Abstract
Natural killer (NK) cells are implicated in the control of metastasis in uveal melanoma, a process that has been ascribed to its cancer stem cell subpopulation. NK cell activation is regulated by specific microRNA (miR). The NK cell sensitivity and regulatory miR production of uveal melanoma cancer stem cells was examined. Cancer stem cells enriched from aggressively metastatic MUM2B uveal melanoma cells by selecting CD271+ cells or propagating as non-adherent spheres in stem-cell supportive were more resistant to NK cell cytolysis than cancer stem cells enriched from less aggressively metastatic OCM1 uveal melanoma cells. Both MUM2B and OCM1 cells expressed and secreted NK cell regulatory miRs, including miR 146a, 181a, 20a, and 223. MUM2B cells expressed and secreted miR-155; OCM1 cells did not. Transfecting MUM2B cells with anti-miR-155 increased NK cell sensitivity. CD271+ cells were identified in the blood of patients with metastatic uveal melanoma and were characterized by low expression of melanocyte differentiation determinants and by the ability to form non-adherent spheres in stem-cell supportive media. These cells also expressed NK cell regulatory miRs, including miR-155. These results indicate that uveal melanoma cancer stem cells can vary in their sensitivity to NK cell lysis and their expression of NK cell regulatory miRs. Circulating CD271+ cells from patients with metastatic uveal melanoma manifest cancer stem cell features and express miRs associated with NK cell suppression, including miR-155, that may contribute to metastatic progression.
Keywords: CD271, melanospheres, miR-155, major histocompatibility complex class I molecules, MHC class I-related chain A, microphthalmia-associated transcription factor
Introduction
Uveal melanoma is a rare cancer with a high mortality rate because of its ability to metastasize hematogenously. There is evidence that this process is inhibited by natural killer (NK) cells, critical components of innate immunity. In intraocular melanoma mouse models, disabling NK cells results in an increase in metastasis, while augmenting NK cell activity with type I interferon (IFN) results in a decrease [1]. In patients with uveal melanoma, tumor expression of major histocompatibility complex (MHC) class I molecules, which is necessary for T-cell recognition but which limits NK cell activation through interactions with NK cell inhibitory receptors, is associated with the development of metastasis [2,3]. Loss of tumor MHC class I-related chain (MIC) A, which activates NK cells through the NK group 2 member D (NKG2D) receptor, also has been associated with a poor prognosis [4], Considerable variation in the susceptibility of human uveal metastatic melanoma cells to NK cell cytolysis not predicted by MHC class I and/or MICA expression has, however, been observed, and the mechanisms involved in regulating the NK cell susceptibility are not established [5-8].
Metastasis has been ascribed to an undifferentiated or less differentiated subpopulation of cells within a tumor, referred to as cancer stem cells (CSC), that can not only invade and circulate but that can also self-renew and differentiate into the cells that comprise metastatic tumors. Cells with CSC characteristics have been identified in uveal melanoma tumors, and a CSC-like phenotype of the primary tumor has been associated with the development of metastasis [9-13]. Whether circulating tumor cells, which are frequently identified in patients with uveal melanoma [14], manifest CSC characteristics has not, however, been demonstrated. CSC are resistant to radiotherapy and chemotherapy, and in several model systems, CSC are also resistant to NK cell cytotoxicity. Expression of the nerve growth factor receptor, CD271, which is found on neural crest cells during embryogenesis, has been used to identify CSC in melanoma, including uveal melanoma [15]. In contrast to CD271− melanoma cells, CD271+ melanoma cells are tumorigenic in nude and NOD/SCID mice. NK cell depletion in these mice restores the capacity of CD271− melanoma cells to form tumors [16]. In other model systems, CSC have been susceptible to NK cell cytotoxicity, significantly more susceptible than their more differentiated tumor counterparts [17,18]. Activated NK cells have been shown to kill cutaneous melanoma CSC isolated by several methods [19]. The NK cell sensitivity of uveal melanoma CSC has not been reported.
Epigenetic mechanisms mediated by microRNA (miR) play a central role in CSC maintenance and metastasis [20]. miRs are increasingly being recognized in the regulation of immune responses, and several miRs have been shown to regulate NK cell development and function. Prominent among these are miR-155 and 181a, which regulate NK cell differentiation and activation [21,22]; miR-146a and 223, which regulate NK cell effector molecules [23,24]; and miR-20a, which regulates MICA/B [25]. miRs are released by cells, and, because of incorporation into microparticles and exosomes, are highly stable in the circulation and can mediate cell-cell signaling via paracrine and endocrine routes [26]. Circulating extracellular miRs have been shown to activate NK cells [27]. Plasma levels of NK cell regulatory miR-146a, 155, 20a, and 223 were observed to increase, and miR-181a, to decrease, in patients with uveal melanoma developing metastasis [28]. To examine a potential mechanism by which metastatic progression in uveal melanoma may be regulated, the expression of NK cell regulatory miRs by uveal melanoma CSC, including those in the circulation of patients, was examined.
Materials and Methods
Cell lines
MUM2B and OCM1 uveal melanoma cell lines were studied [29] as were NK-92, SKMEL-28, Daudi, and K562 cell lines (ATCC, Manassas, VA). NK-92 cells were maintained in RPMI 1640 culture medium supplemented with 10% fetal calf serum (FCS). The other cell lines were maintained in Dulbecco's Modified Essential Medium (DMEM) with 10% heat-inactivated FCS, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were also cultured in human embryonic stem cell culture medium (StemPro hESC SFM, Life Technologies, Grand Island, NY). All cultures were maintained at 37°C in 5% CO2.
Cell preparations
Buffy coats were isolated from whole blood by centrifugation. Contaminating erythrocytes were lysed using isotonic ammonium chloride buffer. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll gradient centrifugation. CD271+ cells were isolated using CD271 Cell Isolation Kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Cells were magnetically labeled with CD271 microbeads and separated on a MACS Separator. Both the magnetically labelled CD271+ cells and the unlabeled CD271− cells were collected. Purities by flow cytometry for CD271+ cells was >90%, and for CD271− cells, >85%. NK cells were isolated from PBMC that were incubated in RPMI 1640 with 10% FCS supplemented with 200 IU/mL recombinant human interleukin 2 (Prometheus Laboratories Inc. San Diego, CA) for 48 hours. The NK Cell Isolation Kit, human (Miltenyi Biotech GmbH) was then applied according to the manufacturer's instructions. Flow cytometry confirmed that >90% of the isolated cells expressed CD56 but not CD3.
