Abstract
Long noncoding (lnc)RNAs modulate gene expression alongside presenting unexpected source of neoantigens. Despite their immense interest, their ability to be transferred and control adjacent cells is unknown. Extracellular Vesicles (EVs) offer a protective environment for nucleic acids, with pro and antitumourigenic functions by controlling the immune response. In contrast to extracellular nonvesicular RNA, few studies have addressed the full RNA content within human fluids’ EVs and have compared them with their tissue of origin. Here, we performed Total RNA‐Sequencing on six Formalin‐Fixed‐Paraffin‐Embedded (FFPE) prostate cancer (PCa) tumour tissues and their paired urinary (u)EVs to provide the first whole transcriptome comparison from the same patients. UEVs contain simplified transcriptome with intron‐free cytoplasmic transcripts and enriched lnc/circular (circ)RNAs, strikingly common to an independent 20 patients’ urinary cohort. Our full cellular and EVs transcriptome comparison within three PCa cell lines identified a set of overlapping 14 uEV‐circRNAs characterized as essential for prostate cell proliferation in vitro and 28 uEV‐lncRNAs belonging to the cancer‐related lncRNA census (CLC2). In addition, we found 15 uEV‐lncRNAs, predicted to encode 768 high‐affinity neoantigens, and for which three of the encoded‐ORF produced detectable unmodified peptides by mass spectrometry. Our dual analysis of EVs‐lnc/circRNAs both in urines’ and in vitro’s EVs provides a fundamental resource for future uEV‐lnc/circRNAs phenotypic characterization involved in PCa.
Keywords: CircRNA, EV, lncRNA, neoantigen, TSA
1. INTRODUCTION
Extracellular Vesicles (EVs) are secreted membrane‐enclosed vesicles ranging from 50 to 1000 nm (Mathieu et al., 2019; Tkach & Théry, 2016). They are composed of a variety of proteins, lipids, metabolites and nucleic acids, and their cargo is controlled by specific molecular sorting machineries (Van Niel et al., 2018). Recent efforts have been conducted toward a better and more rigorous classification of the multiple forms of EVs differing in size and composition (Théry et al., 2018), which led to identification of other particles containing proteins, lipids and nucleic acids, often coisolated with EVs. These particles have been called exomeres, extracellular nanoparticles (ENPs) (Hoshino et al., 2020) or distinct nanoparticles (Jeppesen et al., 2019). EVs and probably ENPs protect biomolecules from potential degradation by nucleases, proteases or other environmental stress present in fluids. Since EVs work as a safe way to transport biological information through the whole body, they are now recognized as an important source of biomarkers in clinics but also as a mechanism of cell‐cell communication within tumour cells and their distant environment (Becker et al., 2016; Tkach & Théry, 2016). Multiple studies in different cancer types have demonstrated that EVs play an important role in cell proliferation, migration, invasion, angiogenesis, epithelial‐to‐mesenchymal transition (EMT), metastasis and immune response (Kalluri & Lebleu, 2020; Pelissier Vatter et al., 2021; Spinelli et al., 2021; Tkach & Théry, 2016). EVs also act as a source of antigenic peptides for the activation of T and B cells (Chaput et al., 2004; Wolfers et al., 2001). Therefore, the role of EVs as potential therapeutic agents is actively considered in addition to their use as diagnostics and prognostic options (Lebleu & Kalluri, 2020; Reiner et al., 2017; Wiklander et al., 2019) .
RNA studies in biological fluids were mostly focused on small ncRNAs (Buschmann et al., 2018; Murillo et al., 2019; Tosar et al., 2020) or captured mRNA/circRNA exomes (Hulstaert et al., 2020; Vo et al., 2019), whenever performed on nonvesicular extracellular (ex)RNAs or EVs/ENPs. Following the early work on urinary microvesicules revealing extensive forms of every types of long RNAs (Miranda et al., 2014), it is only recently that systematic exRNA whole transcriptome approaches started to emerge (Amorim et al., 2017; Barreiro et al., 2020; Everaert et al., 2019; Galvanin et al., 2019; Giraldez et al., 2019; Li et al., 2019). Whole transcriptome analyses assess the numerous types of RNA molecules including protein‐coding RNAs (mRNA), lncRNAs, fusion transcripts, splice variants and RNA modifications. LncRNAs constitute the most prevalent family in human transcriptome and can exert a plethora of functions, such as epigenetic regulation, chromatin remodelling, regulation of proteins’ activity and stability, mRNA stability and translation. They make an intimate part of mechanisms controlling cell/tissue homeostasis as well as various diseases including cancers (Rinn & Chang, 2012) now nicely reported in the CLC2 lncRNA database (Vancura et al., 2021). Although lncRNAs were initially defined as noncoding, several contain open reading frames (ORFs) that are translated into functional peptides and subjected to translation‐dependent decay (Andjus et al., 2021; Chen, 2020). Among lncRNAs, circRNAs are circular molecules with a covalently closed loop structure lacking 5′cap and 3′polyadenylated tail, formed through noncanonical splicing (Kristensen et al., 2019). Due to high stability, circRNAs can be easily found in circulating fluids as exRNAs, especially in cancer and proposed as potent biomarkers (Everaert et al., 2019; Hulstaert et al., 2020; Li et al., 2019). CircRNAs were shown to function as miRNA sponges to prevent mRNA translation or as part of ribonucleoprotein complexes regulating splicing or transcription (Chen, 2020; Hansen et al., 2013; Okholm et al., 2020). Despite their regulatory potential, expression of lncRNAs in circulating EVs/ENPs isolated from cancer patients remains poorly explored. Determination of their identity, origin, and features through comparative transcriptomic studies of liquid and solid biopsies together with existing in vitro cell systems is a challenge for understanding lncRNAs and circRNAs functional significance.
Urine is an excellent source of biomarkers containing EVs secreted from different tissues/organs alongside the urogenital tract and is of high value for extracellular vesicler cargo identification now reaching a consensus on the best methodological practices (Erdbrügger et al., 2021). Prostate cancer (PCa) is the second leading cause of death from cancer in men. During medical digital rectal examination, prostate material, and particularly PCa cells and EVs, can be released from prostatic ducts into the urethra and end up in urine. The identification of two well‐known prostate‐associated RNAs, PCA3, and TMPRSS2:ERG in urinary EVs (Nilsson et al., 2009) was the starting signal for intense efforts to define novel EVs‐RNA markers in urine for urological‐localized cancers (Erdbrügger et al., 2021). To date, only two extensive transcriptome profiling have targeted PCa patient's vesicular or nonvesicular exRNAs in urine and they are excellent bases for future biomarker discovery on larger cohorts. In an elegant benchmarking study, Everaert et al. were pioneers in performing deep whole transcriptome on carefully isolated urinary vesicular exRNA for prostate cancer in one patient (Everaert et al., 2019) but lacked the statistics of a larger cohort. Vo et al. provided the first exome capture in three PCa patients’ urinary samples but limit their study on nonvesicular circRNA (Vo et al., 2019) for biomarkers discovery. Both studies lack key information on the originating tissue RNAs, fundamental for future functional studies. Thus, there is now a need to extend the urine cohort of PCa patients to robustly assess the enrichment of vesicular long exRNA content in comparison with their paired originating tumours, and more importantly to identify putative regulatory lncRNA enrichment within the EVs/ENPs.
Here, we performed the first whole transcriptome analysis of prostate tumour tissues and paired urine‐derived EV‐enriched ultracentrifugation pellets, called thereafter uEVs, to provide an exhaustive comparison of RNA components between uEVs and their tissues of origin from the same patients. We showed that most of the RNAs enriched in the uEVs are intron‐free cytoplasmic RNA but also contain previously identified functional circRNAs and cancer‐related lncRNAs. In addition, we predict lncRNAs encoding potential neoantigens for which some can have ribosome footprint or can produce detectable peptides in immortalized cell lines by mass spectrometry. Finally, to orientate subsequent functional studies, we generated and explored the transcriptomes of three broadly used PCa cell lines and their corresponding EVs. In conclusion, in this study, most of the urine and EVs collection methodologies (urine collection, storage, RNA extraction and quality controls, see methods) were in line with the gold standard very recently described by the urine EVs task force (Erdbrügger et al., 2021). Thus, our study provided an extensive resource for methodologies, tools, and datasets to determine the lncRNA content of uEVs paving a way for future studies of their role in prostate cancer.
2. RESULTS
2.1. CircRNAs are enriched in uEVs
We performed total RNA extraction from matched prostate tumour FFPE tissue and uEVs from six PCa patients, RNA‐sequencing and differential expression analysis. Briefly, after prostatic massage on the patients, urine was collected, depleted from living cells and cell debris with a series of low‐speed centrifugations, followed by ultracentrifugation for 2.5 h, to pellet mostly microvesicles and exosomes with limited presence of ENPs, normally recovered only after an overnight ultracentrifugation (Zhang et al., 2019). Although these preparations can still contain some nonvesicular components (Erdbrügger et al., 2021), we choose to call these pellets uEVs, to simplify reading for the rest of the article (Figure 1a). The size distribution and aspect of uEVs were analysed by, respectively, Nanoparticle tracking analysis and Transmission Electron Microscopy, showing the presence of EVs of sizes ranging from 50 to 300 in diameter (Extended data Figure 1a, b and supplemental text). The presence of transmembrane tetraspanins CD63, CD9 and cytosolic protein Syntenin‐1 and absence of the cellular endoplasmic reticulum protein Calreticulin confirmed the absence of cellular debris in the preparation (Extended Data Figure 1c) (Théry et al., 2018). Then, RNA from FFPE biopsies and from isolated uEVs were extracted, quantified, and used for total RNA library preparation using an ultralow input protocol for RNA‐sequencing (see supplemental text and Extended Data Figure 1e, f, g). The uniquely mapped reads (at least five counts for circRNAs and 20 counts for other RNAs, see methods), corresponding to Gencode v32 annotation of snoRNA, snRNA, Repeat, Pseudogene, mRNA, lncRNA and circRNA, were quantified and compared between uEVs and tissue samples (Extended Data Figure 2a). Quantification of each annotated genomic feature revealed similar RNA content in both sample types but striking differences in distribution for some RNA families (Extended Data Figure 2b). Remarkably, mRNAs and pseudogenes numbers were comparable in both sets, respectively, with 15,490 in FFPE tissues and 13,614 in uEVs, and 1225 in FFPE tissues and 823 in uEVs, showing that EVs transcriptomes are as much as complex as tumour transcriptomes (Extended Data Figure 2b, Extended Data Table 2). Furthermore, a large majority of reads came from mRNAs confirming previous results (Everaert et al., 2019). The specificity of the prostate‐enriched RNAs was further explored by comparing the uEVs transcriptome with bladder and kidney tissue transcriptomes (see supplemental text and Extended Data Figure 3a, b and c) showing statistically significant enrichment of prostatic RNAs (SuperExactTest, p < 10–320), supporting urine as an excellent source of prostate EVs after prostatic massage. Among the ncRNA, premiRNA, repeats and snRNA genes were under‐represented in uEVs’ transcriptome, whereas circRNAs were over‐represented in uEVs with a number of 8796 versus 1240 in tumours. LncRNAs and pseudogenes showed a slight reduction in uEVs with 3776 genes in tumour and 1907 in uEVs for lncRNAs, and 1225 in tumours and 823 in uEVs for pseudogenes. Among circRNAs, 66% of uEVs circRNAs and 52% of tumour circRNAs were found in circBase, with the rest representing novel circRNAs (Extended Data Figure 2b). To further confirm this observation, we compared the number and the level of expression of circRNAs and lncRNAs in pooled tumour tissues versus pooled uEVs (Figure 1b). The global correlation between tumour and uEVs indicate a larger disproportion of circRNA in uEV (R2 = 0.35) in comparison with lncRNAs mostly enriched in tumour (R2 = 0.16). Similar correlations were found in each patient comparing tumours RNA expression and uEVs (Extended Data Figure 4a) supporting a general tendency for enrichment of circRNAs in uEVs observed in previous analyses (Everaert et al., 2019).
FIGURE 1.
