Abstract
The discovery of functional organic–inorganic hybrid nanoflowers (FNFs) consisting of proteins/enzymes as the organic components and Cu(ii) ion as the inorganic component has made an enormous impact on enzyme immobilization studies. The FNFs synthesized by an encouraging and novel approach not only showed high stabilities but also much enhanced catalytic activities as compared to free and conventionally immobilized enzymes. A recent development demonstrated that FNF formation has moved beyond the initial discovery in which enzymes and Cu2+ ions used as the organic and inorganic parts, respectively, are replaced with new organic (chitosan, amino acid and plant extracts) and inorganic (Cu2+ and Fe2+) materials. The new organic materials incorporated into FNFs act as Fenton-like agents and then show peroxidase-like activity owing to the metal ions and the porous structure of FNFs in the presence of hydrogen peroxide (H2O2). All FNFs have been widely utilized in many different scientific and industrial fields due to their greatly enhanced activities and stabilities. This review focuses primarily on the preparation, characterization, and bioanalytical applications of FNFs and explains the mechanisms of their formation and enhanced activities and stabilities.
The discovery of functional organic–inorganic hybrid nanoflowers (FNFs) consisting of proteins/enzymes as the organic components and Cu(ii) ion as the inorganic component has made an enormous impact on enzyme immobilization studies.
1. Introduction
Enzymes have been considered as a versatile biological tool for the synthesis, cleavage and conversion of compounds in biological and industrial systems. The reasons for their extensive use in free form are their intrinsically high catalytic activity, substrate selectivity, low toxicity and water solubility. Typically, they effectively and selectively catalyze substrates with high reaction speed in optimal experimental conditions (typically at room temperature, atmospheric pressure and around neutral pH values). They may catalyze one or few substrates by reacting with either the entire substrate or its certain parts due to substrate specificity.1–3 Despite these advantages, their short lifetimes in aqueous and organic media, the rapid loss of activity (at high temperatures, at acidic or basic pHs and high salt concentration), and lack of separation from the reaction medium as well as the fact that they cannot be reused are potential disadvantages of free enzymes, which make them quite expensive; these disadvantages also highly limit their use in a diverse range of applications.4–7
It is worthy to mention that enzymes in natural systems exist as immobilized enzymes because they are usually bound to the cell membrane and show high stability and activity. For this reason and to overcome the drawbacks of free enzymes, the conjugation of free enzymes to supports called “enzyme immobilization” was offered. Researchers have aimed to simultaneously increase the stability and activity of enzymes by immobilizing them on various materials with different immobilization methods. However, these methods were not successful. While enzymes in free form exhibit high activities and low stabilities as mentioned above, immobilized enzymes usually show lower activities and higher stabilities when compared to free enzymes, owing to three major effects: (1) mass transfer limitations, (2) restriction of enzyme mobility and (3) unfavorable changes of enzyme conformation.8–10 There are only a few specific reports on enhanced catalytic activities after immobilization. For instance, Ackerman and co-workers demonstrated that the organophosphorus hydrolase enzyme immobilized onto mesoporous silica nanomaterials exhibited increased activity (200% activity) as compared to the free form of the corresponding enzyme.11
2. Conventional immobilization
Up to now, many enzymes have been immobilized on various supports such as glass, membrane, polymer, gel beads, sol–gel supports, porous silicon matrix, porous monolithic and nanosized materials with the use of mostly four different conventional immobilization methods including adsorption, covalent binding, entrapment and cross-linking.12–16 The water solubility and/or insolubility, biocompatibility, surface functionality and size and morphology are critical features to consider while choosing supports for immobilization. Using various external supports generally enables the adjustment of the amount of immobilized enzymes and their re-use.2,6 Despite all these developments, most of the immobilized enzymes have shown great stabilities towards environmental conditions (temperature, pH values, etc.), but they inevitably have exhibited reduction in their catalytic activities due to the following effects: (1) restriction in mass transfer between the enzyme and substrate, (2) limited mobility and (3) unfavorable conformation as compared to that of free enzymes.17–35 For instance, Corderio et al., Kumar et al., and Singh et al., successfully immobilized α-amylase onto reactive polymers, amonolithic silica nanohybrid and PAni-PVA materials, respectively. Immobilized α-amylase was highly stable against reaction temperature, and some metal ions were used as inhibitors, and it allowed us to reperatedly use them; however, it showed lower activity as compared to its free form.19–21
