Abstract
Animal and plant species exhibit an astonishing diversity of sexual systems, including environmental and genetic determinants of sex, with the latter including genetic material in the mitochondrial genome. In several hermaphroditic plants for example, sex is determined by an interaction between mitochondrial cytoplasmic male sterility (CMS) genes and nuclear restorer genes. Specifically, CMS involves aberrant mitochondrial genes that prevent pollen development and specific nuclear genes that restore it, leading to a mixture of female (male-sterile) and hermaphroditic individuals in the population (gynodioecy). Such a mitochondrial-nuclear sex determination system is thought to be rare outside plants. Here, we present one possible case of CMS in animals. We hypothesize that the only exception to the strict maternal mtDNA inheritance in animals, the doubly uniparental inheritance (DUI) system in bivalves, might have originated as a mitochondrial-nuclear sex-determination system. We document and explore similarities that exist between DUI and CMS, and we propose various ways to test our hypothesis.
Keywords: bivalves, mitochondria, mitonuclear interactions, plants, sex determination
INTRODUCTION
Although a rich diversity of sexual systems exists in animals and plants, two are most common and evolutionary stable. The first is dioecy or gonochorism, a system in which individuals reproduce in only one sexual role during their lifetime with males producing sperm and females producing eggs. The second is simultaneous hermaphroditism with out-crossing, in which individuals can reproduce through both sperm and eggs in a single breeding season, and mate with other individuals in both the male and female sexual roles.[1] The other, less evolutionary stable sexual systems include environmental sex determination, exclusively selfing hermaphroditism, sequential hermaphroditism, trioecy, gynodioecy and androdioecy[1] (Table 1). Sexual systems can be conserved across a given taxonomic level, but some lineages exhibit a wide diversity among species, or even among populations within the same species.[2] Indeed, it remains enigmatic why some taxa quickly evolve diversity in sexual systems while others remain unchanged over hundreds of millions of years.
TABLE 1.
Sexual systems in plants and animals
| Sexual system | Definition |
|---|---|
| Dioecy or gonochorism | A sexual system in which individuals reproduce in only one sexual role during their lifetime with males producing sperm and females producing eggs |
| Hermaphroditism | A sexual system in which individuals can reproduce through both sperm and eggs in a single breeding season, and mate with other individuals in both the male and female sexual roles |
| Androdioecy | A sexual system which consists of populations composed of simultaneous hermaphrodites and males |
| Gynodioecy | A sexual system which consists of populations composed of simultaneous hermaphrodites and females |
| Trioecy | A sexual system which consists of populations composed of simultaneous hermaphrodites, females, and males |
| Environmental sex determination | A sexual system in which sex is established by non-genetic cues |
| Exclusively selfing hermaphroditism | A sexual system in which individuals reproduce through self-fertilization exclusively |
| Sequential hermaphroditism | A sexual system in which individuals reproduce through eggs during one part of their life and through sperm during another |
Multiple transitions from simultaneous hermaphroditism to dioecy (or the reverse) have been studied in depth in plants,[3,4] but fewer examples are known in animals.[1,5] For angiosperms, two main paths to dioecy from hermaphroditism have been proposed: one is via monoecy, which involves hermaphrodites with separate male and female flowers as opposed to “perfect” hermaphrodites with flowers bearing both male and female parts.[6,7] In this scenario, a hermaphroditic population gradually increases sexual specialization because of disruptive selection (in which extreme values for a trait are favored over intermediate values), such that some individuals produce more male flowers over female ones and vice versa.[7] In these situations, individuals that specialize in being either male or female are more fit than those with both flower types. The second path to dioecy implicates a gynodioecious intermediate.[3,6–8] The underlying genetic transitions have been modeled for this scenario,[3,6–8] in which a male-sterility mutation first occurs in a hermaphroditic population (Figure 1). Once gynodioecy has evolved, hermaphrodites will experience greater selective pressure to produce more pollen and specialize in being male, eventually leading to dioecy if selection ultimately favors those individuals that only produce pollen (Figure 1). For the reverse path, that is, dioecy to simultaneous hermaphroditism, it is assumed that females able to produce small amounts of pollen for self-fertilization might be favored when population density drops and mates are difficult to find,[1,8,9] thus leading to androdioecy (Figure 1). Once androdioecy has evolved, males will experience a relative disadvantage as fewer ova become available for them to fertilize. Selection for more male sex allocation in hermaphrodites can ultimately lead to stochastic loss of males and a simultaneous hermaphroditic sexual system[1,8] (Figure 1).
FIGURE 1.
