Abstract
To overcome the antibiotic resistance mechanism mediated by β-lactamases, small-molecule β-lactamase inhibitors, such as clavulanic acid, have been used. This approach, however, has applied selective pressure for mutations that result in β-lactamases no longer sensitive to β-lactamase inhibitors. On the basis of the structure of β-lactamase inhibitor protein (BLIP), novel peptide inhibitors of β-lactamase have been constructed. BLIP is a 165-amino-acid protein that is a potent inhibitor of TEM-1 β-lactamase (Ki = 0.3 nM). The cocrystal structure of TEM-1 β-lactamase and BLIP indicates that residues 46 to 51 of BLIP make critical interactions with the active site of TEM-1 β-lactamase. A peptide containing this six-residue region of BLIP was found to retain sufficient binding energy to interact with TEM-1 β-lactamase. Inhibition assays with the BLIP peptide reveal that, in addition to inhibiting TEM-1 β-lactamase, the peptide also inhibits a class A β-lactamase and a class C β-lactamase that are not inhibited by BLIP. The crystal structures of class A and C β-lactamases and two penicillin-binding proteins (PBPs) reveal that the enzymes have similar three-dimensional structures in the vicinity of the active site. This similarity suggests that the BLIP peptide inhibitor may have a broad range of activity that can be used to develop novel small-molecule inhibitors of various classes of β-lactamases and PBPs.
β-Lactam antibiotics such as the penicillins and cephalosporins are among the most often used antimicrobial agents. Due to widespread β-lactam antimicrobial use, bacterial resistance has been increasing and now represents a serious threat to the continued use of antibiotic therapy (46). The most common mechanism of bacterial resistance to β-lactam antibiotics is the synthesis of β-lactamases that cleave the amide bond in the β-lactam ring to generate ineffective products (7). On the basis of primary sequence homology, β-lactamases have been grouped into four classes. Classes A, C, and D are active-site serine enzymes that catalyze the hydrolysis of the β-lactam via a serine-bound acyl intermediate (18). Class B enzymes require zinc for activity, and catalysis does not proceed via a covalent intermediate (6, 9, 48). The active-site serine β-lactamases belong to a larger family of penicillin-recognizing enzymes that includes the penicillin-binding proteins (PBPs), which cross-link bacterial cell walls (32). All of these enzymes contain the active-site serine as well as a conserved triad of K(S/T)G between the active-site serine and the C terminus (32). The crystal structures of several class A enzymes, three class C enzymes, and two PBPs show that these enzymes have similar three-dimensional structures, particularly around the active site, suggesting a common evolutionary origin for the penicillin-recognizing enzymes (25). The structures of three class B enzymes confirm the lack of similarity with the serine β-lactamases and PBPs and indicate an independent evolutionary origin for these enzymes (9, 12, 47).
TEM-1 β-lactamase is a class A enzyme encoded by blaTEM-1 (14). Epidemiological studies have shown that TEM-1 is the most common plasmid-encoded β-lactamase in gram-negative bacteria (49). It is able to efficiently hydrolyze penicillins and many cephalosporins; therefore, it is an important source of bacterial resistance to the β-lactam antibiotics (7). The SHV-1 β-lactamase is 68% identical to the TEM-1 β-lactamase and also occurs frequently in gram-negative bacteria (3, 16). The SHV-1 β-lactamase exhibits a substrate hydrolysis profile similar to that of TEM-1 (29). Two approaches have been used to combat TEM-1 and SHV-1 β-lactamase-mediated resistance. First, extended-spectrum cephalosporins such as cefotaxime and ceftazidime were developed, in part, because the TEM-1 and SHV-1 β-lactamases are not able to hydrolyze these antibiotics (10). Second, the coadministration of an inhibitor of β-lactamase, such as clavulanic acid or sulbactam, with a β-lactam antibiotic, such as ampicillin or amoxicillin, has been shown to overcome β-lactamase-mediated resistance (35, 37).
Both of these approaches apply selective pressure for mutations that result in a β-lactamase that either cleaves extended-spectrum cephalosporins or is no longer sensitive to β-lactamase inhibitors (22, 33, 36). Both types of mutations have been found in the genes for TEM-1 and SHV-1 β-lactamases from clinical isolates resistant to these therapies (22, 36). This has led to an increasing problem of resistance to antibiotics and a corresponding diminution of effective therapies for some bacterial infections. The emergence of resistance to β-lactamase inhibitors and the relatively rapid emergence of resistance to new antibiotics imply that the design of new antibiotics must keep pace with the evolution of bacterial resistance.
Naturally occurring antibiotics also include secreted, protein inhibitors of β-lactamase and PBP function. The β-lactamase inhibitor protein (BLIP) is a 165-amino-acid protein produced by the gram-positive soil bacterium Streptomyces clavuligerus (15). S. clavuligerus also produces β-lactam antibiotics such as cephamycins as well as a β-lactamase inhibitor, clavulanic acid (23). BLIP has been shown to bind to and inhibit the TEM-1 β-lactamase with a Ki of 0.1 to 0.6 nM (38, 40, 45). In addition, BLIP binds to and inhibits the class A β-lactamases from Staphylococcus aureus, Bacillus cereus, and Bacillus licheniformis with Ki values of 1 to 3 μM. BLIP does not efficiently bind to class B, C, or D β-lactamases (45).
