Abstract
Fiber-based materials with microchannels have drawn considerable attention in recent years owing to their ability to mimic intrinsic morphologies of living tissues. Folded morphologies, which are common in vivo, such as in skeletal muscle capillaries and intestine luminal endoderm, play important roles in the achievement of tissue functions. Here, microfibers with folded hollow channels are fabricated. Channel morphologies, such as straight-folded, double-folded and double-helical channels, can be regulated by adjusting flow conditions in the microfluidic devices. To further demonstrate the potential to be used in tissue engineering, intestine and skeletal muscle constructs are fabricated using these microfibers as building blocks. Furthermore, the properties of perfusability, permeability, cytocompatibility and weavability of the microfibers are evaluated. The asymmetric molecular distributions in the microfibers provide promising platforms for the study of nutrient exchange and energy supplement between normal and tortuous tissues. The new features of biofibers and proof-of-concept of tissue constructs with folded morphologies may contribute to the development of regenerative medicine and drug screening in the future.
Perfusable microfibers with folded channels are generated to fabricate small intestine and skeletal muscle constructs for tissue engineering.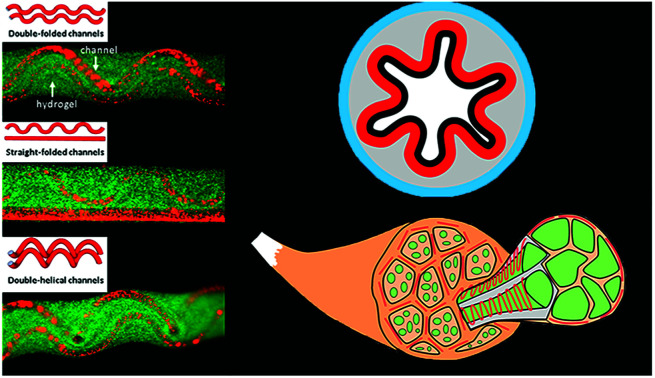
1. Introduction
Fabricating tissue constructs in vitro can alleviate the crisis of organ shortage in transplantation,1–3 and provide platforms for cancer research,4 regenerative medicine and drug screening.5,6 Mimicking three-dimensional tissue morphologies is a key point among processes of construction.7 The folded morphology, which is common in vivo, such as in skeletal muscle capillaries,8 the cerebral cortex,9,10 intestine luminal endoderm and tracheal mucosa,11–14 plays an important role in tissue functionalization, like oxygen flux improvement, information processing, nutrient absorption. Nevertheless, some folded morphologies cause diseases, such as asthma caused by folded mucosa penetrating into airway lumen.15 In addition, blood vessels with folded channels, an anomalous morphology, are associated with various severe diseases like atherosclerosis, diabetes, hypertension, cancer and genetic diseases.16–23 Thus, inherent biomechanical and geometrical mechanisms need to be studied for the purpose of developing new tools for the prevention and treatment of diseases.18,24
Complexity of tissue structures makes it hard to build their folding morphologies.25 At present, many efforts have been paid to construct folding structures in vitro, including by 3D bioprinting, electrospinning and microfluidic techniques.25–32 Current 3D bioprinting methods, based on layer-by-layer stacking, are usually difficult to print complex hollow structures, because layers must be connected and mechanically supported to avoid collapse and imprecise geometric configurations.33 In the sacrificial material-based method, platforms capable of fast bioink switching deposit sacrificial materials into matrix material, occupying channels' positions at beginning and then dislodged after molding.34 As for electrospinning method, nanofibers with several morphologies including porous, solid, core–shell, and hollow structures are produced.35–37 However, the random whipping motion of the electrospun nanofiber makes it a challenge to handle single fiber for constructing complex tissues.38 As additional methods, microfluidic techniques are developed to fabricate on-chip and fiber-based tissue constructs. However, limited by conventional lithography methods,39 channels embedded in PDMS chips usually have rectangle cross-sections, which are inconsistent with consecutive curved structures in vivo. In recent years, fiber-based techniques have been widely used in producing microfibers with various morphologies, including solid,40,41 helical,42,43 grooved,44 spindle,45,46 folded shapes,47 and the channel in microfibers could be straight,48 helical-channel and multi-channel.49,50 Owing to the similarity to hierarchical structures in vivo, these microfibers have the potential to be used as building blocks for tissue engineering.51–53 Nevertheless, to the best of our knowledge, microfibers with folded channels have not yet been reported.
In this study, we propose a microfluidics-based method to fabricate microfibers with double-folded, folded-straight and double-helical channels. Channel morphologies can be regulated by adjusting flow rates in the microfluidic devices. Intestine and skeletal muscle constructs are fabricated using these microfibers for future applications in tissue engineering. Moreover, characteristics of perfusability, permeability, biocompatibility and weavability endow these microfibers with the potential to build up in vitro tortuous vessel models.
