Abstract
To face the coronavirus disease 2019 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) virus, our institute has developed the rVSV‐ΔG‐spike vaccine, in which the glycoprotein of vesicular stomatitis virus (VSV) was replaced by the spike protein of SARS‐CoV‐2. Many process parameters can influence production yield. To maximize virus vaccine yield, each parameter should be tested independently and in combination with others. Here, we report the optimization of the production of the VSV‐ΔG‐spike vaccine in Vero cells using the Ambr15 system. This system facilitates high‐throughput screening of process parameters, as it contains 24 individually controlled, single‐use stirred‐tank minireactors. During optimization, critical parameters were tested. Those parameters included: cell densities; the multiplicity of infection; virus production temperature; medium addition and medium exchange; and supplementation of glucose in the virus production step. Virus production temperature, medium addition, and medium exchange were all found to significantly influence the yield. The optimized parameters were tested in the BioBLU 5p bioreactors production process and those that were found to contribute to the vaccine yield were integrated into the final process. The findings of this study demonstrate that an Ambr15 system is an effective tool for bioprocess optimization of vaccine production using macrocarriers and that the combination of production temperature, rate of medium addition, and medium exchange significantly improved virus yield.
Keywords: Ambr15 system, Fibra‐Cel, optimization, Vero cells, virus vaccine production, VSV‐ΔG‐spike vaccine
Rosen et al. report the optimization of the VSV‐ΔG‐spike vaccine production process in Vero cells using the Ambr15 system. Many process parameters can influence production yield and during optimization, many critical parameters were tested. The findings of this study demonstrate that an Ambr15 system is an effective tool for bioprocess optimization of vaccine production using macrocarriers and that the combination of production temperature, rate of medium addition, and medium exchange significantly improved virus yield. Moreover, the optimized parameters were subsequently tested in the BioBLU 5p vaccine bioreactors production process and most were found to have the same useful effects and were, therefore, integrated into the final production process.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was identified as the causative agent for the novel coronavirus disease 2019 (COVID‐19). Similar to the disease caused by two other family viruses—severe acute respiratory syndrome (SARS) and middle east respiratory syndrome—COVID‐19 illness is accompanied by clinical symptoms of cough fever and lunge pneumonia (Guan et al., 2020). As of January 2022, the ongoing worldwide pandemic has resulted in the infection of 350 million people and 5.6 million deaths, causing severe economic burdens and hindering social development worldwide.
Many researchers worldwide are working around the clock to discover and develop new vaccines against COVID‐19 (Kiesslich & Kamen, 2020). This led to almost 300 vaccine candidates that are based on varying strategies entering clinical trials. One such vaccine being tested in clinical trials was developed by the Israel Institute for Biological Research (IIBR). This vaccine is based on vesicular stomatitis virus (VSV), where the native surface glycoprotein (VSV‐G) has been replaced by the SARS‐CoV‐2 spike protein (rVSV‐ΔG‐spike), creating a recombinant replicating virus (Yahalom‐Ronen et al., 2020). A similar vaccine strategy, where a recombinant vesicular stomatitis virus (rVSV) viral platform expresses the Zaire Ebola virus glycoprotein and is produced in Vero cells, was previously approved for Ebola virus (Monath et al., 2019). Several inactivated SARS‐CoV‐2‐based vaccine candidates produced in Vero cells are in current clinical trials for disrupting the COVID‐19 pandemic (WHO, 2021).
On the basis of the above‐noted advantages, the IIBR vaccine is produced in serum‐free Vero cells. These cells originated from a kidney of an African monkey (Yasumura & Kawakita, 1963) and are adherent. They lack the ability to express interferon, which reduces their virus protection actions (Emeny & Morgan, 1979), making them ideal for virus production. In addition, the use of defined and serum‐free cell culture allows more consistent processes (Aubrit et al., 2015). On the basis of these characteristics, Vero cells are now considered one of the most broadly used cell lines for the manufacture of vaccines for human use, particularly viral vaccine production (Barrett et al., 2017). In many screening experiments, the highest virus productivity was achieved by Vero cells, and those cells also maintained their high virus productivity when transferred to a stirred tank reactor (e.g., Elahi et al., 2019; Grein et al., 2017). The preparation of vaccines in Vero host cells has been approved for the production of polio, smallpox, rabies, Japanese encephalitis, rotavirus, vaccinia, and influenza vaccines. Consistently, the development of new vaccines and vaccine vectors, such as vaccinia, polio, Ross River, West Nile, chikungunya, tick‐borne encephalitis virus, hepatitis A, SARS‐coronavirus, rely on the use of Vero as host cells (Damodharan et al., 2021).
Generally, virus‐based vaccine production occurs in two major phases: cell growth and virus production. The purpose of the first phase is to grow the host cells producing the viral vaccine to high biomass. During their growth, Vero cells adhere to surfaces and require an expanding area for propagation. Such a vast area is eligible for the use of porous Fibra‐Cel carriers. Fibra‐Cels are macrocarriers (each has a 5 cm2 growth area) that allow the growth of up to a few million cells on each carrier (Han & Sha, 2017). This material, made of polyester fibers, adsorb the cells electrostatically and is suitable for pharmaceutical use. Practically, the rVSV‐ΔG‐spike vaccine has been produced in cells grown on Fibra‐Cel carriers in single‐use BioBLU 5p bioreactors (Cino et al., 2011), each containing ∼35,000 Fibra‐Cels (150 g, 180,000 cm2 total growth surface) per bioreactor (Han et al., 2018). Cells are first allowed to adsorb to Fibra‐Cel carriers and then grown for 6 days. After infection, the virus is produced and released into the culture medium, and at the end of the 48 h production process, the medium is harvested.
