Abstract
Bacterial peptidoglycan (PGN) stimulates toll-like receptor 2 (TLR2) on the surface of keratinocytes (KCs), triggering signaling pathways that promote an innate immune response. However, excessive TLR2 activation can lead to inappropriate inflammation, which contributes to skin conditions such as rosacea. To better treat these conditions, there is a need to understand the molecular mechanisms that regulate the cellular response to TLR2 activation in the skin. Aryl hydrocarbon receptor (AhR) is a transcription factor that modulates the immune response in KCs and is a promising therapeutic target for inflammatory skin diseases. Here, we investigated the role of the AhR in regulating the transcriptional response of human KCs to PGN. We performed whole-transcriptome sequencing in wild-type and AhR-depleted KCs after PGN stimulation. AhR depletion altered the expression of 72 genes in response to PGN, leading to increased expression of 48 genes and repression of 24 genes, including interleukin (IL)-1β. Chromatin immunoprecipitation showed that PGN stimulation resulted in AhR binding the promoters of IL-1β and IL-6 to activate them. More broadly, AhR promoted inflammatory gene expression by increasing JNK/mitogen-activated protein kinase signaling and FosB expression. Finally, we observed that AhR depletion increased TLR2 expression itself, raising the hypothesis that AhR may serve to restrain TLR2-mediated inflammation in KCs through negative feedback. Viewed together, our findings demonstrate a significant and complex role for AhR in modulating the expression of inflammatory genes in KCs in response to PGN.
Keywords: Peptidoglycan, Aryl hydrocarbon receptor, Toll-like receptor 2
Introduction
The epidermis is the outermost layer of the skin and comprises predominantly of keratinocytes (KCs) that serve as a primary barrier to the outside environment. KCs sense and respond to harmful microbes. One major mechanism is recognition of pathogen-associated molecular patterns, small molecular motifs conserved across classes of pathogens that activate toll-like receptors (TLRs) on the surface of KCs. Engagement of TLRs triggers signaling pathways that induce an immune response to combat infections [1].
In the ideal response, inflammation is activated by microbes and then is tapered down after pathogens are eliminated. Excessive or prolonged responses can drive states of chronic inflammation [2]. A prominent example is seen in rosacea, a common skin condition that classically manifests as fixed erythema, telangiectasias, and inflammatory papules and pustules on the face. Rosacea can be worsened by dietary or environmental triggers such as alcohol, ultraviolet irradiation, or microbes. This leads to a mixed infiltrate of cells from both the innate and adaptive immune responses that permeate the superficial and deep dermis, leading to edema, dilated vessels, and spongiosis [3].
The excessive innate immune response in rosacea is driven by elevated expression and activity of TLR2 [4]. TLR2 recognizes an array of bacterial elements, including peptidoglycan (PGN), a component of the bacterial cell wall. Activation of TLR2 leads to expression of pro-inflammatory proteins such as interleukin (IL)-1β and IL-6 [5], which recruit additional immune cells and polarize T lymphocytes [6]. What is not fully understood are the mechanisms that regulate the downstream activity of TLR2. Increased insights into these mechanisms would be valuable to understanding the pathogenesis of chronic inflammatory conditions.
One candidate regulator of TLR2-mediated skin inflammation is the AhR. Originally studied in the skin for its involvement in chloracne and response to industrial chemicals, AhR has been subsequently found to have wide range of impacts on skin biology, affecting processes such as skin differentiation, carcinogenesis, and inflammation [7]. AhR is a ligand-bound transcription factor that translocates to the nucleus to regulate gene expression. It is a versatile but complex regulator responding to a wide array of ligands including organic chemicals, products of ultraviolet irradiation, dietary molecules, and microbial elements [8].
The main goal of this work is to characterize the effect of AhR on the transcriptional response of human KCs to PGN. Our hypothesis is that PGN can activate AhR and that AhR modulates the expression of TLR2 gene targets through inflammatory signaling pathways as well as by direct transcriptional activation. To test this hypothesis, we performed PGN stimulation in wild-type versus AhR-depleted primary human KCs and assessed the molecular phenotype of AhR depletion using whole-transcriptome sequencing, chromatin immunoprecipitation, and qRT-PCR of TLR2 downstream gene targets, as well as evaluation of the activity of intracellular signaling pathways.
Materials and Methods
Cell Culture and Stimulation
Primary epidermal KCs were isolated from discarded neonatal foreskins taken from elective circumcisions, collected with informed consent under an institutional review board protocol approved by the University of California San Diego. Cells were cultured in a 50:50 mixture of K-SFM and 154 media (Life Technologies) with recommended supplements and 1× antibiotic-antimycotic (Thermo Fisher Scientific). Cells were seeded as indicated and cultured for 24 h at 37°C in a humidified atmosphere containing 5% CO2. Twenty-four hours later, the medium was changed and supplemented with 1.2 mM calcium. After 24 h, cells were treated with the chemical AhR antagonist CH223191 (10 μM, Sigma-Aldrich, St. Louis, MO, USA) and/or PGN (20 μg/mL, Sigma-Aldrich), as indicated. CH223191 was added to cell medium 1 h prior to PGN in dual treatment experiments. Water was applied as vehicle control for PGN. DMSO was applied as vehicle control for CH223191.
Real-Time Quantitative PCR
KCs were seeded onto 24-well plates (2.5 × 105 cells/well). Total RNA from KCs was extracted using Trizol (Ambion, Foster City, CA, USA) and an RNA Miniprep kit (Zymo Research, Tustin, CA, USA) based on manufacturer's instructions. Quantitative PCR was performed using SYBR Green Supermix (BIO-RAD, Hercules, CA, USA). Human RPL32 was used as an internal reference expression gene. Primer sequences are as shown in Table 1. GeneForward 5′→3′Reverse 5′→3′CXCL8CCTGATTTCTGCAGCTCTGTGCCAGACAGAGCTCTCTTCCATCCL20CAAAGAACTGGGTACTCAACACTTGTATCCAAGACAGCAGTCAAAGTNF-αATGTTGTAGCAAACCCTCAAGCGAGGAGCACATGGGTGGAGIL-1βGATGGCTTATTACAGTGGCAATGACCCTTGCTGTAGTGGTGGTCIL-6AGACAGCCACTCACCTCTTCAGTTCTGCCAGTGCCTCTTTGCTGChIP: IL-6GACTCAGTGGCAATGGGGAGCACCTGCTTCAGCCCACTTAChIP: IL-1βTGGCTTGTTTTGCGCTTTGATGTGAACAAGAGCATGGGTGTChIP: neg ctrlAGGTTCAGGTATGGATGGGTGTTATAGGATTACAGGCATAAGCCACTGRPL32AGGCATTGACAACAGGGTTCGTTGCACATCAGCAGCACTT
Table 1.