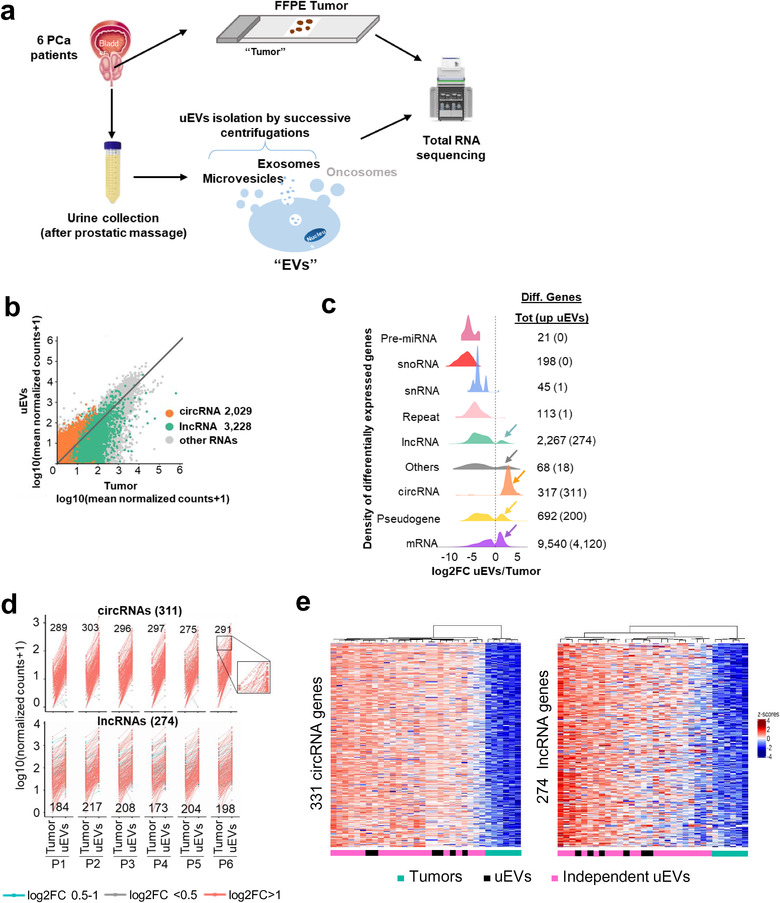
uEVs are enriched in circRNAs and some lncRNAs. (a). Full transcriptome of paired liquid and solid biopsies of prostate cancer patients. Experimental procedure from prostate tumour biopsies and urines collections to RNA sequencing, through FFPE biopsies and uEVs isolation. (b). Mean gene expression of paired Tumour against uEVs (n = 83,980 RNAs if at least one counts, n = 21,397 if at least five counts for cirRNAs and 20 counts for lncRNAs and other RNAs). DEseq2 normalized counts, for each type of RNA are plotted; circRNAs (orange), lncRNAs (green), all other types of RNAs (grey). Each dot represents all transcripts for each gene. R2 = 0.35 and R2 = 0.16 for 2029 circRNAs and 3228 lncRNAs, respectively (> = 5 counts for cirRNAs, > = 20 counts for lncRNAs and other RNAs). (c). Density plot showing the distribution of log2 fold change uEVs/Tumour ratio per gene types. The right side of dotted line correspond to enriched genes in uEVs compared to Tumour tissues. The left side of dotted line correspond to the enriched genes in tumours compared to uEVs. Each colour represents an RNA type. Number of total differentially enriched genes in Tumour and uEVs are indicated on the right with the number of enriched RNA in EVs in bracket. (d). Interaction between the normalized number of counts of the 311 upregulated circRNAs (top) and 274 upregulated lncRNAs (bottom) in paired Tumour and uEVs for each patient (P1 to P6). Each dot corresponds to a circRNA or lncRNA upregulated in uEVs compared to Tumour with Log2 FC = 0,5–1 (blue), Log2 FC < 0,5 (grey), Log2 FC > 1 (red). The number of counts for the same RNA, in Tumour and in uEV are linked together with a line as shown for four circRNAs in the zoom window of patient 6. For each patient are indicated the numbers of circRNA and lncRNA with a log2(FC) > 1. (e). Heatmap display unsupervised hierarchical clustering with euclidean distance (CED) of 311 circRNAs (left) and 274 lncRNAs (right) up regulated in uEVs (FC > = 1.5, at least 20 counts) in each of the individual samples from six Tumour biopsies (green), paired six uEVs (black) and 20 independent uEVs (pink) from prostate cancer patients. Colour scales represent z‐scores of log10(TPM+1)
We next identified the most differentially overrepresented ncRNAs in uEVs and tumours. Differential expression analyses on the whole transcriptome (Extended Data Figure 4b) and hierarchical clustering by Euclidean distance between pooled uEVs and tumour tissues datasets (Extended Data Figure 4c) revealed 4925 enriched RNAs in EVs and 8336 enriched in tumours (Extended Data Table 4). From these differential analyses, very few or none premiRNAs, snoRNAs, snRNAs and repeats were defined as significantly overrepresented in urines. In contrast, a subgroup of circRNAs (N = 311), mRNAs (N = 4120), pseudogenes (N = 200) and lncRNAs (N = 274) were defined as significantly enriched in uEVs compared to tumours (Figure 1c; Extended Data Figure 4b). To explore further to which extent each patient’ urine contained specific noncoding RNAs, we analysed the number of normalized counts of lncRNAs and circRNAs by plotting each value in uEVs and tumours and calculated for each linked pair the enrichment value (Figure 1d). Our result showed that most of the differential circRNAs/lncRNAs were reproducibly differential in all six patients (with a minimum of 88% common circRNAs in patient five and 63% lncRNAs in patient 4, Figure 1d), pointing to a general rule for the enrichment of the two families of ncRNAs in each specimen. To validate this observation on a larger cohort, we extracted uEV‐RNAs independently from 20 additional PCa patients and performed total RNA‐seq experiments. Our data show that the enriched circRNAs and lncRNAs, previously identified, remarkably present similar levels of expression in these 20 novel uEVs (Figure 1e). Principle Component Analysis provided a striking cluster of all 26 uEVs’ samples together, far from the tumour RNAs expression cluster (Extended Data Figure 2c).
We conclude that uEVs cargo all sorts of RNAs in urine with robustly enriched subgroups when compared to their respective tumours. Among the noncoding RNAs, two subfamilies of lncRNAs and circRNAs present the most important differential enrichment in vesicular urines, robustly validated in an independent cohort.
2.2. Mature lncRNAs and mRNAs are enriched in uEVs
Next, to further characterize the uEVs RNA content, we compared the genic structures of transcripts in three types of samples, frozen, FFPE and uEVs, based on read counts across six gencode v32 features: exon, intron, 5′‐UTR, 3′‐UTR, promoter, and intergenic region in tumours and uEVs specimens (Figure 2a, Extended Data Table 5). We found that intergenic and promoter mapped reads were extremely low in uEVs datasets (less than 1% in uEVs vs. 4% in FFPE tumours). This is in striking contrast with 5′, 3′ untranslated regions and exonic regions all enriched in uEVs (3.2% and 8.3% for 3′UTR, 14.2% and 27% for 5′UTR, and 32% to 62% in FFPE tumours and EVs, respectively). Although the three fractions were sequenced using the same total RNA‐seq protocol, intronic counts were very low in uEVs (2.6%) in comparison to FFPE tumours (46%) or frozen tumour (27%). Interestingly, the latter comparison confirms previous report on glioblastoma samples showing the enrichment of intronic tags in FFPE tissues in comparison with frozen tissues, which could be explained by a stabilization of nuclear RNAs (thus intronic ones) being protected by the FFPE treatment (Esteve‐Codina et al., 2017). Nevertheless, despite the prostate FFPE samples showed a higher proportion of intronic tags (0,7 Exon/intron (E/I) ratio) when compared to the 1,78 E/I ratio of frozen prostate tumour samples (Figure 2a), the uEVs E/I ratio (35,7) dramatically increased by 50‐fold or 20‐fold when compared to FFPE's or frozen tumours’ E/I, strongly supporting that uEVs are indeed depleted for intronic RNAs. This observation has been further validated using cellular RNAs and corresponding EVs from three prostate cell lines (Extended Data Figure 5c), where we measured an average E/I ratio of 14.3 in cEVs, while the cells present a lower ratio of 5,2. We concluded from this experiment that uEVs and cEVs were robustly devoid of intronic RNAs, when compared to their originating tissues/cells.
FIGURE 2.
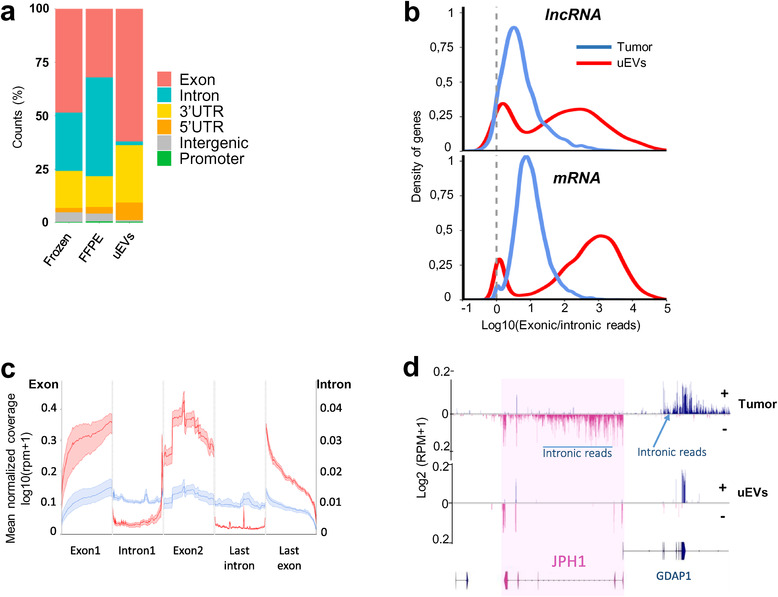
Intronic RNAs are depleted in uEVs compared to Tumour. (a). Genomic read counts distribution by percentage across exon, intron, 3′UTR, 5′UTR, intergenic and promoter in frozen, FFPE and uEVs biopsies. (b). Distribution of log10(exonic read counts/intronic read counts) normalized by length from Tumour (blue) and uEVs (red) samples for lncRNA (top, 2151 in tumours, 1393 in uEVs) and mRNA (bottom, 13,084 in tumours, 11,483 in uEVs) annotations. (c). Metagene of mean coverage for two first exons, last exon, first intron and last intron of 25,166 mRNAs and lncRNAs from Tumour (blue) and uEVs (red) samples. (d). GGbio‐generated RNA read profiling along minus (−; pink) and plus (+; bleu) strands of chr8:74198516–74398516 in Tumour and uEVs specimens. Arrow lines represent introns and rectangles represent exons of GENCODE‐annotated protein‐coding gene JPH1 (pink) and part of GDAP1. The maximum value of coverage, read count is shown in the left panel of read mapping. Some intronic reads are indicated for the two genes
To get a quantitative assessment of the uEVs’ intron loss per RNA biotypes, we then analysed the density distribution of read counts of exonic enrichment, normalized by the lengths of each feature in lncRNAs and mRNAs (Figure 2b). The density plot showed that uEVs have a significantly larger proportion of exonic‐enriched mRNAs (shift of the Exon‐rich peak from log10FC(1) to log10FC(> 3)). The lncRNA distribution was less homogenous than for mRNA, probably reflecting the less accurate annotation for lncRNA exon/intron features (Hezroni et al., 2015). Nevertheless, it also showed a shift in the E/I ratio in uEVs when compared to tumours. We conclude that the higher E/I ratios observed in uEVs corresponded to a group of lncRNAs and mRNA harbouring essentially mature spliced transcripts. Metagene profiling of reads across the two first exons, the first intron and the last exon and intron of all transcripts with a minimum of three exons (N = 25,166 genes), revealed higher coverage of exons in uEVs than in tumours and higher coverage of introns in tumours than in uEVs (Figure 2c). These results suggested that uEVs contain preferentially fully processed mRNAs and lncRNAs, as illustrated for the mRNA GDAP1 and JPH1 (Figure 2d), both harbouring intronic reads in tumours and none in uEVs. We hypothesized that uEVs carry mature RNAs, most probably originating from the cytoplasmic compartment, but less or none from the nucleus.
