Abstract
Purpose of the review:
Immune dysregulation disorders are among the most rapidly growing set of inborn errors of immunity. One particular subset is the category where early-onset inflammatory bowel disease is the most common manifestation. These disorders are being increasingly appreciated although there has been minimal effort to articulate a unified approach to their diagnosis and management. This review will cover current thinking and strategies related to diagnosis and management of very early-onset inflammatory bowel disease.
Recent findings:
There is an expanding set of monogenic causes of early-onset inflammatory bowel disease. In many cases, the precise genetic cause dictates management. Lessons learned from the management of these monogenic conditions can sometimes be extrapolated to other refractory cases of inflammatory bowel disease.
Summary:
An integrated approach to diagnosis, risk analysis and management can include diagnostic approaches not often utilized for traditional inflammatory bowel disease such as whole exome sequencing. Management can also include non-traditional approaches such as targeted biologics or hematopoietic cell transplantation.
Keywords: Monogenic, primary immunodeficiencies, inborn errors of immunity, multidisciplinary clinic
INTRODUCTION
Inflammatory bowel disease (IBD), includes Crohn’s disease, ulcerative colitis and inflammatory bowel disease-unspecified. The phenotype ranges from mild disease to severe refractory disease [1*]. There can be benefits to establishing a multidisciplinary clinic to evaluate and manage patients with very early-onset IBD (VEO IBD), especially when the phenotype is severe or unusual. While not all patients will require an integrated approach, many patients with complex or refractory disease will benefit from it. The simple act of establishing a diagnosis can be impressively challenging as non-inflammatory causes are common (Table 1) [2*]. One of the advances in the past 5 to 10 years has been the increasing recognition of the genetic contribution to VEO IBD. Indeed, nearly all IBD is thought to have a genetic component. The difference is the high frequency of monogenic or Mendelian causes in VEO IBD. This review will cover strategies to integrate care for patients, genetic evaluations, and multimodal approaches.
Table 1:
Differential Diagnosis
| Consideration | Notes | Potential for confusion with VEO-IBD |
|---|---|---|
| Anatomical | ||
| Diversion colitis | The excluded large bowel can show changes similar to IBD. | Medium |
| Diverticular colitis | Sigmoid disease usually in older adults with diverticulitis | Low |
| Ischemic colitis | The anatomic distribution is distinctive although the pathologic appearance can resemble IBD | Low |
| Radiation colitis | Fibrosis and vascular changes are characteristic in addition to chronic inflammation | Low |
| Infection | ||
| HIV related colitis | HIV testing and extensive pathogen testing required | Low |
| Lymphogranuloma venereum | Testing for syphilis can distinguish this feature and is typically seen in the setting of HIV | Low |
| Tuberculosis | Large granulomas | Medium |
| Yersinia | Abundant granulomas with central necrosis | High |
| Medication | ||
| Non-steroidal anti-inflammatory use | Epithelial cell apoptosis prominent | Low |
| Mycophenylate mofitil | Pathology indistinguishable from IBD | Low |
| Immune-mediated | ||
| Autoimmune enteropathy | Distinct autoantibodies. Common in primary immunodeficiencies | Low |
| Behcet’s disease | Lymphoid aggregates | Low |
| Graft versus host disease | Pathologically is nearly indistinguishable from IBD although apoptotic crypt epithelial cells are prominent | Low |
| Microscopic colitis (collagenous colitis or lymphocytic colitis) | These are not uncommon in primary immune deficiencies but respond to alternative therapies | Low |
| Congenital diarrhea | ||
| Congenital diarrhea syndromes | Defects in epithelial nutrient/electrolyte transport, polarity, and entero-endocrine function | Medium |
REVIEW
This review will attempt to address common questions that arise in the consideration of a clinic dedicated to VEO IBD. For the purposes of this review, we will consider VEO IBD to be children with age of onset less than six years of age. This is aligned with a recent effort to standardize the nomenclature [3].
Epidemiology of IBD and VEO IBD
The incidence of IBD is increasing worldwide. A more recent recognition is that industrialized countries have begun to plateau whereas emerging economies continue to have an increasing incidence [4*]. Immigrants rapidly acquire the risk profile of their new country [4]. A seminal study from Canada demonstrated that in an industrialized country early age of onset is the fastest growing subset of patients [5*,6]. One study suggests that early onset IBD is now also beginning to plateau [7] although others found a continued rise [8,9].
In Saudi Arabia, infantile or toddler age onset was strongly associated with a positive family history as well as consanguinity [10]. This supports the concept that earlier onset is associated with a stronger genetic contribution. The first monogenic form of IBD was reported in 2009: defects in the IL-10 receptor [11]. In the Middle East, a genetic approach to early-onset IBD identified enrichment of variants within the IL-10 and IL-12 pathways [12*]. In China, where the economy is more recently matured, diverse mutations in early-onset IBD have been identified, such as in the IL-10 and autoinflammatory pathways [13*,14*]. In a recent study from China, the Han ancestral group was found to have enrichment of the IL10RB R101W and T179T (a relatively common polymorphism that affects splicing) [15]. A total of 93 infantile-onset IBD patients had IL10RA variants identified with a phenotype that included 90% perianal disease, onset at a mean of 10 days of age, and oral ulcers. 28% had a positive family history of VEO IBD.
In adults, asthma has been recently identified as a risk factor for subsequent development of Crohn’s disease [16]. This finding may be confounded by the high rate of undiagnosed pulmonary disease in adults with IBD [17]. One could hypothesize that genetic or environmental factors that impact epithelial barrier function could drive both an increased risk of atopic disease as well as an increased risk of IBD. Somewhat counterintuitively, there are recent studies that suggest that poor oral health protects against IBD [18]. Thus, there is a great need to further explore the environmental contributions to the development of IBD.
Diagnostic delay
There are a number of causes of diarrhea in infants and children that represent potential diagnostic confusion [2]. These include defects in lipid trafficking, solute carrier defects, and epithelial dysplasia. However, the most common cause of diarrhea in childhood is certainly infectious. Therefore, it is perhaps no surprise that the diagnosis of IBD in young children can be delayed. Crohn’s disease and isolated small bowel disease are risk factors for delay [19**]. Nearly 2/3 of patients with Crohn’s disease diagnosed after more than two years of symptoms had at least one complication at the time of diagnosis compared to just one quarter of patients diagnosed within four months [20**]. These studies support efforts to increase awareness of early-onset IBD in the community as the single largest source of delay in diagnosis was the time to referral to a gastroenterologist.
