Abstract
A novel tetracycline resistance gene, designated tet(32), which confers a high level of tetracycline resistance, was identified in the Clostridium-related human colonic anaerobe K10, which also carries tet(W). tet(32) was transmissible in vitro to the rumen anaerobe Butyrivibrio fibrisolvens 2221R. The predicted gene product of tet(32) has 76% amino acid identity with Tet(O). PCR amplification indicated that tet(32) is widely distributed in the ovine rumen and in porcine feces.
The widespread use of antibiotics has resulted in the emergence of antibiotic resistance in both human and veterinary pathogens, and resistance mechanisms exist for all antibiotics currently in clinical use (1). Most resistance genes have been isolated from pathogenic bacteria or from antibiotic producers, and there has been comparatively little research on antibiotic resistance in the commensal flora of the human or animal gut. The dominant microorganisms in gut ecosystems are obligate anaerobes, specifically low-G+C-content gram-positive bacteria, bifidobacteria, and members of the gram-negative Cytophaga-Flavobacter-Bacteroides phylum (11, 16). It is not clear how much genetic exchange occurs between these obligate anaerobes and the smaller populations of facultative anaerobes that include major pathogenic species.
A new ribosome protection (RP) tetracycline resistance (Tcr) gene, tet(W) (GenBank accession no. AJ222769), identified recently in a range of rumen anaerobes from geographically distant locations (3), was transferable in vitro between strains of Butyrivibrio fibrisolvens (13). tet(W) was also identified in a Clostridium-related human fecal anaerobe, K10, and in Bifidobacterium longum isolates (14). We report here a second novel transmissible Tcr gene, designated tet(32) (8), in the commensal anaerobe K10.
The anaerobic bacterial strains B. fibrisolvens 2221R, rifampin-resistant strain 2221 (13), and K10 were cultured in M2GSC broths (10). Transfer of Tcr was investigated in anaerobic filter matings (13) in the absence of tetracycline. Transconjugants were selected on M2GSC plates containing 10 μg of tetracycline/ml and 100 μg of rifampin/ml, incubated for 2 days. The level of Tcr was tested using 16-h cultures to inoculate fresh M2GSC broths containing various tetracycline concentrations. Minimum inhibitory tetracycline concentrations, inhibiting 90% of bacterial growth (MIC90), were estimated and confirmed in broth cultures.
DNA was extracted from overnight cultures using either the Wizard Genomic purification kit or the Wizard Plasmid purification kit (Promega, Southampton, United Kingdom). DNA for genomic sequencing was extracted using the QIAGEN Genomic DNA Buffer set and 100/G tips, and for purification of total DNA from fecal and rumen fluid samples, the QIAamp DNA Stool mini-kit (Qiagen, Crawley, United Kingdom) was used. Restriction digestion, Southern blotting, and hybridization of genomic DNA followed standard procedures.
PCR amplification was done using various primer combinations: tetWfor and Tet2, specific for tet(W) (14); degenerate primers Tet1 and Tet2, which recognize all known RP-type genes (3); or primers specific for tet(32): Tet(32)For (5′ GAACCAGATGCTGCTCTT 3′) and Tet(32)Rev (5′ CATAGCCACGCCCACATGAT 3′). Optimizing the annealing temperature of the latter amplification to 57°C resulted in no amplification of the related RP genes, tet(M), tet(O), tet(Q), or tet(W). PCR products were sequenced using a Taq ABI PRISM kit (Perkin-Elmer, Warrington, United Kingdom), and for direct genomic sequencing (7), the Thermofidelase I enzyme (Fidelity Systems Inc.) was utilized. Sequences were separated on an ABI377 automated sequencer. Sequences were assembled using UWGCG software (6), which is available through the HGMP facility (Human Genome Mapping Project, Cambridge, United Kingdom).
The tet(32) gene was identified during investigations into the transmission of tet(W) from the human isolate K10 to a Rifr mutant of the rumen anaerobe B. fibrisolvens 2221R. Tcr transferred at frequencies of 10−4 per donor cell. Surprisingly, although the donor K10 gave the expected tet(W) PCR product of 1.8 kb, the transconjugants did not. However, both donor and transconjugants gave the expected 1.3-kb product in PCR amplifications using degenerate RP primers. We concluded that the transferable Tcr gene in K10 was not tet(W) but was a second Tcr gene, tet(32).
