Abstract
Sepsis-induced myeloid-derived suppressor cells (MDSCs) increase mortality risk. We previously identified that long non-coding RNA Hotairm1 supports myeloid precursor shifts to Gr1<sup>+</sup>CD11b<sup>+</sup> MDSCs during mouse sepsis. A major unanswered question is what molecular processes control Hotairm1 expression. In this study, we found by a genetic deletion that a specific PU.1-binding site is indispensable in controlling Hotairm1 transcription. We then identified H3K4me3 and H3K27me3 at the PU.1 site on the Hotairm1 promoter. Controlling an epigenetic switch of Hotairm1 transcription by PU.1 was histone KDM6A demethylase for H3K27me3 that derepressed its transcription with possible contributions from Ezh2 methyltransferase for H3K27me3. KDM6A knockdown in MDSCs increased H3K27me3, decreased H3K4me3, and inhibited Hotairm1 transcription activation by PU.1. These results enlighten clinical translation research of PU.1 epigenetic regulation as a potential sepsis immune-checkpoint treatment site.
Keywords: Sepsis, Myeloid-derived suppressor cell, Immune suppression, Hotairm1, Epigenetics
Introduction
Sepsis-induced immunosuppression hampers inflammation resolution and immune homeostasis, leading to the development of protracted sepsis with elevated morbidity and mortality [1, 2, 3, 4]. We [5, 6] and others [7, 8] previously reported that myeloid-derived suppressor cells (MDSCs) − a mixed population of precursors of monocytes, neutrophils, and dendritic cells [9, 10] − contribute to immunosuppression in mice and humans with sepsis. Recently, we reported that S100A9 protein is constitutively expressed in circulating leukocytes [11, 12], secreted from myeloid cells via inflammation-derived signals [13, 14], and promotes protracted sepsis [15]. S100A9 accumulates in the nucleus in MDSCs in mice and patients with protracted sepsis [15]. S100A9 knockout mice do not develop protracted sepsis, and their MDSCs are not immunosuppressive [15].
Long non-coding RNAs (lncRNAs) are protein non-coding transcripts of >200 nucleotides [16, 17] that regulate cellular biology [18, 19, 20]. Immune-related lncRNAs increase in cell- and tissue-specific ways [18], with differential expression found during immune cell responses [20]. LncRNAs regulate innate immune cell development and inflammatory gene expression [19, 20]. LncRNAs interact directly with proteins, RNA, or DNA [19, 21], and most immune-related lncRNAs act through protein binding [22, 23] and may modify protein transport [19, 20, 24, 25]. Using lncRNA expression microarray and real-time PCR, we found expression of lncRNA HOXA transcript antisense RNA myeloid-specific 1 (Hotairm1) − a key regulator of myeloid cell development [26, 27] − increases as sepsis progresses to the protracted state. Hotairm1 levels were significantly increased in MDSCs in mice and patients with post-acute sepsis [28], which correlates with an increase in MDSC numbers [15]. We also found that Hotairm1 binds to S100A9 protein in post-sepsis MDSCs and that S100A9 relocalizes in the cytosol following Hotairm1 knockdown [28].
Thus, Hotairm1 modifies S100A9 biological function and can induce MDSC development and immunosuppression during post-acute sepsis. In the present study, we investigated Hotairm1 transcriptional regulation in MDSCs during murine sepsis. The results showed that Hotairm1 transcription is driven by a proximal promoter containing a consensus binding site for the transcription factor PU.1. Binding of PU.1 at the Hotairm1 promoter was increased as sepsis protracted. The results also revealed that epigenetic modifications of histones around the PU.1 binding site switched Hotairm1 transcription from repression during acute sepsis to activation after acute sepsis. This study provides insights into Hotairm1 expression induction during sepsis and suggests that targeting Hotairm1 transcription provides a molecular-based approach to attenuate MDSC development and associated immunosuppression in sepsis.
Materials and Methods
Mice
Male C57BL/6 mice (8–10 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). The mice were housed in a pathogen-free facility and were acclimated to the new environment for a week before surgery. All experiments were conducted following National Institutes of Health guidelines and were approved by the East Tennessee State University Animal Care and Use Committee (Protocol No. P190603).
Sepsis
Polymicrobial sepsis was induced by cecal ligation and puncture (CLP) in mice as described previously [29]. In brief, a midline abdominal incision was made, and the cecum was ligated distal to the ileocecal valve and punctured twice with a 23-gauge needle. A small amount of fecal matter was extruded into the abdominal cavity, and the abdominal wall and skin were sutured in layers with 3–0 silk. To establish intra-abdominal infection and approximate the clinical conditions of human sepsis [30] and delay in MDSC development, mice received (i.p.) 1 mL lactated Ringers plus 5% dextrose for fluid resuscitation and were subcutaneously administered antibiotic (imipenem; 25 mg/kg body weight) in saline (0.9% sodium chloride) at 8 and 16 h after CLP. These interventions are in line with the recommendations for MQTiPSS [31]. These manipulations result in high mortality (∼60–70%) during a post-acute sepsis phenotype [5]. Survival was followed for 28 days. Mice moribund during acute sepsis (defined as the first 5 days after CLP) or post-acute sepsis (day 7–28) [29] were euthanized and analyzed. A corresponding number of mice from the control/sham group were also analyzed at the same time point.
Several clinical and experimental studies have shown that cell-mediated immune responses are depressed in males while enhanced in females during sepsis [32, 33] and that female mouse is more immunologically competent than the male mouse in surviving CLP insult [34]. And because MDSCs suppress both innate and adaptive immune responses, we used male mice to assess the maximal effect of this immunosuppressive cell population on sepsis outcome.
Gr1+CD11b+ Cells
Gr1+CD11b+ cells were isolated from bone marrow by negative selection using the EasySep mouse MDSC isolation kit according to the manufacturer's protocol (Stemcell Technologies, Cambridge, MA, USA). In brief, the bone marrow was flushed out of the femurs with RPMI-1640 medium under aseptic conditions. A single-cell suspension was made by filtering through a 70-μm mesh nylon strainer, followed by incubation with erythrocyte lysis buffer and washing. The cells were incubated with Gr1+CD11b+ isolation biotinylated antibody cocktail (bind to all non-Gr1+CD11b+ cells) at room temperature for 10 min, followed by adding streptavidin-coated magnetic particles and incubating at room temperature for 5 min. Sample tubes were placed onto a magnet for 3 min, and the enriched cell suspension (supernatant) containing Gr1+CD11b+ cells was transferred to a fresh tube. The cells were >90% Gr1+CD11b+ positive as determined by flow cytometry.
In some experiments, Gr1+CD11b+ cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (all from Hyclone Laboratories, Logan, UT, USA), and 10% FBS (Atlanta Biologicals, Lawrenceville, GA, USA) at 37°C and 5% CO2.
siRNA-Mediated Knockdown
PU.1, Ezh2, and KDM6A were knocked down in Gr1+CD11b+ cells using pools of gene-specific or scrambled (control) siRNAs (Santa Cruz Biotechnology, Dallas, TX, USA). The siRNA mixture was suspended in HiPerFect reagent (Qiagen, Germantown, MD, USA) at a 0.5 μM final concentration. The cells were transfected and incubated with RPMI-1640 medium for 48 h.