Multiple and varied parameters influence the production yields, as each parameter influences the production in a different way. To maximize the production yield, it is, thus, necessary to test the effect of each parameter separately and in combination with the others. Hence, many experiments must be performed in parallel. The need to conduct such a large number of experiments under bioreactor conditions has resulted in the development of miniaturized high‐throughput technologies for process development, including the Ambr15 system (Sartorius) (Fletcher, 2014). This technology is very cost‐effective, high throughput, containing disposable reactor vessels with maximum working volumes of 15 ml, controlled by an automated workstation. The system provides parallel processing with the use of 24–48 disposable reactors with online monitoring of pH, temperature, and dissolved oxygen (DO) for each reactor. Full control of impeller speed and culture temperature is also achievable in this system. For process optimization, we purchased the Ambr15 system with 24 reactors, which allows the simulation of up to 24 different processes. Before optimization, the system was adjusted to perform the vaccine production process (Jayson et al., 2022).
In this study, we present the vaccine production process optimization that was achieved with the Ambr15 system. Many parameters were tested for their effect on virus yield, all of which are associated with the infection and production steps. These include cell densities, the multiplicity of infections (MOIs), production temperatures, medium additions and exchanges, and the influence of glucose. Acquisition of this information deepened our understanding of the production process and assisted in process control and knowledge of factors that influence it. In addition, the values of the parameters that were found in the Ambr15 system to have a positive effect on virus yield were tested in the production process BioBLU 5p bioreactors (3.5 L working volume). Those who were proved to have a favorable effect were incorporated in the final production process in those bioreactors (manuscript in preparation).
2. MATERIALS AND METHODS
2.1. The Ambr15 system
All experiments were performed in the Ambr15 system. The system was adapted beforehand to perform the production process including Fibra‐Cel carriers as described in detail in (Jayson et al. 2022). The agitation speed, DO, and pH were set and maintained at a constant value of 300 rpm (upstir), 50%, and 7.05, respectively. If not stated, 60 Fibra‐Cel carriers were added to each reactor, the working volume was 14 ml, and the medium was Flex‐20.
2.2. Materials
Vero cells, derived from the kidneys of female green monkeys, were obtained from World Health Organization (WHO) (WHO, RCB 10‐87). Vero E6 was purchased from American Type Culture Collection (VERO C1008, Vero 76, clone E6, Vero E6). NutriVero FLEX‐20 and NutriVero Flex‐10 are serum‐free, animal component‐free medium. FLEX‐20 was specially designed for serum‐free adapter WHO, RCB 10‐87 Vero cells, Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), nonessential amino acids (NEAAs), l‐alanine‐l‐glutamine, penicillin/streptomycin (pen/strep) antibiotics, recombinant trypsin‐EDTA solution, trypan blue solution, and phosphate‐buffered saline (PBS) were purchased from Biological Industries, Israel (05‐069‐1A, 05‐068‐1A, 01‐055‐1A, 04‐121‐1A, 01‐340‐1B, 03‐022‐1B, 03‐031‐1C, 03‐079‐1A, 03‐102‐1B, and 02‐020‐1A, respectively). OptiPRO SFM was purchased from Gibco (12309‐019). Fibra‐Cel carriers were purchased from Eppendorf (New Brunswick Scientific; M1292‐9984). Alamar blue was purchased from Promega (CellTiter‐blue, G808). Calcein AM was purchased from Sigma‐Aldrich (C1359). Tragacanth was purchased from Merck (G1128).
2.3. Vero cells cultivation
Vero cells were maintained at 37°C under 5% CO2 in T‐flasks in FLEX‐20 medium supplemented with 2 mM l‐alanine l‐glutamine and 0.1% pen/strep antibiotics. Cells were harvested by washing the flasks with PBS and incubating with recombinant trypsin‐EDTA. The detached cells were centrifuged for 5 min at 1200 rpm. The supernatant was removed, and the pellet was resuspended in a fresh medium. Cells count was performed with cell countessTM (Invitrogen) after 1:1 mixing with trypan blue.
2.4. Preparing reactors with Fibra‐Cel carriers
Fibra‐Cel carriers were weighed and 0.28 g (corresponding to 60 Fibra‐Cel carriers) were added to 50 ml tubes containing 15 ml PBS and autoclaved at 121°C for 20 min. The sterilized Fibra‐Cel carriers in PBS were then inserted into reactors using a specific funnel. All reactors were drained and then filled with 14 ml fresh FLEX‐20 medium. The reactors and Fibra‐Cels were maintained for 24 h at 37°C and 50% DO.
2.5. Adsorption and growth of cells on Fibra‐Cel carriers
On the next day, the reactors were inoculated. Three milliliters of medium were first drained from each reactor and manually replaced with 3 ml containing the desired cell numbers into each reactor. At the end of the adsorption period (2 h), the reactors were drained and 14 ml of fresh medium was added. At this point, the system was fixed at 50% DO, 7.05–7.2 pH, 37°C and agitation speed of 300 rpm (upstir). Cells were allowed to grow for 6 days. Replacement of medium with the fresh medium was done during cell growth phase based on glucose consumption, with a minimum of 1 g/L glucose in the medium, and before infection.
2.6. Virus production
The rVSV‐ΔG‐spike virus was constructed as described in (Yahalom‐Ronen et al., 2020). Briefly, the amplified polymerase chain reaction spike product was digested by restriction enzymes and ligated into the pVSV‐FL+(2) vector (Kerafast), precut by the same enzymes to remove the VSV‐G gene. The rVSV‐ΔG‐spike virus was first grown in Vero E6 cells containing serum and later a virus stock in serum‐free Vero cells was established (stock concentration 1 × 108 PFU/ml). rVSV‐ΔG‐spike virus stock was loaded into the system. Infection was carried out at the desired MOI. The amount to be added was calculated in advance, by multiplying the total number of cells in each reactor (determined by glucose consumption) by 0.1 (for an MOI of 0.1). During the virus production process, a sample was taken every 24 h and analyzed with the plaque‐forming units (PFUs) assay.
The cell‐specific virus yield (CSVY) was calculated by dividing the total active virus particle at harvest by the total number of cells at infection.
