L. Appl. Environ. Microbiol. 2001;67:5219–5224. doi: 10.1128/AEM.67.11.5219-5224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.-S. Yun H. Jang J.-W. Park I. Kim B.-G. Appl. Microbiol. Biotechnol. 2003;61:463–471. doi: 10.1007/s00253-003-1250-6. [DOI] [PubMed] [Google Scholar]
- JoVE Science Education Database, in Basic methods in cellular and molecular biology, Cambridge, MA, 2018 [Google Scholar]
- Zhang Y. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChemAxon, 2017, http://chemaxon.com/products/marvin
- Trott O. Olson A. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, 2018, http://www.schrodinger.com/maestro
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.