Western Blotting
Whole cell lysates were resolved onto SDS-10% polyacrylamide gel (Bio-Rad, Hercules, CA, USA) and transferred to nitrocellulose membranes (Thermo Fisher Scientific, Waltham, MA, USA). The membranes were blocked with 5% milk in Tris-buffered saline/Tween-20 for 1 h at room temperature and then probed overnight at 4°C with anti-PU.1 (Cat #MA5-15064; Invitrogen, Carlsbad, CA, USA), anti-Ezh2 (Cat #166609, Santa Cruz Biotechnology), anti-KDM6A (Cat #33510S), or KDM6B/JMDJ3 (Cat #3457S) (Cell Signaling Technology, Danvers, MA, USA). After washing, blots were incubated with the appropriate HRP-conjugated secondary antibody for 2 h at room temperature. Proteins were detected with the enhanced chemiluminescence detection system (Thermo Fisher Scientific), the bands were visualized using the ChemiDoc XRS System (Bio-Rad), and the images were captured with the Image Lab Software V. 3.0. The membranes were stripped and reprobed with β-actin (Invitrogen).
Luciferase Assay
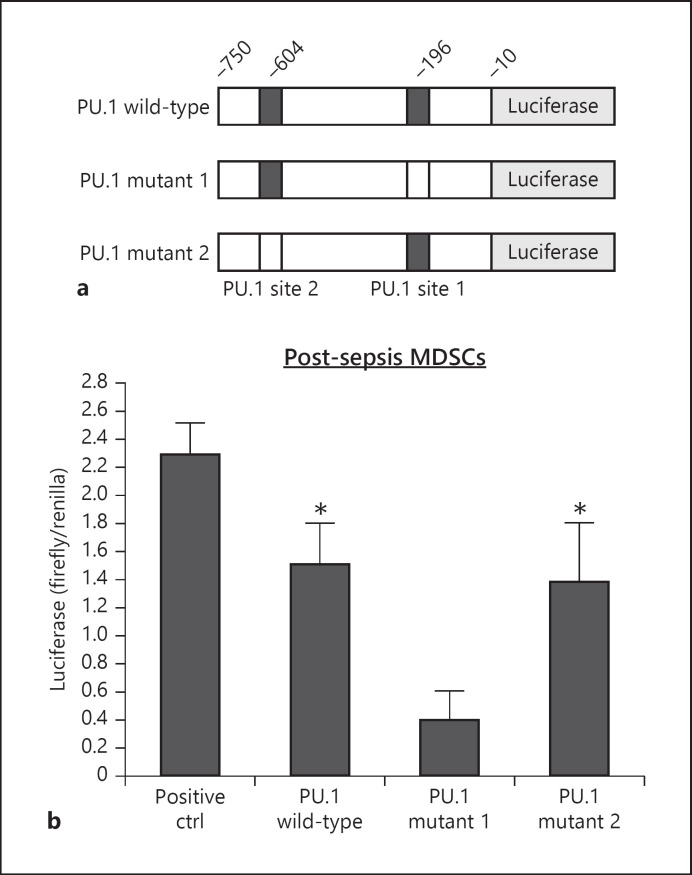
A 740-bp Hotairm1 promoter fragment from −750 to −10 relative to the transcription start site containing 2 putative PU.1 binding sites (Fig. 1a) was amplified from the mouse genomic DNA via PCR. The DNA fragment was cloned upstream of the firefly luciferase reporter via 5′ Spel and 3′ SalI sites in the pEZX-FR01 firefly-Renilla dual-reporter vector (Fig. 3). For PU.1 mutant constructs, the core sequence GGAA on the opposite strand at −196 (PU.1 mutant 1) or −604 (PU.1 mutant 2) was changed to ATCG using site-directed mutagenesis.
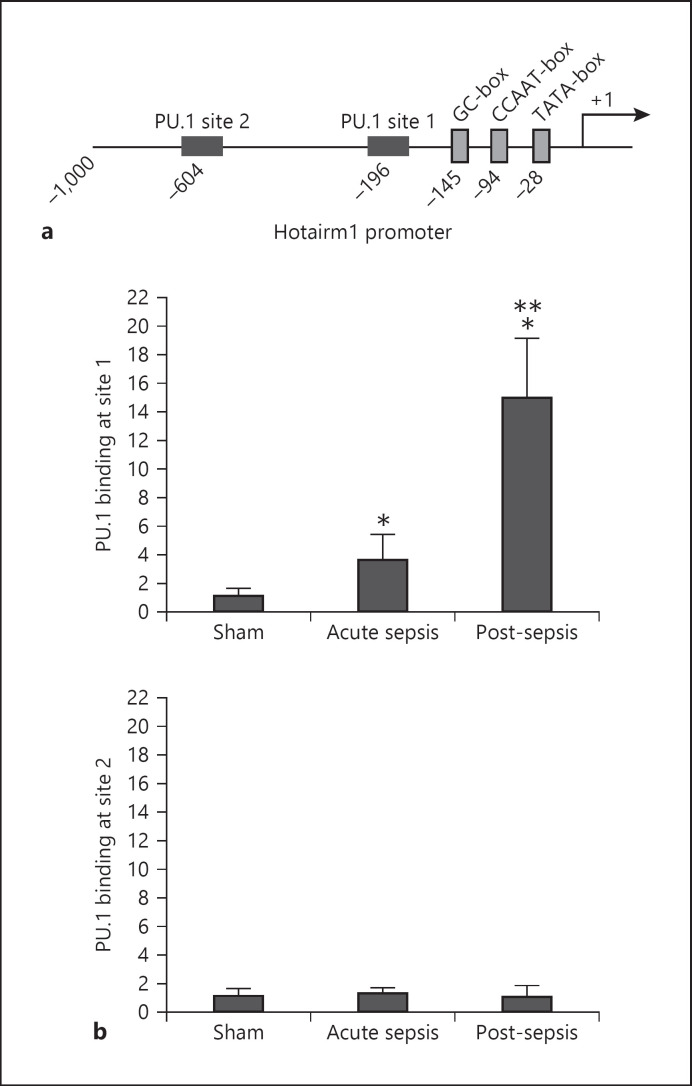
Fig. 1.
PU.1 binds at the Hotairm1 promoter. a A schematic diagram depicting the proximal promoter region of mouse Hotairm1. The PU.1 binding sites and consensus promoter motifs are shown. b ChIP assay for PU.1 binding. Bone marrow cells were harvested from septic mice that were moribund and sacrificed at days 1–5 (representing acute sepsis) and at days 7–28 (representing post-acute sepsis), as well as sham mice. Gr1+CD11b+ cells were then purified by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. The cells were fixed in formaldehyde to cross-link protein-DNA and lysed, and the pelleted nuclei were digested with a chromatin shearing enzymatic cocktail. The sheared chromatin was immunoprecipitated with anti-PU.1 or anti-IgG isotype control antibody. Next, chromatin cross-links were reversed to recover the protein-bound DNA. To measure the amount of PU.1 binding at the promoter, the ChIPed DNA was analyzed by real-time qPCR using primers that amplify the promoter sequences surrounding the PU.1 binding sites 1 and 2 (see Fig. 1a). Sample values were normalized to the “input” DNA (DNA isolated before the immunoprecipitation) and are presented as fold change relative to the IgG-immunoprecipitated samples (1-fold). Data are means ± SD for 5–7 mice per group, from 3 experiments. *p < 0.05 versus sham; **p < 0.05 versus acute sepsis. ChIP, chromatin immunoprecipitation.