Genetics and genomics
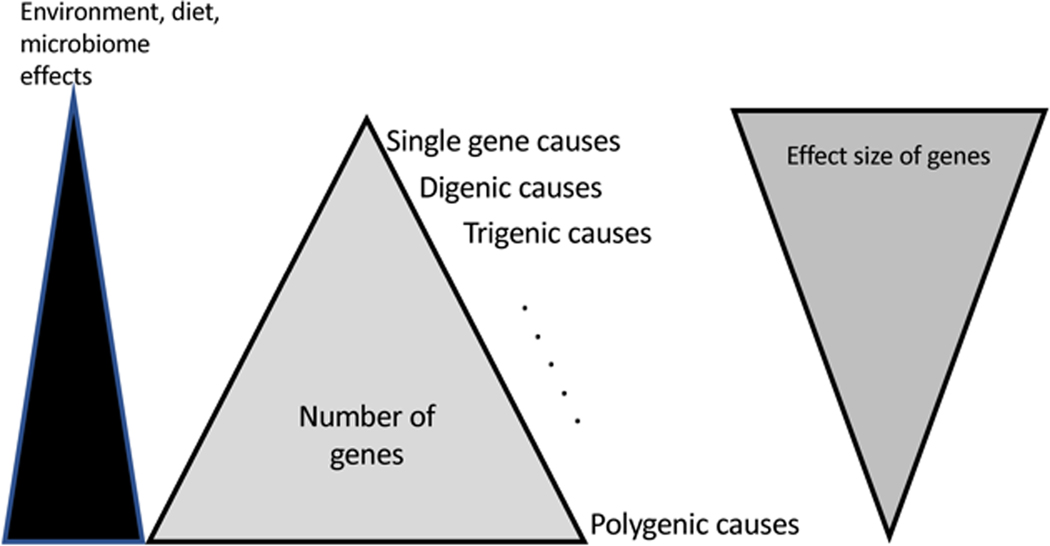
Since the seminal paper that identified age of onset as a predictor of Mendelian causality of IBD [21], there have been diverse and energetic efforts to identify, categorize, and understand the genetic basis of VEO IBD. The specific genes identified have been used to establish gene sequencing panels that are now often used diagnostically, as a targeted exome slice or as part of a larger comprehensive immunodeficiency panel [22*]. Table 2 offers a current listing of the genes clearly identified as associated with VEO IBD. These genes are pathophysiologically nearly identical to the genes identified in genome-wide association studies (GWAS) [23]. An overall model of IBD can be represented as a triangle with highly penetrant single gene defects at the tip and polygenic contributions, where each variant contributes modestly to the overall risk of developing IBD, at the bottom (Figure 1). Environmental contributions, gut microbiome, and epigenetics are envisioned to play significantly larger roles when the genetic risk is lower and are less impactful when the genetic risk is high. Having said that, it is known from murine studies that even highly penetrant genetic foundations of IBD often require a specific bacterial stimulus to manifest disease and can be impacted by other variables such as degradation of the mucus layer [24].
Table 2:
Genetic causes of VEO-IBD*
| Gene (Inheritance pattern) | Frequency of IBD among those with gene defect | History | Physical examination | Immunologic features |
|---|---|---|---|---|
| High likelihood of presenting with VEO-IBD | ||||
| ADAM17 (AR) | >80% | Staphylococcal infections | Psoriasiform erythroderma, pustules, broken hair, abnormal nails | Slightly low TNF response to LPS in PBMCs and T cells. |
| ARPC1B | >80% | Vasculitis prominent | Vasculitis | Microthrombocytes with aberrant spreading |
| FOXP3 (XL) | >80% enteropathy | Onset near birth | Dermatitis | Low regulatory T cells |
| GUCY2C (AD GOF) | >80% diarrhea, IBD less common | Ileal obstruction, adhesions, esophagitis, electrolyte abnormalities | Dilation of small intestine | Not assessed |
| IL10 (AR) | >80% | IBD onset near birth | Folliculitis, perianal disease | Normal |
| IL10RA (AR) | >80% | IBD onset near birth | Folliculitis, perianal disease | Lack of IL-10 suppression of LPS response |
| IL10RB (AR) | >80% | IBD onset near birth | Folliculitis, perianal disease | Lack of IL-10 suppression of LPS response |
| MALT1 | >80% | Respiratory infections, sepsis, CMV | Dermatitis | Poor response to vaccines, poor antigen-specific proliferation, low switched memory B cells |
| PLA2G4A | >80% stenosing enteritis | Early strictures, Platelet dysfunction |
- | None reported |
| SKIV2L (AR) | >80% enteropathy | IUGR, FTT | Trichorrhexis nodosa, frontal bossing | Villous atrophy, low immunoglobulins, low T cells |
| SLC02A1 (AR) |
>80% ileal ulceration (females) | No response to TNF inhibitors | Digital clubbing in males | Normal |
| SLC9A3 | >80% | Prenatal onset diarrhea | Abdominal distension | Normal or Low IgG, high fecal sodium |
| TTC37 (AR) | >80% enteropathy | IUGR, FTT | Trichorrhexis nodosa, frontal bossing | Villous atrophy, low immunoglobulins, low T cells |
| TTC7A (AR) | >80% | Intestinal atresia | Dermatitis, alopecia | Low T cells |
| Intermediate likelihood of presenting as VEO-IBD | ||||
| COL7A1 (AR) | 16% in childhood | Epidermolysis bullosa | Bullous lesions | Normal or low IgG |
| FERMT1 (AR) | 25% in adults | Infections | Bullous lesions, progressive poikiloderma, gingival fragility |
- |
| IKBKG (XL) | >50% by mid-childhood | Infections | Hypodontia, poor sweat, thin hair, frontal bossing | Poor titers, low NK function, abnormal TLR responses |
| IL2RA (AR) | 30–50% | Infections | - | Poor T cell responses |
| IL2RB (AR) | 30–50% | Infections | - | Poor T cell responses |
| POLA1 | 50% in males | FTT, corneal disease | Reticular pigmentation, photophobia | High interferon signature, amyloid in papillary dermis |
| RTEL (AR) | 30–50% | IUGR, FTT, microcephaly | Fine hair | Low NK cells, apoptosis in biopsy |
| SLC37A4 | 30% | Hypoglycemic episodes | Hepatomegaly | Poor neutrophil funciton |
| STXBP2 (AR) | <50% | HLH | - | Poor NK function, low IgG |
| XIAP (XL) | 30–50% | HLH, IBD does not respond to medications | Splenomegaly | Low IgG |
| Lowest likelihood of presenting with VEO-IBD | ||||
| BTK (XL) | <10% in early life | Infections | Small tonsils | Very low B cells and immunoglobulins |
| CTLA4 (AD) | <10% in early childhood | Infections, interstitial pneumonitis | - | Low immunoglobulins, low switched memory B cells, low CD4 T cells |
| CYBA (AR) P22phox |
<10% in early life | Infections | - | Low DHR, reduced switched memory B cells, low T cells |
| CYBB (XL) Gp91phox |
<10% in early life | Infections, maternal discoid lupus | - | Low DHR, reduced switched memory B cells, low T cells |
| DKC1 (XL) | <10% in early childhood | Microcephalic, IUGR | Small, nail dystrophy | Low B cells. NK cells |
| DOCK8 (AR) | ? | Cutaneous viral infections, other infections | Eczema | Low T cells, high eosinophils |
| G6PC3 (AR) | <10% | Cardiac anomalies, urogenital defects, IUGR | Superficial vessels enlarged | Neutropenia, intermittent thrombocytopenia, lymphopenia in severe forms |
| HPS1 (AR) | <10% in early childhood | Pulmonary fibrosis, founder effect in Puerto Rico and Swiss Alps, bleeding | Oculocutaneous albinism | Pigmented macrophages, lymphoid aggregates on biopsy |
| HPS4 (AR) | <10% in early childhood | Pulmonary fibrosis, bleeding | Oculocutaneous albinism | Pigmented macrophages, lymphoid aggregates on biopsy |
| HSP6 (AR) | <10% early childhood | Bleeding | Oculocutaneous albinism | Pigmented macrophages, lymphoid aggregates on biopsy |
| ICOS (AR) | <10% | Infectious enteritis, founder effect along the Danube river | Splenomegaly | Absent class switched memory B cells, low TFH, poor germinal centers in lymph nodes, low IgG, nodular lymphoid hyperplasia of GI tract |
| ITGB2 (AR) | <10% in early childhood | Severe infections, delayed separation of umbilical cord | Gingivitis, scarring (poor wound healing) | High WBC/ANC, low CD18 expression, reduction of factor XIIIa+ DC in lymph node |
| LRBA (AR) | <10% in early childhood | Infections, interstitial pneumonitis | - | Low immunoglobulins, low switched memory B cells, low CD4 T cells |
| MEFV (AR) | <10% | Serositis, founder effect in Mediterranean | - | - |
| MVK | <10% in early life | Nausea and fever episodically, abdominal pain | Adenopathy, oral ulcers, arthritis | - |
| NCF1 (AR) P47phox |
<10% in early life | Infections | - | Low DHR, reduced switched memory B cells, low T cells |
