Abstract
Purpose
The pattern of immune cells infiltrating the corneal stroma has been extensively studied in mice, but data on human tissue have been far less elaborate. To further characterize the number and differentiation state of resident immune cells in organ-cultured human corneal tissue, we employed a comprehensive bioinformatic deconvolution (xCell) of bulk RNA-sequencing (RNA-seq) data, immunohistochemistry (IHC), and flow cytometry (FC).
Methods
A transcriptome-based analysis of immune cell types in human corneal samples was performed. The results were validated by IHC, focusing on the identification of pro-inflammatory (M1) and regulatory (M2) macrophages. A protocol was established to identify these 2 different macrophage populations in human corneal tissue by means of FC. Subsequently, corneal samples in organ culture were differentially stimulated by IL-10, IL-4 & IL-13, or LPS and macrophage populations were evaluated regarding their response to these stimuli. Furthermore, cell survival was analyzed in correlation with time in organ culture.
Results
xCell-based mathematical deconvolution of bulk RNA-seq data revealed the presence of CD8 T cells, Th17 cells, dendritic cells, and macrophages as the predominant immune cell types in organ-cultured human corneal tissue. Furthermore, RNA-seq allowed the detection of different macrophage marker genes in corneal samples, including PTPRC (CD45), ITGAM (CD11b), CD14, and CD74. Our RNA-seq data showed no evidence of a relevant presence of monocytes in human corneal tissue. The presence of different macrophage subtypes was confirmed by IHC. The disintegration and subsequent FC analysis of human corneal samples showed the presence of both M1 (HLA-DR<sup>+</sup>, CD282<sup>+</sup>, CD86<sup>+</sup>, and CD284<sup>+</sup>) and M2 (CD163<sup>+</sup> and CD206<sup>+</sup>) macrophage subtypes. Furthermore, we found that the total number of macrophages in corneal samples decreased more than the total cell count with increasing tissue culture time. Treatment with IL-10 led to higher total cell counts per cornea and to an increased expression of the M2 marker CD163 (p < 0.05) while expression levels of various M1 macrophage markers were not significantly reduced by interleukin treatment.
Conclusions
Regarding different macrophage populations, untreated human corneas showed more M1 than M2 macrophages. With increasing organ culture time, these macrophages decreased. In terms of cell dynamics, adding interleukins to the organ culture medium influenced the phenotype of macrophages within the cornea as detected by FC. Modifying the immunomodulatory properties of human grafts appears a promising approach to further reduce the risk of graft rejection in patients. In this context, treatment with interleukins was more effective in upregulating M2 macrophages than in suppressing M1 macrophages in corneal tissue.
Keywords: Cornea, Macrophages, RNA-sequencing, Flow cytometry, Immunophenotyping, Corneal graft, Corneal graft rejection
Introduction
Despite its transparency, the human cornea contains various cell types, including both differentiated and undifferentiated immune cells. Among these cells, CD8 T cells, dendritic cells (DC), and macrophages take on a special role [1, 2]. Even though the innate immune cells are considered to be tissue resident, especially macrophages and DC can traffic in and out of the cornea in the steady state and translocate to locoregional lymph nodes following corneal transplantation [3, 4]. Especially during corneal graft rejection, donor antigen-presenting cells (APCs) leave the graft and interact with the recipient's immune system (direct pathway), just as recipient APCs can enter the graft, take up and process alloantigens, and induce consecutive immune responses (indirect pathway) [5]. In humans, the role and contribution of the 2 different pathways in corneal graft rejection is not yet fully understood, nor is the role of the phenotype of the cells present in the graft at the time of transplantation.
Identification, localization, and phenotyping of APCs in the human cornea remain an experimental bottleneck in this context. Furthermore, the innate immune cell profile of the human cornea seems to be a moving target − altered not only by corneal graft rejection but also by underlying diseases of the cornea and the ocular surface [6]. Early work describing the cellular composition of the human cornea mainly focused on histology and immunohistochemistry [7] as well as confocal microscopy [8, 9]. However, these approaches do not allow a simultaneous analysis of several parameters of macrophages as a substantial part of the corneal immune system. Recent approaches therefore focus on immunophenotyping using RNA-sequencing (RNA-seq) [2] and flow cytometry (FC) [10].
Human corneas are kept in organ culture for several days to weeks before transplantation. One approach to overcome graft rejection mediated by the direct route may be to add supplements to the cell culture to shape the phenotype of immune cells within the graft. In mice, incubation of donor corneas with IL-10 and TGF-β altered the graft-APC-phenotype and has been shown to improve allograft survival [11]. If this were also true for human APC in corneal tissue, it would offer an additional therapeutic option for patients with increased risk for corneal graft rejection.
Currently, the composition of immune cells in the human cornea is underinvestigated. The present knowledge is mostly restricted to murine data and to adaptive rather than innate immune cells. Moreover, the current FC protocols appear to be little or not at all standardized. In the present study, we therefore performed RNA-seq on formalin-fixed and paraffin-embedded (FFPE) healthy corneas and established a FC protocol for processing and phenotyping human corneal cells. We investigated which macrophage subtypes were present in the human cornea, with special focus on pro-inflammatory (M1) and regulatory (M2) macrophages. We also analyzed the functional response of these cells to different interleukins and assessed the APC differentiation state in organ culture.
Materials and Methods
Ethics
Ethics approval was granted by the local Ethics Committee (Research Ethics Committee of University of Freiburg; Germany; reference numbers: 572/14 and 25/20). This study adheres to the tenets of the Declaration of Helsinki.
RNA Extraction
Corneal samples were dissected from 2 enucleated eyes (due to ciliary body melanoma), as well as from 1 corneal explant (due to corneal endothelial failure), which were FFPE immediately after surgery, as previously described [12]. Samples were fixed immediately after surgery in 4% formalin for 12 h, dehydrated in alcohol, and processed for paraffin embedding. Histological diagnoses were made by 2 experienced ophthalmic pathologists. 10 to 15 FFPE sections (4 µm thick) from each cornea were collected and stored in tubes until RNA isolation, which was performed as previously described [12]. Briefly, total RNA was isolated from FFPE samples using the Quick-RNA FFPE Kit (Zymo Research). Following a DNAse I digestion using the Baseline-ZERO kit (Epicentre), the RNA concentration was measured with the Qubit RNA HS Assay Kit on a Qubit Fluorometer (Life Technologies). The RNA quality was determined with the RNA Pico Sensitivity Assay on a LabChip GXII Touch (PerkinElmer).
RNA-Sequencing
RNA-sequencing was performed using massive analysis of cDNA ends (MACE), a 3′ RNA-sequencing method as previously described [13]. Briefly, 3 barcoded libraries were sequenced simultaneously on the NextSeq 500 (Illumina). PCR bias was removed based on unique molecular identifiers encoded in the library barcodes. The sequencing data are available in the Gene Expression Omnibus Database under the accession number GSE164192.
