Abstract
Bacteria possess the ability to evolve varied and ingenious strategies to outwit the host immune system, instigating an evolutionary arms race. Proteases are amongst the many weapons employed by bacteria, which specifically cleave and neutralize key signalling molecules required for a coordinated immune response. In this article, we focus on a family of S8 subtilisin-like serine proteases expressed as cell-envelope proteases (CEPs) by group A and group B streptococci. Two of these proteases known as Streptococcus pyogenes CEP (SpyCEP) and C5a peptidase cleave the chemokine CXCL8 and the complement fragment C5a, respectively. Both CXCL8 and C5a are potent neutrophil-recruiting chemokines, and by neutralizing their activity, streptococci evade a key defence mechanism of innate immunity. We review the mechanisms by which CXCL8 and C5a recruit neutrophils and the characterization of SpyCEP and C5a peptidase, including both in vitro and in vivo studies. Recently described structural insights into the function of this CEP family are also discussed. We conclude by examining the progress of prototypic vaccines incorporating SpyCEP and C5a peptidase in their preparation. Since streptococci-producing SpyCEP and C5a peptidase are responsible for a considerable global disease burden, targeting these proteases by vaccination strategies or by small-molecule antagonists should provide protection from and promote the resolution of streptococcal infections.
Keywords: Bacterial infection, Cell-envelope protease, Chemoattractants, Complement system, Streptococcus
Introduction
Successful host colonization by bacteria is due in part to the evolution of ingenious strategies to evade the innate immune response. If left unchecked, this can result in the initiation of systemic and severe infections. One common strategy amongst diverse bacterial pathogens is the production and secretion of proteases that degrade key components of the immune system. By degrading molecules pivotal to the host response, bacteria are able to evade detection and clearance with a spectrum of clinical consequences [1]. In this article, we focus on a family of S8 subtilisin-like serine proteases expressed by the Gram-positive bacteria Streptococcus pyogenes (group A streptococcus [GAS]) and Streptococcus agalactiae (group B streptococcus [GBS]) [2]. These proteases are expressed as cell-envelope proteases (CEPs) and selectively abrogate neutrophil recruitment by specifically degrading key chemotactic factors vital for the innate immune response. Severe GAS-related disease presents a considerable human global disease burden that is estimated to result in over 600,000 annual cases of invasive infection, including toxic shock and necrotizing fasciitis [3]. GBS is an emerging human and veterinary pathogen and is the leading cause of early-onset severe infant sepsis. The highly specific activity of this family of streptococcal proteases and their role in combating pathogen clearance underline the critical role that neutrophil chemotactic factors play in response to streptococcal infection.
Neutrophil Recruitment and Activation in the Innate Immune Response
Neutrophils play a major role in the early response of the innate immune system by neutralizing and phagocytosing bacteria. They are guided to sites of microbial infection by molecules known as chemoattractants, typically small proteins up to 10 kDa in size. Chemoattractants function by binding with high affinity to specific G protein-coupled receptors (GPCRs) on the neutrophil surface, coordinating the directional migration (chemotaxis) towards the source of attractant. Neutrophil chemoattractants can be released or secreted by cells in response to infection (e.g., chemokines) or generated by the cleavage of pre-existing soluble factors (e.g., complement fragments) [4]. Both categories of chemoattractants are substrates for the streptococcal CEPs.
Complement-Derived Neutrophil Chemoattractants
The complement pathway is a tightly regulated network of self-perpetuating proteolytic cascades that result in the clearance of pathogens by opsonization, anaphylatoxin release, and formation of the lytic membrane attack complex. Although the membrane attack complex is not known to play a role in the clearance of streptococci, anaphylatoxins C3a and C5a are critical neutrophil chemoattractants, while complement C3b-mediated opsonization is required for neutrophil phagocytosis of streptococci [5]. Both the anaphylatoxins and C3 act as substrates for CEPs. The complement cascade is activated by 3 routes: namely, the classical, lectin, and alternative pathways, which converge on the central complement component and α-macroglobulin family member, C3 [6]. C3 comprises an α chain (111 kDa) and a β chain (75 kDa), linked by multiple disulphide bonds. C3 is modified by the enzyme C3 convertase, which liberates the 9-kDa C3a fragment from the N-terminus of the C3 α-chain, shown in Figure 1. The remaining fragment of C3, known as C3b, contains the 102-kDa α-chain and the 75-kDa β-chain, and is the activated form of C3. The cleavage of C3 into C3b exposes a reactive thioester residue, permitting covalent deposition on the surface of the bacteria. C3b binds the pro-enzyme factor B and is cleaved by factor D to form the enzyme complex C3b2Bb, known as C5a convertase, which cleaves the complement component C5. Cleavage of C5 by C5a convertase releases the N-terminal 10.4-kDa component C5a in a similar manner to the liberation of C3a from C3. Generation of the C3 convertase is controlled by the regulatory protein factor I, a plasma protease that cleaves C3b into the inactive form iC3b. Unlike intact C3b, iC3b is unable to bind factor B and therefore amplify the complement cascade. Both C3b and iC3b function as opsonins, binding to 4 distinct complement receptors (CR1–CR4) expressed by neutrophils and monocytes. Binding to the receptor triggers the phagocytosis of pathogens. CR1 (CD35) binds C3b, while iC3b can bind to CR2 (CD21), CR3 (CD11b/CD18), and CR4 (CD11c/CD18).
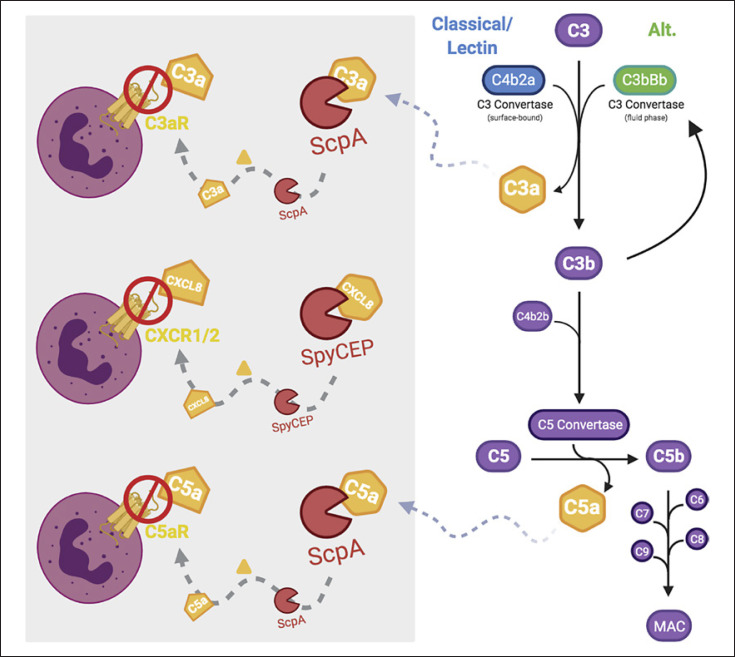
Fig. 1.
Generation of anaphylatoxins C3a and C5a and subsequent degradations by CEPs. A schematic presentation of the common terminal complement system pathway. The production of anaphylatoxins C3a and C5a, depicted in orange, by the C3 convertases and C5 convertase remains central to the pathways, leading to the formation of the MAC. Pictured in the grey box is the cleavage of chemoattractants by CEPs and the resultant inability to bind and activate their respective receptors. CEPs, cell-envelope proteases; MAC, membrane attack complex; SpyCEP, Streptococcus pyogenes CEP. Adapted from Monk et al. [7]. Created with BioRender.com.
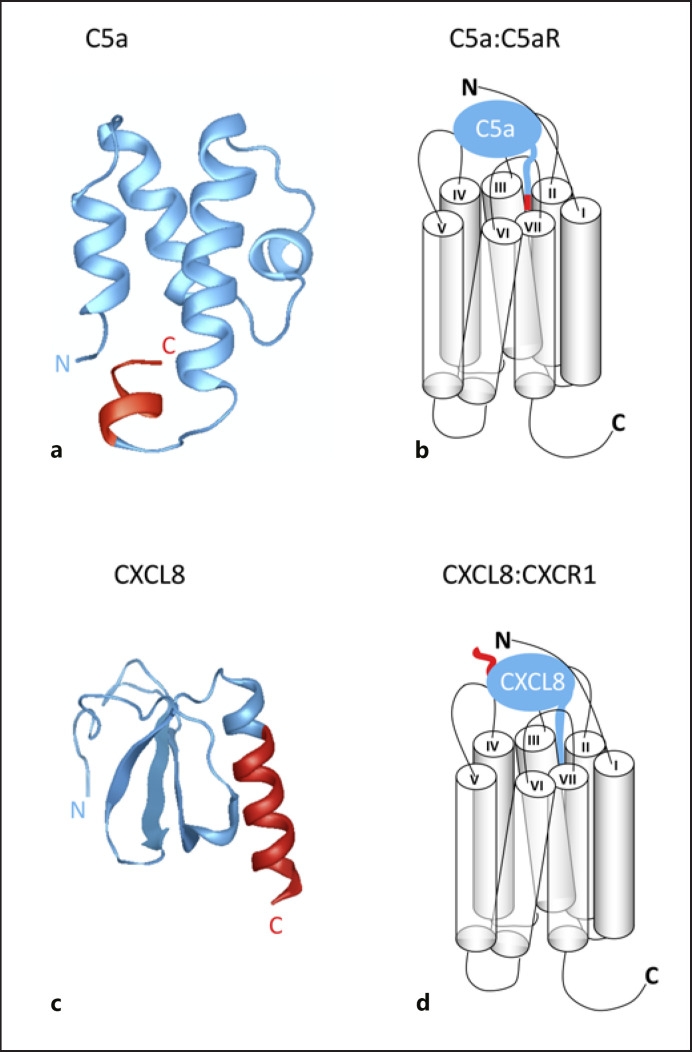
The structures of C3a and C5a anaphylatoxins have been solved by NMR [4] and comprise a 4-α-helical bundle, shown in Figure 2a. C3a and C5a function by binding and activating specific GPCRs known as the C3a receptor (C3aR) and the C5a receptor (C5aR) (reviewed in [7, 8]). Both ligands bind to their receptors in a two-step model in which the N-termini of the receptors tether the anaphylatoxins. This orientates them so that the C-terminal-most residue (arginine) is able to insert into the helical bundle of the GPCR and stabilize an active conformation. This results in the activation of G proteins, as shown in Figure 2b. A recent crystal structure of the C5aR in complex with the small-molecule antagonist PMX53 [9] validated earlier mutagenesis and modelling efforts, suggesting that the C-terminal R75 of C5a contacts D282 of C5aR [10, 11]. It will be seen that the C-termini of both C3a and C5a, which are targets of the streptococcal CEPs, are critical for anaphylatoxin function.
Fig. 2.
C5a and CXCL8 activate their receptors via distinct two-step models. a Cartoon of C5a derived from the PDB file 1KJS with N- and C-termini denoted. b Schematic diagram highlighting how the C-terminal portion of C5a inserts into the C5aR helical bundle to induce signalling. c Cartoon of CXCL8 derived from the PDB file 1IKL with N- and C-termini denoted. d Schematic diagram highlighting how the N-terminal portion of CXCL8 inserts into the C5aR helical bundle to induce signalling. In all panels, the portion of the chemoattractant coloured red marks the piece cleaved from the main body of the chemoattractant (blue) by the CEPs. CEPs, cell-envelope proteases. Generated with Protean 3DTM version 17.0.1, DNASTAR, Madison, WI.
