Abstract
BMS-284756, a novel des-fluoro(6)-quinolone, was used to select for in vitro mutants of Staphylococcus aureus ISP794. Step mutants were obtained, and the quinolone resistance-determining regions of four target genes, gyrA, gyrB, grlA, and grlB, were sequenced. The data suggest that DNA gyrase is the primary target for BMS-284756 in S. aureus.
BMS-284756 is a novel des-fluoro(6)-quinolone which has a fluorine incorporated through a C8 difluoromethyl ether linkage instead of the classical C6 fluorine of fluoroquinolones (1, 3). When compared to five fluoroquinolones (trovafloxacin, moxifloxacin, levofloxacin, ofloxacin, and ciprofloxacin), BMS-284756 was the most active against staphylococci, streptococci, pneumococci, and Enterococcus faecalis (1).
S. aureus ISP794, a group II wild-type strain of NCTC8325 (pig-131), was obtained from David C. Hooper (Massachusetts General Hospital, Boston, Mass.) and was used for the selection of spontaneous step mutants by both BMS-284756 and ciprofloxacin (6). Mutants were isolated from Mueller-Hinton agar plates containing two, four and eight times the MIC determined for the preceding step strain (4). The frequency of resistance emergence (FRE) was calculated by dividing the number of mutants obtained at the highest concentration of drug on which resistant colonies of bacteria emerged in each round of selection by the number of cells plated. Each FRE experiment was repeated twice. All MICs were determined according to the National Committee for Clinical Laboratory Standards microdilution standard protocol (5). The quinolone resistance-determining regions (QRDRs) of the parental and mutant strains were sequenced.
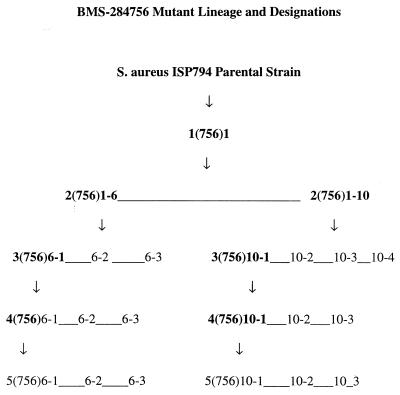
Two microliters of a culture with an optical density at 550 nm of 0.2 was directly lysed in a standard PCR, or isolated chromosomal DNA was used as the template to obtain PCR fragments for sequencing. The following primers were used to amplify the QRDR of each target gene: grlA, 5′ primer, ACTTGAAGATGTTTTAGGTGAT; and 3′ primer, TTAGGAAATCTTGATGGCAA; grlB, 5′ primer, AGACAAATTGCCATTCTATTTAGAAG; and 3′ primer, AAACCTTTGTAACGTTGTAACG; gyrA, 5′ primer, TATTACCAGTGAAATGCGTRAATC; and 3′ primer, ACGAGAACGCATTTGAATTGAACC; and gyrB, 5′ primer, TAGACTTTCTGGTGAAGATACACG; and 3′ primer, ATTTTGGTGTTGGATTCAATTCAG. PCRs were purified for sequencing using the QIAquick PCR Purification Kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Internal primers as well as PCR primers were used to sequence the DNA templates on an ABI Prism 3700 capillary sequencing machine (PE Applied Biosystems, Norwalk, Conn.). Sequencing traces were analyzed using the Lasergene software program. Any mutation was confirmed by sequencing an independently obtained PCR fragment. The mutant strain designation reflects the following information: mutant step, the antibiotic used for mutant selection, and the number of the mutant (Fig. 1).
FIG. 1.
The mutant lineage of BMS-284756-selected strains is shown. Strains in boldface were used to produce the next step mutants.
In all mutants selected by BMS-284756, for which the MIC was ≥0.06 μg/ml, the GyrA 84 (Ser→Leu) mutation was present (Tables 1 and 2). In the case of fourth-step BMS-284756-selected mutants which had an amino acid substitution in both GyrA 84 (Ser→Leu) and GrlA 80 (Ser→Phe) (Table 1), the ciprofloxacin MICs increased four to eightfold when the GrlA 80 (Ser→Phe) mutation was present. In contrast, the BMS-284756 MIC remained the same (Tables 1 and 2). To explore whether mutations outside the QRDRs might be responsible for the increased MICs for BMS-284756 fourth-step mutants, the entire gyrA/gyrB region was sequenced for two mutants.