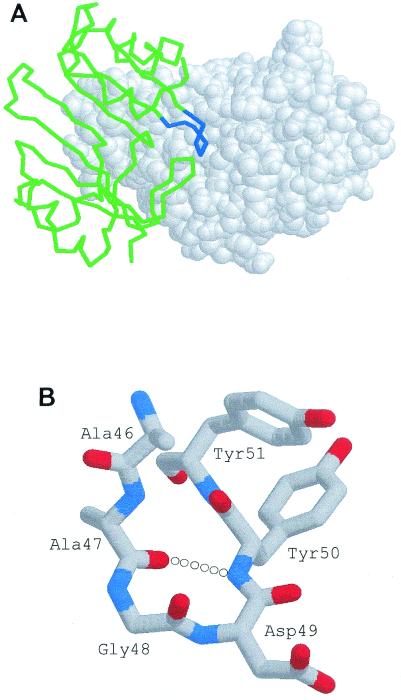
The three-dimensional structures of BLIP alone and BLIP in complex with the TEM-1 β-lactamase have been determined to high resolution (44, 45). The structure of the complex indicates that a type II′ β turn encompassing residues 46 to 51 of BLIP makes critical interactions with the active site of the TEM-1 β-lactamase (Fig. 1) (38, 44). Because of these interactions, it was hypothesized that a peptide that includes turn residues 47 to 50 would retain sufficient binding energy to interact with β-lactamase in the absence of the remaining portion of BLIP (44). If this peptide did inhibit β-lactamase, it could serve as a starting point for the design of peptide analogues that inhibit β-lactamase.
FIG. 1.
(A) Representation of BLIP (green) binding to TEM-1 β-lactamase (white, space-fill model). The region of BLIP from residues 45 to 52 is shown in blue. (B) Structure of the Ala-46 to Tyr-51 peptide extracted from the BLIP structure (44) showing the type II′ β turn generated by residues 47 to 50.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli XL1-Blue [F′::Tn10 proA+B+ lacIq Δ(lacZ) M15/recA1 endA1 gyrA96 (Nalr) thi-1 hsdR17 supE44 relA1 lac; Stratagene, Inc.] (5) was used for transformation of the ligation reaction of plasmids pCM01 and pZZ101. E. coli RB791 (strain W3110 lacIqL8) was used for expression and purification of the TEM-1, SHV-1, IMP-1, and P99 β-lactamases (1, 4). IMP-1 and P99 β-lactamase expression vector pGR32 was constructed as described previously (38).
Peptide synthesis.
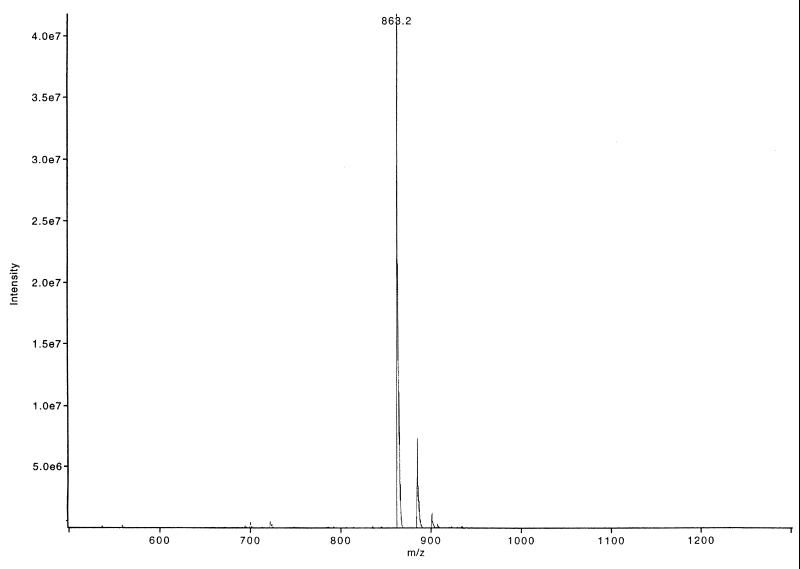
The Biotin-MiniBLIP peptide (N-biotin-Gly-Ser-Gly-Cys-Ala-Ala-Gly-Asp-Tyr-Tyr-Cys-COOH) and the BP46-51 peptide (N-Cys-Ala-Ala-Gly-Asp-Tyr-Tyr-Cys-COOH) were synthesized at the Baylor College of Medicine protein chemistry core with an ABI 433A synthesizer. The synthesized peptides were cyclized by the dropwise addition of ammonium hydroxide to the solution to pH 8.0. The progress of the reaction was monitored by reverse-phase high-pressure liquid chromatography (HPLC), and the final product was purified to >90% homogeneity by reverse-phase HPLC (Fig. 2). The BP41-50 peptide (N-His-Cys-Arg-Gly-His-Ala-Ala-Gly-Asp-Tyr-COOH) was synthesized by Research Genetics, Inc. (Huntsville, Ala.). Disulfide bonds were reduced in the BP46-51 peptide by the addition of 10 mM dithiothreitol (DTT) to the peptide stock prior to performance of the inhibition assays.
FIG. 2.
Mass spectrometry spectrum of HPLC-purified BP46-51. The major peak corresponding to the BP46-51 peptide is labeled with the molecular weight of the peptide.
IMP-1 and P99 cloning.
The blaIMP gene was amplified from pBCAM-52E by PCR (27) with top-strand external primer Imp-SacI0 (5′-GCGGGAGCTCGTGGAAACGGATGAAGGCAC-3′) and bottom-strand external primer Imp-XbaI (5′-GGGCGGGTCTAGAAATTTAGTTGCTTGGTTTTGATGG-3′). The blaP99 gene was amplified from plasmid pHU356 by PCR (17) with top-strand external primer AmpC-1 (5′-CCGCGCGAGCTCCGTTTGTCAGGCACAGTCAAATC-3′) and bottom strand external primer AmpC-2 (5′-CCCCCCTCTAGACCCGGCAATGTTTTACTGTAGCG-3′). A SacI site (underlined) in Imp-SacI0 and AmpC-1 and an XbaI site (underlined) in Imp-XbaI and AmpC-2 allowed the enzyme-digested PCR products to be cloned into SacI- and XbaI-digested pGR32, which contains an inducible Ptrc promoter to control expression of the gene inserts (38). The insertion of each clone was verified by DNA sequence analysis. The resulting IMP-1 expression vector was named pCM01, and the P99 expression vector was named pZZ101.
Purification of β-lactamase proteins.