Neutrophil-Specific Chemokines
The major neutrophil-recruiting chemokines are a subset of the CXC chemokine family possessing an N-terminal glutamate-leucine-arginine (ELR) motif, termed ELR+ chemokines [12]. These are exemplified by CXCL8/interleukin-8, the first chemokine to have its structure solved in 1989 [13]. All chemokines contain a conserved tertiary structure comprising a disordered N-terminal signalling domain and a structured core domain consisting of an N-loop, which contains a 3-stranded antiparallel β-sheet, and a C-terminal helix as shown in Figure 2c. Activation of the chemokine receptors follows a two-step model that is analogous to the anaphylatoxin receptors. However, unlike C3a and C5a, tethering by the receptor is followed by the N-terminus of the chemokine inserting into the helical bundle of the chemokine receptor. This drives a conformational change that induces intracellular signalling, shown in Figure 2d. The two-step model was derived by early mutagenesis studies of CXCL8 and its receptors CXCR1 and CXCR2. These studies identified a role for the CXCL8 N-terminus in receptor activation following ligand binding and recognition [14, 15]. Recently, this was validated by a cryo-EM-derived structure of CXCR2 in complex with CXCL8 and G protein [16]. Unlike the anaphylatoxins, therefore, the C-terminus of chemokines lacks apparent specific activity.
Targeting of Neutrophil Chemoattractants by Streptococcal CEPs
The C5a Peptidases of GAS, GBS, and Other Streptococci
The first streptococcal CEP discovered was C5a peptidase from GAS, known as ScpA. ScpA cleaves anaphylatoxin C5a [17, 18], targeting the His-Lys peptide bond that is not accessible prior to processing of C5 [19]. Lynskey et al. [20] showed that ScpA cleaves C3a at a site analogous to that in C5a, effectively removing the C-terminus. Given the role of the C-terminus in anaphylatoxin signalling (Fig. 2a–b), it is not surprising that cleavage results in the inactivation of both C3a and C5a.
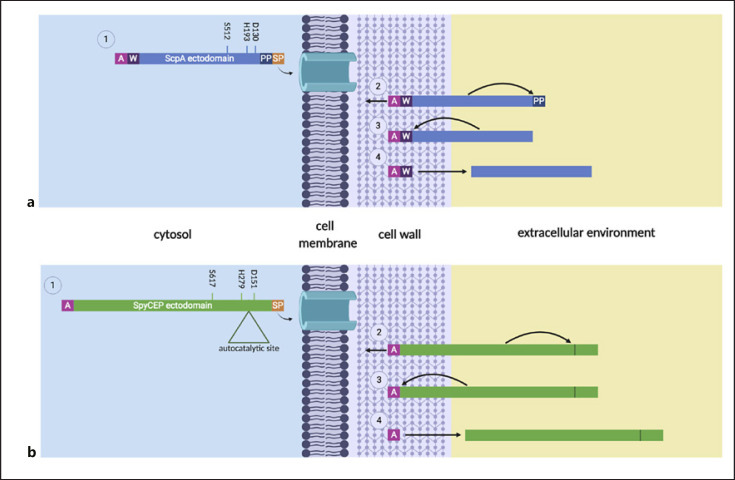
ScpA is 1,167 amino acids in length and is initially produced as a precursor; the N-terminal signal peptide directs ScpA to the bacterial cell wall for secretion into the extracellular environment and is autocatalytically removed during this process, while an LPXTG motif at the C-terminus mediates attachment to the streptococcal cell wall through sortase-mediated processing (Fig. 3a) [18, 19]. Further autocatalytic processing generates catalytically active ScpA through the removal of the pro-sequence [21]. The exact N-terminal residue after cleavage has not been identified, so the length of the pro-sequence remains unclear, but crystallographic analysis has shown that the mature and active form of ScpA starts at residue 97 [22]. The role of ScpA in GAS infection is emerging. Early in vivo experiments showed that the expression of ScpA by GAS strains injected intraperitoneally led to delayed accumulation of neutrophils into the peritoneum of mice but did not affect overall GAS virulence [23]. However, intranasal immunization of mice with ScpA led to delayed GAS clearance, suggesting a greater role for ScpA in noninvasive than invasive infection [24]. There are structural differences between mouse and human anaphylatoxins. ScpA-mediated cleavage of human C5a and C3a is now known to be rapid in contrast to cleavage of murine homologues, potentially offering some explanation for results in mice [20]. Indeed, recent studies using a soft tissue model of infection confirmed that any activity of ScpA in mice might be complement independent. Expression of ScpA reduced clearance of GAS in mice lacking both C3 and C5 [20]. It seems possible that ScpA contributes to virulence by promoting GAS adhesion to epithelial and endothelial cells, in a manner independent of catalytic activity, at least in mice [20]. Notwithstanding findings in mice, ScpA-mediated C3a and C5a cleavage leads to a marked reduction in human neutrophil activation and chemotaxis, while cleavage of C3 leads to reduction in opsonization and phagocytosis of GAS. Taken together, ScpA likely plays a key role in human infection that cannot be fully modelled in mice.
Fig. 3.
Model for ScpA (a) and SpyCEP (b) export, processing, and maturation. a. ScpA: (1) The signal peptide (Sp) of ScpA is recognized, mediating protein translocation across the membrane, and removed. (2) Post-translocation, the anchoring domain (A) mediates covalent cell wall attachment through a sortase-dependent mechanism and ScpA autocatalytically removes its prosequence (PP), activating itself. (3) and (4) ScpA is liberated from the cell wall by either an uncharacterized enzymatic route or autocatalysis of the cell wall spanning region (W). b. SpyCEP: (1) The Sp of SpyCEP is recognized, mediating protein translocation across the membrane, and removed. (2) Post-translocation, the anchoring domain (A) mediates covalent cell wall attachment through a sortase-dependent mechanism and SpyCEP autocatalytically liberates the N- and C-terminal domains, producing the heterodimeric mature enzyme. (3) and (4) SpyCEP is liberated from the cell wall by either autocatalysis or an uncharacterized enzymatic route. SpyCEP, Streptococcus pyogenes cell-envelope protease. Created with BioRender.com.
Consistent with a role as an adhesin, the homologue of ScpA in GBS, ScpB, possesses adhesive properties in addition to C5a cleavage activity. A phage-display approach identified ScpB as binding fibronectin with high affinity, which was confirmed with a GBS mutant deficient in ScpB expression which showed 50% less binding to fibronectin than its wild-type parent strain [25]. A follow-up study showed the affinity of ScpB to be in the nanomolar range, supportive of a role in virulence [26]. Both studies explain an earlier observation that an anti-ScpB serum raised in rabbits could block the invasion of the human lung epithelial cell line A549 in vitro [27]. Although GBS are not known to replicate within lung epithelial cells, they are able to survive within these cells and their entry via attachment to endogenous fibronectin may represent a mechanism through which GBS can traverse mucosal membranes [27]. As such, ScpB has been termed an “invasin.”
In GAS, the gene ScpA is situated within an operon encoding a number of anti-immune response virulence factors such as M protein and the streptococcal inhibitor of complement, the expression of which is positively regulated by the global transcriptional regulator Mga [28]. In contrast, evidence suggests that ScpB is predominantly regulated by the 2-component CsrRS regulator in GBS, which is also known as CovR/S in GAS [29]. Deactivating mutations in both CsrR and CsrS led to de-repression of ScpB transcription and increased expression [29]. However, whether this effect is due to a direct interaction between CsrRS and scpB remains unclear.
In addition to being a leading human pathogen associated with neonatal sepsis and meningitis, GBS is also a major causative agent of bovine mastitis. ScpB and Lmb (laminin-binding protein) are found on a transposon, flanked by insertion sequences in GBS, and these genes show high sequence identity to the genes in GAS, suggesting acquisition by horizontal gene transfer [30]. Intriguingly, this transposon is absent from many bovine isolates of GBS [31]. In vitro analyses of an Scp reporter gene construct found that the expression of ScpB and Lmb is correlated with exposure to increasing levels of human, but not foetal calf serum in strains possessing the transposon [32].
Homologues of C5a peptidase are found in other streptococcal species. A gene encoding a subtilisin-like serine protease with domain architecture similarity to C5a peptidase was recently discovered in the emerging zoonotic pathogen, Streptococcus suis [33]. Increased expression of this gene, SSU1773, was identified in the blood, joints, and hearts of infected pigs that were infected with a highly virulent strain of the pathogen [33] and is implicated in adhesion to porcine cells in vitro as it was identified during a transposon-directed insertion sequencing screen of S. suis genes involved in adhesion and colonization of pig nasal epithelium [34]. Streptococcus equi and Streptococcus zooepidemicus are predicted to encode CEPs named ScpZ whose substrate is C5a [35, 36], while Streptococcus iniae is a pathogen associated with finfish aquaculture and has been shown to encode a C5a peptidase known as ScpI [37]. PCR analysis of virulence factors coupled with emm typing found that the vast majority (99.2%) of 246 isolates of Streptococcus dysgalactiae subspecies equisimilis (SDSE) harboured the gene encoding ScpA [38]. SDSE is a beta-haemolytic streptococcal species that causes similar disease presentations to GAS. Table 1 summarizes functionally related streptococcal CEPs shown to inactivate chemotactic factors.
Table 1.
Summary of functionally related chemotactic factors inactivating streptococcal CEPs
| Species | Protease | Substrate | Location | References |
|---|---|---|---|---|
| Group A streptococcus | SpyCEP | CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8 | Cell surface and released | [45] |
|
|
||||
| ScpA | C5a, C3, and C3a | Cell surface and released | [20] | |
|
| ||||
| Group B streptococcus | CspA | Fibrinogen and other chemokines | Cell surface and released | [75] |
|
|
||||
| ScpB | C5a | Cell surface and released | [27] | |
|
| ||||
| Streptococcus equi | SeCEP | CXCL8 | Cell surface and released | [52] |
|
|
||||
| ScpC | C5a (predicted) | Predicted cell surface | [36] | |
|
| ||||
| Streptococcus zooepidemicus | SzoCEP or ScpC | CXCL8 | Cell surface and released | [52] |
|
|
||||
| ScpZ | C5a (predicted) | Predicted cell surface | [35] | |
|
| ||||
| Streptococcus iniae | ScpI | C5a | Cell surface and released | [37] |
|
|
||||
| CepI | CXCL8 | Cell surface and released | [59] | |
CEPs with characterized function have been included, but it must be noted that there is an abundance of S8 protease sequences that have been inferred from homology with SpyCEP and ScpA/ScpB, which require experimental confirmation. CEP, cell-envelope protease; SpyCEP, Streptococcus pyogenes CEP.
SpyCEP, the CXCL8-Cleaving Serine Protease of GAS
The S. pyogenes CEP (SpyCEP) was originally identified by its ability to cleave human CXCL8, after it was noted that lethal necrotizing GAS infections were characterized by poor neutrophil recruitment to sites of infection despite the significant bacterial burden [39, 40]. SpyCEP is a 180-kDa heterodimer comprising 2 non-covalently linked polypeptide fragments derived from autocatalytic processing [41]. N- and C-terminal fragments, 30 and 150 kDa, respectively, both contribute residues to the catalytic triad (D151, H279, and S617) and can be produced separately and recombined to produce a functional enzyme [42]. Similar to ScpA, SpyCEP is secreted via an N-terminal signal peptide and anchored to the bacterial cell wall by the C-terminal LPXTG cell wall-anchoring motif (Fig. 3b) [40]. SpyCEP is subsequently liberated from the cell wall by an undefined mechanism and found free in culture [43].