Fig. 3.
The proximal PU.1 binding site in the Hotairm1 promoter induces luciferase gene expression in Gr1+CD11b+ cells. a A diagram of wild-type and mutant reporter constructs containing the proximal promoter region of Hotairm1 cloned upstream of the luciferase gene. b Gr1+CD11b+ cells were isolated from the bone marrow of post-acute septic mice. The cells were transfected with the indicated luciferase constructs. After 48 h, the cells were harvested for the measurement of firefly and Renilla luciferase activities. A luciferase vector with the GAPDH promoter served as a positive control for maximum firefly luciferase gene activity. Background reading from transfection with a negative control vector was subtracted, and firefly luciferase values were normalized to Renilla luciferase activity. Data are means ± SD for 6 mice per group, from 3 experiments, and are presented as firefly/Renilla ratio. *p < 0.05 versus PU.1 mutant 1. MDSCs, myeloid-derived suppressor cells.
Gr1+CD11b+ cells were isolated from the bone marrow by positive selection and transfected (∼2 × 106 cells) with 0.5 μg of luciferase plasmid (GeneCopoeia, Rockville, MD, USA) using HiPerFect transfection reagent per the manufacturer's instructions (Qiagen). After 48 h, the cells were harvested, and firefly and Renilla luciferase activities were determined with the dual luciferase reporter assay system (Promega, Madison, WI, USA). The pEZX-MT06 vector, in which the Renilla luciferase gene is controlled by the GAPDH promoter and an SV40 early enhancer controls the firefly luciferase gene, served as a positive control for maximum firefly luciferase gene activity. An empty pEZX-FR01 served as a negative control. Firefly luciferase values were normalized to Renilla luciferase activity and are presented as firefly/Renilla ratio.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed to determine in vivo protein-DNA interactions at the Hotairm1 promoter using the ChIP-IT Express Enzymatic Shearing kit according to the manufacturer's instructions (Active Motif, Carlsbad, CA, USA). In brief, Gr1+CD11b+ cells were harvested and protein-DNA complexes were cross-linked by fixation in 1% formaldehyde in a minimal culture medium at room temperature for 10 min. After washing with cold PBS, cells were resuspended in lysis buffer containing protease inhibitor cocktail and incubated on ice for 1 h. The cell lysate was cleared by centrifugation at 5,000 rpm and 4°C for 10 min. The pelleted nuclei were then resuspended in digestion buffer containing an enzymatic shearing cocktail and incubated at 37°C for 10 min. After stopping the reaction with EDTA, the sheared chromatin (supernatant) was recovered by centrifugation at 15,000 rpm for 10 min at 4°C. Ten microliters of the sheared chromatin solution were reserved as the “input DNA” sample. Next, 5 μg of antibody against PU.1 (Cat #MA5-15064; Invitrogen), H3K4me3 (Cat #MBS9401981), H3K27me3 (Cat #MBS3010330), H3K9Ac (Cat #MBS9126034), H3K27Ac (Cat #MBS3011047) (all from MyBioSource, San Diego, CA, USA), H3K9me3 (Cat #A-4036-025; Epigentek, Farmingdale, NY, USA), Ezh2/ENX-1 (Cat #166609-x, Santa Cruz Biotechnology), KDM6A (Cat #33510S), KDM6B/JMJD3 (Cat #3457S) (Cell Signaling Technology), or isotype control antibody and 25 μL of protein G-coated magnetic beads were added to 150 μL of the sheared chromatin, and the chromatin was then immunoprecipitated at 4°C overnight with rotation. The chromatin/antibody complexes captured on the beads were washed 3 times in ChIP buffer and then eluted by incubation for 15 min in 50 μL elution buffer. To reverse the DNA-protein cross-links, 50 μL of reverse cross-linking buffer was added to the eluted chromatin, and the samples were incubated, along with the “input” DNA samples, at 95°C for 15 min. After treatment with 5 μL of proteinase K at 37°C for 1 h (to degrade proteins), the ChIPed DNA was recovered and stored at −20°C until analyzed by PCR as described below.
Semiquantitative PCR
Standard PCR was used to determine the presence of Hotairm1 promoter DNA in the PU.1-immunoprecipitated (ChIPed) DNA. PCR was performed in a 50-μL volume containing 25 μL HotStarTaq Master Mix (Qiagen), 200 ng of ChIPed DNA, and 5 μL of 2 μM primer mix. The PCR conditions were as follows: 1 cycle at 95°C for 15 min, 30 cycles of 94°C, 58°C, and 72°C for 1 min each, and a final cycle at 72°C for 10 min. Equal amounts of PCR products were run on 1% ethidium bromide-stained agarose gel. The bands were visualized using the ChemiDoc XRS Detection System (Bio-Rad), and the images were captured with the Image Lab Software V. 3.0 (Bio-Rad). The PCR primers were designed to amplify 220-bp and 219-bp promoter sequences that contain PU.1 binding site 1 at −196 and PU.1 site 2 at −604, respectively. The primers were PU.1/site 1 forward 5′-tcccagagtcgccactgccaa-3′; reverse 5′-tagagtcacgtgtcctcccc-3′ and PU.1/site 2 forward 5′-ctggtggtggtggtgatgct-3′; reverse 5′-ccccagacggctacttacca-3′ (Integrated DNA Technologies, Coralville, IA, USA).
Quantitative PCR
Quantitative real-time RT-PCR was used to determine Hotairm1 levels in Gr1+CD11b+ cells. Total RNA was isolated using TRIzol reagent (Invitrogen) and subjected to reverse transcription using the QuantiTect Reverse Transcription kit (Qiagen). The cDNA was amplified by real-time qPCR using the QuantiTect SYBR Green PCR Master Mix kit and QuantiTect qPCR Primer Assay specific to Hotairm1 (Qiagen). The expression level was calculated using the 2−∆∆Ct cycle threshold. Gene expression was normalized to GAPDH, and the results are presented as a fold change or percentage relative to the control samples.
For ChIP analysis, the ChIPed DNA was amplified by real-time qPCR using QuantiTect SYBR Green PCR Master Mix (Qiagen) and the same primer sets used for quantitative PCR. The PCR was performed in duplicate in 50-μL volumes. The cycling conditions were as follows: 1 cycle at 95°C for 15 min, 35 cycles at 94°C, 58°C, and 72°C for 30 s each, and a final cycle at 72°C for 10 min.
Statistical Analysis
Data analysis was performed using Microsoft Excel, V. 3.0. Values are expressed as mean ± SD. Differences between 2 groups were determined by a 2-tailed Student's t test. Differences among multiple groups were determined by a one-way ANOVA. Statistical significance with p values <0.05 is reported.