TABLE 1.
| Strain | Location of mutation for indicated gene
|
|||
|---|---|---|---|---|
| GrlA | GrlB | GyrA | GyrB | |
| Third-step mutant strains | ||||
| 3(756)6-1 | 116 (Ala→Glu) | —c | 84 (Ser→Leu) | — |
| 3(756)6-2 | — | — | 84 (Ser→Leu) | — |
| 3(756)6-3 | 157 (Pro→Leu) | — | — | — |
| 3(756)10-1 | — | — | 84 (Ser→Leu) | — |
| 3(756)10-2 | — | — | 84 (Ser→Leu) | — |
| 3(756)10-3 | — | — | 84 (Ser→Leu) | — |
| Fourth-step mutant strains | ||||
| 4(756)6-1 | — | — | 84 (Ser→Leu) | — |
| 4(756)6-2 | 80 (Ser→Phe) | — | 84 (Ser→Leu) | — |
| 4(756)10-1 | — | — | 84 (Ser→Leu) | — |
| 4(756)10-3 | 80 (Ser→Phe) | — | 84 (Ser→Leu) | — |
| Fifth-step mutant strains | ||||
| 5(756)6-1 | — | 472 (Glu→Lys)d | 84 (Ser→Leu) | — |
| 5(756)6-2 | — | 472 (Glu→Lys) | 84 (Ser→Leu) | — |
| 5(756)10-1 | — | — | 84 (Ser→Leu) | — |
| 5(756)10-3 | — | — | 84 (Ser→Leu) | — |
First- and second-step BMS-284756-selected mutants had no changes in the QRDRs of the four genes sequenced.
Four ciprofloxacin-selected mutants, 1C6(1)-1, 2C6(1)-1, 3C6(1)-1, and 4C6(1)-1, were sequenced. They all had only one QRDR mutation, grlA (A116E).
The designation — indicates no change.
Mutations not previously reported.
TABLE 2.
Summary of antibiotic MICs for S. aureus mutants selected by BMS-284756
| Strain | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|
| BMS-284756 | Ciprofloxacin | Levofloxacin | Moxifloxacin | |
| Parental strain | ||||
| ISP794 | 0.007 | 0.125 | 0.125 | 0.06 |
| First-step strain | ||||
| 1(756)1 | 0.015 | 0.06 | 0.125 | 0.03 |
| Second-step strains | ||||
| 2(756)1-6 | 0.015 | 0.125 | 0.125 | 0.03 |
| 2(756)1-10 | 0.015 | 0.125 | 0.125 | 0.03 |
| Third-step strains | ||||
| 3(756)6-1 | 0.06 | 0.125 | 0.125 | 0.03 |
| 3(756)6-2 | 0.06 | 0.25 | 0.125 | 0.03 |
| 3(756)6-3 | 0.03 | 0.5 | 0.5 | 0.06 |
| 3(756)10-1 | 0.06 | 0.25 | 0.125 | 0.03 |
| 3(756)10-2 | 0.06 | 0.25 | 0.125 | 0.03 |
| 3(756)10-3 | 0.06 | 0.25 | 0.125 | 0.03 |
| 3(756)10-4 | 0.06 | 0.25 | 0.125 | 0.03 |
| Fourth-step strains | ||||
| 4(756)6-1 | 1.0 | 1.0 | 1.0 | 1.0 |
| 4(756)6-2 | 1.0 | 8.0 | 4.0 | 2.0 |
| 4(756)6-3 | 1.0 | 1.0 | 1.0 | 1.0 |
| 4(756)10-1 | 1.0 | 2.0 | 2.0 | 1.0 |
| 4(756)10-2 | 1.0 | 1.0 | 1.0 | 1.0 |
| 4(756)10-3 | 1.0 | 8.0 | 4.0 | 2.0 |
| Fifth-step strains | ||||
| 5(756)6-1 | 2.0 | 2.0 | 1.0 | 2.0 |
| 5(756)6-2 | 2.0 | 2.0 | 1.0 | 2.0 |
| 5(756)6-3 | 2.0 | 2.0 | 1.0 | 2.0 |
| 5(756)10-1 | 2.0 | 2.0 | 2.0 | 2.0 |
| 5(756)10-2 | 2.0 | 2.0 | 2.0 | 1.0 |
| 5(756)10-3 | 2.0 | 2.0 | 2.0 | 2.0 |
Fourth-step mutants 4(756)10-1 and 4(756)10-3 were chosen because the MIC of BMS-284756 was 1 μg/ml against both strains, while ciprofloxacin, levofloxacin, and moxifloxacin MICs were two- to fourfold higher against 4(756)10-3 than against 4(756)10-1 (Table 2). While both mutants had the QRDR mutation GyrA 84 (Ser→Leu), 4(756)10-3 had an additional QRDR mutation, GrlA 80 (Ser→Phe) (Table 1). The only mutation identified outside the QRDRs was found in mutant 4(756)10-1 (GrlA 18 [Phe→Lys]).