Additionally, decreasing the material size to the nano range in enzyme immobilization has become much common owing to much larger surface-to-volume ratio as compared to that of micro-sized materials. Nanomaterials (NMs) effectively increase the number of loading enzymes on their surface and local enzyme concentration. Thus, researchers have predicted that immobilized enzyme on NMs such as plasmonic, magnetic metal and metal oxide and polymeric NMs (Fe3O4 NMs, CdTe dots, ZnO, Au and SiO2 NMs, etc.) may improve their stability and activity. When a significant amount of enzyme was immobilized per unit volume of NMs, higher mechanical and thermal stabilities were observed, but an increase in the activity was not observed and thus, it is not suitable as compared to the free enzyme dissolved in solution.27–35
3. Synthesis of functional organic–inorganic hybrid nanoflowers (FNFs) as nanobiocatalysts
A previously reported enzyme immobilization method that was accidentally discovered by Zare and coworkers demonstrated that immobilized enzymes simultaneously show remarkable activities and stabilities as compared to free and conventionally immobilized enzymes;36 they reported the synthesis of organic–inorganic nanostructures in a flower-shape called functional organic–inorganic hybrid nanoflowers (FNFs) using proteins/enzymes as the organic parts and Cu2+ ion as the inorganic part in a phosphate-buffered saline (PBS) solution. The main principle of the formation of nanoflowers relies on the following effects: (1) Cu2+ ions react with phosphate to create the primary copper phosphate nanocomplex (Cu3(PO4)2) in a nucleation step, (2) the amide groups in the protein backbone preferentially bind to the Cu2+ ions in the Cu3(PO4)2 nanocomplexes to form flower petals in the growth step and (3) the petals adhere to each other to produce a nanoflower in the final completion step. A proposed mechanism of organic–inorganic nanoflower formation is illustrated in Fig. 1.
Fig. 1. Production of organic component-incorporated nanoflowers with potential mechanism including three successive steps (nucleation, growth and completion).
In a typical FNF procedure, a certain volume of CuSO4 stock solution (120 mM) freshly prepared in ultrapure water and 0.02 mg mL−1 organic component were mixed in 10 mM phosphate-buffered saline (PBS) solution (pH 7.4). Then, the resultant mixture was vigorously agitated prior to incubation for 3 days under undisturbed conditions at room temperature (RT = 20 °C). After incubation, the blue colored precipitate, which can be considered as FNFs, was collected via centrifugation at 10 000 rpm for 20 min.
3.1. Single enzyme-incorporated FNFs
The enhanced activity of FNFs can be explained with the following effects: (1) porous structure and high surface area, (2) facilitation of mass transfer between FNFs and the substrate, (3) synergistic effects occurring between the enzyme molecules and the enzyme-metal ions, (4) high local enzyme concentration and (5) favorable conformation of enzyme molecules in FNFs.
Many researchers inspired from the nanoflower concept have developed various enzyme-inorganic hybrid nanoflowers for a variety of applications. In terms of the inorganic component, some metal ions including Cu2+, Fe2+ and Ca2+ have been used as the inorganic part of FNFs.37–47 A wide range of enzymes37–50 including commercially available and isolated enzymes such as HRP,37–39 SBP,40 alfa-chymotrypsin,41 laccase,42,47 urease,43 lactoperoxidase,44 lipase,45 Turkish black radish peroxidase,46 and trypsin47 have been successfully incorporated into FNF synthesis. FNFs have provided a variety of applications such as colorimetric sensors to detect H2O2, phenol,37 dopamine,38,44 epinephrine,42,44 dye decolorization,46,50 and protein digestion.41
3.2. Multi-enzyme-incorporated FNFs
One of the most exciting advances in nanomaterial synthesis is the ability to produce hybrid nanomaterials (HNMs). HNMs consisting of several individual domains (components) usually exhibit enhanced physical and chemical properties or multifunctionalities in one single platform as compared to single-component NMs. The development of HNMs has also opened up numerous new opportunities to use HNMs in different scientific and technical areas.51,52
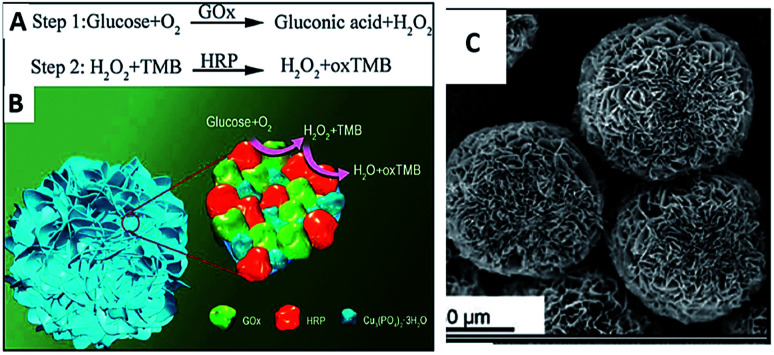
Recently, as examples of multifunctionalities of HNMs, several groups have reported syntheses and applications of multi-enzyme-containing FNFs. Having multi-enzymes in a single FNF allows simultaneous or stepwise catalysis of cascade reactions in one pot. Thus, an enzymatic reaction by an enzyme in an FNF is catalyzed by an adjacent enzyme on the same FNF to perform another reaction. The close proximity between the co-enzymes in the FNF induces cascade reactions to occur rapidly in a short time and also reduces diffusion and decomposition of the product of one of the adjacent enzymes, which can be subsequently catalyzed by the other enzyme. For example, Sun et al. reported the synthesis of glucose oxidase (GOx)-horseradish peroxidase (HRP) FNFs and their calorimetric sensor application for glucose detection,53,54 as shown in Fig. 2.