2.7. Experiments schedule
All experiments were carried out using the following steps:

On Days 6–8, virus titer was monitored by PFU and on Days 0–8 metabolites and waste products were monitored by a chemistry analyzer.
2.8. Glucose consumption
To estimate cell quantity on Fibra‐Cel carrier, a correlation between daily glucose consumption and the number of cells was obtained and used. For that purpose, during the first experiment, the glucose concentration was measured in different time intervals in each reactor. The daily glucose consumption of each reactor was calculated based on: the difference between consecutive results multiplied by the time interval divided by 24 h per day. In parallel, for the same time points, the numbers of cells on Fibra‐Cel carriers were quantified by Alamar blue (Rosen, Jayson, Natan, & Epstein, 2021; Rosen, Jayson, Natan, Monash, et al., 2021). A strong correlation between daily glucose consumption and a number of cells on the Fibra‐Cel carrier was found and was used to estimate the number of cells on the Fibra‐Cel carrier. The results indicated that 1 × 109 cells consumed 1 g glucose in 24 h. In addition, the Alamar blue assay was also used to check and verify cell density on different Fibra‐Cel carriers within a reactor.
2.9. Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) measurements were performed using a NanoSight NS300 (Malvern), consisting of a conventional optical microscope, charged‐coupled device camera, and a sample unit with a laser light source. Samples were diluted to reach a particle concentration suitable for analysis with NTA (1 × 108−1 × 109 particles/ml). Samples were injected with a 1 ml sterile syringe. Each sample was recorded five times for 60 s and analyzed with NanoSight 3.0.
2.10. Plaque‐forming unit
The virus titer was determined with the PFU assay (Dulbecco, 1952). The rVSV‐ΔG‐spike creates a center of destruction on a monolayer cell culture. The cells used for this assay were Vero E6. The cells were grown in 75 cm2 tissue culture flasks with vented caps. For cultivation, the cells were grown in sterile DMEM supplemented with 10% FBS inactivated 30 min at 56°C, 0.1 mM NEAA, 2 mM l‐glutamine, and 2% pen/strep at 37°C under 5% CO2. Cultures were passaged twice weekly until Passage 20 and discharged. Briefly, tested samples withdrawn from reactors were serially diluted in MEM containing 2% fetal calf serum (FCS), 2 mM glutamine, and 1% NEAA. The different diluted samples were implemented (0.2 ml) on E6 Vero cell monolayers in six‐well plates. After 1‐h incubation, a layer of 1 ml 0.8% Tragacanth in MEM containing 2% FCS, 2 mM glutamine, and 1% NEAA was added to each well. Following 3 days of incubation (37°C, 5% CO2) the cells were fixed, stained with crystal violet, and the number of plaques was counted. The virus titer (PFU/ml) was calculated by multiplying the number of plaques formed in a well by the dilution factor and the sow factor volume.
2.11. Monitoring metabolites and waste products
On each day, metabolites and waste products were monitored. Tracking was performed with Cobas Integra 400 plus wet chemistry analyzer (Roche Diagnostics GmbH). Eppendorf tubes containing 100 µl samples were tested for glucose, lactate, glutamate, alanine‐glutamine (GlutaMAX), NH3, and lactate dehydrogenase (LDH) concentrations with commercially available kits.
2.12. Host‐cell protein analysis
Residual proteins of Vero cells were measured using a Vero cell host‐cell protein (HCP) Enzyme‐Linked Immunosorbent Assay Kit (Cygnus Technologies), following the manufacturer's instructions.
3. RESULTS AND DISCUSSION
Production of various VSV‐based vaccines in Vero cells has been reported. However, all reported processes were either in microcarriers (Kiesslich et al., 2020; Mangion et al., 2020) or in cell suspension (Kiesslich et al., 2021) and none were reported in a Fibra‐Cel based production process. Before optimizing the process parameters in the Ambr15 system, a preliminary study was conducted (Jayson et al., 2022). In this preliminary study, special accessories were developed to enable the insertion of tens of Fibra‐Cel carriers into the reactors. In addition, after optimizing system parameters, each of the process steps (cells adsorption, cell growth, infection, and vaccine virus production) was performed by programming the system software and was shown to be similar to the steps in BioBLU 5p bioreactors. The establishment of the process simulation was a basic requirement before the process optimization described here.
During the optimization, many parameters were tested (highlighted in Figure 1). Those include parameters in the infection step (Figure 1, red)—cell density and MOI; and in the virus production step (Figure 1, blue)—virus production temperature; a combination of perfusion and media addition at different times and quantities; and addition of glucose in the virus production step.
Figure 1.
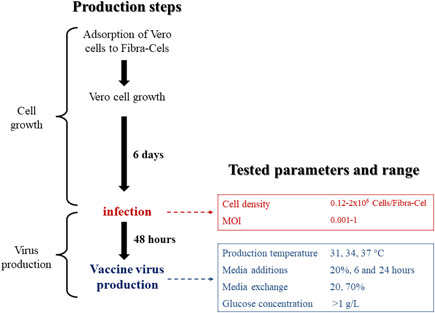
SARS‐CoV‐2 production steps and the parameters that were optimized with the Ambr15 system. The process comprised two major phases, cell growth, and virus production. The parameters that were optimized by the Ambr15 system are all in the virus production phase and they are highlighted in red and blue. MOI, multiplicity of infection; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.1. Cell density impact
Since the virus is produced in cells, it is assumed that one effective way to increase functional titers is to increase cell density at infection. To determine if cell density influences virus titer, two sets of experiments were conducted. In the first, inoculation was performed with the same number of cells, but their growth temperature was varied so they reached different cell densities. In the second experiment, inoculation was performed with different numbers of cells (growth was done under the same conditions); hence the cells reached different cell densities. In both experiments, cells densities were estimated based both on daily glucose consumption (Jayson et al., 2022) and by the Alamar blue assay (Rosen, Jayson, Natan, & Epstein, 2021; Rosen, Jayson, Natan, Monash, et al., 2021). The number of cells on different Fibra‐Cel carriers within a reactor was found to be similar (based on the Alamar blue assay).