2. Experimental
2.1. Materials and chemicals
Alginic acid sodium salt (Sigma-Aldrich, 1.0% w/v, 19.4 cP) dissolved in ddH2O was filtered through filter membranes (0.45 μm, Millex-HV). CaCl2 (Sigma-Aldrich, 100 mM) solution was prepared in ddH2O. Brilliant Blue and Allura Red was purchased from Shanghai Dyestuffs Research Institute Co. Ltd. Blunt needles, stainless steel tube, epoxy adhesive resin glue were purchased from the Taobao website. Glass capillaries were obtained from Sutter Instrument Co. Ltd. Fluorescein isothiocyanate-dextran (70 kDa, Sigma-Aldrich) and rhodamine B isothiocyanate-dextran (70 kDa, Sigma-Aldrich) were both dissolved in 100 mM CaCl2 solution to a final concentration of 25 μM respectively. Phosphate Buffered Saline (PBS 1×) was purchased from Hyclone. Calcein AM and PI were purchased from KeyGEN BioTECH. Endothelial Cell Medium (ECM) was obtained from ScienCell. DMEM and 0.05% trypsin–EDTA (1×) phenol red was purchased from Thermo Fisher Scientific. Fluorescence particles (FluoSpheres carboxylate-modified, 0.2 μm, red 580/605; yellow-green 505/515; blue 365/415) and CellTracker Green (492/517) were purchased from Thermo Fisher Scientific. EDTA was obtained from Beijing Chemical Works.
2.2. Fabrication of microfibers
The microfluidic device used to fabricate microfibers was made up of two CaCl2 inlets, one alginate inlet and one outlet. A 20 G blunt needle was punched as connector and alginate inlet. Two 32 G stainless needles were inserted into the connector in parallel as CaCl2 inlets. A glass capillary (outer diameter, inner diameter and length are 1 mm, 0.75 mm and 2 cm respectively) was inserted from the other side serving as the outlet. All the connections were sealed with epoxy adhesive resin glue. Syringe pumps (Harvard PHD) were used to injecting the solutions into the microfluidic device. The outlet of the device was immersed into CaCl2 bath to form microfibers.
2.3. Perfusion and permeation test of microfibers
A home-made perfusion system was used in the perfusion experiment. The detailed information was introduced in our previous work.49 To be brief, the microfiber was firstly sucked and fixed into a glass capillary. Then fluorescent particles solutions (diluted 500 times) were perfused into hollow channels through a three-way valve and recorded per 100 ms. For the permeation test, fluorescent dextran solutions were perfused into hollow channels through a three-way valve. After dextran filling the channel, perfusion was stopped and the fluorescence intensity was recorded per 10 s to 360 s and then per 60 s.
2.4. Cell culture
Human skeletal muscle cells (HSMCs, purchased from Shanghai and Shanghai Zhen Biotechnology Co., Ltd.) were maintained in DMEM supplemented with 10% fetal bovine serum. Human umbilical vein endothelial cells (HUVEC, purchased from ScienCell) were maintained in complete ECM. Culture medium was changed every 2 days.
2.5. Cell staining
To evaluate cell viability, microfibers with embedded cells were washed with PBS before stained with live/dead cell viability assay. After 20 minutes incubation, cells were released by dissolving the Ca-alginate matrix using 20 mM EDTA and imaged with confocal laser-scanning microscopy (Nikon, A1RSi). Software Image J was used to count cells. To locate HSMCs in skeletal muscle constructs, HSMCs cultured on Petri dishes were firstly stained with 5 μM CellTracker Green in serum-free DMEM and incubated for 30 min. Then the labeled cells were dispensed into microfibers at 5 × 106 ml−1 and observed under microscope.
2.6. Characterization
The generation processes of microfibers were recorded in real-time with the help of Full HD 1080p camera microscope. Bright field and fluorescence images were captured by Olympus IX81 microscopy. 3D fluorescence images were obtained by Carl Zeiss LSM 780 microscopy. Scanning electron microscope (SEM, FEI Quanta 200) was used to illustrate channel positions. Three-dimensional structure of microfibers was snapped by Zeiss Lightsheet Z.1.