Fig. 2. (A) Proposed mechanism of glucose detection using a two-step enzymatic reaction in one pot. (B) The mechanistic illustration of glucose detection with GOx and HRP and Cu(ii)-incorporated nanoflowers. (C) SEM images of GOx and HRP nanoflowers.
In the synthesized FNFs, H2O2 produced via the oxidation of glucose by GOx acts as a cofactor for HRP to oxidize 3,3′,5,5′-tetramethylbenzidine, resulting in a color change. The performance of GOx-HRP FNFs in glucose detection was approximately 4-fold higher than that of the free enzymes and FNFs formed from only GOx or HRP. Glucose detection with the use of a GOx-HRP FNF-deposited microfluidic paper-based analytic device (μPAD) was also developed by Zhu et al.55 In another example, Ye et al. synthesized concanavalin A (Con A)-invertase FNFs with Ca2+ ion as the inorganic part in place of Cu2+ ion.56 Owing to the E. coli O157 : H7 recognition of Con A and the hydrolysis of sucrose to glucose by invertase, they succeeded at the point-of-care detection of E. coli O157 : H7 from milk by measuring the amount of glucose present. The same group also performed a similar detection of a food pathogen by using Con A-GOx FNFs;57 they used GOx as the signal amplification unit to convert glucose to gluconic acid, resulting in the decrease in the pH value, which was measured by a portable pH meter. Another calorimetric nanosensor application of multi-enzyme-incorporated FNFs was obtained with the use of streptavidin (SA)-HRP dual enzyme by Liu et al.58 The designed FNFs were able to detect alpha-fetoprotein (AFP) with much better sensitivity as compared to commercial enzyme-linked immunosorbent assay (ELISA) kits.
3.3. Other organic component-incorporated FNFs
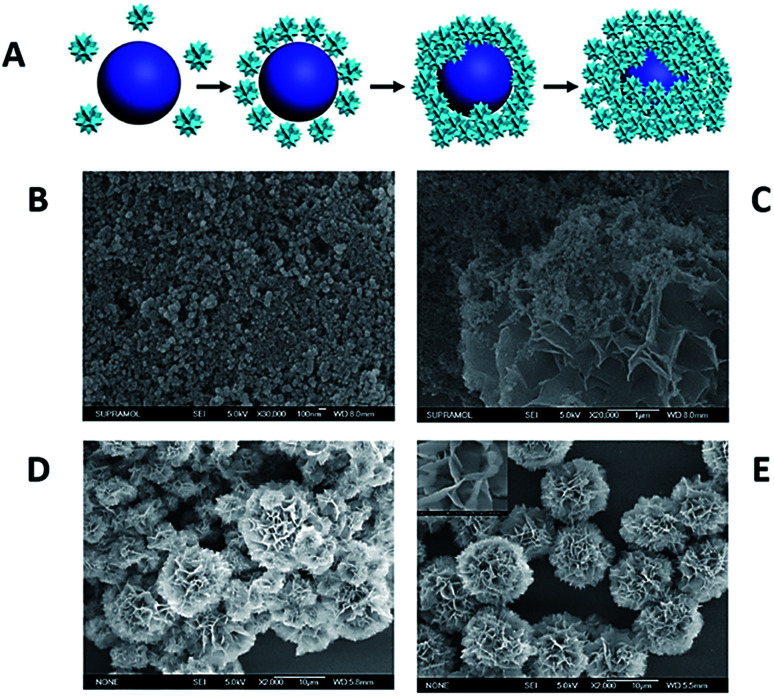
The potential mechanism for the formation of protein-inorganic FNFs relies on the coordination reaction between the accessible amide groups in the protein/enzyme backbone and the Cu2+ ions in PBS solution. In addition to that, a very recent development proved that not only the Cu–N bonds but also the Cu–O bonds can be driving forces in the FNF formation.58–61 For instance, Wu et al. used 20 natural amino acids as the organic component and Cu2+ ion as the inorganic component to form FNFs. The study demonstrated that the Cu2+ ion reacted with both amine and carboxyl groups, and FNFs consisted of Cu–N and Cu–O bonds.62 A schematic for the synthesis of amino acid (AA)-based FNFs and their time-dependent SEM images are presented in Fig. 3.
