Abstract
Plants are indispensable on earth and their improvement in terms of food security is a need of time. The current study has been designed to investigate how biogenic zinc nanoparticles (Zn NPs) can improve the growth and development of Brassica napus L. In this study, Zn NPs were synthesized utilizing Mentha arvensis aqueous extracts, and their morphological and optical properties were assessed using UV-Visible spectrophotometry, scanning electron microscopy (SEM), transmission electron microscopy (TEM), and X-ray diffraction (XRD). The synthesized Zn NPs were irregular in shape, indicating aggregation in pattern, with an average particle size of 30 nm, while XRD analysis revealed the crystalline structure of nanoparticles. The growth and development of B. napus varieties (Faisal canola and Shiralee) were assessed after foliar treatments with different concentrations of biogenic Zn NPs. In B. napus varieties, exposure to 15 mg/L Zn NPs dramatically increased chlorophyll, carotenoid content, and biomass accumulation. Similarly, proteomic analyses, on the other hand, revealed that proteins associated with photosynthesis, transport, glycolysis, and stress response in both Brassica varieties were substantially altered. Such exposure to Zn NPs, differential expression of genes associated with photosynthesis, ribosome structural constituents, and oxidative stress response were considerably upregulated in B. napus var. (Faisal and Shiralee canola). The results of this study revealed that foliar applications of biogenic Zn NPs influence the transcriptome and protein profiling positively, therefore stimulating plant growth and development.
Keywords: nanotechnology, proteomics, Brassica napus, zinc nano fertilizer, green synthesis
Introduction
Plants extracts have the extraordinary potential to transform bulk metals into nanoparticles (NPs). They can reduce bulk material in an eco-friendly reaction into nanostructures. The photometabolites along with the reducing capabilities stabilize and provide surface functionalization attributes to the NPs (Javed and Mashwani, 2020b). The physical and chemical methods of reduction of bulk material involve the utilization of hazardous chemicals, which make nanostructures unsuitable for use in biological applications due to their toxic nature and harmful effects on plants, the environment, and other living organisms (Ahmad et al., 2020; Javed and Mashwani, 2020a). However, biofabrication using plant extracts results in the synthesis of biocompatible nanomaterials. Plants have the natural mechanism to absorb essential nutrients from the soil and reduce them into smaller particles, which latter assimilate and help improve growth and development of the plants (Javed et al., 2020).
Recent advances in nanotechnology allowed the development of NPs that can be widely used in an increasing number of different applications (Allhoff et al., 2010). Nanoparticles are aggregates of atoms or molecules with small size, usually less than 100 nm, and large surface areas, which drastically alter their physicochemical characteristics as compared with bulk materials (Love et al., 2005). The exposure of some plants to some NPs gives rise to reactive oxygen species (ROS) production, which has both positive and negative effects (Mazumder et al., 2020). Moreover, the nature, size, surface area, and composition of metal materials greatly influence the activity of NPs (Jain et al., 2007). The massive production and utilization of NPs are becoming a serious issue regarding their environmental impact (Klaine et al., 2008; Raghib et al., 2020). Plants are vital elements of the ecosystem and can react to NP exposition (Khot et al., 2012; Prasad et al., 2014). Therefore, the release of NPs into the environment, contact of NPs with plants, and their effect on plants, ecosystems, and the entire environment need a serious reconsideration.
Advanced nanotechnology has provided the technological platform to study the effects as well as mechanism of NPs effects in plants (Sahoo et al., 2007; Newman et al., 2009). The unique characteristics of nanosized particles (1–100 nm) have arouse the attention of diverse scientific communities, such as agronomy, technology, biology, and chemistry, because of their unique characteristics in comparison with bulk material (Ejaz et al., 2018). The NPs, which are made up of metal oxides, are used extensively for commercial purposes, such as antimicrobial soaps, topical sunscreens, and coatings for self-cleaning (Javed et al., 2020). The contamination of ecosystems by NPs is becoming a major concern due to their overproduction for use in daily consumables (Liu et al., 2021). The rapid development of the nanomaterial industry and the utilization of nanomaterial in mass products have caused difficulties in controlling the release of engineered NPs into the environment (Piccinno et al., 2012). However, there is very limited knowledge available about NPs effects on the environment and further studies are needed.
Zinc nanoparticles (Zn NPs) are widely used in many fields, e.g., agriculture and medicine. It was observed that Zn NPs can greatly influenced plant growth, yield, and fatty acid profiles of maize (Taheri et al., 2016). An increase in germination percentage and root length of maize plants was observed after exposure to zinc oxide nanoparticles (ZnO NPs) (Meena et al., 2017), whereas increased shoot length was observed in oat and berseem plants (Maity et al., 2018). It was also confirmed that ZnO NPs enhance the plant growth and increased root biomass of the Glycine max plant (De la Rosa et al., 2013). Foliar application of Zn NPs significantly increases the leaf area and dry mass of maize (Taheri et al., 2016). However, ZnO NPs and aluminum oxide NPs prevented root elongation of plants like cucumber, cabbage, corn, soybean, and carrot. The submergence effect on plants can be reduced by improving the duration of thresholds and flooding depth. Hossain et al. (2012) proclaimed that metallic NPs can protect plants from the detrimental effects of flooding by enhancing ATP generation rate and regulating the pathways of secondary metabolism. Zinc oxide nanoparticles can interact with plant roots in a various way. They can release Zn ions (Zn2+) that can be taken up by Zn transporters (Milner et al., 2013). Uptake might also happen through pores that are larger than the ZnO NPs (Fleischer et al., 1999). The rate of uptake and effectiveness of several NPs on growth and metabolism differ among various plants (Singh et al., 2018; Sturikova et al., 2018).
Keeping in mind the importance of zinc as a micronutrient, we present the environmentally friendly bioassisted synthesis of Zn NPs by using aqueous extracts of Mentha as an efficient oxidizing/reducing and capping agent. Although the biosynthesis of Zn NPs has been documented as Brassica oleracea (Awan et al., 2021), Brassica juncea (Mazumder et al., 2020), and Brassica nigra (Zafar et al., 2016), their varied effect on plant physiological and biochemical profiling has received less attention. The purpose of this study was to disclose the biological consequence of green synthesized Zn NPs on Brassica napus proteomics profiling, which was ignored in many published studies on Brassica crop (Mousavi Kouhi et al., 2015; El-Badri et al., 2021). In addition, different physiological and growth attributes have been investigated to strengthen this research. The idea was that Zn NPs would be advantageous at low doses, because higher dose may cause reduction in growth and lead to toxicity in plants. Plants are the foundational elements of the ecosystem, providing the initial energy for the food chain’s major consumers; therefore, Brassica crop has been used as model plant in this study.
Materials and Methods
Plant Extract Preparation
The aqueous leaf extracts of Mentha arvensis were used as a reducing and capping agent to produce Zn NPs. Fresh and healthy leaves were vigorously washed with tap water and distilled water. Notably, 20 g of leaf material were mixed with 100 ml of Milli-Q® water and incubated at 50°C for 4 h. The extract was filtered two times by using Whatman filter paper No. 21 and kept at 4°C for further use (Dobrucka and Długaszewska, 2016).
Green Synthesis of Nanoparticles
The green synthesis of NPs was performed by the reduction of ZnNO3⋅7H2O (zinc nitrate heptahydrate) using plant aqueous extract. In this process, 5-mM solution of Zn salt was prepared using Milli-Q® water and was kept on a magnetic stirrer followed by the continuous addition of 100 ml of Mentha plant aqueous extract dropwise for 40 min. The color of the reaction mixture was changed to light brown, indicating the synthesis of Zn NPs. However, the synthesis of Zn NPs was confirmed by measuring the absorbance of the colloidal suspension of NPs between 200 and 800 nm of the light wavelength using a UV-visible spectrophotometer (Beckman Model DU640). This reaction solution was centrifuged at 1,000 × g for 1 h. The Zn NPs pellet was collected carefully, followed by dispersion in ethanol and re-centrifugation. The procedure was repeated two times to remove impurities. The Zn NPs were placed in the oven at 100°C to completely dry. The dried Zn NPs were stored in airtight vials for future use.