3.1.1. Variations in cell growth temperature
A total of 100,000 cells/Fibra‐Cel carriers were inoculated into eight reactors and the cells were grown for 6 days. Four reactors were incubated at 37°C and the other four at 34°C, leading to a different number of cells/Fibra‐Cel carrier. At the end of the growth phase, based on daily glucose consumption (Jayson et al., 2022, the number of cells/Fibra‐Cel was found to be 750,000 and 1,000,000 per carrier with a total of 45 × 106 and 60 × 106 cells/reactor, respectively.
All reactors were sequentially infected with 0.1 MOI (4.5 × 106 and 6 × 106 PFU/reactor). The influence of different cell densities (differing by 25%) was tested by monitoring the virus titer for 2 days. No significant difference was seen between virus titers (both reached 3 × 108 PFU/ml) at the different cells' densities (Figure 2, gray and green). The different cells' densities, and the modification of cells' growth temperature, led to the same virus titer, concluding that higher cell density did not improve the titer.
Figure 2.
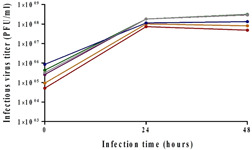
Impact of cell density on virus titer. Reactors were inoculated with either 100,000 cells/Fibra‐Cel and cells were grown for 6 days in different temperatures or with various numbers of cells and cells were grown for 6 days under the same condition. At the end of growing phase, 120,000 (red), 220,000 (orange), 600,000 (purple), 750,000 (gray), 1,000,000 (green), or 2,000,000 (blue) cells/Fibra‐Cel were estimated. Virus titers were monitored for 2 days after infection with 0.1 MOI. MOI, multiplicity of infection; PFU, plaque‐forming unit
3.1.2. Variation in cells inoculation concentration
Based on the results described above (Section 3.1.1), cell density higher than 750,000 cells/Fibra‐Cel seems to have no influence on virus titer. To further elaborate if lower densities could affect virus titers, reactors were inoculated with different cell numbers ranging from 12,500 to 120,000 cells/Fibra‐Cel. After 6 days of growth at 37°C, the number of cells/Fibra‐Cel in each reactor was estimated by daily glucose consumption to be 120,000, 220,000, 600,000, 1,000,000 and 2,000,000 cells/Fibra‐Cel (total of 7.2 × 106, 13.2 × 106, 36 × 106, 60 × 106, and 120 × 106 cells/reactor, respectively). All reactors were infected with 0.1 MOI.
As shown in Figure 2, similar titers (3 × 108 PFU/ml) were reached with the high initial cell densities (600,000 and 1,000,000 cells/Fibra‐Cel, Figure 2 compare green to purple) while there was even a slight decrease in titer (1.5 × 108 PFU/ml) at the highest density (2,000,000 cells/Fibra‐Cel, Figure 2 blue). These results are comparable to the results described in the previous chapter, suggesting that high cells density (>600,000 cells/Fibra‐Cel) did not improve virus titers. The upper titer limit, at high cell densities, could have resulted from another limiting parameter overriding the cell number. On the other hand, this was not the case at lower initial cell densities (220,000 and 120,000 cells/Fibra‐Cel, Figure 2 orange and red). Lower cell densities resulted in significantly lower titers (8 × 107 and 5 × 107 PFU/ml, respectively), which correlated with cell densities. These results demonstrate that below a certain cell number, cell densities have an impact on virus titer.
To further elaborate on the effect of cell density on virus yield, the CSVY was calculated. The results were 97, 85, 117, 93, 70, and 18 for 120,000, 220,000, 600,000, 750,000, 1,000,000, and 2,000,000 cells/Fibra‐Cel, respectively. These results demonstrate that different virus production efficiencies occur at different cell densities and that the best results were achieved with 600,000 cells/Fibra‐Cel. Together with previous results, described in the previous section, it was concluded that cell densities do have an impact on virus titer as initially assumed.
3.2. MOI influence
MOI refers to the number of infectious virions added per cell during infection. This ratio is considered to be a major process parameter since the number of viruses added to cells can influence the virus's ability to infect and multiply. Theoretically, one can presume that higher MOIs are desired, leading to more viruses infecting the cells and a more intense and efficient process. On the other hand, massive amounts of virus infection can impact the future cell viability, growth, and cycle and interfere with the production of the next virus generation. Moreover, it was reported that defective‐interfering particles appear following infection at high MOI, ensuing hampered viral replication, resulting in reduced virus titers (Kibenge & Kibenge, 2016; Linder et al., 2021).
Studies described in the literature, use an MOI value of 0.1 as a point of reference (e.g., Elahi et al., 2019; Gélinas et al., 2019). Hence, all experiments conducted to optimize other parameters than MOI conducted up to this point were performed with an MOI of 0.1. Nevertheless, because some studies had suggested the use of larger or smaller MOI, the impact of MOI was investigated here.
To evaluate the impact of high MOI, rVSV‐spike infections were first performed at MOIs of 1 versus 0.1, and the virus titer was monitored for 2 days. As seen in Figure 3, increasing the virus input did not improve the virus titer (compare yellow to red). The contributing effect to viral yield at low MOI was described by Gélinas et al. (2019) in other VSV‐based production processes. To evaluate the impact of lower MOI in our system, infections were performed with MOI of 0.01 and 0.001 versus 0.1, and virus titer was monitored for 2 days. Despite two log differences between MOIs tested (0.001 vs. 0.1), virus yield (3.5 × 108 PFU/ml) was indistinguishable (Figure 3, red, blue, and purple). These results indicate a large operating window in terms of MOI and compare well with other VSV‐based studies (Elahi et al., 2019; Gélinas et al., 2019).
Figure 3.