2.3. Nuclear lncRNAs are under‐represented in uEVs
To directly prove that uEVs are devoid of nuclear transcripts, we analysed public strand‐specific RNA‐seq data from cytoplasmic and nuclear fractions of 22Rv1 PCa cells (Kaul et al., 2020). Nuclear and cytoplasmic transcripts were defined by differential expression analysis of nuclear versus cytoplasmic datasets (see method). In total, 4518 transcripts were considered as highly enriched in nucleus and 6419 transcripts as enriched in cytoplasm. It is important to note that the public dataset was produced from poly(A) RNA‐seq, thus missing circRNAs and nonpolyadenylated lncRNAs. Nevertheless, the list of 22Rv1 cytoplasmic and nuclear‐enriched RNAs was intersected with those enriched in tumours (n = 8336) and uEVs (n = 4925) (Figure 3a, Extended Data Table 6). The assumed cytoplasmic RNAs represented 58% of uEVs‐enriched RNAs, whereas the nuclear transcripts were mostly depleted (representing only 3%) from uEVs (Figure 3b). In contrast, tumours mainly contained transcripts from both compartments (57%) and nuclear (24%) whereas cytoplasmic RNA fraction was of only 15% (Figure 3b). To distinguish what type of transcripts were specifically depleted or enriched in the cytoplasmic and nuclear compartments in uEVs and tumours, we inferred the subcellular localization of mRNAs, pseudogenes and lncRNAs from 22Rv1 datasets. The density distribution of the ratio cytoplasmic/nuclear reads showed that tumour libraries preferentially contained nuclear lncRNAs and pseudogene transcripts. As expected, the mRNAs were both nuclear and cytoplasmic. On the contrary, in uEVs the majority of enriched lncRNAs, pseudogenes and mRNA were all detected within the cytoplasmic fraction (Figure 3c). Like the exon/intron analysis, we observed similar enrichment of cytoplasmic RNAs within the cEVs when compared to their originating in vitro cells, albeit in a less dramatic proportions (Extended Data Figure 6a, b).
FIGURE 3.
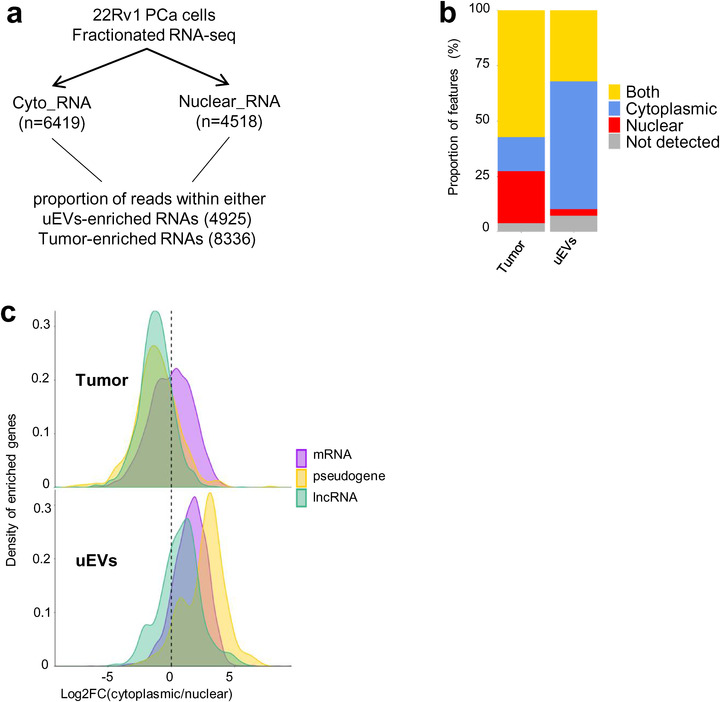
Depleted lncRNAs in uEVs are nuclear. (a). Experimental procedure, starting from 22Rv1 cell line fractionation polyA RNA‐seq, to propose cytoplasmic or nuclear localization of up‐regulated genes in uEVs and up regulated genes in Tumour biopsies. (b). Stacked barplot distribution, by percentage, of cytoplasmic (blue), nuclear (red), both (yellow) or nonpolyA RNAs (grey) of up‐regulated genes in Tumour (8336) and up regulated genes in uEVs (4925). 15.4% and 57.8% upregulated RNAs, respectively, in tumours and uEVs are cytoplasmic; 23.6% and 2.9% are nuclear; 57.2% and 32% are both. (c). Density distribution of log2 (fold change cytoplasmic/nuclear ratio) per RNA types (7732 RNAs from Tumour and 4556 RNAs from uEVs), mRNA (purple), pseudogene (yellow), lncRNA (green) in Tumour (top) and uEVs (bottom). The left side of dotted line in both graphs corresponds to the nuclear RNAs, the right side corresponds to cytoplasmic RNAs
FIGURE 6.
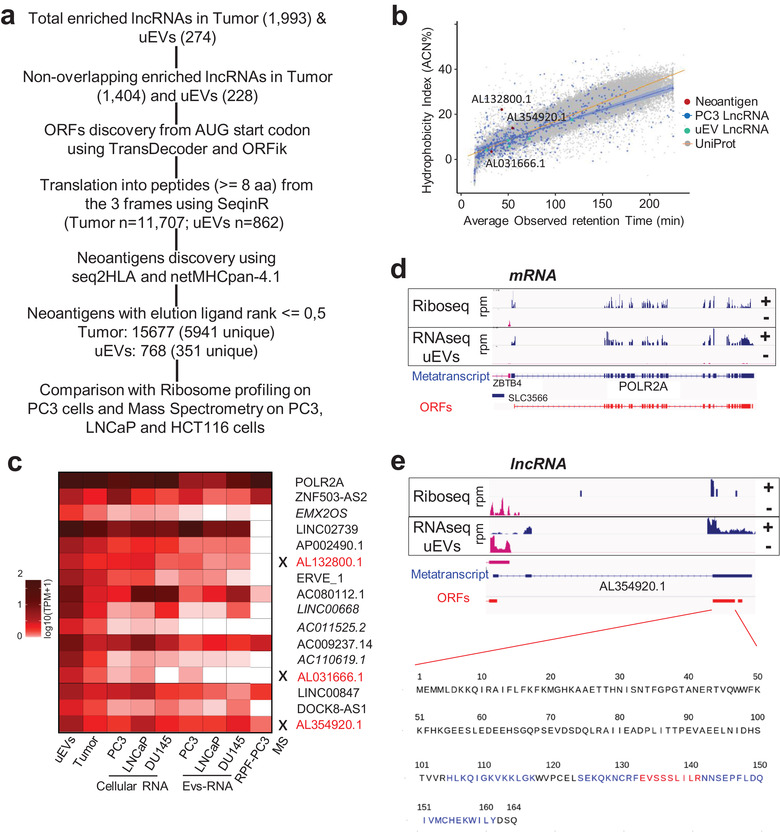
uEVs‐enriched RNAs show lncRNA‐encoding neoantigens shared by prostate cell lines. (a). Workflow of ORFs, peptides and neoantigens prediction from lncRNAs enriched in Tumour (n = 1993) and uEVs (n = 274). (b). Correlation between hydrophobicity and observed retention time for Neoantigen peptides (red dots), PC3 lncRNA peptides (blue dots and regression line with 95% confidence interval, R2 = 0.482), peptides from lncRNAs upregulated in uEVs (cyan) and peptides derived from Uniprot‐annotated human proteins (grey dots and orange regression line, R2 = 0.889) from three cell lines (PC3, LNCAP and HCT116 in N = 5 biological replicate). (c). Heatmap representing the relative expression by log10(TPM+1), of POLR2A mRNA and 15 uEVs‐neoLncRNAs, encoding the 351 strong unique neoantigens within uEVs, Tumour, PC3‐, LNCaP‐ and DU145‐cells and their respective EVs and RPF from PC3 ribosome profiling dataset. Highlighting of peptides by mass spectrometry is indicated by a cross and writing in red. (d). Example of EV‐neoLncRNA. IGV‐generated public PC3 prostate cancer cell line ribosome occupancy (Hsieh et al., 2012) and uEVs RNA‐seq profiling along plus (+, blue) and minus (−, pink) strands of POLR2A mRNA and (e). of AL354920.1 EV‐neoLncRNA. Blue arrow‐lines and rectangles represent introns and exons of metatranscripts, respectively. RPM, reads per million mapped reads. Open reading frames (ORFs, red rectangles), starting from AUG codon, of the most abundant transcripts for POLR2A and AL354920.1, are indicated. The sequence of one AL354920.1 ORF from the frame 1, generated with GGplot2, is presented and the regions of the 20 translated neopeptides are in blue and red. Red sequence is the strongest 9‐mer neoantigen
In conclusion, transcriptome analyses of subcellular fractions confirmed that uEVs are enriched in cytoplasmic RNAs, in agreement with the presence of mostly mature intron‐less RNAs.
2.4. Most uEVs‐circRNAs are present in EVs released by prostate cell lines
EVs‐RNAs have a key role in intercellular communication. While a lot of attention has been focused on miRNAs in the past, little is known on the potential functionality of lncRNAs and circRNAs in this intercellular signalling, raising the need to properly characterize cellular models of EVs cargo. We performed total RNA extraction and sequencing of PC3, DU145, and LNCaP human PCa cell lines and resulting EVs collected from the cell culture media (cEVs,, characterized by NTA, electron microscopy and Western blotting, Extended Data Figure 1a, b, d). We focused our attention on circRNA species as the most abundant type of uEVs‐lncRNAs (Extended Data Table 7). Strikingly, the intersection of circRNAs present in patients uEVs, cell lines and cEVs (Figure 4a and Extended Data Table 7) showed 60% (187) cellular and 95% (296) cEVs‐circRNAs in common with the 311 uEVs‐enriched circRNAs, making these cell lines excellent models for functional studies. Among these uEV‐circRNAs, the androgen‐responsive circ‐SMARCA5 has been reported to present an oncogenic activity (Kong et al., 2017), and is also detected within the three cell lines and in their cEVs. This finding supported the attractive hypothesis that circ‐SMARCA5, shown to control cellular proliferation, could act both within the tumour context and the vesicular‐mediated signalling. Moreover, we also identified several circRNAs already named “essential‐circRNA” required for high proliferation rate in PCa cell lines (Chen et al., 2019). Out of these 171 essential circRNAs, 40 (23%) and 130 (76%) were detected in our own PCa cell lines and cEVs, respectively (Figure 4a and Extended Data Table 7). Among those, 14 were enriched in uEVs as well as in cEVs (Figure 4b and Extended Data Table 7) and 12 of these 14 circRNAs were also expressed in cell models as illustrated for circ‐GOLPH3 (Figure 4c). We conclude that PC3, DU145 and LNCaP PCa cell lines and associated cEVs are excellent cellular models for functional studies of uEV‐enriched circRNAs.
FIGURE 4.
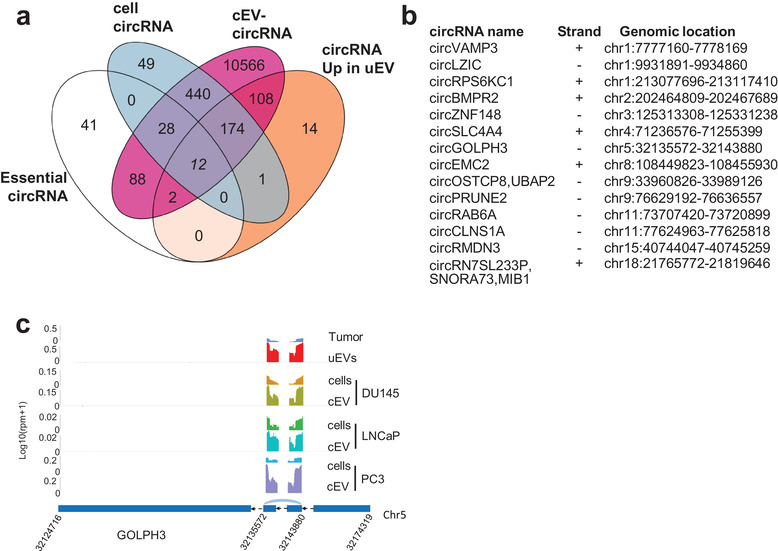
uEVs‐enriched circRNAs contain essential circRNAs and are common to PCa cell lines EVs. (a). Venn diagram showing number of over‐represented circRNAs in prostate cancer uEVs (n = 311), 171 essential circRNAs defined by Chen et al., circRNAs expressed (at least five DEseq normalized counts) in PC3, LNCaP and DU145 PCa cell lines (cell circRNAs, n = 704) and in cell‐EVs (cEVs circRNAs, n = 11,418). (b). List of the 14 essential circRNAs up‐regulated in uEVs c. Sequencing read coverage from back splicing of GOLPH3 circRNA, from chromosome 5:32124716–32174319, is shown using GGplot2 in Tumour, uEVs, PC3, LNCaP and DU145 cEVs and in cells. The maximum value of coverage read count is shown in the left panel of read mapping. Parental transcript ENST000000265070.7 is schematized by blue rectangles representing exons and black arrow lines representing introns (shrunk to 100 nt). Junction of back splicing is indicated in light blue.