Results
Transcription Factor PU.1 Binds at Hotairm1 Promoter in MDSCs during Sepsis
Hotairm1 expression increases following sepsis initiation [28]. To study the mechanism of Hotairm1 transcription, we examined the Hotairm1 proximal promoter for consensus transcription factor binding sites within a 1-kb sequence upstream of the transcription start site. Bioinformatic analysis using the Eukaryotic Promoter Database of the Swiss Institute of Bioinformatics (https://www.expasy.org/resources/epd) revealed 2 consensus sequences with high binding affinity to the transcription factor PU.1, which we designated as PU.1 binding site 1 and PU.1 binding site 2 (Fig. 1a). The analysis also identified consensus GC, CCAAT, and TATA motifs.
PU.1 controls hematopoiesis and its levels change with myeloid cell differentiation [35, 36, 37]. To test whether PU.1 regulates Hotairm1 expression in MDSCs during sepsis, we performed a ChIP assay using a PU.1-specific antibody. The ChIP DNA was analyzed by real-time qPCR using primer pairs designed to amplify the promoter sequences containing the PU.1 binding site 1 or 2. The results showed binding of PU.1 at the most proximal site (site 1), but not site 2 (Fig. 1b). Notably, PU.1 binding increased significantly in MDSCs from mice with post-acute sepsis.
PU.1 Induces Hotairm1 Expression during Sepsis
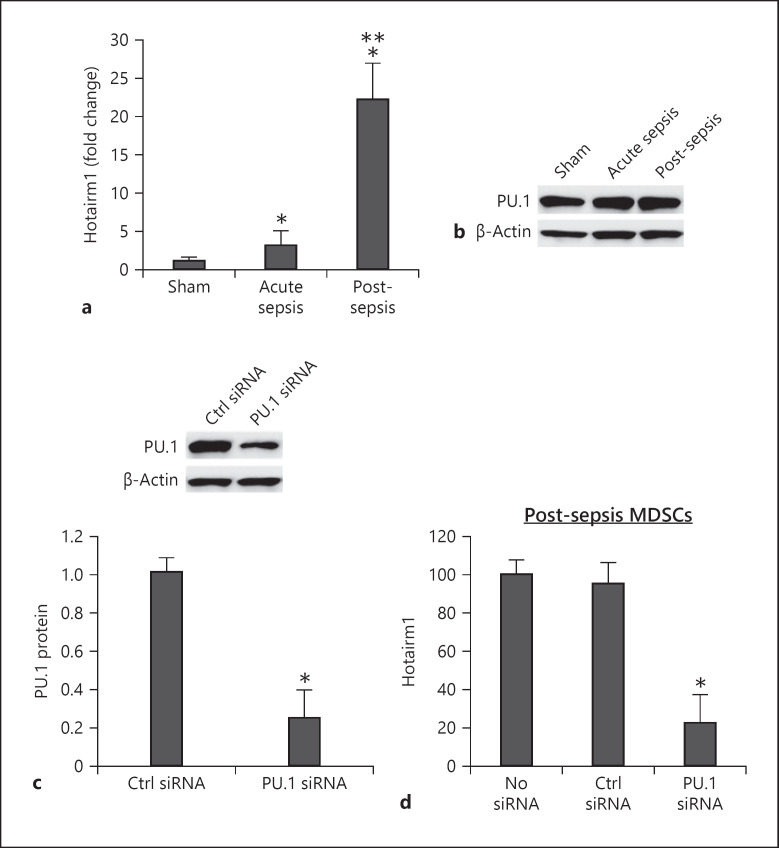
To examine the physiological contribution of PU.1 to Hotairm1 expression, we measured levels of Hotairm1 RNA and PU.1 protein in MDSCs. Hotairm1 level significantly increased in Gr1+CD11b+ MDSCs during post-acute sepsis (Fig. 2a), which correlates with the expansion of Gr1+CD11b+ MDSCs [15]. PU.1 protein was detected at high levels before and after sepsis induction (Fig. 2b). Next, we knocked down PU.1 in post-acute sepsis MDSCs. The knockdown significantly decreased PU.1 protein and Hotairm1 expression (Fig. 2c, d), supporting that PU.1 activates Hotairm1 expression in sepsis.
Fig. 2.
Knockdown of PU.1 reduces Hotairm1 expression. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. a Total RNA was isolated using TRIzol reagent, and Hotairm1 levels were determined by real-time RT-qPCR using QuantiTect qPCR Primer Assay specific to Hotairm1. Sample values were normalized to GAPDH RNA as an internal control. Data are means ± SD for 5–6 mice per group, from 3 experiments, and are presented relative to sham (1-fold). b Western blot of PU.1 protein in Gr1+CD11b+ cells. The results are representative of 2 blots from 2 experiments. c Knockdown of PU.1 in Gr1+CD11b+ cells. The cells were transfected with PU.1-specific or scrambled/control siRNA for 48 h. PU.1 protein levels were determined by Western blotting. Representative blots and densitometric analysis (lower panel) of 3 blots from 2 experiments are shown. Values were normalized to β-actin and are presented relative to control siRNA. *p < 0.05. d Hotairm1 RNA levels were determined by real-time RT-qPCR as in a. Data are means ± SD for 6 mice per group, from 3 experiments, and are presented relative to nontransfected cells. *p < 0.05 versus no siRNA or control (Ctrl) siRNA. MDSCs, myeloid-derived suppressor cells.
Hotairm1 Proximal Promoter Activates Reporter Gene Expression in MDSCs
To determine the functional significance of the putative PU.1 binding sites in the Hotairm1 promoter, we generated reporter constructs, with the luciferase gene transcription under the control of a Hotairm1 promoter fragment containing the 2 PU.1 binding sites (Fig. 3a). These promoter constructs were transfected into post-acute sepsis MDSCs. The promoter construct with the 2 PU.1 binding sites had high luciferase activity but lower than the positive control construct, in which luciferase expression is driven by the GAPDH promoter and SV40 enhancer (Fig. 3b). Site-directed mutagenesis of PU.1 site 1 significantly reduced the luciferase gene expression compared to the wild-type promoter construct, whereas mutation of PU.1 site 2 had no effect (Fig. 6b). These results suggest that the most proximal PU.1 binding site in the Hotairm1 promoter can activate Hotairm1 transcription in MDSCs during sepsis.
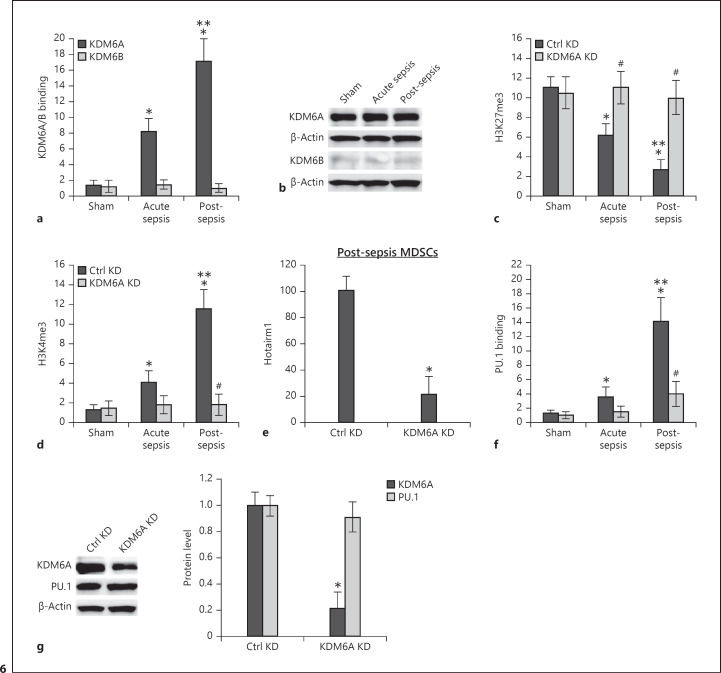
Fig. 6.