3. Results and discussion
3.1. Generation of hollow hydrogel microfibers with folded channels
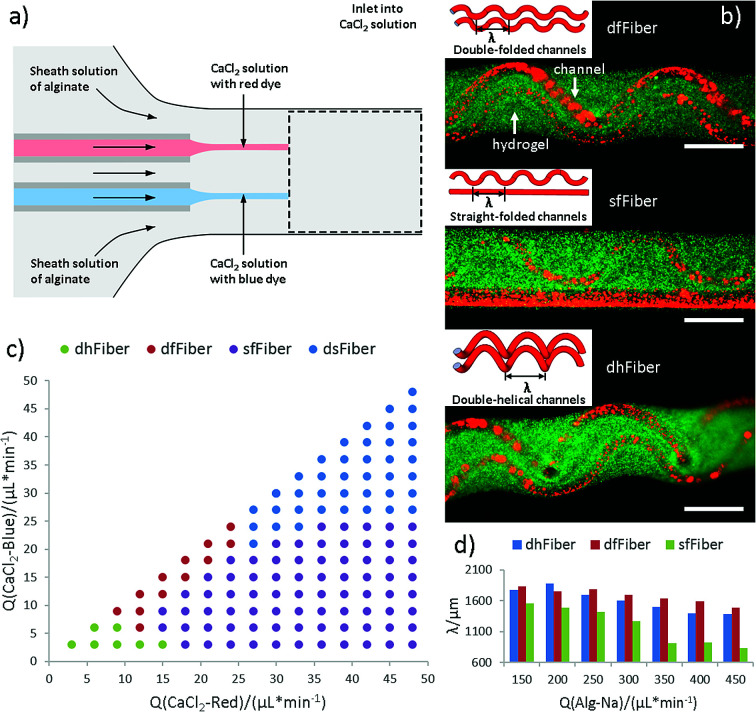
The co-flow device designed for producing microfibers with folded channels are illustrated in Fig. 1. Sodium alginate is chosen as sheath material while calcium chloride solution as core stream. Microfibers with two hollow channels could soon be generated attributed to instantaneous crosslink reaction between alginate and calcium ion at the interface (Fig. 1a and Video S1 in ESI†). Considering that the two aligned inner channels are equivalent in our experiment, microfibers with double-folded channels (dfFiber, Fig. 1b), folded-straight channels (sfFiber, Fig. 1b), double-helical channels (dhFiber, Fig. 1b) and double-straight channels (dsFiber) are produced by adjusting the flow rate of core solution (Qi), keeping sheath flow rates (Qo) constant (200 μl min−1). To explain these phenomena, the relationship between flow patterns and Qi is illustrated in Fig. 1c. With Qi increasing, morphology of inner channels transforms from helical to folded and straight in turn. Demarcation lines of the four patterns vary according to device specifications and fluid viscosity. Aside from the above-mentioned morphologies, numerous patterns can be realized by adding the number of embedded inner channels theoretically, such as microfiber with three-helical channels or three-folded channels (Fig. S1, Video S2 and S3 in ESI†). Moreover, the relationship between period (λ) of df, dh, sf channels and flow rates are also explored. As demonstrated in Fig. 1d, under the circumstance that Qi/Qo is fixed, raising the flow rates of Qi and Qo relates to decreased λ.
Fig. 1. Generation of hollow hydrogel microfibers with folded channels. (a) Scheme of the formation of hollow fiber with double channels. (b) The 3D structure of three fibers, in which green fluorescent microspheres represent area of the alginate hydrogel shell, and red microspheres represent hollow channel. The scale bar is 500 μm. (c) Channel shapes of microfibers at different flow conditions of two Qi with constant Qo (200 μl min−1). (d) The relationship between the period (λ) of folded channels and Qo when Qi/Qo is constant.
3.2. Mechanism of microfiber generation
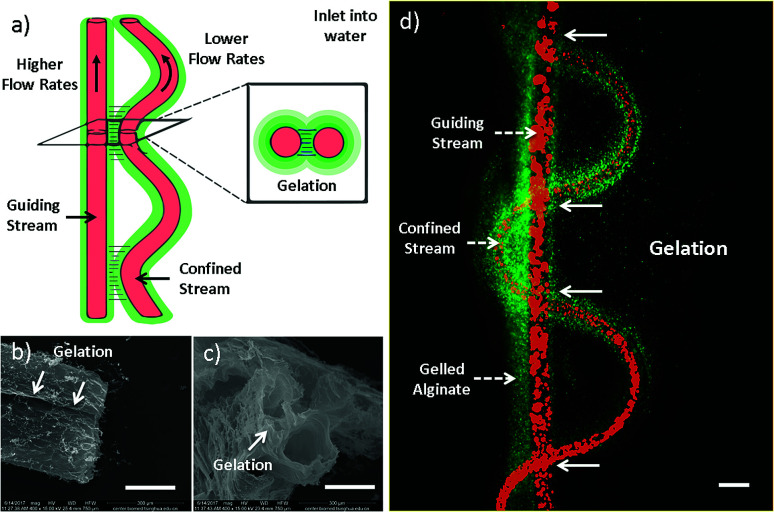
The formation mechanism has also been studied (Fig. 2). As mentioned earlier, inner stream tends to form straight channel when Qi/Qo is high, while helical channel is facilitated when Qi/Qo is low. The formation mechanism of helical channel can be explained by the hetero-generated liquid rope-coil effect.49 To be brief, alginate solution nearby inner channels reacts with Ca2+ firstly, forming a semi-crosslinked Ca-alginate layer with higher viscosity, which afterward causing the coiling phenomenon. However, in the case of sf-microfiber, the two streams are close with each other and thus fluid-flows are not independent (Fig. 2a). The thin layer of alginate sandwiched between the two streams is fully crosslinked owing to rapid and thoroughly Ca2+ diffusion (Fig. 2d). Therefore, the coiling trend is influenced by the nearby straight stream and the resultant flow pattern becomes a vibrating folded channel paralleling to a straight one. We define this mechanism as “gelation-induced guiding effect”. The straight and sine-wave channels serve as “guiding channel” and “confined channel” respectively. This explanation suits for the formation of dfFibers and three-folded fibers as well. To confirm this speculation, microfibers are injected into water instead of CaCl2 solution in order to diffuse uncrosslinked alginate (Fig. 2b and c). The gelation part is clearly demonstrated in the scanning electron microscope images.
