Abstract
The absence of long term, primary untransformed in vitro models that support hepatitis B virus (HBV) infection and replication have hampered HBV pre-clinical research, which was reflected in the absence of a curative therapy until recently. One of the limitations for in vitro HBV research has been the absence of high titer and pure recombinant HBV stocks, which, as we describe here, can be generated using simple, and reproducible protocols. In addition to infection of more conventional in vitro and in vivo liver model systems, recombinant high titer purified HBV stocks can also be used to efficiently infect differentiated human liver organoids, whose generation, maintenance, and infection is discussed in detail in a companion organoid protocol. Here, we also describe the protocols for the detection of specific viral read-outs, including HBV DNA in the supernatant of the cultures, covalently closed circular DNA (cccDNA) from intracellular DNA preparations, and HBV viral proteins and viral RNA, which can be detected within the cells, demonstrating the presence of a complete viral replication cycle in infected liver organoids. Although an evolving platform, the human liver organoid model system presents great potential as an exciting new tool to study HBV infection and progression to hepatocellular carcinoma (HCC) in primary cells, when combined with the use of high-titer and pure recombinant HBV stock for infection.
Keywords: HBV infection, Production of HBV, Heparin purification, Readouts for viral products
Background
HBV is a member of the hepadnaviridae family, a group of viruses harboring a partially double stranded DNA genome, which is replicated through an RNA intermediate. Infectious and replication-competent recombinant hepatitis B virus was generated from HepG2.2.15 cells, a HepG2-derived cell line stably transfected with the full-length HBV genome. Recombinant HBV, released in the supernatant of HepG2.2.15 cells, needs to be concentrated before infection of organoids, to get the desired amount of viral particles in a smaller volume and achieve the desired multiplicity of infection (MOI) per organoid. This can be easily done, using a commercial PEG virus precipitation kit (detailed protocol is given below) (De Crignis et al., 2021 ). To generate a higher-titer virus stock, which is purified by heparin affinity chromatography to remove subviral particles and enrich for complete HBV, we are currently using a modified version of a previously published protocol that increases the infection efficiency of the organoids ( Wettengel et al., 2021 ). In the previously published protocol by Wettengel et al. (2021) , sedimentation centrifugation by the sucrose-gradient system was used to further concentrate the heparin-column purified eluate HBV stock. Therefore, the final HBV stock that is generated via Wettengel et al. (2021) contains a high percentage of sucrose solution, which makes it unsuitable for infection of liver organoids, as the presence of high sucrose concentrations in the culture media can cause changes in osmolarity that result in the disruption of the 3D structure of the liver organoids. In the protocol described below, centrifugal filter units are used to further concentrate the eluate HBV stock purified through a heparin column, instead of sedimentation centrifugation by sucrose-gradient. Therefore, the final HBV stocks generated using this protocol are in a solution of Ad+++ (see Recipes section), which makes them suitable for use to infect human liver organoids. For more information on this, please refer to the companion protocol paper, which describes the generation, maintenance, and infection of the human liver organoids with HBV ( Romal et al., 2022 ).
Upon infection, specific viral products that are produced at specific viral replication steps can be detected, which confirms the presence of a complete viral replication cycle in infected human liver organoids. Infection of liver organoids with recombinant HBV results in the generation of two different forms of HBV DNA, that can be detected using polymerase chain reaction (PCR). These are: 1) relaxed circular DNA (rcDNA), which is the actual genomic DNA of the virus, and can be detected both in DNA preparation from the culture supernatant as well as intracellular DNA; 2) cccDNA, which is generated inside the host cell after successful infection, and can only be detected from the intracellular DNA preparation. Detailed procedures for production of recombinant HBV virus and detection of viral products in human liver organoids are mentioned below.
Materials and Reagents
Sterile filter pipette tips (Greiner Bio-One, catalog numbers: 774288 [P20]; 739288 [P200]; 740288 [P1000])
Low retention sterile pipette tips with filter (Biotix, catalog numbers: M-0010-9FC [P10]; M-0020-9FC [P20]; M-0200-9FC [P200]; M-1000-9FC [P1000])
Disposable sterile serological pipette with filter (VWR, catalog numbers: 89130-910 [10 mL]; 89130-890 [25 mL])
15 mL Falcon tubes (Greiner, catalog number: 188285)
50 mL Falcon tubes (Greiner, catalog number: 227285)
T175 cell culture flask (Thermo Fisher Scientific, catalog number: 159920)
Sterile syringe filter, 0.45 μm (GE Healthcare, Whatman, catalog number: 6896-2504)
Sterile syringe filter, 0.2 μm (GE Healthcare, Whatman, catalog number: 6900-2502)
1.5 mL microcentrifuge tubes, (Biotix, catalog number: MT-0150-BC)
24 well suspension plates (Greiner, catalog number: 662102)
48 well suspension plates (Greiner, catalog number: 677102)
100 mm Cell culture dish (Thermo Fisher Scientific, catalog number: 150464)
HYPERFlask® cell culture vessels (Sigma-Aldrich, Corning, catalog number: CLS10030)
500 mL Vacuum Filter/Storage Bottle System, 0.45 µm Pore (Sigma-Aldrich, Corning, catalog number: CLS430770)
HiTrap® Heparin High Performance, 5 mL (GE Healthcare, catalog number: GE17-0407-01)
Amicon® Ultra-15 Centrifugal Filter Unit, 100 KDa (Sigma-Aldrich, MerckMillipore, catalog number: UFC9100)
CryoTubesTM Vials (Thermo Scientific, Nunc, catalog number: 366656)
HepG2.2.15 cell line (CCTCC, catalog number: CCTCC-GDC0141)
DMEM (Dulbecco's Modified Eagle Medium) (Thermo Fisher Scientific, GibcoTM, catalog number: 41966029)
Fetal Bovine Serum (FBS) (Capricorn Scientific, catalog number: FBS-12A)
Penicillin-Streptomycin (10,000 U/mL) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122)
Advanced DMEM/F-12 (Thermo Fisher Scientific, GibcoTM, catalog number: 12634028)
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 M Buffer Solution in 0.85%
NaCl (Lonza, catalog number: BE17-737E)
UltraGlutamineTM I 200 mM in 0.85% NaCl Solution (Lonza, catalog number: BE17-605E/U1)
Collagen R solution 0.2% (Serva, catalog number: 47254.02)
Trypsin-EDTA solution (Sigma-Aldrich, catalog number: T3924)
PEG Virus Precipitation Kit (Abcam, catalog number: ab102538)
Phosphate Buffered Saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023)
Sodium chloride (Honeywell, catalog number: 71380)
Ethanol absolute (Sigma-Aldrich, MerckMillipore, catalog number: 1009832500)
Cultrex Basement Membrane Extract, Type 2, Pathclear (R&D Systems, catalog number: 3533-010-02)
Phenol:Chloroform:Isoamyl Alcohol 25:24:1, Saturated with 10 mM Tris, pH 8.0, 1 mM EDTA
Chloroform (Sigma, catalog number: p3803)
Isoamyl alcohol (Merck, catalog number: 100979)
Chloroform stabilized with Ethanol (Boom, catalog number: 76025322.2500)
2-propanol (isopropanol) (Honeywell, catalog number: 33539)
QIAamp MinElute Virus Spin Kit (Qiagen, catalog number: 57704)
Proteinase K (Sigma, catalog number: P2308)
Sodium acetate (Honeywell, catalog number: 32319)
Nuclease free water (Promega, catalog number: P1193)
DNeasy Blood & Tissue Kit (Qiagen, catalog number: 69504)
T5 Exonuclease: (New England Biolabs, catalog number: NEB:M0363)
Plasmid-Safe ATP-Dependent DNase (Epicentre, catalog number: E3101K)
Ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA) (Sigma, catalog number: E1644)
QIAquick PCR Purification Kit (Qiagen, catalog number: 28104)
Tris (hydroxymethyl) aminomethane (THAM) hydrochloride (Sigma, catalog number: PHG0002)
Sodium Hydroxide (Sigma, catalog number: S8045)
NP-40 Surfact-AmpsTM Detergent Solution (Thermo Scientific, catalog number: 85124)
Sodium dodecyl sulfate (Sigma, catalog number: 71729)
2-Butanol (Honeywell, catalog number: 19440)
Glycogen (20 mg/mL), molecular biology grade (Thermo Scientific, catalog number: R0561)
Potassium Acetate (Honeywell, catalog number: 236497)
Ammonium acetate (Honeywell, catalog number: 17836)
Platinum Taq DNA Polymerase (Invitrogen, catalog number: 10966034)
100 mM Deoxynucleoside triphosphate (dNTP) set (Invitrogen, catalog number: 10297-117)
2× Light Cycler 480 probe master (Roche, catalog number: 04887301001)