Fig. 3. (A) The potential mechanism of AA-FHN formation. Time-dependent formation of FHNs in (B) 0 hour, (C) 0.5 hours, (D) 2 hours and (E) 24 hours.
As mentioned above, Ca2+ ion-incorporated α-amylase FNFs were reported in a previous study; however, instead of using a protein, Wang et al. fabricated microflowers using chitosan (CS) biomolecule as the organic component and Ca2+ ion as the inorganic component. CS was cross-linked by tripolyphosphate (TPP) through ionotropic gelation to form nanocomplex tripolyphosphate (TPP).58 The CS-TPP microflower production was completed via the mechanism of FNF formation. When the catalase (CAT) enzyme was used as a model enzyme immobilized onto the CS-microflowers, drastic enhancements in the activity and stability were observed.63
It is well-known that plant extracts and their active components have been commonly used as reducing and capping agents for the synthesis of metallic nanomaterials. Plant extracts have several advantages over other chemical agents in nanomaterial synthesis owing to the following factors: (1) they are cost-effective, (2) they are easily obtainable, (3) they do not pose a contamination risk and (4) no expertise is required while using them.64–69
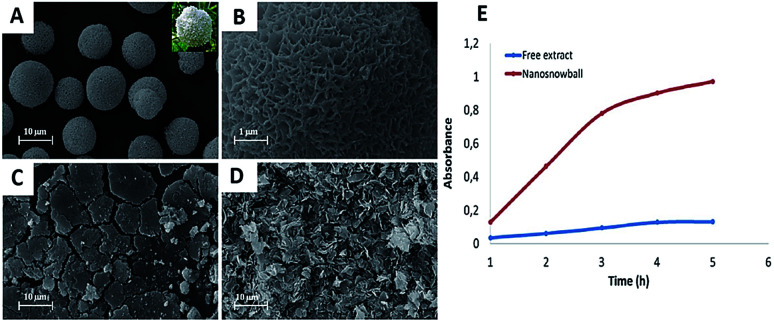
Ildiz et al. and Baldemir et al. benefited from the nanoflower concept and reported for the first time the synthesis of FHNs using whole plant extracts and standard molecules acting as the organic components.70–72 Ildız and coworkers used a Viburnum opulus (VO) extract to form guelder rose-like morphology called “nanosnowball” (NSBs) and investigated how the concentration of the VO extract influenced the formation of NSBs. They demonstrated that NSBs behaved like peroxidase enzyme through a Fenton-like reaction mechanism when guaiacol was used as the model substrate. The study also reported that NSBs showed effective antimicrobial activities against some bacterial pathogens such as Escherichia coli, Salmonella typhi, Enterococcus faecium, Enterococcus faecalis, Bacillus, and Staphylococcus aureus as compared to free VO extract. Morphology and peroxidase-like activity of NSBs are shown in Fig. 4.
Fig. 4. (A) SEM image of NSBs synthesized using 0.1 mg mL−1 VO extract. Inlet: photograph of natural VO flower, (B) the high resolution image of (A), (C) NSB formation was not observed when 0.5 mg mL−1 VO extract was used. (D) SEM image of copper phosphate nanocrystals without VO extract. (E) Catalytic activity of free VO extract (blue line) and NSBs (red line).