Fig. 2. Mechanism of microfiber generation with sine-wave channel. (a) Scheme of the mechanism of microfiber generation with sine-wave channel. Gelation happens primarily between where guiding and confined channels are closer. (b and c) Side-view and cross-section view of double straight channels taken by SEM. (d) Figure of sfFiber inlet into water. The scale bar of (b–d) are 200 μm.
3.3. Intestine construct fabricated using microfibers
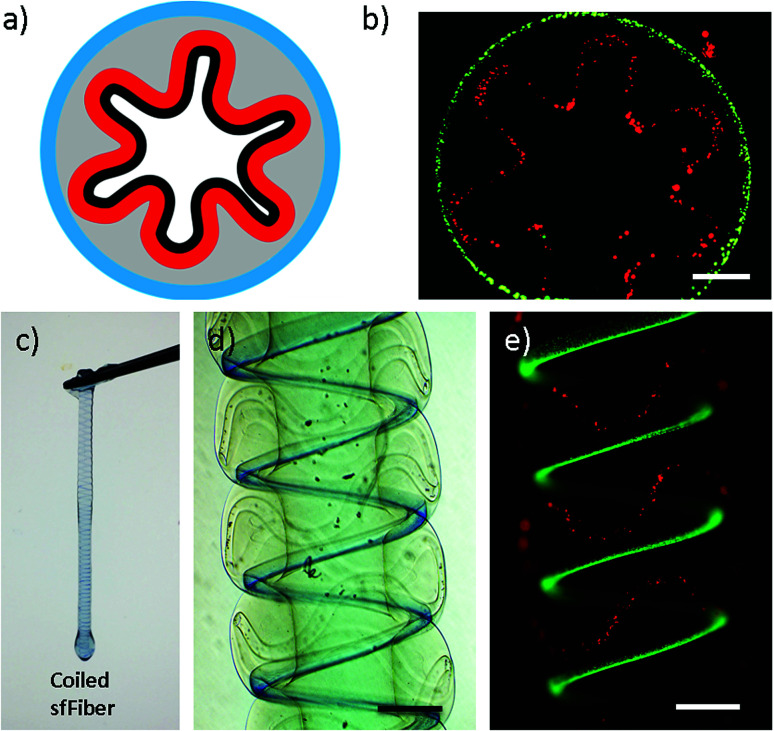
The new features of microfibers enable their potential in fabricating tissue constructs with folded morphologies. Folding morphologies can be found commonly in tissues, such as small intestine. Coiling sfFibers with folded channels inward, small intestine constructs are fabricated in vitro (Fig. 3). Side and cross-section view of intestine constructs are presented in Fig. 3d, e and b respectively. Immersed in CaCl2 solution to crosslink microfibers, a complete intestine construct is built and the ability of being raised by tweezers indicates enough strength for manipulation, as shown in Fig. 3c. This may provide a simple intestine tissue construct for tissue engineering.
Fig. 3. Intestine construct fabricated using sfFiber. (a) Scheme of the cross-section view of small intestine. (b) The sfFiber coiled in a circle. (c) Coiled sfFiber to generate tough hydrogel tube. (d and e) Picture of coiled sfFiber in bright field and fluorescence field. Scale bar: 500 μm.