2.5. Cancer lncRNAs census are found both in uEVs‐enriched lncRNAs and in PCa cell lines EVs
In addition to circRNAs, lncRNAs have been shown to be associated to cancer promoting or suppressing the tumour progression (Huarte & Rinn, 2011). Within the last decade enormous efforts have been made not only to list cancer‐related lncRNAs but to provide their mechanistic functions. Several databases have been released reporting their misregulation but also their functional significance (Mitranscriptome (Iyer et al., 2015) or RNA atlas (Lorenzi et al., 2021) among others). Recently, a nice curated list of lncRNA‐mediating cancer progression has been publicly released as the cancer lncRNA census (CLC2) (Vancura et al., 2021). To define which of the uEV‐enriched lncRNAs could play a role into the hosting cell, we intersected the list of uEVs‐enriched, PCa cellular and cEVs lncRNAs with the 492 CLC2 lncRNAs list (Figure 5a, Extended Data Table 8). Remarkably, 28 CLC2 lncRNAs were found enriched in uEVs (Figure 5b). Among those, PCAT6 is well enriched in uEVS and is present also in PCa cell lines and their EVs (Figure 5c). This lncRNA has been firstly described in mitranscriptome as a Prostate CAncer Transcript (PCAT), and more recently as controlling bladder, and colorectal cancer cell proliferation by sponging miRNA (Ghafouri‐Fard et al., 2021). Our results suggest that in addition to functional circRNAs, uEVs can also transfer functional lncRNAs that could potentially reprogram gene expression into the hosting cell.
FIGURE 5.
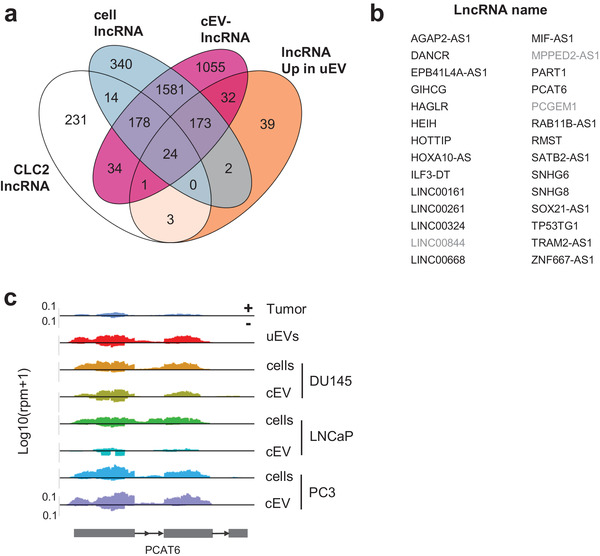
uEVs‐enriched lncRNAs contain cancer lncRNAs and are common to PCa cell lines EVs. (a). Venn diagram showing number of over‐represented lncRNAs in prostate cancer uEVs (n = 274), 485 functional cancer lncRNAs defined in CLC2(Vancura et al., 2021), lncRNAs expressed (at least 20 DEseq normalized counts) in PC3, LNCaP and DU145 PCa cell lines (cell lncRNAs, n = 2312) and in cell‐EVs (cEVs lncRNAs, n = 3078). (b). List of the 28 CLC2 upregulated uEVs lncRNAs including 25 common with cEVs (black) and three exclusive to uEVs (grey). (c). Sequencing read coverage of PCAT6 lncRNA (strand +) is shown using GGplot2 in Tumour, uEVs, DU145‐, LNCaP‐ and PC3‐cEVs and in cells. The maximum value of coverage read count is shown in the left panel of read mapping. Metagene transcript is schematized by grey rectangles representing exons and arrow line representing introns
2.6. uEVS and cEVs are a source of lncRNAs that can be translated into neoantigens
Although lncRNAs are originally defined as unable to code for proteins, some were reported to contain ORF that encodes functional peptides (Chen et al., 2020). Here, we investigated the coding potential of uEV‐enriched lncRNAs, with a specific focus on neoantigenic properties, in comparison with those enriched in tumour tissues. First, to avoid any ambiguity with existing ORFs or proteins, all lncRNA transcripts overlapping annotated protein‐coding genes or pseudogenes from Gencode V32 were filtered out from the list, to obtain 1404 and 228 lncRNAs, respectively, in tumour and uEVs (Figure 6a). We then defined all potential coding sequences starting from AUG codons and finishing with a STOP codon, in the three reading frames (including those that are shorter than 100 codons). For the genes transcribed into several isoforms, the most prevalent was considered for further analysis (see method). We retrieved all peptide sequences with at least eight aminoacids, resulting in 11,707 tumours and 862 uEVs lncRNA‐encoded peptides. Neoantigens were then predicted with NetMHCpan‐4, using MHC typing computed by seq2HLA from the RNA‐seq file of each patient (Jurtz et al., 2017). To best separate ligands from nonligand peptides, only neopeptides with an elution ligand index lower than 0.5 were kept, corresponding to 15,677 (5941 unique) for tumours and 768 (351 unique) for uEVs, derived from 244 and 15 transcripts, respectively (see method, Extended Data Table 9, Figure 6a and Extended Data Figure 7a). From the top 2% predictions, we estimated that the majority of potential lncRNA‐derived neoantigens were 9‐mer peptides (Extended Data Figure 7b), the gold standard size for neoantigen recognition (Dash et al., 2017). This result showed that both EVs‐lncRNAs and tumour‐lncRNAs have similar capacities (and proportion) to encode peptides with high affinity binding to MHC. In conclusion, among the uEVs‐enriched lncRNAs, we identified 15 transcripts (called EVs‐neoLncRNA hereafter) that could potentially serve as templates for the translation of 9‐mer peptides with potent neoantigens properties, if internalized and translated into the host cells.
To determine whether the uEV‐LncRNAs could indeed encode for detectable and stable peptides, we performed proteomic mass spectrometry on PC3, LNCaP and HCT116 cells and compared MS‐detected peptides with the predicted encoded peptides from the ORF of PC3‐LncRNAs, of the upregulated uEVs‐lncRNAs and the UniProt database (see method). The observed retention times for these MS‐detected peptides were correlated with the calculated hydrophobicity of each unmodified MS‐detected peptide (Figure 6b). The results indicated a correlation for 616 PC3 cellular‐lncRNA peptides and eight uEV‐LncRNA peptides (Figure 6b). Strikingly, three MS‐peptides found in both aforementioned groups were derived from ORFs of EVs‐neoLncRNAs for which peptides were predicted to be high affinity MHC classI neoantigens, suggesting that these peptides might be stable enough to be detected (Figure 6b). To confirm that uEV‐neolncRNAs showed active translation features, we first quantified their expression in PC3, DU145 and LNCaP PCa cell lines and corresponding cEVs. As shown in the heatmap, all the 15 EVs‐neolncRNAs are expressed, at least in one of the three cell lines, at lower but comparable levels as the POLR2A mRNA used here as a moderate expressed control (Figure 6c). We then reanalysed a public ribosome profiling dataset for the PC3 cell line, to determine ribosome occupancy on the 15 EVs‐neolncRNAs (Hsieh et al., 2012). We observed that 5, out of 15 neolncRNAs expressed in PC3, were associated with actively translated ribosomes (Figure 6c). Finally, ORFs encoding one of the three MS‐detected neopeptides showed ribosome footprints as a mark of an active translation in PC3 cell line (Figure 6c), indicating that this EV‐lncRNA was translated, produced stable peptides, predicted to contain excellent class1 antigen properties. As illustrated, the POLR2A mRNA showed similar levels (Figure 6d) of Ribosome Protected Fragments (RPFs) as the predicted AL354920.1 EV‐neoLncRNA encoding 20 predicted high affinity neoantigens(Figure 6e), for which a tryptic sequence was detected by Mass spectrometry.
In conclusion, our results showed that uEVs can cargo lncRNAs containing ORFs with a high potential to be translated and produced peptides with excellent predicted features for MHC classI presentation as neoantigen.
3. DISCUSSION
To our knowledge at this date, here we report the first systematic comparison of whole RNA sequencing libraries from paired urinary EV‐enriched pellets and prostate tumour FFPE samples. Our results show that uEVs are enriched in intron‐free, processed cytoplasmic RNAs. Remarkably, this extracellular material is enriched in some lncRNAs and circRNAs. Our whole transcriptome analyses confirmed the presence of these lncRNAs in EV pellets obtained from PCa cell lines and hence the utility of in vitro systems for functional molecular and cellular studies. Recent works suggest that lncRNAs are a largely ignored source of neoantigens (Laumont et al., 2018) and our results point out several of those as circulating templates for neoantigen production if internalized in a recipient cell. Historically, a challenge with cell‐based phenotypic screens is the difficulty in gaining molecular insight that are shared with real tumour expressing tissues. Our results in the present study, however, demonstrate in six paired tumours and urinary EVs, the robustness of such candidates that can be found in in vitro models. The data generated herein should thus be an important resource to further investigate loss or gain of function of the prostate circulating RNAs using cellular models and develop genetic tools to address EVs functional signalling pathways. The power of genome‐scale CRISPR/Cas‐9 or Cas‐13 loss‐of‐function and gain of‐function screens will provide mechanistic clues for such circulating RNAs.
It is conceivable that some of these lncRNAs and circRNAs can be brought in clinics as novel therapeutic strategies to limit communication with the immune response system or the oncogenic transformation of adjacent cells. EVs secreted by PCa cells can alter the transcription of infiltrating T cells (Chen et al., 2021) and their RNA content might be part of their signalling activity controlling key elements in the recipient hosting cells driving their transcription.
We identified here a subset of circRNAs that were previously defined and named “essential”‐circRNA to increase proliferation of prostate cancer cells (Chen et al., 2019). One such oncogenic mechanism could be transferred from tumour to adjacent cells and tissues to initiate perturbation. Such perturbations could provoke uncontrolled expanding proliferation in proximal cells, thus overwhelming antitumoural regulators while being beneficial to the initial tumour in a competitive environment. Since we found that those circRNAs are enriched in circulating EVs, and present in in vitro EVs, it is tempting to further address their role in intercellular communication using epithelial recipient cells in contact with such EVs containing or not these essential circRNAs. In contrast, we note that circ‐ELK4, the most‐enriched circRNA in uEVs and present in cEVs, has been previously shown to be the most abundant PCa tumour circRNA, whose expression is negatively correlated with proliferation markers for prostate cancer (Cuzick et al., 2011; Vo et al., 2019), similarly to other circRNAs found to be anticorrelated with proliferative genes in ovarian normal and cancer cell lines (Bachmayr‐Heyda et al., 2015). Furthermore, in uEVs and cEVs, we also found previously characterized “highly stable” circ‐ARID1A, circ‐FAM13B, circ‐MAN1A2 and circ‐RHOBTB3, detected by rtqPCR independently in nonvesicular urinary samples of PCa patients (Vo et al., 2019) reinforcing the robustness of our approach and the highly significant enrichment of those circRNAs. These observations raised the exciting hypothesis that such amount of circulating vesicular circ‐RNAs such as circ‐ELK4 for instance, could impact proximal normal tissues by contributing to cell cycle arrest and perturbing again the competitive environment of the tumour. An alternative scenario would follow previous hypothesis where acquisition of metastatic traits by tumours could be achieved by eliminating a negative regulator such as miR23 through exosomes (Ostenfeld et al., 2014). Hence, similarly, the disposal of the cell cycle arrest factor circ‐ELK4 in the uEVs, or others, would promote proliferation of the PCa EV‐secreting cells.
Concerning lncRNAs, we similarly identified several of them which might have a functional significance by sponging and titrating cytoplasmic miRNA for instance if internalized in the recipient cell. However, in addition to reprogram gene expression, we propose here that the traveling lncRNAs characterized in this study, might be also susceptible for translation. This hypothesis is even more attractive since these lncRNAs were originally mostly cytoplasmic and mature. We provide a first list of lncRNA candidates that can be translated, predicted to form high affinity neoantigens and even produced stable and detectable peptides. EV‐RNAs can in principle serve as a source of novel proteins in recipient cells, since mRNAs transported by EVs can be actively translated into the recipient cells (Skog et al., 2008; Valadi et al., 2007) even only as short as 1 h after EV uptake during coculture cells (Lai et al., 2015). Thus, one may hypothesize these circulating neolncRNAs could also be translated in the host cells and act as a decoy to the immune system or intracellular signalling peptides. One of the attractive scenarios would predict that some tumour cells not only could silence their own HLA system using epigenetic regulation as recently shown (Griffin et al., 2021; Zhang et al., 2021), as some sort of invisibility blanket, but would transfer information and tumour specific neoantigens lncRNA templates to nontumour adjacent cells. With their fully operational antigen presenting machinery, these cells would then divert the antitumour immune response away from the true tumour. Further work will be required to formally demonstrate the existence of such peptides but also their immunogenicity performance.