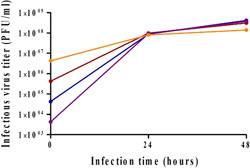
Impact of MOI on virus titer. Reactors were inoculated with 125,000 cells/Fibra‐Cel and cells were grown for 6 days under the same conditions. At the end of the growing phase, 900,000 cells/Fibra‐Cel were estimated. Infections were performed with MOIs of 1 (orange), 0.1 (red), 0.01 (blue), or 0.001 (purple). Virus titers were monitored for 2 days after infection. MOI, multiplicity of infection; PFU, plaque‐forming unit
3.3. Virus production temperatures
Another important parameter that was investigated in the Ambr15 system was production temperature. The cells were grown at 37°C, which is ideal for cell propagation. Virus production, up to this point, was also performed at 37°C. However, it had previously been reported that decreasing the production temperature of VSV‐based vaccines could have a considerable impact on virus yield (Elahi et al., 2019; Gélinas et al., 2019; Paillet et al., 2009). Therefore, to assess the temperature impact, rVSV‐spike infections were performed (in triplicate) at either 34°C or 37°C. Figure 4 shows the resulting virus titers with a threefold increase in titer at 48 h for production at 34°C as compared to 37°C (2.6 × 108 vs. 7.4 × 108 PFU/ml). Our results are in agreement with previous studies conducted by Elahi et al. (2019) and Gélinas et al. (2019) showing that shifting to 34°C (from 37°C) led to a threefold or sixfold increase in virus titer. In addition, the production of a VSV‐based Ebola vaccine showed a preference for production at 34°C (Gélinas et al., 2019, 2020; Kiesslich et al., 2021). These results led us to test an additional lower temperature value of 31°C. However, the virus titer was not improved at this temperature (data not shown), so the temperature for future experiments was set to 34°C. Temperature reduction was later tested in BioBLU 5p bioreactors, and a virus yield improvement was observed in them as well.
Figure 4.
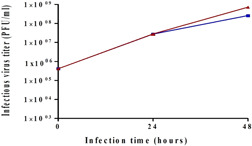
Effect of virus production temperature on virus titer. Reactors were inoculated with 125,000 cells/Fibra‐Cel and cells were grown for 6 days under the same conditions. At the end of the growing phase, 900,000 cells/Fibra‐Cel were estimated. Infections were performed with 0.1 MOI. Virus titers were monitored for 2 days after infection. Bars represent the mean of samples from five different replicates ± standard deviation. MOI, multiplicity of infection; PFU, plaque‐forming unit
Throughout the virus production process (at 34°C and 37°C), samples were taken at 24 and 48 h and sent for HCP and LDH analysis. The HCP proteins are process‐related protein impurities produced by the host organism and the majority of HCPs (>99%) are removed from the final product during the purification process. LDH is an inner cell enzyme that typically is released into the medium during cell lysis. As such, it has long been used as a marker of cell death. The average HCP and LDH values at 24 and 48 h at 37°C versus 34°C in three independent experiments are shown in Table 1. Much lower HCP and LDH values were found when production was done at 34°C, although a threefold virus titer increase was observed, indicating yet another advantage for production at 34°C. Both analyses suggest that less cell damage occurred at 34°C.
Table 1.
HCP and LDH values during virus production
| Temperature | |||
|---|---|---|---|
| Hours | 37°C | 34°C | |
| HCP, μg/ml | 24 | 33 ± 4 | 17 ± 4 |
| 48 | 62 ± 8 | 37 ± 4 | |
| LDH, U/L | 24 | 123 ± 5 | 56 ± 2 |
| 48 | 537 ± 12 | 166 ± 9 | |
Note: Results are presented as mean ± SD from three different experiments.
Abbreviations: HCP, host‐cell protein; LDH, lactate dehydrogenase.
Another analysis that was done at the end of virus production is NTA which characterize nanoparticles (NP) from 10 to 1000 nm in solution. Each particle is individually but simultaneously analyzed by direct observation and measurement of diffusion events. The particle‐by‐particle methodology produces high‐resolution results for NP size distribution and concentration. Examination of the NTA results for virus produced at 34°C versus 37°C yielded interesting results: size distribution of particles was similar but the concentration was much higher (almost one log) for virus produced at 37°C (data not shown). Since the values for active particles (as measured by PFU/ml) were higher for 34°C, these results suggest a different ratio between total particles to an active particle at the different temperatures (1: a few tens vs. 1: a few hundred for 34°C vs. 37°C, respectively). These ratios suggest different efficiencies for active virus production at the different temperatures and can indicate more efficient production or better stability at the lower temperature.
3.4. Medium exchange, addition, and selection
During cell growth, medium exchanges were performed daily, based on glucose consumption. These exchanges were done to supply essential medium components for the cells' growth and to remove undesirable metabolic byproducts that accumulate in the medium during cell growth. At the end of the growth step, and just before infection, the medium is also exchanged to remove impurities and optimize infection conditions. After infection, supplementing medium components is important, but since the produced virions are secreted into the media, media exchanges might take out the newly produced virions as well. Therefore, several strategies were suggested and tested in the Ambr15 system for supplementing medium:
-
1.
Twenty‐percent medium exchange 24 h after infection.
-
2.
Medium addition at different time points after infection. As part of this experiment, different medium additions were examined.
-
3.
Seventy‐percent medium exchange a short time (8 h) after infection.
3.4.1. Twenty‐percent medium exchange
To assess the impact of medium exchange, a 20% exchange was implemented 24 h after infection. The rationale for the exchange (removing and adding medium) was as follows: after 24 h metabolic byproducts may need to be removed so taking out and replacing 20% of the reactor volume can assist. On the other hand, the virus has begun to accumulate in the medium, though, by far the majority of virus production takes place after 24 h. Taking 20% of already produced virions can influence the titer, but the benefits of fresh medium might be greater. Virus titers after 48 h were compared between reactors with and without medium exchange (2.5 × 108 PFU/ml). Figure 5a indicates that exchanging 20% of the medium 24 h postinfection had no effect on virus titer.