TRI Reagent (Trizol) (Sigma, catalog number: T9424)
RealiaPrep RNA cell miniprep system (Promega, catalog number: Z6012)
DNase I (ThermoFisher, Invitrogen, catalog number: 18047019)
Superscript II Reverse Transcriptase (ThermoFisher, Invitrogen, catalog number:18064022)
Random Primers (ThermoFisher, Invitrogen, catalog number:48190011)
Formaldehyde, 16%, methanol free, Ultra-Pure (Polysciences, catalog number: 18814)
Acetone (Honeywell, catalog number: 00585)
Glycine (Sigma, catalog number: G7126)
Monolisa Hepatitis B surface Antigen (HBsAg) ULTRA (Bio-Rad, catalog number: 72346)
Human Hepatitis B e Antigen (HBeAg) ELISA Kit (Cusabio, catalog number: CSB-E13557h)
Hoechst 33342 trihydrochloride, trihydrate (Invitrogen, catalog number: H3570)
Anti-Hepatitis B Virus Core Antigen antibody, (Abcam, catalog number: ab115992)
Alexa FluorTM 488 Phalloidin (actin stain) (Invitrogen, catalog number: A12379)
TrypLE Express (1×), no Phenol Red (Invitrogen, catalog number: 12604-013)
Triton X-100 (Sigma aldrich, catalog number: 10789704001)
Dimethyl sulfoxide, DMSO (Sigma aldrich, catalog number: D9170)
Sodium phosphate dibasic dihydrate, Na2HPO4·2H2O (Sigma aldrich, catalog number: 71643)
Potassium phosphate monobasic, KH2PO4 (Sigma aldrich, catalog number: P9791)
Potassium chloride, KCl (Sigma aldrich, catalog number: P9541)
Sodium Bicarbonate, NaHCO3 (Fisher Scientific, catalog number: S233-500)
DMEM ++ culture medium (500 mL) (see Recipes)
Ad+++ culture medium (500 mL) (see Recipes)
70% Ethanol solution (100 mL) (see Recipes)
Wash solution (see Recipes)
Elution buffer (100 mL) (see Recipes)
Column wash buffer (100 mL) (see Recipes)
Heparin column storage buffer (100 mL) (see Recipes)
Cell lysis buffer (50 mL) (see Recipes)
Alkali lysis buffer (50 mL) (see Recipes)
PBSTD (50 mL) (see Recipes)
10× PBS solution (see Recipes)
Equipment
Calibrated micropipettes (VWR, Gilson, catalog numbers: 613-5946 [P20]; 613-5948 [P200]; 613-5949 [P1000])
Biosafety cabinet (Clean Air by Baker, Model name: BioVanguard Biological Safety cabinet-Class ll)
Cell culture incubator with 5% CO2, 37°C (Panasonic catalog number: MCO-170AICUVH-PE)
Eppendorf Centrifuge 5810 R for 15 mL Falcon tubes and cell culture plates (Eppendorf, model: A-4-62)
Eppendorf Centrifuge 5417 R for 1.5 mL tubes (Eppendorf, catalog number: F45-30-11)
Water bath 37°C (Grant, VFP)
-80°C freezer (Panasonic, catalog number: MDF-794-PE)
Freezing container (NalgeneTM Cryo, catalog number: 5100-0001)
Refrigerator (Liebherr, catalog number: CP3523-22)
Peristaltic Pump (Ismatec, model: ECOLINE VC-380)
Eppendorf Thermomixer 5436 (Eppendorf, catalog number: T1442-1EA)
Milli-Q® IQ 7000 Ultrapure Lab Water System (Merck)
NanoDrop 2000/2000c spectrophotometers (Thermo Fisher Scientific, catalog number: ND2000LAPTOP)
Bright field microscope: Leica DMIL microscope and a DFC420C camera
Leica SP5 confocal
Software
Leica LAS AF Lite software
Procedure
-
Production of virus for PEG precipitation
Day 1:
Add 10 mL of 0.01% Collagen-R solution in sterile water to each 10-cm cell culture plate.
-
Perform incubation at room temperature overnight.
Day 2:
Remove the collagen solution with a 10-mL pipette.
Wash twice with 10 mL of PBS.
Seed 3 × 106 HepG2.2.15 cells (up to passage 25) in 10 mL of DMEM ++ (see Recipes) per plate.
-
Culture in the incubator at 37°C with 5% CO2 for 3-4 days (until the cells are 90–100% confluent).
Day 5 (based on 90–100% confluency):
Replace the culture medium to 10 mL of Ad+++ (see Recipes).
-
Culture in the incubator at 37°C with 5% CO2 for 4 days.
# Concentrating virus with PEG precipitation kit
Day 8 (The complete procedure should be performed on ice):
Collect the HBV-rich culture supernatant in a 50-mL Falcon tube.
Centrifuge the culture supernatant at 3,200 × g and 4°C for 15 min, to remove cell-debris.
Collect the supernatant in a new 50-mL Falcon tube.
Add 2.5 mL of 5× PEG solution per 10 mL of HBV-rich culture supernatant and mix properly.
-
Refrigerate the PEG-HBV culture supernatant mix overnight.
Critical step: PEG-HBV is stable at 4°C for up to 2 days.
Day 9 (The complete procedure should be performed on ice):
Centrifuge the PEG-HBV culture supernatant mix at 3,200 × g and 4°C for 30 min.
Carefully remove the supernatant with a 10-mL pipette. Do not touch the beige/white virus pellet.
Resuspend the virus pellet that comes from 10 mL of culture supernatant in 100 μL of Ad+++, or proportionally adjust the volumes according to the initial volume of HBV-rich culture supernatant (e.g., if 50 mL of HBV-rich culture supernatant is used, then the final volume of Ad+++ to be used to resuspend the virus pellet will be 500 μL).
-
Make small aliquots (100–500 μL) of virus stock and store at -80°C.
Critical step: Avoid freeze/thaw cycles to maximize virus recovery.
-
Production of high-titer purified and concentrated viral stock from HBV-rich HYPERFlask supernatant
# Production of high-titer HBV-rich supernatant in HYPERFlask (The following protocol is designed for one HYPERFlask)
Day 1:
Add 20 mL of 0.01% Collagen-R solution in sterile water to each T175 cell culture flask.
Prepare five T175 flasks for one HYPERFlask.
-
Perform incubation at room temperature overnight.
Day 2:
Remove the collagen solution with a 10 mL pipette.
Wash twice with 20 mL of PBS.
Seed approximately 6 × 106 of HepG2.2.15 cells in 35 mL of DMEM++ per T175 flask.
-
Culture in the incubator at 37°C with 5% CO2 until day 5.
Day 4:
Add 550 mL of 0.01% Collagen-R solution in sterile water to a 10-layer HYPERFlask cell culture vessel.
-
Perform incubation at room temperature overnight.
Day 5:
# Preparation of HYPERFlask for cell seeding
-
Remove the collagen solution by pouring it into a bottle.
Optional step: Collect 0.01% Collagen R solution and store at 4°C for future HYPERflask collagen-coatings. The solution can be reused approximately five times.
Wash twice with 300 mL of PBS.
-
Label the two sides of the flask as A and B (Figure 1).
# Seeding of cells in the HYPERFlask
Remove the culture media from the T175 flasks with a 10-mL pipette.
Wash gently once with 20 mL of PBS.
Add 10 mL of Trypsin-EDTA to each T175 flask and place it in the incubator, until all the cells are detached from the surface (the average time needed for the detachment of all the cells is 7–10 min).
Stop the trypsin digestion by adding 20 mL of pre-warmed (at 37°C) DMEM++ culture media per T175 flask, which makes the final volume of liquid in each flask 30 mL.
Collect 27 mL of cell suspension from each flask in a separate sterile bottle, and leave 3 mL.
The remaining 3 mL of cell suspension can be cultured again. Add 27 mL of DMEM++ and culture in the incubator until day 8.
Collect the total volume from step 17 for five T175 flasks (27 × 5 =135 mL) in a sterile bottle.
Add 415 mL of pre-warmed DMEM++ media, to make the final volume 550 mL.
-
Add the whole 550 mL of cell suspension (approximately 100 × 106 HepG2.2.15 cells) into the collagen coated HYPERFlask.
Critical step: Remove all remaining air by applying pressure to the center of the HYPERFlask, enabling homogenous cell spreading, and guaranteeing space for medium extension during the 37°C incubation, to avoid a pressure burst of the HYPERFlask.
-
Culture in the incubator at 37°C with 5% CO2 until day 8, with the A-labeled (step 12, Figure 1) side of the flask up.
Day 8:
Follow steps 13–16 again (# Seeding of cells in the HYPERFlask) for the five T175 flasks prepared at day 5, with the remaining 3 mL of cell suspension (step 17).
Collect 30 mL of cell suspension from each flask in a separate sterile bottle.
Collect the total volume from step 24 for five T175 flasks (30 × 5 =150 mL) in a sterile bottle.
Add 400 mL of pre-warmed DMEM++ media, to make the final volume 550 mL.
Remove the culture media from the HYPERFlask that was seeded at day 5.
Wash gently once with 300 mL of PBS.
Add 550 mL of cell suspension (from step 26) to the HYPERFlask (approximately 100 × 106 HepG2.2.15 cells).
Culture in the incubator with the B-labeled (step 12, Figure 1) side up, which allows attachment and growth of cells on opposite sides of the HYPERFlask layers.
-
Harvest the supernatant for purification in 4-day intervals (3–4 harvest cycles per HYPERFlask; see Notes).
Critical step: Due to the presence of a high number of dead and non-attached cells, discard the first supernatant collection after each cell plating round.
Notes:
Do not harvest supernatant earlier than 2 days after changing the culture medium, to ensure that enough mature HBV virus particles are secreted in the supernatant. Cells will also start to detach and die after 5–6 days without media change. It is recommended to harvest supernatant and add fresh media in alternating 3–4 days interval.