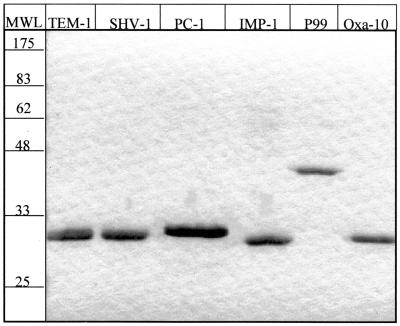
The TEM-1 and SHV-1 β-lactamases were purified to >90% homogeneity with a zinc-chelating Sepharose (fast flow) column (Pharmacia) and by Sephadex G-75 gel filtration chromatography as described previously (8). Fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to estimate the purity of each protein (Fig. 3). The PC-1 β-lactamase was a gift from Osnat Herzberg at the University of Maryland Biotechnology Institute, and the OXA-10 β-lactamase was a gift from Shariar Mobashery of Wayne State University.
FIG. 3.
Purified β-lactamase proteins. The six β-lactamase proteins were purified to >90% homogeneity, as determined by SDS-PAGE. The values on the molecular weight ladder (MWL) are expressed in units of kilodaltons.
For IMP-1 β-lactamase purification, plasmid pCM01 was transformed into E. coli RB791 by electroporation. The resulting strain was grown overnight with shaking at 37°C in 25 ml of Luria-Bertani (LB) medium containing 12.5 μg of chloramphenicol per ml. The overnight culture was diluted 1:500 into 1 liter of LB medium containing 12.5 μg of chloramphenicol per ml and was grown at 37°C to an optical density at 600 nm (OD600) of approximately 0.6. The β-lactamase gene was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and further incubation at 25°C for 4 h.
Following induction the cells were pelleted and the supernatant containing the secreted, soluble β-lactamase was concentrated to 100 ml with an Amicon Centriprep-10 concentrator (Millipore Corp.). The concentrated solution was dialyzed overnight against 4 liters of buffer H (50 mM HEPES [pH 7.5], 50 μM ZnSO4). The dialyzed supernatant was filtered and loaded onto a HiTrap SP Sepharose cation-exchange column (Amersham Pharmacia Biotech) equilibrated with buffer H. The enzyme was eluted with a linear gradient of 0 to 0.5 M NaCl in buffer H. Fractions containing β-lactamase activity were identified by nitrocefin hydrolysis. Active fractions were pooled and concentrated to a 2-ml volume with an Amicon Centriprep-10 concentrator, followed by dialysis against 4 liters of buffer H. The β-lactamase was further purified by Sephadex G-75 filtration chromatography with buffer H. The purified active enzyme was pooled and concentrated with an Amicon Centriprep-10 concentrator. The purity of the enzyme was verified by SDS-PAGE (Fig. 3). All preparations were >90% pure and were stored at −80°C.
For purification of the P99 β-lactamase, plasmid pZZ101 was transformed into E. coli RB791 by electroporation. An overnight culture was grown by shaking at 37°C in 25 ml of LB medium containing 12.5 μg of chloramphenicol per ml. The overnight culture was diluted 1:1,000 into 1 liter of LB medium containing 12.5 μg of chloramphenicol per ml and was grown at 37°C to an OD600 of approximately 0.4. The β-lactamase gene was induced by the addition of IPTG to a final concentration of 0.2 mM and incubation at 25°C overnight.
Following overnight induction, β-lactamase was isolated by an osmotic shock procedure (34). The solution obtained by osmotic shock was dialyzed against 4 liters of 25 mM morpholineethanesulfonic acid (MES) buffer (pH 6.1) at 4°C overnight. The dialyzed supernatant was filtered, and the β-lactamase was concentrated as described above for the IMP-1 β-lactamase. The enzyme was purified by ion-exchange chromatography in 25 mM MES buffer (pH 6.1). The enzyme was further purified by gel filtration in 25 mM phosphate buffer (pH 7.6), and the purity was verified by SDS-PAGE (Fig. 3).
Biotin-MiniBLIP ELISA.
Enzyme-linked immunosorbent assay (ELISA) experiments with TEM β-lactamase and the Biotin-MiniBLIP peptide were performed in 96-well microtiter plates precoated with the biotin binding protein NeutrAvidin (Pierce). A total of 0.2 ml of Biotin-MiniBLIP peptide in 1× Tris-buffered saline (TBS; pH 7.5) (42) was added to coated wells at a final peptide concentration of 10 μg/ml. The plates were gently agitated at 25°C for 15 min, followed by four washes with 0.2 ml of wash buffer (1× TBS [pH 7.5], 1 mg of bovine serum albumin [BSA] per ml, 0.5 g of Tween 20 per liter) to remove unbound peptide. The wells were blocked for nonspecific binding with 0.2 g of dry-milk powder per liter in wash buffer for 1 h at 25°C, followed by six washes with wash buffer. A total of 100 μg of the TEM-1 β-lactamase per ml or 80 μg of the control protein, maltose-binding protein (MBP), per ml in 0.2 ml of wash buffer was added to the Biotin-MiniBLIP-coated wells in duplicate; and the plates were incubated with slight agitation for 2 h at 25°C. The wells were washed six times with wash buffer to remove unbound protein. Bound TEM-1 β-lactamase and MBP were detected with anti-β-lactamase polyclonal antibodies and anti-MBP polyclonal antibodies (New England Biolabs, Inc.), respectively. Bound antibodies were detected with antirabbit antibodies conjugated to horseradish peroxidase (Amersham).
The sera with anti-β-lactamase and anti-MBP antibodies were tested for reactivity by coating microtiter wells with 0.2 ml of a 10-μg/ml solution of TEM-1 β-lactamase, MBP, Biotin-MiniBLIP, or BSA in 0.05 M Na2CO3 (pH 9.6) overnight at 4°C. The wells were blocked as described above, followed by 10 washes with 0.2 ml of wash buffer. Immobilized proteins were detected as described above by using sera with either anti-β-lactamase or anti-MBP antibodies.
BLIP inhibition assays.