| NCF2 (AR) P67phox |
<10% in early life | Infections | - | Low DHR, reduced switched memory B cells, low T cells |
| NPC1 | <10% in early life | Airway infections, developmental delay | Splenomegaly, ataxia | Foam cell macrophages on biopsy |
| PIK3CD (AD GOF) P100 |
<10% in early life | Infections, PSC, herpes, lymphoma | Bronchiectasis, adenopathy, HSM | High IgM, low IgG, low CD4/CD45RA, nodular lymphoid hyperplasia, EBV viremia |
| PIK3R1 (AD LOF) P85 |
<10% in early life | Insulin resistance | Short stature | High IgM, low IgG, low CD4/CD45RA, nodular lymphoid hyperplasia, |
| PLCG2 (AD) | <10% in early childhood | Pleomorphic inflammation, may be cold induced | Dermatitis, evaporative cooling may induce urticaria | Low immunoglobulins, low switched memory B cells |
| PTEN (AD) | <10% in early life | Thyroiditis, autoimmune hemolytic anemia, hamartomas | Adenopathy, large tonsils, macrocephaly, developmental delay | Nodular lymphoid hyperplasia, low IgG |
| SLC26A3 (AR) | <10% in early life, but all have diarrhea | Onset of secretary diarrhea at birth | - | Inflammation occurs later in life |
| SLC37A4 (AR) | <10% in early life | Hypoglycemic episodes | Hepatomegaly | Neutropenia |
| STAT1 (AD GOF) | <10% in early life but enteropathy common | Pleomorphic autoimmunity, candida, other infections | Poor growth | Low NK cells, low IgA |
| STAT3 (AD GOF) | <10% in early life but enteropathy common | Pleomorphic autoimmunity: diabetes, thyroid | Poor growth, eczema | Low regulatory T cells, low IgG |
| TGFBR1 (AD) | <10% early in life | Aneurysms, | Cleft palate/uvula, hypertelorism, arachnodactyly, pectus, joint laxity | Eosinophilic colitis, high IgE, high eosinophils |
| TGFBR2 (AD) | <10% early in life | Aneurysms, | Cleft palate/uvula, hypertelorism, arachnodactyly, pectus, joint laxity | Eosinophilic colitis, high IgE, high eosinophils |
| WAS (XL) | <10% early in life | Thrombocytopenia | Eczema | Poor T/B/NK function |
| ZBTB24 (AR) | <10% | Developmental delay | Frontal bossing, short stature | Progressive decline in antibodies and T cells |
| Unknown frequency of IBD | ||||
| ALPI (AR) | ? | None noted | - | - |
| ANKZF1 (AR) | ? | Infantile onset IBD | Perioral and oral inflammation | Lymphocytic colon infiltrates, T/B/NK lymphopenia (some) |
| CD55 | ? | Protein losing enteropathy, thrombosis | Complement deposition on biopsy | |
| DUOX2 | ? | Hypothyroidism | - | High lymphocyte count |
| IL21R (AR) | ? | Cryptosporidia, cholangitis | - | Low switched memory B cells |
| IL21 (AR) | ? | Infections | - | Low B cells, low switched memory B cells, low IgG |
| NCF4 (AR) P40phox |
? | Infections | - | DHR slightly low |
| NLRC4 (AD) | ? | HLH, episodic inflammation | - | - |
| TGFB1 | ? | Encephalopathy, eosinophilic esophagitis | Posterior leukoencephalopathy | High IgG and IgE, poor proliferative responses |
| TRIM22 | ? | Early infancy onset, infections | Severe perianal disease | - |
Excluding leaky severe combined immunodeficiency genes- all of which could theoretically be associated with VEO-IBD.
Abbreviations: DHR= dihydrorhodamine, HLH= hemophagocytic lymphohistiocytosis, IUGR= intrauterine growth retardation, FTT= failure to thrive, HSM= hepatosplenomegaly, and PSC= primary sclerosing cholangitis. Inheritance is given as AR= autosomal recessive, AD= autosomal dominant, XL= X-linked, GOF= gain of function mutation.
Figure 1. The genetic basis of VEO IBD.
A schematic diagram illustrating the contributions to the development of IBD. Environmental effects are most pronounced in polygenic disease where the effect size of any individual gene is low and collectively, the genetic contribution to disease is less than what is seen for single gene causality. Environment and microbiome are nearly always a contribution, in the monogenic cases of VEO IBD, their relative contribution is less than what is seen in polygenic disease. Age of onset somewhat tracks with single gene causality but increasingly adults are being recognized with significant inborn errors of immunity causing IBD.
Genome-wide association studies
Genome wide association studies (GWAS) paved the way for a functional understanding of the genomic architecture of IBD. GWAS continue to be performed, refining risk variant gene assignment and adding additional loci. There are now over 230 individual loci associated with IBD. Recent studies have highlighted the role of integrin-associated variants [25], a concept aligned with our current understanding of IBD pathogenesis and highlighted by the success of integrin-directed therapy (vedolizumab). An additional area for fertile study is the recognition that some loci may include variants that lie within microRNAs (miRNAs) [26]. MiRNAs regulate the transcriptome by binding to messenger RNA and modulating either translation or stability. The recent identification of variants that impact miRNAs suggests that this may be a future area of emphasis. Less frequently investigated have been studies evaluating the epigenome in IBD. One study identified differential DNA methylation at inflammatory genes in pediatric IBD that were associated with dysregulated expression of miRNAs [27]. In another study, the authors compared intestinal epithelial cells in newly diagnosed pediatric patients with IBD and found changes in DNA methylation and transcriptome patterns [28**]. These studies suggest that in spite of the maturation of the GWAS studies for IBD that additional mechanisms of disease will be revealed with additional refinement. The recently developed Human disease methylation database (DiseaseMeth) has provided a comprehensive DNA methylation repository of human disease and will serve to expand the study of epigenetics in IBD [29].
New genes identified as associated with early onset IBD
Multiple new genes have been identified in the past two years [30-33*, 34-35*, 36*, 37*, 38*, 39* 40*, 41]. Additional studies defining phenotypic differences and therapeutic interventions directed at gene-stratified patient populations are eagerly awaited. The gene variants currently implicated in VEO IBD are highlighted in Table 1. Many classical primary immunodeficiencies are associated with IBD or enteropathy [42*]. The list in Table 1 is restricted to genes where VEO IBD has been identified in multiple cases.
Mechanisms of disease
The two areas most intensively investigated over the past two years are the microbiome and the role of T cells in IBD. None of these studies specifically target VEO IBD but the lessons are likely extrapolatable. The interactions of the maturing microbiome and T cells in childhood is just beginning to reveal the critical interfaces. An excellent recent review portrays the pathologic pathways in VEO IBD graphically [43*].