3.4. Skeletal muscle constructs fabricated using microfibers
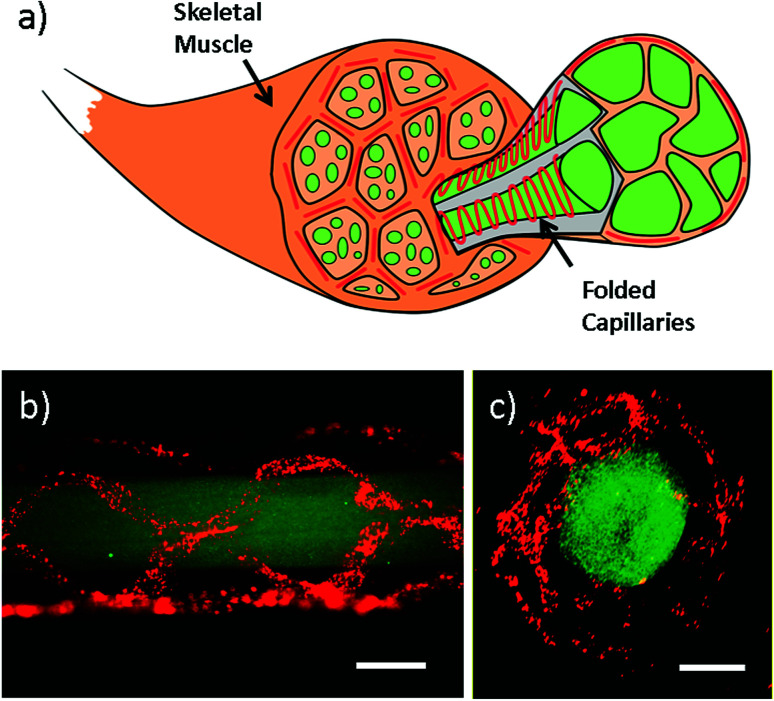
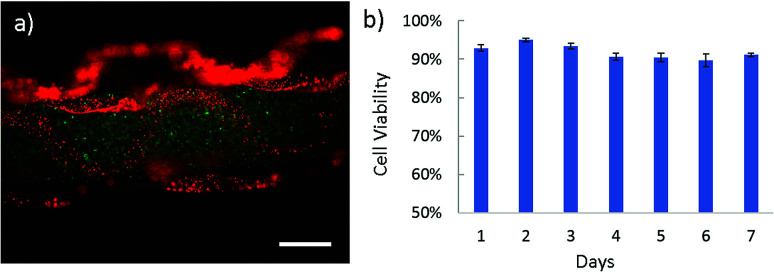
Apart from intestine constructs, human skeletal muscle fiber is constructed (Fig. 4 and Video S4 in ESI†). In contracted state, muscle fibers are positioned into the polygonal regions encircled by convoluted capillaries and anastomotic vessels. Using our microfiber-based strategy, capillaries between fascicles are structurally simulated by folded channels. Thus a muscle fascicle unit is established by embedding one straight microfiber into several surrounded dfFibers (Fig. 4b and c). In addition, human skeletal muscle cells (HSMCs) are cultured in the fascicle unit in order to test the biocompatibility (Fig. 5a). Live/dead staining results reveal that high viability (over 90%) still remains after 7 days culture indicating that this construct possesses the possibility for tissue engineering (Fig. 5b).
Fig. 4. Skeletal muscle constructs fabricated using dfFiber. (a) Scheme of skeletal muscle constructs. Skeletal muscle fibers are surrounded by sine-wave blood capillaries. (b and c) Side view and cross-section of skeletal muscle constructs marked by green and red fluorescent microspheres. Scale bar: 500 μm.
Fig. 5. The biocompatibility of skeletal muscle constructs fabricated using dfFiber. (a) Picture of HSMCs embedded in microfiber, which are marked by CellTracker dye as green color. (b) HSMC cell viability collected every single day for 7 days. Scale bar: 500 μm.
3.5. Properties of microfibers with folded channels
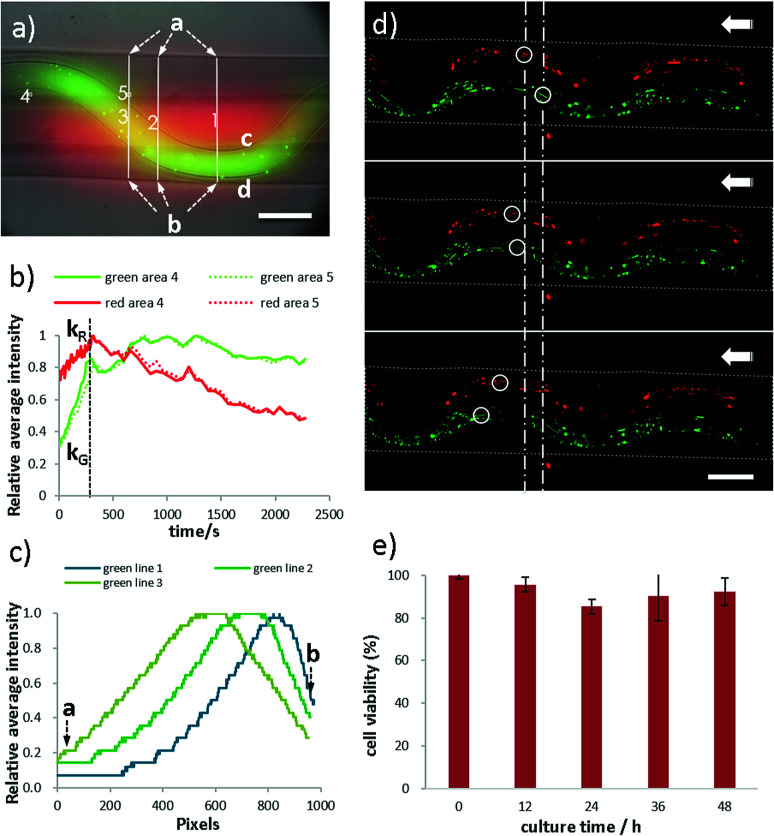
Moreover, properties of perfusability, permeability, biocompatibility and weavability of the microfiber have been investigated (Fig. 6). In the permeability test, FITC-dextran (green, 70 kDa), TRITC-dextran (red, 70 kDa) are dissolved in CaCl2 and perfused in the sfFiber simultaneously (Fig. 6a). The fluorescent intensity of two square areas with same distance to the two inner channels and three lines across the microfiber is measured. The relationship between relative fluorescent intensity and time is depicted in Fig. 6b. The fluorescent intensity increases rapidly in the beginning and then slowly decreases. The diffuse rate of the folded channel is calculated to be 2.83 times greater than the straight channel (ESI†), presenting faster material exchange capacity. The possible explanation of this phenomenon is that the channel with folded morphology has a higher surface area than straight channel in same length of microfiber. An asymmetric concentration distribution is observed by folded channel in microfiber due to its asymmetric position. The c side has the same concentration gradient, while the d side differs about 2.71 times (Fig. 6c, ESI†). This phenomenon could be explained that the fluorescent molecules diffuse faster in outer solution than in microfibers. The distance between folded channel and the edge of microfiber is near at line 1 position and far at line 3 position from d side, thus the different concentration gradient is observed.