BLIP inhibition assays were performed as described previously (38). Briefly, various concentrations of BLIP, BP46-51, BP41-50, or the control peptide, protein kinase C substrate (N-Pro-Ser-Arg-Thr-Leu-Ser-Val-Ala-Ala-Lys-Lys-COOH; Sigma Chemical Co.), were incubated in the presence of 1 nM TEM-1 β-lactamase, 1 nM SHV-1 β-lactamase, 3 nM PC-1 β-lactamase, 3 nM OXA-10 β-lactamase, 0.1 nM IMP-1 β-lactamase, or 0.1 nM P99 β-lactamase for 2 h at 25°C. For all enzymes except IMP-1 the enzyme-inhibitor incubation was in 0.05 M phosphate buffer (pH 7.0) containing 1 mg of BSA per ml. For the IMP-1 β-lactamase the incubation was in buffer H containing 1 mg of BSA per ml. Following the 2-h incubation, a β-lactam substrate was added at a concentration at least 10-fold lower than the Km of the substrate for the β-lactamase being tested. For the TEM-1 and SHV-1 β-lactamases the β-lactam substrate cephaloridine was used. Nitrocefin was used to assay the activities of the remaining β-lactamase enzymes. Hydrolysis of the β-lactam substrate was monitored at A260 for cephaloridine and A500 for nitrocefin. Equilibrium dissociation constants (Ki) were determined for each enzyme as described previously (38).
RESULTS
Peptide binding to TEM-1 β-lactamase.
To test the hypothesis that the β turn from BLIP residues 47 to 50 can bind to β-lactamase, a peptide, Biotin-MiniBLIP, with the sequence N-biotin-Gly-Ser-Gly-Cys-Ala-Ala-Gly-Asp-Tyr-Tyr-Cys-COOH was synthesized. This sequence consists of BLIP residues Ala-46 to Tyr-51 flanked on either side by cysteine residues. After synthesis the peptide was oxidized to form a disulfide bond between the cysteines to create a cyclic peptide. The N terminus of the peptide contains two glycine residues, a serine residue, and biotin. The biotin was included to facilitate characterization of the binding properties of the peptide. The two glycine residues serve as a flexible spacer between the cyclic peptide and the biotin, while serine was included to enhance solubility.
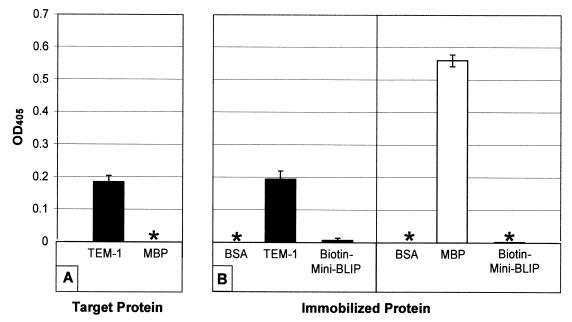
Biotin-MiniBLIP was initially tested for binding to TEM-1 β-lactamase by ELISA. The peptide was immobilized in a NeutrAvidin-coated microtiter well by taking advantage of the biotin moiety on the peptide. Soluble TEM-1 β-lactamase was then added to the well and allowed to bind, and the wells were washed extensively. Bound β-lactamase was detected with sera containing anti-TEM-1 β-lactamase polyclonal antibody. As a control, soluble MBP was added to the immobilized peptide, washed, and detected with sera containing an anti-MBP polyclonal antibody. As seen in Fig. 4A, the TEM-1 β-lactamase was retained in the microtiter well, while the MBP was not. As a further control, TEM-1 β-lactamase and MBP were immobilized directly into microtiter wells and detected with either the anti-TEM-1 β-lactamase or anti-MBP polyclonal sera (Fig. 4B). In both cases a strong signal was obtained, indicating that the sera are active and, therefore, that the failure to detect MBP in the peptide binding experiment was not due to a failure of the anti-MBP detection antibody. In addition, the failure to detect a signal in ELISA wells coated with BSA or Biotin-MiniBLIP with sera with either antibody indicates that the reactivity of each antibody is specific for the target protein. These results suggest that the TEM-1 β-lactamase binds to the cyclic biotin-peptide.
FIG. 4.
(A) ELISA of TEM-1 β-lactamase binding to immobilized Biotin-MiniBLIP peptide. A total of 10 μg of Biotin-MiniBLIP per ml was immobilized on NeutrAvidin-coated microtiter wells. After extensive washing to remove unbound protein, TEM-1 β-lactamase and MBP were tested for their levels of binding to the immobilized peptide. Bound TEM-1 β-lactamase was assayed with a polyclonal sera directed toward the TEM-1 β-lactamase. Bound MBP was assayed with a polyclonal sera directed toward MBP. (B) As a control, the sera with anti-β-lactamase and anti-MBP antibodies were tested for their reactivities against the indicated proteins. A total of 10 μg of each protein per ml was immobilized in microtiter wells and was then probed with polyclonal sera directed toward either the TEM-1 β-lactamase (B, left panel) or MBP (B, right panel). All datum points are the averages for two samples. An asterisk indicates that no detectible ELISA signal was observed for the samples.