Morphological and Optical Characterization
The biosynthesized Zn NPs were subjected to different material characterization techniques in order to determine their morphological, optical, and biochemical attributes (Almessiere et al., 2020). The scanning electron microscopy (SEM) SIGMA model (MIRA3; TESCAN Brno) and transmission electron microscopy (TEM, JEM-2200FS) were used to collect the micrographic images (Sohail et al., 2019). The drop-coating method was used to prepare the samples, and the images were collected at different voltages and laser intensities. The crystalline nature of Zn NPs was confirmed by using the X-ray diffraction (XRD, Bruker D2 Phaser) analysis with the monochromatic Cu-Kα1 radiation at a 2θ angle between 10° and 80° (Sohail et al., 2020).
Plant Materials and Growth Conditions
Locally available seeds of B. napus varieties (e.g., Faisal canola and Shiralee) collected from the National Agricultural Research Center, Islamabad, Pakistan, were grown directly in 5.5 cm × 5.5 cm pots with TS-1 white peat bedding substrate and were kept in dark for 2 days at 4°C. Later, the plant pots were transferred to a climate chamber at a temperature of 22°C with light intensity (photosynthetic active radiation) of approximately 120 mol/m2s and a light/dark photoperiod of 16 h/8 h.
Zinc Nanoparticles Treatment
For proteomics analysis, seeds of canola varieties were sown in pots, and two consecutive sprays of Zn NPs (i.e., 5, 15, and 25 mg/L) were made after seedling emergence. One-week-old plants were treated with 5, 15, and 25 mg/L of biosynthesized Zn NPs by foliar spraying, followed by a second spray at day 3 after the first treatment.
Biomass and Water Content Measurement
Four-week-old plants from each pot of three biological replicates were harvested and washed thoroughly with tap water and distilled water. The moisture was removed by using a paper towel. The roots and shoots were separated, and fresh weight (FW) was measured. The sample was then dried in an oven at 80°C for 20 min, followed by vacuum dry at 40°C to constant mass before recording dry weight (DW). The plant water contents were calculated using the following formula (Whetherley, 1950):
Chlorophyll and Carotenoid Content Measurement
To determine the chlorophyll and carotenoid contents, rosette leaves of treated plants were harvested and transported in polythene zipper bags. The leaves were washed with tap and distilled water and homogenized with 80% acetone followed by centrifugation for 10 min at 14,000 rpm. The supernatant was collected and subjected to UV-visible spectrophotometry analysis. The chlorophyll a (Chl a), chlorophyll b (Chl b), total chlorophyll (Chl T), and carotenoid (Car) contents were measured using the methodology described by Arnon (1949). The chlorophyll content was determined using methodology described by Lichtenthaler and Wellburn (1983).
Protein Extraction
For the preparation of GTEN buffer, the buffer was mixed in the distilled water and kept at 4°C. The extraction buffer was prepared in a prechilled tube by equipping GTEN buffer with 10 mM dithiothreitol (DTT), 0.2% Nonidet-40 (Igepal), antiprotease tablet (Sigma-Aldrich) (1 tablet per 10 ml), and 2% polyvinylpyrrolidone (PVPP). About 100 mg of fresh leaves were powdered in liquid nitrogen using a pestle and mortar. The powder was mixed with 150 mM GTEN buffer (10% glycerol, Tris–HCl, pH 7.5), 1 mM ethylenediaminetetraacetic acid (EDTA), 150 mM NaCl, 10 mM DTT, 0.2% Nonidet-40 (Igepal), antiprotease tablet complete™ Proteasehemmer-Cocktail (1 tablet for 10 ml), and 2% PVPP. The thawed material was incubated on a rotator for 20 min at 4°C. Each sample was then centrifuged at 5,000 × g at 4°C for 20 min to remove cell debris. The supernatant was collected carefully and shifted to a new prechilled tube on ice. A second centrifugation at 5,000 × g at 4°C for 5 min of each sample was carried out and the collected supernatant was shifted to a prechilled tube by filtering through Miracloth. Total protein concentration in the sample was investigated by using Bradford’s (1976) assays.
Digestion of Protein for Mass Spectrometry Analysis
For proteomics analysis, proteins were obtained from Center of Proteome, University of Tübingen, Tübingen, Germany. The proteins were separated on a short SDS page and visualized by Coomassie staining. Gel pieces were in-gel digested with trypsin (Borchert et al., 2010). The extracted peptides were desalted and then labeled using C18 Stage Tips (Rappsilber et al., 2007; Boersema et al., 2009). Dimethyl labeling is a technique that used a specific reagent (i.e., cyanoborohydride and formaldehyde in unlabeled and stable isotope-labeled forms) in order to tag the primary amine, i.e., N-terminus and the ε-amino group of lysine in peptides, in proteins (Yan et al., 2020). Finally, the samples were labeled with dimethyl “light” (CH3)2 and dimethyl “intermediate” (CH1D2)2. Saturating incorporation levels of the dimethyl labels were achieved in all cases.
Nano-Liquid Chromatography-Tandem Mass Spectrometry Analysis
Nano-liquid chromatography-tandem MS analysis was carried out, and eluted peptides were mixed in a 1:1 ratio conferring to measured protein amount. The analysis of the peptide mixture was achieved on an Easy-nLC 1200 System (Thermo Fisher Scientific) coupled with LTQ Orbit Rap Elite MS (Thermo Fisher Scientific). The peptides were separated with solvent A (0.1% of formic acid) at a flow rate of 500 nl/min and subsequently eluted with a 230 min gradient of (10-33-50-90%) HPLC solvent B (80% acetonitrile in 0.1% of formic acid) with a constant flow rate of 200 nl/min. The 15 most intense precursor ions were sequentially fragmented in each scan cycle (Boersema et al., 2009).
Protein Identification From the Mass Spectrometry Data
The MS data were processed by using MaxQuant software suite version 1.5.2.8 (Cox and Mann, 2008), while database search was performed by using the Andromeda search engine (Cox et al., 2011), which is a module of the MaxQuant software. The MS/MS spectra were searched against protein entries from B. napus, and a database consisting of 285 commonly observed contaminants and reverse decoy matches were removed from the protein identification list. In a database search, full tryptic specificity was required and up to two missed cleavages were allowed. The protein N-terminal acetylation and oxidation of methionine were set as variable modifications. The peptide, protein, and modification-site identifications were filtered using a target-decoy approach at a false discovery rate (FDR) set to 0.01 (Elias and Gygi, 2007). A minimum of two quantified peptides were registered for protein group quantification. The Perseus software (version 1.5.0.15), a module from the MaxQuant suite (Tyanova et al., 2016), was used for the calculation of the significance B (p sig. B) for each protein ratio with respect to the distance of the median of the distribution of all protein ratios as well as its intensities. All proteins with p sig. B < 0.1 in a pairwise comparison were considered differentially expressed.
RNA Extraction
For total RNA extraction (100 mg), leaf tissue was powdered in liquid nitrogen using mortar and pestle. Total RNA extraction was carried out by using MACHEREY-NAGEL KIT Germany and treated with RNase-free DNase I during extraction. The concentration was checked using NanoDrop.
Reverse Transcription/cDNA Synthesis
To synthesized cDNA from the extracted RNA, synthesis was carried out using the protocol of Cello et al. (2002). The extracted RNA was equipped with 2 μl of oligo dT primer 0.5 mM and kept incubated for 10 min at 70°C followed by incubation for 2 min on ice. Precise amount of 8 μl of Reveres Transcription (RT) Buffer, 4 μl of dNTP, 2 μl of Reverse aid RT (company), and 1 μl of Ribo lock (Thermo Fisher Scientific) was added to the reaction mixture. The mixture was then incubated for 90 min at 42°C. The incubated mixture (cDNA) was stored at −20°C for future use (Table 1).
TABLE 1.
Details of selected genes and primers designed by using the NCBI primer-BLAST tool for the quantitative real-time PCR (qRT-PCR) analysis of the plants treated with 15 mg/L of Zn NPs.
| Plants | Gene/Accession No | 5′ Primer | 3′ Primer |
| Faisal canola | BnaC09g00220D/PsaD | TGCTTGAATTC CTAAGTTTGC |
AATATGCCCA TTCCCATCAA |
| BnaC03g44500D | ATTGTCAAGG CTTCTGCTTA |
AGAATGAAAT GCTCTCACCT |
|
| BnaA01g14450D/FSD1 | GGTCCACTAA GGAAGAAACA |
TCCTAGCTTCG GCTATATCA |
|
| Shiralee | BnaC09g00220D/PsaD | TTGTCGTTG TCGTTAAAACC |
TTGCTTAAAC CATGCTACGA |
| BnaA04g15980D | GAGTTGATTG CTGTTGGAAG |
CCCATGTCCT GCAAATTAAC |
|
| BnaAnng33440D | TTGGTTTGTC CATGTTGTGA |
CCGAGAATGAG TAACGAGTA |
Three biological replicates were used for gene expression.