Bioinformatics
Sequencing data (fastq files) were uploaded to and analyzed on the Galaxy web platform (https://usegalaxy.eu/) [14]. Quality control was performed with FastQC Galaxy Version 0.72 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ last access on December, 21 2020). Reads were mapped to the human reference genome (Gencode, release 36, hg38) with RNA STAR Galaxy Version 2.7.6a [15] with default parameters using the Gencode annotation file (Gencode, release 36, https://www.gencodegenes.org/human/releases.html). Reads mapped to the human reference genome were counted using featureCounts Galaxy Version 2.0.1 [16] with default parameters using the aforementioned annotation file. The output of featureCounts was imported to RStudio (Version 1.2.1335, R Version 3.5.3). Gene symbols were determined based on ENSEMBL release 101 (Human genes, GRCh38.p13, downloaded on November 23, 2020) [17]. Transcripts per million were calculated based on the output of featureCounts (assigned reads and feature length), as previously described [18]. Immune cell types enriched in corneal samples were determined using bioinformatic deconvolution analysis. For this purpose the tool xCell [19] was applied, using the implemented Bindea cell signature. The results were filtered for immune cell types. Heatmaps were created with the R package ComplexHeatmap 1.20.0 [20]. Other data visualization was performed using the ggplot2 package [21].
Immunofluorescence
For immunofluorescence staining, 3 macroscopically normal eyes from 2 female and 1 male body donors were analyzed. The eyes were enucleated in accordance with the informed consent of body donors, which was contractually secured during lifetime. In accordance with ethical requirements, no data other than age, sex, body weight, and cause of death were recorded. Enucleation, tissue preparation, and staining were performed at the Institute of Anatomy, Leipzig University. Eyes were fixed in 4% PFA for 16 h at 4°C and dissected under a binocular microscope. Corneas were harvested, dehydrated in 30% sucrose and embedded in Tissue-Tek® O.C.T.TM Compound (Sakura Finetek Germany GmbH) for cryosections. 12 µm sections were blocked with 2% bovine serum albumin (BSA) and 2% normal donkey serum (NDS) in PBS Triton-X 0.1% for 60 min at room temperature. Primary antibodies were added at a dilution of 1:100 for CD206 (ab64693, Abcam), 1:200 for HLA-DP, DQ, DR (M0775, DAKO), and 1:500 for IBA1 (ab5076, Abcam) in PBS containing 2% BSA and 2% NDS in PBS Triton-X 0.1% overnight at 4°C. Following washing with 2% BSA and 0.2% NDS in PBS Triton-X 0.1%, secondary antibodies were added at a dilution of 1:500 (Alexa Fluor® 568 and Alexa Fluor® 647, Thermo Fisher Scientific) in PBS Triton-X 0.1% at room temperature for 90 min in the dark. After washing at least 3 times with 2% BSA and 0.2% NDS in PBS Triton-X 0.1%, the slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) 1:10,000 for 10 min, washed 3 times with PBS followed by autofluorescence quenching with TrueBlack® Lipofuscin Autofluorescence Quencher (Biotium) according to the manufacturers' instructions. Slides were imaged using a Fluoview FV 1000 confocal microscope (Olympus) equipped with a 20 × 0.75 NA U Plan S Apo and 40 × 0.95 NA U Plan S (Olympus).
Corneal Samples Acquisition for FC
Human corneas were received from the Lions Cornea Bank Baden-Württemberg (Freiburg im Breisgau, Germany). Corneas not suitable for transplantation due to a reduced corneal endothelial cell count (endothelial cell count between 1,000 and 2,000 cells/m2) or cornea remnants following Descemet's membrane endothelial keratoplasty implant preparation were used as previously described [22]. Informed consent by the donor or next of kin for scientific use was prerequisite for study enrollment. Corneas from donors with transmissible diseases were excluded from the experiments according to the list of contraindications of infectious origin endorsed by the European Eye bank Association (https://www.eeba.eu/minimum-medical-standards-revision-5.html).
After collection, corneas were stored in the eye bank in Tissue-C medium (reference TIS 001-00, ALCHIMIA, Italy) with 2% FCS at 37°C and 5% CO2. In general, the organ medium was changed weekly. An exception to this are the experiments related to the cell content of the culture medium. In this case, the objective was to achieve maximum culture duration. The manufacturer indicates a theoretical culture duration of up to 6 weeks; in the respective experiments, the corneas were maintained for 45 days ± 5.9 (mean ± SD). The culture medium supernatants as well as the pH were checked regularly to exclude contamination. For experiments involving medium containing dextran, the corneas were cut in half and one half was incubated in the aforementioned Tissue-C medium, the other half in Carry-C medium (containing 5% dextran, reference CAR 001-00, ALCHIMIA, Italy) for 24 h in the incubator as under the above conditions.
Stimulation
Central corneas were trephined with a 9.5-mm trephine; corneo-scleral rims were not used. Corneal buttons were flattened by 4 radial cuts toward the center. Corneas were immersed in a 12-well plate (Thermo Fischer Scientific, Ref. 150628) with 3 mL tissue culture medium (CARRY-C, Alchimia, Ref. CAR 001-00) and the appropriate stimulus (Table 1) at 37°C and 5% CO2 for 24 h, respectively. LPS and IL-10 were each used separately as stimuli, whereas IL-4 and IL-13 were used in combination. Untreated corneas were incubated without a stimulus and served as controls.
Table 1.
Stimuli
| Stimulus | Stock solution | Working solution, ng/mL | Company |
|---|---|---|---|
| LPS | 1 mg/mL | 100 | Sigma Ref. L4391 |
| IL-13 | 2 µg/mL | 20 | Peprotech Ref. 200-13 |
| IL-4 | 5 µg/mL | 40 | Peprotech Ref. 200-04 |
| IL-10 | 2 µg/mL | 30 | Peprotech Ref. 200-10 |
Processing of Human Corneal Samples for FC
After stimulation corneas were removed from the well and rinsed with PBS (Gibco, Ref. 10010023). All corneas were de-epithelialized before cutting. Descemet's membrane of the cornea of samples from the eye bank were cautiously peeled off and completely removed, similar to the preparation for DMEK. Corneas were cut into 1 × 1 cm2 fragments using a sterile scalpel. Fragments were placed into a 12-well plate and covered with 1.1 mg/mL collagenase D (Roche, Ref. 11088866001) in HBSS w/o CaCl2 and MgCl2 (Gibco, Ref. 14170-088) and incubated overnight at 37°C and 5% CO2. The following day, fragments were resuspended vigorously and sheared through a cell strainer (70-µm nylon mesh, Falcon, Ref. 352350) into a 50 mL FALCON tube; the strainer was again rinsed with 10 mM EDTA (Serva, Cat.11280) in PBS. The resulting cell suspension was centrifuged at 4°C and 450 g for 15 min. The supernatant was discarded and the cells were transferred to a FC tube. The cell pellet was washed twice with 300 µL DPBS (Biotech, Ref. P04-35500) and resuspended in a FC tube for further use.
Flow Cytometry
To discriminate dead cells, 1 μL viability dye (Fixable viability stain 780, BD Horizon, Ref. 565388) was used according to the manufacturer's instructions. Afterward the cell suspension was washed with 2 mL FC buffer (2% FBS with 10 mM EDTA in DPBS) once, centrifuged for 7 min at 450 g at 4°C.
Before antibody staining, 20 µL blocking solution (FcR blocking reagent (human), MACS, Ref. 130-059-901) was added. The sample was incubated for 5 min at room temperature. In a next step, the antibodies were added to each tube (all diluted in a 1:100 dilution in FC buffer (2% FBS with 10 mM EDTA in DPBS; Table 2). Samples were vortexed and covered for 20 min on ice. After incubation the cell pellet was washed in 2 mL FC buffer, centrifuged for 7 min at 450 g, the supernatant was discarded. For fixation, 100 µL intracellular fixation buffer (eBioscienceTM IC Fixation Buffer, Formaldehyde, Invitrogen, Ref. 00-8222-49) was added and incubated for 40 min at room temperature.