Microbiome
Significant differences in the microbiome have been recognized in patients with IBD compared to healthy controls [44]. These differences have been attributed to both a cause of IBD and an effect of IBD. The microbiome develops between birth and 3 years of life suggesting a vulnerable window of time. In IBD, decreased relative abundance of bacteria within the phyla, Firmicutes and Bacteroidetes, and enrichment of Proteobacteria and Actinobacteria have been seen. Host genetics and environmental factors (smoking, diet, air pollution, and antibiotic use) have been shown to influence the microbiome in IBD. A recent study in mice demonstrated that antibiotics administered at the time of delivery can alter the offspring’s intestinal microbiome and leads to a long-term increased susceptibility to colitis [45*]. Given the high rate of antibiotic use to prevent group B streptococcal sepsis in newborns, this represents a significant finding. A second murine study was directed at defining trajectories of change as a strategy to understand the relationship between alterations to the microbiome and the interacting T cells. Increased responses to bacterial ligands preceded the clinical development of IBD [46*]. At the time of clear disease activity, increased T cell activation was identified [47].
The microbiome is also a living community and metabolites also impact the immune response. Commensal bacterial products stimulate cytokines and release of antimicrobial peptides, such as RegIIIƔ [48,49]. For example Bacteroides fragilis produces short chain fatty acids, inhibiting activity of histone deacetylases in colonocytes and immune cells leading to down-regulation of pro-inflammatory cytokines and induction of Treg cells [50]. Perhaps pertinent to the rise in incidence in VEO IBD, recent studies suggest that metabolites can be maternally transferred to the offspring and affect the developing immune response [51*].
One of the confounding aspects in the study of microbiota and IBD is the vast diversity of species and genera observed. The microbial dysbiosis index collapses this diversity into a manageable variable, a strategy that may enhance analyses [52]. Vedolizumab response, a monoclonal antibody to T cell integrins required for gastrointestinal migration was associated with high bacterial diversity at baseline [53*]. Branched-chain amino acid synthesis was significantly higher at baseline in those achieving remission as well. Another metabolic feature, bacterial urease production and nitrogen flux, have been implicated in the development of intestinal dysbiosis and subsequent immune-mediated colitis [54*]. The importance of these findings is that microbial metabolic pathways can be targeted through medical management [47].
One additional study on the microbiome highlights the systemic effects of Crohn’s disease. Pediatric patients with Crohn’s disease were found to have a completely different subgingival microbial structure in children with Crohn’s disease compared to controls [55]. Patients who responded to therapy normalized their subgingival microbiota suggesting that subclinical inflammation throughout the gastrointestinal tract impacts the microbial community.
T cells
T cell immunodeficiencies have a high rate of IBD [56*]. Recent data have focused on atypical T cells as well as conventional T cells and their functional responses. The gastrointestinal tract has been termed the largest immune organ in the body. The richness of the cell types and their intracellular location have posed methodologic problems and only recently have advances been made in understanding the diversity and function. Long recognized are the innate lymphoid cells (ILC). ILC have been demonstrated to mold responses to commensal bacteria by killing highly responsive T cells [57]. The highly responsive T cells are largely polarized to produce γ-interferon, IL-17, and IL-22 [58*]. In the setting of VEO IBD, patients with mutations in the IL-10 receptor have enhanced production of IL-17 [59*]. This is paired with increased macrophage expression of IL-1β [60]. IL-17 inhibition is not felt to be therapeutic and in fact can be harmful by disrupting T cell interactions with barrier function, however, IL-1β inhibition has been used therapeutically and was found to be specifically useful in patients with IL-10 receptor deficiency [60].
In murine models, the effect of the glycosylation pathway has been recently investigated. Mucosal T cells in patients with ulcerative colitis have a deficiency in branched chain glycosylation. This was modeled in mice to show that the branched chain glycosylation pathway is critical for T cell function [61**]. Indeed, supplementation with glycosylation precursors was able to restore T cell function. This study directly relates to VEO IBD because oral GlcNAc is available for human utilization and has been described as promoting mucus production in children with treatment-resistant IBD [62]. A second glycosylation defect in epithelial cells was identified in mice suggesting that glycosylation is a growth area for understanding IBD [63].
An additional study in mice identified an atypical T cell utilizing the γδ T cell receptor [64*]. The location of the γδT cells at the tips of villi is dependent on the microbiota and they exhibit rapid movements in response to changes in the microbial community. These T cells regulate the intestinal barrier via epithelial tight junction proteins. Therefore, the concept of a highly dynamic interplay between the microbial community, conventional T cells, and unconventional T cells at the epithelial surface and within the lamina propria is developing.
Providing integrated care to patients with VEO IBD
An integrated program can have significant benefits to the patient by simplifying and unifying the approach. As is true for all conditions, patient care is improved at centers with high volume and expertise. Physicians also benefit by having real time interactions regarding patient care. Nevertheless, there are some challenges to developing an integrated care approach. This section will attempt to address common hurdles for the development and implementation of an integrated approach.
Which patients should be invited to participate in an integrated clinic?
There are three conceptual approaches to identify the constituency. The selection can be according to age criteria, severity, or geographic constraints. The VEO IBD clinic at The Children’s Hospital of Philadelphia is based largely on age of onset (<6 years of age at onset) but older patients with refractory or unusual disease can petition for evaluation. Approximately 500 patients are followed at this time and >90% had their onset at <6 years of age.
Which providers will be involved in the structure of the clinic?
The VEO IBD clinic at The Children’s Hospital of Philadelphia includes specialists in Gastroenterology, Immunology, Nutrition, and Social work at a minimum. Extensive nursing involvement supports the complex care and the nurses act as liaisons for the patients. The addition of Genetics and Rheumatology is patient-specific. The holistic approach ensures that patients receive coordinated care, have the best support through their difficult journey and receive a consistent message. Patients also appreciate the one-stop approach to care.
People who support the clinic but are not directly involved in patient care include a Rheumatologist, dedicated Geneticist, Bioinformatician, Study Coordinators, Study Researchers and research laboratory staff who perform flow cytometry, functional analyses and examine biomarkers and microbiome. This component is not essential for patient care but is often critical for variant validation.
What laboratory evaluation is critical to identify an inborn error of immunity?
In our clinic, very low switched memory B cells has the highest sensitivity for the identification of Mendelian inborn errors of immunity. Other commercially available studies have a low yield when used in an unselected fashion but are useful in specific settings. Infantile-onset disease with fistulas suggests an IL-10 pathway defect and an assay that detects lack of IL-10 inhibition of responses to lipopolysaccharide will detect receptor mutations (but not IL-10 ligand defects). The sensitivity of widely available flow cytometry assays, often the standard evaluation for inborn errors of immunity, is surprisingly low. For this reason, sequencing often represents a cost-effective approach.