Figure 5.
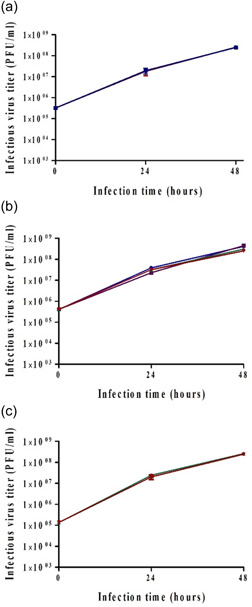
Effect of 20% medium exchange (a), medium additions (b), and 70% medium exchange (c) on virus titer. Reactors were inoculated with 125,000 cells/Fibra‐Cel and cells were grown for 6 days under the same conditions. At the end of the growing phase, 900,000 cells/Fibra‐Cel were estimated. Infections were performed with 0.1 MOI. Virus titers were monitored for 2 days after infection. Bars represent the mean of samples from three different replicates ± standard deviation. MOI, multiplicity of infection; PFU, plaque‐forming unit
3.4.2. Medium addition at different time points
Next, the benefit of adding 20% medium was tested. The advantage of adding medium (vs. exchanging) is that the cells receive new substances needed for their growth but already produced virions are not discarded. Two medium additions were tested, each of 10%, one at 6 h after infection and the other at 24 h. 1. Flex‐20 which is the normal growth medium; 2. Flex‐10 which is another serum‐free medium from the same company (Biological Industries); and 3. Serum‐free medium from another company (Gibco). All these media are suitable for Vero cells adapted to serum‐free growth. As a control, a reactor with no medium additions was included (the additional 20% media volume and the dilution effect were considered in titer calculations). The results (Figure 5b) indicate a positive influence for Flex media additions of about twofold on virus titer (purple and blue [4.5 × 108 PFU/ml]). There was no significant effect of Gibco medium addition compared to control (2.5 × 108 PFU/ml). In light of these results, and the fact that Flex‐20 is the usual medium in a cell growth phase, Flex‐20 medium additions at 6 and 24 h postinfection were tested in the BioBLU 5p process bioreactors and were found to have a similar positive effect.
3.4.3. Seventy‐percent medium exchange
Previous experiments had shown that exchanging 20% of the media 24 h postinfection did not improve the virus titer but the addition of a total of 20% of the medium volume (10% at each time point) had an improvement effect. The next experiment examined the effect of 70% medium exchange a few hours postinfection. Since the virions are secreted to the medium, it is important to exchange the medium in an early stage, to avoid virion loss. The virus does not enter cells immediately, and our results suggested that 6–8 h postinfection, most of the virions were inside cells (data not shown). During that time, some metabolic byproducts from cell growth had already accumulated in the medium so medium exchange could be beneficial without losing virions
To test the impact of high volume exchange, 70% of the medium was exchanged 8 h postinfection, and virus titers were compared to reactors with no medium exchange. The results show there is no difference between titers (Figure 5c, 2.5 × 108 PFU/ml). These results support previous findings suggesting that at 6–8 h postinfection, most of the produced virions are inside cells. The encouraging results have led us to conduct the same experiments in the process of BioBLU 5p bioreactors. Comparing virus titers in bioreactors after 70% medium exchange 8 h postinfection to those with no exchange indicated no effect on virus titers. To assess the beneficial effect of medium exchange on the removal of waste products, the HCP levels were compared. Lower HCP values were found in BioBLU 5p bioreactors after 70% medium exchange, supporting the hypothesis that contaminant removal was beneficial for such exchange. On the basis of these results, 70% of medium exchanges were incorporated in the final process of virus vaccine production.
3.5. Does glucose have an effect on virus titer?
Glucose metabolism in glycolysis is the source of energy. Cells utilize glucose as their primary source for metabolism and they consume it constantly from the medium. Consequently, during the growth phase, samples of medium were taken and glucose consumption was tracked as an indicator to cell growth and cell numbers. Depending on the measured glucose concentration, during cell growth, medium exchange was performed to maintain a glucose concentration above 1 g/L. However, after infection, it was not clear that keeping glucose level >1 g/L was essential for virus production. Previous studies of VSV‐based production had reported contrary information regarding the importance of glucose levels during the production phase (Gélinas et al., 2019).
Thus, the prominence of glucose levels was tested in the following experiment: reactors with the same number of cells were infected at 0.1 MOI. For some reactors, the level of glucose was maintained >1 g/L by adding a suitable amount of glucose from a stock of 200 g/L, while the others were left untouched. In the latter reactors, glucose levels decreased to <1 g/L and totally consumed. The resulting virus titers are shown in Figure 6. Similar titers (2.5 × 108 PFU/ml) were obtained, indicating that during the virus production phase, it is not essential to keep glucose level >1 g/L.
Figure 6.
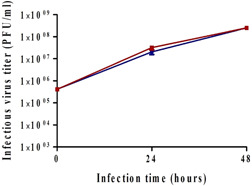
Effect of glucose concentration on virus titer. Reactors were inoculated with 125,000 cells/Fibra‐Cel and cells were grown for 6 days under the same conditions. At the end of the growing phase, 900,000 cells/Fibra‐Cel were estimated. Infections were performed with 0.1 MOI. The level of glucose was monitored during the virus production phase. In two reactors, the level of glucose was maintained >1 g/L by adding glucose (from a 200 g/L stock). No additions of glucose were made to the other two reactors. Virus titers were monitored for 2 days after infection. MOI, multiplicity of infection; PFU, plaque‐forming unit
4. CONCLUSION
In the current study, we demonstrated for the first time the amenability of the Ambr15 cell culture reactor system for the development and optimization of a scalable SARS‐CoV‐2 vaccine production process that involves macrocarriers. The very promising results generated in this study provided the basis for process optimization of the rVSV‐spike Fibra‐Cel‐based platform in our process BioBLU 5p bioreactors.