Quantitative Real-Time PCR (qRT-PCR) for Gene Amplification
Real-time quantitative PCR analysis was performed with Bio-Rad iQ5 Thermo Fisher. The reaction mixture was prepared by mixing SYBR® Green dye, 10 μM of forward primer and reverse primer, and 1.5 μl of RNA-free water. Later, 1 μl of cDNA of B. napus cultivars were added to each well. All steps were performed essentially according to the manufacturer’s protocol. Relative gene expression calculation was performed with the Bio-Rad CFX-Manager Software version 1.6 using the DDC(t)method and ACTIN as reference.
Statistical Analysis
All the experiments were performed in triplicates using three different biological replicates. The data were analyzed statically using SPSS version 20 software for the analysis of variance (ANOVA) and the mean significant differences were separated using Duncan’s multiple range test (DMRT).
Availability of Data and Materials
The proteome data set of this study is available as supplementary material. The biogenic Zn NPs and plants material are available from the corresponding authors on reasonable request.
Ethics Statement
This material is the authors’ own original work, which has not been previously published elsewhere. This study does not involve any human or animal trials and was purely based on the use of green synthesized Zn NPs for the growth and development of rapeseed plant.
Results and Discussion
Morphological and Optical Characterization of the Photosynthesized Zinc Nanoparticles
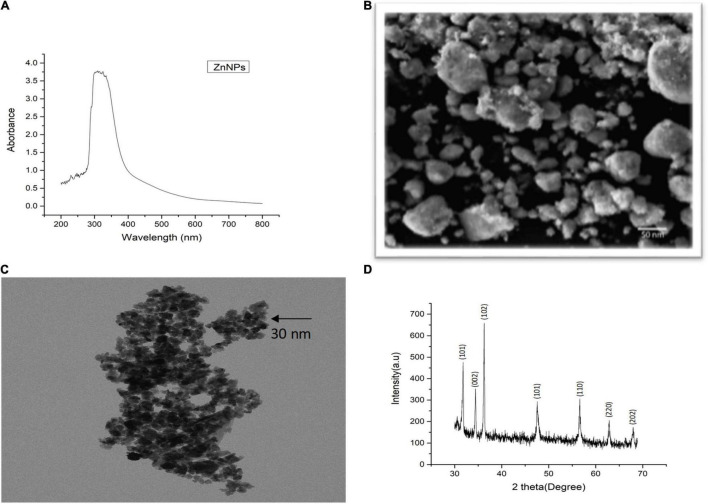
Plant-mediated green synthesis is considered an eco-friendly approach for the synthesis of biocompatible NPs. In this study, Zn NPs were synthesized by using M. arvensis aqueous extract as a reducing and stabilizing agent (Sohail et al., 2019). A surface plasmon resonance (SPR) band characteristic of Zn NPs was observed at 292 nm wavelength (Figure 1A). The SPR band is a response of the interaction of the oscillating electromagnetic light waves with the NPs resonance (Yasmeen et al., 2017). The SEM images visualize mostly the surface of the Zn NPs. Most of the Zn NPs are visible as aggregates (Figure 1B). The TEM images revealed that most of the NPs exist in the size range of 30–50 nm. The morphological evaluation of the Zn NPs photosynthesized by M. arvensis exhibits an irregular shape and a size of the NPs that is suitable for plant exposition experiments (Figure 1C). The NPs appear aggregated, confirming the SEM analysis results obtained for these NPs. Transmission electron microscopy reveals the irregular shape of Zn NPs with a similar size range as revealed by SEM. The XRD analysis confirmed the crystalline nature of the Zn NPs, while different peaks represent the pattern and phase angle of the synthesized NPs (Figure 1D). The diffraction peaks were observed at the angle of 14.3°, 17.6°, 27.6°, 47.8°, 51.6°, 65.7°, 73.4°, and 80.2° with their corresponding lattice plane. These angels are analogous to (101), (104), (002), (110), (111), (220), and (202) Miller indices, respectively. The XRD analysis exhibits sharp peaks that are referenced according to the Joint Committee Powdered Diffraction (JCPD) number, proving the presence of Zn NPs in the sample (Lu and Yeh, 2000).
FIGURE 1.
Characterization of green synthesized Zn NPs using Mentha arvensis leaves with (A) UV-visible spectroscopy, (B) scanning electron microscopy (SEM), (C) transmission electron microscopy (TEM), and (D) X-ray diffraction analysis (XRD). SEM analysis showed aggregated nature of Zn NPs in the solution used, indicating a gel-like appurtenance, while clear morphology of the green synthesized NPs can be seen in TEM image, revealing an irregular shape. The XRD peaks are in reference to the standard present in the library data of the XRD machine, which confirmed the presence of Zn NPs in the sample.
Chlorophyll and Carotenoid Content of the Brassica napus Varieties Treated With Zinc Nanoparticles
To investigate the effects of these Zn NPs on the physiological responses of the canola varieties, i.e., Faisal and Shiralee, the 4-week-old plants were treated with 5, 15, and 25 mg/L of Zn NPs, while untreated plants were considered controls. Chlorophyll and carotenoids play an indispensable role in the photosynthetic process largely determining the photosynthetic capacity of the plants. Chlorophyll and carotenoid contents can alter depending on the growth conditions, health status, and other environmental factors (Pinto et al., 2020).
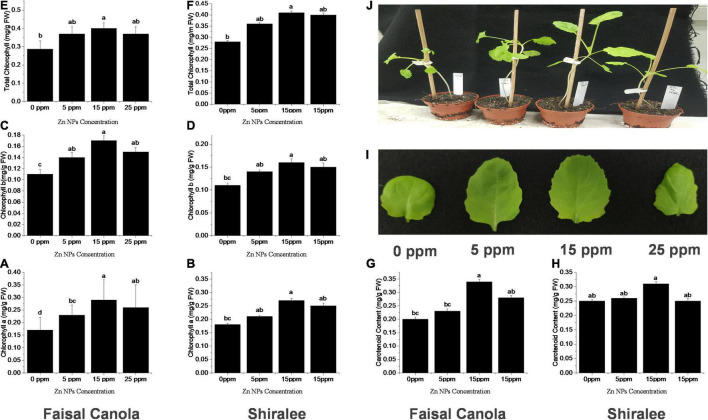
Zinc nanoparticles treatment had a positive effect on chlorophyll contents in both Faisal and Shiralee B. napus varieties. Plants treated with Zn NPs had a significantly increased level of Chl a, Chl b, and Chl T content after Zn NPs treatment with all concentrations applied. The highest increase was obtained with 15 mg/L Zn NPs. The Chl a content was enhanced by 47 and 50% in both canola varieties, respectively, after foliar application of 15 mg/L of Zn NPs (Figures 2A,B). The Chl b content was 54 and 46% higher in both canola varieties, respectively, with this treatment (Figures 2C,D). The Chl T content with 15 mg/L of Zn NPs application showed 46% increase in Shiralee and 50% increase in Faisal (Figures 2E,F).
FIGURE 2.
Effects of Zn NPs on chlorophylls and carotenoids contents in Brassica napus rosette leaves. (A,B) Chlorophyll a, (C,D) Chlorophyll b, (E,F) total chlorophylls, and (G,H) carotenoids in rosette leaves of 5-week-old B. napus (Faisal canola and Shiralee), (I) leaf size, and (J) phenotype of plants treated with 0, 5, 15, and 25 ppm of Zn NPs. Chlorophylls and carotenoids were extracted from rosette leaves of 5-week-old B. napus plants using 80% acetone and measured using a spectrophotometer. Data represent the mean ± SD of three replicates. Different letters indicate significantly different (p < 0.05).
The carotenoid content was increased by 65% in Faisal canola and 34% in Shiralee with 15 mg/L of Zn NPs (Figures 2G,H). There has been a considerable rise in leaf size on the same treatment, and the phenotypic also correlates with this effect of Zn NPs (Figures 2I,J).