Fig. 6. Properties of microfibers with folded channels. (a) HUVEC cell viability in sfFiber. (b–d) Permeability of sfFiber. (b) The relationship between relative average intensity to diffusion time in area 4 and area 5. (c) The fluorescence intensity distribution of diffused fluorescent dye in line 1–3 (red in straight channel and green in folded channel) at the time of 10 seconds. (d) Perfusability of dfFiber. Green and red microparticles flow leftwards and their flow rates are independent. (e) The cell viability of HUVEC in sfFiber. Scale bar: 200 μm.
Furthermore, perfusability of microfibers is studied using dfFiber (Fig. 6d and Video S5† in ESI†). Green and red fluorescence particles are perfused through two channels separately, and their flow rates can be independent. Moreover, the biocompatibility has also been studied (Fig. 6e). Human umbilical vein endothelial cells (HUVECs) are embedded in microfibers and cultured at 37 °C and viability data are collected every 12 hours. As shown in Fig. 6e, the viability of embedded HUVECs is 92% after 48 hours culture, indicating that the proposed method holds great biocompatibility. In addition, weavability of microfibers with folded channels is explored by knotting (Fig. S2, ESI†). Moreover, microfibers with folded channels may provide platforms for pathological study. For instance, the physiological environment of blood vessel, such as vessel tortuosity, blood pressure, viscosity and shear force, plays a vital role in the pathology of atherosclerosis. Despite tremendous efforts have been paid, the interplay between them remains unclear.54 The models proposed in this work can be used as platforms for the evaluation of endothelial cell responses to atherogenic factors, including ROS and lipoprotein permeability, under specific blood and vessel tortuosity, which benefits further researches on the prevention of atherosclerosis. This model may also be used in fields of drug screening and pathophysiology, exploring the difference of drug diffusion, materials exchange and energy supplement between normal and tortuous tissues in future studies.
4. Conclusions
In summary, we propose a microfluidics-based method to fabricate microfibers with folded hollow channels, which can be used as building blocks for tissue constructs in vitro. By adjusting the flow rates, microfibers with various channel morphologies including double-folded, folded-straight and double-helical are produced. The gelation-induced guiding effect is used to understand the intrinsic mechanism. The proof-of-concept in vitro intestine and skeletal muscle constructs are produced using the above-mentioned microfibers. Moreover, characteristics of perfusability, permeability and asymmetric molecular distributions in microfibers make it possible to be used as platforms for the study of nutrient exchange and energy supplement between normal and tortuous tissues.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the Ministry of Science and Technology (Nos. 2017YFC0906902 and 2017ZX09301032) and National Natural Science Foundation of China (No. 21621003).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04192j
References
- Petersen T. H. Calle E. A. Zhao L. Lee E. J. Gui L. Raredon M. B. Gavrilov K. Yi T. Zhuang Z. W. Breuer C. Herzog E. Niklason L. E. Science. 2010;329:538. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby B. Science. 2012;338:921. doi: 10.1126/science.1226340. [DOI] [PubMed] [Google Scholar]
- Badylak S. F. Weiss D. J. Caplan A. Macchiarini P. Lancet. 2012;379:943. doi: 10.1016/S0140-6736(12)60073-7. [DOI] [PubMed] [Google Scholar]
- Loessner D. Stok K. S. Lutolf M. P. Hutmacher D. W. Clements J. A. Rizzi S. C. Biomaterials. 2010;31:8494. doi: 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- Murphy S. V. Atala A. Nat. Biotechnol. 2014;32:773. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Pati F. Gantelius J. Svahn H. A. Angew. Chem., Int. Ed. 2016;55:4650. doi: 10.1002/anie.201505062. [DOI] [PubMed] [Google Scholar]