Various process parameters were found to optimize the production process and maximize virus yield. First, cell density and MOI were evaluated and their working ranges were determined. The results indicated that for optimal process, a biomass >600,000 cells/Fibra‐Cel was needed and MOI <0.1 (with at least two logs flexibility). Then, optimal conditions were established: two medium additions of 10% each at 6 and 24 h postinfection; 70% media exchange 8 h postinfection; and a temperature shift to 34°C at the virus production phase. These results deepen our knowledge of the process and the effect of each parameter on the outcome. All conditions that were found beneficial in the Ambr15 system were subsequently tested in the BioBLU 5p vaccine production process bioreactors and most were found to have the same useful effects, emphasizing the benefit of the simulation in our modified Ambr15 system.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENT
The authors would like to thank Dr. Sandy Livnat for editorial assistance.
Rosen, O. , Jayson, A. , Goldvaser, M. , Dor, E. , Monash, A. , Levin, L. , Cherry, L. , Lupu, E. , Natan, N. , Girshengorn, M. , & Epstein, E. (2022). Optimization of VSV‐ΔG‐spike production process with the Ambr15 system for a SARS‐COV‐2 vaccine. Biotechnology and Bioengineering, 119, 1839–1848. 10.1002/bit.28088
Contributor Information
Osnat Rosen, Email: osnatr@iibr.gov.il.
Eyal Epstein, Email: eyale@iibr.gov.il.
REFERENCES
- Aubrit, F. , Perugi, F. , Léon, A. , Guéhenneux, F. , Champion‐Arnaud, P. , Lahmar, M. , & Schwamborn, K. (2015). Cell substrates for the production of viral vaccines. Vaccine, 33(44), 5905–5912. 10.1016/j.vaccine.2015.06.110 [DOI] [PubMed] [Google Scholar]
- Barrett, P. N. , Terpening, S. J. , Snow, D. , Cobb, R. R. , & Kistner, O. (2017). Vero cell technology for rapid development of inactivated whole virus vaccines for emerging viral diseases. Expert Review of Vaccines, 16(9), 883–894. 10.1080/14760584.2017.1357471 [DOI] [PubMed] [Google Scholar]
- Cino, J. , Mirro, R. , & Kedzierski, S. (2011). An update on the advantages of Fibra‐Cel® disks for cell culture. https://www.eppendorf.com/uploads/media/Application_bioprocess_shakers_incubators_Application-Note-Boo.pdf
- Damodharan, K. , Arumugam, G. S. , Ganesan, S. , Doble, M. , & Thennarasu, S. (2021). A comprehensive overview of vaccines developed for pandemic viral pathogens over the past two decades including those in clinical trials for the current novel SARS‐CoV‐2. RSC Advances, 11(33), 20006–20035. 10.1039/D0RA09668G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco, R. (1952). Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proceedings of the National Academy of Sciences of the United States of America, 38(8), 747–752. 10.1073/pnas.38.8.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi, S. M. , Shen, C. F. , & Gilbert, R. (2019). Optimization of production of vesicular stomatitis virus (VSV) in suspension serum‐free culture medium at high cell density. Journal of Biotechnology, 289, 144–149. 10.1016/j.jbiotec.2018.11.023 [DOI] [PubMed] [Google Scholar]
- Emeny, J. M. , & Morgan, M. J. (1979). Regulation of the interferon system: Evidence that Vero cells have a genetic defect in interferon production. Journal of General Virology, 43(1), 247–252. 10.1099/0022-1317-43-1-247 [DOI] [PubMed] [Google Scholar]
- Fletcher, T. (2014). Using a microscale bioreactor to develop cell culture media explicitly for steady state processes. https://www.researchgate.net/profile/Tom-Fletcher-6/publication/304676645_Using_a_microscale_bioreactor_to_develop_cell_culture_media_explicitly_for_steady_state_processes/links/5776e0c408ae4645d60d81a6/Using-a-microscale-bioreactor-to-develop-cell-cultur
- Gélinas, J.‐F. , Azizi, H. , Kiesslich, S. , Lanthier, S. , Perdersen, J. , Chahal, P. S. , Ansorge, S. , Kobinger, G. , Gilbert, R. , & Kamen, A. A. (2019). Production of rVSV‐ZEBOV in serum‐free suspension culture of HEK 293SF cells. Vaccine, 37(44), 6624–6632. 10.1016/j.vaccine.2019.09.044 [DOI] [PubMed] [Google Scholar]
- Gélinas, J.‐F. , Kiesslich, S. , Gilbert, R. , & Kamen, A. A. (2020). Titration methods for rVSV‐based vaccine manufacturing. MethodsX, 7, 100806. 10.1016/j.mex.2020.100806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein, T. A. , Schwebel, F. , Kress, M. , Loewe, D. , Dieken, H. , Salzig, D. , Weidner, T. , & Czermak, P. (2017). Screening different host cell lines for the dynamic production of measles virus. Biotechnology Progress, 33(4), 989–997. 10.1002/btpr.2432 [DOI] [PubMed] [Google Scholar]
- Guan, W.‐J. , Ni, Z.‐Y. , Hu, Y. , Liang, W.‐H. , Ou, C.‐Q. , He, J.‐X. , Liu, L. , Shan, H. , Lei, C.‐L. , Hui, D. , Du, B. , Li, L.‐J. , Zeng, G. , Yuen, K.‐Y. , Chen, R.‐C. , Tang, C.‐L. , Wang, T. , Chen, P.‐Y. , Xiang, J. , … Zhong, N.‐S. , China Medical Treatment Expert Group for Covid‐19 . (2020). Clinical characteristics of coronavirus disease 2019 in China. The New England Journal of Medicine, 382(18), 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. (K.) , Becken, U. , & Sha, M. (2018). Vero perfusion, packed‐bed vessels intensify vaccine production. Genetic Engineering & Biotechnology News, 38(15), S16–S18. 10.1089/gen.38.15.13 [DOI] [Google Scholar]