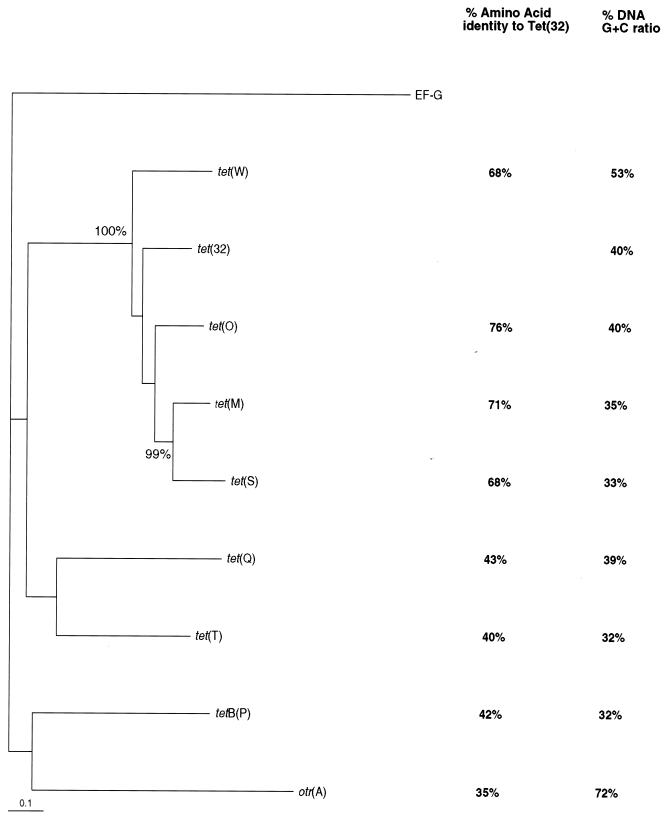
The complete sequence of tet(32) contains an ORF whose product of 594 amino acids has 76% identity to Tet(O), 71% to Tet(M), and 68% to Tet(W) and Tet(S) (Fig. 1). tet(32) has a G+C content of only 40%, which is considerably lower than that of tet(W) (53%) but is similar to that of tet(M) (35%) and tet(O) (40%). The sequence upstream of the tet(32) start codon contains a GGAGG ribosome binding site (+7 nt) and two sets of inverted repeats forming secondary stem-loop structures with ΔG values of −20.6 and −14.0 kcal (17). The sequence of this upstream region is virtually identical to that of Campylobacter jejuni tet(O) (GenBank accession no. M18896; 4 in 150 nt differences), including the transcription initiation sites and promoter regions (18).
FIG. 1.
Phylogenetic tree showing the evolutionary relationships of RP-type Tcr proteins. The amino acid sequence of the Aquifex aeolicus fusA gene (accession no. AE000657) for translation factor EF-G was used to root the tree. GenBank accession numbers are as follows: Tet(O), Y07780; Tet(M), U58986; Tet(S), X92946; Tet(W), AJ222769; Tet(Q), X58717; Tet(T), L42544; TetB(P), L20800; and Otr(A), X53401. Figures beside nodes indicate bootstrap values when greater than 95% (based on 500 trials). Percent G+C content and percent amino acid sequence identity for each sequence relative to tet(32) are indicated.
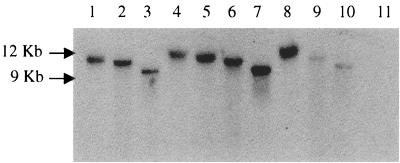
Additional plasmid DNA was not detected in transconjugants, and hybridization of genomic DNA to tet(32) identified bands from 9 to 12 kb in size (Fig. 2), implying that tet(32) is chromosomally encoded. Hybridization of the same 12-kb SmaI fragment in the donor and transconjugants implicated an element of at least this size in tet(32) transfer. There was no cross-hybridization to a B. fibrisolvens 2221R transconjugant containing tet(W) (3). Transconjugant DNA failed to hybridize to probes specific to regions of conjugative transposons Tn916 (15) or TnB1230 (13), indicating that tet(32) transfer does not involve similar mobile elements.