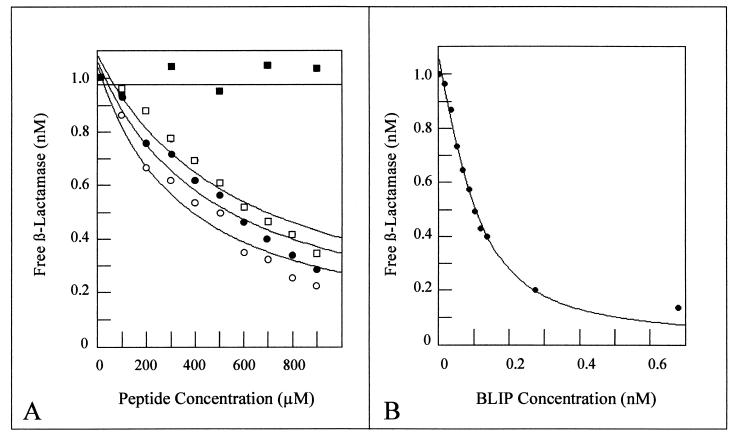
The BLIP peptide was tested for binding and inhibition of TEM-1 β-lactamase in solution by a quantitative inhibition assay developed previously (38). Because the biotin moiety was not required for these assays, the cyclic BLIP peptide, BP46-51, with the sequence N-Cys-Ala-Ala-Gly-Asp-Tyr-Tyr-Cys-COOH, was used. This peptide lacks the N-terminal biotin and peptide linker of Biotin-MiniBLIP but retains the two cysteine residues for generation of a cyclized peptide. For the inhibition assay, BP46-51 was incubated with the TEM-1 β-lactamase for 2 h to achieve binding equilibrium. After the incubation, the β-lactam antibiotic cephaloridine was added and its rate of hydrolysis was monitored. The concentration of free β-lactamase was calculated from the rate of cephaloridine hydrolysis in the presence of a given quantity of peptide. Fitting of the data obtained when various concentrations of peptide were incubated with 1 nM TEM-1 β-lactamase resulted in a Ki of 603 μM (Table 1). As a control, the same assay was performed with a peptide with the sequence N-Pro-Ser-Arg-Thr-Leu-Ser-Val-Ala-Ala-Lys-Lys-COOH. This peptide is a protein kinase C substrate, and its sequence has no homology to the BLIP peptide or to any sequence within the BLIP protein. As seen in Fig. 5, the control peptide does not inhibit the TEM-1 β-lactamase. This result indicates that peptide-mediated inhibition of TEM-1 β-lactamase is specific to the BLIP peptide sequence. Thus, two independent assays have shown that the cyclic peptide from residues 46 to 51 can bind to the TEM-1 β-lactamase. However, the binding is approximately 106-fold weaker than the binding of the TEM-1 β-lactamase by the wild-type BLIP molecule. The weaker binding presumably reflects the loss of a large number of protein-protein contacts that are present in the wild-type BLIP–β-lactamase complex (44). Nevertheless, the binding is experimentally significant and could be further optimized by cycles of mutagenesis and selection.
TABLE 1.
BLIP and BLIP peptide Kis for various β-lactamases
| β-Lactamase |
Ki (μM)
|
||||
|---|---|---|---|---|---|
| BLIP | Oxidized BP46-51 | Reduced BP46-51 | BP41-50 | Protein kinase C substrate | |
| TEM-1 | 0.0003 | 603 | 488 | 359 | — |
| SHV-1 | 1.0 | 680 | 420 | 458 | — |
| PC-1 | —a | — | 1,400 | — | — |
| IMP-1 | — | — | NDb | — | — |
| P99 | — | — | 135 | — | — |
| OXA-10 | — | — | — | — | — |
—, for those peptide-enzyme combinations for which there was no detectable inhibition, peptide was tested up to 1,000 μM.
ND, not determined.
FIG. 5.
(A) Inhibition assays of TEM-1 β-lactamase using various BLIP-derived peptides. Peptide binding and inhibition of β-lactamase were determined by measuring the amount of free β-lactamase at various peptide concentrations. β-Lactamase concentrations were 1.0 nM in all assays. For each set of datum points a nonlinear regression fit was calculated as described previously (38). Closed squares, protein kinase C peptide; open squares, cyclic BP46-51 peptide; closed circles, reduced BP46-51 peptide; open circles, BP41-50 peptide. Each datum point is the average for two independent experiments. (B) Inhibition assay of TEM-1 β-lactamase by wild-type BLIP. A Ki of 0.23 nM was determined by using a nonlinear regression fit as described above.
Peptide binding to other β-lactamases.
The SHV-1 β-lactamase is a class A enzyme that is 68% identical to the TEM-1 β-lactamase. The crystal structures of both enzymes have been solved, and the active-site regions are superimposable (26, 45). Nevertheless, BLIP binds to and inhibits the TEM-1 β-lactamase with a Ki of 0.23 nM (Fig. 5B), while it inhibits SHV-1 10,000-fold less efficiently, with a Ki of 1 μM (38). Therefore, it was of interest to determine if the SHV-1 β-lactamase is inhibited by the BP46-51 peptide. By using the quantitative assay described above, the peptide was found to inhibit the SHV-1 β-lactamase with a Ki of 680 μM (Table 1). This value is similar to that obtained for inhibition of TEM-1 β-lactamase. The high degree of similarity between the SHV-1 and the TEM-1 active-site pockets predicts that a peptide that binds within the active-site pocket would exhibit similar affinities for both enzymes. The result also suggests that the large difference in binding affinity of wild-type BLIP for SHV-1 versus TEM-1 is due to a steric clash between BLIP and SHV-1 somewhere outside of the region of the enzyme bound by the peptide, i.e., outside the active-site pocket. It has been suggested that the weaker inhibition of SHV-1 by BLIP compared to that of TEM-1 is due to sequence differences between the enzymes at SHV-1 residues Ala-124, Gln-100, Asp-104, and Arg-215 (26). All of these positions are outside the active-site pocket of the enzyme and may result in the weaker binding of the enzyme-inhibitor complex by generating multiple perturbations along the interface of the enzyme-inhibitor complex (26). These changes would not affect the binding of the BP46-51 peptide since the residues implicated in the disruption of BLIP binding to SHV-1 are found outside the active-site pocket region where BP46-51 is expected to bind.
The BP46-51 peptide was also tested for binding and inhibition of a number of other β-lactamases. The S. aureus PC-1 β-lactamase is a class A β-lactamase that is approximately 40% identical to the TEM-1 β-lactamase (2). As seen in Table 1, neither wild-type BLIP nor the cyclic peptide inhibited the PC-1 enzyme. This could be due to the conformational differences observed between the TEM-1 and PC-1 β-lactamase active-site pockets (20, 43). In addition, neither wild-type BLIP nor the cyclic peptide inhibited the P99 β-lactamase of class C, the OXA-10 β-lactamase of class D, or the IMP-1 metallo-β-lactamase of class B (Table 1). The lack of binding and inhibition is presumably due to the sequence and structural divergence of these enzymes compared to the sequence and the structure of the TEM-1 β-lactamase (11, 19, 31).