In addition to their regulatory roles, circRNAs received considerable attention as potential liquid biopsy biomarkers (reviewed in (Li & Han, 2019; Bach et al., 2019)), being much more stable than their linear mRNAs isoforms (Jeck et al., 2013) (48h vs. 20h), and are released to the cytoplasm during mitosis, where they show extraordinary stability. Here we showed that circRNAs are an important fraction of urinary vesicular‐enriched lncRNAs, independently of their parental RNA enrichment. Indeed, by comparing the list of the 4614 enriched RNAs in urine (excluding circRNAs) with the 311 uEV‐enriched circRNAs, we observed that only 130 corresponded to parental linear RNAs leaving 61% enriched circRNAs without a corresponding enrichment of their parental RNA. This suggests that the mechanism of circRNA enrichment in uEVs could be independent of their parental linear RNA expression and instead may be due to the regulation of splicing mechanisms or to the circRNA lifetime itself. Further experiments will determine if specific circRNA processing is linked to their externalization.
Several causes could explain the increase of circRNA or lncRNA concentration in urine as for any RNA release in the stream: (1) cell death induced by stresses like hypoxia, (2) inflammation, (3) antitumour therapies, (4) tumour invasion and (5) metastasis process. In addition, several reports identified specific motifs within miRNA sequences that could drive their release/retention in EVs (Garcia‐Martin et al., 2022; Temoche‐Diaz et al., 2019; Villarroya‐Beltri et al., 2013), and it is tempting to extrapolate similar mechanisms in controlling circRNAs and lncRNAs into EVs. Altogether, these might be the reasons for the disproportion between circRNA and some lncRNA levels in urine versus tumours. EV heterogeneity and subfamily classification are currently the target of large research efforts (Théry et al., 2018). Our work did not address the specific contents of the different members of the EV family, nor did we formally demonstrate that the RNA analysed here were specifically incorporated inside EVs: we cannot exclude that some of the RNA sequences may be nonspecifically associated to EVs secondarily after EV secretion. Further work properly distinguishing EVs from nonvesicular circulating factors by size exclusion chromatography for instance (Royo et al., 2020), will address whether some of these extracellular entities contain distinct types of noncoding RNAs. Nevertheless, our results come as further support of considering these noncoding RNAs as surrogate circulating markers and regulatory elements for prostate cancer progression (Tucker et al., 2020).
4. MATERIAL AND METHODS
4.1. Sample collection and processing
Six urine and paired Formalin‐Fixed Paraffin‐Embedded (FFPE) tissue, 20 independent urine were collected from prostate cancer patients by Henri Mondor hospital, Créteil, France with written consent and approval by ethical Comité de Protection des Personnes (CPP) Ile‐de‐France V, N° ID‐RCB:2016‐A00789‐42. All patients were newly diagnosed and had not received treatment for prostate cancer before biospecimen collection.
Following digital rectal exam (DRE) performed by the attending oncologist, first‐catch urine samples were collected in 50 ml Falcon tubes without adding protease, nuclease inhibitors. Urines were placed at 4°C, centrifuged within 72 h of collection at 2000 g for 12 min followed by 3500 g for 17 min at 4°C to remove cell debris. A total of 30 ml of supernatant was used for EVs enrichment by ultracentrifugation at 160,000 g for 2.5 h at 4°C using a SW32‐Ti swing bucket rotor in an Optima L‐80 XP ultracentrifuge (Beckman Coulter, USA). The resultant pellets were resuspended in 100 μl cold PBS and then directly stored at ‐80°C for EVs characterization or vortexed 1 min with 700 μL of Qiazol (Qiagen) then keep 5 min at room temperature before storage at ‐80°C, for RNA extraction.
Absence of bacteria in urine supernatant was confirmed by inoculation on Lennox Broth media plates.
The prostate biopsies were fixed with 10% neutral buffered formalin solution (Sigma‐Aldrich, Merck) at room temperature for 8 h but no longer than 24 h. Tissues were dehydrated through ascending grades of alcohol and cleared in xylene and embedded into paraffin blocks. Biopsy specimens were stored at room temperature at Henri Mondor hospital.
PC3, DU145 and LNCaP human prostate cancer cell lines, were obtained from American Type Culture Collection (Manassas, VA, USA), and cultured, in triplicate for EVs analyses and in five replicates for mass spectrometry, in RPMI 1640 media supplemented with 10% Foetal Bovine Serum (FBS), GlutaMAX (Gibco‐Invitrogen Corporation, Carlsbad, CA, USA). Five replicates of human colon cancer cell line HCT‐116 were cultured with Dulbecco's Modified Eagle Medium (DMEM) with High Glucose, 4 mM of L‐Glutamine, 4500 mg/L Glucose and sodium pyruvate (HyClone) supplemented with 10% FBS (Eurobio) and 1% antibiotics (v/v) (penicillin and streptomycin (Gibco)). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. When prostate cells reached 70% confluence, they were washed twice in PBS and cultured for an additional 48h reaching a maximum of 80–90% confluence, in the same media composition without FBS, to eliminate vesicles coming from the serum. LNCaP growth media were supplemented in 2 nM dihydrotestosterone. Conditioned medium (3 × 30 ml from a total of 10 to 24 million of cells) was then processed for EV enrichment as described for urine samples. Briefly, medium was centrifuged at 2000 g for 12 min followed by 3500 g for 17 min at 4°C and an ultracentrifugation at 160,000 g for 2.5 h at 4°C using a SW32‐Ti swing bucket rotor in an Optima L‐80 XP ultracentrifuge (Beckman Coulter, USA). The resultant pellets were resuspended in 100 μl cold PBS and then directly stored at ‐80°C for EVs characterization or vortexed 1 min with 700 μL of Qiazol (Qiagen) then keep 5 min at room temperature before storage at ‐80°C, for RNA extraction.
4.2. Extracellular vesicles characterization
Characterization of particle number and size, of the EVs‐enriched pellets was performed using nanoparticle tracking analysis (NTA) using a ZetaView PMX‐120 video microscope (Particle Metrix) equipped with a 488 nm laser and the software Zeta View version 8.05.10. For optimal measurements, samples were diluted with PBS until particle concentration was within the optimal concentration range for particle analysis. Experiments were performed briefly as follows: the instrument was set at 25°C, sensitivity of 70 and shutter of 75. Measurements were done at 11 different positions (five cycles per position) and frame rate of 30 frames per second. 55 s videos were recorded with a resolution of 0.714μm/px). Then, the software tracks the brownian motion of individual vesicles, visualized by light scattering, and calculates their size and total concentration with corresponding standard error.
Electron microscopy (EM) was performed on pellets stored at ‐80°C and never unfrozen. EV suspension in PBS was deposited on formvar‐carbon‐coated cooper/palladium grids for whole‐mount analysis as described previously in Chapter 3.Unit3‐22 (Théry et al., 2006)). Images were acquired with a digital camera (Gatan Orius) mounted on a JEM‐1010 transmission electron microscope (JEOL Inc., USA) operated at 80 kV.
4.3. Western blot to measure proteins content in EVs and cellular extracts
Cell lysates for Western blot were obtained by incubating HEK293 (for uEV Western blot) and PC3 or DU145 (for cEV Western blot) cell pellets at a concentration of 2 × 10 (Hoshino et al., 2020) cells in 25 ul of lysis buffer (50 mM Tris, pH 7.5, 0.15 M NaCl, 1% Triton X‐100) with 2% complete protease inhibitor (Roche) for 20 min on ice, followed by an 18,516×g centrifugation for 15 min at 4 °C to recover the supernatant. Both cell lysates and EV pellets (10 (Spinelli et al., 2021) particles) were mixed with Laemmli sample buffer (BioRad), without reducing agent. After boiling for 5 min at 95 °C, samples were loaded on a 4–15% Miniprotean TGX stain‐free gels (BioRad). Total proteins were imaged from the stain‐free gels with the ChemiDoc Touch Imager (BioRad). Transfer was performed on Immuno–Blot PVDF membranes (BioRad), with the Trans–Blot Turbo Transfer System (BioRad) during 7 min. Blocking was performed during 30 min with Blocking Reagent (Roche) in TBS 0.1% Tween. Primary antibodies were incubated overnight at 4 °C and secondary antibodies during 1 h at room temperature (RT). Development was performed using either the Clarity Western ECL Substrate (BioRad) or the Immobilon Forte Western HRP substrate (Millipore), and the ChemiDoc Touch Imager (BioRad). Intensity of the bands was quantified using ImageJ.
4.4. Antibodies and reagents
Primary antibodies for Western blot were mouse antihuman CD63 (BD Bioscience, clone H5C6, 1/1000), ‐human CD9 (Millipore, clone MM2/57, 1/1000), rabbit antihuman Syntenin (Abcam, clone EPR8102, 1/2000). Mouse anti human Calreticulin (Abcam, clone FMC 75, 1/1000), rabbit antihuman Calnexin (Abcam, clone EPR3633(2), 1/1000).
Secondary antibodies HRP‐conjugated goat antirabbit IgG (H + L) and HRP conjugated goat antimouse IgG (H + L) was purchased from Jackson Immuno–Research and used 1/10000.
4.5. RNA extraction
Extraction of total RNA including miRNA from urinary and cell samples was performed by using the miRNeasy micro kit (Qiagen) with modified on‐column digestion of DNA recommended by Qiagen for less than 1 μg of RNA. The Modified procedure consists of reapplying to the membrane the flow‐through after digestion and washing and to prepare RWT buffer with isopropanol instead of ethanol. RNA was eluted with 14 μL nuclease free water.
RNA from FFPE tissues was purified using RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Life technologies) from 10 slices of 5 μm. RNA concentration has been evaluated by with Qubit RNA HS Assay Kit (Life technologies). The extracted RNA was checked for size distribution on the Agilent Bioanalyzer using RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA, USA).
RNA from frozen tumour prostate tissue was extracted as described previously (Pinskaya et al., 2019) using the TRizol reagent (Thermo Fisher Scientific), according to the manufacturer's procedure.
The Extracted RNA was checked for concentration and contaminants on NanoDrop spectrophotometer (Thermo Fisher Scientific) and Qubit fluorometer using Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and for size distribution on the Agilent Bioanalyzer using RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA, USA).
4.6. Preparation of libraries and Illumina sequencing
For each sample, 10 ng of total RNA was used to construct a strand‐specific library using SMARTer Stranded Total RNA‐Seq Kit v2 ‐ Pico Input Mammalian kit which incorporates a technology that enables removal of ribosomal cDNA following cDNA synthesis (Takara Bio, Europe). The libraries were sequenced on NovaSeq2 sequencing system from Next–Generation Sequencing platform of the Institut Curie as 2 × 100 nucleotides paired‐end reads to obtain about 50 million reads per sample.
4.7. Bioinformatics analysis
4.7.1. RNA‐seq reads quality control and alignment for FFPE and uEVs
The Curie bioinformatic platform performed the quality control of the RNA‐seq data using MultiQC, and the alignment of the reads on the human genome hg38 was done using STAR 2.6.1a, with the following parameters : –outMultimapperOrder Random –outSAMtype BAM Unsorted –outSAMattributes All –outSAMprimaryFlagOneBestScore –outSAMmultNmax 1 –outFilterTypeBySJout –outFilterMultimapNmax 20 –outFilterMismatchNmax 999 –outFilterMismatchNoverLmax 0.04 –alignIntronMin 20 –alignIntronMax 1000000 –alignMatesGapMax 1000000 –alignSJoverhangMin 8 –alignSJDBoverhangMin 1 (Esteve‐Codina et al. 2017; Zhang et al. 2019).
4.7.2. Differential expression analysis of FFPE and uEVs
Read counting was performed for each sample on the human gene annotation gencode v32 (https://www.gencodegenes.org/human/release_32.html) and on the human repeats (http://www.repeatmasker.org/species/hg.html), using Kallisto 0.46.1 with the parameter –rf‐stranded (Hezroni et al. 2015). The Kallisto index was built with the extracted sequences from the human genome hg38, using the annotations and the getfasta command of BEDTools 2.29 (for gencode 32, before the extraction, the exons of all the transcripts of each gene were merged by location, in order to obtain directly the counts at the gene level) (Kaul et al., 2020).