Immunofluroescence
About 50,000 to 100,000 cells were seeded per slide using a Shandon cytospin III centrifuge (Thermo Fisher Scientific, Waltham, MA). Immunofluorescence staining was performed with the following antibodies: anti-CD271 (rabbit polyclonal; Abeam, San Francisco, CA), anti-CD45 pre-conjugated to Alexa 488 (Molecular Probes, Thermo Fisher Scientific) to identify leukocytes, and anti-Melan-A (rabbit monoclonal, Novus Biologicals, Littleton, CO; or mouse clone A103, Dako, Carpinteria, CA). Cells on the cytospin slides were permeabilized with 0.1% Triton X100 (Sigma Aldrich, St. Louis, MO) for 10 minutes, and blocked using a buffer containing 0.5% Bovine Serum Albumin (Sigma Aldrich), 1% human serum in phosphate buffered saline (PBS), for 1 hour at room temperature in a humid chamber. The primary and secondary antibodies were diluted independently and incubated sequentially for 1 hour or 45 minutes, respectively. Slides were washed three times for 10 minutes each, in PBS containing 0.01% Tween 20 (Sigma Aldrich) after incubation. The slides were mounted on coverslips set with Vectashield containing DAPI (Vector Laboratories, Burlingame, CA) for nuclear staining.
Imaging was done either on a Leica DM-5500 fluorescent microscope system with a controller CTR5500, and a halogen fluorescent lamp and QCapture Pro acquisition software (Qlmaging, Surrey, BC) was used to acquire images in three (red, blue and green) channels, using specific excitation and emission narrow band pass filters for DAPI, Alexa 568 and Alexa 488, or a Leica SP2 confocal imaging system with Leica confocal Software v.2.61 (Leica Microsystems, Inc., Buffalo Grove, IL). Bandwidths for excitation and emission for confocal imaging were selected such that they were in mutually non-overlapping regions of the spectra, eliminating bleed-through signal contaminations.
Flow cytometry
Cell surface staining was accomplished using phycoerythrin labeled CD271, fluroescein isothiocynate labeled CD3, allophycocyanin labeled CD56; APC-conjugated anti-human HLA-ABC (clone G46-2.6,) and phycoerythrin labeled MICA antibodies (BD Biosciences, San Jose, CA). Standard methodologies and appropriate isotype controls were used in all experiments. All samples were analyzed using an Accuri 6 flow cytometer (Beckman Coulter, Fullerton, CA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA from cells was obtained using the RNeasy method (Qiagen, Valencia, CA) according to the manufacturer's direction. qRT-PCR was performed on an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). Pre-standardized primers and TaqMan probes for microphthalmia-associated transcription factor (MITF), dopachrome tautomerase (DCT), tyrosinase (TYR), Melan-A/MART-1 (MLANA), vimentin (VIM), and E-cadherin (CDH1) mRNA were used (Applied Biosystems). The reverse transcription and PCR was accomplished using a one-step protocol and TaqMan Universal Master Mix (Applied Biosystems). Ct values were determined in duplicate, averaged, and the relative number of copies (RQ) of mRNA was calculated using the ΔΔCt method. miR levels were similarly assessed using TaqMan MicroRNA Assay kits for human miRs 146a, 155, 181a, 20a, and 223 (Applied Biosystems) in a two-step process. Reverse transcription reactions were first performed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The product obtained was then used in PCR reactions using TaqMan universal master mix as above. RQ was determined using the ΔΔCT method. miR data were normalized to a C. elegans synthetic miR sequence, cel-miR-39 (Qiagen), which was spiked in as a control during RNA isolation. Factors with Ct values ≥ 37 were considered “undetectable.”
NK cell cytotoxicity
NK cell cytotoxicity was determined using CytoTox 96 Assay (Promega, Madison, WI), which measures lactate dehydrogenase release by the enzymatic reduction of a tetrazolium salt into a red formazan product. A cytotoxicity experiment consisted of NK cells as effectors combined with 2 × 103 tumor cells as targets at specific ratios (E:T) for measuring experimental release, tumor cells and NK cells alone for measuring spontaneous release, and tumor cells treated with lysing reagent for assessing total release, all in triplicate in a 96-well plate. The plates were centrifuged at 50 g for 5 minutes and incubated at 37°C in 5% CO2 for 4 hours. After incubation, the plates were centrifuged at 500 g for 5 minutes and 50 μl of the supernatant from each well was transferred to a fresh 96-well plate. The supernatants were incubated with the reagents from CytoTox 96 Kit according to the manufacturer's protocol. Relative light units were determined by measuring the optical density colorimetrically at 490 nm using an ELISA reader and averaged. Percent lysis was calculated using the following formula: 100% × [experimental − spontaneous (tumor + NK cell) release/(total − spontaneous (tumor) release]. Cytotoxicity experiments were repeated at least three times.
Transfection
Anti-miR transfections were performed by electroporation (Nucleofector 4D System; Lonza Cologne AG, Cologne, Germany). MUM2B and OCM1 cells (5 × 106) were suspended in transfection medium consisting of 100 ul DMEM with 5 mM KCl, 15 mM MgCl2, 15 mM HEPES, and 150 mM Na2HPO4/NaH2PO4 (pH 7.2), and anti-miR-155 and anti-miR-146a (Ambion, Austin, TX) were added (1 ul of 5 uM solution). Transfection was accomplished with the FF-120 pulse. Cells electroporated without anti-miRs were used as controls. Cells were then incubated in DMEM with 10% heat-inactivated fetal calf serum cells for 1 hour prior to testing. Transfection efficiency was monitored using a co-transfected GFP-expressing plasmid (Pmax GFP, Lonza).
Statistical analysis
Data are presented as means ± SD. Differences between specific groups of replicates were analyzed by two-sided Student's t test. P < 0.05 was considered significant. Differences in RQ had also to be >0.5 log to be considered significant.