In addition to CXCL8, SpyCEP cleaves all neutrophil-specific chemokines that possess an ELR motif, namely, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL7, thereby inhibiting the recruitment of neutrophils to sites of infection and inflammation [44, 45]. Evolution and conservation of the gene encoding SpyCEP are consistent with a strong selective pressure that favours subversion of neutrophil-specific chemokines. As the major neutrophil-specific chemokine, it seems likely that CXCL8 is the main substrate of SpyCEP, while other ELR+ chemokines serve as inadvertent substrates due to shared structural homology [45]. In the case of CXCL8, SpyCEP cleaves the peptide bond between Q59 and R60, resulting in the production of a 59-amino acid N-terminal fragment, and a 13-amino acid C-terminal fragment that corresponds to the C-terminal α-helix. Despite the high level of amino acid sequence identity between ELR+ CXC chemokines, the cleavage sites of CXCL1, CXCL2, CXCL6, and CXCL8 vary in their primary sequence but exhibit high secondary structural homology, wherein a C-terminal α-helix is liberated by SpyCEP in each case [46]. This is perhaps curious since the C-terminus of the CXC chemokines has not been considered to play a major role in chemokine signalling and might not, alone, be predicted to influence receptor ligation and chemokine activity.
Chemokine Inactivation by SpyCEP
Cell membranes are typically decorated with a negatively charged glycocalyx comprising glycosaminoglycans (GAGs), repeating negatively charged disaccharide units. The C-termini of chemokines are typically positively charged and have been defined in many instances as a GAG-binding site. This is certainly true for CXCL8, which possesses a cluster of positively charged residues within the C-terminus (notably R60 and K67) that are critical for supporting the electrostatic interactions, which drive GAG binding [45, 46, 47, 48, 49]. GAGs are essential for the proper functioning of chemokines in vivo [47] and exhibit changes in their structure and localization following tissue injury and inflammation (reviewed in Ref. [48]). While GAGs display diverse physiological roles that support tissue homeostasis, their interactions with chemokines on the vascular endothelium support haptotactic cell migration by establishing stable cell-surface chemokine gradients that are detected by leukocytes in the peripheral circulation [49]. For example, chemokines produced in response to a tissue infection have to be translocated to the endothelial surface to be visible to neutrophils in the periphery. Without the physical interaction with endothelial GAGs, the chemokine would be washed away by the blood flow within the vessel, rather acting being concentrated close to the site of infection. Furthermore, the ability of chemokines to form oligomers on GAGs allows concentrated “depots” of chemokines to be generated to enhance leukocyte recruitment. Although mutant obligate monomeric forms of chemokines have been shown to be active in in vitro chemotaxis assays, such forms are often inactive in vivo, highlighting the importance of oligomer formation [47]. In the case of CXCL8, GAG binding via the C-terminal α-helix facilitates both the translocation of the chemokine to the luminal endothelial surface [50] and the oligomerization of CXCL8 into dimeric and higher order species [51]. Since SpyCEP cleaves the C-terminal GAG-binding motif from CXCL8, it was originally postulated that the lack of neutrophils observed in post-mortem tissues from a patient with invasive GAS infection was due to a lack of CXCL8 on the endothelial surfaces, able to induce their arrest and transmigration from the peripheral circulation [40, 52]. However, this hypothesis did not explain the manifest loss of chemotactic activity exhibited in vitro [40].
Recent work from Goldblatt et al. [45] has extended these observations to put forward a role for neutrophil GAGs in CXCL8 signalling. Cleavage of CXCL8 by SpyCEP was shown to render the chemokine unable to bind CXCR1 and CXCR2 in vitro, with a subsequent loss of all downstream signalling events including chemotaxis. SpyCEP cleavage of CXCL8 was also shown to ablate heparin binding, and subsequent glycanase treatment to remove cell-surface GAGs from neutrophils was shown to abolish CXCL8-induced activation of neutrophils in vitro. Collectively, these data support the hypothesis that chemokine binding to neutrophil-bound GAGs is required for the subsequent activation of CXCR1 and CXCR2. These interactions expose an inherent vulnerability in the initiation of innate immunity, which is exploited by SpyCEP to inactivate CXCL8 in vivo [52].
The removal of the GAG-binding region of CXCL8 by SpyCEP also introduces the possibility that SpyCEP subverts host immunity by interfering with the process of chemokine oligomerization. Neutrophil pericellular GAGs comprising the glycocalyx sequester local soluble chemokines in multiple degrees of oligomerization, to sample the chemokine gradients present at the leading edge of the neutrophil. In situ, CXCL8 can form monomeric and dimeric variants, with the dimer exhibiting reduced activation of CXCR1 in chemotaxis assays relative to the monomeric form [53]. While both monomeric and dimeric CXCL8 bind GAGs, they are unable to activate CXCR1 or CXCR2 while bound [54]. It is thought that neutrophil cell surface GAGs serve to increase local concentrations of chemokines in the vicinity of their cognate receptors to improve the chemokine gradient sampling efficiency. Therefore, in addition to its impact on reducing CXCR1/CXCR2 signalling, SpyCEP may also disrupt CXCL8 oligomerization and in turn, reduce CXCL8 dimer dissociation into its receptor-activating monomeric subunits.
SpyCEP Impact on Pathogenesis
In addition to human ELR+ CXC chemokines, SpyCEP cleaves the murine CXCL1 and CXCL2 homologues MIP-2 and KC, which signal via mCXCR2 and play a significant role in recruiting neutrophils to the sites of infection in mice [40, 55, 56, 57]. Several in vivo experimental studies in mice using strains that differed in SpyCEP gene expression have demonstrated a role for SpyCEP in soft tissue dermonecrosis [44, 46, 55, 58, 59]. Although some studies have demonstrated paradoxical results [46, 58], discrepancies may arise due to differences in models, strains, and the possibility of regulatory gene mutations. Several in vivo studies have demonstrated that SpyCEP expression reduces the levels of murine chemokines in either tissue or serum while retarding neutrophil influx. Interestingly, SpyCEP expression confers a lethal phenotype to the otherwise innocuous Lactococcus lactis [46], with an increased bacterial burden and dissemination observed in a SpyCEP-dependent manner. A recent report showed that S. pyogenes can utilize draining lymph nodes to metastasize to distant sites following intramuscular injection of S. pyogenes into mouse hind limbs [60]. The same study also found a role for SpyCEP in limiting neutrophil recruitment to lymph nodes, thereby promoting survival of GAS and underlining a role for CXC chemokines in leukocyte recruitment to lymph nodes. The implication of the findings from both heterologous expression studies and mutagenesis in S. pyogenes is that SpyCEP not only retards neutrophil recruitment but also impairs neutrophil killing of bacteria, which is consistent with previous reports demonstrating that SpyCEP protects S. pyogenes from neutrophil killing, in part due to the inhibition of chemokine-induced neutrophil extracellular traps [59].
The SpyCEP gene, cepA (also annotated as scpC/prtS), is found ubiquitously in all isolates of GAS; however, the expression of SpyCEP is highly variable between clinical isolates, with invasive blood isolates expressing higher levels of SpyCEP than pharyngeal isolates [40, 61]. In a clinical invasive disease, high SpyCEP expression is associated with disease severity and poor patient outcome [61]. SpyCEP is under the repression of CovR/S (also known as csrRS), the two-component gene regulator that controls approximately 15% of the GAS genome [61, 62, 63]. Naturally occurring mutations at this regulatory locus have been reported to dramatically upregulate SpyCEP expression and contribute to disease severity [40, 61]. Mutations in CovR/S are proposed to occur at the time S. pyogenes transitions from a pharyngeal to a more invasive state and may therefore underlie the observed dichotomy in SpyCEP production between pharyngeal and blood culture isolates.
Other CEPs of Pathogenic Streptococci
The family of CEPs in pathogenic streptococci has orthologues in other lactic bacteria, namely, the lactococci and Streptococcus thermophilus (prtS), where almost all have a nutritional role in cleavage of casein [64, 65, 66]. Along with orthologues to SpyCEP (e.g., SpyCEP in S. dysgalactiae subsp. equisimilis, SeCEP in S. equi, and SzoCEP in S. zooepidemicus) and C5a peptidase, other CEPs have been discovered in pathogenic streptococci. The functions of some of these proteases are yet to be elucidated; however, like SpyCEP and C5a peptidase, most target effectors of the host innate immune system.
PrtA
PrtA was discovered in Streptococcus pneumoniae by screening convalescent-phase serum and was identified as a serine protease containing the histidine, aspartate, and serine catalytic triad [64]. Unlike SpyCEP and C5a peptidase, which maintain a high degree of sequence identity between strains of the same species, PrtA displays a surprising amount of diversity between clinical strains of S. pneumoniae, particularly within the central region of the protein [67]. The exact function of PrtA has not been established, but in vivo evidence is emerging of its role as a virulence factor. Loss of the gene does not impact colonization of the murine nasopharynx, although it does result in attenuated virulence and lower recovery of bacteria from the lung and blood of infected mice [68]. Furthermore, mice infected intraperitoneally with a prtA knockout S. pneumoniae strain showed greater survival compared to mice infected with the prtA-expressing WT parent strain [67]. Interestingly, it was shown that PrtA cleaves human apolactoferrin, releasing lactoferrin-like peptides with bactericidal activity. Therefore, somewhat paradoxically, expression of prtA may lead to decreased bacterial survival [69].
PrtA is transcriptionally regulated by PsaR in S. pneumoniae [70], and while the function of PrtA is unknown, its regulation indicates a role in the progression from nasopharyngeal carriage to more invasive disease. The PsaR regulon is highly upregulated in the presence of nickel, cobalt, and zinc ions and repressed in the presence of magnesium ions [71, 72, 73]. Responding to changes in metal ions is thought to be important for adapting to the change in environment from the nasopharynx to lungs. Interestingly, in vivo expression of prtA is increased in blood compared to the nasopharynx of infected mice [68]. Clearly, there is much more to learn about PrtA, notably its main function and the role, if any, of diversity within Prt and virulence.
CspA and SspA
CspA was first identified in a GBS blood isolate from a case of neonatal sepsis. While it was found to share some homology with C5a peptidase and caseinases from lactic acid bacteria, it does not cleave either C5a or casein [74]. At first, fibrinogen was the only known substrate of CspA, but its similarity to other immunomodulating proteases, coupled with the attenuated virulence of a CspA knockout GBS strain in a neonatal rat infection model, suggested that CspA may also serve a role in pathogenesis [74]. CspA was subsequently found in in vitro assays to cleave the ELR+ CXC chemokines CXCL1, CXCL2, CXCL3, CXCL6, and CXCL7, but not CXCL8 [75]. No cleavage of CC chemokines was shown, although it was not possible for the authors to demonstrate cleavage was not occurring at the extreme C- or N-termini of the proteins [75].
Investigation into the MtaR regulator of GBS, which is required for virulence, showed that CspA was one of the 11 genes downregulated in an MtaR mutant [76], further suggesting a role in GBS infection. Although initially thought to only be located on the cell wall [74], it was later shown that CspA includes a second site for autocatalytic cleavage near the cell wall-anchoring domain, which leads to the release of a mature peptide from the cell wall [75, 77]; however, immunofluorescence staining of GBS has established that CspA maintains a punctate distribution on the cell wall [78].