The effects of different concentrations of ZnO-ZnO-NPs on the photosynthetic pigments (i.e., chlorophyll and carotenoids) were investigated in peanut (Prasad et al., 2012), pearl millet (Tarafdar et al., 2014), green pea (Mukherjee et al., 2014), cluster bean (Raliya and Tarafdar, 2013), and tomato (Raliya et al., 2015) and found a positive effect. These studies support our results in that Zn NPs are growth promoting and do not cause stress or damage to plants. Zinc plays an important role in the synthesis of chlorophyll, DNA replication, nitrogen fixation, and electron transport chain (e.g., chloroplast and mitochondria) (Yruela, 2005; Nouet et al., 2011), indicating that it is essential for plant health and that supplementing with Zn may have growth-promoting effects. It also plays a protective role in photo-oxidative stress; for example, plants growing in zinc-deficient environments are more vulnerable to drought-induced oxidative stress (Cakmak, 2000). Marusenko et al. (2013) have explained experimentally that ZnSO4 (salt) showed a smaller increase in chlorophyll content compared with plants treated with ZnO NPs, suggesting that the NPs can offer advantages over traditional bulk Zn applications (Faizan et al., 2021).
Zinc Nanoparticles Influence the Growth and Biomass Accumulation in the Brassica napus Varieties
To evaluate the effect of NPs on plant performance, morphological characteristics such as shoot FW and DW were measured.
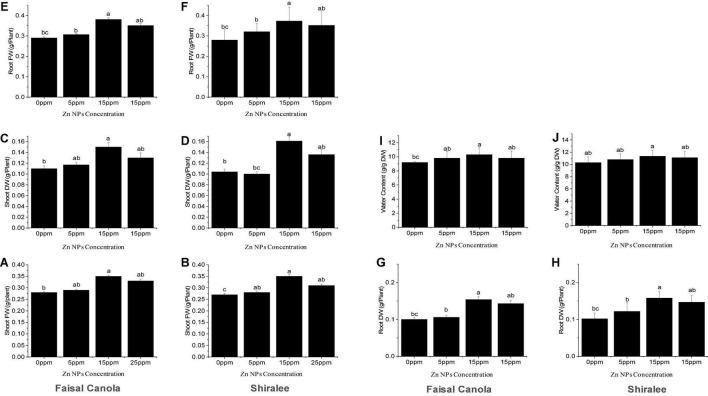
After exposure to Zn NPs, the FW and DW of the shoots increased significantly in the rapeseed varieties. The shoot FW was significantly higher at 15 mg/L of Zn NPs, with an increase in 25% in Faisal canola and 29% in Shiralee as compared with the untreated control plants (Figures 3A,B).
FIGURE 3.
Effect of zinc nanoparticle significantly influences biomass accumulation shoot fresh (A,B) and dry weight (C,D), root fresh (E,F) and dry weight (G,H), and relative water content (I,J) in Brassica napus varieties, i.e., Faisal canola and Shiralee. Data were expressed in mean ± SD. Bar with different letters indicates significance, while bar with similar letters indicates non-significance. Three different biological replicates were used in the study.
In contrast, the Zn NPs showed a slight increase in the FW of the shoot after treatment with 5 mg/L, with 3.5% in Faisal canola and 4.7% in Shiralee.
None of the concentrations tested resulted in decreased biomass, showing that Zn NP treatment enhanced growth and has no detrimental but positive effects on canola FW. This means that the canola variety Faisal responds more strongly to the treatment than do Shiralee. The shoot DW was significantly higher at 15 mg/L of Zn NPs, with an increase in 36% in Faisal canola and 51% in the Shiralee variety, indicating that the water content of Faisal contributed stronger to the FW gain than do Shiralee that shows higher DW and therefore higher biomass increases (Figures 3C,D). Generally, Zn NPs application significantly improved the FW and DW in both the study canola varieties.
The roots of the canola plants showed a similar effect upon treatment with 15 mg/L of Zn NPs. The root FW showed an increase by 31% for Faisal canola and 33% for Shiralee as compared with control plants (Figures 3E,F). In the case of Zn NPs, a dose-dependent tendency was observed at different concentrations, i.e., 5 mg/L of Zn NPs caused an increase by 5% in Faisal canola and 14% in Shiralee canola whereas 25 mg/L of Zn NPs caused 10% in Faisal and 20% in Shiralee (Figures 3G,H). The root DW was increased by 54% in Faisal canola and 58% in Shiralee canola at 15 mg/L of Zn NPs. Therefore, the variety Shiralee responded significantly higher than Faisal canola.
Positive effects of Zn NPs on plant growth have been reported previously in other plant species, supporting our findings. Tarafdar et al. (2014) reported improved wheat plant height, root length, and root weight with the foliar application of Zn NPs. Similarly, an increase in the root/shoot biomass of Vigna radiata was observed (Dhoke et al., 2013; Taheri et al., 2016). Enhanced root/shoot DW was reported in the sunflower by ZnO NPs exposure (Torabian et al., 2016). ZnO NPs promote onion growth and reduce the flowering period at 20 and 30 mg/L concentrations (Laware and Raskar, 2014). Burman et al. (2013) reported an improved growth in the chickpea with foliar application of Zn, whereas Prasad et al. (2012) reported increased root/shoot growth in peanut plant.
Plant Relative Water Content Measurement in Response to Zinc Nanoparticles
The relative water content (RWC) acts as a valuable physiological indicator to plants. The foliar application of 15 mg/L of Zn NPs significantly increased the RWC of both canola varieties (i.e., 11% for the Faisal canola and 10% for the Shiralee) (Figures 3I,J). A slight increase in the RWC was observed in relation to the seeds treated with 5 mg/L of Zn NPs, which showed an increase in 6% in Faisal canola and 4% in Shiralee. These findings resulted that the RWC of both canola varieties showed a significant increase after Zn NPs applications. Many researchers have indicated the importance of Zn in plant growth, development, reproduction, and yield (Camp and Fudge, 1945; Chapman, 1966; Anderson, 1975; Brown et al., 1993; Marschner, 1993; Stampoulis et al., 2009). Khan et al. (1998) reported that Zn can improve the water content of plants. They observed that the addition of Zn increased the water content of leaf and stomatal conductance in chickpea plants. Deficiency of Zn leads to water stress and reduced stomatal conductance and the water-use efficiency of plants (Khan et al., 2003). Thus, as Zn NPs can improve the photosynthetic pigment content, growth, and water content of plants, they should be used to improve the quality of rapeseed and other crop plants.
Effect of Zinc Nanoparticles on Proteomic Profiling of Brassica napus
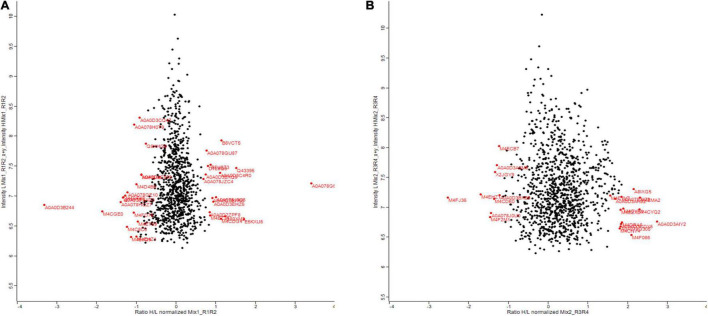
Zinc nanoparticles have a positive effect on plant biomass. To understand how this is achieved, we performed proteomic analysis. Also, to find out which proteins are changed as a result of Zn NP treatment and whether this can explain why the plants perform better. Proteins were extracted from the leaves of a 30-days-old canola plant treated with 15 mg/L of Zn NPs and analyzed by gel-free/label proteomics technique. Both canola varieties showed a significant alteration in their protein profiles. A total of 1,904 proteins were identified from protein extracts by nano-LC/MS-MS, with 35 of the proteins from Faisal canola and 23 proteins from Shiralee being significantly altered (Figures 4A,B).
FIGURE 4.
Summary of the results comparing deep-scale proteome analysis data of Brassica napus variety (A) Faisal canola and (B) Shiralee using nano-flow LC-MS/MS data and data obtained by micro-flow LC-MS/MS data in this study. Red color indicates significantly altered proteins. The red on the right side of the scale starting from 0 shows an increase in protein expression while the red on the left side of 0 indicates the decrease in protein expression. Three different biological replicates were used in the study.
Proteomic profiling of the B. napus variety Faisal canola revealed that various proteins involved in the photosynthesis, carotenoid biosynthetic pathways, light-harvesting protein-chromophore linkage, signal transduction, glutamine metabolic process, cysteine biosynthetic process from serine, heme-binding proteins, iron ion-binding proteins, defense-related protein, NAD(P)H dehydrogenase (quinone) activity, and water-soluble chlorophyll protein (WSCP) were significantly altered in zinc-treated plants (Table 2). Metallic NPs such as the ZnO NPs mainly target the cellular organization mechanism and hormonal and protein-related secondary metabolism under overflowing stress (Hossain et al., 2012). In contrast, B. napus variety Shiralee protein related to photosynthesis, protein metabolic process, defense response, glycolytic process, transport, carbohydrate metabolic process, translation, defense response to biotic stimulus, and carbon fixation was altered in plants treated with Zn NPs (Table 3).