- Zorlutuna P. Annabi N. Unal G. C. Nikkhah M. Cha J. M. Nichol J. W. Manbachi A. Bae H. Chen S. Khademhosseini A. Adv. Mater. 2012;24:1782. doi: 10.1002/adma.201104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter R. F. Groom A. C. Microvasc. Res. 1983;25:68. doi: 10.1016/0026-2862(83)90044-4. [DOI] [PubMed] [Google Scholar]
- Tallinen T. Chung J. Y. Rousseau F. Girard N. Lefèvre J. Mahadevan L. Nat. Phys. 2016;12:588. [Google Scholar]
- Duyn J. H. Gelderen P. V. Li T. Q. Zwart J. A. Koretsky A. P. Fukunaga M. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11796. doi: 10.1073/pnas.0610821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savin T. Kurpios N. A. Shyer A. E. Florescu P. Liang H. Mahadevan L. Tabin C. J. Nature. 2011;476:57. doi: 10.1038/nature10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons B. D. Science. 2013;342:203. doi: 10.1126/science.1245288. [DOI] [PubMed] [Google Scholar]
- Shyer A. E. Tallinen T. Nerurkar N. L. Wei Z. Gil E. S. Kaplan D. L. Tabin C. J. Mahadevan L. Science. 2013;342:212. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A. Eidelman D. H. Hogg J. C. James A. L. Lambert R. K. Ludwig M. S. Martin J. McDonald D. M. Mitzner W. A. Okazawa M. Pack R. J. Pare P. D. Schellenburg R. R. Tiddens H. A. W. M. Wagner E. M. Yager D. J. Appl. Physiol. 1994;77:1011. doi: 10.1152/jappl.1994.77.2.1011. [DOI] [PubMed] [Google Scholar]
- Wiggs B. R. Hrousis C. A. Drazen J. M. Kamm R. D. J. Appl. Phys. 1997;83:1814. doi: 10.1152/jappl.1997.83.6.1814. [DOI] [PubMed] [Google Scholar]
- Nakamura T. Lozano P. R. Ikeda Y. Iwanaga Y. Hinek A. Minamisawa S. Cheng C. F. Kobuke K. Dalton N. Takada Y. Tashirok K. J. Ross Jr Honjo T. Chien K. R. Nature. 2002;415:171. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- Coucke P. J. Willaert A. Wessels M. W. Callewaert B. Zoppi N. Backer J. D. Fox J. E. Mancini G. M. S. Kambouris M. Gardella R. Facchetti F. Willems P. J. Forsyth R. Dietz H. C. Barlati S. Colombi M. Loeys B. Paepe A. D. Nat. Genet. 2006;38:452. doi: 10.1038/ng1764. [DOI] [PubMed] [Google Scholar]
- Zheng Y. Chen J. Lopez J. A. Nat. Commun. 2015;6:7858. doi: 10.1038/ncomms8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H. C. J. Vasc. Res. 2012;49:185. doi: 10.1159/000335123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S. Duda D. G. Xu L. Munn L. L. Boucher Y. Fukumura D. Jain R. K. Physiol. Rev. 2011;91:1071. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avolio A. Jones D. Shadpour M. T. Hypertension. 1998;32:170. doi: 10.1161/01.HYP.32.1.170. [DOI] [PubMed] [Google Scholar]
- Elis S. G. Vandormael M. G. Cowley M. J. DiSciascio G. Deligonul U. Topol E. J. Bulle T. M. Circulation. 1990;82:1193. doi: 10.1161/01.CIR.82.4.1193. [DOI] [PubMed] [Google Scholar]
- Baluk P. Hashizume H. McDonald D. M. Curr. Opin. Genet. Dev. 2005;15:102. doi: 10.1016/j.gde.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Lee A. Y. Han B. Lamm S. D. Fierro C. A. Han H. C. Am. J. Physiol.: Heart Circ. Physiol. 2012;302:H873. doi: 10.1152/ajpheart.00463.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. S. Stevens K. R. Yang M. T. Baker B. M. Nguyen D. T. Cohen D. M. Toro E. Chen A. A. Galie P. A. Yu X. Chaturvedi R. Bhatia S. N. Chen C. S. Nat. Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Zhang F. Yan Z. Ma Q. Li X. Huang Y. Rogers J. A. Nat. Rev. Mater. 2017;2:17019. doi: 10.1038/natrevmats.2017.19. [DOI] [Google Scholar]
- Xu S. Yan Z. Jang K. I. Huang W. Fu H. Kim J. Wei Z. Flavin M. McCracken J. Wang R. Badea A. Liu Y. Xiao D. Zhou G. Lee J. Chung H. U. Cheng H. Ren W. Banks A. Li X. Paik U. Nuzzo R. G. Huang Y. Zhang Y. Rogers J. A. Science. 2015;347:154. doi: 10.1126/science.1260960. [DOI] [PubMed] [Google Scholar]