Results
CSC from uveal melanoma cell lines differ in NK cell sensitivity
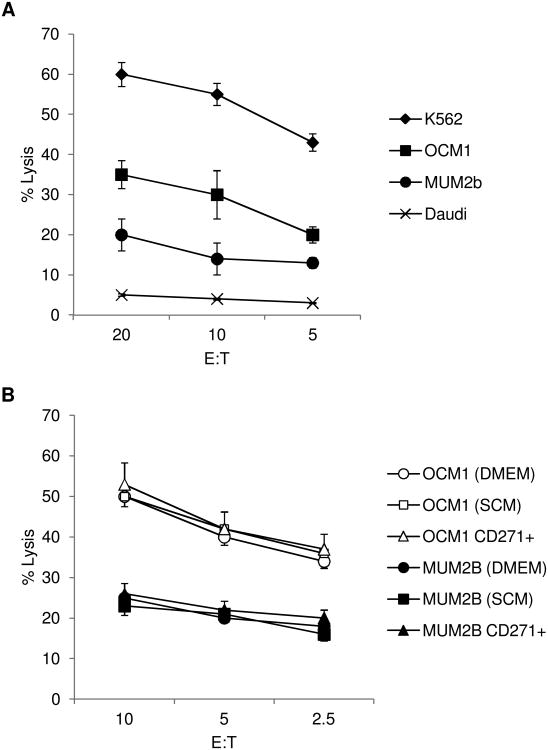
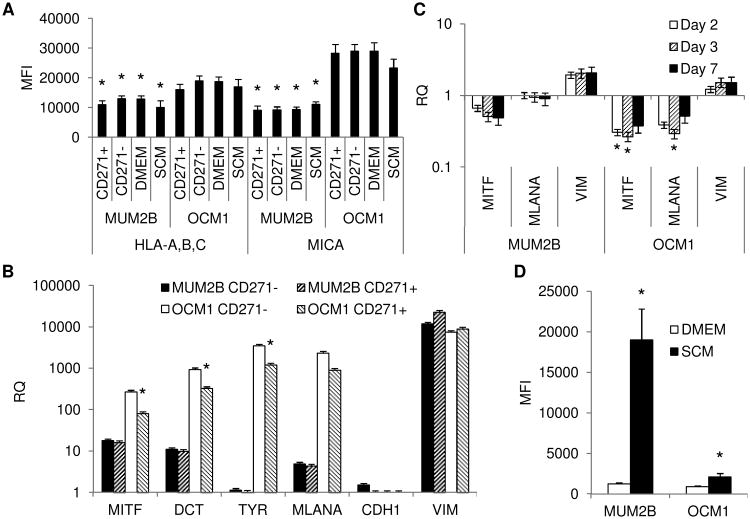
The NK cell susceptibility of the more invasive and metastatic MUM2B and the less invasive and metastatic OCM1 uveal melanoma cell lines when propagated in DMEM as monolayers was examined initially [30]. Susceptibility was compared to that of NK cell sensitive K562 and NK cell resistant Daudi cells. OCM1 cells manifested susceptibility to cytolysis mediated by NK cells isolated from the blood of normal donors and mediated by the human NK cytotoxic cell line NK-92; MUM2B cells were more highly resistant (Figure 1A and IB). Tumor factors associated with NK cell sensitivity were examined. Both cell lines expressed the NK cell inhibitory F1LA-A,B,C (MHC class I) and stimulatory MICA; expression of both of these regulatory molecules was greater by OCM1 cells (Figure 2A). MUM2B cells expressed less melanocyte differentiation determinants, MITF, DCT, MLANA, and TYR, than OCM1 cells, a characteristic of melanoma CSC (Figure 2B). Furthermore, MUM2B expressed more VIM, which is associated with a CSC phenotype in uveal melanoma; only very low levels of CDH1, which is inversely associated with a CSC phenotype, were detectable (Figure 2B) [10,31].
Figure 1.
(A) Cytolytic activity of IL-2 activated CD56+CD3− NK cells isolated from normal donors against MUM2B, OCM1, K562, and Daudi cells. Data represent mean ± SD, n = 3. (B) Cytolytic activity of NK-92 cells against CD271+ MUM2B and OCM1 cells enriched by magnetic cell sorting and against MUM2B and OCM1 cells propagated as monolayers in DMEM (DMEM) or as melanospheres in stem-cell supportive media (SCM). Data represent mean ± SD, n = 4.
Figure 2.
(A) Expression of HLA-A,B,C and MICA by MUM2B and OCM1 cells enriched for CD271+ and CD271− fractions by magnetic cell sorting and when propagated as monolayers in DMEM (DMEM) or as melanospheres in stem-cell supportive media (SCM) for 3 days. Data represent mean fluorescent intensity (MFI) ± SD, n = 3. *, P <0.05 MUM2B vs. OCM1. (B) Levels of melanocyte differentiation determinants of MUM2B and OCM1 cells enriched for CD271+ and CD271− fractions by magnetic cell sorting. Data represent mean ± SD, n = 3. *, P <0.05 CD271+ vs. CD271−. (C) Levels of melanocyte differentiation determinants of MUM2B and OCM1 cells cultured in SCM for 2, 3, of 7 days compared to culture as monolayers in DMEM. Data represent mean ± SD, n = 3. *, P <0.05 SCM vs. DMEM. (D) Expression of CD271 by OCM1 and MUM2B propagated either as monolayers in DMEM or as melanospheres in SCM for 3 days. Data represent mean MFI ± SD, n = 3. *, P <0.05 SCM vs. DMEM.
CSC from each cell line were first enriched by selecting CD271+ cells and tested for susceptibility to NK cell cytolysis. CD271+ cells derived from the same melanoma cell line were equally susceptible to NK cell mediated lysis when compared to their CD271− counterpart (Figure 1B). Relative expression of melanocyte determinants was lower in the CD271+ fraction of OCM1 cells to the CD271− fraction consistent with less differentiation (Figure 2B). Levels of HLA-A,B,C and MICA were similar (Figure 2A). NK cell sensitivity of CSC enriched by another established method, namely culturing in stem-cell supportive media to produce non-adherent spheres, referred to as melanospheres, was also examined. MUM2B and OCM1 cells formed typical melanospheres (Supplementary Figure S1). The susceptibility of the melanospheres to NK cell cytolysis mediated by NK-92 cells did not vary from the monolayer counterpart (Figure 1B). Melanospheres that developed in both cell lines were characterized by decreases in MITF and MLANA and increases in VIM consistent with de-differentiation (Figure 2C). The decreases in MITF and MLANA in OCM1 cells were >0.5 log. CD271 expression increased when the cell lines were propagated as melanospheres (Figure 2D) compared to propagation as monolayers, whereas levels of HLA-A,B,C and MICA were similar (Figure 2A).