A homologue of CspA, the serine protease SspA, was discovered in S. suis, by screening both the convalescent-phase serum [79] and a transposon mutant library for isolates deficient in cleavage of a chromogenic substrate specific for chymotrypsin-like proteases [80]. SspA possesses a His-Asp-Ser catalytic triad and shares the greatest degree of sequence identity with PrtS of S. thermophilus (95.9%) and CspA (49.5%) [80]. Mutants deficient in SspA exhibit delayed growth and decreased survival in whole human blood, while inducing milder clinical signs and significantly decreased mortality in mice following intraperitoneal infection [80]. SspA degrades gelatin [81] and, unlike CspA, degrades CCL5 [82]. A further immunomodulatory role for SspA was uncovered following incubation of stimulated THP-1-derived macrophages with active and heat-inactivated recombinant SspA. This in vitro study showed that regardless of proteolytic activity, SspA induced IL-1β, IL-6, TNF-α, CXCL8, and CCL5 release [82]. An inverse relationship between SspA and CCL5 concentration was observed, as although SspA induced the secretion of CCL5, high concentrations of exogenous recombinant SspA resulted in the degradation of de novo secreted CCL5, while, conversely, low concentrations of SspA led to increased concentrations of CCL5 [82]. Induction of pro-inflammatory cytokines points to another strategy of immunomodulation by bacterial serine proteases. Unlike the secretion of other streptococcal CEPs, evidence suggests that SspA secretion is dependent on a type-4 secretion system (T4SS) encoded by the 89K pathogenicity island of S. suis [83]. However, detectable amounts of SspA were found in the growth supernatant of the T4SS knockout strain, and previous work showed that SspA was only present on the cell wall of S. suis but not in the supernatant [79]. Another CspA homologue, SFP (subtilase-family protein), was identified in S. pneumoniae; however, its role in virulence has yet to be determined [84]. Given that S. suis is a major pathogen of pigs worldwide, we expect that this CEP will continue to be the focus of future research.
Sequence and Structural Homology within Bacterial CEPs
Subtilisin-like serine proteases, termed subtilases, are a diverse family found in bacteria, archaea, yeast, fungi, and some eukaryotes [2, 64]. Subtilases are typically extracellular and exhibit low sequence homology and diverse functionality, spanning nutrient scavenging and precursor processing for immune evasion [65]. These proteins are characterized by a multi-domain architecture extending over 1,000 residues, typically comprising an N-terminal signal peptide for export and a complementary C-terminal-anchoring motif, pre-propeptide (removed by autocatalysis), subtilisin-like catalytic domain with an inserted protease-associated (PA) domain, with heterogeneity observed in the composition and number of C-terminal domains. The protease domain encompasses the aspartate-histidine-serine catalytic triad, with the highest sequence conservation observed proximal to the active site, but variation in these residues can be observed [66].
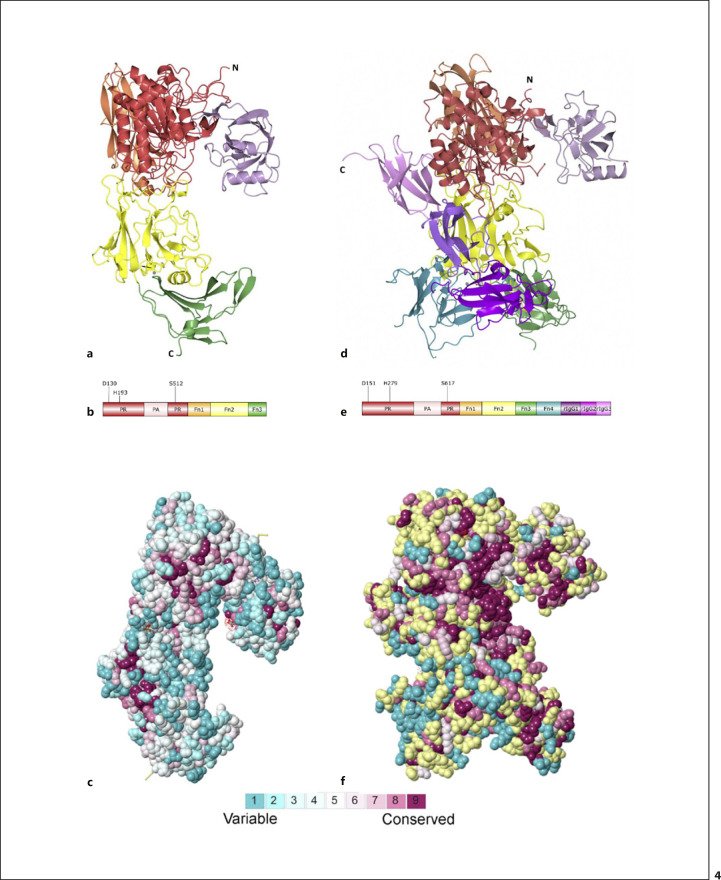
Streptococcal CEPs are members of the S8 family of subtilases and share a common architecture. This comprises a YSIRK signal peptide and a C-terminal LPXTG-anchoring motif, essential for sortase-dependent anchoring and functionality of the protease [85] consistent with cell wall-anchored proteins from Gram-positive bacteria. Bacterial CEPs also contain an S8 catalytic domain interrupted by a PA domain, fibronectin, and immunoglobin-like C-terminal domains and are defined by autocatalytic processing during maturation. Crystal structures have been solved for 3 streptococcal CEPs, SpyCEP, ScpA, and ScpB. The mature and active form of ScpA spans residues 97–1,032, encompassing the catalytic domain with the inserted PA domain followed by 3 fibronectin type III domains (Fn1–Fn3 domains) (Fig. 4a) [22]. The catalytic triad (D130, H193, and S512) is localized in the catalytic domain (Fig. 4b) and is consistent with other characterized S8 subtilases [21, 86]. SpyCEP comprises 9 distinct domains (Fig. 4d, e), where the first 5 domains, catalytic domain with the inserted PA followed by 3 fibronectin type III domains (Fn1–Fn3 domains), are vital for catalytic activity and homologous with the domain organization in ScpA [87]. The remaining 4 domains extend beyond the core and encompass a fibronectin/Indian hedgehog protein (Fn4) domain and 3 reverse-Ig folds.
Fig. 4.
Structures of ScpA and SpyCEP. Structures of wild-type ScpA (3EIF) (a) and SpyCEP (5XYR) (d), coloured according to the domain architecture shown in (b, e), respectively. ConSurf analysis of ScpA (c) and SpyCEP (f). Sequences of the most closely related C5a and chemokine degrading CEPs were analysed to highlight regions of sequence conservation, then mapped onto the structures, and visualized according to the key. Regions that could not be analysed due to insufficient homologous data are shown in yellow. SpyCEP, Streptococcus pyogenes cell-envelope protease. Created with PyMol.
ScpA and ScpB share 98% sequence identity and domain organization but exhibit a surprising level of structural variance. Alignment of all Cα positions on ScpA and ScpB produces a root mean squared difference of 4.7 Å, owing to differences in the orientation but not fold of the PA and Fn1–Fn3 domains and regions within the catalytic domain [22]. However, the ScpB structure is catalytically inactive as it was produced as a shorter construct, impeding autocatalytic processing vital for protein maturation. This was proposed as the cause of the structural differences by Kagawa et al. [22] and is in line with structural variance observed in wild-type and mutants of SpyCEP [87, 88].
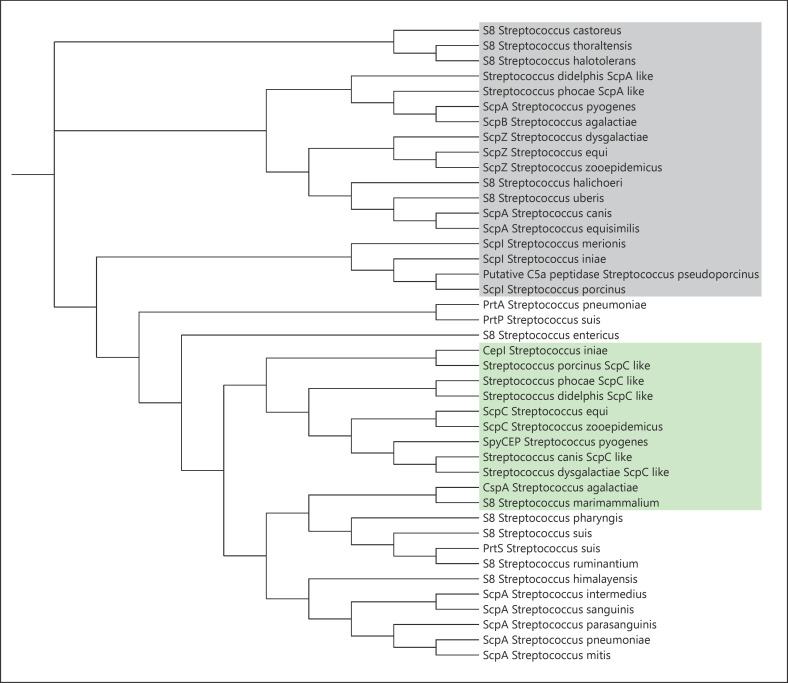
Large-scale genome sequencing has highlighted the prevalence and wide distribution of streptococcal S8 subtilases, whereby function is inferred from the annotation of the S8 catalytic region and homology with ScpA/B or SpyCEP. We carried out phylogenetic analysis of streptococcal CEPs [89, 90, 91, 92], most closely related to SpyCEP and ScpA/B, and found that there is a clustering of sequences with related and experimentally characterized function, where 2 distinct subtrees, related to C5a and CXC chemokine-degrading functions, are observed (Fig. 5). Although functional assignments cannot be made without experimental confirmation, sequences with homology greater than 45% to either ScpA/ScpB or SpyCEP cluster around these branches and have been highlighted. The retention of CEPs across a wide array of pathogenic streptococci, displayed in only the most closely related subset here, indicates the importance of the function played by these proteins in the establishment of infection.
Fig. 5.
Phylogenetic tree of streptococcal CEPs. ConSurf analysis generated a scored list of related sequences for SpyCEP (5XYR), ScpA (3EIF), and ScpB (1XF1) [22, 87]. The 15 most related sequences were taken, with duplicates removed, for each protein and a phylogenetic tree generated using Clustal Omega [85]. Phylogenetic data were visualized using the Interactive Tree Of Life [88]. Sequences were blasted and denoted ScpA (grey shading) or ScpC (green shading) like owing to their homology (>50%) with C5 peptidase (ScpA) or SpyCEP (ScpC), respectively. CEPs, cell-envelope proteases; SpyCEP, Streptococcus pyogenes CEP.
Sequence homology within the streptococcal CEPs is low, but functionally related proteins exhibit greater homology. SeCEP and SzoCEP from S. equi and S. zooepidemicus, respectively, share 96% sequence identity and 61–62% with SpyCEP (Table 2). Experimental characterization has shown that they can cleave human and equine CXCL8 [52]. This sequence identity is concentrated in the protease-related region of the sequences, which is unsurprising when shared substrate specificity is considered. ConSurf, a bioinformatic server that calculates the evolutionary conservation of each amino acid in a protein based on phylogenetic links between homologous sequences, was used to analyse the SpyCEP sequence, with highest sequence conservation corelating with an annotated chemokine degradation function [90, 91]. Highest sequence conservation, when mapped onto the structure of SpyCEP, is observed in the core catalytic domains with least conservation observed at sites most distal to the core of the protein (Fig. 4f). Interestingly, the PA domain exhibits patches of high sequence conservation but has been shown to be mobile in molecular dynamics simulations. It has been hypothesized that the PA domain plays a role in substrate recruitment, with this sequence conservation observation indicative of a functional relevance [88].