Table 2.
FC antibodies
| Antibody | Aliases | Description | Company | Ref. ID |
|---|---|---|---|---|
| Anti-CD11b | Integrin α-M | BV711 rat anti-CD11b | BD Horizon | 563168 |
| Anti-CD14 | LPS receptor | BV711 mouse anti-human CD14 | BD Horizon | 563373 |
| Anti-CD45 | PTPRC | BV510 mouse anti-human CD45 | BD Horizon | 563204 |
| Anti-CD68 | GP110 | BV711 mouse anti-human CD68 | BD Horizon | 565594 |
| Anti-CD86 | B7-2/ B70 | APC mouse anti-human CD86 | BD Horizon | 560956 |
| Anti-CD163 | M130 | PE-CF594 mouse anti-human CD163 | BD Horizon | 562670 |
| Anti-CD206 | MMR | BB515 mouse anti-human CD206 | BD Horizon | 564668 |
| Anti-CD282 | TLR2 | BV421 mouse anti-human CD282 | BD Horizon | 565350 |
| Anti-CD284 | TLR4 | PE mouse anti-human TLR4 CD284 | BD Horizon | 564215 |
| Anti-HLA-DR | MHC class II receptor | BB700 mouse anti-human HLA-DR | BD Horizon | 745782 |
| FC, flow cytometry. |
CD68 requires an intracellular staining protocol. Therefore, the cell pellet was washed after fixation with 2 mL 1x permeabilization buffer (eBioscienceTM Permeabilization Buffer (10X), Invitrogen, Ref. 00-8333-56), centrifuged at 400 g for 5 min at 4°C. Thereafter anti-CD68 was added in 100 µL permeabilization buffer and incubated for 30 min at room temperature and protected from light. After the incubation, cells were washed with 2 mL 1× permeabilization buffer once and the pellet was resuspended in 150 µL FC buffer for analysis with CytoFLEX S FC (Beckman Coulter, Brea, CA, USA).
Gating Strategy
Cells were harvested from enzymatically digested human corneas as mentioned above. Cells were identified using the FCS-A and SSC-A; singlets were then detected in an FSC-H and FSC-W combined gate. Alive cells were gated by means of viability dye. Leukocytes were identified according to CD45 expression. Macrophages were identified by means of CD11b, CD14, and CD68 expression. A sequential gating strategy was used to identify macrophage subsets based on their surface markers, HLA-DR+, CD282+, CD86+, and CD284+ cells were identified as M1 macrophages, whereas CD163+ and CD206+ cells were identified as M2 macrophages. After CD45+ myeloid cells were identified, fluorescence minus one (FMO) staining was applied to identify positive cells for the respective aforementioned surface markers.
Data Analysis and Plotting
FC data analysis was carried out using both FlowJo software (Version 10.6.2) and GraphPad Prism (GraphPad Software, Version 6.0; La Jolla, USA). Data were analyzed descriptively using the R system [23], package ggplot2 [21], and Hmisc [24]. FC data are presented as exact cell count, as percentage (the percentage of positive cells from its parent gate), or per scale (per 50,000 alive cells). For data plotting the R platform, FlowJo software and GraphPad Prism were used. Box-whisker plots are plotted as median with quartiles. For statistical analysis the respective tests are mentioned in the figure legend.
Results
RNA-Seq and Immunofluorescence of Human Corneal Tissue
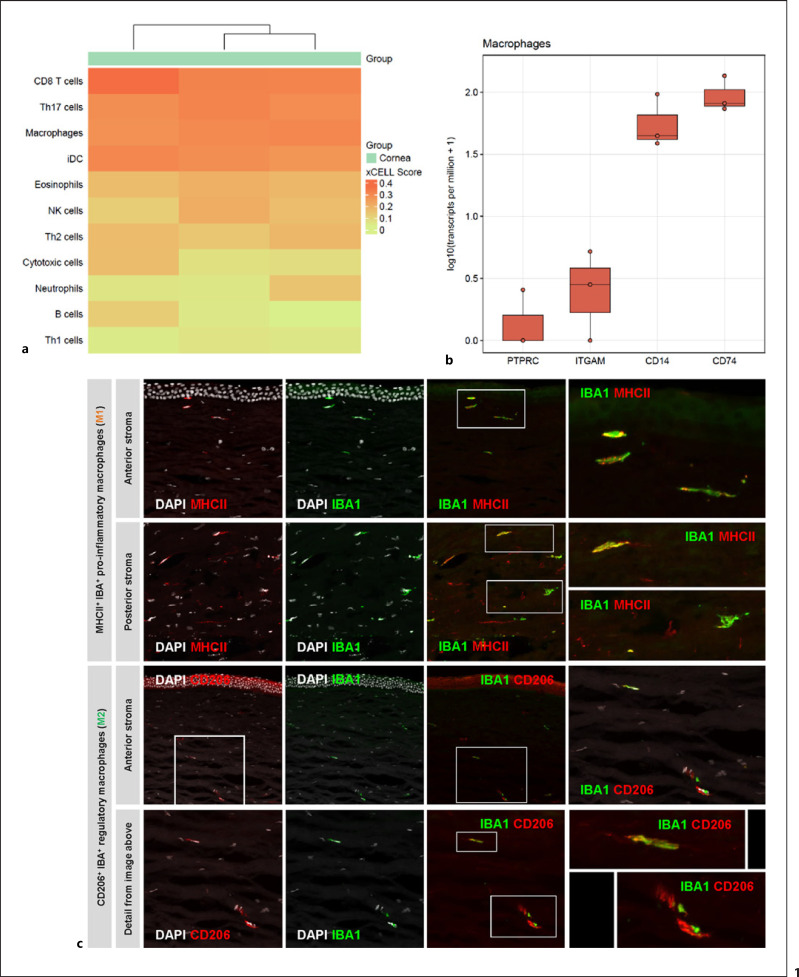
Bulk RNA-seq (online suppl. Fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000516669) revealed various genes being strongly expressed in corneal tissue, including genes that are known to be relevant in corneal cell signaling, such as S100A6 and S100A4 [25], KERA, coding for keratocan, a protein essential for maintaining corneal shape and transparency [26], TGFBI, mutations of which are involved in various corneal dystrophies [27], and ANGPTL7, which maintains corneal avascularity [28]. Profiles of different adaptive and innate immune cell types were identified in the transcriptional profile of 3 healthy corneas using the xCell deconvolution tool. A particularly high xCell score was found for CD8+ T cells and Th17 cells, as well as for DCs and macrophages (Fig. 1a). In line with this, we identified several genes by RNA-seq that are typically expressed in macrophages, including PTPRC (CD45), ITGAM (CD11b), CD14 − which were later applied for FACS-based identification of macrophages − and CD74 (Fig. 1b).
Fig. 1.
a RNA-seq and immunofluorescence. RNA-seq indicated the presence of various immune cells in the human cornea; notably CD8+ T cells, Th17 cells, dendritic cells, and macrophages (n = 3 individual corneas). b Besides PTPRC (CD45), macrophage-specific marker genes, such as ITGAM (CD11b), CD14, and CD74, can be detected by RNA-seq. c Immunofluorescence confirms the presence of M1 and M2 macrophages, with M1 macrophages appearing to be more abundant than M2 macrophages (representative images from 3 individual human corneas). RNA-seq, RNA-sequencing; M1, pro-inflammatory macrophages; M2, regulatory macrophages.