Uveal melanoma CSC express and secrete NK cell regulatory miRs
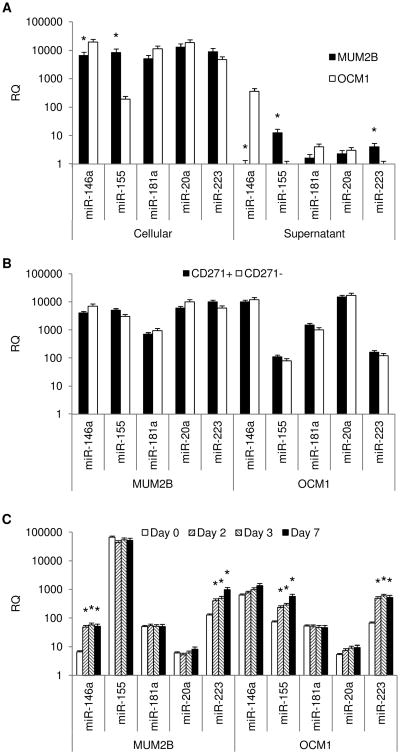
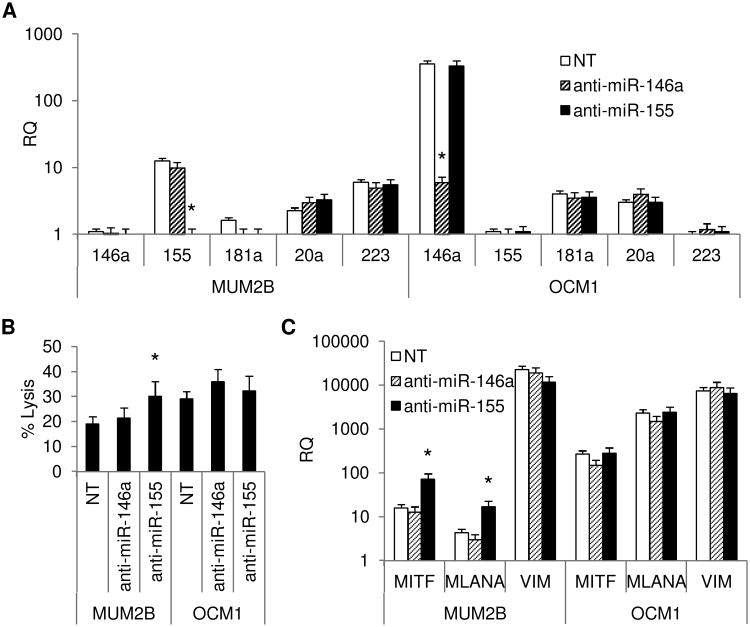
The production of NK cell regulatory miRs by OCM1 and MUM2B cells was determined (Figure 3A). Cellular expression of all miRs tested was found in both MUM2B and OCM1 cells. Significant expression of miR-155 was observed by MUM2B cells but not by OCM1. In contrast, miR-146a expression was greater in OCM1 cells. Expression of miRs 20a, 181a, and 223 was not significantly different. NK cell regulatory miR expression in supernatants paralleled cellular expression with miR-155 expression only measureable in MUM2B supernatants, and miR-146a, higher in OCM1 supernatants. CD271+ cells isolated from both cell lines expressed similar levels of miRs (Figure 3B). Culturing in stem-cell supportive medium was associated with increases in miR-155 in OCM1 cells, though the levels achieved were still 2-log less than that of MUM2B cells. Increases in miR-146a were observed in MUM2B cells. Levels of miR-146a achieved in MUM2B were 1-log less than those of OCM1 cells. miR-223 increased significantly in both cell lines (Figure 3C). MUM2B and OCM1 cells transfected with anti-miR-155 and anti-miR-146a were tested for NK cell sensitivity. Transfection significantly decreased the levels of the corresponding miR in cell supernatants (Figure 4A). Transfecting MUM2B cells with anti-miR-155 increased their NK cell sensitivity; OCM1 cell sensitivity was not altered. Transfecting MUM2B cells with anti-miR-146a had no effect on their NK cell sensitivity; OCM1 sensitivity tended to increase (Figure 4B). The increased NK cell sensitivity produced by anti-miR-155 in MUM2B cells was associated with increases in melanocyte differentiation markers, MITF and MLANA (Figure 4C).
Figure 3.
(A) Cellular and supernatant NK cell regulatory miR expression of MUM2B and OCM1 cells. Data represent mean ± SD, n = 4. *, P <0.05 MUM2B vs. OCM1. (B) Cellular expression of NK cell regulatory miR by MUM2B and OCM1 cells when enriched for CD271+ and CD271− fractions by magnetic cell sorting. Data represent mean ± SD, n = 3. (C) Changes in cellular NK cell regulatory miR expression of MUM2B and OCM1 cells cultured in stem-cell supportive media for 2, 3, or 7 days compared to culture as monolayers in DMEM (Day 0). Data represent mean ± SD, n = 3. *, P <0.05 vs. Day 0.
Figure 4.
(A) Supernatant NK cell regulatory miR production of MUM2B and OCM1 cells transfected by electroporation with anti-miR-155 and anti-miR 146a compared to cells electroporated without anti-miRs (NT). Data represent mean ± SD, n = 3. *, P <0.05 vs. NT. (B) Effect of anti-miR-155 and anti-miR-146a on sensitivity of MUM2B and OCM1 cells to NK cell lysis mediated by NK-92 cells at an E:T ratio of 5:1 compared to cells electroporated without anti-miRs (NT). Data represent mean ± SD, n = 3. *, P <0.05 vs. NT. (C) Levels of melanocyte differentiation determinants of MUM2B and OCM1 cells transfected by electroporation with anti-miR-155 and anti-miR 146a compared to cells electroporated without anti-miRs (NT) cultured for 3 days in DMEM. Data represent mean ± SD, n = 3. *, P <0.05 vs. NT.
Circulating CD271+ cells have CSC characteristics and express NK cell regulatory miRs
That circulating melanoma cells in patients expressed CD271 was first examined by immunofluorescence staining. Cell staining methods were first optimized using huffy coats from normal volunteers spiked with SKMEL-28 cells, cutaneous melanoma cells known to express CD271 (Supplementary Figure S2 and S3) [32], These studies also confirmed a lack of expression of CD271 by peripheral blood leukocytes from normal volunteers. Cytospins of huffy coats were prepared from patients with metastatic uveal melanoma and stained with antibodies to CD271, Melan-A, and CD45 (Figure 5 and Supplementary Figure S4). Approximately 1 in every 50,000 to 100,000 cells among the patient huffy coats stained with CD271 alone. There was heterogeneity in the size and shape of these CD271+ cells. Most were slightly larger than leukocytes with elongated or irregular shapes (Figure 5A), while some were similar in size to leukocytes and more rounded (Figure 5B). Confocal imaging revealed that some of the CD271+ cells were also positive for melan A, indicating melanocyte derivation (Figure 5C). In contrast to the normal volunteer preparations, isolated cells that stained for both CD271 and CD45 were observed in the patient preparations. Cells positive for both Melan-A and CD45 were not observed. The expression of melanocyte differentiation determinants by CD271+ cells isolated by positive selection from patients with metastatic uveal melanoma were then assessed (Figure 6A). Levels were compared to those of the less differentiated MUM2B and the more differentiated OCM1 uveal melanoma cells. CD271+ cells isolated from all patients expressed MITF, the key regulatory of melanocyte differentiation. DCT, MLANA, and TYR, melanocyte differentiation genes downstream of MITF, were not highly expressed, paralleling the less differentiated expression pattern apparent in the NK cell resistant MUM2B cells. CD271 cells isolated from patients with metastatic uveal melanoma were also cultured in stem-cell supportive media. Melanospheres consistent with a CSC phenotype, formed (Figure 5D). These results are consistent with a CSC-like phenotype of circulating CD271+ cells in patients with metastatic uveal melanoma. Figure 6B displays the NK cell regulatory miR expression of CD271 cells isolated from patients with metastatic uveal melanoma. Similar to the uveal melanoma cell lines, NK cell regulatory miRs were measurable in all samples. Notably, significant expression of miR-155 was observed.