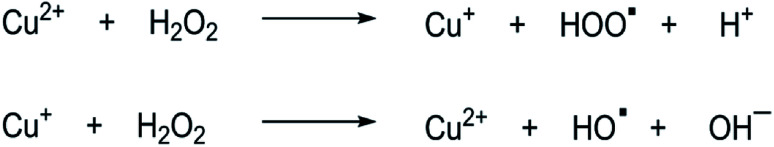
In terms of peroxidase-like activity, the Cu(ii)-integrated FHNs act as a Fenton reagent and exhibit peroxidase-type activity in the presence of hydrogen peroxide (H2O2) due to the intrinsic property of copper compounds. The mechanism of the Fenton-like reaction, as shown in Fig. 5, presents that Cu(ii) ions in the structure of FHNs react with hydrogen peroxide to form Cu(i) ions. The reaction of Cu(i) ions with H2O2 results in a highly reactive hydroxyl radical. This free hydroxyl radical initiates the oxidation of the substrate, and the reaction proceeds through this mechanism. The peroxidase-like activities of FHNs are significantly increased by increasing the concentrations of H2O2 and catalyst to the optimal point.
Fig. 5. Fenton reaction mechanism.
As another example, Baldemir and coworkers extended their study on plant extract-incorporated FHNs. They reported Camellia sinensis (L.) Kuntze extracts as the main components and Laurocerausus officinalis Roemer extract Cu(ii)-assembled nanoflowers and showed their antimicrobial and antiparasitic properties.71,72 Interesting results were obtained: while the green tea (GT) extract (extracted in ethanol) and catechin produced FHNs with very similar morphology, green tea (GT) extract (extracted in water) and caffeine-based FHNs were also quite similar in shape and size. It can be concluded that the main components of GT extracted in ethanol and water can be catechin and caffeine molecules.
4. Summary
The development of functional organic–inorganic hybrid nanoflowers (FNFs) including enzyme- and organic molecule-inorganic hybrid nanoflowers has made an enormous impact on the stability and catalytic activities as compared to that of free enzymes and organic molecules. For instance, single enzyme or multi enzyme-incorporated nanoflowers have overcome major drawbacks (instability, lack of reusability and loss of activity) exhibited by free and/or conventionally immobilized enzymes.
In addition to enzyme-based FNFs, the plant extract-incorporated FNFs were introduced in this review as a first report to show Fenton activity of plant extract-based FNFs; they showed peroxidase-like activity through Fenton-like reaction. The catalytic and antimicrobial activities towards guaiacol and microorganisms were evaluated in the presence of hydrogen peroxide (H2O2). We believe that all FNFs with their unique properties can be used in a variety of technical and scientific fields.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Erciyes University Scientific Research Office (TSA08-590).
References
- Horton H. R., Moran L. A. and Ochs R. S., Principles of biochemistry, Prentice Hall International, Inc., New Jersey, 1996 [Google Scholar]
- Ansari S. A. Husain Q. Biotechnol. Adv. 2012;30:512–523. doi: 10.1016/j.biotechadv.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Tran D. N. Balkus K. J. ACS Catal. 2011;1:956–968. doi: 10.1021/cs200124a. [DOI] [Google Scholar]
- Ahmad R. Sardar M. Biochem. Anal. 2015;4:1–8. [Google Scholar]
- Mateo C. Palomo J. M. Fernandez-Lorente G. Guisan J. M. Fernandez-Lafuente R. Enzyme Microb. Technol. 2007;40:1451–1463. doi: 10.1016/j.enzmictec.2007.01.018. [DOI] [Google Scholar]
- Zhang Y. Ge J. Liu Z. ACS Catal. 2015;5:4503–4513. doi: 10.1021/acscatal.5b00996. [DOI] [Google Scholar]