Primer sequences
| Gene | Forward 5′→3′ | Reverse 5′→3′ |
|---|---|---|
| CXCL8 | CCTGATTTCTGCAGCTCTGTG | CCAGACAGAGCTCTCTTCCAT |
| CCL20 | CAAAGAACTGGGTACTCAACACT | TGTATCCAAGACAGCAGTCAAAG |
| TNF-α | ATGTTGTAGCAAACCCTCAAGC | GAGGAGCACATGGGTGGAG |
| IL-1β | GATGGCTTATTACAGTGGCAATGA | CCCTTGCTGTAGTGGTGGTC |
| IL-6 | AGACAGCCACTCACCTCTTCAG | TTCTGCCAGTGCCTCTTTGCTG |
| ChIP: IL-6 | GACTCAGTGGCAATGGGGAG | CACCTGCTTCAGCCCACTTA |
| ChIP: IL-1β | TGGCTTGTTTTGCGCTTTGA | TGTGAACAAGAGCATGGGTGT |
| ChIP: neg ctrl | AGGTTCAGGTATGGATGGGTGTTA | TAGGATTACAGGCATAAGCCACTG |
| RPL32 | AGGCATTGACAACAGGGTTC | GTTGCACATCAGCAGCACTT |
Immunoblotting
KCs were seeded onto 12-well plates (5 × 105 cells/well). After 24 h, medium was supplemented with calcium (1.2 mM) and incubated for 24 h. Then cells were treated with stimuli as indicated. For harvesting, cells were washed with PBS and lysed in RIPA buffer containing protease inhibitors and phosphatase inhibitors. Protein concentration was determined using a BCA assay (ThermoFisher Scientific, Chicago, IL, USA). Lysates were resolved on SDS-PAGE and transferred to nitrocellulose membranes. Blots were blocked in TBS-Tween containing 5% BSA. The membranes were incubated with antibodies against phospho-p44/42 mitogen-activated protein kinase (MAPK) (Erk1/2) (Thr202/Tyr204) (#9101S), phospho-nuclear factor kappa B (NF-κB) p65 (Ser536) (#3033), phospho-p38 MAPK (Thr180/Tyr182) (#4511), phospho-SAPK/JNK (Thr183/Tyr185) (#4668), NF-κB p65 (#6956), AHR (#83200), FOSB (#2251), or β-tubulin (#2128) (dilution 1:1,000) overnight at 4°C. All antibodies were from Cell Signaling (Danvers, MA, USA). Blots were visualized on an Odyssey imager (Li-Cor).
ELISA
KCs were seeded onto 96-well plates (4 × 104 cells/well) in culture medium treated with stimuli as indicated. Supernatants were assayed to determine the expression of IL-6 and IL-1β with an ELISA kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions.
RNA Interference
KCs were seeded in 6-well plates at 3 × 105 cells/well for 24 h. KCs were transfected with 2 μL short interfering RNA (siRNA) (10 μM) using 5 μL RNAiMAX (Invitrogen). After 48 h, cells were treated as indicated. AhR siRNA (s1199) and control siRNA (4390844) were from Life Technologies (Carlsbad, CA, USA).
RNA-Seq
KCs treated with control or AhR siRNAs were harvested 3 h after PGN stimulation. Two independent biological replicates were used for each condition. Total RNA was isolated using the RNA Miniprep Kit (Zymo Research). Quality and quantity of RNA were assessed using a NanoDrop spectrophotometer (Thermo Scientific). RNA samples were poly-A selected and sequenced on Illumina HiSeq using 150-nucleotide paired-end reads to a depth of >50 million reads per sample. Reads alignment was performed with STAR and transcript quantitation using a standard Partek model (St. Louis, MO, USA). Differentially expressed genes were defined as those displaying >1.5-fold expression difference, p value <0.05 based on a 2-sided t test, and a minimum threshold of 5 read counts.
Chromatin Immunoprecipitation
KCs were seeded onto a 15-cm dish (4 × 106cells/dish), cultured to 70–80% confluence, and then treated with PGN (20 μg/mL) for 1 h. Eight million cells and 5 μg of antibody were used for each antibody pulldown. The following antibodies were used: AhR (#83200, Cell Signaling Technology) and rabbit IgG (#12–370, Millipore, Bedford, MA, USA). Cells were fixed in both formaldehyde (1%, #28908, ThermoFisher) and disuccinimidyl glutarate (2 mM, #20593, ThermoFisher). The chromatin fraction was immunoprecipitated overnight at 4°C. The percentage input method was used to normalize chromatin immunoprecipitation data.
Immunofluorescence
KCs were seeded to confluence in 4-well chamber slides (2.5 × 105 cells/well, Thermo Scientific #177437). After 24 h, medium was supplemented with 1.2 mM calcium and incubated for another 24 h. As indicated, cells were then treated with vehicle control, 20 μg/mL PGN, and/or 10 μM CH223191. CH223191 was added to cell medium 1 h prior to PGN in the dual treatment condition. After 72 h, cells were fixed with ice-cold acetone for 10 min, washed in PBS, and incubated with a TLR2 (TL1.2) antibody (Santa Cruz Biotechnology, sc-21759) at 1:50 dilution overnight at 4°C. Slides were washed in PBS then incubated with a goat anti-mouse Alexa Fluor 488 secondary antibody (Thermo Fisher, A28175) at 1:500 dilution for 1 h. After washing in PBS, nuclei were stained with Hoescht 33342 (Thermo Fisher), rinsed with water, then mounted with Prolong Gold Antifade Mountant (Thermo Fisher), and visualized at ×60 objective magnification using an EVOS M5000 microscope (Thermo Fisher) using 2-channel capture and overlay of GFP (470/525 nm ex/em) and DAPI (357/447 nm ex/em) filters.