FIG. 2.
Southern blot of genomic DNA from B. fibrisolvens 2221R transconjugants Tcm1 (lanes 1 to 4) and Tcm8 (lanes 5 to 8) digested with BamHI, EcoRI, HindIII, or SmaI, in that order, and hybridized to a PCR probe specific to tet(32). Controls of the tet(32) parent strain K10 digested with BamHI (lane 9) and EcoRI (lane 10) and a representative B. fibrisolvens 2221R transconjugant containing only tet(W) (lane 11) are included.
The presence of tet(32) in other gut environments was tested by PCR amplification of total bacterial DNA using specific tet(32) primers. Six out of 9 rumen samples from cannulated sheep and 8 out of 11 pig fecal samples, all from different animals, gave PCR products. Selected products were sequenced and confirmed to be tet(32), suggesting that tet(32) is abundant in farm animals. There were 25 in 584 nt differences between the porcine amplicon sequences and the K10 tet(32) gene, corresponding to a 4% sequence divergence.
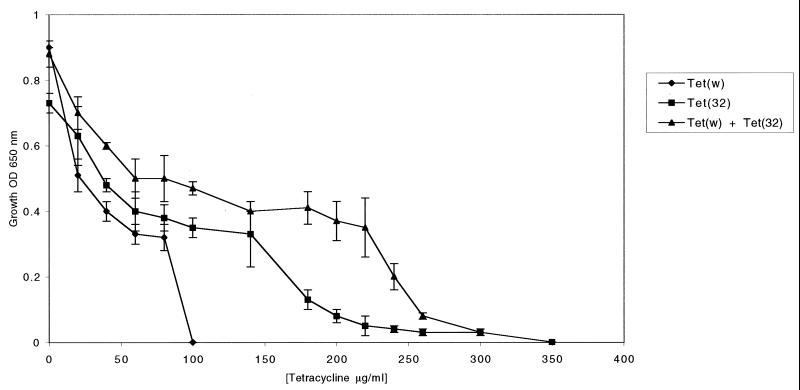
The resistance profiles of B. fibrisolvens 2221R transconjugants expressing Tet(W) or Tet(32) were compared. Tetracycline concentrations above 20 μg/ml gave a progressive reduction in growth (Fig. 3). Broth cultures based on growth data (Fig. 3) confirmed that the MIC90 of tetracycline for bacteria expressing Tet(W) was much lower (90 μg/ml) than that for bacteria expressing Tet(32) (200 μg/ml). For strain K10, which encodes both genes, the MIC90 of tetracycline was even higher (270 μg/ml). As with all RP proteins, Tet(32) also confers resistance to minocycline (10 μg/ml).
FIG. 3.
Levels of tetracycline resistance expressed by Tet(W) and Tet(32) individually following transfer into the host strain B. fibrisolvens 2221R and by Tet(W) and Tet(32) together in the bacterial host K10. Bacteria were grown anaerobically for 16 h in M2GSC broth at 37°C in the concentrations of tetracycline shown. The graph was used to estimate the MIC90 for each strain, which was then confirmed in broth culture. OD 600 nm, optical density at 600 nm.
The identification of a second novel Tcr gene, tet(32), from an anaerobic commensal gut bacterium following the identification of tet(W) (3, 14) demonstrates that the gut microflora harbors novel antibiotic resistance genes. It is important to determine the distribution of these novel resistance genes, in both obligate and facultative anaerobes from different gut and nongut habitats. We have shown that tet(32) is present among the gut microflora of human, ruminant, and porcine hosts and we know that tet(W) has a similar distribution (2, 14). These novel genes could contribute significantly to tetracycline resistance among clinical pathogens where specific resistance genes are currently unidentified (12).
This work provides a third case in which two RP-type Tcr genes are present in the same bacterium. B. fibrisolvens 1.230 carries a mobile tet(W) gene and a nonmobile tet(O) gene (3); strain K10 carries a mobile tet(32) gene and a nonmobile tet(W) gene; Streptococcus pneumoniae contains both tet(M) and tet(O) genes (9). This phenomenon could result from intense selection pressure during the evolution of tetracycline resistance and may contribute to higher resistance levels.