An important question is whether the disulfide bond in the cyclic peptide is critical for the binding and inhibition of β-lactamase. To answer this question, the cyclic peptide was reduced with 10 mM DTT and tested for inhibition of β-lactamase as described above. As seen in Table 1, the disulfide bond is not required for inhibition and may in fact be somewhat detrimental. For example, the reduced peptide binds to TEM-1 and SHV-1 somewhat more tightly than the cyclic peptide does, with Ki values of 488 and 420 μM, respectively (Fig. 5A). In addition, the S. aureus PC-1 enzyme and the class C P99 enzyme are inhibited by the reduced peptide but not the cyclic peptide. As a control, each of the β-lactamases assayed was tested for inhibition by DTT. Since the addition of DTT resulted in the inactivation of only the zinc metallo-β-lactamase, IMP-1, but had no inhibitory activity for the serine β-lactamases assayed (data not shown), inhibition of the serine β-lactamases was due to the BP46-51 peptide and was not due to the addition of DTT. Taken together, these results suggest the disulfide bond restrains the peptide in a conformation that is not optimal for binding to the β-lactamases assayed.
Design of a peptide inhibitor based on contact residues.
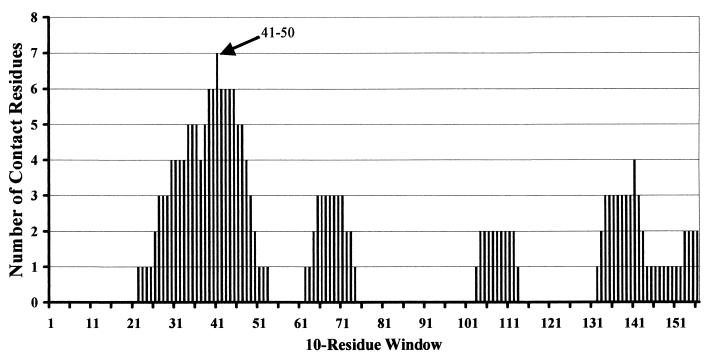
The X-ray structure of BLIP in complex with the TEM-1 β-lactamase is known and indicates that several BLIP residues make direct contact with the TEM-1 β-lactamase (44). We next wanted to assess whether a peptide could be designed to bind to and inhibit the TEM-1 β-lactamase on the basis of maximization of the number of contact residues known from the crystal structure in the peptide sequence. To implement this strategy, a sliding window of 10 amino acids was moved through the BLIP sequence. For each of the 158 independent windows, the number of contact residues was calculated on the basis of the crystal structure. It is apparent from this analysis that of the 23 residues of BLIP that make contact with the TEM-1 β-lactamase, most contact residues are concentrated in the region of the β turn from residues 46 to 51 that inserts into the active-site pocket of the TEM-1 β-lactamase (Fig. 6). The highest percentage of contact residues is found in the window that encompasses residues 41 to 50, which has the sequence N-His-Cys-Arg-Gly-His-Ala-Ala-Gly-Asp-Tyr-COOH. Seven of the 10 residues in the window from residues 41 to 50 are directly in contact with the TEM-1 β-lactamase in the BLIP–TEM-1 β-lactamase cocrystal structure (44).
FIG. 6.
Sliding amino acid window. A window of 10 contiguous BLIP residues was scanned across the BLIP sequence starting at position 1. Each set of 10 contiguous residues was scored for the number of residues that make contact with the TEM-1 β-lactamase on the basis of the cocrystal structure of TEM-1 β-lactamase and BLIP (44). The positions of BLIP amino acid residues that come into contact with the TEM-1 β-lactamase were obtained from the X-ray structure of the BLIP-TEM-1 β-lactamase complex (44). An arrow indicates the 10-residue window representing the BP41-50 peptide. The starting residue for every 10th window is indicated below the graph.
A 10-residue peptide containing residues 41 to 50 of BLIP was synthesized and tested for binding to the TEM-1 β-lactamase by the inhibitor assay described above. The BP41-50 peptide inhibits the TEM-1 β-lactamase with a Ki of 359 μM, which is slightly lower than the Ki for the reduced BP46-51 peptide (Fig. 5 and Table 1). BP41-50 was also found to inhibit the SHV-1 β-lactamase, with a Ki of 458 μM. In contrast, the peptide did not inhibit the S. aureus PC-1 enzyme or the class B, C, or D enzymes in Table 1. These results indicate that a peptide designed to bind to β-lactamase on the basis of the cocrystal structure of the complex does bind to and inhibit β-lactamase, albeit weakly. The relatively weak binding compared to the level of binding of the wild-type BLIP–β-lactamase complex is not surprising, in that the majority of the contacts found in the wild-type complex are not possible with the 10-amino-acid peptide.
DISCUSSION
The reduction of proteins to short peptides that retain binding function is an important intermediate step toward the development of novel small-molecule drugs that disrupt protein-protein interactions (13). Over the past decade, several protein inhibitors have been converted into small-molecule inhibitors that bind to their target substrates with high affinities (28, 30, 50). An example is an inhibitor of the interaction between fibrinogen and the glycoprotein IIb/IIIa (GPIIb/IIIa) that has been developed. GPIIb/IIIa is a membrane protein that mediates aggregation of platelets (39). In response to stimulation by agonists such as thrombin, this protein undergoes a conformational change that allows it to bind to fibrinogen (39). Binding is mediated by β-turn regions within fibrinogen that contain the sequence Arg-Gly-Asp (RGD). A cyclic peptide with the sequence RGDS flanked by cyclic disulfide analogues binds to GPIIb/IIIa and is a potent inhibitor of platelet aggregation (41).
Importantly, the Arg-Gly-Asp sequence motif of fibrinogen is equivalent to the BLIP 47-Ala-Gly-Asp-49 sequences. The cocrystal structure of TEM-1 β-lactamase and BLIP shows that the loop in domain 1 of BLIP that contains the 47-Ala-Gly-Asp-49 sequence forms a type II′ β turn (Fig. 1) (44). This β turn appears to be essential for a proper fit of the loop in the active-site pocket of the TEM-1 β-lactamase by preventing steric clashes with residues that line the enzyme's active site. In addition, the β turn allows the proper positioning of Asp-49 in the active site, which binds to four conserved residues important for substrate binding and hydrolysis by the TEM-1 β-lactamase (44).