Therefore, it appears that the GrlA 80 (Ser→Phe) mutation in 4(756)10-3 is sufficient to increase ciprofloxacin, levofloxacin, and moxifloxacin MICs while having no effect on BMS-284756 MICs. This may be of clinical significance, as a survey of 110 clinical isolates determined that all GyrA Ser84 mutants also had GrlA Ser80 or Glu84 mutations (7). The lack of effect of GrlA 80 (Ser→Leu) mutations on the MIC of BMS-284756 suggests the potential of this drug to cover quinolone-resistant pathogens in the clinic.
At multiples of the MIC, the FRE of ciprofloxacin in first-step mutants was compared to that of BMS-284756 (Table 3). The FREs of ciprofloxacin for the parental S. aureus strain ISP794, FREs were 1.5 × 10−7 to 6.8 × 10−7 at four times the MIC of ciprofloxacin and higher at two times the MIC. There were no mutants at eight times the MIC.
TABLE 3.
FRE in S. aureus ISP794 selected by BMS-284756 and ciprofloxacin
| Multiple of MIC used | FRE for BMS-284756 or ciprofloxacina
|
|||
|---|---|---|---|---|
| BMS-284756 | Ciprofloxacin | |||
| For first-step mutants | ||||
| 2× MIC for parental strain | >8.0 × 10−10 | 5 × 10−10 | >1.5 × 10−7 | >6.8 × 10−7 |
| 4× MIC for parental strain | 8.0 × 10−10 | NDc | 1.5 × 10−7 | 6.8 × 10−7 |
| 8× MIC for parental strain | ND | ND | ND | None |
| For second-step mutants | ||||
| 2× MIC for first-step mutant | 1.4 × 10−9 | 1.2 × 10−9 | 1.3 × 10−8 | 1.7 × 10−8 |
| 4× MIC for first-step mutant | None | None | None | None |
| 8× MIC for first-step mutant | None | ND | ND | None |
| For third-step mutantsb | ||||
| 2× MIC for second-step mutant | >1.3 × 10−9, >8.3 × 10−9 | >4.5 × 10−9, >1.3 × 10−8 | 1.2 × 10−8 | 2.2 × 10−7 |
| 4× MIC for second-step mutant | 1.3 × 10−9, 8.3 × 10−9 | 4.5 × 10−9, 1.3 × 10−8 | None | None |
| 8× MIC for second-step mutant | None | None | ND | ND |
| For fourth-step mutants | ||||
| 2× MIC for third-step mutant | >2.6 × 10−7, >1.3 × 10−8 | >2.5 × 10−7, >6.25 × 10−9 | 5.8 × 10−6 | 1.2 × 10−6 |
| 4× MIC for third-step mutant | >2.6 × 10−7, >1.3 × 10−8 | >2.5 × 10−7, >6.25 × 10−9 | None | None |
| 8× MIC for third-step mutant | 2.6 × 10−7, >1.3 × 10−8 | 2.5 × 10−7, >6.25 × 10−9 | ND | None |
| 16× MIC for third-step mutant | ND, 1.3 × 10−8 | ND, 6.25 × 10−9 | ND | ND |
FREs were determined in two separate experiments for each drug; results from both trials are shown in the two columns.