Table 2.
Percentage sequence identity between functionally characterized CEPs
| CspA | − | ||||||||
| CepI | 42% | − | |||||||
| SpyCEP | 39% | 56% | − | ||||||
| SeCEP | 39% | 56% | 61% | − | |||||
| SzoCEP | 39% | 56% | 62% | 96% | − | ||||
| ScpC | 32% | 29% | 30% | 29% | 29% | − | |||
| ScpZ | 31% | 29% | 29% | 29% | 29% | 95% | − | ||
| ScpI | 32% | 31% | 32% | 31% | 31% | 32% | 32% | − | |
| ScpA | 34% | 33% | 34% | 34% | 33% | 38% | 38% | 38% | − |
| ScpB | 33% | 31% | 32% | 31% | 31% | 36% | 36% | 37% | 98% − |
CEP, cell-envelope protease; SpyCEP, Streptococcus pyogenes CEP.
Comparison of ScpA with closely related C5a degrading CEPs elicits a similar result, with the highest sequence conservation observed in the residues comprising the core of the catalytic domain (Fig. 4c). However, on average, ScpA exhibits less overall sequence conservation than SpyCEP. For example, only the core of the more ordered PA domain is highly conserved in ScpA in comparison with the entire PA domain of SpyCEP. Interestingly, ConSurf identified more related sequences for ScpA than SpyCEP, which could have given rise to the lower overall sequence conservation score, with a wider evolutionary space sampled for ScpA. Moreover, SpyCEP has a higher sequence identity with related CEPs (Table 2), further contributing to the high sequence conservation observed.
Intrinsically disordered regions (IDRs) are a common feature at the extreme N- and C-termini of CEPs, imparting flexibility required for autocatalysis-mediated maturation and a high degree of freedom to a cell wall-anchored protease. These regions are typically missing from crystallographic analysis but have been characterized for SpyCEP using NMR [88]. The N- and C-terminal IDRs present in SpyCEP (residues 34–115 and 1,575–1,613, respectively) are characterized by distinct subsets of disorder, highlighting the functional plasticity imparted by these sequences particularly within the context of a structured protein. The C-terminal IDR is directly upstream of the LPXTG-anchoring motif, exhibits limited structural propensity, and is hypothesized to function as a flexible linker. However, the N-terminal IDR exhibits helical propensity indicative of functional relevance. Moreover, truncation of this region impedes crystallization [93] and inhibits the heterodimerization of autoprocessed SpyCEP. Interestingly, IUPRED analysis of the most closely related sequences to SpyCEP, SeCEP and SzoCEP, highlights the consistent presence of disorder at the extreme N-termini but not the C-termini [94, 95, 96]. The predicted retention of the N-terminal IDR indicates the importance of this region within the CEPs. Further analysis is required to determine functional relevance and the role of disorder within the CEPs, potentially broadening or confining the observations made on SpyCEP.
Vaccine Development Targeting Pathogenic CEPs
There is a global demand for robust vaccines against pathogenic streptococci. Worldwide S. pneumoniae and S. pyogenes infections alone account for over 2.1 million deaths per year [97, 98], highlighting the significant healthcare burden and requirement for accessible global vaccines. Developments over the last 2 decades in reverse vaccinology and cell surface proteomics have revealed many novel and immunogenic streptococcal proteins as potential vaccine targets. In a field that is increasingly searching for highly conserved pan-serotype antigens, the CEPs of pathogenic streptococci represent ideal candidates for inclusion in both inter- and intraspecies vaccine designs. In this section, we will discuss the use of CEPs as vaccine components (summarized in Table 3).
Table 3.
Summary of studies using CEPs as vaccine antigens for immunization against streptococcal species
| Target | Vaccine | Model | Findings | Reference |
|---|---|---|---|---|
| SpyCEP | Recombinant SpyCEP with alum adjuvant | IP immunisation with M1 GAS IN challenge | Increased survival after challenge | [88] |
|
|
||||
| Recombinant SpyCEP with Freund's adjuvant | IP immunization followed by intranasal lethal challenge with M23 GAS of CD1 mice | Increased survival to 70% | [99] | |
|
|
||||
| Recombinant SpyCEP with Freund's adjuvant | BALB/c mice immunized IM, followed by IM and IN challenge with GAS and S. equi | No reduction in bacterial burden at the site of infection. Decreased dissemination of GAS and S. equi following both IM and IN challenge | [52] | |
|
|
||||
| Combo (SpyCEP, SpyAD, and streptolysin O) adjuvanted with Freund's or alum hydroxide | CD1 mice immunized IP, challenged with Ml, M6, M12, and M23 GAS | Cross serotype protection against GAS 50-80% survival following IN and IM challenge Decreased bacterial burden in subcutaneous air pouch infection Induced antibodies that mediated opsonophagocytic killing in whole blood assay | [101] | |
|
|
||||
| SpyCEP with Freund's adjuvant | FVB/n mice immunized IM and IN challenge | Reduced signal of bioluminescent M75 GAS, but no difference in bacterial burden in the nasopharynx | [112] | |
|
|
||||
| SpyCEP with J8-DT (M protein conserved epitope) | BALB/c mice immunized SC, GAS skin challenge following scarification | Protection against pyoderma and bacteraemia Protection against GAS hypervirulent CovRS mutants | [105, 106] | |
|
| ||||
| ScpA | Recombinant ScpA unadjuvanted | Intranasal immunization of CD1 mice, intranasal challenge with M1, M2, M6, M11, and M49 GAS | Reduced nasopharyngeal colonization | [24] |
|
|
||||
| Recombinant ScpA adjuvanted with alum or monophosphoryl lipid A | SC immunization of CD1 mice, IN challenge with M49 and M1 GAS | ScpA-specific antibodies neutralized protease activity of ScpA and ScpB Increased clearance of GAS from lungs and nasal mucosa | [109] | |
|
|
||||
| ScpA adjuvanted with cholera toxin | Intranasal immunization of BALB/c mice, intranasal challenge with M49 GAS | Reduction of colonization of nasal-associated lymphoid tissue Immunized sera gave passive protection against GAS IN infection in naïve mice | [110] | |
|
| ||||
| SpyCEP and ScpA | Spy7 (ScpA, and 6 other highly conserved surface antigens) with Freund's adjuvant vs SpyCEP | FVB/n mice immunized IM, IM challenge with M1 and M3 GAS | Reduction in systemic dissemination | [102] |
|
|
||||
| Combo 5 (ScpA, SpyCEP, arginine deaminase, streptolysin O, and trigger factor) with Alum adjuvant | IM immunization of rhesus macaques, M1 GAS IN challenge | Robust antigen-specific antibody response and reduction in both pharyngitis and tonsillitis following an M1 intranasal challenge | [100] | |
|
| ||||
| ScpB | ScpB adjuvanted with alum and mycobacterial phospholipid, SC immunisation | CD1 mice, GBS serotype VI IN challenge | Reduced lung bacterial burden Conjugation of recombinant ScpB to a nonhomologous polysaccharide (III) can increase the immunogenicity of the polysaccharide when challenged with another serotype (VI) and reduced inflammatory damage to the lungs | [111] |
|
|
||||
| ScpB encapsulated in biodegradable polymer | ICR mice immunized IN or IM, vaginal challenge | Strong IgG response after administration of ScpB by both intranasal and intramuscular routes, with an increased response when encapsulated Intranasal administration elicited a secretory IgA in the vaginal mucosa Mice immunized with encapsulated ScpB fully resisted vaginal colonization after exposure to serotype III GBS. Parental immunization conferred neonate protection | [114] | |
|
| ||||
| SspA | SspA with ScpCL and 3 other S. suis virulence factors adjuvanted with CpG | IN immunization of C57BL6/6JCnc mice, followed by IN, IP, or IV challenge with SS2, SS3, and SS7 serotype S. suis | Significantly reduced NALT bacterial burden Serotype-dependent protection against systemic dissemination Reduced mortality in mice given a lethal IV dose of SS2 S. suis | [120] |
|
| ||||
| PrtA | PrtA with Freund's adjuvant | C3h/HeJ mice, SC immunisation and IP challenge | Mice were protected from lethal challenge of serotypes 6A and 6B but not 4 S. pneumoniae strains Serum from immunized mice was able to protect naive mice from lethal challenge in a serotype- specific manner | [121] |
|
|
||||
| PrtA with ISCOMATRIX | IM immunization of BALB/c and CBA/n mice, followed by IN challenge with serotype 3 or 8 S. pneumoniae | Reduced serotype 3 bacterial burden in the lungs of BALB/c mice Protected CBA/n mice from a lethal intranasal dose of serotype 3 Protective effect against non-lethal serotype 8 infection | [122] | |
|
|
||||
| PrtA with Curdlan adjuvant | IN immunization of BALB/c mice, followed by IN challenge with serotype 2 S. pneumoniae | Increased PrtA-specific IgG and IgA levels in BALF and increased IgA levels in saliva and nasal washes Did not protect against acute pneumonia and systemic dissemination; no significant reduction in lung bacterial burden or blood multiplication | [123] | |
CEPs, cell-envelope proteases; GAS, group A streptococcus; SpyCEP, Streptococcus pyogenes CEP.
SpyCEP as a Target for GAS Vaccination
SpyCEP has been the target of several GAS vaccine designs. It was initially identified through “reverse vaccinology” as a potentially protective cell wall-associated antigen [99]. Immunization with recombinant SpyCEP conferred a 70% survival rate to CD1 mice challenged with a 90% lethal intranasal dose of a serotype M23 S. pyogenes. Subsequently, vaccine studies have shown SpyCEP immunization to induce protease-neutralizing antibodies in mice [52, 100], raising the possibility that vaccine-induced protection resides in the neutralization of virulence as much as opsonic activity.
Turner et al. [52] demonstrated that SpyCEP immunization enhanced protection against S. pyogenes dissemination in both intranasal and intramuscular infection models in mice and highlighted the potential of SpyCEP to induce cross-species protection against S. equi intramuscular infection. More recently, the combination of SpyCEP with 2 additional conserved, highly expressed, and immunogenic proteins, arginine deaminase and streptolysin O, demonstrated cross-serotype (M1, M6, M12, and M23) protection in CD1 mice, conferring 50–80% survival following intranasal and intramuscular challenge and significantly reducing bacterial growth in a subcutaneous air pouch infection model [101]. This combination vaccine is currently in commercial development, while SpyCEP has also been used in a number of other combination vaccines [100, 102, 103].
SpyCEP has also been used to augment the efficacy of M protein-based vaccines, the historical target of S. pyogenes vaccines. An M protein-based, minimal B-cell epitope vaccine conjugated to the diphtheria toxoid, named J8-DT, was shown to be effective against pyoderma in mice, but ineffective against hypervirulent S. pyogenes strains that had mutations in covRS and therefore expressed an abundance of virulence factors [104]. The inclusion of SpyCEP, or a 20-amino acid (aa 205–224) minimal epitope, in the J8-DT vaccine effectively protected against these hypervirulent strains [105, 106].