Results from RNA-seq were confirmed using immunofluorescence. In addition to IBA1 (AIF-1), co-staining with either MHC II (HLA-DRA) for identification of M1 macrophages or CD206 (MRC1) for identification of M2 macrophages was used (Fig. 1c). Staining with IBA1 revealed staining of cells in both the anterior and posterior stroma in all 3 corneas. This was confirmed by co-staining with MHC II, which also showed double positive staining of cells in all parts of the stroma. As in the staining with MHC II, positive cells were also found in the double staining of IBA1 with CD206.
FC Analysis of Corneal Tissue
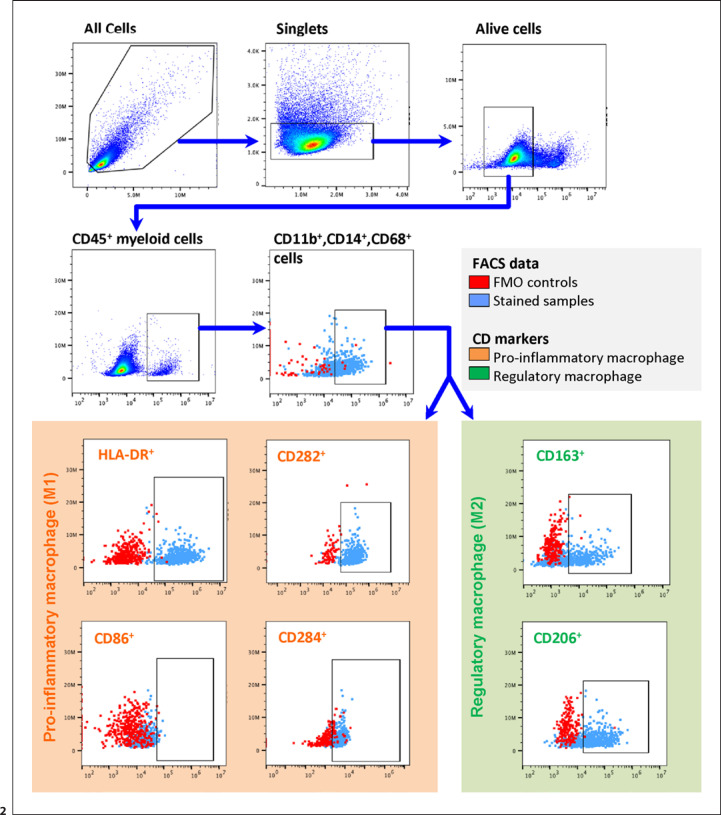
In addition to qualitative analysis, FC allows quantitative ex vivo analysis. In a further approach, RNA-seq and immunofluorescence were therefore extended by FC analysis. For this purpose, we established a FACS protocol for human corneal tissue (Fig. 2). After identification of cells, duplicates and doublets were excluded. Subsequently, identification of living cells was performed with the aid of viability dye. Leukocytes were gated using CD45 staining followed by the corresponding receptors CD11b and CD14 as well as CD68, based on our RNA-seq analysis, to identify cornea macrophages. To further subdivide the macrophages, HLA-DR, CD282, CD284, and CD86 were then used to identify M1 macrophages, whereas CD206 and CD163 were used for M2 macrophages (Fig. 2). In order to increase the precision of our gating strategy, FMO control was performed (red dots in Fig. 2). FMO controls showed a clear separation of the individual populations for most fluorochromes, except for anti-CD86. No distinct signal could be detected for CD86 in most of the samples, even when using the FMO control to set the baseline. Unstimulated corneal samples showed a ratio of M1 to M2 macrophages within CD45+ leukocytes of approximately two-thirds to one-thirds (online suppl. Fig. 4).
Fig. 2.
Gating strategy to identify macrophage subsets in human corneas. Cells were harvested from enzymatically digested human corneas. CD45+ was used to identify leukocytes after exclusion of debris, doublets, and dead cells. Macrophages were identified by means of CD11b, CD14, and CD68 expression. A sequential gating strategy was used to identify macrophage subsets based on their surface markers: HLA-DR+, CD282+, and CD284+ cells were identified as M1 macrophages, whereas CD163+ and CD206+ cells were identified as M2 macrophages. CD86+ was only rarely detected. FMO controls (in red) of the corresponding gate (cells in light blue) were included to determine the baseline for positive events in the particular channels. FMO, fluorescence minus one; M1, pro-inflammatory macrophages; M2, regulatory macrophages.
Given the cellular composition of the cornea, we aimed to explore if additional staining with CD45 as a pan-leukocyte marker would lead to a higher quality of the FACS data or whether this marker can be omitted. We therefore compared our FMO controls without (A in online suppl. Fig. 2) and with CD45 staining (B in online suppl. Fig. 2). We were furthermore interested to see if a CD45 staining would help to exclude other cells such as keratocytes. When staining with an anti-CD45 antibody, all FMO controls showed a more distinct differentiation from the positive population (online suppl. Fig. 2b) than samples without anti-CD45 antibody staining (online suppl. Fig. 2a). Thus, CD45 proved to be an advantageous marker for the identification of leukocytes in the human cornea.
Macrophages in Organ-Cultured Human Corneas
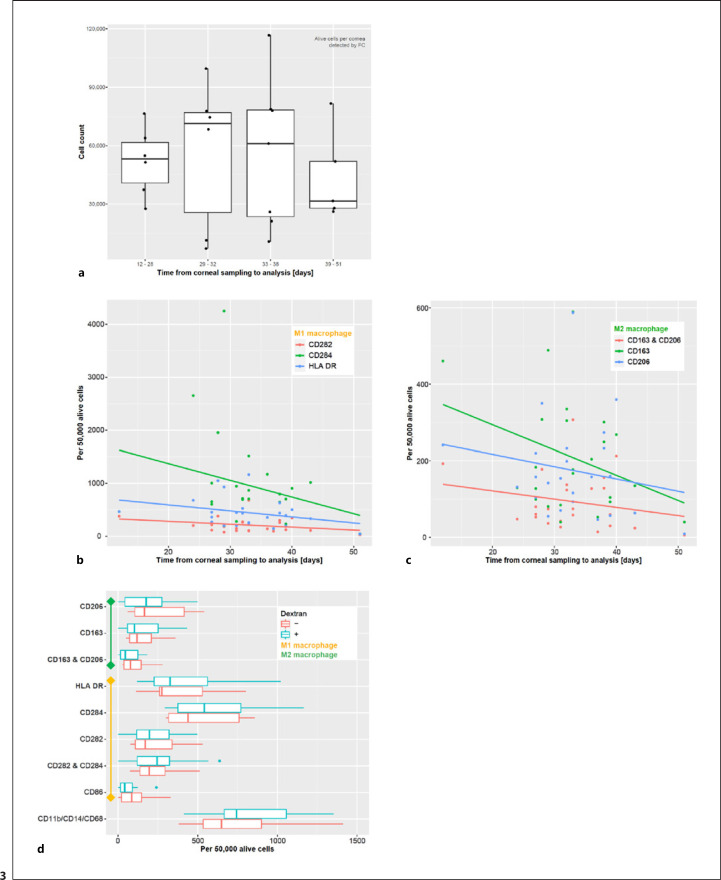
It has been reported that the number of APCs within the cornea decreases with increasing time in organ culture [29]. To investigate this phenomenon in more detail, we analyzed the number of macrophages per cornea in correlation with culture time (Fig. 3): 24 corneas with different incubation times were digested without any other intervention and analyzed by FC according to the protocol mentioned above. Analysis confirmed a slight decrease in total cell count per cornea with increasing incubation time (Fig. 3a). To verify whether this decrease is accompanied by an altered polarization of macrophages, we further analyzed the expression of the individual markers for M1 and M2 macrophages (Fig. 3b, c). We found that the relative cell loss of both M1 as well as M2 macrophages over time was higher than the total cell loss.