The titer of HBV in the HYPERFlask supernatant should be ≥107 copies/mL, to achieve the desired final concentration.
Store cell culture supernatants at 4°C overnight, before purification to precipitate serum lipoproteins and cell debris.
Do not store supernatant at 4°C for more than one day, as HBV infectivity drops with increasing storage time.
Harvest supernatant 3–4 times (depending on cell attachment) from each HYPERFlask.
After harvesting the supernatant four times, the HYPERFlask can be treated with Trypsin-EDTA, washed with PBS, and kept in a sterile environment, to reuse for production of subsequent rounds of HBV-rich supernatant. It is recommended to incubate the HYPERFlask with collagen-R solution before each round of cell seeding.
-
After seeding the HYPERFlask, the HepG2.2.15 cells are cultured in DMEM++ medium, as the HepG2.2.15 cell growth is optimal in DMEM++. However, Ad+++ and not DMEM++ medium is optimal for culturing organoids. Therefore, in section A (Production of virus for PEG precipitation) step 7, the culture medium is changed into Ad+++, to ensure the absence of any residual DMEM in the concentrated virus stock, as the HBV-rich supernatant is directly used to precipitate virus with PEG. However, in the HYPERFlask, while HepG2.2.15 cells are cultured in DMEM++ to ensure their optimum growth, the HBV-rich supernatant from the HYPERFlask is passed through the heparin column after collection, to purify the virus stock. The purified virus is eluted with a high salt-containing buffer, the eluate is diluted four times with Ad+++, and it is then further concentrated using a centrifugal filter unit. Following these steps ensures the absence of residual DMEM++ in the purified and concentrated virus stock from the HYPERFlask.
# Purification and concentration of high titer HBV stock (The complete procedure should be performed on ice or at 4°C)
Cold HYPERFlask supernatant is centrifuged at 500 × g and 4°C for 5 min, to precipitate cell debris.
Clear supernatant from the top is filtered through a 0.45 µm sterile filter, to remove remaining cell debris (see Notes).
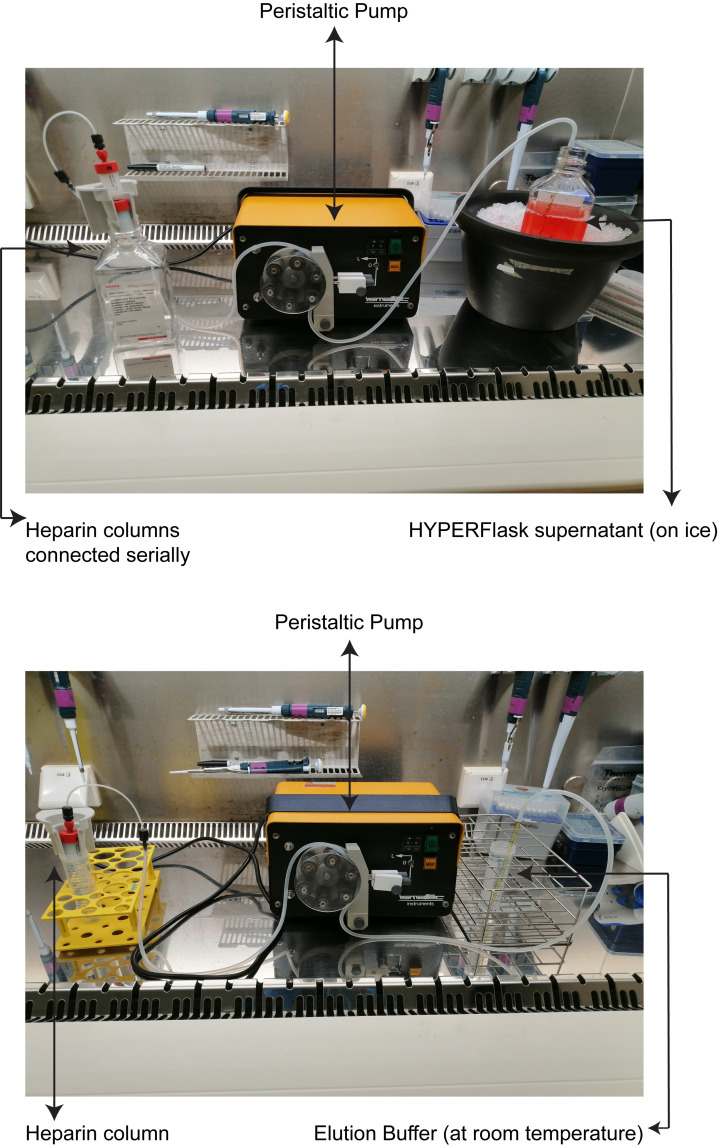
The silicone tubing is attached to a peristaltic pump (Figure 2). One end of the silicone tubing is connected to the heparin column, using a connector which is provided with the column. The remaining end of the silicone tubing is submerged in buffers or supernatant when required. Ensure that the flow direction of the peristaltic pump is from the buffers or supernatant end of the tubing to where the heparin column is attached.
The whole tubing is flushed with 50 mL of 70% ethanol solution.
-
Afterward, the tubing is washed with 50 mL of 1× PBS.
Critical step: Ensure that all the bubbles are removed and the tubing is completely filled with PBS, to prevent damage of the heparin columns.
-
Serially connect two heparin columns (HiTrap heparin 5 mL) for every 550 mL of supernatant (Figure 2).
Critical step: The heparin columns can also be connected in parallel using a stopcock. If connected in parallel, then the flow speed in step 7 can be 20 mL/min for two columns.
-
Keep the filtered supernatant on ice, and perfuse it through the heparin columns at a flow rate of 10 mL/min.
Critical step: A higher perfusion flow rate might reduce the lifetime of the columns.
-
After a complete perfusion of the supernatant, remove the columns and flush the system with elution buffer.
Critical step: Ensure that all the bubbles are removed and the tubing is completely filled with elution buffer.
-
Re-attach one column at a time, and elute with elusion buffer with a 2 mL/min flow rate.
Critical step: Before re-attaching the column, ensure that there is no air left in the system. Release a little bit of elution buffer in the connection point of the column, and then re-attach the column with the tubing.
Critical Step: Two columns can be re-attached together in a serial connection. If re-attached serially, then discard the first 4 mL of the eluent, due to the dead volume, and collect the next 40 mL.
-
Collect 20 mL of eluent per column in a sterile tube (40 mL from two columns).
Critical Step: Keep the elution buffer at room temperature, and also collect the eluent and store at room temperature until step 11. Storing eluent on ice or using ice cold elution buffer may result in precipitation of HBV particles, due to the high salt concentration in the elution buffer.
Immediately add 120 mL of Ad+++ to 40 mL of eluent (final volume =160 mL), to dilute the high salt concentration present in the elution buffer.
Add 12–15 mL of Ad+++ to one Amicon® Ultra-15 Centrifugal Filter Unit of 100 KDa cutoff, and centrifuge at 3,000 × g and 4°C for 10 min, to wash the filter.
Discard the flow through medium.
-
Add 12–15 mL of the purified viral solution to one Amicon® Ultra-15 Centrifugal Filter Unit.
Optional: Using multiple filter units will reduce the processing time.
Centrifuge at a maximum speed of 3,000 × g and 4°C for 10 min.
Discard the flow through filtrate from the bottom of the tube, and top up the remaining residual volume again to 12–15 mL with purified viral solution (step 11).
-
Follow steps 13 and 14 again until the whole 160 mL (step 11) is filtered, and the remaining final volume of purified and concentrated HBV stock is 2 mL (starting from 550 mL of HYPERFlask supernatant).
Critical Step: After 2–3 centrifugation rounds, the remaining viral solution starts to become sticky and might block the filter of the centrifugal unit. Pipetting up and down with a 1,000 μL micropipette during the topping up the centrifugal unit (step 16) helps to homogenize the solution and prevents blocking of the filter.
-
Make 100–500 µL aliquots of virus stock and store it at -80°C for future use.
Critical step: Avoid freeze/thaw cycles to maximize virus recovery and infectivity.
Take 5 µL of purified and concentrated virus, and bring the volume up to 200 µL.
-
Titer the virus stock after isolation of HBV DNA followed by qPCR (see section E).
Note: In the HYPERFlask cell culturing system, a large number of cells are cultured in a single HYPERFlask, resulting in a generous amount of cell debris and proteins present in the supernatant, which can block the heparin column if not removed. To ensure the complete removal of cell debris and proteins, centrifugation of the HYPERFlask supernatant alone is not enough, and filtering with a 0.45 µm sterile filter is essential. However, in section A (Production of virus for PEG precipitation), only 10 mL of supernatant from a single 10 cm cell culture plate is processed in a single 50 mL Falcon tube, which contains limited amounts of cell debris for which centrifugation alone is sufficient to remove.
# Maintenance of purification apparatus and heparin columns- Flush the column with 20 mL of 10× PBS solution per heparin column, at a 5 mL/min flow rate.
- Flush again with 20 mL of column storage buffer, and store the columns in an air-tight manner (by tightening the screw caps provided with the column) at 4°C.
- Clean the tubing with 70% ethanol, to inactivate any remaining HBV.
-
Production of heat-inactivated HBV
Thaw the pre-tittered active recombinant HBV on ice.
Transfer the virus to a 1.5-mL microcentrifuge tube.