- Altinkaynaka C. Tavlasogluc S. Özdemirc N. Ocsoy İ. A. Enzyme Microb. Technol. 2016;93:105–112. doi: 10.1016/j.enzmictec.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Kim J. Grate J. W. Wang P. Trends Biotechnol. 2008;26:639–646. doi: 10.1016/j.tibtech.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Luckarift H. R. Spain J. C. Naik R. R. Stone M. O. Nat. Biotechnol. 2004;22:211–213. doi: 10.1038/nbt931. [DOI] [PubMed] [Google Scholar]
- Mateo C. Grazu V. Palomo J. M. LopezGallego F. Fernandez-Lafuente R. Guisan J. M. Nat. Protoc. 2007;2:1022–1027. doi: 10.1038/nprot.2007.133. [DOI] [PubMed] [Google Scholar]
- Lei C. Shin Y. Liu J. Ackerman E. J. J. Am. Chem. Soc. 2002;124:11242–11243. doi: 10.1021/ja026855o. [DOI] [PubMed] [Google Scholar]
- Kim J. Grate J. W. Wang P. Chem. Eng. Sci. 2006;61:1017–1026. doi: 10.1016/j.ces.2005.05.067. [DOI] [Google Scholar]
- Sheldon R. A. Pelt S. V. Chem. Soc. Rev. 2013;42:6223–6235. doi: 10.1039/C3CS60075K. [DOI] [PubMed] [Google Scholar]
- Zaborsky O., Immobilized Enzymes, CRC Press, Boca Raton, FL, 1973, pp. 117–126 [Google Scholar]
- Sheldon R. A. Biochem. Soc. Trans. 2007;35:1583–1587. doi: 10.1042/BST0351583. [DOI] [PubMed] [Google Scholar]
- Lei C. Shin Y. Magnuson J. K. Fryxell G. E. Lasure L. L. Elliott D. C. Liu J. Ackerman E. J. Nanotechnology. 2006;17:5531–5538. doi: 10.1088/0957-4484/17/22/001. [DOI] [PubMed] [Google Scholar]
- Hanefeld U. Cao L. Chem. Soc. Rev. 2013;42:15. doi: 10.1039/C2CS35286A. [DOI] [PubMed] [Google Scholar]
- Gupta M. Mattiasson B. Methods Biochem. Anal. 1992:1–34. doi: 10.1002/9780470110577.ch1. [DOI] [PubMed] [Google Scholar]
- Cordeiro A. L. Lenk T. Werner C. J. Biotechnol. 2011;154:216–221. doi: 10.1016/j.jbiotec.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Singh V. Kumar P. J. Mol. Catal. B: Enzym. 2011;70:67–73. doi: 10.1016/j.molcatb.2011.02.006. [DOI] [Google Scholar]
- Singh S. Saikia J. P. Buragohain A. K. Colloids Surf., B. 2013;106:46–50. doi: 10.1016/j.colsurfb.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Li B. Chen Y. Chen X. Liu D. Niu H. Xiong J. Wu J. Xie J. Bai J. Ying H. Process Biochem. 2012;47:665–670. doi: 10.1016/j.procbio.2012.01.008. [DOI] [Google Scholar]
- Abd El-Ghaffar M. A. Hashem M. S. J. Appl. Polym. Sci. 2009;112:805–814. doi: 10.1002/app.29292. [DOI] [Google Scholar]
- Konsula Z. Liakopoulou-Kyriakides M. Process Biochem. 2004;39:1745–1749. doi: 10.1016/j.procbio.2003.07.003. [DOI] [Google Scholar]
- Pascoal A. M. Mitidieri S. Fernandes K. F. Food Bioprod. Process. 2011;89:300–306. doi: 10.1016/j.fbp.2010.07.002. [DOI] [Google Scholar]
- Nwagua T. N. Aoyagia H. Okolob B. N. Yoshida S. J. Mol. Catal. B: Enzym. 2012;78:1–8. doi: 10.1016/j.molcatb.2012.01.019. [DOI] [Google Scholar]
- Chen Y. H. Chi M. C. Wang T. F. Chen J. C. Lin L. L. Appl. Biochem. Biotechnol. 2012;166:1711–1722. doi: 10.1007/s12010-012-9575-1. [DOI] [PubMed] [Google Scholar]
- Jiang H. Wang X. Analyst. 2012;137:2582. doi: 10.1039/C2AN00038E. [DOI] [PubMed] [Google Scholar]
- Sahoo B. Sahu S. K. Pramanik P. J. Mol. Catal. B: Enzym. 2011;69:95–102. doi: 10.1016/j.molcatb.2011.01.001. [DOI] [Google Scholar]