Stratification of patients for sequencing
The “hit” rate varies across studies but overall about 20% of patients with VEO IBD have been found to have an inborn error of immunity through sequencing [65–67]. Using a gene panel and looking at a cohort of general pediatric IBD, only 4% had a clear causal gene variant (s) [41]. Using age of onset <10 years, a German study found 7% frequency of Mendelian gene variants [68*]. Conversely a cohort of infantile-onset IBD cases found 31% of cases had Mendelian disease [69]. These studies suggest that early age of onset enriches for the finding of monogenic causality but even adults with Mendelian causality have been described and CGD has a rate of IBD approaching 50% by adulthood [70*,71]. Due to the expense it is common to be asked if there is an approach to risk stratify other than age of onset. The most common inborn error of immunity in our cohort is CGD and we therefore screen all patients with a DHR. In some other cohorts, IL-10 receptor defects have been the most common and the screening test for this is available. Enrichment by selecting patients with infantile onset, from consanguineous parents, and growth failure has been observed in our clinic and has been reported in the literature [69,72*]. Our data suggest that low switched memory B cells may select out a group enriched for inborn errors of immunity. These findings have led to our current approach of opting for sequencing for most patients. There is a caution, however. In our cohort, trisomy 21 and Turner syndrome were found with a collective frequency of 19% among those with an identified Mendelian cause. Furthermore, 33% of those sequenced where a causal variant(s) was found had a variant in a gene not included in IBD panels. Lastly, nearly a third of our patients with causal or potentially causal variants require additional high-level validation approaches. Thus, sequencing should be undertaken with a vision of what is possible at each center and the patients should be educated regarding expectations.
Sequencing methodology
There are four options for sequencing and the costs, yield and turnaround time are different, not to mention the difficulty in getting insurance approval (Table 3). Labs with an interest in VEO-IBD will occasionally do the sequencing on a research basis. There are a number of potential pitfalls here. Data are typically delivered with little analysis requiring time and effort to analyze. Findings most often cannot be used for clinical decision making and any variants must therefore be validated through clinical-grade sequencing.
Table 3:
CLIA approved sequencing options
| Sanger sequencing | Whole exome “slice” or panel | Whole exome sequencing | |
|---|---|---|---|
| Strengths | • High accuracy • Can include splice junctions • Often reflexes to deletion/duplication testing |
• Affordable • Often more timely than WES • No off target findings |
• Most comprehensive |
| Weaknesses | • Limited number of genes sequenced (i.e. best used from his pre-test probability) • Some genes are incompletely sequenced and only “hot spots” are sequenced |
• Same weaknesses as WES for technical approach although coverage is typically higher | • Usually takes the longest to run and analyze • Some exons are missed • Not sensitive (at this time) for deletions and duplications • Regulatory (non-coding) mutations missed • Off target findings common |
| Insurance coverage/cost | • The easiest of the three • Cost is highly variable but single genes are typically less than WES |
• Mid-range difficulty • Less expensive than WES |
• Most difficult • Most expensive generally |
| Notes | • Panel content highly variable • Some panels use WES data and targeted analysis, others use specific gene capture which leads to higher coverage |
• Bioinformatic analysis variable • Strength of variant association can be difficult to discern |
Interpretation of sequencing results
There are three conceptual categories of findings for each patient who has sequencing. There may be a clear variant identified that explains the condition because it has been previously reported in VEO IBD. This type of result is gratifying but well less than half the sequenced patients have this type of result. There may be a variant in a gene that is known to be associated with the condition but the specific variant has not been previously reported. It is tempting to call the new variant causal and report the result to the patient and indeed there are cases where that may be appropriate, however, this type of result generally requires some type of validation. A standard lab study may reveal findings compatible with that genetic diagnosis. An example would be a new mutation in the CYBB, associated with X-linked CGD. The variant may be easily validated using a dihydrorhodamine (DHR) assay. For some genes, there are no clear laboratory or clinical features that would allow validation, and in those cases, a research lab must be sought to perform analyses demonstrating that the variant affects protein function. Finally, in many cases either no gene variant is identified or a gene variant labelled as variant of uncertain significance (VUS) is found. For genes not previously reported to be disease-associated, GeneMatcher (https://genematcher.org) or ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) may be used to see if other patients have been identified in other settings. Lacking a finding of other similar patients, the road in this circumstance is long and arduous. A research lab must be identified with time, interest and funding to support a functional validation and possibly an animal model of the gene variant.
How can the patients be managed?
At our center, we strongly feel that identification of a variant can drive therapeutic decisions. For patients with VEO IBD, the gene often suggests a specific therapy (Table 4). In some cases, knowing the pathway can suggest a treatment. In the cases, where there is no variant identified, we individually determine whether additional studies should be pursued. When there is no guidance from sequencing, experience can be useful. While we have strategies that seem more successful to us, in the end, patients often need a unique therapeutic approach. While the goal is to prevent surgery, diversion and colectomy represent options with clear expectations set [73*,74*].
Table 4:
Genetic stratification for therapy
| Gene defect | Therapeutic considerations |
|---|---|
| CYBB and other etiologies of CGD | IL-1 blockade. TNF inhibitors should not be used due to risk of fungal infections |
| CTLA4 | Abatacept, sirolimus, HSCT |
| FOXP3 | Sirolimus, HSCT |
| IL-10, IL-10RA, IL10RB | HSCT |
| LRBA | Abatacept, sirolimus, colchicine, HSCT |
| MEFV | IL-1 blockade, colchicine |
| MVK | IL-1, TNF blockade |
| NLRC4 | IL-18 binding protein |
| TTC7A | HSCT can be considered, however, success has been limited |
| XIAP | HSCT, IL-18 binding protein study underway |
HSCT: Hematopoietic stem cell transplant
CONCLUSIONS
Multidisciplinary approaches to VEO IBD can provide coordinated and comprehensive care for patients. The genetic underpinnings of VEO IBD represent an opportunity to advance therapeutic decision making beyond trial and error. Strategies that are successful in cases with causal genetic variants can also be applied for those without an identified genetic cause.
A minority of patients with VEO IBD have a single gene cause, although these patients often benefit from specific targeted therapeutic approaches
A multidisciplinary clinic can bring value to the diagnosis and care of patients with VEO IBD
Traditional immunologic studies have limited value but sequencing can often identify patients with monogenic causality
ACKNOWLEDGEMENTS
The authors would like to thank Dawn Westerfer for administrative support, Rawan Shraim and Audrey Merz for expertise in study coordination.
Financial support: This work was supported by the Department of Pediatrics, The Children’s Hospital of Philadelphia and the Wallace Chair of Pediatrics.
Funding:
KES is supported by PCORI and NIH grants U01HG010219, R21AI130967, U24AI086037, and U54AI082973.
JRK is supported by NIH K23DK100461–01A1
Footnotes
Conflicts of interest: The authors declare that they have no conflicts of interest.
REFERENCES
- 1. Dore M, Triana Junco P, Sanchez Galan A, Prieto G, Ramos E, Munoz Romo M, Gomez Cervantes M, Hernandez F, Martinez L, Lopez Santamaria M: Pitfalls in Diagnosis of Early-Onset Inflammatory Bowel Disease. Eur J Pediatr Surg 2018, 28:39–43. *This paper is a nice overview of the types of patients often seen in a clinic dedicated to VEO IBD.
- 2. Ensari A, Kelsen J, Russo P: Newcomers in paediatric GI pathology: childhood enteropathies including very early onset monogenic IBD. Virchows Arch 2018, 472:111–123. *This review covers the VEO IBD mimics and the key pathologic distinctions.
- 3.Turner D, Muise AM: Very Early Onset IBD: How Very Different ‘on Average’? J Crohns Colitis 2017, 11:517–518. [DOI] [PubMed] [Google Scholar]
- 4. Sykora J, Pomahacova R, Kreslova M, Cvalinova D, Stych P, Schwarz J: Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol 2018, 24:2741–2763. *The changing epidemiology of IBD poses a challenge for all efforts to identify patients with Mendelian causes of VEO IBD because the overall incidence of VEO IBD is increasing and the cases with Mendelian causes are harder to identify.