To help maintain the proper fit of the constructed BP46-51 peptide in the active-site pocket of the TEM-1 β-lactamase and to correctly position Asp-49 in the enzyme, we introduced two cysteine residues to the NH2 and COOH termini of the peptide. This feature allowed cyclization of the BP46-51 peptide to potentially generate the type II′ β turn similar to that found in BLIP and the RGD peptide inhibitor of GPIIb/IIIa, with Gly and Asp occupying positions i + 1 and i + 2 of the β turns, respectively. Although the cyclized BP46-51 peptide inhibited the TEM-1 and SHV-1 β-lactamases, it was found that the reduced form of the peptide was a slightly better inhibitor of these enzymes. The reduced peptide also inhibited the PC-1 and P99 β-lactamases, which were not inhibited by the cyclized peptide. Although it cannot be determined from these data if a type II′ β turn is generated in the cyclic or reduced peptides, the data suggest that the formation of the disulfide bond in the BP46-51 peptide generates a conformation that is not optimal for binding of the peptide in the active sites of the β-lactamase enzymes.
In aqueous solution peptides may assume multiple conformations, some of which may be favorable for binding to the target substrate. On the basis of nuclear magnetic resonance analysis, linear RGD peptides have been found to form type II β turns in solution, although at a low efficiency (24). Similarly, a subset of the reduced BP46-51 peptides may adopt a conformation in solution that is compatible with binding to various β-lactamases. Presumably, this structure is different from that of the cyclic peptide.
Constraining the BP46-51 peptide in a more optimal conformation than the cyclized peptide generated here may greatly increase the inhibitory activity of the BP46-51 peptide for β-lactamase. One excellent example has been with the tripeptide RGD sequence, in which the cyclized peptide was constrained in a rigid β-turn structure (21). This was accomplished by the addition of nonpeptidic constituents that mimic the structural properties of the cyclic peptide and the substitution of methyl groups for hydrogen atoms in the peptide backbones. The result is a dramatic decrease in the rotational freedom of the peptide in solution with a corresponding increase in affinity for GPIIb/IIIa of 3 orders of magnitude (21). An additional advantage of replacing residues with a nonpeptidic substitute is an increased stability to proteolysis (21).
Previous studies have shown that the binding between the TEM-1 β-lactamase and BLIP is not optimal and can be improved (38, 40). Phage display has been shown to be an effective means of optimizing protein-protein interactions and therefore could be used to improve the binding between the BP46-51 peptide and various β-lactamases. Further binding improvements can also be achieved by restricting the selected peptides to preferred binding conformations, as described above, to generate potent tightly binding β-lactamase inhibitors. These methods may generate inhibitors of not only class A β-lactamases but also β-lactamases of other classes and PBPs that are not inhibited by the current repertoire of β-lactamase inhibitors.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI32956 to Timothy Palzkill.
We thank Christina Materon and Zhen Zhang for purifying and supplying β-lactamase enzymes for kinetic assays and Hiram Gilbert for assisting with peptide inhibition data analysis. We also thank Osnat Herzberg and Shahriar Mobashery for supplying the PC-1 β-lactamase and the OXA-10 β-lactamase, respectively, and Dasantila Golemi for providing information on OXA-10 kinetics.
REFERENCES
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P, Coulson A F, Frere J M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthelemy M, Peduzzi J, Ben Yaghlane H, Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988;231:217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- 4.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 6.Bush K. Metallo-beta-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantu C, III, Huang W, Palzkill T. Selection and characterization of amino acid substitutions at residues 237–240 of TEM-1 beta-lactamase with altered substrate specificity for aztreonam and ceftazidime. J Biol Chem. 1996;271:22538–22545. doi: 10.1074/jbc.271.37.22538. [DOI] [PubMed] [Google Scholar]
- 9.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere J M, Dideberg O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmine A A, Brogden R N, Heel R C, Speight T M, Avery G S. Cefotaxime. A review of its antibacterial activity, pharmacological properties and therapeutic use. Drugs. 1983;25:223–289. doi: 10.2165/00003495-198325030-00001. [DOI] [PubMed] [Google Scholar]
- 11.Concha N O, Janson C A, Rowling P, Pearson S, Cheever C A, Clarke B P, Lewis C, Galleni M, Frere J M, Payne D J, Bateson J H, Abdel-Meguid S S. Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry. 2000;39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- 12.Concha N O, Rasmussen B A, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham B C, Wells J A. Minimized proteins. Curr Opin Struct Biol. 1997;7:457–462. doi: 10.1016/s0959-440x(97)80107-8. [DOI] [PubMed] [Google Scholar]
- 14.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965;208:239–241. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 15.Doran J L, Leskiw B K, Aippersbach S, Jensen S E. Isolation and characterization of a beta-lactamase-inhibitory protein from Streptomyces clavuligerus and cloning and analysis of the corresponding gene. J Bacteriol. 1990;172:4909–4918. doi: 10.1128/jb.172.9.4909-4918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Bois S K, Marriott M S, Amyes S G. TEM- and SHV-derived extended-spectrum beta-lactamases: relationship between selection, structure and function. J Antimicrob Chemother. 1995;35:7–22. doi: 10.1093/jac/35.1.7. [DOI] [PubMed] [Google Scholar]