Centrifuge at maximum speed and 4°C for 10 min.
Transfer the virus to a new 1.5-mL microcentrifuge tube.
Boil the virus at 100°C for 30 min.
Centrifuge at maximum speed and 4°C for 10 min.
Use the supernatant as heat-inactivated virus.
-
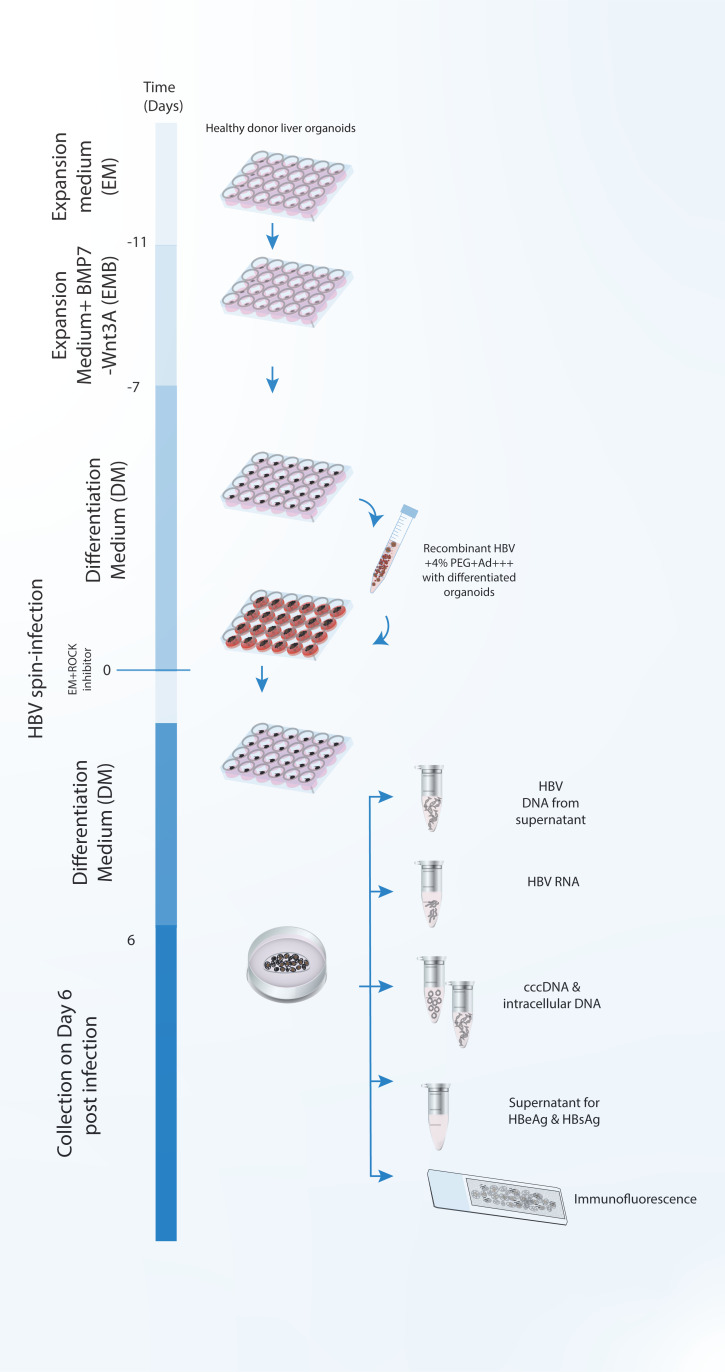
Brief description of the generation, maintenance, and HBV infection of human liver organoids
There is a companion protocol paper that describes the culture method for human liver organoids, and their infection with recombinant HBV ( Romal et al., 2022 ). A very brief description of that protocol paper is given below.
Liver biopsies are dissociated into single cell level, mixed with BME/Ad+++, and cultured as a dome shaped drop, using a specialized culture medium that enables the growth of LGR5+ adult hepatic stem cells and formation of organoids.
Organoids are cultured in an expansion medium that allows the continuous growth of human liver organoids.
Culture medium is changed into differentiation medium, which halts the proliferation of the organoids and induces their differentiation into hepatocytes, as well as expression of differentiated hepatocyte markers and the HBV receptor NTCP, which is necessary for the infection of the organoids with HBV.
Differentiated organoids are mixed with the appropriate amount of HBV and spin infected.
After spin infection, infected organoids are cultured again in 24-well cell culture plates in dome shaped drops containing BME/Ad+++ (Section D, step 31 in Romal et al., 2022 ).
-
Isolation and detection of HBV DNA from the supernatant
HBV DNA from the supernatant can be isolated by either using available commercial kits or using the phenol-chloroform isolation method. A detailed protocol is described below:
# Isolation of HBV DNA from supernatant with a commercial isolation kit:
Take 200 µL of the culture supernatant (see section D, step 5).
-
The HBV DNA from the supernatant is isolated using the QIAamp MinElute Virus Spin Kit, following the manufacturer’s instructions.
# Isolation of HBV DNA from the supernatant using the phenol-chloroform isolation method:
Take 200 µL of the supernatant.
Add 300 µL of 1% SDS+ 0.1M NaHCO3 in MilliQ-water supplemented with 100 µg/mL proteinase K.
Incubate the samples at 55°C for 1 h.
Briefly spin down to collect all the components in the bottom of the tube.
Add 500 µL of phenol-chloroform-isoamyl alcohol (PCI), and vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the top aqueous phase to a new 1.5-mL microcentrifuge tube.
Add 500 µL of 24:1 chloroform-isoamyl alcohol (CI), and vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the top aqueous phase into a new 1.5-mL microcentrifuge tube, add 1 µL of glycogen from the stock (20 mg/mL), and 30 µL of 3M NaOAc pH 5.2. Vortex for 10 s, add 1 mL of 100% ethanol, and vortex again thoroughly for 10 s.
Snap-freeze the microcentrifuge tubes in liquid nitrogen or freeze at -80°C for 30 min or overnight.
Spin the DNA down at maximum speed and 4°C for 30 min.
Wash the pellet with 500 µL of 70% ethanol.
Spin down at maximum speed and 4°C for 15 min.
Carefully remove the supernatant.
Resuspend the DNA pellet in 30 µL of nuclease free-water.
-
Store the DNA at -20°C.
# Detection of HBV DNA
Component Final concentration 2× LightCycler480 Probes Master (Roche) 1× (12.5 µL) Forward Primer (100 µM) 0.5 µM Reverse Primer (100 µM) 0.5 µM Probe (50 µM) 0.1 µM DNA template 4 µL Nuclease free water x µL Total reaction volume 25 µL
* Primer and probe sequences: Forward (5’-GCAACTTTTTCACCTCTGCCTA-3’)
Reverse (5’-AGTAACTCCACAGTAGCTCCAAATT-3’)
Probe (FAM-TTCAAGCCTCCAAGCTGTGCCTTGGGTGGC-BHQ1)
Prepare the reaction mix and briefly centrifuge to collect all the components at the bottom of the PCR plate.
-
Run the amplification, starting with 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s.