TABLE 2.
Quantitative proteomics table of proteins with differential abundance (ratio below 0.8 or above 3.00 relative to the Zn NPs-treated plants).
| Protein IDs | Species | GOMF name | GOBP name | Gene name | Ratio H/L normalized |
| M4CGE0 | B. rapa | Hydrolase activity, hydrolyzing o-glycosyl compounds | Carbohydrate metabolic process | Glu1 | −1.857835 |
| E5KXU6 | B. campestris | Copper ion binding; metal ion binding; protein domain specific binding; superoxide dismutase activity | Circadian rhythm; response to cadmium ion; response to copper ion; response to light intensity; response to ozone | FSD1 | 1.709247 |
| A0A0D3C833 | B. oleracea v | ATP binding; metallo-endopeptidase activity; zinc ion binding | 106292516 | −0.8753542 | |
| A0A0D3BYR6 | B. oleracea | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; oxidoreductase activity | Carotenoid biosynthetic process | PDS | 0.9167823 |
| M4FDA0 | B. rapa | Chlorophyll binding | Photosynthesis | −0.8749309 | |
| Q9XHG8 | B. napus | Channel activity | Transport | gamma-TIP2 | −0.7562578 |
| M4FD78 | B. rapa | Proton transmembrane transporter activity | Transport | −1.066609 | |
| A0A078G6S6 | B. napus | ADP binding | Signal transduction | BnaA01g33720D | 3.414677 |
| A0A078GAD3 | B. napus | Cysteine synthase activity | Cysteine biosynthetic process from serine | BnaC04g49280D | −1.329051 |
| M4EZJ4 | B. rapa | Heme binding; iron ion binding; monooxygenase activity; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | BnaAnng14260D | 0.8665519 | |
| A0A078GE10 | B. napus | Carbohydrate binding | Lectin Protein | BnaA06g01990D | −1.226274 |
| Q93XM2 | B. carinata | Defense related protein | Glutamine metabolic process | CJAS1 | 1.232845 |
| A0A0D2ZPF8 | B. oleracea | Uncharacterized protein | Uncharacterized protein | BnaC02g36330D | 0.8458312 |
| A0A078GU87 | B. napus | Carbonate dehydratase activity; zinc ion binding | Carbon utilization | BnaCnng05580D | 0.7713785 |
| A0A078H0T9 | B. napus | Cysteine synthase activity | Cysteine biosynthetic process from serine | BnaA04g25390D | −1.053224 |
| A0A078HQZ7 | B. napus | Hydrolase activity, hydrolyzing o-glycosyl compounds; xyloglucan: xyloglucans transferase activity | Cell wall biogenesis | BnaA03g50050D | −1.396688 |
| M4CDG4 | B. rapa | L-aspartate:2-oxoglutarate aminotransferase activity; pyridoxal phosphate binding | Biosynthetic process; cellular amino acid metabolic process | 106320409 | 1.128755 |
| M4CFK0 | B. rapa | FMN binding; NAD(P)H dehydrogenase (quinone) activity | Oxidation-reduction process | BnaCnng38420D | −1.283878 |
| M4EJQ9 | B. rapa | FMN binding; NAD(P)H dehydrogenase (quinone) activity | Oxidation-reduction process | 106331476 | −1.138569 |
| B6VCT5 | B. rapa | Epithiospecifier protein | Post-translational modifications | BnaAnng10080D | 1.142087 |
| A0A078J0C6 | B. napus | Electron transfer activity | Post-translational modifications | 106305373 | 1.004106 |
| Q7X737 | B. juncea | Transferase activity | Detoxification | GSTF2 | −1.337922 |
| A0A078JZC4 | B. napus | Hydrolase activity, hydrolyzing o-glycosyl compounds | Carbohydrate metabolic process | BnaA09g52790D | 0.6890308 |
| A0A0D3B244 | B. oleracea | Manganese ion binding; nutrient reservoir activity | 106329205 | −3.31919 | |
| M4C8C6 | Brassica rapa | Isomerase activity | Cell redox homeostasis | 106328378 | −1.231617 |
| A0A0D3BXQ9 | Brassica oleracea | Triose-phosphate isomerase activity | Glycolytic process; metabolic process | 106342295 | 0.7464848 |
| A0A0D3C4R0 | Brassica oleracea | RNA binding | Uncharacterized protein | 106342759 | 1.1143 |
| A0A0D3CQ47 | Brassica oleracea | Carbohydrate binding | Uncharacterized protein | 106298242 | −0.9183059 |
| A0A0D3EHZ6 | Brassica oleracea | Glutamate synthase activity | Glutamate biosynthetic process | 106317861 | 0.9386246 |
| D1L8Q3 | Brassica napus | Photosystem ii reaction center protein l | Photosynthesis | psbL | 0.8029789 |
| B5U8Z3 | Brassica rapa | Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | Nitrogen compound metabolic process | BrNIT2 | 0.8696346 |
| M4D4B0 | Brassica rapa | ATP binding; l-aspartate:2-oxoglutarate aminotransferase activity; protein serine/threonine kinase activity; pyridoxal phosphate binding; transaminase activity | Cellular amino acid metabolic process; | −0.9932066 | |
| M4EHZ1 | Brassica rapa | Monooxygenase activity; ribulose-bisphosphate carboxylase activity | Carbon fixation; photorespiration; photosynthesis | −0.9936661 | |
| M4ET89 | Brassica rapa | Oxidoreductase activity | Oxidation-reduction process | BnaA05g32330D | −0.96508 |
| Q43395 | Brassica napus | Endopeptidase inhibitor activity | Uncharacterized protein | WSCP | 1.518787 |
GOMF, gene ontology molecular function; GOBP, gene ontology biological function.
TABLE 3.
Quantitative proteomics table of proteins with differential abundance (ratio below 0.8 or above 3.00 relative to the Zn NPs-treated plants).
| Protein IDs | Species | GOMF name | GOBP name | Gene name | Ratio H/L normalized |
| M4EHZ1 | B. rapa | carbon fixation; photorespiration | Photosynthesis | −1.70524 | |
| A0A0D3AHJ2 | B. oleracea | Structural constituent of ribosome | Translation | 1.688717 | |
| M4EYV7 | B. rapa | Structural constituent of ribosome; ubiquitin-like modifier Activating enzyme activity | Translation | 106300706 | 1.558219 |
| A0A0D3A5M6 | B. oleracea | Apoplast; vacuole | Cellular amino acid metabolic process | BnaA01g13280D/106313559 | −1.29138 |
| A0A078GI73 | B. napus | Structural constituent of ribosome | Translation | BnaA06g12670D | 1.842456 |
| A0A078G305 | B. napus | ND | Ribosome biogenesis | BnaA09g03590D | 1.829159 |
| M4F2M1 | B. rapa | Allene oxide synthase activity; heme binding; heme binding; iron ion binding; monooxygenase activity; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | Defense response to fungus; jasmonic acid biosynthetic process; oxylipin metabolic process; response to jasmonic acid; response to wounding | AOS | −1.45815 |
| M4EZX0 | B. rapa | Structural constituent of ribosome | Translational elongation | BnaA04g15980D | 1.821955 |
| A0A0D3BXB6 | B. oleracea | Glucan endo-1,3-beta-D-glucosidase activity; hydrolase activity, hydrolyzing O-glycosyl compounds | Carbohydrate metabolic process; defense response | BnaC04g24330D/106341843 | −1.22014 |
| X2JGY9 | B. rapa | Calcium ion binding; calcium-dependent phospholipid binding | 106295437 | −1.34015 | |
| M4F066 | B. rapa | Hydrolase activity, acting on ester bonds | Uncharacterized protein | 106292567 | 2.09285 |
| A0A078J0U0 | B. napus | Serine-type endopeptidase activity | Hydrolase, Protease, Serine protease | BnaCnng28690D/106313216 | −1.43511 |
| M4EMA2 | B. rapa | ATP binding | Protein metabolic process | 106311155 | 2.309933 |
| A0A0D3AIY2 | B. oleracea | Integral component of membrane | Transport | 106319205 | 2.740755 |
| M4CNY4 | B. rapa | Aspartic-type endopeptidase activity | Protein catabolic process | 106328540 | 1.804962 |
| A0A0D3CDX6 | B. oleracea | DNA binding; DNA-directed 5′-3′ RNA polymerase activity; protein dimerization activity; structural constituent of ribosome | Transcription, DNA-templated; transcription, DNA-templated; translation | rpoA | 1.832607 |
| A8IXG5 | B. campestris | Signaling receptor activity | Defense response; response to biotic stimulus | BnaAnng33440D | 2.154389 |
| M4CDE0 | B. rapa | ATP binding; glucose-1-phosphate adenylyl transferase activity | Photosynthesis | 106319363 | −1.35657 |
| M4CYK1 | B. rapa | Aspartic-type endopeptidase activity | Protein catabolic process | 106319255 | 1.89309 |
| M4CYQ2 | B. rapa | Uncharacterized protein | Uncharacterized protein | BnaC09g47440D | 2.296604 |
| M4DRA6 | B. rapa | Photosynthesis, metal ion binding; protein domain specific binding; protochlorophyllide reductase activity | Photosynthesis | BnaA03g48610D | 1.858459 |
| M4ECB7 | B. rapa | Fructose-bisphosphate aldolase activity | Glycolytic process | −1.23016 | |
| M4FJ36 | B. rapa | Ribulose-bisphosphate carboxylase activity | Carbon fixation | −2.53191 |
Protein alterations in the plant after exposure to NPs could provide a novel biomarker by using advanced bioinformatics tools (Roy et al., 2017). Studies on green synthesized NPs affecting plant morphology and proteomics profiling are limited. Yasmeen et al. (2016) observed that aluminum oxide NPs significantly altered the number of proteins in the soybean plant, which leads to the discovery of a protein involved in signaling and protein metabolism process. Similarly, silver NPs were reported to altering the proteins involved in the cellular metabolism and stress signaling responses (Mustafa et al., 2015).