Fig. 3.
Macrophages in organ-cultured human corneas. a The total cell count per cornea varied over a wide range and decreased with increasing time in the organ culture (n = 24). To assess whether the macrophage populations decrease over time, the number of macrophages per 50,000 alive cells was analyzed at different time points. Corneas with different tissue culture times were digested and examined for cell marker expression (n = 24). Both, cells expressing pro-inflammatory (b) and regulatory markers (c) decreased with increasing time in organ culture. d To assess whether the cell culture conditions affected macrophages in human corneas, media with and without dextran were compared. Cornea halves were kept in the 2 different media for 24 h, followed by tissue processing and FC analysis (n = 10 per group). The presence or absence of dextran in tissue organ culture medium did not significantly alter the marker expression patterns (d). FC, flow cytometry.
There are different culture media used in cornea banks worldwide for various reasons. The main differences between the media used include the dextran content, with dextran leading to less swelling of the grafts or being used to de-swell grafts. We therefore aimed to explore whether the observed cell loss might be caused by culture media conditions. Therefore, we compared cell numbers in corneal culture medium with and without dextran (Fig. 3d). Corneas were cut in half and one half was incubated for 24 h in tissue-C (without dextran), the other half in carry-C medium (containing 5% dextran). Corneal macrophages were then analyzed by FC as above. The expression of all macrophage markers mentioned above showed no statistically significant difference between the tissue-C group and the carry-C group (pairwise t-test, Fig. 3d).
To explore if the reduced number of cells − especially macrophages − in the cornea was caused by cells leaving the graft and floating in the organ culture medium, we investigated the culture medium in which the corneas were stored by FC (online suppl. Fig. 3). It turned out that although living macrophages were detected in the culture medium, the cell count of these cells alone in the medium cannot account for the cell loss detected in corneal tissue.
Differentiation of Human Corneal Macrophages in Organ Culture
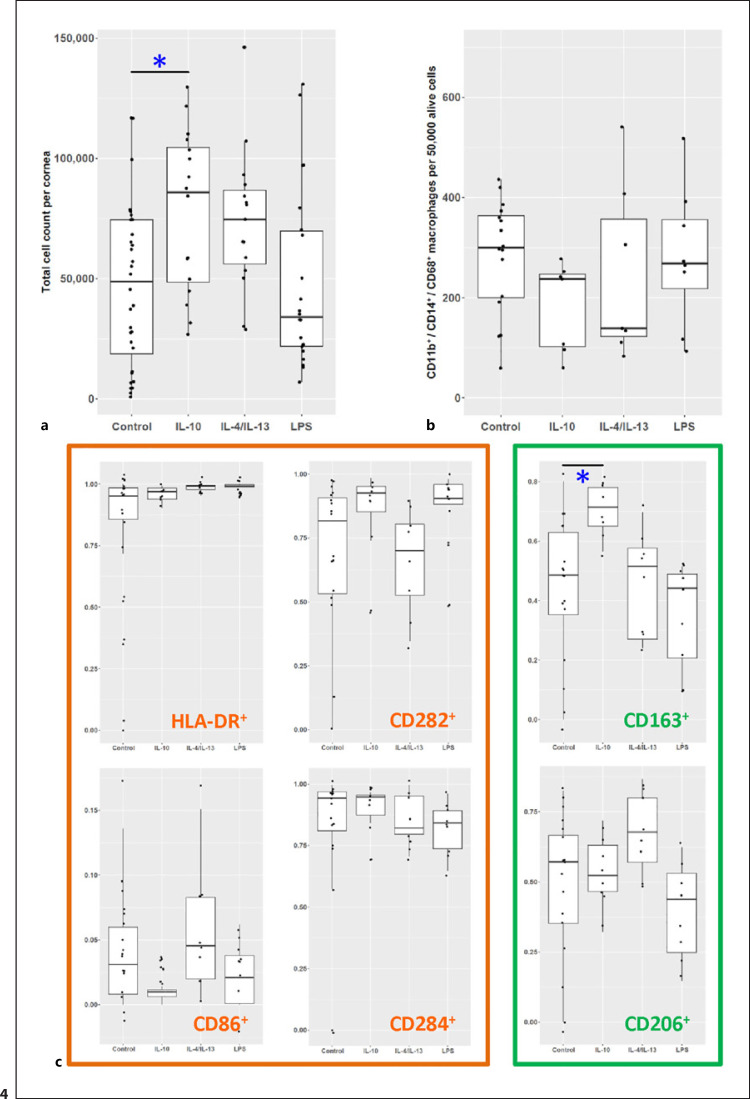
To see if the phenotype of macrophages in the human cornea can be altered, in a next step either IL-10, a combination of IL-4 with IL-13, or LPS were added to the organ culture medium (Fig. 4). LPS is a native and well-characterized stimulus for Toll-like receptor signaling in macrophages which served as a positive control [30]. Prior to the analysis of individual markers before and after stimulation, we first tested for possible toxic effects of the stimulation. Regarding the exact number of alive cells, the IL-10 group interestingly showed significantly more living cells per cornea compared to the group of untreated corneal samples (p < 0.05, one-way ANOVA; Fig. 4a), suggesting that supplementation of IL-10 to organ culture medium either prolongs the survival of cells in the cornea or leads to proliferation of cells in the corneal tissue. There was no evidence for cytotoxic effects caused by the addition of interleukins or LPS to the organ culture medium.
Fig. 4.
Polarization of human corneal macrophages especially monocyte-derived cells. a Corneal cell counts after the respective treatment (each point represents an individual cornea) showed statistically significant increased cell counts for corneas treated with IL-10 (one-way ANOVA p < 0.05). For analyzing macrophages, the cell marker expression was normalized to 50,000 live cells. b Macrophage cell count showed no statistical differences among the different groups. c The percentage of CD45+/CD11b+/CD14+/CD68+ macrophages after treatment revealed an altered macrophage phenotype for CD163+ cells after treatment with IL-10 (*p < 0.05, one-way ANOVA).
In addition to total cell numbers per cornea, we also measured the number of macrophages, especially monocyte-derived macrophages, as it might be influenced by individual treatments. To exclude any confounders in this context, first all CD45+ cells were gated as described above, macrophages, especially monocyte-derived macrophages, were then defined as CD11b+CD14+CD68+ cells (Fig. 4b). Statistics were performed using one-way ANOVA. Since the total cell count varied between corneas, macrophage counts were normalized to the total corneal cell counts (scaled per 50,000 viable cells). In the IL-10-treated group, a lower relative number of macrophages/ monocyte-derived macrophages were observed, although this difference was not statistically significant.