- 17.Dubus A, Wilkin J M, Raquet X, Normark S, Frere J M. Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the Lys-Thr(Ser)-Gly triad. Biochem J. 1994;301:485–494. doi: 10.1042/bj3010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghuysen J M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 19.Golemi D, Maveyraud L, Vakulenko S, Tranier S, Ishiwata A, Kotra L P, Samama J-P, Mobashery S. The first structural and mechanistic insights for class D β-lactamases: evidence for a novel catalytic process for turnover of β-lactam antibiotics. J Am Chem Soc. 2000;122:6132–6133. [Google Scholar]
- 20.Herzberg O, Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science. 1987;236:694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- 21.Jackson S, DeGrado W, Dwivedi A, Parthasarathy A, Higley A, Krywko J, Rockwell A, Markwalder J, Wells G, Wexler R, Mousa S, Harlow R. Template-constrained cyclic peptides: design of high-affinity ligands for GPIIb/IIIa. J Am Chem Soc. 1994;116:3220–3230. [Google Scholar]
- 22.Jacoby G A, Medeiros A A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen S E. Biosynthesis of cephalosporins. Crit Rev Biotechnol. 1986;3:277–301. [Google Scholar]
- 24.Johnson W C, Jr, Pagano T G, Basson C T, Madri J A, Gooley P, Armitage I M. Biologically active Arg-Gly-Asp oligopeptides assume a type II beta-turn in solution. Biochemistry. 1993;32:268–273. doi: 10.1021/bi00052a034. [DOI] [PubMed] [Google Scholar]
- 25.Knox J R, Moews P C, Frere J M. Molecular evolution of bacterial beta-lactam resistance. Chem Biol. 1996;3:937–947. doi: 10.1016/s1074-5521(96)90182-9. [DOI] [PubMed] [Google Scholar]
- 26.Kuzin A P, Nukaga M, Nukaga Y, Hujer A M, Bonomo R A, Knox J R. Structure of the SHV-1 beta-lactamase. Biochemistry. 1999;18:5720–5727. doi: 10.1021/bi990136d. [DOI] [PubMed] [Google Scholar]
- 27.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frere J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leatherbarrow R J, Salacinski H J. Design of a small peptide-based proteinase inhibitor by modeling the active-site region of barley chymotrypsin inhibitor 2. Biochemistry. 1991;30:10717–10721. doi: 10.1021/bi00108a016. [DOI] [PubMed] [Google Scholar]
- 29.Lee K Y, Hopkins J D, O'Brien T F, Syvanen M. Gly-238-Ser substitution changes the substrate specificity of the SHV class A beta-lactamases. Proteins. 1991;11:45–51. doi: 10.1002/prot.340110106. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Tom J Y, Oare D, Yen R, Fairbrother W J, Wells J A, Cunningham B C. Minimization of a polypeptide hormone. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 31.Lobkovsky E, Moews P C, Liu H, Zhao H, Frere J M, Knox J R. Evolution of an enzyme activity: crystallographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc Natl Acad Sci USA. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medeiros A A. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 34.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 35.Page M I, Laws A P. The mechanism of catalysis and the inhibition of β-lactamases. Chem Commun. 1998;1998:1609–1617. [Google Scholar]
- 36.Palzkill T. β-Lactamases are changing their activity spectrums. ASM News. 1998;64:90–95. [Google Scholar]
- 37.Parker R H, Eggleston M. Beta-lactamase inhibitors: another approach to overcoming antimicrobial resistance. Infect Control. 1987;8:36–40. doi: 10.1017/s0195941700066972. [DOI] [PubMed] [Google Scholar]
- 38.Petrosino J, Rudgers G, Gilbert H, Palzkill T. Contributions of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of beta-lactamases. J Biol Chem. 1999;274:2394–2400. doi: 10.1074/jbc.274.4.2394. [DOI] [PubMed] [Google Scholar]
- 39.Phillips D R, Charo I F, Scarborough R M. GPIIb-IIIa: the responsive integrin. Cell. 1991;65:359–362. doi: 10.1016/0092-8674(91)90451-4. [DOI] [PubMed] [Google Scholar]
- 40.Rudgers G W, Palzkill T. Identification of residues in beta-lactamase critical for binding beta-lactamase inhibitory protein. J Biol Chem. 1999;274:6963–6971. doi: 10.1074/jbc.274.11.6963. [DOI] [PubMed] [Google Scholar]
- 41.Samanen J, Ali F, Romoff T, Calvo R, Sorenson E, Vasko J, Storer B, Berry D, Bennett D M, Strohsacker M, et al. Development of a small RGD peptide fibrinogen receptor antagonist with potent antiaggregatory activity in vitro. J Med Chem. 1991;34:3114–3125. doi: 10.1021/jm00114a022. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Strynadka N C, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 44.Strynadka N C, Jensen S E, Alzari P M, James M N. A potent new mode of beta-lactamase inhibition revealed by the 1.7 A X-ray crystallographic structure of the TEM-1-BLIP complex. Nat Struct Biol. 1996;3:290–297. doi: 10.1038/nsb0396-290. [DOI] [PubMed] [Google Scholar]
- 45.Strynadka N C, Jensen S E, Johns K, Blanchard H, Page M, Matagne A, Frere J M, James M N. Structural and kinetic characterization of a beta-lactamase-inhibitor protein. Nature. 1994;368:657–660. doi: 10.1038/368657a0. [DOI] [PubMed] [Google Scholar]
- 46.Tenover F C, Hughes J M. The challenges of emerging infectious diseases. Development and spread of multiply-resistant bacterial pathogens. JAMA. 1996;275:300–304. [PubMed] [Google Scholar]
- 47.Ullah J H, Walsh T R, Taylor I A, Emery D C, Verma C S, Gamblin S J, Spencer J. The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J Mol Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Fast W, Valentine A M, Benkovic S J. Metallo-beta-lactamase: structure and mechanism. Curr Opin Chem Biol. 1999;3:614–622. doi: 10.1016/s1367-5931(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 49.Wiedemann B, Kliebe C, Kresken M. The epidemiology of beta-lactamases. J Antimicrob Chemother. 1989;24(Suppl. B):1–22. doi: 10.1093/jac/24.suppl_b.1. [DOI] [PubMed] [Google Scholar]
- 50.Yanofsky S D, Baldwin D N, Butler J H, Holden F R, Jacobs J W, Balasubramanian P, Chinn J P, Cwirla S E, Peters-Bhatt E, Whitehorn E A, Tate E H, Akeson A, Bowlin T L, Dower W J, Barrett R W. High affinity type I interleukin 1 receptor antagonists discovered by screening recombinant peptide libraries. Proc Natl Acad Sci USA. 1996;93:7381–7386. doi: 10.1073/pnas.93.14.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]