Figure 5.
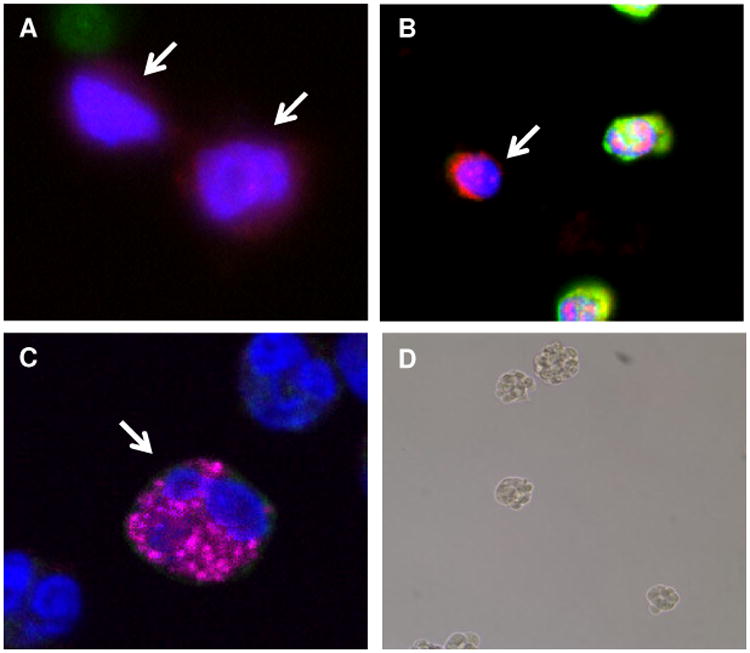
(A and B) Examples of blood nucleated cells from patients with metastatic uveal melanoma that were positive for CD271 and negative for CD45 (arrows) and (C) virtual confocal imaged slice cells of cells positive for CD271 and Melan-A (arrow). Nuclei are visualized with DAPI (blue color). (D) Example of the formation of non-adherent spheres by CD271+ circulating cells from a patient with metastatic uveal melanoma cultured for 4 days in stem-cell supportive media.
Figure 6.
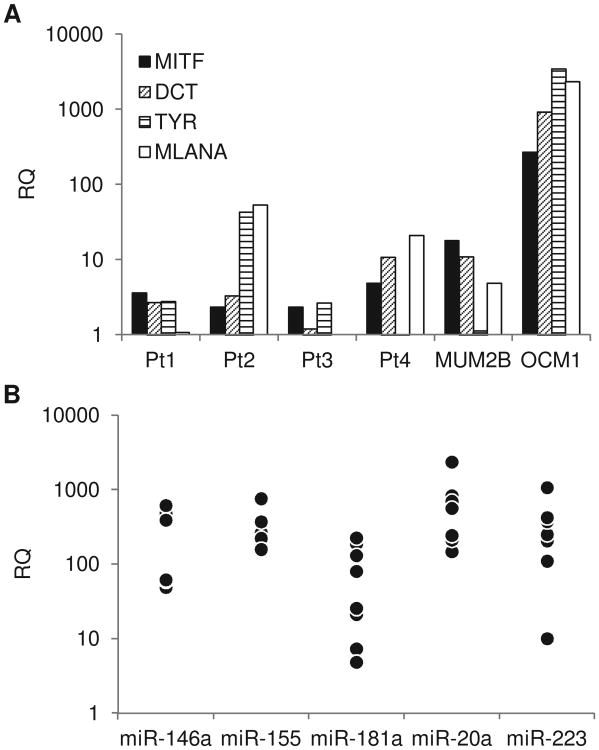
(A) Levels of melanocyte differentiation determinants of CD271+ cells isolated from the blood of four patients with metastatic uveal melanoma (Pt1-4) compared with MUM2B and OCM1 cells. Data represent means of duplicate determinations. (B) NK cell regulatory miR expression of CD271+ cells isolated from the blood of nine patients with metastatic uveal melanoma. Data represent means of duplicate determinations.
Discussion
NK cells and CSC are considered to play central roles in the metastatic progression of uveal melanoma. That CSC would be protected from immune surveillance, as are embryonic stem cells, would be of obvious survival benefit and necessary for the development of metastasis. In several models, however, CSC have been shown to be sensitive to NK cell cytolysis [17-19]. We examined CSC derived from the more aggressively metastatic MUM2B and the less aggressively metastatic OCM1 uveal melanoma cell lines. Differential NK cell sensitivity was observed. CSC isolated from MUM2B cells either by CD271+ selection or by propagation as melanospheres in stemcell supportive media, which increased CD271 expression, were relatively resistant. CSC similarly isolated from OCM1 cells were relatively sensitive. That CSC isolated from cell lines by propagation in stem-cell supportive media or by positive selection are similarly sensitive to NK cell cytolysis than the non-CSC counterpart has been observed in cutaneous melanoma models [19]. In other models, NK cell lysis of CSC but not their differentiated progeny has been demonstrated, suggesting that de-differentiation enhances NK cell sensitivity [18]. In the uveal melanoma models studied, de-differentiation was not associated with increased NK cell sensitivity. Not only were the poorly differentiated MUM2B cells resistant to NK cytolysis, the de-differentiation effected by propagating cells as melanospheres did not increase sensitivity. CSC from both cell lines expressed NK-cell inhibitory MHC class I and stimulatory MICA molecules, levels of which were not substantially affected by CSC enrichment. Collectively, these data provide evidence that NK cells can lyse some uveal melanoma CSC more efficiently than others. These data also underscore the complexity of the interactions involved in conferring NK cell sensitivity.