In the next step, the aforementioned pro-inflammatory and regulatory markers of macrophages were analyzed after stimulation (Fig. 4c). A potential goal of an immunomodulatory intervention would be, for example, depletion of M1 or enrichment of M2 macrophages in corneal grafts. In CD45+CD11b+CD14+CD68+ macrophages, IL-10 treatment upregulated the expression of the M2 marker CD163 (one-way ANOVA, p < 0.05). LPS reduced CD206+ and CD163+ cells compared to all other groups, even though this effect was not statistically significant. For the other 4 pro-inflammatory markers, there was no statistically significant suppression by the interleukins and no significant upregulation by stimulation with LPS. From our experimental design, M1 macrophages appear to be less sensitive to modification than M2 macrophages.
Discussion
Despite its transparency, the cornea contains large numbers of immune cells which contribute to rejection following corneal transplantation [31]. Yet, there are no comprehensive studies characterizing these immune cells in human corneal tissue. In the present study, human corneal tissue was assessed by RNA-seq revealing the presence of CD8+ T cells, Th17 cells, DCs, and macrophages. A detailed immunohistochemical analysis of these macrophages confirmed the presence of M1 macrophages and M2 macrophages. Based on FC, our study substantiates a suspected loss of these APCs with increasing duration of organ culture. Furthermore, we demonstrate that cytokine treatment of human cornea organ cultures can alter the phenotype of resident macrophages as has been suggested in mice [11], whereby M1 macrophages appear to be less sensitive to modifications than M2 macrophages. FC analysis of individual corneas may thus facilitate further development of tissue-priming strategies in human organ culture to diminish transplant rejection risks, as has been suggested based on murine data [11].
Sequencing of corneal tissue has only been performed in a few individual studies to date. These studies are mainly focusing on corneal diseases, such as Fuchs endothelial dystrophy [32], keratoconus [33], and herpetic infections [34], rather than a systemic analysis of cellular components of the cornea. A current study is the first to use a complex single-cell RNA-seq approach to investigate the most diverse cell populations of the cornea and the adjacent limbus in humans [2]. In this study, similar to our results, predominantly monocyte-derived macrophages and DCs were found, as well as CD8+ T cells and various macrophage subpopulations. This is in line with another study using single-cell and bulk RNA-seq paired with turnover studies in mice showing a steady replacement of resident tissue macrophages in the cornea by peripheral blood monocytes [35]. Overall, these most recent RNA-seq studies, as well as our work, suggest an important role for native immune cells in the cornea, although further details need to be elucidated.
Similar to RNA-seq, there are only a few studies on FC analysis of human corneal tissue or whole corneal grafts. The first protocols for a respective FC analysis were established in mice which have a much thinner corneal stroma layer than humans, allowing for easier cell harvesting [10]. Two more recent studies, which examined human tissue, focused on herpetic infections of the cornea [34] and on subtypes of dendritic cells [36], whereby only the first study analyses the dynamics of APCs over time and both studies contain only few samples. The small number of previous studies on human cornea bespeaks the complexity of preparing corneal tissue for FC analysis although FC analysis is actually ideally suited to investigate the dynamics of APC − especially DCs and macrophages.
Macrophages take on a special position in the rejection of corneal grafts. In addition to infiltration of the graft, an accumulation of monocyte-derived macrophages develops in the anterior chamber of the eye [37]. This recruitment of macrophages to the anterior chamber of the eye occurs in response to the recognition of allogeneic tissue by the recipient's immune system. In vitro studies using human macrophages suggest that these cells not only contribute to the destruction of the transplanted tissue [37], but that they also contribute to the recognition of allogeneic corneal tissue in the very early stages of graft rejection [22]. Furthermore, animal studies in different mouse strains show that the phenotype of these macrophages determines the course of transplant rejection, whereby a more aggressive M1 macrophage phenotype seems to be associated with a worse outcome [38].
There is evidence in the literature for a decrease in APCs within human corneas as organ culture time increases [39, 40]. Our data are in line with these observations and clearly show a time-dependent loss of macrophages as a fraction of total live cells, whereas the total cell numbers were largely stable. Previous studies suggest a correlation between APC loss and increased graft survival [29]. However, a prolonged organ culture time also tends to diminish the endothelial cell count, thus increasing the risk of graft failure [41]. Therefore, a drastic extension of organ culture time does not seem to be reasonable in order to prevent graft rejection. Eliminating APCs in corneal grafts therefore seems to be another rational approach to reduce the risk of rejection after transplantation. Zhang et al. [42] depleted corneal CD45+ cells from corneal buttons in mice using ex vivo depletion strategies including a complement-dependent cytotoxicity approach. In their experiments, the authors were able to reduce cells expressing CD45 by up to 70%. However, the depletion of CD45+ cells in grafts did not improve graft survival rates in a murine keratoplasty model. Therefore, it was suggested that the donor APCs participate in tolerance induction [43, 44]. Radical elimination of donor APCs from the graft may have affected tolerance induction and led to increased inflammation. Therefore, changing the phenotype from a pro-inflammatory to an anti-inflammatory regulatory profile, rather than eliminating these cells, could be a valid alternative approach.
Due to its dense arrangement of fibrils and its greater corneal thickness compared to rabbits and mice [45, 46], the human cornea was inaccessible for a long time for obtaining single-cell suspensions for FC analyses. FC analyses of native DCs in human corneal single-cell suspensions have been reported before [34, 36]. To gain more insights into the myeloid cell composition of the cornea we addressed macrophage subtypes in human corneas and the possibility of a therapeutic modulation of their phenotype. Such treatment of donor corneas was shown to reduce corneal transplant rejection rates in animal models [31, 47], but respective data on human tissue are lacking.
In FC analysis of corneal tissue, we observed an increase in the total number of cells per cornea on stimulation with IL-10. This could be explained by an antiapoptotic effect as well as by the proliferation of cells in response to stimulation with IL-10 [48, 49]. In our study, no clear cell population emerged that would explain this effect, quite to the contrary CD45+ CD11b+ CD14+ CD68+ macrophages tended to be reduced in the IL-10 group. In addition to an investigation of other APC populations such as DCs, it would also be important to analyze adaptive immune cells, especially CD8+ T cells − in future studies. Ambarus and coworkers polarized human peripheral blood monocytes (PBMC) in vitro with IFN-γ, IL-4, and IL-10, and concluded that IL-4-polarized macrophages upregulate CD206, whereas IL-10-polarized macrophages upregulate CD163 [50]. Interestingly, our experiments also revealed increased expression of CD163 in macrophages within the human cornea upon stimulation with IL-10. According to a previous selection of CD45+ cells, M1 macrophages seem to be less sensitive to changes than M2 macrophages. However, this observation requires further investigation in more advanced studies. For the sake of completeness, it should be noted that without prior CD45 selection, cells did show reduced expression of pro-inflammatory markers upon stimulation with IL-10 or IL-4/IL-13. This illustrates that interactions of interleukins with corneal immune cells seem to be rather complex. Further studies investigating different immune cells in this context will therefore be necessary. It will also be essential to clarify the biological relevance of modifying individual cell types in preventing rejection. Certainly, a multiarm approach targeting different immune cell types is preferable to the modification of single cell types.
We identified only a small population of CD86+ cells in a few, but not in all, samples. This is consistent with a study by Lužnik, reporting on only a very small percentage of CD86+ cells in the central area of the human cornea [36]. However, in our study the discrimination of CD86-positive and negative populations was vague. CD86 should therefore probably only be used with caution as a marker for the differentiation of various macrophage subtypes in the context of FC studies in corneal tissue.