# Analysis and expected results:
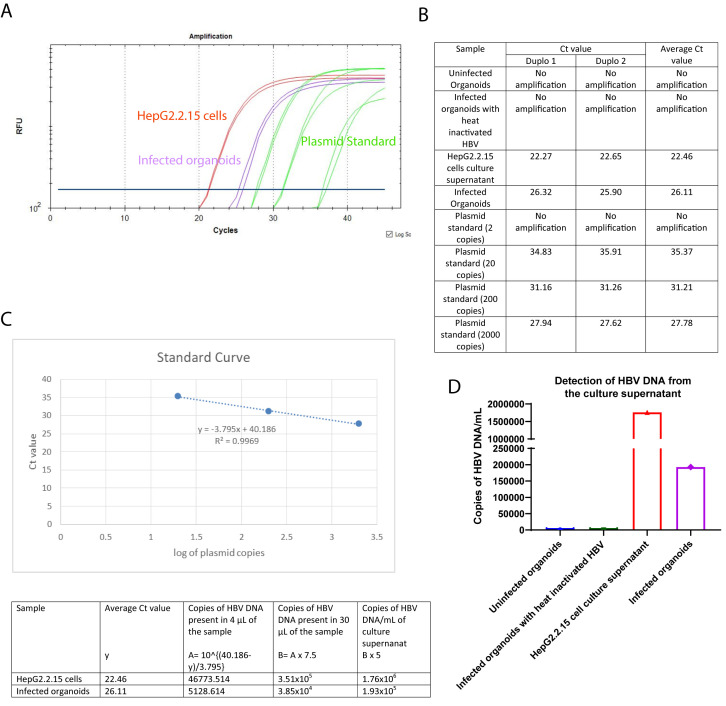
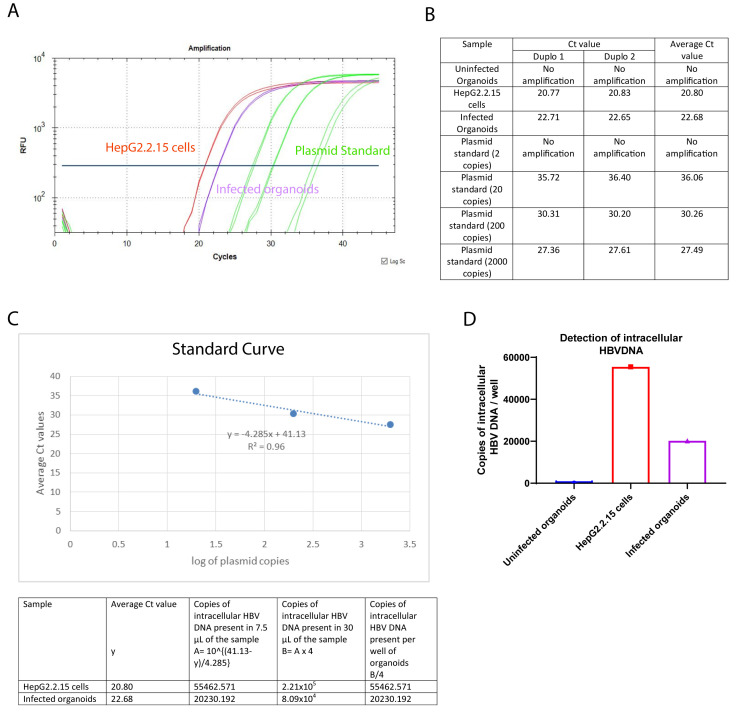
The qPCR reaction to detect HBV DNA from the supernatant included a standard curve made from dilutions of a plasmid containing the lenti-HBV construct with the 1.3mer HBV genome, ranging from 2 to 2 × 104 copies of plasmid. The number of HBV DNA copies is calculated using the standard curve. Figure 3A represents a PCR curve of different samples with the HBV specific primers and probe. For each PCR reaction, 4 µL of the DNA template is used and the starting volume of the DNA preparation of each sample condition is 30 µL. The culture supernatants from the uninfected organoids and the organoids infected with heat-inactivated HBV are used as negative control. The culture supernatant from the HepG2.2.15 cells is used as positive control. A standard curve is generated with 2, 20, 200, and 2000 copies of lenti-HBV plasmid construct containing 1.3 mer HBV genome. There are no amplifications observed in the uninfected organoids or the organoids infected with heat-inactivated HBV. The fluorescent signal curves for the infected organoids, HepG2.2.15 cells, and the standard plasmid dilutions are depicted in purple, red, and green, respectively. Figure 3B represents the individual and average (cycle threshold) Ct values for the different sample conditions. The standard curve (Figure 3C) is prepared by plotting the Ct values of the different standard plasmid dilutions in the y-axis, and the corresponding logarithm of the standard plasmid copies in the x-axis. The r2 value and the equation for the standard line are also mentioned in Figure 3C. Using the average Ct values for the infected organoids and HepG2.2.15 cells, and the standard line equation, the number of HBV DNA copies present per mililiter of the corresponding culture supernatants are calculated (Figure 3C). The bar graph in Figure 3D depicts the HBV DNA copy numbers present in the different sample conditions.
-
Isolation and detection of intracellular HBV DNA and cccDNA
All the available techniques to detect cccDNA have their own sets of limitations. Therefore, using multiple techniques simultaneously is recommended. In our study, we have performed both treatment of whole genomic DNA with T5 exonuclease, and alkali lysis plasmid DNA isolation, followed by plasmid-safe digestion to differentiate cccDNA from rcDNA. Detailed protocols are described below:
Prepare a 15-mL Falcon tube containing 10 mL of Ad+++.
Remove the medium from each well of organoids (see section D, step 5).
Add 1 mL of Ad+++ to each well of organoids, and collect 4–8 wells of a 24-well plate of organoids in each 15-mL Falcon tube.
Incubate on ice for 30 min.
Centrifuge at 200 × g and 4°C for 5 min.
Remove the supernatant.
Add 60–100 µL of TrypLE (a substitute for trypsin that is used to dissociate adherent cells and primary human cell cultures) to the sample, and mechanically digest for 30–60 s at room temperature.
Wash the organoids with ice-cold PBS.
Centrifuge at 200 × g and 4°C for 5 min.
-
Remove the supernatant, leaving the pellet of digested organoids.
# Whole genomic DNA isolation followed by T5 exonuclease digestion
T5 exonuclease degrades all the linear and partially circular dsDNA. However, the enzyme does not degrade completely circular supercoiled dsDNA. T5 exonuclease is used to remove the rcDNA present in the whole genomic DNA preparation, which is partially double stranded, but will not degrade the completely circular and double stranded cccDNA.
The pellet (section F, step 10) is used for whole genomic DNA isolation using the DNeasy Blood and Tissue kit.
Continue the isolation, following the manufacturer’s instructions.
Elute the final DNA from the column in 50 µL of nuclease free-water.
Use 25 µL for T5 exonuclease digestion, and the remaining 25 µL for detection of intracellular HBV DNA.
Follow the manufacturer’s instructions, to continue the digestion with T5 exonuclease.
Inactivate the digestion by adding EDTA, to a final concentration of 11 mM.
Purify the complete inactivated reaction volume using the QIAquick PCR Purification Kit.
Continue the purification, following the manufacturer’s instructions.
-
Elute the purified DNA from the column in 30 µL of nuclease free-water.
# For the alkali lysis plasmid DNA-isolation followed by plasmid safe digestion
Resuspend the pellet (section F, step 10) in 800 µL of ice-cold cell lysis buffer.
Incubate on ice for 10 min.
Add equal volume of alkali lysis buffer.
Incubate the samples at 37°C for 30 min.
Neutralize the DNA by adding 3 M potassium acetate (CH3COOK) (pH 5.0), to a final concentration of 0.6 M.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the supernatant to two new 1.5 mL microcentrifuge tubes, and add ~900 µL of phenol-chloroform-isoamyl alcohol (PCI). Vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the top aqueous phase to a new 1.5 mL microcentrifuge tube.
Repeat steps 7–9.
Centrifuge at maximum speed and room temperature for 5 min.
Add 500 µL of butanol:isopropanol (7:3), and vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the bottom layer of each tube into two new 1.5 mL microcentrifuge tubes. (The bottom layers from two tubes will be distributed equally into four tubes.)
Add 200 µL of 7.5 M Ammonium Acetate (CH3COONH4), 1 µL of glycogen stock (20 mg/mL), and 1 mL of 100% ethanol.
Snap-freeze the microcentrifuge tubes in liquid nitrogen, or freeze at -80°C for 30 min or overnight.
Spin the DNA down at maximum speed and 4°C for 30 min.
Wash the pellet with 1 mL of 70% ethanol.
Spin down at maximum speed and 4°C for 15min.
Carefully remove the supernatant.
-
Resuspend the DNA pellet in 50 µL of nuclease free-water.
Pause step: DNA can be stored at -20°C for one year, until processing for further digestion.
Take 25 µL of the DNA.
Digest the samples using plasmid safe, following the manufacturer’s protocol.
After digestion, top the volume up with PBS to 400 µL.
Add 400 µL of phenol-chloroform-isoamyl alcohol (PCI). Vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Take the top aqueous phase and transfer to a new 1.5 mL microcentrifuge tube.
Add 400 µL of 24:1 Chloroform-isoamyl alcohol (CI). Vortex thoroughly for 10 s.
Centrifuge at maximum speed and room temperature for 5 min.
Transfer the top aqueous phase into a new 1.5 mL microcentrifuge tube, add 1 µL of glycogen from the stock (20 mg/mL), and 20 µL of 3M NaOAc pH 5.2. Vortex for 10 s, add 1 mL of 100% ethanol, and vortex thoroughly for 10 s.
Snap-freeze the microcentrifuge tubes in liquid nitrogen, or freeze at -80°C for 30 min or overnight.
Spin the DNA down at maximum speed and 4°C for 30 min.
Wash the pellet with 500 µL of 70% ethanol.
Spin down at maximum speed and 4°C for 15 min.
Carefully remove the supernatant.
-
Resuspend the DNA pellet in 30 µL of nuclease free-water.
# Detection of intracellular HBV DNA
Component Final concentration 10× PCR Buffer 1× (2.5 µL) 50 mM MgCl2 1.75 mM 10 mM dNTP mix 400 µM Forward Primer (100 µM) 0.5 µM Reverse Primer (100 µM) 0.5 µM Probe (50 µM) 0.15 µM Platinum Taq DNA Polymerase 0.04 units/µL DNA template 7.5 µL Nuclease free water x µL Total reaction volume 25 µL * Primer and probe sequences: Forward (5’-GCAACTTTTTCACCTCTGCCTA-3’)
Reverse (5’-AGTAACTCCACAGTAGCTCCAAATT-3’)
Probe (FAM-TTCAAGCCTCCAAGCTGTGCCTTGGGTGGC-BHQ1)
Prepare the reaction mix and briefly centrifuge, to collect all the components at the bottom of the PCR plate.
-
Run the amplification starting with 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s.
# Detection of cccDNA
Component Final concentration 2× LightCycler480 Probes Master (Roche) 1× (10 µL) Forward Primer (100 µM) 1 µM Reverse Primer (100 µM) 1 µM Probe (50 µM) 0.2 µM DMSO 4% cccDNA template 4.2 µL Nuclease free water x µL Total reaction volume 20 µL * Primer and probe sequences: Forward (5’-GTCTGTGCCTTCTCATCTGC-3’)
Reverse (5’-AGTAACTCCACAGTAGCTCCAAATT-3’)
Probe (FAM-TTCAAGCCTCCAAGCTGTGCCTTGGGTGGC-BHQ1)
Prepare the reaction mix and briefly centrifuge, to collect all the components at the bottom of the PCR plate.