Statistical Analysis
Normal distribution data were presented as mean ± SD. Statistical analysis was performed using a paired t test or 1-way ANOVA for normal distribution data. p value of less than 0.05 was considered significant. The p values are designated as p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). Statistical analysis was conducted using SPSS software 10.1.
Results
AhR Affects the Transcriptional Response of KCs to PGN
To systematically identify the genes activated by PGN stimulation, we performed whole-transcriptome sequencing in biological replicates of primary human KCs following 3 h of PGN treatment. To understand how AhR affects this response, we performed PGN stimulation in both wild-type KCs and in KCs depleted of AhR using siRNA. siRNA-mediated depletion reduced AhR mRNA and protein levels by >75% (shown in online suppl. Figure 1; for all online suppl. material, see www.karger.com/doi/10.1159/000517627). Differentially expressed genes (DEGs) between 2 conditions were defined as those displaying >1.5-fold expression change, p value <0.05, and a minimum threshold of 5 read counts. The full list of DEGs is shown in the online suppl. Table.
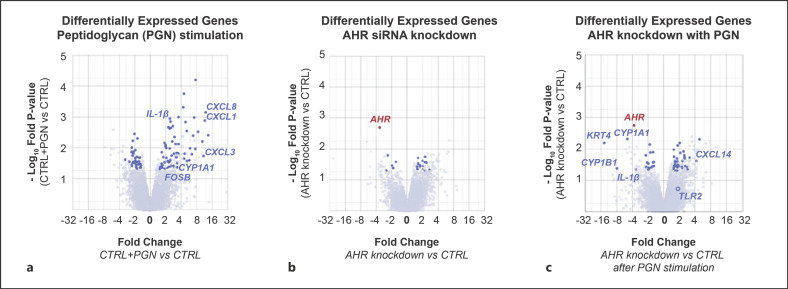
We first compared control, untreated KCs (CTRL) to PGN-treated KCs (CTRL + PGN). Treatment with PGN resulted in upregulation of 80 genes including tumor necrosis factor (TNF), interleukin-1β (IL-1β), and a set of chemokines (CXCL1, 3, 8), all of which are known downstream targets of activated TLR2 (shown in Fig. 1a) [9]. Gene ontology analysis of the upregulated gene set identified enrichment for terms associated with an inflammatory response (p value 4.42 × 10−11). These results affirmed that our experimental conditions captured the expected inflammatory transcriptional response to PGN.
Fig. 1.
AhR regulates the transcriptional response of human KCs to PGN. a Volcano plot displaying DEGs in KCs treated with PGN. Dark blue circles represent gene transcripts meeting criteria of at least 1.5-fold differential expression compared to untreated control (CTRL), minimum read count >5, and p value <0.05. Representative genes of interest are labeled. b Volcano plot displaying differentially expressed genes in KCs after AhR knockdown using siRNA. c Volcano plot displaying DEGs in KCs treated with PGN, comparing AhR-depleted KCs against control. Key genes of interest are labeled. Full gene sets for each pairwise comparison are shown in the online suppl. Table. PGN, peptidoglycan; AhR, aryl hydrocarbon receptor; KC, keratinocyte; DEG, differentially expressed genes; siRNA, short interfering RNA.
We next turned our attention to comparing unstimulated control KCs (CTRL) to unstimulated AhR-depleted KCs. Globally, AhR depletion in unperturbed KCs did not result in clear major patterns of transcriptional changes (shown in Fig. 1b), with gene ontology analysis of DEGs yielding no significantly enriched terms (p > 0.05). Only 6 gene transcripts were downregulated by AhR depletion in unstimulated KCs, one of which was AhR itself.
Using these 2 initial states as experimental controls, we compared the transcriptional response of wild-type versus AhR-depleted KCs to PGN stimulation (shown in Fig. 1c). Twenty-four genes were repressed in AhR-depleted KCs compared to control, representing a set of genes whose expression depends on AhR. In addition to AhR itself, 2 of the most downregulated transcripts were cytochrome P450 1A1 and 1B1 (CYP1A1 and CYP1B1). Members of the cytochrome P450 family metabolize xenobiotic compounds and are established AhR targets [10]. However, we also noted that IL-1β induction by PGN was significantly inhibited in AhR-depleted KCs. IL-1β is a transcriptional target of TLR2 and is a critical cytokine that stimulates leukocyte activation and T-lymphocyte polarization [5].