- Han, X. (K.) , & Sha, M. (2017). High‐density Vero cell perfusion culture in BioBLU® 5p single‐use vessels. https://www.eppendorf.com/product-media/doc/en/308208/Fermentors-Bioreactors_Application-Note_359_BioBLU-5p_High-Density-Vero-Cell-Perfusion-Culture-BioBLU-5p-Single-Vessels.pdf
- Jayson, A. , Goldvaser, M. , Dor, E. , Monash, A. , Levin, L. , Cherry, L. , Lupu, E. , Natan, N. , Girshengorn, M. , Epstein, E. , & Rosen, O. (2022). Application of Ambr15 system for simulation of entire SARS‐CoV‐2 vaccine production process involving macro‐carriers. Biotechnology progress. [DOI] [PMC free article] [PubMed]
- Kibenge, F. S. B. , & Kibenge, M. J. T. (2016). In Kibenge F. S. B., & Godoy M. G. (Eds.), Orthomyxoviruses of fish (Ch. 19, pp. 299–326). Academic Press. 10.1016/B978-0-12-801573-5.00019-X [DOI] [Google Scholar]
- Kiesslich, S. , & Kamen, A. A. (2020). Vero cell upstream bioprocess development for the production of viral vectors and vaccines. Biotechnology Advances, 44, 107608. 10.1016/j.biotechadv.2020.107608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesslich, S. , Kim, G. N. , Shen, C. F. , Kang, C. Y. , & Kamen, A. A. (2021). Bioreactor production of rVSV‐based vectors in Vero cell suspension cultures. Biotechnology and Bioengineering, 118(7), 2649–2659. 10.1002/bit.27785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesslich, S. , Vila‐Chã Losa, J. P. , Gélinas, J.‐F. , & Kamen, A. A. (2020). Serum‐free production of rVSV‐ZEBOV in Vero cells: Microcarrier bioreactor versus scale‐XTM hydro fixed‐bed. Journal of Biotechnology, 310, 32–39. 10.1016/j.jbiotec.2020.01.015 [DOI] [PubMed] [Google Scholar]
- Linder, A. , Bothe, V. , Linder, N. , Schwarzlmueller, P. , Dahlström, F. , Bartenhagen, C. , Dugas, M. , Pandey, D. , Thorn‐Seshold, J. , Boehmer, D. F. R. , Koenig, L. M. , Kobold, S. , Schnurr, M. , Raedler, J. , Spielmann, G. , Karimzadeh, H. , Schmidt, A. , Endres, S. , & Rothenfusser, S. (2021). Defective interfering genomes and the full‐length viral genome trigger RIG‐I after infection with vesicular stomatitis virus in a replication dependent manner. Frontiers in Immunology, 12, 1302. 10.3389/fimmu.2021.595390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangion, M. , Gélinas, J. ‐F. , Bakhshi Zadeh Gashti, A. , Azizi, H. , Kiesslich, S. , Nassoury, N. , Chahal, P. S. , Kobinger, G. , Gilbert, R. , Garnier, A. , Gaillet, B. , & Kamen, A. (2020). Evaluation of novel HIV vaccine candidates using recombinant vesicular stomatitis virus vector produced in serum‐free Vero cell cultures. Vaccine, 38(50), 7949–7955. 10.1016/j.vaccine.2020.10.058 [DOI] [PubMed] [Google Scholar]
- Monath, T. P. , Fast, P. E. , Modjarrad, K. , Clarke, D. K. , Martin, B. K. , Fusco, J. , Nichols, R. , Heppner, D. G. , Simon, J. K. , Dubey, S. , Troth, S. P. , Wolf, J. , Singh, V. , Coller, B.‐A. , & Robertson, J. S. , Brighton Collaboration Viral Vector Vaccines Safety Working Group . (2019). rVSVΔG‐ZEBOV‐GP (also designated V920) recombinant vesicular stomatitis virus pseudotyped with Ebola Zaire glycoprotein: Standardized template with key considerations for a risk/benefit assessment. Vaccine X, 1, 100009. 10.1016/j.jvacx.2019.100009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillet, C. , Forno, G. , Kratje, R. , & Etcheverrigaray, M. (2009). Suspension‐Vero cell cultures as a platform for viral vaccine production. Vaccine, 27(46), 6464–6467. 10.1016/j.vaccine.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Rosen, O. , Jayson, A. , Natan, N. , & Epstein, E. (2021). A simple method for in situ quantification of cells on carriers. Bio‐Protocol, 11(23), e4254. 10.21769/BioProtoc.4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, O. , Jayson, A. , Natan, N. , Monash, A. , Girshengorn, M. , Goldvaser, M. , Levin, L. , & Epstein, E. (2021). Novel method for quantifying cells on carriers and its demonstration during SARS‐2 vaccine development. Biotechnology and Bioengineering, 137, 197–202. 10.1002/bit.27856 [DOI] [PubMed] [Google Scholar]
- WHO . (2021). The COVID‐19 candidate vaccine landscape and tracker. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- Yahalom‐Ronen, Y. , Tamir, H. , Melamed, S. , Politi, B. , Shifman, O. , Achdout, H. , Vitner, E. B. , Israeli, O. , Milrot, E. , Stein, D. , Cohen‐Gihon, I. , Lazar, S. , Gutman, H. , Glinert, I. , Cherry, L. , Vagima, Y. , Lazar, S. , Weiss, S. , Ben‐Shmuel, A. , … Israely, T. (2020). A single dose of recombinant VSV‐∆G‐spike vaccine provides protection against SARS‐CoV‐2 challenge. Nature Communications, 11(1):6402. 10.1038/s41467-020-20228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura, Y. , & Kawakita, Y. (1963). Studies on SV40 in tissue culture‐preliminary step for cancer research in vitro. Nihon Rinsho, 21, 1201–1215. [Google Scholar]