The tool CIRIquant 1.1 was used on the samples with default parameters to discover the location of circRNAs and to quantify them (Zhang et al., 2020). The results (counts from human genes, repeats and circRNAs) were concatenated. The conditions FFPE and uEVs were compared using DESeq2, with the following parameters: betaPrior = FALSE, independentFiltering = F, cooksCutoff = F. Only the features with adjusted p‐value < = 0.05,abs(log2FoldChange ) > = 0.585, and normalized counts > = 20 in at least one sample were retained as differentially expressed. Heatmaps of expression were obtained using the R package ComplexHeatmap from bioconductor.
4.7.3. Read count genomic distribution
We created from the human gene annotation gencode32 (https://www.gencodegenes.org/human/release_32.html) six classes of genomic features (exon, intron, 5′‐UTR, 3′‐UTR, promoter, intergenic), as follows: The exons from all the transcripts of each gene were merged by location, in order to have nonredundant segments of exons, using the merge command of BEDTools 2.29. Introns were inferred from these exons using the R package GenomicFeatures from bioconductor. The 5′ and 3′ UTRs from gencode32 were merged and were subtracted from the previous features. Promoters were inferred by using 1 kb upstream of the first exon (when possible), in respect of the strand. Intergenic parts were constructed after the concatenation of all the previous features, using the complement command of bedtools. Strand‐specific counting from the alignment files was performed on these features using featureCounts of the Subread package (https://sourceforge.net/projects/subread/files/) (Chen et al., 2019). The priority order when the counts were on several features was the following: exon > UTRs > intron > promoter > intergenic.
4.7.4. Exon‐intron read count ratio
The exons from all the transcripts of each gene of the human gene annotation gencode32 (https://www.gencodegenes.org/human/release_32.html) were merged by location using the merge command of BEDTools 2.29, in order to have nonredundant segments of exons. Introns were inferred from these exons using the R package GenomicFeatures from bioconductor. Genes with no introns were discarded.
The alignment files were converted to BEDPE format and were intersected with the new formed annotation to determine their status (exonic or intronic), using the count command of BEDTools 2.29. The number of exonic and intronic counts for each gene was then determined, using the same command. The counts were normalized using the length of each feature, and the ratio exonic count over intronic count was computed (genes that had 0 count on numerator and denominator before calculation were discarded. When just a part is equal to 0, +1 is added to both parts to avoid division by 0). The average ratio for each gene was computed for each condition, and the results were plotted using the R package ggplot2.
4.7.5. Metagenes
To select the exons and introns features, we have created firstly metatrancripts as follows:
The exons from all the transcripts of each gene were merged by location using the merge command of BEDTools 2.29, to have nonredundant segments of exons, and these exons were numbered; Introns were inferred from these exons.
The first exon, first intron, second exon, last intron and last exon were selected for each gene having at least one raw read count (read count from Kallisto).
The alignment files were converted in BigWig files with RPM normalization (reads per million of mapped reads), using UCSC tools (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/), and for each part and each gene, the read coverage was extracted.
The obtained signal was scaled on 100 positions using the R base approx function (allows to compute interpolations), with the following parameters: method = “linearties = ,”“ordered”
The average, minimum, and maximum values were computed at each of the 100 positions, and the result was plotted using the R package ggplot2.
4.7.6. Neoantigen analysis
HLA allotypes from FFPE and uEVs samples were determined using the tool seq2HLA. Upregulated lncRNAs from FFPE &uEVs were selected, and a research of the ORFs (start codon = AUG) of the most abundant transcript for each gene was performed using TransDecoder (https://transdecoder.github.io/) and the R bioconductor package ORFik https://github.com/Roleren/ORFik. The translation into peptides from the three frames was performed using the SeqinR R package (Charif & Jean, 2007), and only the ones with a length > = 8 amino acids were kept. The list of peptides for FFPE and uEVs was given to the tool netMHCpan‐4.1 in order to find neoantigens. Only the peptides with an elution ligand rank < = 2 were kept for downstream analysis.
4.7.7. Ribosome profiling analysis
Raw data were extracted from (Hsieh et al., 2012). The Ribosome protect Fragments were defined by counting reads on the defined putative ORF of the 15 Evs‐lncRNA containing, encoding high affinity binding scores neoantigens, and on the ORF of POLR2A, normalized with the size (TPM).
4.8. Proteomics and mass spectrometry analyses
4.8.1. Sample preparation
Cell samples were lysed in a buffer containing 8 M urea (Euromedex), 200 mM ammonium bicarbonate (ABC, FisherScientific) for 30 min at room temperature. Lysates were sonicated to decrease viscosity and centrifuged at 20,000 x g for 10 min. The protein concentration was measured using the BCA assay (Pierce). 60 μg of total protein was reduced by 5 mM dithiothreitol (DTT, Sigma) for 30 min at 55°C, alkylated with 10 mM iodoacetamide (IAM, Sigma) for 30 min in the dark. Samples were then diluted 10‐fold with 200 mM ABC to obtain a final concentration of urea of 1 M before overnight digestion with Trypsin‐LysC (Promega) at a 1:50 ratio at 37°C. Digested samples were acidified with 1% trifluoroacetic acid (TFA, Thermo) for 15 min on ice and centrifuged at 2000 x g for 15 min and purified using 50 mg Sep‐Pak C18 cartridge (Waters). Peptides were eluted using 40/60 MeCN/H2O + 0.1% formic acid and 1/6 of the starting material was vacuum concentrated to dryness and reconstituted in 10 μl injection buffer (0.3% TFA) before nano‐LC‐MS/MS analysis.
4.8.2. LC‐MS/MS analysis
Chromatography was performed with an RSLCnano system (Ultimate 3000, Thermo Scientific) coupled online to an Orbitrap Exploris 480 mass spectrometer (Thermo Scientific). Peptides were trapped on a C18 column (75 μm inner diameter × 2 cm; nanoViper Acclaim PepMapTM 100, Thermo Scientific) with buffer A (2/98 MeCN/H2O in 0.1% formic acid) at a flow rate of 3.0 μL/min over 4 min. Separation was performed on a 50 cm x 75 μm C18 column (nanoViper Acclaim PepMapTM RSLC, 2 μm, 100Å, Thermo Scientific) regulated to a temperature of 40°C with a linear gradient of 3% to 32% buffer B (100% MeCN in 0.1% formic acid) at a flow rate of 300 nl/min over 211 min. MS full scans were performed in the ultrahigh‐field Orbitrap mass analyser in ranges m/z 375–1500 with a resolution of 120 000 at m/z 200. The top 20 most intense ions were subjected to Orbitrap for further fragmentation via high‐energy collision dissociation (HCD) activation and a resolution of 15 000 with the automatic gain control (AGC) target set to 100%. We selected ions with charge state from 2+ to 6+ for screening. Normalized collision energy (NCE) was set at 30 and the dynamic exclusion to 40s.
4.8.3. Data analysis
LncRNA and Neoantigen data base prediction. For identification, the data were searched against the Homo Sapiens (UP000005640_9606) UniProt database (https://www.expasy.org/resources/uniprotkb‐swiss‐prot) and our two predicted databases of PC3 LncRNA ORF encoded peptides and uEV‐lncRNA ORF encoded peptides, using Sequest HT through proteome discoverer (PD version 2.4). These two databases have been built, respectively, from 12,815 lncRNAs with at least one count in PC3 cell line by RNA sequencing, and from the 228 enriched uEV‐lncRNAs. The selection of the ORFs (start codon = AUG) of the most abundant transcript for each lncRNA was performed using TransDecoder (https://transdecoder.github.io/) and the R bioconductor package ORFik https://github.com/Roleren/ORFik. We identified 7033 (from PC3) and 167 (from uEVs) lncRNAs containing ORFs. Translation of these ORFs into peptides from the three frames were performed using the SeqinR R package (Charif & Jean, 2007), and only the ones with a length > = 8 amino acids were kept. We then obtained 43,387 putative peptides for the PC3 LncRNAs and 862 putative peptides for enriched uEV‐lncRNAs (Extended Data Table 10).
PD enzyme specificity was set to trypsin and a maximum of two miss cleavages sites were allowed. Oxidized methionine, Met‐loss, Met‐loss‐Acetyl and N‐terminal acetylation were set as variable modifications. Carbamidomethylation of cysteins was set as fixed modification. Maximum allowed mass deviation was set to 10 ppm for monoisotopic precursor ions and 0.02 Da for MS/MS peaks. The resulting files were further processed using myProMS v3.9.3 (Poullet et al., 2007) (https://github.com/bioinfo‐pf‐curie/myproms). FDR calculation used Percolator (The et al., 2016) and was set to 1% at the peptide level for the whole study. Observed retention times from nonmodified peptide (excepted for Carbamidomethylation of cysteins) were averaged across all samples and plotted against the predicted hydrophobicity index calculated with an SSRCalc‐based algorithm (Krokhin et al., 2004) using the R package ggplot2.
AUTHORS CONTRIBUTIONS
Anna Almeida: Supervision, validation, investigation, methodology, project administration, funding and article writing—original draft, review, and editing. Marc Gabriel: Resources, data curation, software, formal analysis, validation, investigation, and visualization. Virginie Firlej: Resources, methodologies and data curation. Lorena Martin‐Jaular: Formal analysis, methodology, and investigation. Mathieu Lejars: Methodology, investigation. Rocco Cippola: Resources, investigation. Floriane Petit: Methodology. Nicolas Vogt: Investigation. Florent Dingli: methodology, resources Damarys Loew: Supervision, data curation. Damien Destouches: Resources. Francis Vacherot: Resources. Alexandre De La Taille: Resources and data curation. Clotilde Théry: Supervision, methodology, validation, formal analysis Antonin Morillon: conceptualization, supervision, funding acquisition, project administration, and writing—original draft, review, and editing.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
We deeply thank all the members of our labs for discussions and critical reading of the manuscript. Daniel Gautheret for his constant encouragement and critical readings. Pascale Soyeux for technical help on FFPE manipulation. The platform of biological resources (PRB) of Henri Mondor hospital for tissues collection. We also would like to thank Benoît Albaud, Sonia Lameiras, Patricia Legoix‐Né, Virginie Raynal, and Sylvain Baulande from the Next Generation Sequencing (NGS) platform of Institut Curie for discussions and expertise. RNA‐seq sequencing efforts were supported by the grant from the ICGex program of Institut Curie attributed to A.M. Marc Gabriel and Matthieu Lejars and this project were supported by the European Research Council (875532‐PROSTATOR‐ERC‐2019‐PoC), CARNOT‐2018‐PROSTATOR, (PSL‐Qlife cirdark‐Marker (Q‐life ANR‐17‐CONV‐0005) 17 MECENAT and structurating funds by Institut Curie attributed to A.M. and C.T. This work was supported by “Région Ile‐de‐France” and Fondation pour la Recherche Médicale grants (to D.L.), and by Fondation ARC (PGA1 RF20180206962) and a Subagreement from the Johns Hopkins University with funds provided by Grant No. UG3CA241694 from the National Cancer Institute to CT. The LSMP thanks Patrick Poullet from the bioinformatics platform of the Institut Curie U900 and Victor Laigle from the LSMP for the continuous development of myProMS and bioinformatics support. This work was also funded by the Ligue contre le cancer 94 / Val‐de‐Marne (grant #A.P. 2020) and the Groupement d'Entreprises Françaises dans la Lutte contre le Cancer, GEFLUC (grant #A.P. 2020).
Almeida, A. , Gabriel, M. , Firlej, V. , Martin‐Jaular, L. , Lejars, M. , Cipolla, R. , Petit, F. , Vogt, N. , San‐Roman, M. , Dingli, F. , Loew, D. , Destouches, D. , Vacherot, F. , de la Taille, A. , Théry, C. , & Morillon, A. (2022). Urinary extracellular vesicles contain mature transcriptome enriched in circular & long noncoding RNAs with functional significance in prostate cancer. Journal of Extracellular Vesicles, 11, e12210. 10.1002/jev2.12210
Anna Almeida and Marc Gabriel contributed equally to this work.