Our study is limited by the fact that our FACS protocol does not allow for differentiation between resident macrophages and infiltrating monocyte-derived macrophages. Signs of the presence of a major monocyte population did not emerge in our RNA-seq data, which appears to be in line with other published data in this field [2, 35].
In summary, our data clearly illustrate the feasibility of FC-based quantitative analysis and characterization of innate immune cell phenotypes in human corneas. This technique allows the evaluation of therapeutic priming of innate immune cells in cultured human corneal tissue. Finally, we provide a strategy to translate a stimulus-dependent fine-tuning of the myeloid compartment into a reduced transplant rejection rate in patients.
Statement of Ethics
Ethics approval was granted by the local Ethics Committee (Research Ethics Committee of University of Freiburg; Germany; reference numbers: 572/14 and 25/20). Corneal samples for immunohistochemical analyses were obtained from eyes of body donors; these eyes were enucleated in accordance with the informed consent of the body donors, which was contractually secured during lifetime. In accordance with ethical requirements, no data other than age, sex, body weight, and cause of death were collected. Enucleation, tissue preparation, and staining were performed at the Institute of Anatomy, University of Leipzig. This study adheres to the tenets of the Declaration of Helsinki.
Conflict of Interest Statement
The authors have no conflicts of interest to disclose.
Funding Sources
T.B. is supported by a research grant of the Geschwister-Freter-Foundation (Germany). T.L. is supported by a research grant from the Ernst-und-Berta-Grimmke-Foundation (Germany) and the Gertrud-Kusen-Foundation (Germany). The funding organizations had no role in the design and conduct of this study. The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg in the funding programme Open Access Publishing.
Author Contributions
Conceptualization: G.S., T.B., P.W., C.L., T.R., and T.L. Investigation: X.Z., J.W., D.D.R., T.B., R.L., and T.L. Data curation: X.Z., J.W., D.D.R., T.B., R.L., D.B., and T.L. Formal analysis: G.S., J.W., D.B., P.W., C.L., T.R., and T.L. Administration and supervision: G.S., D.B., P.W., C.L., T.R., and T.L. Funding acquisition: T.B. and T.L. Visualization: X.Z., G.S., J.W., D.B., P.W., C.L., and T.L. Writing − original draft: X.Z., G.S., J.W., P.W., C.L., and T.L. Writing − reviewing and editing: all authors.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We would like to thank Prof. Dr. Philip Maier and his team from the Lions Cornea Bank Baden-Württemberg for supporting this study and providing us with corneal tissue. We would also like to thank Prof. Dr. Gabriele Niedermann and her team for kindly providing the CytoFLEX S flow cytometer. Last but not least, we would like to especially thank Dr. Anja Schlecht and Ms. Gabriele Prinz for their excellent technical support during much of the work in the laboratory.
References
- 1.Novak N, Siepmann K, Zierhut M, Bieber T. The good, the bad and the ugly: APCs of the eye. Trends Immunol. 2003;24((11)):570–4. doi: 10.1016/j.it.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Collin J, Queen R, Zerti D, Bojic S, Moyse N, Molina MM, et al. A single cell atlas of human cornea that defines its development, limbal stem and progenitor cells and the interactions with the limbal niche. bioRxiv. 2020 Jul; doi: 10.1016/j.jtos.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamagami S, Dana MR. The critical role of lymph nodes in corneal alloimmunization and graft rejection. Invest Ophthalmol Vis Sci. 2001 May;42((6)):1293–8. [PubMed] [Google Scholar]
- 4.Amouzegar A, Chauhan SK, Dana R. Alloimmunity and tolerance in corneal transplantation. J Immunol. 2016 May;196((10)):3983–91. doi: 10.4049/jimmunol.1600251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan A, Fu H, Tan LA, Harper JE, Beutelspacher SC, Larkin DFP, et al. Dendritic cell modification as a route to inhibiting corneal graft rejection by the indirect pathway of allorecognition. Eur J Immunol. 2013;43((3)):734–46. doi: 10.1002/eji.201242914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palomar APd, Montolío A, Cegoñino J, Dhanda SK, Lio CT, Bose T. The innate immune cell profile of the cornea predicts the onset of ocular surface inflammatory disorders. J Clin Med. 2019 Dec 2;8((12)):2110. doi: 10.3390/jcm8122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagami S, Usui T, Amano S, Ebihara N. Bone marrow-derived cells in mouse and human cornea. Cornea. Cornea. 2005 Nov;24((8 Suppl)):S71–4. doi: 10.1097/01.ico.0000178732.42921.05. [DOI] [PubMed] [Google Scholar]
- 8.Yamagami S, Yokoo S, Usui T, Yamagami H, Amano S, Ebihara N. Distinct populations of dendritic cells in the normal human donor corneal epithelium. Invest Ophthalmol Vis Sci. 2005 Dec;46((12)):4489–94. doi: 10.1167/iovs.05-0054. [DOI] [PubMed] [Google Scholar]
- 9.Hau S, Clarke B, Thaung C, Larkin DFP. Longitudinal changes in corneal leucocyte density in vivo following transplantation. Br J Ophthalmol. 2019 Aug;103((8)):1035–41. doi: 10.1136/bjophthalmol-2018-312386. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa M, Inomata T, Shiang T, Tsubota K, Murakami A. Method for selective quantification of immune and inflammatory cells in the cornea using flow cytometry. J Biol Methods. 2018 Nov 22;5((4)):e102. doi: 10.14440/jbm.2018.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahvildari M, Emami-Naeini P, Omoto M, Mashaghi A, Chauhan SK, Dana R. Treatment of donor corneal tissue with immunomodulatory cytokines: A novel strategy to promote graft survival in high-risk corneal transplantation. Sci Rep. 2017 Dec;7((1)):971. doi: 10.1038/s41598-017-01065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf J, Auw-Haedrich C, Schlecht A, Boneva S, Mittelviefhaus H, Lapp T, et al. Transcriptional characterization of conjunctival melanoma identifies the cellular tumor microenvironment and prognostic gene signatures. Sci Rep. 2020 Dec;10((1)):17022. doi: 10.1038/s41598-020-72864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boneva S, Schlecht A, Böhringer D, Mittelviefhaus H, Reinhard T, Agostini H, et al. 3′ MACE RNA-sequencing allows for transcriptome profiling in human tissue samples after long-term storage. Lab Invest. 2020 Oct;100((10)):1345–55. doi: 10.1038/s41374-020-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020 Jul;48((W1)):W395–402. doi: 10.1093/nar/gkaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 Jan;29((1)):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014 Apr;30((7)):923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 17.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2020 Jan;48((D1)):D682–8. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012 Dec;131((4)):281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 19.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017 15;18((1)):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016 Sep;32((18)):2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 21.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2009. [Google Scholar]
- 22.Wolf J, Zhuang X, Hildebrand A, Boneva S, Schwämmle M, Kammrath Betancor P, et al. Corneal tissue induces transcription of metallothioneins in monocyte-derived human macrophages. Mol Immunol. 2020 Nov;128:188–94. doi: 10.1016/j.molimm.2020.10.016. [DOI] [PubMed] [Google Scholar]
- 23.R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. Available from: http://www.R-project.org. [Google Scholar]
- 24.Harrell F. Hmisc: Harrell Miscellaneous [Internet]. Httpscranr-Proj. [cited 2020 Aug 16]. Available from: http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc.