AhR Affects the Expression of TLR2 Downstream Transcriptional Targets
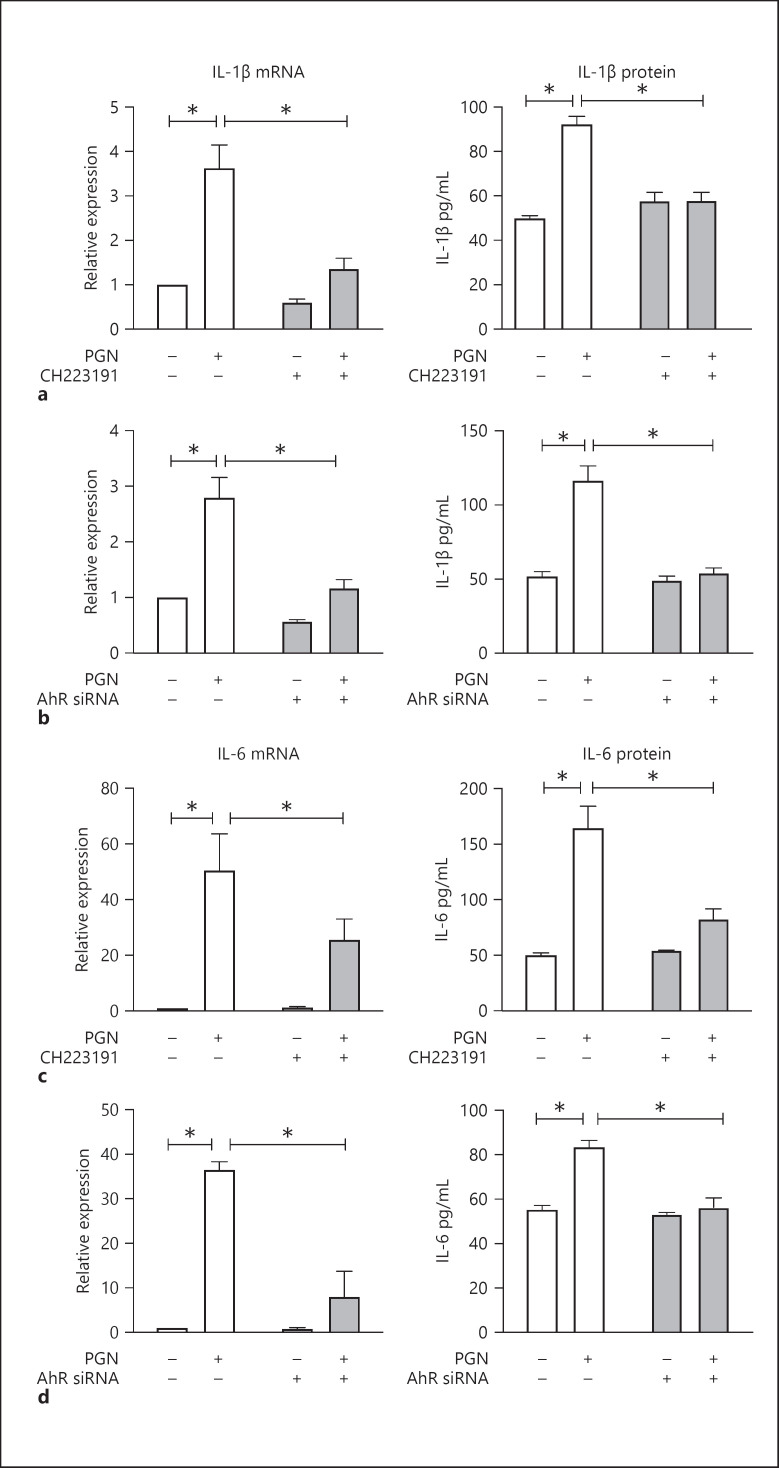
The results from transcriptome sequencing led us to further focus our attention on TLR2-related inflammatory mediators IL-1β and IL-6 [3]. To affirm our transcriptome results using an orthogonal method, we evaluated the impact of a chemical inhibitor of AhR, CH223191, on the transcription of these key genes. PGN treatment led to an increase in IL-1β and IL-6, as observed previously. This induction was attenuated by CH223191, mirroring the results from genetic AhR inhibition (shown in Fig. 2a–d). To corroborate that the transcriptional changes that we observed were also reflected in the levels of protein expression, we performed ELISA to measure protein levels of these targets. The results confirmed that AhR is required for induction of IL-1β and IL-6 after PGN stimulation (shown in Fig. 2a–d).
Fig. 2.
AhR is required for upregulation of IL-1β and IL-6 in response to PGN. a Quantitative RT-PCR and ELISA for IL-1β mRNA and protein. KCs were pretreated with AhR chemical antagonist CH223191 (10 μM) or DMSO vehicle control for 1 h, then stimulated by PGN (20 μg/mL) or water control for 3 h for qPCR or 24 h for ELISA. b Quantitative RT-PCR and ELISA for IL-1β mRNA and protein. KCs transfected with control siRNA or AhR siRNA were stimulated by PGN (20 μg/mL) or water control for 3 h for qPCR or 24 h for ELISA. c Quantitative RT-PCR and ELISA for IL-6 mRNA and protein under similar conditions as in a. d Quantitative RT-PCR and ELISA for IL-6 mRNA and protein under similar conditions as b. For all experiments: n = 3; 2-sided t test with error bars = SEM; *p < 0.05. PGN, peptidoglycan; AhR, aryl hydrocarbon receptor; KC, keratinocyte; siRNA, short interfering RNA.
However, the results from whole-transcriptome sequencing also showed that AhR depletion did not uniformly inhibit expression of TLR2-associated downstream target genes. The expression of CCL20 and IL-36G was not affected by AhR chemical inhibition (shown in online suppl. Fig. 2a). Furthermore, PGN-mediated induction of TNFα and the chemokine CXCL8 increased further with AhR inhibition (shown in online suppl. Fig. S2a, b). Viewed together, these results suggested that AhR and TLR2 both contribute to the transcriptional response to PGN stimulation, but their effects on specific genes can be cooperative or antagonizing.
PGN Activates AhR to Directly Target Promoters of IL-1β and IL-6
To better understand how AhR influences the transcriptional response to PGN stimulation, we sought to gain more insights into its molecular mechanism. AhR is a basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor. In its inactive state, AhR resides in the cytoplasm. Upon binding to a ligand, AhR undergoes a conformational change and translocates to the nucleus where it binds xenobiotic response element motifs in the promoter of target genes to induce gene expression [8].