Our data on the differential expression of proteins provide a valuable data set that can be used to study specific physiological effects of Zn NPs on canola and may be also used to extrapolate on other plants.
Differential Expression Analysis of Candidate Genes
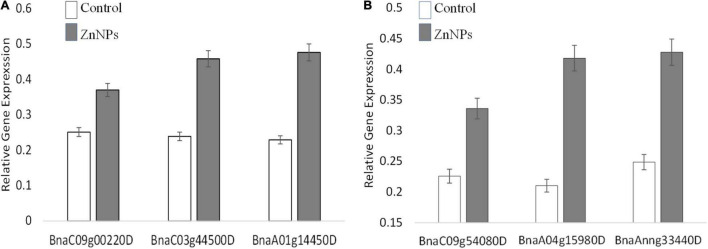
Proteomic analyses revealed numerous proteins were influenced differently after exposure to Zn NP. In this study, three genes in each variety were selected and their expressions were checked at the transcriptional level. The selection of genes was based on the most altered expression of their proteins in proteomic analyses. Actin (ACT7) was used as a reference gene, and three biological replicates were analyzed on the transcriptional level. The gene responsible for the photosynthesis, superoxide dismutase (SOD), and ribosome structural components (Protein ID/Gene: BnaC09g00220D/PsaD, BnaA01g14450D/FSD1, and BnaC03g44500D) in Faisal canola and (BnaC09g00220D/PsaD, BnaAnng33440D, and BnaA04g15980D) in Shiralee were significantly upregulated in canola varieties on exposure to Zn NPs at 15 mg/L as compared with control plants (Figure 5A). The gene associated with photosynthesis showed more expression/upregulation, followed by gene involved in SOD activity as compared with control plants (Figure 5B).
FIGURE 5.
Expression of the randomly selected gene from Brassica napus varieties (A) Faisal canola and (B) Shiralee were assisted using quantitative real-time PCR Bio-Rad iQ5 Thermo Fisher. Four-week-old Zn NPs treated (15 mg/L) plants were harvested, RNA was extracted, and cDNA was synthesized. Represented values are the mean ± SD obtained from three individual experiments performed for each sample.
To date, only a few studies have been carried out to identify differentially expressed genes after Zn-NP exposure. Landa et al. (2012) reported differential expression upon Zn NPs exposure including a putative carbonic anhydrase family protein (At1g58180), Naringenin 3-dioxygenase (At3g51240), 2-oxoisovalerate dehydrogenase (At1g21400), and an acidic phosphatase class B family protein (At4g29270). Numerous genes were upregulated and downregulated in Arabidopsis thaliana upon exposure to Ag NPs (Xu et al., 2011). Due to very limited literature, the function of Zn NP in the gene expression is not very cleared.
Conclusion
This study reports the effects of biosynthesized and therefore biocompatible and non-toxic Zn NPs on two different canola varieties. Multiple characterization results revealed that the Zn NPs are 30 nm in size and showed aggregation with irregular shape and morphology, which makes them suitable for various biological applications. All tested concentrations of the Zn NPs have positive effects on both canola varieties, with 15 mg/L giving the strongest effects. Chlorophyll and carotenoid content, growth, and water content were enhanced in all tested conditions and both varieties, i.e., Faisal and Shiralee. Proteomic and gene expression analysis of the Zn NPs-treated plants revealed that the NPs have the ability to influence protein levels and gene transcription, which may explain the alteration of the morphological and physiological parameters. The selected genes expression analysis showed that genes related to photosynthetic pathways, SOD activity, defense, response to the biotic stress, and the structural constituents of the ribosomes. The findings of this study endorse the foliar applications of the Zn NPs to induce gene expression and alter the protein level of certain genes/proteins of the plant to increase biomass, RWC, and plant chlorophyll and carotenoid contents. Due to very limited literature, the role of Zn NP in the gene expression related to growth and production is not very cleared. Future studies are needed to be carried out to understand the molecular role of Zn NPs on plant physiological process.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
Sohail: writing, review, analysis, investigation, methods, and research. LS, EF, Y-DS, and BK: editing and review. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are highly thankful to the Transmission and Electron Microscopy and Center of Proteomics, University of Tübingen, Tübingen, Germany, for helping in this microscopy and proteomics profiling.