DATA AVAILABILITY STATEMENT
The bioinformatic codes, intermediate data and Extended Data Tables are available at: https://github.com/MorillonLab/Prostate_6_tumor_FFPE_vs_6_tumor_uEVs_analysis. The raw data are available under GEO accession GSE183070. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Perez‐Riverol et al., 2019)
REFERENCES
- Amorim, M. G. , Valieris, R. , Drummond, R. D. , Pizzi, M. P. , Freitas, V. M. , Sinigaglia‐Coimbra, R. , Calin, G. A. , Pasqualini, R. , Arap, W. , Silva, I. T. , Dias‐Neto, E. , & Nunes, D. N. (2017). A total transcriptome profiling method for plasma‐derived extracellular vesicles: Applications for liquid biopsies. Scientific Reports, 7, 14395. 10.1038/s41598-017-14264-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjus, S. , Morillon, A. , & Wery, M. (2021). From yeast to mammals, the nonsense‐mediated mRNA decay as a master regulator of long non‐coding RNAs functional trajectory. Noncoding RNA, 44, 7. 10.3390/ncrna7030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, D. ‐. H. , Lee, S. K. , & Sood, A. K. (2019). Circular RNAs in cancer. Molecular Therapy. Nucleic Acids, 16, 118–129. 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmayr‐Heyda, A. , Reiner, A. T. , Auer, K. , Sukhbaatar, N. , Aust, S. , Bachleitner‐Hofmann, T. , Mesteri, I. , Grunt, T. W. , Zeillinger, R. , & Pils, D. (2015). Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific Reports, 5, 8057. 10.1038/srep08057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , Dwivedi, Om P. , Leparc, G. , Rolser, M. , Delic, D. , Forsblom, C. , Groop, P. ‐. H. , Groop, L. , Huber, T. B. , Puhka, M. , & Holthofer, H. (2020). Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. Journal of Extracellular Vesicles, 10, e12038. 10.1002/jev2.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A. , Thakur, B. K. , Weiss, J. M. , Kim, H. S. , Peinado, H. , & Lyden, D. (2016). Extracellular vesicles in cancer: Cell‐to‐cell mediators of metastasis. Cancer Cell, 30, 836–848. 10.1016/j.ccell.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann, D. , Kirchner, B. , Hermann, S. , Märte, M. , Wurmser, C. , Brandes, F. , Kotschote, S. , Bonin, M. , Steinlein, O K. , Pfaffl, M W. , Schelling, G. , Reithmair, M. (2018) Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next‐generation sequencing. Journal of Extracellular Vesicles, 7, 1481321. 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput, N. , Schartz, N. E. C. , André, F. , Taïeb, J. , Novault, S. , Bonnaventure, P. , Aubert, N. , Bernard, J. , Lemonnier, F. , Merad, M. , Adema, G. , Adams, M. , Ferrantini, M. , Carpentier, A. F. , Escudier, B. , Tursz, T. , Angevin, E. , & Zitvogel, L. (2004). Exosomes as potent cell‐free peptide‐based vaccine. II. Exosomes in CpG adjuvants efficiently prime naive Tc1 lymphocytes leading to tumor rejection. Journal of Immunolgy, 172, 2137–2146. 10.4049/jimmunol.172.4.2137 [DOI] [PubMed] [Google Scholar]
- Charif, D. , & Jean R. L.. (2007). SeqinR 1.0‐2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. 207–232 (Springer Verlag; ). [Google Scholar]
- Chen, J. , Brunner, A. ‐. D. , Cogan, J. Z. , Nuñez, J. K. , Fields, A. P. , Adamson, B. , Itzhak, D. N. , Li, J. Y. , Mann, M. , Leonetti, M. D. , & Weissman, J. S. (2020). Pervasive functional translation of noncanonical human open reading frames. Science, 367, 1140–1146. 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nature Reviews. Molecular Cell Biology, 21, 475–490. 10.1038/s41580-020-0243-y [DOI] [PubMed] [Google Scholar]
- Chen, S. , Huang, V. , Xu, X. , Livingstone, J. , Soares, F. , Jeon, J. , Zeng, Y. , Hua, J. T. , Petricca, J. , Guo, H. , Wang, M. , Yousif, F. , Zhang, Y. , Donmez, N. , Ahmed, M. , Volik, S. , Lapuk, A. , Chua, M. L.K. , Heisler, L. E. … He, H. H. (2019). Widespread and functional RNA circularization in localized prostate cancer. Cell, 176, 831–843.e22. 10.1016/j.cell.2019.01.025 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Zhu, G. , Yang, Y. , Wang, F. , Xiao, Yu‐T. , Zhang, Na , Bian, X. , Zhu, Y. , Yu, Y. , Liu, F. , Dong, K. , Mariscal, J. , Liu, Y. , Soares, F. , Loo Yau, H. , Zhang, Bo , Chen, W. , Wang, C. , Chen, D. … Ren, S. (2021). Single‐cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nature Cell Biology, 23, 87–98. 10.1038/s41556-020-00613-6 [DOI] [PubMed] [Google Scholar]
- Cuzick, J. et al. (2011). Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. The Lancet. Oncology, 12, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, P. , Fiore‐Gartland, A. J. , Hertz, T. , Wang, G. C. , Sharma, S. , Souquette, A. , Crawford, J. C. , Clemens, E. B. , Nguyen, T. H. O. , Kedzierska, K. , La Gruta, N. L. , Bradley, P. , & Thomas, P. G. (2017). Quantifiable predictive features define epitope‐specific T cell receptor repertoires. Nature, 547, 89–93. 10.1038/nature22383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A. … Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles, 10, e12093. 10.1002/jev2.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve‐Codina, A. , Arpi, O. , Martinez‐García, M. , Pineda, E. , Mallo, M. , Gut, M. , Carrato, C. , Rovira, A. , Lopez, R. , Tortosa, A. , Dabad, M. , Del Barco, S. , Heath, S. , Bagué, S. , Ribalta, T. , Alameda, F. , De La Iglesia, N. , Balaña, C. (2017). A comparison of RNA‐Seq results from paired formalin‐fixed paraffin‐embedded and fresh‐frozen glioblastoma tissue samples. PloS One, 12, e0170632. 10.1371/journal.pone.0170632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaert, C. , Helsmoortel, H. , Decock, A. , Hulstaert, E. , Van Paemel, R. , Verniers, K. , Nuytens, J. , Anckaert, J. , Nijs, N. , Tulkens, J. , Dhondt, B. , Hendrix, An , Mestdagh, P. , & Vandesompele, Jo (2019). Performance assessment of total RNA sequencing of human biofluids and extracellular vesicles. Scientific Reports, 9, 17574. 10.1038/s41598-019-53892-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvanin, A. , Dostert, G. , Ayadi, L. , Marchand, V. , Velot, É. , Motorin, Y. (2019). Diversity and heterogeneity of extracellular RNA in human plasma. Biochimie, 164, 22–36. 10.1016/j.biochi.2019.05.011 [DOI] [PubMed] [Google Scholar]
- Garcia‐Martin, R. , Wang, G. , Brandão, B. B. , Zanotto, T. M. , Shah, S. , Kumar Patel, S. , Schilling, B. , & Kahn, C. R. (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature, 601, 446–451. 10.1038/s41586-021-04234-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri‐Fard, S. , Khoshbakht, T. , Taheri, M. & Ebrahimzadeh, K. A review on the role of PCAT6 lncRNA in tumorigenesis. Biomedicine & Pharmacotherapy = Biomedecine & pharmacotherapie, 142, 112010. 10.1016/j.biopha.2021.112010 (2021). [DOI] [PubMed] [Google Scholar]
- Giraldez, M. D. , Spengler, R. M. , Etheridge, A. , Goicochea, A. J. , Tuck, M. , Choi, S. W. , Galas, D. J. , & Tewari, M. (2019). Phospho‐RNA‐seq: A modified small RNA‐seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO Journal, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, G K. , Wu, J. , Iracheta‐Vellve, A. , Patti, J C. , Hsu, J. , Davis, T. , Dele‐Oni, D. , Du, P P. , Halawi, A G. , Ishizuka, J J. , Kim, S Y. , Klaeger, S. , Knudsen, N H. , Miller, B C. , Nguyen, T H. , Olander, K E. , Papanastasiou, M. , Rachimi, S. , Robitschek, E J. … Bernstein, B E. (2021). Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature, 595, 309–314. 10.1038/s41586-021-03520-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, T B. , Jensen, T I. , Clausen, B H. , Bramsen, J B. , Finsen, B. , Damgaard, C K. , Kjems, J. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hezroni, H. , Koppstein, D. , Schwartz, M. G. , Avrutin, A. , Bartel, D. P. , & Ulitsky, I. (2015). Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Reports, 11, 1110–1122. 10.1016/j.celrep.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. … Lyden, D. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell, 182, 1044–1061.e18. 10.1016/j.cell.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, A C. , Liu, Yi , Edlind, M P. , Ingolia, N T. , Janes, M R. , Sher, A. , Shi, E Y. , Stumpf, C R. , Christensen, C. , Bonham, M J. , Wang, S. , Ren, P. , Martin, M. , Jessen, K. , Feldman, M E. , Weissman, J S. , Shokat, K M. , Rommel, C. , Ruggero, D. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature, 485, 55–61. 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte, M. , Rinn, J. L. Large non‐coding RNAs: Missing links in cancer? Human Molecular Genetics, 19, R152–R161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert, E. , Morlion, A. , Avila Cobos, F. , Verniers, K. , Nuytens, J. , Vanden Eynde, E. , Yigit, N. , Anckaert, J. , Geerts, A. , Hindryckx, P. , Jacques, P. , Brusselle, G. , Bracke, K. R. , Maes, T. , Malfait, T. , Derveaux, T. , Ninclaus, V. , Van Cauwenbergh, C. , Roelens, K. … Mestdagh, P. (2020). Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Reports, 33(13), 108552. 10.1016/j.celrep.2020.108552 [DOI] [PubMed] [Google Scholar]
- Iyer, M. K. , Niknafs, Y. S. , Malik, R. , Singhal, U. , Sahu, A. , Hosono, Y. , Barrette, T. R. , Prensner, J. R. , Evans, J. R. , Zhao, S. , Poliakov, A. , Cao, X. , Dhanasekaran, S. M. , Wu, Yi‐Mi , Robinson, D. R. , Beer, D. G. , Feng, F. Y. , Iyer, H. K. , & Chinnaiyan, A. M. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics, 47, 199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck, W. R. , Sorrentino, J. A. , Wang, K. , Slevin, M. K. , Burd, C. E. , Liu, J. , Marzluff, W. F. , & Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna, 19, 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Qi , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , Coffey, R. J. (2019) Reassessment of exosome composition. Cell, 177, 428–445.e18. 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtz, V. , Paul, S. , Andreatta, M. , Marcatili, P. , Peters, B. , Nielsen, M. (2017) NetMHCpan‐4.0: Improved peptide‐MHC Class I interaction predictions integrating eluted ligand and peptide binding affinity data. Journal of Immunology, 199, 3360–3368. 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. , & Lebleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science, 367(6478), eaau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, T. , Morales, M. E. , Sartor, A. O. , Belancio, V. P. , & Deininger, P. (2020). Comparative analysis on the expression of L1 loci using various RNA‐Seq preparations. Mobile DNA, 11, 2. 10.1186/s13100-019-0194-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Z. , Wan, X. , Zhang, Y. , Zhang, Pu , Zhang, Y. , Zhang, X. , Qi, X. , Wu, H. , Huang, J. , & Li, Y. (2017). Androgen‐responsive circular RNA circSMARCA5 is up‐regulated and promotes cell proliferation in prostate cancer. Biochemical and Biophysical Research Communications, 493, 1217–1223. 10.1016/j.bbrc.2017.07.162 [DOI] [PubMed] [Google Scholar]
- Kristensen, L. S. , Andersen, M. S. , Stagsted, L. V. W. , Ebbesen, K. K. , Hansen, T. B. , & Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics, 20, 675–691. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- Krokhin, O. V. et al. (2004). An improved model for prediction of retention times of tryptic peptides in ion pair reversed‐phase HPLC: Its application to protein peptide mapping by off‐line HPLC‐MALDI MS. Molecular & Cellular Proteomics : MCP, 3, 908–919. 10.1074/mcp.M400031-MCP20 [DOI] [PubMed] [Google Scholar]