- Chan H. N. Chen Y. Shu Y. Chen Y. Tian Q. Wu H. Microfluid. Nanofluid. 2015;19:9. doi: 10.1007/s10404-014-1542-4. [DOI] [Google Scholar]
- Song H. Bringer M. R. Tice J. D. Gerdts C. J. Ismagilova R. F. Appl. Phys. Lett. 2003;83:4664. doi: 10.1063/1.1630378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. Tice J. D. Ismagilov R. F. Angew. Chem., Int. Ed. 2003;115:792. doi: 10.1002/ange.200390172. [DOI] [PubMed] [Google Scholar]
- He H. Wang L. Yan X. Zhang L. Yu M. Yu G. Dong R. Xia L. Ramakrishna S. Long Y. RSC Adv. 2016;6:29423. doi: 10.1039/C6RA04566A. [DOI] [Google Scholar]
- He X. Wang W. Deng K. Xie R. Ju X. Liu Z. Chu L. RSC Adv. 2015;5:928. doi: 10.1039/C4RA10696B. [DOI] [Google Scholar]
- Mandrycky C. Wang Z. Kim K. Kim D. Biotechnol. Adv. 2016;34:422. doi: 10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertassoni L. E. Cecconi M. Manoharan V. Nikkhah M. Hjortnaes J. Cristino A. L. Barabaschi G. Demarchi D. Dokmeci M. R. Yange Y. Khademhosseini A. Lab Chip. 2014;14:2202. doi: 10.1039/C4LC00030G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. Jun Y. Qin J. Lee S. Biomaterials. 2017;114:121. doi: 10.1016/j.biomaterials.2016.10.040. [DOI] [PubMed] [Google Scholar]
- Bosworth L. A. Turner L. Cartmell S. H. Nanomedicine. 2013;9:322. doi: 10.1016/j.nano.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Chang G. Shen J. Macromol. Mater. Eng. 2011;296:1071. doi: 10.1002/mame.201100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- France K. Xu F. Hoare T. Adv. Healthcare Mater. 2018;7:1700927. doi: 10.1002/adhm.201700927. [DOI] [PubMed] [Google Scholar]
- Wilson M. E. Kota N. Kim Y. Wang Y. Stolz D. B. LeDuc P. R. Ozdoganlar O. B. Lab Chip. 2011;11:1550. doi: 10.1039/C0LC00561D. [DOI] [PubMed] [Google Scholar]
- Onoe H. Okitsu T. Itou A. Kato-Negishi M. Gojo R. Kiriya D. Sato K. Miura S. Iwanaga S. Kuribayashi-Shigetomi K. Matsunaga Y. T. Shimoyama Y. Takeuchi S. Nat. Mater. 2013;12:584. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- Liang Z. Liu C. Li L. Xu P. Luo G. Ding M. Liang Q. Sci. Rep. 2016;6:33462. doi: 10.1038/srep33462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. Fu F. Shang L. Cheng Y. Gu Z. Zhao Y. Adv. Mater. 2017;29:1605765. doi: 10.1002/adma.201605765. [DOI] [PubMed] [Google Scholar]
- Tottoriab S. Takeuchi S. RSC Adv. 2015;5:33691. doi: 10.1039/C5RA01037C. [DOI] [Google Scholar]
- Shi X. Ostrovidov S. Zhao Y. Liang X. Kasuya M. Kurihara K. Nakajima K. Bae H. Wu H. Khademhosseini A. Adv. Funct. Mater. 2015;25:2250. doi: 10.1002/adfm.201404531. [DOI] [Google Scholar]
- Yu Y. Wei W. Wang Y. Xu C. Guo Y. Qin J. Adv. Mater. 2016;28:6649. doi: 10.1002/adma.201601504. [DOI] [PubMed] [Google Scholar]
- Ji X. Guo S. Zeng C. Wang C. Zhang L. RSC Adv. 2015;5:2517. doi: 10.1039/C4RA10389K. [DOI] [Google Scholar]
- Nunes J. K. Constantin H. Stone H. A. Soft Matter. 2013;9:4227. doi: 10.1039/C3SM27579E. [DOI] [Google Scholar]
- Lee K. H. Shin S. J. Park Y. Lee S. H. Small. 2009;5:1264. doi: 10.1002/smll.200801667. [DOI] [PubMed] [Google Scholar]
- Xu P. Xie R. Liu Y. Luo G. Ding M. Liang Q. Adv. Mater. 2017;29:2701664. doi: 10.1002/adma.201701664. [DOI] [PubMed] [Google Scholar]
- Xie R. Xu P. Liu Y. Li L. Luo G. Ding M. Liang Q. Adv. Mater. 2018;30:1705082. doi: 10.1002/adma.201705082. [DOI] [PubMed] [Google Scholar]
- Colosi C. Shin S. R. Manoharan V. Massa S. Costantini M. Barbetta A. Dokmeci M. R. Dentini M. Khademhosseini A. Adv. Mater. 2016;28:677. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. Zheng F. Lu J. Shang L. Xie Z. Zhao Y. Chen Y. Gu Z. Adv. Mater. 2014;26:5184. doi: 10.1002/adma.201400798. [DOI] [PubMed] [Google Scholar]
- Nunes J. K. Wu C. Y. Amini H. Owsley K. Carlo D. D. Stone H. A. Adv. Mater. 2014;26:3712. doi: 10.1002/adma.201400268. [DOI] [PubMed] [Google Scholar]
- Zheng W. Huang R. Jiang B. Zhao Y. Zhang W. Jiang X. Small. 2016;12:1961. doi: 10.1002/smll.201670073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.