- 25.Wang Y, Gao G, Wu Y, Wang Y, Wu X, Zhou Q. S100A4 silencing facilitates corneal wound healing after alkali burns by promoting autophagy via blocking the PI3K/Akt/mTOR signaling pathway. Invest Ophthalmol Vis Sci. 2020 Sep;61((11)):19. doi: 10.1167/iovs.61.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao WW-Y, Liu C-Y. Roles of lumican and keratocan on corneal transparency. Glycoconj J. 2002 Jun;19((4–5)):275–85. doi: 10.1023/A:1025396316169. [DOI] [PubMed] [Google Scholar]
- 27.Han KE, Choi S, Kim T, Maeng Y, Stulting RD, Ji YW, et al. Pathogenesis and treatments of TGFBI corneal dystrophies. Prog Retin Eye Res. 2016 Jan;50:67–88. doi: 10.1016/j.preteyeres.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Toyono T, Usui T, Yokoo S, Taketani Y, Nakagawa S, Kuroda M, et al. Angiopoietin-like 7 is an anti-angiogenic protein required to prevent vascularization of the cornea. PLoS One. 2015;10((1)):e0116838. doi: 10.1371/journal.pone.0116838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier P, Heinzelmann S, Böhringer D, Reinhard T. Prolonged organ culture reduces the incidence of endothelial immune reactions. Eye (Lond) 2016 Jan;30((1)):127–32. doi: 10.1038/eye.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang RB, Mark MR, Gray A, Huang A, Xie MH, Zhang M, et al. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998 Sep;395((6699)):284–8. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 31.Tahvildari M, Amouzegar A, Foulsham W, Dana R. Therapeutic approaches for induction of tolerance and immune quiescence in corneal allotransplantation. Cell Mol Life Sci. 2018 May;75((9)):1509–20. doi: 10.1007/s00018-017-2739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirp A, Leite K, Tuvikene J, Nurm K, Sepp M, Timmusk T. The Fuchs corneal dystrophy-associated CTG repeat expansion in the TCF4 gene affects transcription from its alternative promoters. Sci Rep. 2020 Oct;10((1)):18424. doi: 10.1038/s41598-020-75437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karolak JA, Gambin T, Rydzanicz M, Polakowski P, Ploski R, Szaflik JP, et al. Accumulation of sequence variants in genes of Wnt signaling and focal adhesion pathways in human corneas further explains their involvement in keratoconus. PeerJ. 2020;8:e8982. doi: 10.7717/peerj.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knickelbein JE, Buela KA, Hendricks RL. Antigen-presenting cells are stratified within normal human corneas and are rapidly mobilized during ex vivo viral infection. Invest Ophthalmol Vis Sci. 2014 Feb;55((2)):1118–23. doi: 10.1167/iovs.13-13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieghofer P, Hagemeyer N, Sankowski R, Amann L, Gruber M, Koch J, et al. Mapping the origin and fate of myeloid cells in distinct compartments of the eye by single-cell profiling. EMBO J. 2021 Mar;40((6)):e105123. doi: 10.15252/embj.2020105123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lužnik Z, Kopitar AN, Lapajne L, Pižem J, Ferrari S, Ihan A, et al. Identification and characterization of dendritic cell subtypes in preserved and cultured cadaveric human corneolimbal tissue on amniotic membrane. Acta Ophthalmol (Copenh) 2019 Mar;97((2)):e184–93. doi: 10.1111/aos.13854. [DOI] [PubMed] [Google Scholar]
- 37.Lapp T, Zaher SS, Haas CT, Becker DL, Thrasivoulou C, Chain BM, et al. Identification of therapeutic targets of inflammatory monocyte recruitment to modulate the allogeneic injury to donor cornea. Invest Ophthalmol Vis Sci. 2015 Nov;56((12)):7250–9. doi: 10.1167/iovs.15-16941. [DOI] [PubMed] [Google Scholar]
- 38.Bleul T, Zhuang X, Hildebrand A, Lange C, Böhringer D, Schlunck G, et al. Different innate immune responses in BALB/c and C57BL/6 strains following corneal transplantation. J Innate Immun. 2020 Sep;:1–11. doi: 10.1159/000509716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer WJ, Irschick UM, Moser P, Wurm M, Huemer HP, Romani N, et al. Characterization of antigen-presenting cells in fresh and cultured human corneas using novel dendritic cell markers. Invest Ophthalmol Vis Sci. 2007 Oct;48((10)):4459–67. doi: 10.1167/iovs.06-1184. [DOI] [PubMed] [Google Scholar]
- 40.Al-Fakih A, Faltus V, Jirsova K. A decrease in the density of HLA-DR-positive cells occurs faster in corneas stored in organ culture than under hypothermic conditions. Ophthalmic Res. 2011 Dec;47((1)):39–46. doi: 10.1159/000322801. [DOI] [PubMed] [Google Scholar]
- 41.Cohen EJ, Holland EJ, Mannis MJ, Matoba AY, Meisler DM, Udell IJ, et al. Corneal Endothelial Photography: Three-year Revision. 1997 [Google Scholar]
- 42.Zhang X, Shen L, Jin Y, Saban DR, Chauhan SK, Dana R. Depletion of passenger leukocytes from corneal grafts: An effective means of promoting transplant survival? Invest Ophthalmol Vis Sci. 2009;50((7)):3137–44. doi: 10.1167/iovs.08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ezzelarab M, Thomson AW. Tolerogenic dendritic cells and their role in transplantation. Semin Immunol. 2011 Aug;23((4)):252–63. doi: 10.1016/j.smim.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrod KR, Chang CK, Liu F-C, Brennan TV, Foster RD, Kang S-M. Targeted lymphoid homing of dendritic cells is required for prolongation of allograft survival. J Immunol. 2006 Jul 15;177((2)):863–8. doi: 10.4049/jimmunol.177.2.863. [DOI] [PubMed] [Google Scholar]
- 45.Kaye GI, Pappas GD. Studies on the cornea. I. The fine structure of the rabbit cornea and the uptake and transport of colloidal particles by the cornea in vivo. J Cell Biol. 1962 Mar;12((3)):457–79. doi: 10.1083/jcb.12.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henriksson JT, McDermott AM, Bergmanson JPG. Dimensions and morphology of the cornea in three strains of mice. Invest Ophthalmol Vis Sci. 2009;50((8)):3648–54. doi: 10.1167/iovs.08-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klebe S, Sykes PJ, Coster DJ, Krishnan R, Williams KA. Prolongation of sheep corneal allograft survival by ex vivo transfer of the gene encoding interleukin-10. 2001 doi: 10.1097/00007890-200105150-00006. [DOI] [PubMed] [Google Scholar]
- 48.Rowbottom AW, Lepper MW, Garland RJ, Cox CV, Corley EG, Oakhill A, et al. Interleukin-10-induced CD8 cell proliferation. Immunology. 1999 Sep;98((1)):80–9. doi: 10.1046/j.1365-2567.1999.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng G, Li K, Chen S, Chen P, Zheng H, Yu B, et al. Interleukin10 promotes proliferation and migration, and inhibits tendon differentiation via the JAK/Stat3 pathway in tendonderived stem cells in vitro. Mol Med Rep. 2018 Dec;18((6)):5044–52. doi: 10.3892/mmr.2018.9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TRDJ, Reedquist KA, et al. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J Immunol Methods. 2012 Jan 31;375((1-2)):196–206. doi: 10.1016/j.jim.2011.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data