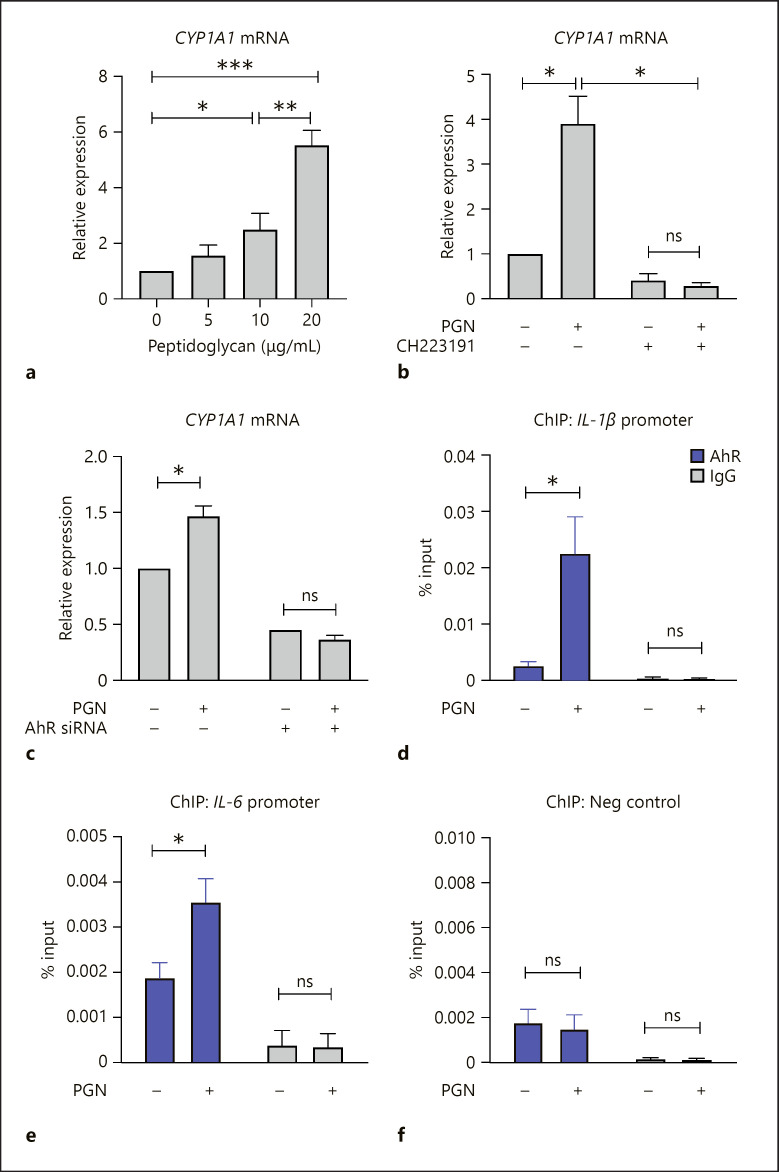
We assessed if PGN stimulation results in AhR activation in KCs. CYP1A1 and CYP1B1 are well-characterized AhR-responsive genes that are directly induced by activated AhR [8]. Our RNA-seq results indicated that PGN increased the expression of CYP1A1 and CYP1B1, and that this upregulation was repressed by AhR knockdown (shown in Fig. 1c and online suppl. Table). We confirmed the induction of CYP1A1 using a dose concentration series of PGN. We found a dose-dependent increase in CYP1A1 expression (shown in Fig. 3a) that was blocked by depletion of AhR using a chemical inhibitor (shown in Fig. 3b) and also by siRNA depletion (shown in Fig. 3c). These results indicated that PGN leads to activated AhR, which can directly bind and induce target genes. We did not assume that PGN directly activates AhR but may result in the indirect stimulation of endogenous AhR ligands [8].
Fig. 3.
PGN results in AhR activation to induce gene targets in KCs. a–c Quantitative RT-PCR for CYP1A1 mRNA. a KCs were treated with a range of PGN concentrations for 3 h. b KCs were pretreated with CH223191 (10 μM) for 1 h, then stimulated with PGN (20 μg/mL) for 3 h. c KCs transfected with control siRNA or AhR siRNA were treated with PGN (20 μg/mL) for 3 h. d–f ChIP to assess binding of AhR at IL-1β promoter (d) IL-6 promoter (e) and negative genomic control (f), performed before and after PGN treatment. Statistical analysis by ANOVA for a and 2-sided t test for b−f. N = 3; error bars = SEM; *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant. PGN, peptidoglycan; AhR, aryl hydrocarbon receptor; KC, keratinocyte; siRNA, short interfering RNA; ChIP, chromatin immunoprecipitation.
RNA-seq (shown in Fig. 1) and confirmatory experiments (shown in Fig. 2) had indicated that IL-1β and IL-6 induction by PGN was dependent, at least in part, on AhR. We therefore examined if AhR directly bound the promoter of these genes to activate them. To do so, we performed chromatin immunoprecipitation in primary human KCs before and after PGN stimulation. We observed enrichment of AhR in both the IL-1β and IL-6 promoters that was specifically induced by PGN stimulation (shown in Fig. 3d–f). These data showed that the full transcriptional induction of some genes in response to PGN occurs by activation and binding of AhR to gene targets.
AhR Promotes PGN-Stimulated JNK/MAPK and Fosb
In addition to directly binding and activating target genes, AhR can also influence inflammation and gene transcription through cross-regulation of signaling pathways [11, 12]. We therefore turned our attention to NF-κB and the MAPKs, signaling pathways downstream of TLR2 [2]. Activation of NF-κB and MAPKs promote production of pro-inflammatory mediators including IL-1β and IL-6 [9].