-
Run the amplification starting with 95°C for 10 min, followed by 50 cycles of 95°C for 15 s, and 61°C for 1 min, as mentioned in a previously published study ( Winer et al., 2017 ).
# Analysis and the expected results:
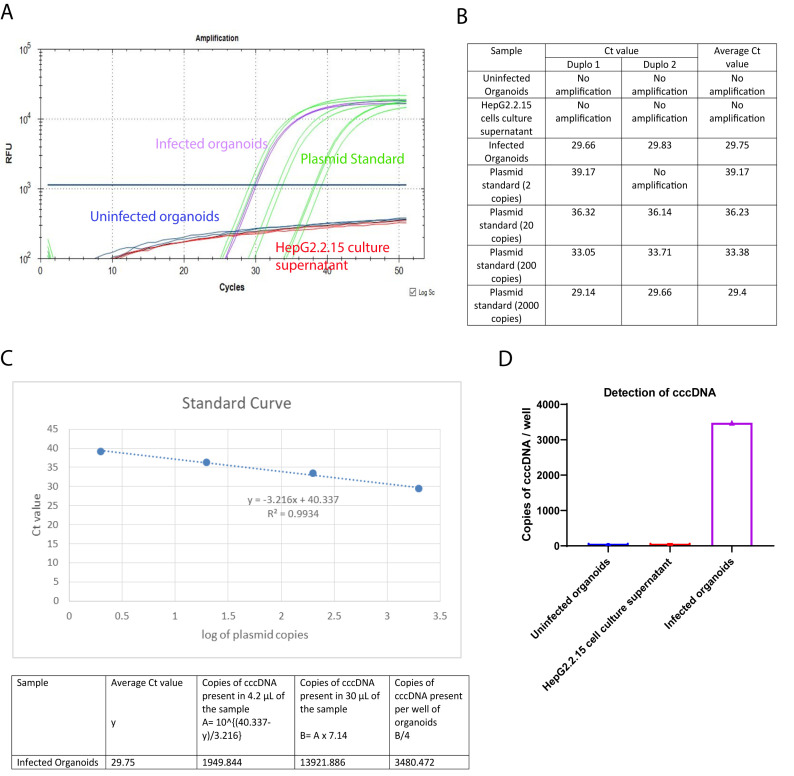
Both PCRs to detect intracellular HBV DNA and cccDNA included a standard curve made from dilutions of a plasmid containing the lenti-HBV plasmid construct with 1.3mer HBV genome, ranging from 2 to 2 × 104 copies of plasmid. The number of intracellular HBV DNA and cccDNA copies were calculated using the standard curve. Figure 4A represents a PCR curve of the different samples, with the cccDNA specific primers and probe. For each PCR reaction, 4.2 µL of the DNA template is used, and the starting volume of the DNA preparation of each sample condition is 30 µL. DNA preparations from uninfected organoids and HepG2.2.15 cell culture supernatant, which should only contain rcDNA, are used as negative control. To generate a standard curve, 2, 20, 200, and 2000 copies of lenti-HBV plasmid construct containing 1.3 mer HBV genome are used. There is no amplification observed in the uninfected organoids (red) or HepG2.2.15 cell culture supernatant (navy blue). The fluorescent signal curves for the infected organoids and the standard plasmid dilutions are depicted in purple and green, respectively. Figure 4B represents the individual and average Ct values for the different sample conditions. The standard curve (Figure 4C) is prepared by plotting the Ct values of the different standard plasmid dilutions in the y-axis, and the corresponding logarithm of the standard plasmid copies in the x-axis. The r2 value and the equation for the standard line are also mentioned in Figure 4C. Using the average Ct value for the infected organoids and the standard line equation, the number of cccDNA copies present in the infected organoid sample is calculated (Figure 4C). The bar graph in Figure 4D depicts the cccDNA copy number present in the different samples.
Figure 5A represents a PCR curve of different samples with the HBV specific primers and probe. For each PCR reaction, 7.5 µL of the DNA template is used, and the starting volume of the DNA preparation for each sample is 30 µL. The uninfected organoids are used as negative control. The HepG2.2.15 cells are used as a positive control. To generate a standard curve, 2, 20, 200, and 2000 copies of lenti-HBV plasmid construct containing 1.3 mer HBV genome are used. There is no amplification observed in the uninfected organoids sample. The fluorescent signal curves for the infected organoids, HepG2.2.15 cells, and the standard plasmid dilutions are depicted in purple, red, and green, respectively. Figure 5B represents the individual and average Ct values for the different samples. The standard curve (Figure 5C) is prepared by plotting the Ct values of the different standard plasmid dilutions in the y-axis, and the corresponding logarithm of the standard plasmid copies in the x-axis. The r2 value and the equation for the standard line are also mentioned in Figure 5C. Using the average Ct values for the infected organoids and HepG2.2.15 cells, and the standard line equation, the number of intracellular HBV DNA copies present are calculated (Figure 5C). The bar graph in Figure 5D depicts the intracellular HBV DNA copy numbers present in the different samples.
-
Isolation and detection of HBV RNA
Isolation of the HBV RNA can be performed using both a commercial kit and Trizol RNA isolation procedure. Detailed protocols are described below:
Remove the medium from the organoids.
-
Collect 1–2 wells of a 24-well plate of organoids (see section D; step 5) with 1 mL of Ad+++, mix by pipetting 4–5 times, and transfer to a 1.5-mL microcentrifuge tube.
Critical Step: One well of a 24-well plate with 90% density, or if the density is low use two wells of a 24-well plate. The samples can be stored in -80°C for six months, or can be directly isolated.
Incubate on ice for 10–20 min.
Centrifuge the samples at 200 × g and 4°C for 5 min.
-
Remove the supernatant, leaving the organoids pellet behind.
# Isolation of RNA using commercial kit
Resuspend the organoids pellet (see section G, step 5) with 500 µL of cell lysis buffer provided in the kit (RealiaPrep RNA Cell Miniprep System, Promega).
Continue the isolation, following the manufacturer’s instructions.
Elute the RNA in 40 µL of nuclease free-water.
-
Store the RNA at -80°C, until further processing.
# Isolation of RNA using Trizol RNA isolation protocol
Lyse the organoid pellet (see section G, step 5) with 1 mL of Trizol, by pipetting up and down, and vortexing vigorously for 20–40 s.
Incubate the sample at room temperature for 5 min.
Add 200 μL of chloroform, vortex the samples vigorously for 20 s, and incubate at room temperature for 2–3 min.
Centrifuge the samples at 12,000 × g and 4°C for 15 min.
Carefully transfer the upper aqueous phase, without disturbing the interphase, into a new 1.5-mL microcentrifuge tube.
Precipitate the RNA by adding 500 μL of isopropyl alcohol, inverting the tubes four to five times, and incubating the samples at room temperature for 10 min.
Centrifuge the samples at 12,000 × g and 4°C for 10 min.
-
Remove the supernatant.
Critical Step: To avoid losing the RNA pellet, especially when the pellet is not visible or is very small, leave ~50 μL of the supernatant.
-
Add 1 mL of 75% ethanol, and centrifuge at 7,500 × g and 4°C for 5 min.
Critical Step: Do not vortex the RNA pellet.
-
Repeat step 9, and remove the wash buffer completely.
Critical Step: To make sure that the wash buffer is completely removed, briefly centrifuge the samples, to get rid of the remaining wash buffer from the edges of the tube.
Air-dry the pellet for 10 min.
Resuspend the pellet in 40 μL of nuclease free water.
-
Store the RNA samples at -80°C.
#DNase l treatment
Use 1 μL of the sample to measure the RNA concentration using a NanoDrop 2000/2000c spectrophotometer.
Use 300–1,000 ng of RNA for DNase l treatment.
-
Complete the DNase treatment, following the manufacturer’s instruction.
# cDNA synthesis
Use the DNase l treated samples for cDNA synthesis with Superscript II Reverse Transcriptase kit.
-
Complete the cDNA synthesis, following the manufacturer’s instructions, and using random primers.
# Detection of HBV RNA
Component Final concentration 10× PCR Buffer 1× (2.5 µL) 50 mM MgCl2 1.75 mM 10 mM dNTP mix 400 µM Forward Primer (100 µM) 0.5 µM Reverse Primer (100 µM) 0.5 µM Probe (50 µM) 0.15 µM Platinum Taq DNA Polymerase 0.04units/µL cDNA template (diluted 1:2.5 or 1:5 in water, based on the starting amount of RNA used for cDNA synthesis) 4 µL Nuclease free water x µL Total reaction volume 25 µL * Primer and probe sequences (HBV): Forward (5’-GCAACTTTTTCACCTCTGCCTA-3’)
Reverse (5’-AGTAACTCCACAGTAGCTCCAAATT-3’)
Probe (FAM-TTCAAGCCTCCAAGCTGTGCCTTGGGTGGC-BHQ1)
* Primer and probe sequences (Beta-2-microglobulin):
Forward (5’- AGCGTACTCCAAAGATTCAGGTT-3’)
Reverse (5’- ATGATGCTGCTTACATGTCTCGAT-3’)
Probe (FAM- TCCATCCGACATTGAAGTTGACTTACTG-BHQ1)
Prepare the reaction mix, and briefly centrifuge to collect all the components at the bottom of the PCR plate.