- Lai, C. P. , Kim, E. Y. , Badr, C. E. , Weissleder, R. , Mempel, T. R. , Tannous, B. A. , & Breakefield, X. O. (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications, 6, 7029. 10.1038/ncomms8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumont, C. M. , Vincent, K. , Hesnard, L. , Audemard, É. , Bonneil, É. , Laverdure, J. ‐. P. , Gendron, P. , Courcelles, M. , Hardy, M. ‐. P. , Côté, C. , Durette, C. , St‐Pierre, C. , Benhammadi, M. , Lanoix, J. , Vobecky, S. , Haddad, E. , Lemieux, S. , Thibault, P. , & Perreault, C. (2018). Noncoding regions are the main source of targetable tumor‐specific antigens. Science Translational Medicine, 10. 10.1126/scitranslmed.aau5516 [DOI] [PubMed] [Google Scholar]
- Lebleu, V. S. , & Kalluri, R. (2020). Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer, 6, 767–774. 10.1016/j.trecan.2020.03.007 [DOI] [PubMed] [Google Scholar]
- Li, S. , Han, L. Circular RNAs as promising biomarkers in cancer: Detection, function, and beyond. Genome Medicine, 11, 15. 10.1186/s13073-019-0629-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhao, J. , Yu, S. , Wang, Z. , He, X. , Su, Y. , Guo, T. , Sheng, H. , Chen, J. , Zheng, Q. , Li, Y. , Guo, W. , Cai, X. , Shi, G. , Wu, J. , Wang, Lu , Wang, P. , He, X. , & Huang, S. (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clinical Chemistry, 65, 798–808. 10.1373/clinchem.2018.301291 [DOI] [PubMed] [Google Scholar]
- Lorenzi, L. , Chiu, H. ‐. S. , Avila Cobos, F. , Gross, S. , Volders, P. ‐. J. , Cannoodt, R. , Nuytens, J. , Vanderheyden, K. , Anckaert, J. , Lefever, S. , Tay, A. P. , De Bony, E. J. , Trypsteen, W. , Gysens, F. , Vromman, M. , Goovaerts, T. , Hansen, T. B. , Kuersten, S. , Nijs, N. … Mestdagh, P. (2021). The RNA Atlas expands the catalog of human non‐coding RNAs. Nature Biotechnology, 39, 1453–1465. 10.1038/s41587-021-00936-1 [DOI] [PubMed] [Google Scholar]
- Mathieu, M. , Martin‐Jaular, L. , Lavieu, G. , & Théry, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nature Cell Biology, 21, 9–17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- Miranda, K C. , Bond, D T. , Levin, J Z. , Adiconis, X. , Sivachenko, A. , Russ, C. , Brown, D. , Nusbaum, C. , Russo, L M. (2014). Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non‐coding RNA. PloS One, 9, e96094. 10.1371/journal.pone.0096094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo, O. D. , Thistlethwaite, W. , Rozowsky, J. , Subramanian, S. L. , Lucero, R. , Shah, N. , Jackson, A. R. , Srinivasan, S. , Chung, A. , Laurent, C. D. , Kitchen, R. R. , Galeev, T. , Warrell, J. , Diao, J. A. , Welsh, J. A. , Hanspers, K. , Riutta, A. , Burgstaller‐Muehlbacher, S. , Shah, R. V. … Milosavljevic, A. (2019). exRNA Atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell, 177, 463–477.e15. 10.1016/j.cell.2019.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, J. , Skog, J. , Nordstrand, A. , Baranov, V. , Mincheva‐Nilsson, L. , Breakefield, X. O. , Widmark, A. Prostate cancer‐derived urine exosomes: a novel approach to biomarkers for prostate cancer. British Journal of Cancer, 100, 1603–1607. 10.1038/sj.bjc.6605058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okholm, T. L. H. , Sathe, S. , Park, S. S. , Kamstrup, A. B. , Rasmussen, A. M. , Shankar, A. , Chua, Z. M. , Fristrup, N. , Nielsen, M. M. , Vang, S. , Dyrskjøt, L. , Aigner, S. , Damgaard, C. K. , Yeo, G. W. , & Pedersen, J. S. (2020). Transcriptome‐wide profiles of circular RNA and RNA‐binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Medicine, 12, 112. 10.1186/s13073-020-00812-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostenfeld, M. S. , Jeppesen, D. K. , Laurberg, J. R. , Boysen, A. T. , Bramsen, J. B. , Primdal‐Bengtson, B. , Hendrix, An , Lamy, P. , Dagnaes‐Hansen, F. , Rasmussen, M. H. , Bui, K. H. , Fristrup, N. , Christensen, E. I. , Nordentoft, I. , Morth, J. P. , Jensen, J. B. , Pedersen, J. S. , Beck, M. , Theodorescu, D. … Ørntoft, T. F. (2014). Cellular disposal of miR23b by RAB27‐dependent exosome release is linked to acquisition of metastatic properties. Cancer Research, 74, 5758–5771. 10.1158/0008-5472.CAN-13-3512 [DOI] [PubMed] [Google Scholar]
- Pelissier Vatter, F. A. , Cioffi, M. , Hanna, S. J. , Castarede, I. , Caielli, S. , Pascual, V. , Matei, I. , & Lyden, D. (2021). Extracellular vesicle‐ and particle‐mediated communication shapes innate and adaptive immune responses. The Journal of Experimental Medicine, 218(8), e20202579. 10.1084/jem.20202579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Riverol, Y. , Csordas, A. , Bai, J. , Bernal‐Llinares, M. , Hewapathirana, S. , Kundu, D. J. , Inuganti, A. , Griss, J. , Mayer, G. , Eisenacher, M. , Pérez, E. , Uszkoreit, J. , Pfeuffer, J. , Sachsenberg, T. , Yılmaz, Ş. , Tiwary, S. , Cox, J. , Audain, E. , Walzer, M. … Vizcaíno, J. A. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research, 47, D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya, M. , Saci, Z. , Gallopin, M. , Gabriel, M. , Nguyen, Ha Tn , Firlej, V. , Descrimes, M. , Rapinat, A. , Gentien, D. , Taille, A. D.e La , Londoño‐Vallejo, A. , Allory, Y. , Gautheret, D. , & Morillon, A. (2019). Reference‐free transcriptome exploration reveals novel RNAs for prostate cancer diagnosis. Life Science Alliance, e201900449, 2. 10.26508/lsa.201900449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poullet, P. , Carpentier, S. , & Barillot, E. (2007). myProMS, a web server for management and validation of mass spectrometry‐based proteomic data. Proteomics, 7, 2553–2556. 10.1002/pmic.200600784 [DOI] [PubMed] [Google Scholar]
- Reiner, A. T. , Witwer, K. W. , Van Balkom, B. W.M. , De Beer, J. , Brodie, C. , Corteling, R. L. , Gabrielsson, S. , Gimona, M. , Ibrahim, A. G. , De Kleijn, D. , Lai, C. P. , Lötvall, J. , Del Portillo, H. A. , Reischl, I. G. , Riazifar, M. , Salomon, C. , Tahara, H. , Toh, W. S. , Wauben, M. H.M. … Lim, S. K. (2017). Concise review: Developing best‐practice models for the therapeutic use of extracellular vesicles. Stem Cells Translational Medicine, 6, 1730–1739. 10.1002/sctm.17-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn, J. L. , & Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annual Reviews in Biochemistry, 81, 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo, F. , Théry, C. , Falcón‐Pérez, J. M. , Nieuwland, R. , & Witwer, K. W. (2020). Methods for separation and characterization of extracellular vesicles: Results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells, 1955, 9. 10.3390/cells9091955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , Van Rijn, S. , Meijer, D H. , Gainche, L. , Curry, W T. , Carter, B S. , Krichevsky, A M. , Breakefield, X O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10, 1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, C. , Tawil, N. , Adnani, L. , Rak, J. , & Choi, D. (2021). Extracellular vesicle mediated vascular pathology in glioblastoma. Subcellular Biochemistry, 97, 247–273. 10.1007/978-3-030-67171-6_10 [DOI] [PubMed] [Google Scholar]
- Temoche‐Diaz, M. M. , Shurtleff, M. J. , Nottingham, R. M. , Yao, J. , Fadadu, R. P. , Lambowitz, A. M. , & Schekman, R. (2019). Distinct mechanisms of microRNA sorting into cancer cell‐derived extracellular vesicle subtypes. eLife, 8. 10.7554/eLife.47544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The, M. , Maccoss, M. J. , Noble, W. S. , & Käll, L. (2016). Fast and accurate protein false discovery rates on large‐scale proteomics data sets with Percolator 3.0. Journal of the American Society for Mass Spectrometry, 27, 1719–1727. 10.1007/s13361-016-1460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, , 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. 10.1080/20013078.2018.153575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach, M. , & Théry, C. (2016). Communication by extracellular vesicles: Where we are and where we need to go. Cell, 164, 1226–1232. 10.1016/j.cell.2016.01.043 [DOI] [PubMed] [Google Scholar]
- Tosar, J. P. , Segovia, M. , Castellano, M. , Gámbaro, F. , Akiyama, Y. , Fagúndez, P. , Olivera, Á. , Costa, B. , Possi, T. , Hill, M. , Ivanov, P. , Cayota, A. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Research, 48, 12874–12888. 10.1093/nar/gkaa674 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, D. , Zheng, W. , Zhang, Da‐H. , & Dong, X. (2020). Circular RNA and its potential as prostate cancer biomarkers. World Journal of Clinical Oncology, 11, 563–572. 10.5306/wjco.v11.i8.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Vancura, A. , Lanzós, A. , Bosch‐Guiteras, N. , Esteban, M. T. , Gutierrez, A. H. , Haefliger, S. , & Johnson, R. (2021). Cancer LncRNA Census 2 (CLC2): An enhanced resource reveals clinical features of cancer lncRNAs. NAR Cancer, 3, zcab013. 10.1093/narcan/zcab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Niel, G. , D'angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology, 19, 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Villarroya‐Beltri, C. , Gutiérrez‐Vázquez, C. , Sánchez‐Cabo, F. , Pérez‐Hernández, D. , Vázquez, J. , Martin‐Cofreces, N. , Martinez‐Herrera, D. J. , Pascual‐Montano, A. , Mittelbrunn, M. , Sánchez‐Madrid, F. (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications, 4, 2980. 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, J. N. , Cieslik, M. , Zhang, Y. , Shukla, S. , Xiao, L. , Zhang, Y. , Wu, Yi‐Mi , Dhanasekaran, S. M. , Engelke, C. G. , Cao, X. , Robinson, D. R. , Nesvizhskii, A. I. , & Chinnaiyan, A. M. (2019). The landscape of circular RNA in cancer. Cell, 176, 869–881.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklander, O. P. B. , Brennan, M. Á. , Lötvall, J. , Breakefield, X. O. , & El Andaloussi, S. (2019). Advances in therapeutic applications of extracellular vesicles. Science Translational Medicine, 11(452), eaav8521. 10.1126/scitranslmed.aav8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers, J. , Lozier, A. , Raposo, G. , Regnault, A. , Théry, C. , Masurier, C. , Flament, C. , Pouzieux, S. , Faure, F. , Tursz, T. , Angevin, E. , Amigorena, S. , & Zitvogel, L. (2001). Tumor‐derived exosomes are a source of shared tumor rejection antigens for CTL cross‐priming. Nature Medicine, 7, 297–303. 10.1038/85438 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Chen, S. , Yang, J. , & Zhao, F. (2020). Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nature Communications, 11(1), 90. 10.1038/s41467-019-13840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Higginbotham, J. N. , Jeppesen, D. K. , Yang, Yu‐P. , Li, W. , Mckinley, E. T. , Graves‐Deal, R. , Ping, J. , Britain, C. M. , Dorsett, K. A. , Hartman, C. L. , Ford, D. A. , Allen, R. M. , Vickers, K. C. , Liu, Qi , Franklin, J. L. , Bellis, S. L. , & Coffey, R. J. (2019). Transfer of functional cargo in exomeres. Cell Reports, 27, 940–954.e6. 10.1016/j.celrep.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.‐M. , Cai, W. L. , Liu, X. , Thakral, D. , Luo, J. , Chan, L. H. , Mcgeary, M. K. , Song, E. , Blenman, K. R. M. , Micevic, G. , Jessel, S. , Zhang, Y. , Yin, M. , Booth, C. J. , Jilaveanu, L. B. , Damsky, W. , Sznol, M. , Kluger, H. M. , Iwasaki, A. … Yan, Q. (2021). KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature, 598, 682–687. 10.1038/s41586-021-03994-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The bioinformatic codes, intermediate data and Extended Data Tables are available at: https://github.com/MorillonLab/Prostate_6_tumor_FFPE_vs_6_tumor_uEVs_analysis. The raw data are available under GEO accession GSE183070. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Perez‐Riverol et al., 2019)