References
- Ahmad P., Alyemeni M. N., Al-Huqail A. A., Alqahtani M. A., Wijaya L., Ashraf M., et al. (2020). Zinc oxide nanoparticles application alleviates arsenic (As) toxicity in soybean plants by restricting the uptake of as and modulating key biochemical attributes, antioxidant enzymes, ascorbate-glutathione cycle and glyoxalase system. Plants 9:825. 10.3390/plants9070825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allhoff F., Lin P., Moore D. (2010). What is Nanotechnology and Why Does it Matter?: From Science to Ethics. Oxford: Wiley-Blackwell, 30. [Google Scholar]
- Almessiere M. A., Slimani Y., Baykal A. (2020). Synthesis and characterization of Co1–2xNixMnxCeyFe2–yO4 nanoparticles. J. Rare Earths 38 188–194. [Google Scholar]
- Anderson J. W. (1975). “Function of sulphur in plant growth and metabolism,” in Sulphur in Australasian Agriculture, ed. McLachlan K. D. (Sydney, NSW: University Press; ). [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan S., Shahzadi K., Javad S., Tariq A., Ahmad A., Ilyas S. (2021). A preliminary study of influence of zinc oxide nanoparticles on growth parameters of Brassica oleracea var italic. J. Saudi Soc. Agric. Sci. 20 18–24. 10.1016/j.jssas.2020.10.003 [DOI] [Google Scholar]
- Boersema P. J., Mohammed S., Heck A. J. (2009). Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 44 861–878. 10.1002/jms.1599 [DOI] [PubMed] [Google Scholar]
- Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., et al. (2010). Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20 837–846. 10.1101/gr.103119.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- Brown P. H., Cakmak I., Zhang Q. (1993). “Form and function of zinc plants,” in Zinc in Soils and Plants, ed. Robson A. D. (Dordrecht: Springer; ), 93–106. 10.1007/978-94-011-0878-2_7 [DOI] [Google Scholar]
- Burman U., Saini M., Kumar P. (2013). Effect of zinc oxide nanoparticles on growth and antioxidant system of chickpea seedlings. Toxicol. Environ. Chem. 95 605–612. 10.1080/02772248.2013.803796 [DOI] [Google Scholar]
- Cakmak I. (2000). Tansley review no. 111 possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 146 185–205. 10.1046/j.1469-8137.2000.00630.x [DOI] [PubMed] [Google Scholar]
- Camp A. F., Fudge B. R. (1945). Zinc as a nutrient in plant growth. Soil Sci. 60 157–164. 10.1097/00010694-194508000-00009 [DOI] [Google Scholar]
- Cello J., Paul A. V., Wimmer E. (2002). Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297 1016–1018. 10.1126/science.1072266 [DOI] [PubMed] [Google Scholar]
- Chapman H. E. (1966). Diagnostic criteria for plants and soils. Diagn. Crit. Plants Soils 137 594–600. [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10 1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- De la Rosa G., Lopez-Moreno M. L., de Haro D., Botez C. E., Peralta-Videa J. R., Gardea-Torresdey J. L. (2013). Effects of ZnO nanoparticles in alfalfa, tomato, and cucumber at the germination stage: root development and X-ray absorption spectroscopy studies. Pure Appl. Chem. 85 2161–2174. 10.1351/pac-con-12-09-05 [DOI] [Google Scholar]
- Dhoke S. K., Mahajan P., Kamble R., Khanna A. (2013). Effect of nanoparticles suspension on the growth of mung (Vigna radiata) seedlings by foliar spray method. Nanotechnol. Dev. 3 111–123. [Google Scholar]
- Dobrucka R., Długaszewska J. (2016). Biosynthesis and antibacterial activity of ZnO nanoparticles using Trifolium pratense flower extract. Saudi J. Biol. Sci. 23 517–523. 10.1016/j.sjbs.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz M., Raja N. I., Mashwani Z. U., Ahmad M. S., Hussain M., Iqbal M. (2018). Effect of silver nanoparticles and silver nitrate on growth of rice under biotic stress. IET Nanobiotechnol. 12 927–932. 10.1049/iet-nbt.2018.0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Badri A. M., Batool M., Mohamed I. A., Khatab A., Sherif A., Wang Z., et al. (2021). Modulation of salinity impact on early seedling stage via nano-priming application of zinc oxide on rapeseed (Brassica napus L.). Plant Physiol. Biochem. 166 376–392. 10.1016/j.plaphy.2021.05.040 [DOI] [PubMed] [Google Scholar]
- Elias J. E., Gygi S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4 207–214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Faizan M., Bhat J. A., Chen C., Alyemeni M. N., Wijaya L., Ahmad P., et al. (2021). Zinc oxide nanoparticles (ZnO-NPs) induce salt tolerance by improving the antioxidant system and photosynthetic machinery in tomato. Plant Physiol. Biochem. 161 122–130. 10.1016/j.plaphy.2021.02.002 [DOI] [PubMed] [Google Scholar]
- Fleischer A., O’Neill M. A., Ehwald R. (1999). The pore size of non-graminaceous plant cell walls is rapidly decreased by borate ester cross-linking of the pectic polysaccharide rhamnogalacturonan II. Plant Physiol. 121 829–838. 10.1104/pp.121.3.829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z., Hajika M., Komatsu S. (2012). Comparative proteome analysis of high and low cadmium accumulating soybeans under cadmium stress. Amino Acids 43 2393–2416. 10.1007/s00726-012-1319-6 [DOI] [PubMed] [Google Scholar]
- Jain P. K., Huang X., El-Sayed I. H., El-Sayed M. A. (2007). Review of some interesting surface plasmon resonance-enhanced properties of noble metal nanoparticles and their applications to biosystems. Plasmonics 2 107–118. 10.1007/s11468-007-9031-1 [DOI] [Google Scholar]
- Javed B., Mashwani Z. U. (2020a). Synergistic effects of physicochemical parameters on bio-fabrication of mint silver nanoparticles : structural evaluation and action against HCT116 colon cancer cells. Int. J. Nanomed. 15 3621–3637. 10.2147/IJN.S254402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed B., Mashwani Z. U. (2020b). Phytosynthesis of colloidal nanosilver from Mentha longifolia and Mentha arvensis: comparative morphological and optical characterization. Microsc. Res. Tech. 83 1299–1307. 10.1002/jemt.23518 [DOI] [PubMed] [Google Scholar]
- Javed B., Nadhman A., Razzaq A., Mashwani Z. (2020). One-pot phytosynthesis of nano-silver from Mentha longifolia L.: their characterization and evaluation of photodynamic potential. Mater. Res. Express 7:055401. 10.1088/2053-1591/ab903b [DOI] [Google Scholar]
- Khan H. R., McDonald G. K., Rengel Z. (1998). Chickpea genotypes differ in their sensitivity to Zn deficiency. Plant Soil 198 11–18. [Google Scholar]
- Khan H. R., McDonald G. K., Rengel Z. (2003). Zn fertilization improves water use efficiency, grain yield and seed Zn content in chickpea. Plant Soil 249 389–400. [Google Scholar]
- Khot L. R., Sankaran S., Maja J. M., Ehsani R., Schuster E. W. (2012). Applications of nanomaterials in agricultural production and crop protection: a review. Crop Prot. 35 64–70. 10.1016/j.cropro.2012.01.007 [DOI] [Google Scholar]
- Klaine S. J., Alvarez P. J., Batley G. E., Fernandes T. F., Handy R. D., Lyon D. Y., et al. (2008). Nanomaterials in the environment: behavior, fate, bioavailability, and effects. Environ. Toxicol. Chem. 27 1825–1851. 10.1897/08-090.1 [DOI] [PubMed] [Google Scholar]
- Landa P., Vankova R., Andrlova J., Hodek J., Marsik P., Storchova H., et al. (2012). Nanoparticle-specific changes in Arabidopsis thaliana gene expression after exposure to ZnO, TiO2, and fullerene soot. J. Hazard. Mater. 241 55–62. 10.1016/j.jhazmat.2012.08.059 [DOI] [PubMed] [Google Scholar]
- Laware S. L., Raskar S. (2014). Influence of zinc oxide nanoparticles on growth, flowering and seed productivity in onion. Int. J. Curr. Microbiol. Sci. 3 874–881. [Google Scholar]
- Lichtenthaler H. K., Wellburn A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 603 591–592. 10.1042/bst0110591 [DOI] [Google Scholar]
- Liu Q., Chen Z., Chen Y., Yang F., Yao W., Xie Y. (2021). Microplastics and nanoplastics: emerging contaminants in food. J. Agric. Food Chem. 69 10450–10468. 10.1021/acs.jafc.1c04199 [DOI] [PubMed] [Google Scholar]
- Love J. C., Estroff L. A., Kriebel J. K., Nuzzo R. G., Whitesides G. M. (2005). Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 105 1103–1170. 10.1021/cr0300789 [DOI] [PubMed] [Google Scholar]
- Lu C. H., Yeh C. H. (2000). Influence of hydrothermal conditions on the morphology and particle size of zinc oxide powder. Ceram. Int. 26 351–357. 10.1016/j.jes.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Maity A., Natarajan N., Vijay D., Srinivasan R., Pastor M., Malaviya D. R. (2018). Influence of metal nanoparticles (nanoparticles) on germination and yield of oat (Avena sativa) and berseem (Trifolium alexandrinum). Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 88 595–607. 10.1007/s40011-016-0796-x [DOI] [Google Scholar]