Two BMS-284756 mutants were carried forward (designated no. 6 and 10) for third- and fourth-step mutants. Each frequency was repeated twice. The first column under BMS-284756 represents mutants 6A and 6B, and the second column represents mutants 10A and 10B.
ND, not determined.
In contrast, the FRE for BMS-284756 was 8.0 × 10−10 at four times the MIC and higher at two times the MIC, and no resistant mutants were selected at eight times the MIC, representing approximately a 3-log difference in FRE between BMS-284756 and ciprofloxacin (Table 3). In all other steps, BMS-284756 showed a 1- to 2-log-lower FRE than did ciprofloxacin (Table 3).
Differences in the susceptibility of these resistant mutants to the four quinolones were observed. MICs of ciprofloxacin for the parental strain and the first-, second-, third-, and fourth-step ciprofloxacin-selected mutants were 0.125, 1, 4, 4 to 8, and 4 to 8 μg/ml, respectively. In contrast, levofloxacin MICs for the same mutants were 0.125, 0.5, 1, 1, and 1 to 2 μg/ml, respectively, and moxifloxacin MICs for the same mutants were 0.06, 0.06, 0.25 to 0.5, 0.25, and 0.25 μg/ml, respectively. However, BMS-284756 MICs were 0.015, 0.03, 0.03 to 0.06, 0.06, and 0.06 μg/ml, respectively. The emergence of resistant mutants selected by BMS-284756 resulted in cross-resistance to the other three quinolones (Table 2). In addition, one step was required using ciprofloxacin selection for the MICs for the S. aureus mutants to reach the ciprofloxacin breakpoint of 1 μg/ml, just as with Streptococcus pneumoniae (2), while four steps were required by BMS-284756 for MICs to increase to 1 μg/ml (still within the anticipated breakpoint of 2 to 4 μg/ml [J. Barrett, Bristol-Myers Squibb, unpublished]). These data agree with results for S. pneumoniae which showed that BMS-284756-selected mutants were isolated at a relatively low rate and that mutations were initially found in the GyrA protein within the QRDR (2). BMS-284756 is primarily selecting for the mutations present in DNA gyrase in gram-positive bacteria. The low FRE for BMS-284756 in S. aureus ISP794 and the lack of effect of GrlA 80 (Ser→Leu) mutations on its MIC indicate the potential of this drug to have reduced resistance development and to cover quinolone-resistant pathogens in the clinic.
Acknowledgments
We thank the Bristol-Myers Squibb DNA Sequencing Core Facility, Hopewell, N.J., for the DNA sequencing. We also thank Michael J. Pucci for his critical reading of the manuscript.
REFERENCES
- 1.Fung-Tomc J C, Minassian B, Kolek B, Huczko E, Aleksunes L, Stickle T, Washo T, Gradelski E, Valera L, Bonner D P. Antibacterial spectrum of a novel des-fluoro(6) quinolone, BMS-284756. Antimicrob Agents Chemother. 2000;44:3351–3356. doi: 10.1128/aac.44.12.3351-3356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartman-Neumann, S., K. DenBleyker, L. A. Pelosi, L. E. Lawrence, J. F. Barrett, and T. J. Dougherty. Selection and genetic characterization of Streptococcus pneumoniae mutants resistant to the des-F(6)-quinolone BMS-284756. Antimicrob. Agents Chemother, in press. [DOI] [PMC free article] [PubMed]
- 3.Hosaka M, Yasue T, Fukuda H, Tomizawa H, Aoyama H, Hirai K. In vitro and in vivo antibacterial activities of Am-1155, a new 6-fluoro-8-methoxy quinolone. Antimicrob Agents Chemother. 1992;36:2108–2117. doi: 10.1128/aac.36.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ince D, Hooper D C. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob Agents Chemother. 2000;44:3344–3350. doi: 10.1128/aac.44.12.3344-3350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Committee for Clinical Library Standards. National Committee for Clinical Laboratory Standards, Wayne, Pa. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. M7–A4. [Google Scholar]
- 6.Stahl M, Pattee P A. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983;154:406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi H, Kikuchi T, Shoji S, Fujimura S, Lutfor A B, Tokue Y, Nukiwa T, Watanabe A. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:49–57. doi: 10.1093/jac/41.1.49. [DOI] [PubMed] [Google Scholar]