- Sharma B. Mandani S. Sarma T. K. Sci. Rep. 2013;3:2601. doi: 10.1038/srep02601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorilyi R. P. Melnyk I. V. Zub Y. L. Seisenbaevab G. A. Kessler V. G. J. Mater. Chem. B. 2014;2:2694. doi: 10.1039/C4TB00018H. [DOI] [PubMed] [Google Scholar]
- Garg S. De A. Mozumdar S. J. Biomed. Mater. Res. 2015;103:1771–1783. doi: 10.1002/jbm.a.35314. [DOI] [PubMed] [Google Scholar]
- Ali A. AlSalhi M. S. Atif M. Ansari A. A. Israr M. Q. Sadaf J. R. Ahmed E. Nur O. J. Phys.: Conf. Ser. 2013;414:012024. doi: 10.1088/1742-6596/414/1/012024. [DOI] [Google Scholar]
- Zhou J. Cao J. Huang W. Huang L. Wang Y. Zhang S. Yuan Y. Hua D. Chem. Biochem. Eng. Q. 2013;27(4):431–437. [Google Scholar]
- Itabaiana Jr I. Sutili F. K. Leite S. G. F. Gonçalves K. M. Cordeiro Y. Leal I. C. R. Miranda L. S. M. Ojeda M. Luqued R. de Souza R. O. M. A. Green Chem. 2013;15:518. doi: 10.1039/C2GC36674F. [DOI] [Google Scholar]
- Ge J. Lei J. Zare R. N. Nat. Nanotechnol. 2012;7(7):428. doi: 10.1038/nnano.2012.80. [DOI] [PubMed] [Google Scholar]
- Lin Z. Xiao Y. Yin Y. Hu W. Liu W. Yang H. ACS Appl. Mater. Interfaces. 2014;6(13):10775–10782. doi: 10.1021/am502757e. [DOI] [PubMed] [Google Scholar]
- Somturk B. Hancer M. Ocsoy I. Özdemir N. Dalton Trans. 2015;44(31):13845–13852. doi: 10.1039/C5DT01250C. [DOI] [PubMed] [Google Scholar]
- Wang L.-B. Wang Y.-. He R. Zhuang A. Wang X. Zeng J. Hou J. G. J. Am. Chem. Soc. 2013;135(4):1272–1275. doi: 10.1021/ja3120136. [DOI] [PubMed] [Google Scholar]
- Ocsoy I. Dogru E. Usta S. Enzyme Microb. Technol. 2015;75:25–29. doi: 10.1016/j.enzmictec.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Yu Y. Fei X. Tian J. Xu L. Wang X. Wang Y. Colloids Surf., B. 2015;130:299–304. doi: 10.1016/j.colsurfb.2015.04.033. [DOI] [PubMed] [Google Scholar]
- Yin Y. Xiao Y. Lin G. Xiao Q. Lin Z. Cai Z. J. Mater. Chem. 2015;3(11):2295–2300. doi: 10.1039/C4TB01697A. [DOI] [PubMed] [Google Scholar]
- Batule B. S. Park K. S. Kim M. I. Park H. G. Int. J. Nanomed. 2015;10:137. doi: 10.2147/IJN.S90274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somturk B. Yilmaz I. Altinkaynak C. Karatepe A. Özdemir N. Ocsoy I. Enzyme Microb. Technol. 2016;86:134–142. doi: 10.1016/j.enzmictec.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Altinkaynak C. Yilmaz I. Koksal Z. Özdemir H. Ocsoy I. Özdemir N. Int. J. Biol. Macromol. 2016;84:402–409. doi: 10.1016/j.ijbiomac.2015.12.018. [DOI] [PubMed] [Google Scholar]
- Ke C. Fan Y. Chen Y. Xu L. Yan Y. RSC Adv. 2016;6(23):19413–19416. doi: 10.1039/C6RA01564F. [DOI] [Google Scholar]
- Lin Z. Xiao Y. Wang L. Yin Y. Zheng J. Yang H. Chena G. RSC Adv. 2014;4:13888–13891. doi: 10.1039/C4RA00268G. [DOI] [Google Scholar]
- Wu X. Hou M. Ge M. Catal. Sci. Technol. 2015;5:5077–5085. doi: 10.1039/C5CY01181G. [DOI] [Google Scholar]
- Lei Z. Gao C. Chen L. He Y. Ma W. Lin Z. J. Mater. Chem. B. 2018;6:1581–1594. doi: 10.1039/C7TB03310A. [DOI] [PubMed] [Google Scholar]
- Altinkaynak C. Tavlasoglu S. Kalin R. Sadeghian N. Ozdemir H. Ocsoy I. Özdemir N. Chemosphere. 2017;182:122–128. doi: 10.1016/j.chemosphere.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Li H. Hou J. Duan L. Ji C. Zhang Y. Chen V. J. Hazard. Mater. 2017;338:93–101. doi: 10.1016/j.jhazmat.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Lee D. C. Smith D. K. Heitsch A. T. Korgel B. A. Annu. Rep. Prog. Chem., Sect. C: Phys. Chem. 2007;103:351–402. doi: 10.1039/B605630J. [DOI] [Google Scholar]