The mechanism of transfer of tet(32) from strain K10 is unknown but may involve a mobile chromosomal element, as shown for many RP-type Tcr genes, including tet(W) (3), tet(Q), and tet(M) (12). Although it was first identified as Fusobacterium prausnitzii, full-length 16S ribosomal DNA sequencing shows that K10 has less than 95% identity with known species but belongs to the Cluster XIVa Clostridium subphylum of low-G+C-content gram-positive bacteria (5). This bacterial cluster includes B. fibrisolvens and many other abundant colonizers of the human colon and rumen (4, 19).
This work provides the first direct experimental evidence that genetic exchange can occur between gram-positive obligate anaerobes from the human colon and those from the rumen. Evidence for transfer between commensal anaerobes and pathogenic gut bacteria is limited, but identical tet(O) genes are present in the rumen anaerobe B. fibrisolvens and the human pathogen S. pneumoniae (3). The species distribution and sequence diversity of the novel tet(32) and tet(W) genes, and of tet(O), should contribute significantly to our understanding of gene flow within and between gut microbial communities.
Nucleotide sequence accession number.
The complete sequence of the tet(32) gene was submitted to the GenBank database (accession no. AJ295238).
Acknowledgments
We thank SEERAD (Scottish Executive Environmental Rural Affairs Department) and FSA (Food Standards Agency) for their financial support.
We thank Pauline Young for the automated sequencing.
REFERENCES
- 1.Aarestrup F M. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int J Antimicrob Agents. 1999;12:279–285. doi: 10.1016/s0924-8579(99)90059-6. [DOI] [PubMed] [Google Scholar]
- 2.Aminov R I, Garrigues-Jeanjean N, Mackie R I. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Antimicrob Agents Chemother. 2001;67:22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa T M, Scott K P, Flint H J. Evidence for recent intergeneric transfer of a new tetracycline resistance gene tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ Microbiol. 1999;1:53–64. doi: 10.1046/j.1462-2920.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 4.Barcenilla A, Pryde S E, Martin J C, Duncan S H, Stewart C S, Henderson C, Flint H J. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiner C R, Hunkapiller K L, Chen S M, Glass J I, Chen E Y. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 1998;8:557–561. doi: 10.1101/gr.8.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy S B, McMurry L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luna V A, Roberts M C. The presence of the tet(O) gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. Int J Antimicrob Chemother. 1998;42:613–619. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki K, Martin J C, Marinsek-Logar R, Flint H J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 11.Pryde S E, Richardson A J, Stewart C S, Flint H J. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl Environ Microbiol. 1999;65:5372–5377. doi: 10.1128/aem.65.12.5372-5377.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts M C. Tetracycline resistance determinants: mechanism of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 13.Scott K P, Barbosa T M, Forbes K J, Flint H J. High frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1997;63:3405–3411. doi: 10.1128/aem.63.9.3405-3411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott K P, Melville C M, Barbosa T M, Flint H J. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–777. doi: 10.1128/aac.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senghas E, Jones J M, Jamamoto M, Gawron-Burke C, Clewell D B. Genetic organisation of the bacterial conjugative transposon Tn916. J Bacteriol. 1988;170:245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suau A, Bonnet R, Sutren M, Godon J, Gibson G, Collins M, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;66:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinoco I, Borer P N, Dengler B, Levine M D. Improved estimation of secondary structure in ribonucleic acids. Nature. 1973;14:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Taylor D E. A DNA sequence upstream of the tet(O) gene is required for full expression of tetracycline resistance. Antimicrob Agents Chemother. 1991;35:2020–2025. doi: 10.1128/aac.35.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willems A, Amat-Marco M, Collins M D. Phylogenetic analysis of Butyrivibrio strains reveals three distinct groups of species within the Clostridium subphylum of the Gram-positive bacteria. Int J Syst Bacteriol. 1996;46:195–199. doi: 10.1099/00207713-46-1-195. [DOI] [PubMed] [Google Scholar]