-
Run the amplification starting with 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s.
# Analysis and the expected results:
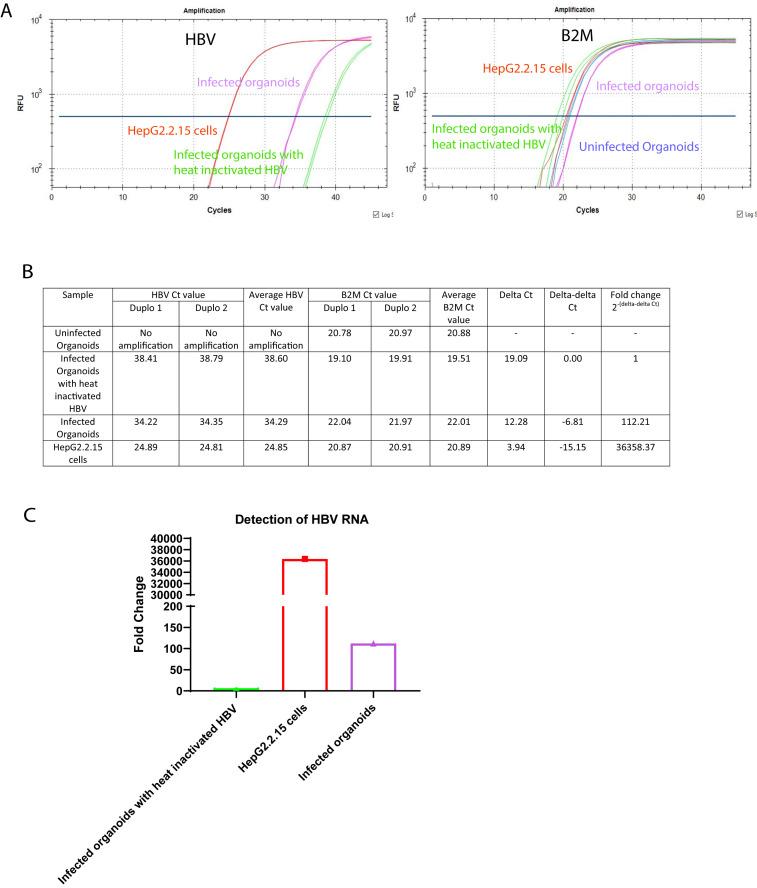
Beta-2-microglobulin (B2M) is used as a housekeeping control for analysis of cDNA sample expression. Fold increase is calculated using the 2-ΔΔCt method. Figure 6A represents PCR curves of different samples with the HBV specific primers and probe, and the B2M specific primers and probe. For each PCR reaction, 4 µl of the cDNA template is used. The uninfected organoids and the organoids infected with heat-inactivated HBV are used as negative control. The HepG2.2.15 cells are used as positive control. The fluorescent signal curves for the uninfected organoids, infected organoids, HepG2.2.15 cells, and the organoids infected with heat-inactivated HBV are depicted in blue, purple, red, and green, respectively. Figure 6B represents the individual and average Ct values for the different samples, as well as the calculation to determine the fold changes of the HBV RNA expression in the infected organoids and the HepG2.2.15 cells compared to the organoids infected with heat-inactivated HBV. There is no expression of HBV RNA in the uninfected organoids. Therefore, the uninfected organoids sample is excluded from the fold change calculation. The bar graph in Figure 6C depicts the fold changes of the HBV RNA expression in the different samples.
-
Immunofluorescence
Critical step: Use an ultra-low retention pipette tip to reduce sample loss while triturating.
First, pre-wet the pipette with the wash medium or buffer solution, and then resuspend the sample.
Prepare a 15-mL Falcon tube with 10 mL of ice-cold Ad+++, and place it on ice.
Remove the medium from all the wells (see section D, step 5).
Add ~1 mL of ice-cold Ad+++ to each well.
-
Collect the drops containing the organoids with a 10 mL pipette, by scrapping gently.
Critical step: Avoid breaking the organoids.
Mix three to four times by triturating with a 10-mL pipette.
-
Incubate on ice for 30 min to 1 h.
Critical step: Mix the organoids by inverting the Falcon tubes four to five times every 5 min. This will help the BME to dissolve efficiently.
Pellet the organoids by centrifuging at 100 × g at 4°C for 5 min.
Wash 1× with 10 mL of ice-cold Ad+++, mix by pipetting up and down four to five times with a 10 mL pipette.
Centrifuge at 100 × g and 4°C for 5 min.
Remove the medium and wash once with 10 mL of ice-cold PBS.
-
Remove the medium and gently resuspend the pellet with 200 µL of freshly prepared fixative solution [either 4% PFA for HBcAg, Hepatocyte Nuclear Factor 4 Alpha (HNF4α), sodium taurocholate co-transporting polypeptide (NTCP), and Albumin, or 100% acetone for HBsAg].
Critical step: Fixative reagent and concentration can differ depending on the antibodies.
Incubate on ice for 30 min.
Remove fixative solution and add 10 mL of ice-cold PBS.
Centrifuge at 100 × g and 4°C for 5 min.
Remove the PBS.
Add 1 mL of freshly made 0.1 M glycine in PBS.
-
Incubate at room temperature for 15–30 min.
Critical step: Treating organoids with glycine after fixation will work as a quencher. This can help in blockage of unreacted aldehydes, which may can cause an increase in background fluorescence.
-
Add 10 mL of PBS, and centrifuge at 100g × g for 5 min.
Pause step: Fixed organoids can be stored at 4°C for 3–4 weeks.
Remove the PBS.
Permeabilize the organoids by adding 1 mL of 0.3% Triton in PBS (HBcAg, HNF4α, and NTCP), 1% Triton X-100 in PBS (Albumin), or 100% acetone (HBsAg).
Incubate at room temperature for 30 min.
Centrifuge at 100 × g for 5 min, and remove the permeabilizing solution.
Wash once with 1 mL of 0.5% FBS in PBS at room temperature.
Centrifuge at 100 × g and room temperature for 5 min.
-
Add 1 mL of blocking buffer (PBSTD; PBS + 0.5% serum + 0.3% triton+ 1% DMSO for HBcAg, HNF4α, Albumin, and NTCP, or 10% BSA+ 0.5% serum in PBS for HBsAg), and incubate at room temperature for 2 h.
Optional: Use goat serum for the blocking step, in case all the secondary antibodies are from goat.
Centrifuge at 100 × g and room temperature for 5 min.
Remove the blocking buffer.
Incubate the organoids with primary antibody diluted in PBS + 10% blocking buffer (check the dilutions as recommended by the manufacturers).
Incubate in an orbital shaker at 4°C overnight.
Centrifuge at 100 × g and 4°C for 5 min.
Remove the primary antibody mix.
Add 10 mL of 0.5% FBS in PBS. Incubate in an orbital shaker at room temperature for 10 min. Remove the wash buffer and repeat this step at least three times.
Add 200 μL of 0.5% FBS in PBS containing the appropriate secondary antibody, and incubate in an orbital shaker in the dark at room temperature for 2 h.
Remove the secondary antibody.
Wash by adding 10 mL of 0.5% FBS in PBS. Incubate in an orbital shaker at room temperature for 10 min.
Centrifuge at 100 × g and room temperature for 5 min.
Repeat steps 35–36 at least four to five times.
Add 200 μL of 0.5% FBS in PBS containing 5 μg/mL of Hoechst.
Incubate in an orbital shaker in the dark at room temperature for 15–20 min.
Remove the Hoechst and wash with 1 mL of 0.5% FBS in PBS.
Incubate in an orbital shaker in the dark at room temperature for 10 min.
Centrifuge at 100 × g and room temperature for 5 min.
Remove the supernatant using a plastic Pasteur pipette, and transfer the organoids to the slide.
Add one drop of anti-fade solution, and apply the coverslip.
Store the slides in the dark at 4°C.
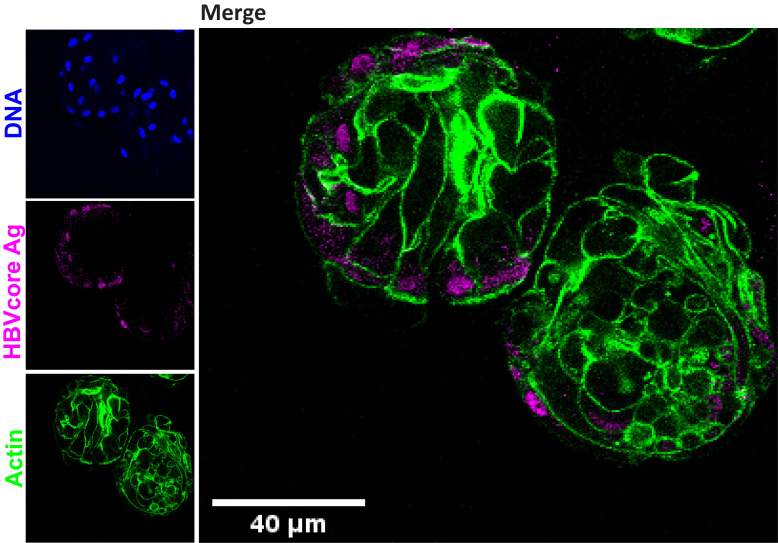
Images are taken and processed using Leica LAS AF Lite software (Leica SP5 confocal) (Figure 7).