Increasing evidence support a role for miRs in immune regulation. We found that uveal melanoma CSC express and secrete NK cell regulatory miRs. miR-155 was the most highly and differentially expressed in the NK cell resistant MUM2B cells, increased the sensitivity of MUM2B cells to NK cell cytolysis when inhibited, and was highly expressed in circulating CD271+ cells from patients with metastatic uveal melanoma. miR-155 has been shown to regulate both the threshold and extent of NK cell activation by affecting multiple signaling pathways. miR-155 regulates NK cell IFN-γ and granzyme B production and NKG2D expression [33,34], NK cell IFN-γ production has also been shown to increase with miR-155 overexpression [33]. IFN-γ renders uveal melanoma cells resistant to granule mediated lysis by cytotoxic lymphocytes [35]. Several other non-immunologic roles in oncogenesis for miR-155 and the other miRs studied have been demonstrated. miR-155 has been implicated in melanoma development [36]. miR-155 has also been shown to promote CSC phenotypes and to regulate tumor adhesion, epithelial-mesenchymal transition, invasion, migration, and angiogenesis [37-40]. CD271 has been used to identify CSC not only in melanoma but also in neuroblastoma, oral, hypopharyngeal, and esophageal squamous cell, and breast carcinomas [41]. The metastatic potential of CD271+ melanoma cells has been shown to vary [42], Our results indicate that expression of CD271 does not confer NK cell resistance/sensitivity. Studies of the characteristics of CD271+ cells in uveal melanoma have been limited to cell lines in vitro [12]. This is the first report characterizing circulating CD271+ cells in patients with uveal melanoma. The circulating CD271+ cells isolated manifested decreased expression of melanocyte differentiation determinants characteristic of uveal melanoma CSC [43,44], Circulating CSC are considered to be central to the metastatic process, the cellular subset that is not only be capable of evading from the primary tumor, but also escaping from immune surveillance, surviving in the circulation, and subsequently forming metastases in distant organs. CD271 expression is not neural or tumor specific. It can be expressed by non-neural, normal circulating cells, including a subset of B cells [45]. Other markers have been used to identify uveal melanoma CSC, such as the transmembrane glycoprotein CD133 (prominin-1) [11]. CD133 is expressed by several non-neural circulating cells, not only hematopoietic but also endothelial in origin [46,47].
That miRs play a role in regulating the metastatic properties of CSC is supported by several studies [20]. Our results suggest the possibility that CSC miRs may play a role in regulating immune surveillance of uveal melanoma. Molecular profiling of tumors can identify patients with uveal melanoma at high risk for metastasis, but the mechanisms regulating the clinical manifestation of metastasis, which can occur 10 to 15 years after primary therapy, are not established [48,49]. Whether the sensitivity of uveal melanoma CSC to NK cell cytolysis varies over time and the development of metastasis and the role of immune regulatory miRs will require further study. Highly stable in the circulation, miRs hold great promise as a new class of blood biomarkers of the metastatic process [50]. miRs are also potentially therapeutic targets. That supplementing miRs in vivo can stimulate protective antitumor immunity has been demonstrated in mouse models [51]. Methods of directly modifying miR expression are under investigation [52],
Supplementary Material
Supplementary Figure S1. Melanosphere formation. (A) MUM2B cells and (B) OCM1 cells cultured as adherent monolayers in DMEM. (C) MUM2B and (D) OCM1 cells cultured as non-adherent spheres in stem cell supportive media for 3 days.
Supplementary Figure S2. Staining optimization. (A) Normal blood nucleated cells (huffy coat) were stained with CD271, and DAPI. Not the absence of signal greater than background on these cells. (B) SKMEL-28 melanoma cells show robust staining with CD271. (C) Mixed sample containing normal blood nucleated cells spiked with a few SKMEL-28 cells and stained with CD271, CD45 and DAPI. Red arrows point to CD271 expression on SKMEL-28 cells, which are detected by their relatively larger size compared with the blood cells.
Supplementary Figure S3. Staining controls. (A) A representative image of a secondary antibody control which were part of all optimizations for all antibodies. (B) Mixed sample of cells containing normal blood nucleated cells spiked with SKMEL-28 cells and stained with Melan-A. Red arrows indicate Melan-A stained cells, and green arrows indicate CD45 stained nucleated blood leukocytes.
Supplementary Figure S4. Circulating cells from patients. Blood nucleated cells (Buffy coat) from patients with metastatic uveal melanoma were stained with CD271, CD45, Melan-A antibodies and with DAPI. They were imaged with Leica SP2 confocal imaging system using LASERs: Argon UV for DAPI at wavelengths 351-364nm, Argon-krypton at 488 nm, LED diode at 361 nm, and Helium-Neon at 633 nm. Non-overlapping, narrow excitation and emission bandwidths were selected to eliminate signal contamination between channels. Images presented are from a virtual confocal imaged slice. (A-D) and (F-I) Nuclei stained with DAPI (blue), CD45 (green), CD271 (red), and Melan-A (magenta). (E and J) Composite image of channels from AD and F-I respectively. (E1, E2, J3, J4) Enlarged images of inset cells are CD271 + are also Melan-A+ but not CD45+ (magenta arrows).
Acknowledgments
This work was supported in part by R21CA175671 from the National Cancer Institute, National Institutes of Health, Bethesda, MD.
Footnotes
Compliance with Ethical Standards: The authors declare that they do not have any competing or financial interests.