ScpA as a Target for GAS Vaccination
ScpA was proposed as a universal candidate GAS vaccine almost 30 years ago, to circumvent the problems inherent in vaccinating against GAS, a pathogen with multiple “M” serotypes [19, 107]. Immunization with recombinant ScpA is highly immunogenic in rabbits and mice and can induce cross-serotype (M1, M2, M6, M11, and M49)-neutralizing antibodies and reduce cross-serotype streptococcal colonization [24]. Modern immunoproteomic techniques, including reverse vaccinology and approaches to uncover the anti-GAS-protective antigenic targets of human intravenous immunoglobulin, have identified and confirmed ScpA to be a key protective antigenic target in GAS infection [99, 102]. More recent work has therefore included ScpA in various combination vaccines, such as Spy7 [102] and Combo5 [100], all of which have shown protective effects in different animal models with Combo5 showing protection in rhesus macaques.
Given the ability of ScpA to modulate human neutrophil recruitment and opsonization, it is unsurprising that it has emerged as a key target in S. pyogenes vaccine design. Anti-ScpA-specific IgG and secretory IgA immunoglobulins are detectable from convalescent patient samples, with antibodies purified from sera effectively neutralizing ScpA activity [108]. Immunization studies have additionally highlighted ScpA as a pan species vaccine target, demonstrating that specific ScpA-induced antibodies can inhibit protease activity of C5a peptidase from both GAS and GBS [109]. Intranasal immunization with ScpA adjuvanted with cholera toxin significantly reduced M49 S. pyogenes colonization of mouse nasal-associated lymphoid tissue [110]. Additionally, intranasal administration of immunized sera containing specific anti-ScpA immunoglobulin provided passive protection in naive mice. Given the reduced enzymatic functionality of ScpA in mice, it seems likely that the protection afforded may reside in opsonic or anti-adhesin activity; the mechanisms by which immunization confers protection require elucidation.
ScpB as a Target for GBS Vaccination
As described above, immunization of mice with either ScpA or ScpB induces neutralizing antibodies and can reduce the bacterial burden following a subsequent intranasal challenge with GAS [109]. Similarly, adjuvanted ScpB immunization can reduce lung bacterial burden following a serotype VI GBS intranasal challenge, while ScpB immune antiserum can additionally provide passive cross-protection from both GAS and GBS infections [109, 111].
Vaccine initiatives against GBS are primarily aimed at reducing the risk of infant infections, which are associated with maternal colonization at the time of birth. As GBS can be a normal part of the enteric flora in up to 30% of healthy women, induction of long-term immunity is challenging. A number of strategies have evolved to elicit effective and longer lasting anti-GBS immunity including encapsulation of ScpB in a biodegradable polymer and combination with a GBS surface-exposed lipoprotein [113, 114, 115]. While all have shown some promise in different models of infection, none have yet reached clinical evaluation.
Streptococcus suis
Streptococcus suis is a global swine pathogen and a potent zoonotic agent. While zoonotic infections are rare, S. suis can cause meningitis, sepsis, and death in humans. This burden is particularly high in Asia, where over 90% of human cases occur [116]. In Southern Vietnam, S. suis is the leading causative agent of adult meningitis [117] and responsible for a mortality rate of up to 6% [118]. Historically vaccine designs for S. suis have focused on bacterins (attenuated bacteria); however, the field is now shifting towards conserved proteins including the CEP SspA and other proteins to develop cross-serotype protection [119, 120]. Despite these efforts, there is currently no effective commercially available vaccine developed against S. suis.
Streptococcus pneumoniae
Despite the introduction of both the 23-valent polysaccharide (PPV23) vaccine and the 10/13-valent conjugate vaccine (PCV10/13), there is still a large global health burden associated with S. pneumoniae infection. Vaccine coverage remains a challenge, however, due to a need to combat non-vaccine serotypes and capsule switching; development of the next generation of protein-based vaccines may require the use of pan-serotype antigenic targets including virulence factors such as the CEPs. The S. pneumoniae CEP PrtA, known to cleave apo lactoferrin, can evoke protective immunity in some animal models of infection and could, in combination with other antigens, provide a solution to the problems of serotype specificity in pneumococcal vaccinology as demonstrated in a number of murine models of pneumonia [121, 122, 123].
Conclusion − Looking Ahead
The CEPs of pathogenic streptococci have an exclusive relationship with their substrates; no other group of enzymes targets the entire family of neutrophil-active CXC chemokines or neutrophil families of chemotactic agents. Beyond the fascinating role that the CEPs play in circumventing host innate immunity, this family of serine proteases may yet have additional functions related to adhesin activity and play an important role in future vaccine development strategies.
The knowledge gained by understanding GAG-chemokine interactions and their interdiction by proteases, such as SpyCEP, raises possibilities for therapeutic development. Potential SpyCEP antagonists modelled on CXCL8 but with reduced CXCR1 and CXCR2 activity have previously been described [14, 15] and could in theory provide adjuvant therapies for more severe invasive streptococcal infections that fail to respond to antibacterial agents alone. However, the ability of CEPs to exhibit class action against entire families of neutrophil chemoattractants raises the possibility that their activities might be harnessed in the treatment of inflammatory disorders − particularly those where CXC chemokines or anaphylatoxins are proven deleterious mediators. Examples of disorders known to be driven by CXCL8 might include inflammatory disorders such as Crohn's disease, COPD, and ARDS; however, caution would be required to ensure impedance of neutrophil recruitment did not render subjects prone to bacterial infections. ScpA has already been developed as a potential anti-inflammatory immune modulator, although clinical application is yet to be demonstrated. As humans often develop antibodies to bacterial enzymes such as ScpA and SpyCEP, such therapies may require better understanding of enzyme activity and development of novel agents that emulate the native bacterial enzymatic action. Of more recent relevance, it has been reported that CXCL8 and C5a are present in abundance in hospitalized patients with COVID-19 [124, 125]. If confirmed to be deleterious, then either SpyCEP or ScpA analogues might be of value either as locally or as systemically delivered reagents in the more severe cases.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
Our work in this area is supported by the Wellcome Trust Collaborative Award 215539: “Understanding and exploiting Group A streptococcal anti-chemotactic proteases in vaccines for infection” awarded to S.S., S.M., and J.E.P.
Author Contributions
S.McK., K.K.H., S.G., M.P., M.S.M.A.S., S.S., S.M., and J.E.P. all contributed to the writing of the initial draft of the manuscript and subsequent edits.
References
- 1.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1((2)):70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osowicki J, Vekemans J, Kaslow DC, Friede MH, Kim JH, Steer AC. WHO/IVI global stakeholder consultation on group A Streptococcus vaccine development: report from a meeting held on 12–13 December 2016. Vaccine. 2018;36((24)):3397–405. doi: 10.1016/j.vaccine.2018.02.068. [DOI] [PubMed] [Google Scholar]
- 4.Jarva H, Jokiranta TS, Würzner R, Meri S. Complement resistance mechanisms of streptococci. Mol Immunol. 2003;40((2)):95–107. doi: 10.1016/s0161-5890(03)00108-1. [DOI] [PubMed] [Google Scholar]
- 5.Yuste J, Sen A, Truedsson L, Jönsson G, Tay LS, Hyams C, et al. Impaired opsonization with C3b and phagocytosis of Streptococcus pneumoniae in sera from subjects with defects in the classical complement pathway. Infect Immun. 2008;76((8)):3761–70. doi: 10.1128/IAI.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437((7058)):505–11. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 7.Monk PN, Scola AM, Madala P, Fairlie DP. Function, structure and therapeutic potential of complement C5a receptors. Br J Pharmacol. 2007;152((4)):429–48. doi: 10.1038/sj.bjp.0707332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klos A, Wende E, Wareham KJ, Monk PN. International union of basic and clinical pharmacology. [corrected]. LXXXVII. Complement peptide C5a, C4a, and C3a receptors. Pharmacol Rev. 2013;65((1)):500–43. doi: 10.1124/pr.111.005223. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Kim HR, Deepak RNVK, Wang L, Chung KY, Fan H, et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat Struct Mol Biol. 2018;25((6)):472–81. doi: 10.1038/s41594-018-0067-z. [DOI] [PubMed] [Google Scholar]
- 10.Cain SA, Monk PN. The orphan receptor C5L2 has high affinity binding sites for complement fragments C5a and C5a des-Arg(74) J Biol Chem. 2002;277((9)):7165–9. doi: 10.1074/jbc.C100714200. [DOI] [PubMed] [Google Scholar]
- 11.Nikiforovich GV, Marshall GR, Baranski TJ. Modeling molecular mechanisms of binding of the anaphylatoxin C5a to the C5a receptor. Biochemistry. 2008;47((10)):3117–30. doi: 10.1021/bi702321a. [DOI] [PubMed] [Google Scholar]
- 12.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36((5)):705–16. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Determination of the secondary structure of interleukin-8 by nuclear magnetic resonance spectroscopy. J Biol Chem. 1989;264((32)):18907–11. [PubMed] [Google Scholar]
- 14.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J Biol Chem. 1991;266((34)):23128–34. [PubMed] [Google Scholar]
- 15.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J Biol Chem. 1994;269((23)):16075–81. [PubMed] [Google Scholar]
- 16.Liu K, Wu L, Yuan S, Wu M, Xu Y, Sun Q, et al. Structural basis of CXC chemokine receptor 2 activation and signalling. Nature. 2020;585:135–40. doi: 10.1038/s41586-020-2492-5. [DOI] [PubMed] [Google Scholar]
- 17.Wexler DE, Nelson RD, Cleary PP. Human neutrophil chemotactic response to group A streptococci: bacteria-mediated interference with complement-derived chemotactic factors. Infect Immun. 1983;39((1)):239–46. doi: 10.1128/iai.39.1.239-246.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor SP, Cleary PP. Localization of the streptococcal C5a peptidase to the surface of group A streptococci. Infect Immun. 1986;53((2)):432–4. doi: 10.1128/iai.53.2.432-434.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleary PP, Prahbu U, Dale JB, Wexler DE, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60((12)):5219–23. doi: 10.1128/iai.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynskey NN, Reglinski M, Calay D, Siggins MK, Mason JC, Botto M, et al. Multi-functional mechanisms of immune evasion by the streptococcal complement inhibitor C5a peptidase. PLoS Pathog. 2017;13((8)):e1006493. doi: 10.1371/journal.ppat.1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson ET, Wetherell MG, Winter LA, Olmsted SB, Cleary PP, Matsuka YV. Processing, stability, and kinetic parameters of C5a peptidase from Streptococcus pyogenes. Eur J Biochem. 2002;269((19)):4839–51. doi: 10.1046/j.1432-1033.2002.03183.x. [DOI] [PubMed] [Google Scholar]
- 22.Kagawa TF, O'Connell MR, Mouat P, Paoli M, O'Toole PW, Cooney JC. Model for substrate interactions in C5a peptidase from Streptococcus pyogenes: a 1.9 A crystal structure of the active form of ScpA. J Mol Biol. 2009;386((3)):754–72. doi: 10.1016/j.jmb.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor SP, Cleary PP. In vivo Streptococcus pyogenes C5a peptidase activity: analysis using transposon- and nitrosoguanidine-induced mutants. J Infect Dis. 1987;156((3)):495–504. doi: 10.1093/infdis/156.3.495. [DOI] [PubMed] [Google Scholar]