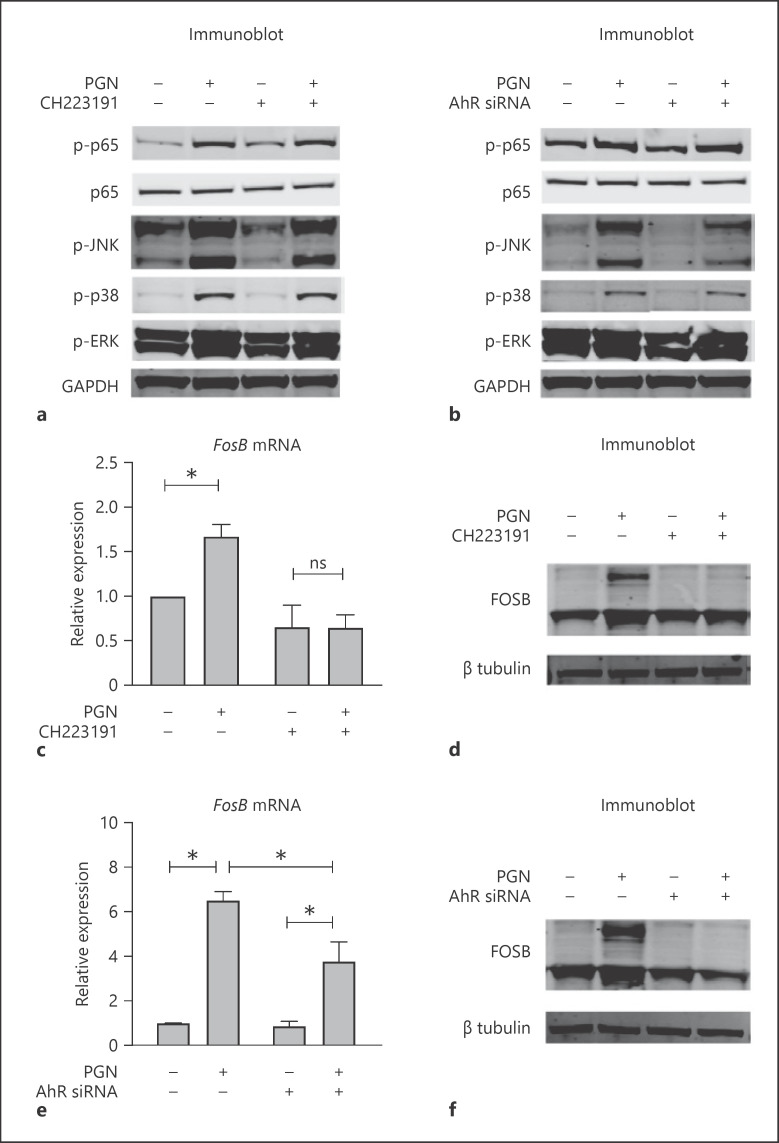
We evaluated if AhR affected activity of NF-κB and MAPK pathways by immunoblotting to assess the phosphorylation of sentinel proteins of each signaling cascade. As expected, stimulation of KCs by PGN promoted phosphorylation of p65/NF-κB, JNK/MAPK, p38/MAPK, and ERK/MAPK (shown in Fig. 4a). To evaluate if AhR influenced the activation of these pathways, we performed PGN stimulation in KCs where AhR was depleted by either chemical CH223191 inhibition (shown in Fig. 4a) or siRNA (shown in Fig. 4b). AhR inhibition resulted in a reduction of phospho-JNK levels. No consistent effects were appreciated on the other signaling pathways.
Fig. 4.
AhR promotes JNK/MAPK and FosB in response to PGN. a, b Immunoblot for canonical signaling proteins of the NF-κB (p65) and MAPKs pathways (JNK, p38, ERK). Where indicated in a, KCs were pretreated with CH223191 (10 μM) for 1 h, then stimulated with PGN (20 μg/mL) for 15 min. Where indicated in b, KCs were previously transfected with control or AhR siRNA. c, d Quantitative RT-PCR and immunoblot for FosB. KCs were pretreated with CH223191 (10 μM) or vehicle, then stimulated with PGN or vehicle control. e, f Quantitative RT-PCR and immunoblot for FosB. KCs were transfected with control or AhR siRNA, then stimulated with PGN or vehicle control. For all experiments: n = 3; 2-sided t test with error bars = SEM; *p < 0.05; ns, not significant. PGN, peptidoglycan; AhR, aryl hydrocarbon receptor; KC, keratinocyte; MAPK, mitogen-activated protein kinase; siRNA, short interfering RNA.
To extend this finding, we evaluated the expression of activator protein-1 (AP-1) transcription factors, which function downstream of JNK/MAPK signaling. The transcription factor AP-1 consists of dimers composed of members of the Jun and Fos protein families, and are linked to inflammation, wounding, and stress [13]. We performed qPCR to assess if the expression of Jun or Fos was altered by PGN stimulation. Among all AP-1 protein family members (shown in online suppl. Fig. 3), only FosB increased reproducibly upon stimulation with PGN (shown in Fig. 1a, 4c–e). Both chemical and genetic inhibition of AhR suppressed the induction of FosB mRNA and protein (shown in Fig. 4c–f), confirming the requirement of AhR for PGN-mediated FosB upregulation. Viewed together, our results suggest that PGN stimulation of KCs stimulates NF-κB and multiple MAPK pathways, and that AhR is required to support the full activation of JNK/MAPK and its downstream effectors.
AhR Attenuates PGN Induction of TLR2
These results delineate how AhR can influence the expression of TLR2-related inflammatory genes and activity of signaling pathways. However, although their downstream effects clearly intersect with each other, we also wanted to consider if AhR activity affected TLR2 expression itself. Examining our transcriptome results, we observed that the expression of TLR2 did not change notably if AhR was depleted in unstimulated KCs. However, upon PGN stimulation, TLR2 increased ∼1.6-fold in AhR-depleted KCs based on RNA-seq data (shown in Fig. 1c). This result provided an initial indication that AhR might have a repressive effect on TLR2 expression.