KDM6A binds at the Hotairm1 proximal promoter, and its knockdown reduces H3K4me3 and Hotairm1 expression. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. Chromatin was prepared as described in Figure 1. a The chromatin was immunoprecipitated with an antibody specific to KDM6A, KDM6B, or IgG isotype control antibody. The ChIPed DNA was analyzed by real-time qPCR using primers that amplify the promoter sequences surrounding PU.1 binding site 1. b Western blot analysis of KDM6A and KDM6B proteins in Gr1+CD11b+ cells. The results are representative of 2 blots from 2 experiments. c, d KD of KDM6A increases H3K27me3 and decreases H3K4me3. The Gr1+CD11b+ cells were transfected with KDM6A-specific or scrambled/control siRNA for 48 h. Chromatin was prepared and immunoprecipitated with antibody specific to H3K27me3 or H3K4me3, and the ChIPed DNA was analyzed by real-time PCR as in a. e Knockdown of KDM6A decreases Hotairm1 expression. Gr1+CD11b+ cells were purified from bone marrow cells from post-acute septic mice and transfected with KDM6A specific or scrambled/control siRNA for 48 h. The cells were harvested, and levels of Hotairm1 were determined by real-time RT-qPCR using QuantiTect Primer Assay specific to Hotairm1. Sample values were normalized to GAPDH RNA as an internal control. Data are means ± SD for 5–6 mice per group, from 3 experiments, and are presented relative to control KD. *p < 0.05. f Knockdown of KDM6A disrupts PU.1 binding. The Gr1+CD11b+ cells were transfected as in e, and the chromatin was immunoprecipitated with anti-PU.1 or anti-IgG isotype control antibody. The ChIPed DNA was analyzed by real-time PCR as in a. In a, c, d, f, samples values were normalized to the “input” DNA and are presented as fold change relative to the IgG-immunoprecipitated samples (1-fold). Data are means ± SD for 5–7 mice per group, from 3 experiments. *p < 0.05 versus sham; **p < 0.05 versus acute sepsis; #p < 0.05 versus Ctrl KD. g Western blotting of KDM6A and PU.1 proteins in Gr1+CD11b+ cells after KDM6A knockdown. Representative blots and densitometric analysis (right panel) of 3 blots from 2 experiments are shown. Values were normalized to β-actin and are presented relative to control KD. *p < 0.05 versus Ctrl KD. KD, knockdown; MDSCs, myeloid-derived suppressor cells.
Sepsis Induces Histone H3K4 Trimethylation and H3K9 Acetylation at Hotairm1 Promoter in MDSCs
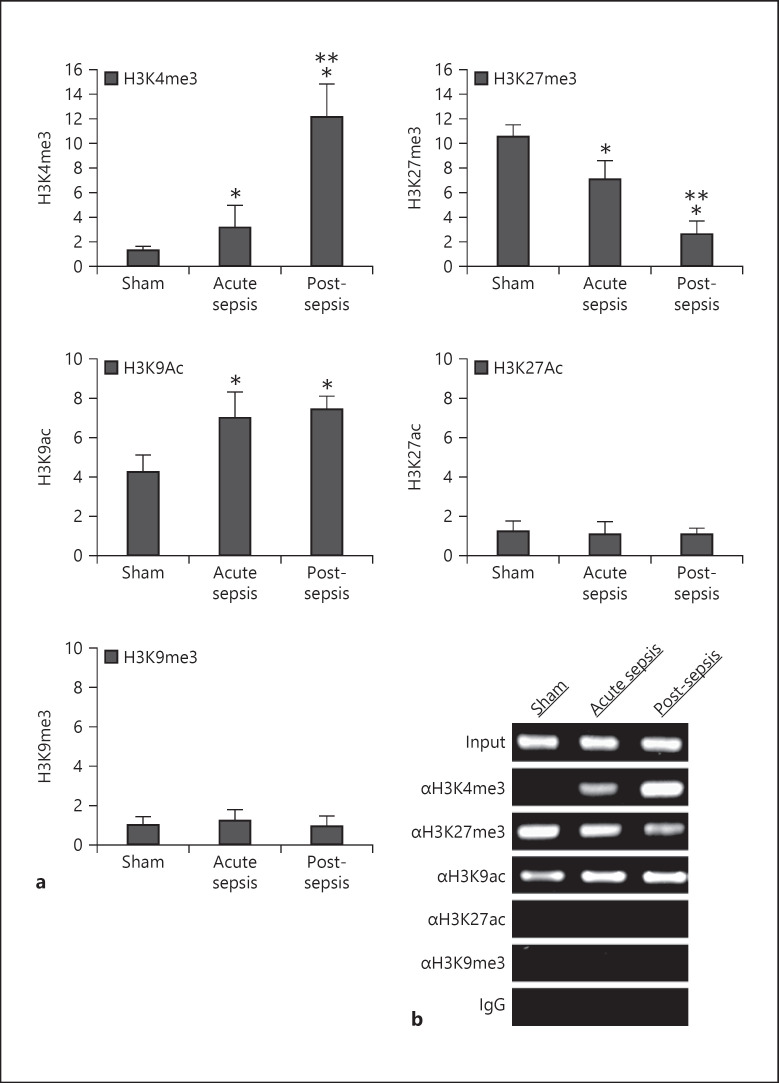
LncRNAs are regulated by epigenetic mechanisms involving histone modifications [38, 39, 40]. We examined histone methylation and acetylation marks around the PU.1 binding site 1. We focused on known histone modifications that support transcription activation and/or repression [41, 42, 43, 44]. ChIP, followed by a real-time qPCR analysis of the ChIPed DNA using a primer set that amplifies the sequence around the PU.1 binding site 1, showed that this sequence was enriched for H3K4me3, H3K27me3, and H3K9ac (Fig. 4a). Specifically, H3K4me3 increased in MDSCs during acute sepsis and further increased during post-acute sepsis, whereas H3K27me3 was already high in control/sham MDSCs and gradually declined after sepsis initiation. The H3K9ac level was high in MDSCs from sham mice and increased further during sepsis, with no significant difference between acute and post-acute sepsis (Fig. 4a). We did not detect significant changes in H3K27ac or H3K9me3 occupancy on the promoter after sepsis initiation. In addition, semiquantitative PCR showed almost similar changes in these histone marks (Fig. 4b). These results demonstrate that sepsis induces H3K4 trimethylation and H3K9 acetylation while inhibiting H3K27 trimethylation at the Hotairm1 proximal promoter.
Fig. 4.
Detection of histone modifications at the Hotairm1 proximal promoter. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. Chromatin was prepared as described in Figure 1 and immunoprecipitated with antibodies specific to the indicated histone marks or anti-IgG isotype control antibody. a The ChIPed DNA was analyzed by real-time qPCR using primers that amplify the promoter sequences surrounding PU.1 binding site 1. Sample values were normalized to the “input” DNA and are presented as fold change relative to the IgG-immunoprecipitated samples (1-fold). Data are means ± SD for 6–7 mice per group, from 3 experiments. *p < 0.05 versus sham; **p < 0.05 versus acute sepsis. b The ChIPed DNA was amplified by semiquantitative PCR using the same primers as in a. The PCR products (220 bp) were separated by electrophoresis using 1% agarose gel.