- Marschner H. (1993). “Zinc uptake from soils,” in Zinc in Soils and Plants, ed. Robson A. D. (Dordrecht: Springer; ), 59–77. 10.1007/978-94-011-0878-2_5 [DOI] [Google Scholar]
- Marusenko Y., Shipp J., Hamilton G. A., Morgan J. L., Keebaugh M., Hill H., et al. (2013). Bioavailability of nanoparticulate hematite to Arabidopsis thaliana. Environ. Pollut. 174 150–156. 10.1016/j.envpol.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Mazumder J. A., Khan E., Perwez M., Gupta M., Kumar S., Raza K., et al. (2020). Exposure of biosynthesized nanoscale ZnO to Brassica juncea crop plant: morphological, biochemical and molecular aspects. Sci. Rep. 10:8531. 10.1038/s41598-020-65271-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena D. S., Jayadeva H. M., Gautam C., Meena H. M. (2017). Effects of nano zinc oxide (ZnO) particles on germination of maize (Zea mays L.) seeds. Int. J. Plant Soil Sci. 16 1–5. 10.9734/ijpss/2017/33687 [DOI] [Google Scholar]
- Milner M. J., Seamon J., Craft E., Kochian L. V. (2013). Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 64 369–381. 10.1093/jxb/ers315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi Kouhi S. M., Lahouti M., Ganjeali A., Entezari M. H. (2015). Comparative effects of ZnO nanoparticles, ZnO bulk particles, and Zn2+ on Brassica napus after long-term exposure: changes in growth, biochemical compounds, antioxidant enzyme activities, and Zn bioaccumulation. Water Air Soil Pollut. 226:364. [Google Scholar]
- Mukherjee A., Pokhrel S., Bandyopadhyay S., Madler L., Peralta-Videa J. R., Gardea-Torresdey J. L. (2014). A soil mediated phyto-toxicological study of iron doped zinc oxide nanoparticles (Fe@ ZnO) in green peas (Pisum sativum L.). Chem. Eng. J. 258 394–401. 10.1016/j.cej.2014.06.112 [DOI] [Google Scholar]
- Mustafa G., Sakata K., Hossain Z., Komatsu S. (2015). Proteomic study on the effects of silver nanoparticles on soybean under flooding stress. J. Proteom. 122 100–118. 10.1016/j.jprot.2015.03.030 [DOI] [PubMed] [Google Scholar]
- Newman M. D., Stotland M., Ellis J. I. (2009). The safety of nanosized particles in titanium dioxide–and zinc oxide–based sunscreens. J. Am. Acad. Dermatol. 61 685–692. 10.1016/j.jaad.2009.02.051 [DOI] [PubMed] [Google Scholar]
- Nouet C., Motte P., Hanikenne M. (2011). Chloroplastic and mitochondrial metal homeostasis. Trends Plant Sci. 16 395–404. 10.1016/j.tplants.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Piccinno F., Gottschalk F., Seeger S., Nowack B. (2012). Industrial production quantities and uses of ten engineered nanomaterials in Europe and the world. J. Nanopart. Res. 14:1109. [Google Scholar]
- Pinto F., Celesti M., Acebron K., Alberti G., Cogliati S., Colombo R., et al. (2020). Dynamics of sun-induced chlorophyll fluorescence and reflectance to detect stress-induced variations in canopy photosynthesis. Plant Cell Environ. 43 1637–1654. 10.1111/pce.13754 [DOI] [PubMed] [Google Scholar]
- Prasad R., Kumar V., Prasad K. S. (2014). Nanotechnology in sustainable agriculture: present concerns and future aspects. Afr. J. Biotechnol. 13 705–713. 10.5897/ajbx2013.13554 [DOI] [Google Scholar]
- Prasad T. N. V. K. V., Sudhakar P., Sreenivasulu Y., Latha P., Munaswamy V., Reddy K. R., et al. (2012). Effect of nanoscale zinc oxide particles on the germination, growth and yield of peanut. J. Plant Nutr. 35 905–927. 10.1080/01904167.2012.663443 [DOI] [Google Scholar]
- Raghib F., Naikoo M. I., Khan F. A., Alyemeni M. N., Ahmad P. (2020). Interaction of ZnO nanoparticle and AM fungi mitigates Pb toxicity in wheat by upregulating antioxidants and restricted uptake of Pb. J. Biotechnol. 323 254–263. 10.1016/j.jbiotec.2020.09.003 [DOI] [PubMed] [Google Scholar]
- Raliya R., Tarafdar J. C. (2013). ZnO nanoparticle biosynthesis and its effect on phosphorous-mobilizing enzyme secretion and gum contents in clusterbean (Cyamopsis tetragonoloba L.). Agric. Res. 2, 48–57. 10.3390/plants10091808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raliya R., Nair R., Chavalmane S., Wang W. N., Biswas P. (2015). Mechanistic evaluation of translocation and physiological impact of titanium dioxide and zinc oxide nanoparticles on the tomato (Solanum lycopersicum L.) plant. Metallomics 7 1584–1594. 10.1039/c5mt00168d [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Roy D. N., Goswami R., Pal A. (2017). Nanomaterial and toxicity: what can proteomics tell us about the nanotoxicology? Xenobiotica 47 632–643. 10.1080/00498254.2016.1205762 [DOI] [PubMed] [Google Scholar]
- Sahoo S., Maiti M., Ganguly A., Jacob George J., Bhowmick A. K. (2007). Effect of zinc oxide nanoparticles as cure activator on the properties of natural rubber and nitrile rubber. J. Appl. Polym. Sci. 105 2407–2415. 10.1002/app.26296 [DOI] [Google Scholar]
- Singh A., Singh N. Á, Afzal S., Singh T., Hussain I. (2018). Zinc oxide nanoparticles: a review of their biological synthesis, antimicrobial activity, uptake, translocation and biotransformation in plants. J. Mater. Sci. 53 185–201. 10.1007/s10853-017-1544-1 [DOI] [Google Scholar]
- Sohail, Amara U., Shad S., Ilyas N., Manaf A., Raja N. I., et al. (2019). In vitro germination and biochemical profiling of B.napus L. in response to biosynthesised zinc nanoparticles. IET Nanobiotechnol. 13 46–51. 10.1049/iet-nbt.2018.5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail, Kamran K., Kemmerling B., Shutaywi M., Mashwani Z. U. R. (2020). Nano zinc elicited biochemical characterization, nutritional assessment, antioxidant enzymes and fatty acid profiling of rapeseed. PLoS One 15:e0241568. 10.1371/journal.pone.0241568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampoulis D., Sinha S. K., White J. C. (2009). Assay-dependent phytotoxicity of nanoparticles to plants. Environ. Sci. Technol. 43 9473–9479. 10.1021/es901695c [DOI] [PubMed] [Google Scholar]
- Sturikova H., Krystofova O., Huska D., Adam V. (2018). Zinc, zinc nanoparticles and plants. J. Hazard. Mater. 349 101–110. 10.1016/j.jhazmat.2018.01.040 [DOI] [PubMed] [Google Scholar]
- Taheri M., Qarache H. A., Qarache A. A., Yoosefi M. (2016). The effects of zinc-oxide nanoparticles on growth parameters of corn (SC704). STEM Fellowship J. 1 17–20. 10.17975/sfj-2015-011 [DOI] [Google Scholar]
- Tarafdar J. C., Raliya R., Mahawar H., Rathore I. (2014). Development of zinc nanofertilizer to enhance crop production in pearl millet (Pennisetum americanum). Agric. Res. 3 257–262. 10.1007/s40003-014-0113-y [DOI] [Google Scholar]
- Torabian S., Zahedi M., Khoshgoftar A. H. (2016). Effects of foliar spray of two kinds of zinc oxide on the growth and ion concentration of sunflower cultivars under salt stress. J. Plant Nutr. 39 172–180. 10.1080/01904167.2015.1009107 [DOI] [Google Scholar]
- Tyanova S., Temu T., Cox J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11 2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Whetherley P. E. (1950). Studies in the water relations of cotton plants. I. The field measurement of water deficit in leaves. New Phytol. 49 81–87. 10.1111/j.1469-8137.1950.tb05146.x [DOI] [Google Scholar]
- Xu L., Takemura T., Xu M., Hanagata N. (2011). Toxicity of silver nanoparticles as assessed by global gene expression analysis. Mater. Express 1 74–79. 10.1166/mex.2011.1010 [DOI] [Google Scholar]
- Yan X., Sun L., Dovichi N. J., Champion M. M. (2020). Minimal deuterium isotope effects in quantitation of dimethyl-labeled complex proteomes analyzed with capillary zone electrophoresis/mass spectrometry. Electrophoresis 41 1374–1378. 10.1002/elps.202000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmeen F., Raja N. I., Mustafa G., Sakata K., Komatsu S. (2016). Quantitative proteomic analysis of post-flooding recovery in soybean root exposed to aluminum oxide nanoparticles. J. Proteomics 143, 136–150. 10.1016/j.jprot.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Yasmeen F., Raja N. I., Razzaq A., Komatsu S. (2017). Proteomic and physiological analyses of wheat seeds exposed to copper and iron nanoparticles. Biochim. Biophys. Acta Proteins Proteom. 1865 28–42. 10.1016/j.bbapap.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Yruela I. (2005). Copper in plants. Braz. J. Plant Physiol. 17 145–156. [Google Scholar]
- Zafar H., Ali A., Ali J. S., Haq I. U., Zia M. (2016). Effect of ZnO nanoparticles on Brassica nigra seedlings and stem explants: growth dynamics and antioxidative response. Front. Plant Sci. 7:535. 10.3389/fpls.2016.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The proteome data set of this study is available as supplementary material. The biogenic Zn NPs and plants material are available from the corresponding authors on reasonable request.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.