Ethical standards: Blood samples were collected from patients with metastatic uveal melanoma and from normal subjects according to protocols approved by the Cleveland Clinic and Wake Forest University Institutional Review Boards. All subjects gave informed consent prior to inclusion in the study. This research was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
References
- 1.Dithmar S, Rusciano D, Lynn MJ, Lawson DH, Armstrong CA, Grossniklaus HE. Neoadjuvant interferon alfa-2b treatment in a murine model for metastatic ocular melanoma: a preliminary study. Arch Ophthalmol. 2000;118:1085–9. doi: 10.1001/archopht.118.8.1085. [DOI] [PubMed] [Google Scholar]
- 2.Blom DJ, Luyten GP, Mooy C, Kerkvliet S, Zwinderman AH, Jager MJ. Human leukocyte antigen class I expression: marker of poor prognosis in uveal melanoma. Invest Ophthalmol Vis Sci. 1997;38:1865–72. [PubMed] [Google Scholar]
- 3.Ericsson C, Seregard S, Bartolazzi A, Levitskaya E, Ferrone S, Kiessling R, Larsson O. Association of HLA class I and class II antigen expression and mortality in uveal melanoma. Invest Ophthalmol Vis Sci. 2001;42:2153–6. [PubMed] [Google Scholar]
- 4.Vetter CS, Lieb W, Bröcker EB, Becker JC. Loss of nonclassical MHC molecules MIC-A/B expression during progression of uveal melanoma. Br J Cancer. 2004;91:1495–9. doi: 10.1038/sj.bjc.6602123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma D, Luyten GP, Luider TM, Niederkom JY. Relationship between natural killer cell susceptibility and metastasis of human uveal melanoma cells in a murine model. Invest Ophthalmol Vis Sci. 1995;36:435–41. [PubMed] [Google Scholar]
- 6.Blom DJ, De Waard-Siebinga I, Apte RS, Luyten GP, Niederkom JY, Jager MJ. Effect of hyperthermia on expression of histocompatibility antigens and heat-shock protein molecules on three human ocular melanoma cell lines. Melanoma Res. 1997;7:103–9. doi: 10.1097/00008390-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Repp AC, Mayhew ES, Apte S, Niederkom JY. Human uveal melanoma cells produce macrophage migration-inhibitory factor to prevent lysis by NK cells. J Immunol. 2000;165:710–5. doi: 10.4049/jimmunol.165.2.710. [DOI] [PubMed] [Google Scholar]
- 8.Jager MJ, Hurks HM, Levitskaya J, Kiessling R. HLA expression in uveal melanoma: there is no rule without some exception. Hum Immunol. 2002;63:444–51. doi: 10.1016/s0198-8859(02)00389-0. [DOI] [PubMed] [Google Scholar]
- 9.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- 10.Kalirai H, Damato BE, Coupland SE. Uveal melanoma cell lines contain stem-like cells that self-renew, produce differentiated progeny, and survive chemotherapy. Invest Ophthalmol Vis Sci. 2011;52:8458–66. doi: 10.1167/iovs.11-7379. [DOI] [PubMed] [Google Scholar]
- 11.Thill M, Berna MJ, Grierson R, Reinhart I, Voelkel T, Piechaczek C, Galambos P, Jager MJ, Richard G, Lange C, Gehling UM. Expression of CD133 and other putative stem cell markers in uveal melanoma. Melanoma Res. 2011;21:405–16. doi: 10.1097/CMR.0b013e328348db10. [DOI] [PubMed] [Google Scholar]
- 12.Valyi-Nagy K, Kormos B, Ali M, Shukla D, Valyi-Nagy T. Stem cell marker CD271 is expressed by vasculogenic mimicry-forming uveal melanoma cells in three-dimensional cultures. Mol Vis. 2012;18:588–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013;13:371. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres V, Triozzi P, Eng C, Tubbs R, Schoenfiled L, Crabb JW, Saunthararajah Y, Singh AD. Circulating tumor cells in uveal melanoma. Future Oncol. 2011;7:101–9. doi: 10.2217/fon.10.143. [DOI] [PubMed] [Google Scholar]
- 15.Murphy GF, Wilson BJ, Girouard SD, Frank NY, Frank MH. Stem cells and targeted approaches to melanoma cure. Mol Aspects Med. 2014;39:33–49. doi: 10.1016/j.mam.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, Seifert B, Moch H, Dummer R, van den Broek M, Sommer L. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71:3098–109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 17.Tseng HC, Arasteh A, Paranjpe A, Teruel A, Yang W, Behel A, Alva JA, Walter G, Head C, Ishikawa TO, Herschman HR, Cacalano N, Pyle AD, Park NH, Jewett A. Increased lysis of stem cells but not their differentiated cells by natural killer cells; de-differentiation or reprogramming activates NK cells. PLoS One. 2010;5:e11590. doi: 10.1371/journal.pone.0011590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng HC, Inagaki A, Bui VT, Cacalano N, Kasahara N, Man YG, Jewett A. Differential Targeting of Stem Cells and Differentiated Glioblastomas by NK Cells. J Cancer. 2015;6:866–76. doi: 10.7150/jca.11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietra G, Manzini C, Vitale M, Balsamo M, Ognio E, Boitano M, Queirolo P, Moretta L, Mingari MC. Natural killer cells kill human melanoma cells with characteristics of cancer stem cells. Int Immunol. 2009;21:793–801. doi: 10.1093/intimm/dxp047. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–4. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–75. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trotta R, Chen L, Ciarlariello D, Josyula S, Mao C, Costinean S, Yu L, Butchar JP, Tridandapani S, Croce CM, Caligiuri MA. miR-155 regulates IFN-γ production in natural killer cells. Blood. 2012;119:3478–85. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, Heinz ME, Crosby SD, Nagarajan R, Ramsingh G, Link DC, Ley TJ, Mardis ER. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Zhang J, Hou Z, Han Q, Zhang C, Tian Z. miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression. Cell Cycle. 2015;14:243–52. doi: 10.4161/15384101.2014.977112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stem-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 26.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome- mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.He S, Chu J, Wu LC, Mao H, Peng Y, Alvarez-Breckenridge CA, Hughes T, Wei M, Zhang J, Yuan S, Sandhu S, Vasu S, Benson DM, Jr, Hofmeister CC, He X, Ghoshal K, Devine SM, Caligiuri MA, Yu J. MicroRNAs activate natural killer cells through Toll-like receptor signaling. Blood. 2013;121:4663–71. doi: 10.1182/blood-2012-07-441360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Achberger S, Aldrich W, Tubbs R, Crabb JW, Singh AD, Triozzi PL. Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease. Mol Immunol. 2014;58:182–6. doi: 10.1016/j.molimm.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folberg R, Kadkol SS, Frenkel S, Valyi-Nagy K, Jager MJ, Pe'er J, Maniotis AJ. Authenticating cell lines in ophthalmic research laboratories. Invest Ophthal Vis Sci. 2008;49:4697–701. doi: 10.1167/iovs.08-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folberg R, Leach L, Valyi-Nagy K, Lin AY, Apushkin MA, Ai Z, Barak V, Majumdar D, Pe'er J, Maniotis AJ. Modeling the behavior of uveal melanoma in the liver. Invest Ophthalmol Vis Sci. 2007;48:2967–74. doi: 10.1167/iovs.06-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan F, Lin M, Li C, Ding X, Qian G, Zhang H, Ge S, Fan X, Li J. Effects of inhibition of hedgehog signaling on cell growth and migration of uveal melanoma cells. Cancer Biol Ther. 2014;15:544–59. doi: 10.4161/cbt.28157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truzzi F, Marconi A, Lotti R, Dallaglio K, French LE, Hempstead BL, Pincelli C. Neurotrophins and their receptors stimulate melanoma cell proliferation and migration. J Invest Dermatol. 