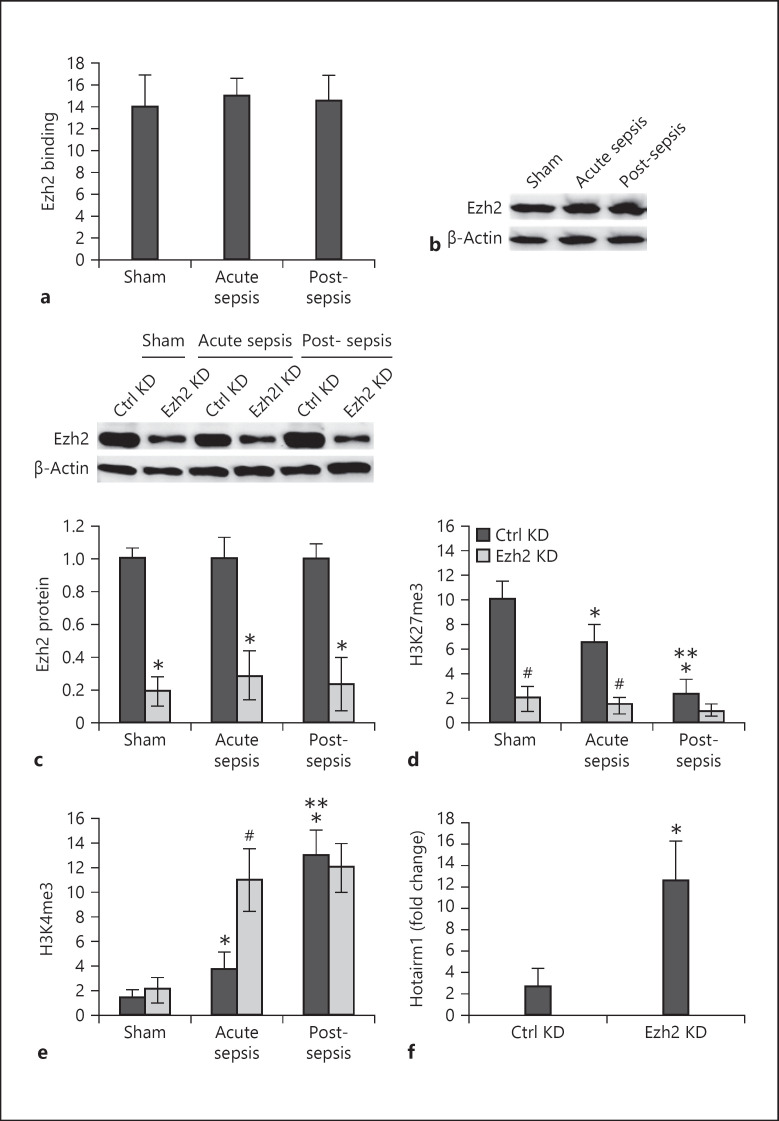
Histone Methyltransferase Ezh2 Binds at Hotairm1 Promoter and Induces H3K27 Trimethylation in MDSCs
Enrichment of H3K4me3 and H3K27me3 at target gene promoters is associated with expression activation and repression, respectively [42, 44, 45]. Because these 2 histone marks significanlty changed as sepsis progressed to the post-acute sepsis stage, they were carried forward for further analysis.
Ezh2 is the catalytic subunit of the polycomb repressor complex 2 (PRC2) as it induces H3K27 trimethylation [46]. We determined whether Ezh2 is involved in H3K27 trimethylation in MDSCs during sepsis. ChIP analysis showed a high amount of Ezh2 binding at the Hotairm1 proximal promoter in MDSCs from sham and septic mice, and there was no significant difference between acute and post-acute sepsis (Fig. 5a). In addition, Western blot showed Ezh2 protein expression in sham and sepsis MDSCs, with no marked change throughout sepsis (Fig. 5b). Next, we measured H3K27me3 and H3K4me3 levels after Ezh2 knockdown. The knockdown reduced Ezh2 protein level to a large extent (Fig. 5c). In addition, Ezh2 knockdown significantly decreased H3K27me3 (Fig. 5d). Notably, the knockdown resulted in significant increases in H3K4me3 levels after sepsis initiations, with no marked difference between acute and post-acute sepsis (Fig. 5e). Since Ezh2 knockdown in MDSCs from acute sepsis resulted in significant changes in H3K27me3 and H3K4me3 levels, we measured Hotairm1 levels in these cells. As shown in Figure 5f, Hotairm1 expression significantly increased after the Ezh2 knockdown. These results suggest that Ezh2 deposits H3K27me3 at the Hotairm1 proximal promoter in MDSCs and attenuates its transcription and that the presence of H3K27me3 precludes H3K4me3.
Fig. 5.
Ezh2 binds at the Hotairm1 proximal promoter and induces H3K27 trimethylation. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. Chromatin was prepared as described in Figure 1. a The chromatin solution was immunoprecipitated with anti-Ezh2 or anti-IgG isotype control antibody. The ChIPed DNA was analyzed by real-time qPCR using primers that amplify the promoter sequences surrounding PU.1 binding site 1. b Western blot analysis of Ezh2 protein in Gr1+CD11b+ cells. The results are representative of 3 blots from 2 experiments. c–e Ezh2 knockdown decreases H3K27me3 and increases H3K4me3. The Gr1+CD11b+ cells were transfected with Ezh2-specific or scrambled/control siRNA for 48 h. c Western blotting of Ezh2 protein in Gr1+CD11b+ cells after Ezh2 knockdown. Representative blots and densitometric analysis (lower panel) of 3 blots from 2 experiments are shown. Values were normalized to β-actin and are presented relative to control KD. *p < 0.05 versus control KD. Chromatin was prepared and immunoprecipitated with antibodies specific to H3K27me3 (d) or H3K4me3 (e), and the ChIPed DNA was analyzed by real-time qPCR as in a. In a, d, e, samples values were normalized to the “input” DNA and are presented as fold change relative to the IgG-immunoprecipitated samples (1-fold). Data are means ± SD for 6 mice per group, from 3 experiments. *p < 0.05 versus sham; **p < 0.05 versus acute sepsis; ♯p < 0.05 versus Ctrl KD. f Levels of Hotairm1 in Gr1+CD11b+ cells from acute sepsis were determined by real-time RT-qPCR. Sample values were normalized to GAPDH RNA as an internal control. Data are means ± SD for 5 mice per group, from 3 experiments, and are presented relative to cells from sham mice with Ctrl KD (1-fold). *p < 0.05 versus Ctrl KD. KD, knockdown.