- 5. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. : Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018, 390:2769–2778. *This systematic review of studies provides a very clear picture of the changing landscape of IBD and highlights potential environmental factors that may play a role in the increasing incidence.
- 6.Benchimol EI, Mack DR, Nguyen GC, Snapper SB, Li W, Mojaverian N, Quach P, Muise AM: Incidence, outcomes, and health services burden of very early onset inflammatory bowel disease. Gastroenterology 2014, 147:803–813 e807; quiz e814–805. [DOI] [PubMed] [Google Scholar]
- 7.Bequet E, Sarter H, Fumery M, Vasseur F, Armengol-Debeir L, Pariente B, Ley D, Spyckerelle C, Coevoet H, Laberenne JE, et al. : Incidence and Phenotype at Diagnosis of Very-early-onset Compared with Later-onset Paediatric Inflammatory Bowel Disease: A Population-based Study [1988–2011]. J Crohns Colitis 2017, 11:519–526. [DOI] [PubMed] [Google Scholar]
- 8.Benchimol EI, Bernstein CN, Bitton A, Carroll MW, Singh H, Otley AR, Vutcovici M, El-Matary W, Nguyen GC, Griffiths AM, et al. : Trends in Epidemiology of Pediatric Inflammatory Bowel Disease in Canada: Distributed Network Analysis of Multiple Population-Based Provincial Health Administrative Databases. Am J Gastroenterol 2017. [DOI] [PMC free article] [PubMed]
- 9.Coughlan A, Wylde R, Lafferty L, Quinn S, Broderick A, Bourke B, Hussey S, Study D: A rising incidence and poorer male outcomes characterise early onset paediatric inflammatory bowel disease. Aliment Pharmacol Ther 2017, 45:1534–1541. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hussaini A, El Mouzan M, Hasosah M, Al-Mehaidib A, K AL, Saadah OI, Al-Edreesi M: Clinical Pattern of Early-Onset Inflammatory Bowel Disease in Saudi Arabia: A Multicenter National Study. Inflamm Bowel Dis 2016, 22:1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. : Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009, 361:2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nemati S, Teimourian S, Tabrizi M, Najafi M, Dara N, Imanzadeh F, Ahmadi M, Aghdam MK, Tavassoli M, Rohani P, et al. : Very early onset inflammatory bowel disease: Investigation of the IL-10 signaling pathway in Iranian children. Eur J Med Genet 2017, 60:643–649. *This is one of the few reports to define the frequency of a specific gene defect in a population.
- 13. Fang YH, Luo YY, Yu JD, Lou JG, Chen J: Phenotypic and genotypic characterization of inflammatory bowel disease in children under six years of age in China. World J Gastroenterol 2018, 24:1035–1045. *This study defined the frequency of different gene defects in an unselected cohort of patients. A key finding is that early age of onset enriched for monogenic conditions.
- 14. Ye Z, Zhou Y, Huang Y, Wang Y, Lu J, Tang Z, Miao S, Dong K, Jiang Z: Phenotype and Management of Infantile-onset Inflammatory Bowel Disease: Experience from a Tertiary Care Center in China. Inflamm Bowel Dis 2017, 23:2154–2164. *This study from China evaluated Mendelian causes and offers a comprehensive clinical picture of the patients.
- 15.Huang Z, Peng K, Li X, Zhao R, You J, Cheng X, Wang Z, Wang Y, Wu B, Wang H, et al. : Mutations in Interleukin-10 Receptor and Clinical Phenotypes in Patients with Very Early Onset Inflammatory Bowel Disease: A Chinese VEO-IBD Collaboration Group Survey. Inflamm Bowel Dis 2017, 23:578–590. [DOI] [PubMed] [Google Scholar]
- 16.Kuenzig ME, Barnabe C, Seow CH, Eksteen B, Negron ME, Rezaie A, Panaccione R, Benchimol EI, Sadatsafavi M, Avina-Zubieta JA, et al. : Asthma Is Associated With Subsequent Development of Inflammatory Bowel Disease: A Population-based Case-Control Study. Clin Gastroenterol Hepatol 2017, 15:1405–1412 e1403. [DOI] [PubMed] [Google Scholar]
- 17.Casella G, D’Inca R, Oliva L, Daperno M, Saladino V, Zoli G, Annese V, Fries W, Cortellezzi C, Italian Group IBD: Prevalence of celiac disease in inflammatory bowel diseases: An IG-IBD multicentre study. Dig Liver Dis 2010, 42:175–178. [DOI] [PubMed] [Google Scholar]
- 18.Yin W, Ludvigsson JF, Liu Z, Roosaar A, Axell T, Ye W: Inverse Association Between Poor Oral Health and Inflammatory Bowel Diseases. Clin Gastroenterol Hepatol 2017, 15:525–531. [DOI] [PubMed] [Google Scholar]
- 19. Ricciuto A, Fish JR, Tomalty DE, Carman N, Crowley E, Popalis C, Muise A, Walters TD, Griffiths AM, Church PC: Diagnostic delay in Canadian children with inflammatory bowel disease is more common in Crohn’s disease and associated with decreased height. Arch Dis Child 2018, 103:319–326. **Diagnostic delay was found to be common in IBD and this study finds pivotal features that increase the likelihood of delay. Diagnostic delay was associated with worse height at later years
- 20. Nguyen VQ, Jiang D, Hoffman SN, Guntaka S, Mays JL, Wang A, Gomes J, Sorrentino D: Impact of Diagnostic Delay and Associated Factors on Clinical Outcomes in a U.S. Inflammatory Bowel Disease Cohort. Inflamm Bowel Dis 2017, 23:1825–1831. **Diagnostic delay represents a signficant burden for the child and family. This study found that serious complications are also increased with diagnostic delay.
- 21.Uhlig HH, Schwerd T, Koletzko S, Shah N, Kammermeier J, Elkadri A, Ouahed J, Wilson DC, Travis SP, Turner D, et al. : The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 2014, 147:990–1007 e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Picard C, Bobby Gaspar H, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Crow YJ, Cunningham-Rundles C, Etzioni A, Franco JL, et al. : International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J Clin Immunol 2018, 38:96–128. *The inborn errors of immunity are tabulated in this publication. Those strongly associated with early onset IBD are identified for easy reference.
- 23.de Lange KM, Barrett JC: Understanding inflammatory bowel disease via immunogenetics. J Autoimmun 2015, 64:91–100. [DOI] [PubMed] [Google Scholar]
- 24.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT: Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji SG, et al. : Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017, 49:256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccacci C, Politi C, Biancone L, Latini A, Novelli G, Calabrese E, Borgiani P: Polymorphisms in MIR122, MIR196A2, and MIR124A Genes are Associated with Clinical Phenotypes in Inflammatory Bowel Diseases. Mol Diagn Ther 2017, 21:107–114. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Song P, Timofeeva M, Meng X, Rudan I, Little J, Satsangi J, Campbell H, Theodoratou E: Systematic meta-analyses and field synopsis of genetic and epigenetic studies in paediatric inflammatory bowel disease. Sci Rep 2016, 6:34076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howell KJ, Kraiczy J, Nayak KM, Gasparetto M, Ross A, Lee C, Mak TN, Koo BK, Kumar N, Lawley T, et al. : DNA Methylation and Transcription Patterns in Intestinal Epithelial Cells From Pediatric Patients With Inflammatory Bowel Diseases Differentiate Disease Subtypes and Associate With Outcome. Gastroenterology 2018, 154:585–598. **Intestinal epithelial cells have substantial changes to their epigenome in IBD as measured by DNA methylation at pivotal biological pathways. Impressively, DNA methylation changes predicted disease severity and were partly stable over time, suggesting that treatment will need to elicit durable changes to epigenome to reverse the disease effects.