2008;128:2031–40. doi: 10.1038/jid.2008.21. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan RP, Fogel LA, Leong JW, Schneider SE, Wong R, Romee R, Thai TH, Sexl V, Matkovich SJ, Dorn GW, 2nd, French AR, Fehniger TA. MicroRNA-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J Immunol. 2013;191:5904–13. doi: 10.4049/jimmunol.1301950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elemam NM, Mekky RY, El-Ekiaby NM, El Sobky SA, El Din MA, Esmat G, Abdelaziz AI. Repressing PU.1 by miR-29a* in NK cells of HCV patients, diminishes its cytolytic effect on HCV infected cell models. Hum Immunol. 2015;76:687–94. doi: 10.1016/j.humimm.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Hallermalm K, Seki K, De Geer A, Motyka B, Bleackley RC, Jager MJ, Froelich CJ, Kiessling R, Levitsky V, Levitskaya J. Modulation of the tumor cell phenotype by IFN-gamma results in resistance of uveal melanoma cells to granule-mediated lysis by cytotoxic lymphocytes. J Immunol. 2008;180:3766–74. doi: 10.4049/jimmunol.180.6.3766. [DOI] [PubMed] [Google Scholar]
- 36.Levati L, Pagani E, Romani S, Castiglia D, Piccinni E, Covaciu C, Caporaso P, Bondanza S, Antonetti FR, Bonmassar E, Martelli F, Alvino E, D'Atri S. MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24:538–50. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Kong X, Lv L, Gao J. TGF-β1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015;359:288–98. doi: 10.1016/j.canlet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 38.Ji J, Zheng X, Forgues M, Yamashita T, Wauthier EL, Reid LM, Wen X, Song Y, Wei JS, Khan J, Thorgeirsson SS, Wang XW. Identification of microRNAs specific for epithelial cell adhesion molecule-positive tumor cells in hepatocellular carcinoma. Hepatology. 2015;62:829–40. doi: 10.1002/hep.27886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375–80. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 40.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–89. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomellini E, Lagadec C, Polakowska R, Le Bourhis X. Role of p75 neurotrophin receptor in stem cell biology: more than just a marker. Cell Mol Life Sci. 2014;71:2467–81. doi: 10.1007/s00018-014-1564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheli Y, Bonnazi VF, Jacquel A, Allegra M, De Donatis GM, Bahadoran P, Bertolotto C, Ballotti R. CD271 is an imperfect marker for melanoma initiating cells. Oncotarget. 2014;5:5272–83.39. doi: 10.18632/oncotarget.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergeron MA, Champagne S, Gaudreault M, Deschambeault A, Landreville S. Repression of genes involved in melanocyte differentiation in uveal melanoma. Mol Vis. 2012;18:1813–22. [PMC free article] [PubMed] [Google Scholar]
- 44.Landreville S, Lupien CB, Vigneault F, Gaudreault M, Mathieu M, Rousseau AP, Guerin SL, Salesse C. Identification of differentially expressed genes in uveal melanoma using suppressive subtractive hybridization. Mol Vis. 2011;17:1324–33. [PMC free article] [PubMed] [Google Scholar]
- 45.Melamed I, Kelleher CA, Franklin RA, Brodie C, Hempstead B, Kaplan D, Gelfand EW. Nerve growth factor signal transduction in human B lymphocytes is mediated by gpl40trk. Eur J Immunol. 1996;26:1985–92. doi: 10.1002/eji.1830260903. [DOI] [PubMed] [Google Scholar]
- 46.Dome B, Timar J, Dobos J, Meszaros L, Raso E, Paku S, Kenessey I, Ostoros G, Magyar M, Ladanyi A, Bogos K, Tovari J. Identification and clinical significance of circulating endothelial progenitor cells in human non-small cell lung cancer. Cancer Res. 2006;66:7341–7. doi: 10.1158/0008-5472.CAN-05-4654. [DOI] [PubMed] [Google Scholar]
- 47.Vroling L, Lind JS, de Haas RR, Verheul HM, van Hinsbergh VW, Broxterman HJ, Smit EF. CD 133+ circulating haematopoietic progenitor cells predict for response to sorafenib plus erlotinib in non-small cell lung cancer patients. Br J Cancer. 2010;102:268–75. doi: 10.1038/sj.bjc.6605477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damato B, Coupland SE. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol. 2009;127:423–9. doi: 10.1001/archophthalmol.2009.40. [DOI] [PubMed] [Google Scholar]
- 49.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 51.Cubillos-Ruiz JR, Baird JR, Tesone AJ, Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, Anadon-Arnillas J, Harwood NM, Korc M, Fiering SN, Sempere LF, Conejo-Garcia JR. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–93. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson A, Linsley PS. The therapeutic potential of microRNA modulation. Discov Med. 2010;9:311–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Melanosphere formation. (A) MUM2B cells and (B) OCM1 cells cultured as adherent monolayers in DMEM. (C) MUM2B and (D) OCM1 cells cultured as non-adherent spheres in stem cell supportive media for 3 days.
Supplementary Figure S2. Staining optimization. (A) Normal blood nucleated cells (huffy coat) were stained with CD271, and DAPI. Not the absence of signal greater than background on these cells. (B) SKMEL-28 melanoma cells show robust staining with CD271. (C) Mixed sample containing normal blood nucleated cells spiked with a few SKMEL-28 cells and stained with CD271, CD45 and DAPI. Red arrows point to CD271 expression on SKMEL-28 cells, which are detected by their relatively larger size compared with the blood cells.
Supplementary Figure S3. Staining controls. (A) A representative image of a secondary antibody control which were part of all optimizations for all antibodies. (B) Mixed sample of cells containing normal blood nucleated cells spiked with SKMEL-28 cells and stained with Melan-A. Red arrows indicate Melan-A stained cells, and green arrows indicate CD45 stained nucleated blood leukocytes.
Supplementary Figure S4. Circulating cells from patients. Blood nucleated cells (Buffy coat) from patients with metastatic uveal melanoma were stained with CD271, CD45, Melan-A antibodies and with DAPI. They were imaged with Leica SP2 confocal imaging system using LASERs: Argon UV for DAPI at wavelengths 351-364nm, Argon-krypton at 488 nm, LED diode at 361 nm, and Helium-Neon at 633 nm. Non-overlapping, narrow excitation and emission bandwidths were selected to eliminate signal contamination between channels. Images presented are from a virtual confocal imaged slice. (A-D) and (F-I) Nuclei stained with DAPI (blue), CD45 (green), CD271 (red), and Melan-A (magenta). (E and J) Composite image of channels from AD and F-I respectively. (E1, E2, J3, J4) Enlarged images of inset cells are CD271 + are also Melan-A+ but not CD45+ (magenta arrows).