Histone Demethylase KDM6A Reduces H3K27 Trimethylation and Facilitates PU.1 Binding at Hotairm1 Promoter in MDSCs
The decrease in H3K27me3 at the Hotairm1 proximal promoter after sepsis initiation, despite normal levels of Ezh2 protein, suggested that the H3K27me3 level may be regulated by a histone demethylase activity. Histone lysine demethylases KDM6A (also known as UTX) and KDM6B (also known as JMJD3) catalyze the demethylation of H3K27me3 [47]. ChIP assay showed KDM6A but not KDM6B binding at the Hotairm1 proximal promoter after sepsis initiation, and KDM6A binding increased further in MDSCs from post-acute septic mice (Fig. 6a). In addition, Western blotting showed that KDM6A protein was expressed in Gr1+CD11b+ cells at high levels before and during sepsis, whereas KDM6B was barely detected (Fig. 6b).
Next, we investigated the effect of KDM6A on H3K27 and H3K4 trimethylation using siRNA-mediated knockdown. As shown in Figure 6c, KDM6A knockdown increased H3K27me3 levels before and after sepsis initiation. In addition, KDM6A knockdown resulted in a significant decrease in H3K4 trimethylation (Fig. 6d) and significantly inhibited Hotairm1 transcription (Fig. 6e). Notably, KDM6A knockdown significantly decreased the binding of PU.1 at the Hotairm1 proximal promoter (Fig. 6f). Western blotting showed that PU.1 protein level was not affected by the knockdown of KDM6A (Fig. 6g). These results suggest that KDM6A demethylates H3K27me3 in MDSCs during sepsis, resulting in an increase in H3K4 trimethylation and transcription activation of the Hotairm1 promoter. These results also suggest that H3K4 trimethylation is required for PU.1 binding at the Hotairm1 proximal promoter.
Discussion
Elevated expression of Hotairm1 increases S100A9 protein-mediated induction of MDSC expansion and sepsis-associated immunosuppression [15, 28]. In the present study, we identified transcription factor PU.1 as a pivotal regulator of Hotairm1 transcription in MDSCs during sepsis. We show that PU.1 binds to and activates the Hotairm1 promoter in MDSCs as they become more immunosuppressive. We demonstrate an epigenetic mechanism that controls Hotairm1 transcription by a specific PU.1 binding site. The results thus uncover a molecular path that modifies S100A9 function as a leading contributor to MDSC development in sepsis.
PU.1/SPI1 is a hematopoietic-specific Ets family transcription factor that plays an essential role in myeloid cell development and differentiation [37]. It does so by controlling the expression of genes required for myeloid cell differentiation and maturation, including c-Jun and C/EBPα [48], macrophage colony-stimulating factor, granulocyte colony-stimulating factor, and GM-CSF, as well as CD11b and CD18 [49, 50]. In the steady-state, PU.1 levels are high in mouse myeloid cells, with lower expression in mature compared to bone marrow myeloid progenitors [36]. Our results showed that PU.1 protein levels increased in bone marrow Gr1+CD11b+ MDSCs before and after sepsis induction. Importantly, PU.1 binding at the Hotairm1 promoter increased during post-acute sepsis and PU.1 knockdown reduced Hotairm1 levels. This is compatible with the notion that PU.1 binding at the Hotairm1 promoter is a rate-limiting step in MDSC development during sepsis, which is supported by luciferase reporter expression in the MDSCs. PU.1 controls all-trans retinoic acid-induced expression of human Hotairm1 during granulocytic differentiation of acute promyelocytic leukemia-derived NB4 cells [51]. In that study, however, PU.1 binding at Hotairm1 DNA occurred distal to putative DNA binding sites, downstream of the transcription start site.
Posttranscriptional modifications on histone proteins control gene transcription by modifying the chromatin structure [43]. Modified histone residues recruit chromatin-modifying cofactors and nonhistone proteins that dynamically condense or open chromatin, leading to transcription repression or activation [41, 43]. H3K27me3 represses gene transcription, whereas H3K4me3 usually activates transcription [41]. Following sepsis induction, we observed increases in H3K4me3 at the Hotairm1 proximal promoter with a gradual decrease in H3K27me3, as a putative transcription switch to increase Hotairm1 in MDSCS during post-acute sepsis. H3K27me3 might repress Hotairm1 transcription in the steady-state or during sepsis initiation. In support of our findings of increased H3K4me3 with transcription activation of Hotairm1 promoter is that H3K4me3 identifies other transcriptionally active lncRNAs [38, 40].
H3K27me3 is regulated by the polycomb repressor complex core subunit, methyltransferase Ezh2 [46]. In this study, Ezh2 protein bound the Hotairm1 promoter in MDSCs before and throughout sepsis, and its knockdown decreased H3K27me3 concurrent with an increase in H3K4me3 at the Hotairm1 promoter. This suggests an epigenetic crosstalk mechanism. Decreased expression of human Hotairm1 during differentiation of blood monocytes into dendritic cells correlates with decreases in H3K27me3 around Hotairm1 genomic regions [52]. Enrichment of the H3K9me1 mark at the human Hotairm1 promoter in liver cancer cells reduces Hotairm1 expression [53], but we did not detect H3K9me1 at the mouse Hotairm1 promoter in MDSCs (data not shown).
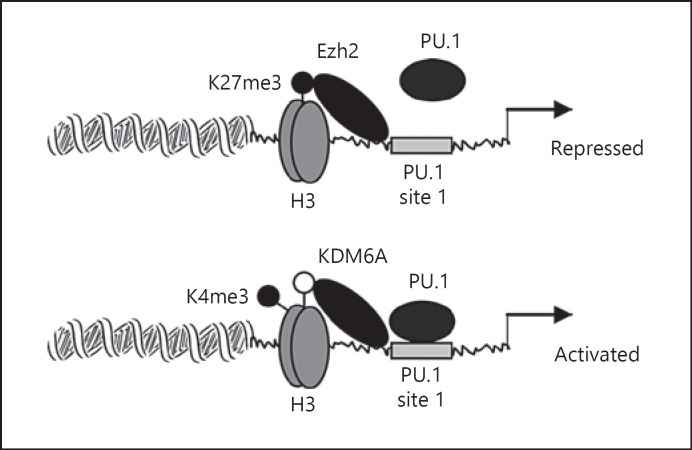
KDM6A specifically demethylates H3K27me3 [54]. KDM6A knockdown in this study increased H3K27me3, decreased H3K4me3 with concomitant inhibition of PU.1 binding, and decreased Hotairm1 transcripts. As such, a dynamic change in the H3K27me3 and H3K4me3 axis may direct Hotairm1 promoter transcription in mouse MDSCs. The current study, however, does not identify the signal or path that induces KDM6A binding at the promoter. This study detected high levels of H3K9ac at the Hotairm1 promoter before sepsis initiation, and its levels increased during sepsis. Histone acetylation activates transcription [43], but histone acetylation unlike methylation is unstable, and it is unclear whether this acetylation mark supports Hotairm1 transcription. Figure 7 models how epigenetic changes at the mouse Hotairm1 promoter may regulate PU.1 binding and Hotairm1 transcription to direct MDSC development in sepsis. Studies of concept translation into humans are warranted.
Fig. 7.
A diagram depicting the putative role of PU.1 in the transcription regulation of the Hotairm1 promoter during sepsis. In the steady-state, Ezh2 binds at the Hotairm1 proximal promoter and catalyzes trimethylation of histone 3 on lysine 27 (H3K27me3), which prevents binding of transcription factor PU.1 at its consensus sequence and thus represses the promoter transcription. After sepsis initiation, H3K27me3 is gradually decreased due to demethylation by KDM6A, leading to an increase in trimethylation of histone 3 on lysine 4 (H3K4me3). This epigenetic modification allows binding of PU.1 at the Hotairm1 promoter and subsequently activates Hotairm1 transcription. H3, histone 3; K, lysine; me3, trimethylation.