- 24.Ji Y, Carlson B, Kondagunta A, Cleary PP. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect Immun. 1997;65((6)):2080–7. doi: 10.1128/iai.65.6.2080-2087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckmann C, Waggoner JD, Harris TO, Tamura GS, Rubens CE. Identification of novel adhesins from Group B Streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect Immun. 2002;70((6)):2869. doi: 10.1128/IAI.70.6.2869-2876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura GS, Hull JR, Oberg MD, Castner DG. High-affinity interaction between fibronectin and the Group B Streptococcal C5a peptidase is unaffected by a naturally occurring four-amino-acid deletion that eliminates peptidase activity. Infect Immun. 2006;74((10)):5739. doi: 10.1128/IAI.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Q, Stafslien D, Purushothaman SS, Cleary P. The Group B Streptococcal C5a peptidase is both a specific protease and an invasin. Infect Immun. 2002;70((5)):2408. doi: 10.1128/IAI.70.5.2408-2413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol Microbiol. 2006;62((2)):491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in Group b Streptococcus. J Bacteriol. 2005;187((3)):1105. doi: 10.1128/JB.187.3.1105-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franken C, Haase G, Brandt C, Weber-Heynemann J, Martin S, Lämmler C, et al. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol Microbiol. 2001;41((4)):925–35. doi: 10.1046/j.1365-2958.2001.02563.x. [DOI] [PubMed] [Google Scholar]
- 31.Emaneini M, Khoramian B, Jabalameli F, Abani S, Dabiri H, Beigverdi R. Comparison of virulence factors and capsular types of Streptococcus agalactiae isolated from human and bovine infections. Microb Pathog. 2016;91:1–4. doi: 10.1016/j.micpath.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Gleich-Theurer U, Aymanns S, Haas G, Mauerer S, Vogt J, Spellerberg B. Human serum induces Streptococcal C5a peptidase expression. Infect Immun. 2009;77((9)):3817. doi: 10.1128/IAI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arenas J, Bossers-de Vries R, Harders-Westerveen J, Buys H, Ruuls-van Stalle LMF, Stockhofe-Zurwieden N, et al. In vivo transcriptomes of Streptococcus suis reveal genes required for niche-specific adaptation and pathogenesis. Virulence. 2019;10((1)):334–51. doi: 10.1080/21505594.2019.1599669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockmeier SL, Loving CL, Nicholson TL, Wang J, Peters SE, Weinert L, et al. Use of proteins identified through a functional genomic screen to develop a protein subunit vaccine that provides significant protection against virulent Streptococcus suis in pigs. Infect Immun. 2018;86((3)):e00559–17. doi: 10.1128/IAI.00559-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Z, Fu Q, Chen Y, Li M, Cong P, Mo D, et al. Streptococcus equi ssp. zooepidemicus C5a peptidase, a putative invasin, induces protective immune response in mice. Res Vet Sci. 2013;95((2)):444–50. doi: 10.1016/j.rvsc.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 36.Turner CE, Bubba L, Efstratiou A. Pathogenicity factors in group C and G Streptococci. Microbiol Spectr. 2019;7((3)) doi: 10.1128/microbiolspec.gpp3-0020-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locke JB, Aziz RK, Vicknair MR, Nizet V, Buchanan JT. Streptococcus iniae M-like protein contributes to virulence in fish and is a target for live attenuated vaccine development. PLoS One. 2008;3((7)):e2824. doi: 10.1371/journal.pone.0002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo HH, Cheng WS. Distribution of virulence factors and association with emm polymorphism or isolation site among beta-hemolytic group G Streptococcus dysgalactiae subspecies equisimilis. APMIS. 2015;123((1)):45–52. doi: 10.1111/apm.12305. [DOI] [PubMed] [Google Scholar]
- 39.Hidalgo-Grass C, Dan-Goor M, Maly A, Eran Y, Kwinn LA, Nizet V, et al. Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet. 2004;363((9410)):696–703. doi: 10.1016/S0140-6736(04)15643-2. [DOI] [PubMed] [Google Scholar]
- 40.Edwards RJ, Taylor GW, Ferguson M, Murray S, Rendell N, Wrigley A, et al. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J Infect Dis. 2005;192((5)):783–90. doi: 10.1086/432485. [DOI] [PubMed] [Google Scholar]
- 41.Zingaretti C, Falugi F, Nardi-Dei V, Pietrocola G, Mariani M, Liberatori S, et al. Streptococcus pyogenes SpyCEP: a chemokine-inactivating protease with unique structural and biochemical features. FASEB J. 2010;24((8)):2839–48. doi: 10.1096/fj.09-145631. [DOI] [PubMed] [Google Scholar]
- 42.Fritzer A, Noiges B, Schweiger D, Rek A, Kungl AJ, von Gabain A, et al. Chemokine degradation by the Group A streptococcal serine proteinase ScpC can be reconstituted in vitro and requires two separate domains. Biochem J. 2009;422((3)):533–42. doi: 10.1042/BJ20090278. [DOI] [PubMed] [Google Scholar]
- 43.Lei B, Mackie S, Lukomski S, Musser JM. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect Immun. 2000;68((12)):6807–18. doi: 10.1128/iai.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumby P, Zhang S, Whitney AR, Falugi F, Grandi G, Graviss EA, et al. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect Immun. 2008;76((3)):978–85. doi: 10.1128/IAI.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldblatt J, Lawrenson RA, Muir L, Dattani S, Hoffland A, Tsuchiya T, et al. A requirement for neutrophil glycosaminoglycans in chemokine: receptor interactions is revealed by the Streptococcal protease SpyCEP. J Immunol. 2019;202((11)):3246–55. doi: 10.4049/jimmunol.1801688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurupati P, Turner CE, Tziona I, Lawrenson RA, Alam FM, Nohadani M, et al. Chemokine-cleaving Streptococcus pyogenes protease SpyCEP is necessary and sufficient for bacterial dissemination within soft tissues and the respiratory tract. Mol Microbiol. 2010;76((6)):1387–97. doi: 10.1111/j.1365-2958.2010.07065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proudfoot AEI, Johnson Z, Bonvin P, Handel TM. Glycosaminoglycan interactions with chemokines add complexity to a complex system. Pharmaceuticals. 2017;10((3)):70. doi: 10.3390/ph10030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proudfoot AE. Chemokines and glycosaminoglycans. Front Immunol. 2015;6:246. doi: 10.3389/fimmu.2015.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100((4)):1885–90. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, et al. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 1997;91((3)):385–95. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 51.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, et al. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36((44)):13570–8. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 52.Turner CE, Kurupati P, Wiles S, Edwards RJ, Sriskandan S. Impact of immunization against SpyCEP during invasive disease with two streptococcal species: Streptococcus pyogenes and Streptococcus equi. Vaccine. 2009;27((36)):4923–9. doi: 10.1016/j.vaccine.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasser MW, Raghuwanshi SK, Grant DJ, Jala VR, Rajarathnam K, Richardson RM. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 2009;183((5)):3425–32. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joseph PRB, Sawant KV, Rajarathnam K. Heparin-bound chemokine CXCL8 monomer and dimer are impaired for CXCR1 and CXCR2 activation: implications for gradients and neutrophil trafficking. Open Biol. 2017;7((11)):170168. doi: 10.1098/rsob.170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hidalgo-Grass C, Mishalian I, Dan-Goor M, Belotserkovsky I, Eran Y, Nizet V, et al. A Streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 2006;25((19)):4628–37. doi: 10.1038/sj.emboj.7601327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180((6)):4308–15. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 57.Ansaldi F, Canepa P, de Florentiis D, Bandettini R, Durando P, Icardi G. Increasing incidence of Streptococcus pneumoniae serotype 19A and emergence of two vaccine escape recombinant ST695 strains in Liguria, Italy, 7 years after implementation of the 7-valent conjugated vaccine. Clin Vaccine Immunol. 2011;18((2)):343–5. doi: 10.1128/CVI.00383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sjölinder H, Lövkvist L, Plant L, Eriksson J, Aro H, Jones A, et al. The ScpC protease of Streptococcus pyogenes affects the outcome of sepsis in a murine model. Infect Immun. 2008;76((9)):3959–66. doi: 10.1128/IAI.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, et al. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe. 2008;4((2)):170–8. doi: 10.1016/j.chom.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siggins MK, Lynskey NN, Lamb LE, Johnson LA, Huse KK, Pearson M, et al. Extracellular bacterial lymphatic metastasis drives Streptococcus pyogenes systemic infection. Nat Commun. 2020;11((1)):4697. doi: 10.1038/s41467-020-18454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner CE, Kurupati P, Jones MD, Edwards RJ, Sriskandan S. Emerging role of the interleukin-8 cleaving enzyme SpyCEP in clinical Streptococcus pyogenes infection. J Infect Dis. 2009;200((4)):555–63. doi: 10.1086/603541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 2002;99((21)):13855–60. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2((1)):e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siezen RJ, de Vos WM, Leunissen JA, Dijkstra BW. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991;4((7)):719–37. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- 65.Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6((3)):501–23. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siezen RJ, Renckens B, Boekhorst J. Evolution of prokaryotic subtilases: genome-wide analysis reveals novel subfamilies with different catalytic residues. Proteins. 2007;67((3)):681–94. doi: 10.1002/prot.21290. [DOI] [PubMed] [Google Scholar]
- 67.Bethe G, Nau R, Wellmer A, Hakenbeck R, Reinert RR, Heinz HP, et al. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol Lett. 2001;205((1)):99–104. doi: 10.1111/j.1574-6968.2001.tb10931.x. [DOI] [PubMed] [Google Scholar]
- 68.Mahdi LK, Van der Hoek MB, Ebrahimie E, Paton JC, Ogunniyi AD. Characterization of pneumococcal genes involved in bloodstream invasion in a mouse model. PLoS One. 2015;10((11)):e0141816. doi: 10.1371/journal.pone.0141816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirza S, Wilson L, Benjamin WH, Jr, Novak J, Barnes S, Hollingshead SK, et al. Serine protease PrtA from Streptococcus pneumoniae plays a role in the killing of S. pneumoniae by apolactoferrin. Infect Immun. 2011;79((6)):2440–50. doi: 10.1128/IAI.00489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hendriksen WT, Bootsma HJ, van Diepen A, Estevão S, Kuipers OP, de Groot R, et al. Strain-specific impact of PsaR of Streptococcus pneumoniae on global gene expression and virulence. Microbiology (Reading) 2009;155((Pt 5)):1569–79. doi: 10.1099/mic.0.025072-0. [DOI] [PubMed] [Google Scholar]