- 29.Xiong Y, Wei Y, Gu Y, Zhang S, Lyu J, Zhang B, Chen C, Zhu J, Wang Y, Liu H, et al. : DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res 2017, 45:D888–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuang LS, Villaverde N, Hui KY, Mortha A, Rahman A, Levine AP, Haritunians T, Evelyn Ng SM, Zhang W, Hsu NY, et al. : A Frameshift in CSF2RB Predominant Among Ashkenazi Jews Increases Risk for Crohn’s Disease and Reduces Monocyte Signaling via GM-CSF. Gastroenterology 2016, 151:710–723 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Haaften-Visser DY, Harakalova M, Mocholi E, van Montfrans JM, Elkadri A, Rieter E, Fiedler K, van Hasselt PM, Triffaux EMM, van Haelst MM, et al. : Ankyrin repeat and zinc-finger domain-containing 1 mutations are associated with infantile-onset inflammatory bowel disease. J Biol Chem 2017, 292:7904–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahr WH, Pluthero FG, Elkadri A, Warner N, Drobac M, Chen CH, Lo RW, Li L, Li R, Li Q, et al. : Loss of the Arp2/3 complex component ARPC1B causes platelet abnormalities and predisposes to inflammatory disease. Nat Commun 2017, 8:14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parlato M, Charbit-Henrion F, Pan J, Romano C, Duclaux-Loras R, Le Du MH, Warner N, Francalanci P, Bruneau J, Bras M, et al. : Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol Med 2018, 10. *ALPI defiency represents one of the newest recognized Mendelain causes of VEO IBD.
- 34.Guerrerio AL, Frischmeyer-Guerrerio PA, Huang C, Wu Y, Haritunians T, McGovern DPB, MacCarrick GL, Brant SR, Dietz HC: Increased Prevalence of Inflammatory Bowel Disease in Patients with Mutations in Genes Encoding the Receptor Subunits for TGFbeta. Inflamm Bowel Dis 2016, 22:2058–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parlato M, Charbit-Henrion F, Hayes P, Tiberti A, Aloi M, Cucchiara S, Begue B, Bras M, Pouliet A, Rakotobe S, et al. : First Identification of Biallelic Inherited DUOX2 Inactivating Mutations as a Cause of Very Early Onset Inflammatory Bowel Disease. Gastroenterology 2017, 153:609–611 e603. *Reactive oxygen species (ROS) defects have been recognized as associated with IBD for many years. The focus has been on ROS produced by phagocytic cells such as neutrophils. DUOX2 is expressed primarily in the GI tract and deficiencies are associated with susceptibility to IBD.
- 36. Conrad MA, Dawany N, Sullivan KE, Devoto M, Kelsen JR: Novel ZBTB24 Mutation Associated with Immunodeficiency, Centromere Instability, and Facial Anomalies Type-2 Syndrome Identified in a Patient with Very Early Onset Inflammatory Bowel Disease. Inflamm Bowel Dis 2017, 23:2252–2255. *This immunodeficiency has been reported as associated with diarrhea but this is the first study to clearly associate centromeric instability with IBD.
- 37. Schwerd T, Pandey S, Yang HT, Bagola K, Jameson E, Jung J, Lachmann RH, Shah N, Patel SY, Booth C, et al. : Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut 2017, 66:1060–1073. *Niemann-Pick disease is a devastating neurologic condition. This is the first descriptio of an assocaited with IBD and the authors pursue a mechanistic understanding, finding an effect on autophagy.
- 38. Neves JF, Afonso I, Borrego L, Martins C, Cordeiro AI, Neves C, Lacoste C, Badens C, Fabre A: Missense mutation of TTC7A mimicking tricho-hepato-enteric (SD/THE) syndrome in a patient with very-early onset inflammatory bowel disease. Eur J Med Genet 2018, 61:185–188. *TTC7A mutations have been identified in patients with multiple intestinal atresia and severe combined immunodeficiency, IBD, and now this paper reports a new phenotype. They found a picture of enteropathy with villous blunting, autoimmunity and hepatic dysfunction.
- 39. Schwerd T, Bryant RV, Pandey S, Capitani M, Meran L, Cazier JB, Jung J, Mondal K, Parkes M, Mathew CG, et al. : NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol 2018, 11:562–574. *This study represents anotehr example of the increasing evidence that reactive oxygen species require meticulous regulation.
- 40. Kotlarz D, Marquardt B, Baroy T, Lee WS, Konnikova L, Hollizeck S, Magg T, Lehle AS, Walz C, Borggraefe I, et al. : Human TGF-beta1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat Genet 2018, 50:344–348. *The TGFβ receptor deficiencies are associated with Loeys Dietz syndrome, itself associated with IBD. This is the first report of the TGFβ ligand being associated with IBD.
- 41.Ashton JJ, Andreoletti G, Coelho T, Haggarty R, Batra A, Afzal NA, Beattie RM, Ennis S: Identification of Variants in Genes Associated with Single-gene Inflammatory Bowel Disease by Whole-exome Sequencing. Inflamm Bowel Dis 2016, 22:2317–2327. [DOI] [PubMed] [Google Scholar]
- 42. Kelsen JR, Baldassano RN: The role of monogenic disease in children with very early onset inflammatory bowel disease. Curr Opin Pediatr 2017, 29:566–571. *This review is a recent report of VEO IBD diagnosis, mechanism of disease and management.
- 43. Uhlig HH, Powrie F: Translating Immunology into Therapeutic Concepts for Inflammatory Bowel Disease. Annu Rev Immunol 2018, 36:755–781. **This review has impressive graphics defining the mechanisms of disease.
- 44.Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD: Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm Bowel Dis 2015, 21:1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyoshi J, Bobe AM, Miyoshi S, Huang Y, Hubert N, Delmont TO, Eren AM, Leone V, Chang EB: Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep 2017, 20:491–504. *Antibiotics have been recognized as a risk factor for IBD. This study found that peripartum antibiotics to the mother had long-lasting effects on the pups’ microbiome.
- 46. Sharpton T, Lyalina S, Luong J, Pham J, Deal EM, Armour C, Gaulke C, Sanjabi S, Pollard KS: Development of Inflammatory Bowel Disease Is Linked to a Longitudinal Restructuring of the Gut Metagenome in Mice. mSystems 2017, 2. *To date, nearly all microbiome studies have been cross-sectional snapshots. This study clearly defined trajectories of change as a key variable in IBD in mice.
- 47.Halfvarson J, Brislawn CJ, Lamendella R, Vazquez-Baeza Y, Walters WA, Bramer LM, D’Amato M, Bonfiglio F, McDonald D, Gonzalez A, et al. : Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017, 2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R: Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118:229–241. [DOI] [PubMed] [Google Scholar]
- 49.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV: The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larmonier CB, Shehab KW, Ghishan FK, Kiela PR: T Lymphocyte Dynamics in Inflammatory Bowel Diseases: Role of the Microbiome. Biomed Res Int 2015, 2015:504638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakajima A, Kaga N, Nakanishi Y, Ohno H, Miyamoto J, Kimura I, Hori S, Sasaki T, Hiramatsu K, Okumura K, et al. : Maternal High Fiber Diet during Pregnancy and Lactation Influences Regulatory T Cell Differentiation in Offspring in Mice. J Immunol 2017, 199:3516–3524. *Fiber was found to promote Treg development, a key cell type for the prevention of autoimmunity.