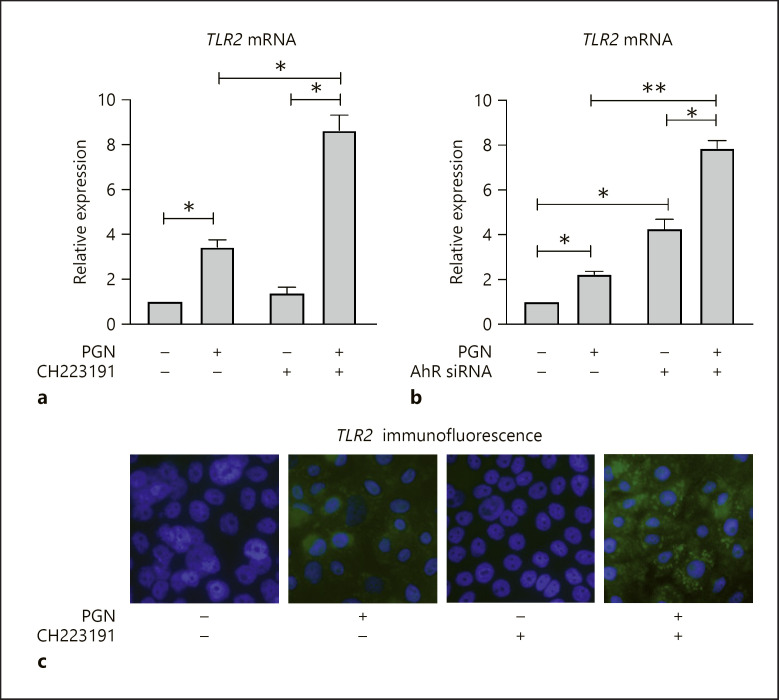
To further examine this possibility, we treated KCs with PGN and measured the expression of TLR2 using quantitative RT-PCR. We found that TLR2 expression increased ∼2–3-fold after PGN stimulation (shown in Fig. 5a). If KCs were pre-treated with the AhR inhibitor CH223191 prior to PGN stimulation, TLR2 expression was amplified further, up to ∼8–9-fold above untreated baseline levels. We observed concordant findings if we depleted AhR using siRNA (shown in Fig. 5b). To corroborate if the transcriptional changes in TLR2 expression were concordant with protein expression, we performed immunocytochemical staining of TLR2 in KCs (shown in Fig. 5c). In untreated and CH223191-treated KCs, we did not detect TLR2. However, treatment with PGN resulted in faint TLR2 staining and combination treatment with CH223191 and PGN led to the most prominent TLR2 expression. Viewed together, these data support that loss of AhR magnifies TLR2 expression in response to PGN. By inference, in a normal state, AhR may restrain TLR2 expression in response to PGN stimulation. This relationship could prevent overexuberant TLR2 signaling and/or contribute to a negative feedback loop to terminate TLR2-related inflammation.
Fig. 5.
TLR2 induction by PGN is inhibited by AhR. Quantitative RT-PCR for TLR2 mRNA. a KCs were pre-treated with CH223191 (10 μM) or vehicle control for 1 h and then stimulated with PGN (20 μg/mL) for 24 h. b KCs were transfected with control or AhR siRNA, then stimulated with PGN (20 μg/mL) or vehicle control for 24 h. For all experiments: n = 3; 2-sided t test with error bars = SEM; *p < 0.05, **p < 0.01. c Immunofluorescence for TLR2 in cultured KCs. KCs were pre-treated with CH223191 (10 μM) or vehicle control for 1 h and then stimulated with PGN (20 μg/mL). TLR2 is stained by Alexa Fluor 488 (green) and nuclei stained with Hoescht 33342 (blue). Images were captured at ×60 magnification. PGN, peptidoglycan; TLR2, toll-like receptor 2; AhR, aryl hydrocarbon receptor; KC, keratinocyte; NF-κB, nuclear factor kappa B; siRNA, short interfering RNA.
Discussion/Conclusion
The skin senses and responds to harmful microbes by recognizing common molecular patterns such as bacterial PGN through surface TLRs. Although TLR-mediated immunological responses are important to host defense, excessive inflammation can be detrimental and contribute to chronic inflammatory skin conditions such as rosacea. The mechanisms that regulate the TLR2 inflammatory response to PGN in the skin remain incompletely understood. Identification of the molecules and pathways that control TLR2 inflammatory gene expression will facilitate better understanding of the innate immune response and can help identify molecular targets to treat overexuberant inflammation.
In this study, we used whole transcriptome sequencing to systematically evaluate the role of AhR on the transcriptional response of primary human KCs to PGN. We found that AhR modulates PGN induction of the inflammatory cytokines IL-1β and IL-6, which we further confirmed using an AhR chemical inhibitor, quantitative RT-PCR, and ELISA. We also observed that PGN led to increased expression of CYP1A1, a classical AhR direct transcriptional target. This raises the possibility that AhR may broadly participate in host-microbe interactions. We do not assume that PGN activates AhR directly but may function through stimulated production of endogenous AhR ligands. Upon PGN stimulation, AhR and TLR2 can be activated simultaneously. Through binding as a transcription factor, AhR increases the expression of cytokines IL-1β and IL-6, further amplifying the induction of these cytokines by TLR2 signaling. In addition to its direct transcriptional effects, we showed that AhR promotes JNK/MAPK signaling that correlates with the upregulation of FosB, which also ultimately regulates inflammatory gene expression. Finally, we observed that expression of TLR2, a key receptor for PGN, is upregulated upon AhR depletion. This raises the possibility that AhR participates in a negative feedback loop to prevent excessive TLR2-mediated inflammation.
Our results largely concur with the conclusions of related studies. A recent report indicated that AhR is activated by Staphylococcus epidermidis [14], supporting the notion that PGN can promote AhR activation in KCs. However, multiple other studies emphasize that the role of AhR depends on the specific ligand and cell context [10]. Imiquimod is a TLR7 agonist that can induce psoriasis-like skin inflammation in mice [15]. In AhR knockout mice, imiquimod treatment amplified IL-1β and inflammation, suggesting that AhR has an immune suppressive role in this system [16]. However, by contrast, AhR activation by the ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in cultured sebocytes led to increased TLR2-mediated induction of TNFα and IL-8, suggesting that it promoted inflammation in this context [17].
Our finding that AhR induces some PGN-related inflammatory genes while suppressing others further depicts a complex role of AhR in inflammatory gene expression. AhR mechanisms may engage different signaling pathways depending on ligand and/or cell type. For instance, TCDD activates AhR, which induces IL-1β expression and represses IL-12 in human dendritic cells. This depends on activated NF-κB/RelB and caudal type homeobox 2, respectively [18]. The results shown here and elsewhere indicate that a detailed understanding of AhR mechanisms in the skin will be needed to rationally pursue its potential as a therapeutic target for rosacea and other skin diseases. AhR-based therapeutics have shown promise in the treatment of skin inflammation in mouse models [19]. The results shown here can guide the investigation of specific AhR-associated inflammatory genes during the course of therapeutic development of AhR-targeted therapies in the skin.