- Ocsoy I. Gulbakan B. Shukoor M. I. Xiong X. Chen T. Powell D. H. Weihong T. ACS Nano. 2013;7(1):417–427. doi: 10.1021/nn304458m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. Ge J. Liu W. Lan M. Zhang H. Wang P. Wang Y. Niu Z. Nanoscale. 2014;6(1):255–262. doi: 10.1039/C3NR04425D. [DOI] [PubMed] [Google Scholar]
- Zhu X. Huang J. Liu J. Zhang H. Jiang J. Yu R. A. Nanoscale. 2017;9(17):5658–5663. doi: 10.1039/C7NR00958E. [DOI] [PubMed] [Google Scholar]
- Li Z. Zhang Y. Su Y. Ouyang P. Ge J. Liu Z. Chem. Commun. 2014;50:12465–12468. doi: 10.1039/C4CC05478D. [DOI] [PubMed] [Google Scholar]
- Sun J. Ge J. Liu W. Lan M. Zhang H. Wang P. Niu Z. Nanoscale. 2014;6(1):255–262. doi: 10.1039/C3NR04425D. [DOI] [PubMed] [Google Scholar]
- Zhu X. Huang J. Liu J. Zhang H. Jiang J. Yu R. A. Nanoscale. 2017;9(17):5658–5663. doi: 10.1039/C7NR00958E. [DOI] [PubMed] [Google Scholar]
- Ye R. Zhu C. Song Y. Song J. Fu S. Lu Q. Lin Y. Nanoscale. 2016;8:18980–18986. doi: 10.1039/C6NR06870G. [DOI] [PubMed] [Google Scholar]
- Ye R. Zhu C. Song Y. Lu Q. Ge X. Yang X. Lin Y. Bioinspired Synthesis of All-in-One Organic–Inorganic Hybrid Nanoflowers Combined with a Handheld pH Meter for On-Site Detection of Food Pathogen. Small. 2016;12:3094–3100. doi: 10.1002/smll.201600273. [DOI] [PubMed] [Google Scholar]
- Liu Y. Chen J. Du M. Wang X. Ji X. He Z. Biosens. Bioelectron. 2017;92:68–73. doi: 10.1016/j.bios.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Wu Z. F. Wang Z. Zhang Y. Ma Y. L. He C. Y. Li H. Chen L. Huo Q. S. Wang L. Li Z. Q. Sci. Rep. 2016;6:22412. doi: 10.1038/srep22412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Shi J. Li Z. Zhang S. Wu H. Jiang Z. ACS Appl. Mater. Interfaces. 2014;6:14522–14532. doi: 10.1021/am503787h. [DOI] [PubMed] [Google Scholar]
- Ocsoy I. Tasdemir D. Mazicioglu S. Celik C. Katı A. Ulgen F. Mater. Lett. 2017;212:45–50. doi: 10.1016/j.matlet.2017.10.068. [DOI] [Google Scholar]
- Ocsoy I., Tasdemir D., Mazicioglu S. and Tan W., Nanotechnology in Plants, Springer, Berlin, Heidelberg, 2018, pp. 1–13 [DOI] [PubMed] [Google Scholar]
- Akhtar M.-S. Panwar J. Yun Y.-S. ACS Sustainable Chem. Eng. 2013;1:591–602. doi: 10.1021/sc300118u. [DOI] [Google Scholar]
- Ocsoy I. Temiz M. Celik C. Altinsoy B. Yilmaz V. Duman F. J. Mol. Liq. 2017;227:147–152. doi: 10.1016/j.molliq.2016.12.015. [DOI] [Google Scholar]
- Karatoprak G.-Ş. Aydin G. Altinsoy B. Altinkaynak C. Koşar M. Ocsoy I. Enzyme Microb. Technol. 2017;97:21–26. doi: 10.1016/j.enzmictec.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Ocsoy I. Demirbas A. McLamore E. S. Altinsoy B. Ildiz N. Baldemir A. J. Mol. Liq. 2017;238:263–269. doi: 10.1016/j.molliq.2017.05.012. [DOI] [Google Scholar]
- Ildiz N. Baldemir A. Altinkaynak C. Özdemir N. Yilmaz V. Ocsoy I. Enzyme Microb. Technol. 2017;102:60–66. doi: 10.1016/j.enzmictec.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Baldemir A. Kose N. B. Ildız N. Ilgun S. Yusufbeyoglu S. Yilmaz V. Ocsoy I. RSC Adv. 2017;7:44303. doi: 10.1039/C7RA07618E. [DOI] [Google Scholar]
- Baldemir A. Karaman Ü. Yusufbeyoğlu S. Eken A. Ildız N. İlgün S. Çolak C. Kaçmaz G. Öçsoy İ. Çankaya S. Mikrobiyol. Bul. 2018;52:56–71. doi: 10.5578/mb.66400. [DOI] [PubMed] [Google Scholar]