- 71.Kloosterman TG, Witwicki RM, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae. J Bacteriol. 2008;190((15)):5382–93. doi: 10.1128/JB.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manzoor I, Shafeeq S, Kloosterman TG, Kuipers OP. Co(2+)-dependent gene expression in Streptococcus pneumoniae: opposite effect of Mn(2+) and Co(2+) on the expression of the virulence genes psaBCA, pcpA, and prtA. Front Microbiol. 2015;6:748. doi: 10.3389/fmicb.2015.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manzoor I, Shafeeq S, Kuipers OP. Ni2+-dependent and PsaR-mediated regulation of the virulence genes pcpA, psaBCA, and prtA in Streptococcus pneumoniae. PLoS One. 2015;10((11)):e0142839. doi: 10.1371/journal.pone.0142839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris TO, Shelver DW, Bohnsack JF, Rubens CE. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J Clin Invest. 2003;111((1)):61–70. doi: 10.1172/JCI16270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bryan JD, Shelver DW. Streptococcus agalactiae CspA is a serine protease that inactivates chemokines. J Bacteriol. 2009;191((6)):1847–54. doi: 10.1128/JB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryan JD, Liles R, Cvek U, Trutschl M, Shelver D. Global transcriptional profiling reveals Streptococcus agalactiae genes controlled by the MtaR transcription factor. BMC Genomics. 2008;9((1)):607. doi: 10.1186/1471-2164-9-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shelver D, Bryan JD. Expression of the Streptococcus agalactiae virulence-associated protease CspA in a soluble, active form utilizing the Gram-positive host, Lactococcus lactis. J Biotechnol. 2008;136((3–4)):129–34. doi: 10.1016/j.jbiotec.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 78.Brega S, Caliot E, Trieu-Cuot P, Dramsi S. SecA localization and SecA-dependent secretion occurs at new division septa in group B Streptococcus. PLoS One. 2013;8((6)):e65832. doi: 10.1371/journal.pone.0065832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Q, Liu P, Yu Z, Zhao G, Li J, Teng L, et al. Identification of a cell wall-associated subtilisin-like serine protease involved in the pathogenesis of Streptococcus suis serotype 2. Microb Pathog. 2010;48((3–4)):103–9. doi: 10.1016/j.micpath.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Bonifait L, de la Cruz Dominguez-Punaro M, Vaillancourt K, Bart C, Slater J, Frenette M, et al. The cell envelope subtilisin-like proteinase is a virulence determinant for Streptococcus suis. BMC Microbiol. 2010;10:42. doi: 10.1186/1471-2180-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonifait L, Vaillancourt K, Gottschalk M, Frenette M, Grenier D. Purification and characterization of the subtilisin-like protease of Streptococcus suis that contributes to its virulence. Vet Microbiol. 2011;148((2–4)):333–40. doi: 10.1016/j.vetmic.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 82.Bonifait L, Grenier D. The SspA subtilisin-like protease of Streptococcus suis triggers a pro-inflammatory response in macrophages through a non-proteolytic mechanism. BMC Microbiol. 2011;11:47. doi: 10.1186/1471-2180-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin S, Li M, Rao X, Yao X, Zhong Q, Wang M, et al. Subtilisin-like protease-1 secreted through type IV secretion system contributes to high virulence of Streptococcus suis 2. Sci Rep. 2016;6:27369. doi: 10.1038/srep27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Stoppelaar SF, Bootsma HJ, Zomer A, Roelofs JJ, Hermans PW, van 't Veer C, et al. Streptococcus pneumoniae serine protease HtrA, but not SFP or PrtA, is a major virulence factor in pneumonia. PLoS One. 2013;8((11)):e80062. doi: 10.1371/journal.pone.0080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneewind O, Missiakas D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim Biophys Acta. 2014;1843((8)):1687–97. doi: 10.1016/j.bbamcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stafslien DK, Cleary PP. Characterization of the streptococcal C5a peptidase using a C5a-green fluorescent protein fusion protein substrate. J Bacteriol. 2000;182((11)):3254–8. doi: 10.1128/jb.182.11.3254-3258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jobichen C, Tan YC, Prabhakar MT, Nayak D, Biswas D, Pannu NS, et al. Structure of ScpC, a virulence protease from Streptococcus pyogenes, reveals the functional domains and maturation mechanism. Biochem J. 2018;475((17)):2847–60. doi: 10.1042/BCJ20180145. [DOI] [PubMed] [Google Scholar]
- 88.McKenna S, Malito E, Rouse SL, Abate F, Bensi G, Chiarot E, et al. Structure, dynamics and immunogenicity of a catalytically inactive CXC chemokine-degrading protease SpyCEP from Streptococcus pyogenes. Comput Struct Biotechnol J. 2020;18:650–60. doi: 10.1016/j.csbj.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, et al. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr J Chem. 2013;53((3–4)):199–206. [Google Scholar]
- 91.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44((W1)):W344–50. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47((W1)):W256–w59. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abate F, Malito E, Falugi F, Margarit Y Ros I, Bottomley MJ. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of SpyCEP, a candidate antigen for a vaccine against Streptococcus pyogenes. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69((Pt 10)):1103–6. doi: 10.1107/S1744309113024871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21((16)):3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 95.Mészáros B, Erdos G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46((W1)):W329–w37. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Erdős G, Dosztányi Z. Analyzing protein disorder with IUPred2A. Curr Protoc Bioinformatics. 2020;70((1)):e99. doi: 10.1002/cpbi.99. [DOI] [PubMed] [Google Scholar]
- 97.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5((11)):685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 98.WHO . Pneumoccocal disease. 2005. [Google Scholar]
- 99.Rodríguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. 2006;24((2)):191–7. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- 100.Rivera-Hernandez T, Carnathan DG, Jones S, Cork AJ, Davies MR, Moyle PM, et al. An experimental group A streptococcus vaccine that reduces pharyngitis and tonsillitis in a nonhuman primate model. mBio. 2019;10((2)):e00693–19. doi: 10.1128/mBio.00693-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bensi G, Mora M, Tuscano G, Biagini M, Chiarot E, Bombaci M, et al. Multi high-throughput approach for highly selective identification of vaccine candidates: the Group A Streptococcus case. Mol Cell Proteomics. 2012;11((6)):M111015693. doi: 10.1074/mcp.M111.015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reglinski M, Lynskey NN, Choi YJ, Edwards RJ, Sriskandan S. Development of a multicomponent vaccine for Streptococcus pyogenes based on the antigenic targets of IVIG. J Infect. 2016;72((4)):450–9. doi: 10.1016/j.jinf.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu Z, Rivera-Hernandez T, Moyle PM. Development of an enzyme-mediated, site-specific method to conjugate toll-like Receptor 2 agonists onto protein antigens: toward a broadly protective, four component, Group A Streptococcal self-adjuvanting lipoprotein-fusion combination vaccine. ACS Infect Dis. 2020;6((7)):1770–82. doi: 10.1021/acsinfecdis.0c00047. [DOI] [PubMed] [Google Scholar]
- 104.Batzloff MR, Hayman WA, Davies MR, Zeng M, Pruksakorn S, Brandt ER, et al. Protection against group A streptococcus by immunization with J8-diphtheria toxoid: contribution of J8- and diphtheria toxoid-specific antibodies to protection. J Infect Dis. 2003;187((10)):1598–608. doi: 10.1086/374800. [DOI] [PubMed] [Google Scholar]
- 105.Pandey M, Langshaw E, Hartas J, Lam A, Batzloff MR, Good MF. A synthetic M protein peptide synergizes with a CXC chemokine protease to induce vaccine-mediated protection against virulent streptococcal pyoderma and bacteremia. J Immunol. 2015;194((12)):5915–25. doi: 10.4049/jimmunol.1500157. [DOI] [PubMed] [Google Scholar]
- 106.Pandey M, Mortensen R, Calcutt A, Powell J, Batzloff MR, Dietrich J, et al. Combinatorial synthetic peptide vaccine strategy protects against hypervirulent CovR/S mutant Streptococci. J Immunol. 2016;196((8)):3364–74. doi: 10.4049/jimmunol.1501994. [DOI] [PubMed] [Google Scholar]
- 107.Cleary PP, Peterson J, Chen C, Nelson C. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect Immun. 1991;59((7)):2305–10. doi: 10.1128/iai.59.7.2305-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O'Connor SP, Darip D, Fraley K, Nelson CM, Kaplan EL, Cleary PP. The human antibody response to streptococcal C5a peptidase. J Infect Dis. 1991;163((1)):109–16. doi: 10.1093/infdis/163.1.109. [DOI] [PubMed] [Google Scholar]
- 109.Cleary PP, Matsuka YV, Huynh T, Lam H, Olmsted SB. Immunization with C5a peptidase from either group A or B streptococci enhances clearance of group A streptococci from intranasally infected mice. Vaccine. 2004;22((31–32)):4332–41. doi: 10.1016/j.vaccine.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 110.Park HS, Cleary PP. Active and passive intranasal immunizations with streptococcal surface protein C5a peptidase prevent infection of murine nasal mucosa-associated lymphoid tissue, a functional homologue of human tonsils. Infect Immun. 2005;73((12)):7878–86. doi: 10.1128/IAI.73.12.7878-7886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheng Q, Debol S, Lam H, Eby R, Edwards L, Matsuka Y, et al. Immunization with C5a peptidase or peptidase-type III polysaccharide conjugate vaccines enhances clearance of group B Streptococci from lungs of infected mice. Infect Immun. 2002;70((11)):6409–15. doi: 10.1128/IAI.70.11.6409-6415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suvorov AN, Grabovskaia KB, Leont'eva GF, Meringova LF, Koroleva IN, Duplik NV, et al. [Recombinant fragments of conservative proteins of group B streptococci as a basis of specific vaccine] Zh Mikrobiol Epidemiol Immunobiol. 2010;((2)):44–50. [PubMed] [Google Scholar]
- 113.Xue G, Yu L, Li S, Shen X. Intranasal immunization with GBS surface protein Sip and ScpB induces specific mucosal and systemic immune responses in mice. FEMS Immunol Med Microbiol. 2010;58((2)):202–10. doi: 10.1111/j.1574-695X.2009.00623.x. [DOI] [PubMed] [Google Scholar]
- 114.Santillan DA, Rai KK, Santillan MK, Krishnamachari Y, Salem AK, Hunter SK. Efficacy of polymeric encapsulated C5a peptidase-based group B streptococcus vaccines in a murine model. Am J Obstet Gynecol. 2011;205((3)):249. doi: 10.1016/j.ajog.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pietrocola G, Arciola CR, Rindi S, Montanaro L, Speziale P. Streptococcus agalactiae non-pilus, cell wall-anchored proteins: involvement in colonization and pathogenesis and potential as vaccine candidates. Front Immunol. 2018;9:602. doi: 10.3389/fimmu.2018.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect. 2014;3((6)):e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT, Sinh DX, et al. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46((5)):659–67. doi: 10.1086/527385. [DOI] [PubMed] [Google Scholar]
- 118.Huong VTL, Turner HC, Kinh NV, Thai PQ, Hoa NT, Horby P, et al. Burden of disease and economic impact of human Streptococcus suis infection in Viet Nam. Trans R Soc Trop Med Hyg. 2019;113((6)):341–50. doi: 10.1093/trstmh/trz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jobin MC, Grenier D. Identification and characterization of four proteases produced by Streptococcus suis. FEMS Microbiol Lett. 2003;220((1)):113–9. doi: 10.1016/S0378-1097(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 120.Xing X, Bi S, Fan X, Jin M, Liu W, Wang B. Intranasal vaccination with multiple virulence factors promotes mucosal cof Streptococcus suis across serotypes and protects against meningitis in mice. J Infect Dis. 2019;220((10)):1679–87. doi: 10.1093/infdis/jiz352. [DOI] [PubMed] [Google Scholar]
- 121.Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect Immun. 2001;69((3)):1593–8. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anderson RJ, Guru S, Weeratna R, Makinen S, Falconer DJ, Sheppard NC, et al. In vivo screen of genetically conserved Streptococcus pneumoniae proteins for protective immunogenicity. Vaccine. 2016;34((50)):6292–300. doi: 10.1016/j.vaccine.2016.10.061. [DOI] [PubMed] [Google Scholar]
- 123.Hsu CF, Hsiao CH, Tseng SF, Chen JR, Liao YJ, Chen SJ, et al. PrtA immunization fails to protect against pulmonary and invasive infection by Streptococcus pneumoniae. Respir Res. 2018;19((1)):187. doi: 10.1186/s12931-018-0895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carvelli J, Demaria O, Vély F, Batista L, Chouaki Benmansour N, Fares J, et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588((588)):146–50. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park JH, Lee HK. Re-analysis of single cell transcriptome reveals that the NR3C1-CXCL8-neutrophil axis determines the severity of COVID-19. Front Immunol. 2020;11:2145. doi: 10.3389/fimmu.2020.02145. [DOI] [PMC free article] [PubMed] [Google Scholar]