One of the limitations of this work is that it is restricted to a snapshot of the KC transcriptional response to PGN. The immunological response to PGN may evolve over hours to days, and a more exhaustive follow-up study could reveal additional genes and mechanisms that are only apparent during earlier or later time points. Another limitation is that our experiments were performed exclusively on cultured KCs, which might not fully recapitulate the response in vivo. Nonetheless, the controlled culture and treatment conditions provide a useful basis to gain insight to KC cell-intrinsic responses to PGN.
Our future directions will include deeper investigation into the relationship between TLR2 and AhR. How does AhR regulate TLR2 expression and TLR2-related inflammation over a dynamic time course? What is the possible mechanism by which AhR activation could provide negative signaling feedback to suppress TLR2 activity? Finally, although AhR agonists have been shown to be promising to treat some forms of skin inflammation, it will be interesting to evaluate if it is similarly useful for modifying PGN/TLR2-mediated responses.
In summary, we have defined a role for AhR in modifying the expression of inflammatory genes following PGN stimulation of human KCs and shown that PGN leads to activated AhR that both directly binds the genomic loci of target genes while also promoting JNK/MAPK signaling. These results provide new insights into the complex role of AhR in moderating the innate immune response in human skin.
Statement of Ethics
Primary cells were collected and used in accordance with a protocol approved by the University of California San Diego Institutional Review Board (#161332).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was supported by the National Natural Science Foundation of China (No. 81502718). The funders had no role in the preparation of data or the manuscript.
Author Contributions
L.W. and B.K.S. designed the study. L.W., B.C., and B.K.S. performed the experiments and analyzed the data. L.W., B.C., Q.J., and B.K.S. contributed reagents, wrote, and revised the manuscript.
Data Availability Statement
All data analyzed during this study are included in the article and online suppl. files. Primary sequencing files available by inquiry to the corresponding author.
Supplementary Material
Supplementary data
Acknowledgement
The authors thank N. Kulkarni and J. Li for generous technical support.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140((6)):805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2017 Dec;9((6)):7204–18. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buddenkotte J, Steinhoff M. Recent advances in understanding and managing rosacea. F1000Res. 2018 Dec;7:1885. doi: 10.12688/f1000research.16537.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamasaki K, Kanada K, Macleod DT, Borkowski AW, Morizane S, Nakatsuji T, et al. TLR2 expression is increased in rosacea and stimulates enhanced serine protease production by keratinocytes. J Invest Dermatol. 2011;131((3)):688–97. doi: 10.1038/jid.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luan H, Zhang Q, Wang L, Wang C, Zhang M, Xu X, et al. OM85-BV induced the productions of IL-1b, IL-6, and TNF-a via TLR4- and TLR2-mediated ERK1/2/NF-kB pathway in RAW264.7 Cells. J Interferon Cytokine Res. 2014;34:526–36. doi: 10.1089/jir.2013.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen NT, Hanieh H, Nakahama T, Kishimoto T. The roles of aryl hydrocarbon receptor in immune responses. Int Immunol. 2013;25((6)):335–43. doi: 10.1093/intimm/dxt011. [DOI] [PubMed] [Google Scholar]
- 8.Stockinger B, Meglio PD, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32((1)):403–32. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 9.Grassin-Delyle S, Abrial C, Salvator H, Brollo M, Naline E, Devillier P. The role of toll-like receptors in the production of cytokines by human lung macrophages. J Innate Immun. 2020;12:63–73. doi: 10.1159/000494463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haarmann-Stemmann T, Esser C, Krutmann J. The janus-faced role of aryl hydrocarbon receptor signaling in the skin: consequences for prevention and treatment of skin disorders. J Invest Dermatol. 2015;135:2572–6. doi: 10.1038/jid.2015.285. [DOI] [PubMed] [Google Scholar]
- 11.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–6. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huai W, Zhao R, Song H, Zhao J, Zhang L, Zhang L, et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun. 2014;5:4738–9. doi: 10.1038/ncomms5738. [DOI] [PubMed] [Google Scholar]
- 13.Rezzonico R, Yeow K, Loubat A, Ferrua B, Lenegrate G, Turchi L, et al. Dynamic characterization of the molecular events during in vitro epidermal wound healing. J Invest Dermatol. 2002;119((1)):56–63. doi: 10.1046/j.1523-1747.2002.01805.x. [DOI] [PubMed] [Google Scholar]
- 14.Rademacher F, Simanski M, Hesse B, Dombrowsky G, Vent N, Gläser R, et al. Staphylococcus epidermidis activates aryl Hydrocarbon receptor signaling in human keratinocytes: implications for cutaneous defense. J Innate Immun. 2019;11((2)):125–35. doi: 10.1159/000492162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 Axis. J Immunol. 2009 May;182((9)):5836–45. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 16.Meglio PD. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity. 2014;40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou XX, Chen G, Hossini AM, Hu T, Wang L, Pan Z, et al. Aryl hydrocarbon receptor modulates the expression of TNF-α and IL-8 in human sebocytes via the MyD88-p65NF-κB/p38MAPK signaling pathways. J Innate Immun. 2019;11:41–51. doi: 10.1159/000491029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kado S, Chang WLW, Chi AN, Wolny M, Shepherd DM, Vogel CFA. Aryl hydrocarbon receptor signaling modifies toll-like receptor-regulated responses in human dendritic cells. Arch Toxicol. 2017 May;91((5)):2209–21. doi: 10.1007/s00204-016-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SH, Jayawickreme C, Rickard DJ, Nicodeme E, Bui T, Simmons C, et al. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J Invest Dermatol. 2017;137:2110–9. doi: 10.1016/j.jid.2017.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
All data analyzed during this study are included in the article and online suppl. files. Primary sequencing files available by inquiry to the corresponding author.