-
Detection of HBsAg and HBeAg from the supernatant
Collect the organoid supernatant at 3–6 days post infection (see section D, step 5).
Spin the samples down at 200 × g and 4°C for 5 min.
Use 50 μL of the supernatant for detection of HBsAg or HBeAg.
-
Use the HBeAg (Human Hepatitis Be Antigen ELISA Kit, Cusabio) or HBsAg (Monolisa HBs Ag ULTRA, Bio-Rad) specific kits for the detection of HBeAg and HBsAg, respectively.
# Analysis and the expected results:
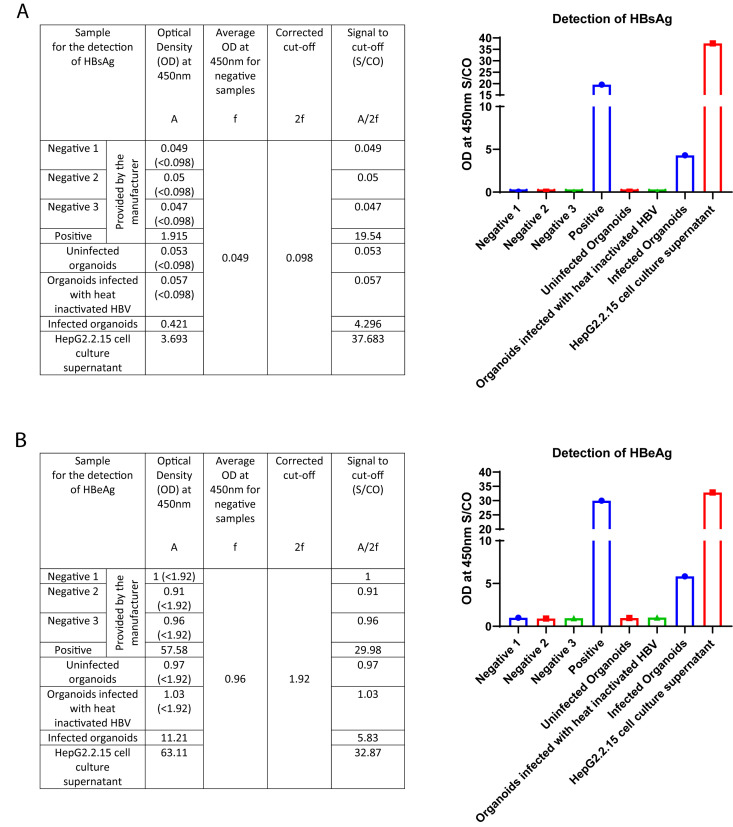
Commercially available ELISA kits are used to detect HBsAg and HBeAg from the culture supernatants. Figure 8A represents the absorbances to detect HBsAg, measured as the optical densities (OD) at a wavelength of 450 nm for the different samples, calculation of the corrected cut-off value using absorbances of the negative samples provided by the kit manufacturer, and the signal to cut-off values for the different samples, as well as the bar diagram depicting the expressions of HBsAg in the different samples. Figure 8B represents the absorbances to detect HBeAg measured as OD450 for the different samples, calculation of the corrected cut-off value using absorbances of the negative samples provided by the kit manufacturer, and the signal to cut-off values for the different samples, as well as the bar diagram depicting the expressions of HBeAg in the different samples.
Figure 1. Labelling of HYPERFlask.
Figure 2. Assembly of purification apparatus for perfusion of HYPERFlask supernatant (up) and elution of HBV particle (down).
Figure 3. Expected results for the detection of HBV DNA from the culture supernatants.
A) PCR Curve of the different sample conditions. B) Observed Ct values and average Ct values for the different sample conditions, C) Generation of the standard curve using the Ct values of different dilutions of the standard plasmids, and calculation of the copies of HBV DNA present in the infected organoids and the HepG2.2.15 cells. D) Bar graph of the HBV DNA copy numbers present in the different samples. The y-axis represents the HBV DNA copy numbers, and the x-axis represents different sample conditions.
Figure 4. Expected results for the detection of cccDNA.
A) PCR Curve of the different sample conditions. B) Observed Ct values and average Ct values for the different sample conditions, C) Generation of the standard curve using the Ct values of different dilutions of the standard plasmids, and calculation of the copies of cccDNA present in the infected organoids sample. D) Bar graph of the cccDNA copy numbers present in the different samples. The y-axis represents the cccDNA copy numbers, and the x-axis represents the different samples.
Figure 5. Expected results for the detection of intracellular HBV DNA from the culture supernatants.
A) PCR Curve of the different sample conditions. B) Observed Ct values and average Ct values for the different sample conditions, C) Generation of the standard curve using the Ct values of different dilutions of the standard plasmids, and calculation of the copies of intracellular HBV DNA present in the infected organoids and the HepG2.2.15 cells. D) Bar graph of the intracellular HBV DNA copy numbers present in the different samples. The y-axis represents the intracellular HBV DNA copy numbers, and the x-axis represents the different sample conditions.
Figure 6. Expected results for the detection of HBV RNA.
A) PCR Curve of the different sample conditions with HBV specific (left) and B2M specific (right) primers and probes. B) Observed Ct values and average Ct values, as well as the HBV RNA fold change calculations for the different samples compared to the organoids infected with heat-inactivated HBV. C) Bar graph of the HBV RNA fold changes present in the different samples. The y-axis represents the fold changes, and the x-axis represents the different sample conditions.
Figure 7. Immunofluorescence staining showing the expression of HBV core antigen (HBcAg) (magenta) together with actin (green), performed in healthy donor liver organoids six days post HBV infection.
Figure 8. Expected results for the detection of HBsAg and HBeAg.
A) Absorbances to detect HBsAg, measured as OD450 for the different samples, calculation of the corrected cut-off value using absorbances of the negative samples provided by the kit manufacturer, and the signal to cut-off values for the different samples, as well as the bar diagram depicting the expressions of HBsAg in the different samples. The y-axis represents the OD at 450 nm S/CO and the x-axis represents the different samples. B) Absorbances to detect HBeAg measured as OD450 for the different samples, calculation of the corrected cut-off value using absorbances of the negative samples provided by the kit manufacturer, and the signal to cut-off values for the different samples, as well as the bar diagram depicting the expressions of HBeAg in the different samples. The y-axis represents the OD at 450 nm S/CO, and the x-axis represents the different samples.
Recipes
-
DMEM ++ culture medium (500 mL)
445 mL of DMEM
50 mL of FBS
5 mL of Penicillin-Streptomycin
-
Ad+++ culture medium (500 mL)
485 mL of DMEM
5 mL of HEPES
5 mL of Ultraglutamine
5 mL of Penicillin-Streptomycin
-
70% Ethanol solution (100 mL)
70 mL of Ethanol absolute
30 mL of Milli-Q treated Water
-
Wash solution
500 mL of 1× PBS, pH 7.4
-
Elution buffer (100 mL)
25 mL of 10× PBS, pH 7.4
75 mL of Milli-Q treated Water
-
Column wash buffer (100 mL)
40 mL of 5M NaCl
60 mL of Milli-Q treated Water
-
Heparin column storage buffer (100 mL)
13 mL of 10× PBS, pH 7.4
20 mL of Ethanol absolute
67 mL of Milli-Q treated Water
-
Cell lysis buffer (50 mL)
100 μL of 0.5M EDTA (pH 8.0)
250 μL of 1 M Tris HCl (pH 7.5)
250 μL of 10% Nonidet P-40
49.4 mL of Milli-Q treated Water
-
Alkali lysis buffer (50 mL)
5 mL of 1 M NaOH
30 mL of 10% SDS
15 mL of Milli-Q treated Water
-
PBSTD (50 mL)
250 μL of FBS
150 μL of Triton X-100
500 μL of DMSO
49.1 mL of PBS
-
10× PBS (500 mL)
8.9 g of Na2HPO4·2H2O
1.2 g of KH2PO4
40 g of NaCl
1 g of KCl
Adjust the final volume to 500 mL with Milli-Q treated Water.
Acknowledgments
Protocol for the generation of high-titer HBV stock has been adapted from a recently published protocol by Wettengel et al. (2021) and necessary modifications have been added to make the virus stock suitable to infect the human liver organoids. Protocol for the detection of the different HBV specific viral products upon infection of the human liver organoids with recombinant HBV has been adapted from the previously published work by the authors (De Crignis et al., 2021 ).
Competing interests
The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. De Crignis E., Hossain T., Romal S., Carofiglio F., Moulos P., Khalid M. M., Rao S., Bazrafshan A., Verstegen M. M. and Pourfarzad F.(2021). Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife 10: e60747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wettengel J. M., Linden B., Esser K., Laue M., Burwitz B. J. and Protzer U.(2021). Rapid and Robust Continuous Purification of High-Titer Hepatitis B Virus for In Vitro and In Vivo Applications . Viruses 13(8): 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winer B. Y., Huang T. S., Pludwinski E., Heller B., Wojcik F., Lipkowitz G. E., Parekh A., Cho C., Shrirao A., Muir T. W., et al.(2017). Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat Commun 8(1): 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Romal S., Hossain T. and Mahmoudi T.(2022). Generation, Maintenance and HBV Infection of Human Liver Organoids. Bio-protocol 12(6): e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]