- 52.Gevers D, Kugathasan S, Knights D, Kostic AD, Knight R, Xavier RJ: A Microbiome Foundation for the Study of Crohn’s Disease. Cell Host Microbe 2017, 21:301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ: Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 2017, 21:603–610 e603. *This sudy demonstrates the promise of microbiome studies for treatment stratification.
- 54. Ni J, Shen TD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, et al. : A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 2017, 9. **Nitrogen flux was identified as a key pathologic pathway in this study. The careful dissection of the role of urease revealed a potential bacterial metabolic target.
- 55.Kelsen J, Bittinger K, Pauly-Hubbard H, Posivak L, Grunberg S, Baldassano R, Lewis JD, Wu GD, Bushman FD: Alterations of the Subgingival Microbiota in Pediatric Crohn’s Disease Studied Longitudinally in Discovery and Validation Cohorts. Inflamm Bowel Dis 2015, 21:2797–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kelsen JR, Sullivan KE: Inflammatory Bowel Disease in Primary Immunodeficiencies. Curr Allergy Asthma Rep 2017, 17:57. *This review analyzes the overall occurrence of IBD in primary immunodeficiencies.
- 57.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, et al. : Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science 2015, 348:1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. : Circulating and Tissue-Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology 2017, 153:1320–1337 e1316. *This study advances our understanding of T cell interactions with enterocytes and the microbial community.
- 59. Shouval DS, Konnikova L, Griffith AE, Wall SM, Biswas A, Werner L, Nunberg M, Kammermeier J, Goettel JA, Anand R, et al. : Enhanced TH17 Responses in Patients with IL10 Receptor Deficiency and Infantile-onset IBD. Inflamm Bowel Dis 2017, 23:1950–1961. *The mechanism of IL-10 receptor deficiency is at least in part due to increased IL-17 production.
- 60.Shouval DS, Biswas A, Kang YH, Griffith AE, Konnikova L, Mascanfroni ID, Redhu NS, Frei SM, Field M, Doty AL, et al. : Interleukin 1beta Mediates Intestinal Inflammation in Mice and Patients With Interleukin 10 Receptor Deficiency. Gastroenterology 2016, 151:1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dias AM, Correia A, Pereira MS, Almeida CR, Alves I, Pinto V, Catarino TA, Mendes N, Leander M, Oliva-Teles MT, et al. : Metabolic control of T cell immune response through glycans in inflammatory bowel disease. Proc Natl Acad Sci U S A 2018, 115:E4651–E4660. **This study is a fascinating look at T cell glycans. Importantly, there are potential therapeutic options.
- 62.Salvatore S, Heuschkel R, Tomlin S, Davies SE, Edwards S, Walker-Smith JA, French I, Murch SH: A pilot study of N-acetyl glucosamine, a nutritional substrate for glycosaminoglycan synthesis, in paediatric chronic inflammatory bowel disease. Aliment Pharmacol Ther 2000, 14:1567–1579. [DOI] [PubMed] [Google Scholar]
- 63.Kudelka MR, Hinrichs BH, Darby T, Moreno CS, Nishio H, Cutler CE, Wang J, Wu H, Zeng J, Wang Y, et al. : Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk. Proc Natl Acad Sci U S A 2016, 113:14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D: Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell 2017, 171:783–794 e713. *This multimodal study defined metabolic changes, motility changes and localization changes related to microbial stimulation.
- 65.Uhlig HH, Schwerd T: From Genes to Mechanisms: The Expanding Spectrum of Monogenic Disorders Associated with Inflammatory Bowel Disease. Inflamm Bowel Dis 2016, 22:202–212. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki T, Sasahara Y, Kikuchi A, Kakuta H, Kashiwabara T, Ishige T, Nakayama Y, Tanaka M, Hoshino A, Kanegane H, et al. : Targeted Sequencing and Immunological Analysis Reveal the Involvement of Primary Immunodeficiency Genes in Pediatric IBD: a Japanese Multicenter Study. J Clin Immunol 2017, 37:67–79. [DOI] [PubMed] [Google Scholar]
- 67.Kammermeier J, Drury S, James CT, Dziubak R, Ocaka L, Elawad M, Beales P, Lench N, Uhlig HH, Bacchelli C, et al. : Targeted gene panel sequencing in children with very early onset inflammatory bowel disease--evaluation and prospective analysis. J Med Genet 2014, 51:748–755. [DOI] [PubMed] [Google Scholar]
- 68. Petersen BS, August D, Abt R, Alddafari M, Atarod L, Baris S, Bhavsar H, Brinkert F, Buchta M, Bulashevska A, et al. : Targeted Gene Panel Sequencing for Early-onset Inflammatory Bowel Disease and Chronic Diarrhea. Inflamm Bowel Dis 2017, 23:2109–2120. *This study represents an overview of genetic findings and reveals the steps they took to validate the identified variants. It represents a case study for a new VEO IBD clinic.
- 69.Kammermeier J, Dziubak R, Pescarin M, Drury S, Godwin H, Reeve K, Chadokufa S, Huggett B, Sider S, James C, et al. : Phenotypic and Genotypic Characterisation of Inflammatory Bowel Disease Presenting Before the Age of 2 years. J Crohns Colitis 2017, 11:60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Angelino G, De Angelis P, Faraci S, Rea F, Romeo EF, Torroni F, Tambucci R, Claps A, Francalanci P, Chiriaco M, et al. : Inflammatory bowel disease in chronic granulomatous disease: An emerging problem over a twenty years’ experience. Pediatr Allergy Immunol 2017, 28:801–809. *IBD occuring in patients with CGD has long been recognized, however, there have been few studies of outcome or treatment. This study rectifies that lack.
- 71.Yu JE, De Ravin SS, Uzel G, Landers C, Targan S, Malech HL, Holland SM, Cao W, Harpaz N, Mayer L, et al. : High levels of Crohn’s disease-associated anti-microbial antibodies are present and independent of colitis in chronic granulomatous disease. Clin Immunol 2011, 138:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim KY, Lee EJ, Kim JW, Moon JS, Jang JY, Yang HR, Ko JS: Higher Morbidity of Monogenic Inflammatory Bowel Disease Compared to the Adolescent Onset Inflammatory Bowel Disease. Pediatr Gastroenterol Hepatol Nutr 2018, 21:34–42. *This study found growth failure was a marker for monogenic disease and also found that the patients had more complications with monogenic disease.
- 73. Rialon KL, Crowley E, Seemann NM, Fahy AS, Muise A, Langer JC: Long-term outcomes for children with very early-onset colitis: Implications for surgical management. J Pediatr Surg 2018, 53:964–967. *These authors found that VEO IBD thought to be unclassified or ulcerative colitis can become reclassified as Crohn’s disease and caution that surgery should be undertaken with an eye to this eventuality.
- 74. Maxwell EC, Dawany N, Baldassano RN, Mamula P, Mattei P, Albenberg L, Kelsen JR: Diverting Ileostomy for the Treatment of Severe, Refractory, Pediatric Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr 2017, 65:299–305. *In contrast to the previous study, these authors found that diversion can be a successful strategy to induce stability of not remission in very sick children.