Statement of Ethics
All experiments were conducted in accordance with National Institutes of Health guidelines and were approved by the East Tennessee State University Animal Care and Use Committee (Protocol No. P190603).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was planned and supported mostly by National Institutes of Health Grant R35GM131692 (to M.E.). Dr. McCall as a contributing interpreter is also supported by NIGMS R35GM126922.
Author Contributions
I.B. and T.A. conducted the experiments. D.Y. and Z.Q.Y. provided critical input and edits. C.E.M. provided insights regarding data interpretation and reviewed the manuscript. M.E. designed the study, interpreted the data, and wrote the manuscript.
References
- 1.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306:2594–605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efron PA, Mohr AM, Bihorac A, Horiguchi H, Hollen MK, Segal MS, et al. Persistent inflammation, immunosuppression, and catabolism and the development of chronic critical illness after surgery. Surgery. 2018;164:178–84. doi: 10.1016/j.surg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13:862–74. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrijver IT, Théroude C, Roger T. Myeloid-derived suppressor cells in sepsis. Front Immunol. 2019;10:327. doi: 10.3389/fimmu.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect Immun. 2012;80:2026–34. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McPeak MB, Youssef D, Williams DA, Pritchett CL, Yao ZQ, McCall CE, et al. Frontline science: myeloid cell-specific deletion of Cebpb decreases sepsis-induced immunosuppression in mice. J Leukoc Biol. 2017;102:191–200. doi: 10.1189/jlb.4HI1216-537R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janols H, Bergenfelz C, Allaoui R, Larsson AM, Rydén L, Björnsson S, et al. A high frequency of MDSCs in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J Leukoc Biol. 2014;96:685–93. doi: 10.1189/jlb.5HI0214-074R. [DOI] [PubMed] [Google Scholar]
- 8.Mathias B, Delmas AL, Ozrazgat-Baslanti T, Vanzant EL, Szpila BE, Mohr AM, et al. Human myeloid-derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock. Ann Surg. 2017;265:827–34. doi: 10.1097/SLA.0000000000001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrand-Rosenberg S, Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J Immunol. 2018;200:422–31. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–66. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 12.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin Chim Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Hsu K, Champaiboon C, Guenther BD, Sorenson BS, Khammanivong A, Ross KF, et al. Anti-infective protective properties of S100 calgranulins. Antiinflamm Antiallergy Agents Med Chem. 2009;8:290–305. doi: 10.2174/187152309789838975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X. S100 proteins as an important regulator of macrophage inflammation. Front Immunol. 2017;8:1908. doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Kumbhare A, Youssef D, McCall CE, El Gazzar M. Intracellular S100A9 promotes myeloid-derived suppressor cells during late sepsis. Front Immunol. 2017;8:1565. doi: 10.3389/fimmu.2017.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20:623–31. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol. 2016;46:504–12. doi: 10.1002/eji.201444558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–19. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. 2011;108:11381–6. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–34. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–87. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkhateeb T, Bah I, Kumbhare A, Youssef D, Yao ZQ, McCall CE, et al. Long non-coding RNA Hotairm1 promotes S100A9 support of MDSC expansion during sepsis. J Clin Cell Immunol. 2020;11((6)):11. [PMC free article] [PubMed] [Google Scholar]
- 29.Brudecki L, Ferguson DA, Yin D, Lesage GD, McCall CE, El Gazzar M. Hematopoietic stem-progenitor cells restore immunoreactivity and improve survival in late sepsis. Infect Immun. 2012;80:602–11. doi: 10.1128/IAI.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazuski JE, Sawyer RG, Nathens AB, DiPiro JT, Schein M, Kudsk KA, et al. The surgical infection society guidelines on antimicrobial therapy for intra-abdominal infections: an executive summary. Surg Infect. 2002;3:161–73. doi: 10.1089/109629602761624171. [DOI] [PubMed] [Google Scholar]
- 31.Osuchowski MF, Ayala A, Bahrami S, Bauer M, Boros M, Cavaillon JM, et al. Minimum quality threshold in pre-clinical sepsis studies (MQTiPSS): an international expert consensus initiative for improvement of animal modeling in sepsis. Shock. 2018;50:377–80. doi: 10.1097/SHK.0000000000001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angele MK, Schwacha MG, Ayala A, Chaudry IH. Effect of gender and sex hormones on immune responses following shock. Shock. 2000;14:81–90. doi: 10.1097/00024382-200014020-00001. [DOI] [PubMed] [Google Scholar]
- 33.De MA, Torres MB, Reeves RH. Genetic determinants influencing the response to injury, inflammation, and sepsis. Shock. 2005;23:11–7. doi: 10.1097/01.shk.0000144134.03598.c5. [DOI] [PubMed] [Google Scholar]
- 34.Zellweger R, Wichmann MW, Ayala A, Stein S, DeMaso CM, Chaudry IH. Females in proestrus state maintain splenic immune functions and tolerate sepsis better than males. Crit Care Med. 1997;25:106–10. doi: 10.1097/00003246-199701000-00021. [DOI] [PubMed] [Google Scholar]
- 35.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel G, DeKoter RP, Wang Q, Hexley P, Ogle CK. Optimization and application of a flow cytometric PU.1 assay for murine immune cells. J Immunol Methods. 2012;382:81–92. doi: 10.1016/j.jim.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–31. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 43.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chittock EC, Latwiel S, Miller TC, Müller CW. Molecular architecture of polycomb repressive complexes. Biochem Soc Trans. 2017;45:193–205. doi: 10.1042/BST20160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marecki S, Fenton MJ. PU. 1/interferon regulatory factor interactions: mechanisms of transcriptional regulation. Cell Biochem Biophys. 2000;33:127–48. doi: 10.1385/CBB:33:2:127. [DOI] [PubMed] [Google Scholar]
- 49.Pahl HL, Scheibe RJ, Zhang DE, Chen HM, Galson DL, Maki RA, et al. The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J Biol Chem. 1993;268:5014–20. [PubMed] [Google Scholar]
- 50.Rosmarin AG, Caprio D, Levy R, Simkevich C. CD18 (beta 2 leukocyte integrin) promoter requires PU.1 transcription factor for myeloid activity. Proc Natl Acad Sci U S A. 1995;92:801–5. doi: 10.1073/pnas.92.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei S, Zhao M, Wang X, Li Y, Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9:44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin J, Li J, Feng Y, Wang L, Zhang Y, Yang R. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. Onco Targets Ther. 2017;10:1307–15. doi: 10.2147/OTT.S124201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li YQ, Sun N, Zhang CS, Li N, Wu B, Zhang JL. Inactivation of lncRNA HOTAIRM1 caused by histone methyltransferase RIZ1 accelerated the proliferation and invasion of liver cancer. Eur Rev Med Pharmacol Sci. 2020;24:8767–77. doi: 10.26355/eurrev_202009_22815. [DOI] [PubMed] [Google Scholar]
- 54.D'Oto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1:34–40. [PMC free article] [PubMed] [Google Scholar]