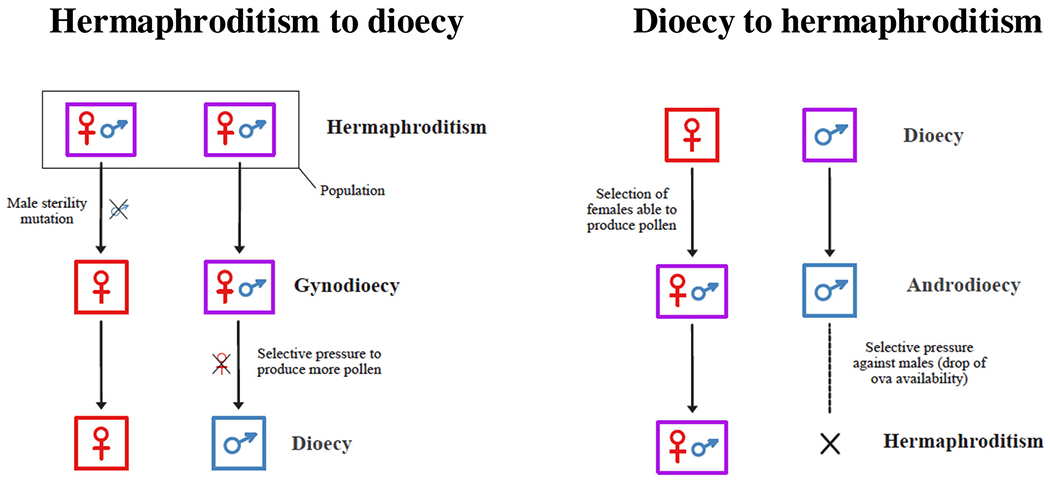
Simultaneous hermaphroditism to dioecy (with a gynodioecious intermediate) and dioecy to simultaneous hermaphroditism (with an androdioecious intermediate)
In the majority of gynodioecious plant species, sex is determined by an interaction between cytoplasmic male-sterility (CMS) genes encoded in the mitochondrial genome that disrupt viable pollen production (leading to female individuals), and nuclear restorer-of-fertility alleles (restorer genes or Rf genes) that counteract the CMS factor when they occur in the same individual (leading to hermaphroditic individuals).[7,10,11] CMS genes are often chimeric and can contain pieces of multiple mitochondrial genes fused together. They are thought to be created by recombination and rearrangement of mtDNA, which is rampant in plants[10,12] (see also next section). Compared to plant mtDNAs (200–2000 kb),[13] the extremely compact nature of bilaterian animal mtDNAs (typically ~16–17 kb) and putatively limited role for mtDNA recombination in animals suggests that there is little room for evolutionary novelties such as the emergence of chimeric or novel genes. This, together with the rarity of gynodioecious animals,[5] suggests that CMS might be a rather uncommon phenomenon outside plants.
However, CMS in bivalve mollusks might be possible. Bivalves display relatively large variation in the size of their mitochondrial genomes (< 14 to > 67 kb), their gene order, and even gene content.[14,15] Moreover, gynodioecy, androdioecy and trioecy have all been reported in bivalves[16–19]; indeed, bivalves, and molluscs more generally, exhibit diverse sexual systems.[20] Mitochondrial genes have also been hypothesized to influence sex-determination pathways in bivalves with doubly uniparental inheritance (DUI) of mtDNA[21,22] (see also the DUI section below). DUI is a bivalve-specific mitochondrial transmission system discovered in the early 1990s[23–26] that involves two divergent sex-linked mtDNAs that are transmitted through eggs or sperm.[27–30] The association between a male-transmitted genome and maleness, which has been observed in more than a hundred species of freshwater mussels, marine mussels and marine clams representing five bivalve orders,[31–33] has led some to hypothesize a role for mitochondria and mtDNA in sex-determination mechanisms in bivalves.[21,22,27,29,34–36] DUI bivalves also contain novel, mitochondrial protein-coding genes that are specific to either the male-transmitted or the female-transmitted genomes in addition to the typical set of 13 oxidative phosphorylation (OXPHOS) genes found across animals.[21,37–41]
Although many parallels exist between DUI and CMS, previous studies have only cursorily explored the hypothesis that DUI had its origins as a cytoplasmic sex-determination system.[14,21,42,43] In this paper, we document and further explore similarities that exist between DUI and CMS. We hypothesize that DUI originated as a nuclear-cytoplasmic sex determining system and that mitonuclear sex determining mechanisms might either still be operational in extant species or be resurrected in certain contexts in contemporary bivalves. We also propose various ways to test this hypothesis.
CMS: SELFISH MITOCHONDRIA, THEIR NUCLEAR EQUALIZERS, AND SEX DETERMINATION
As mentioned above, mitochondrial-encoded genes in plants can cause CMS.[44–46] Because mtDNA is primarily maternally inherited, any mitochondrial mutations that increase female fitness might spread, even if they decrease male function.[47–50] In other words, mutations that negatively affect only males might accumulate in populations because deleterious male-specific fitness effects are “invisible” to natural selection of the maternally transmitted mitochondria. This process has been termed the “mother’s curse”.[49] CMS is a classic example of such selfish mitonuclear conflict: mtDNA variants that cause male sterility can also cause increased female fitness through reallocation of resources to female reproduction or increased offspring fitness through forced outcrossing.[51–53]
The genetic bases of CMS systems are beginning to be described in detail. The key mitochondrial genes responsible for CMS have been identified in several lineages and show remarkable variability. Two main types of CMS genes have been documented: (i) chimeric genes that contain parts of typical mitochondrial genes (e.g., OXPHOS genes), and (ii) novel genes which do not have known homology with other species (ORFans).[44,46,54] CMS genes have arisen independently many times and likely relatively quickly. For example, in different populations of Lobelia siphilitica, several different mitotypes (mitochondrial or mtDNA haplotypes) were found to be segregating at different frequencies, likely representing many novel CMS types.[55] Because plant mtDNA undergoes intragenomic recombination regularly, structural variation can be rapidly generated in plant mitochondrial genomes,[56] which in combination with gene duplication can provide the raw genomic fuel to generate new CMS genes.[57] Also, novel mtDNA can often be acquired in plant mitogenomes via horizontal gene transfer from other lineages.[58,59] Plant mtDNA is therefore a hotspot for generating the chimeric genes and ORFans underlying CMS.
When new CMS genes arise, there should be strong selection on nuclear-encoded genes to counteract them and restore fertility, as nuclear encoded genes benefit from male transmission. Such nuclear restorer of fertility (Rf) genes have been documented in many species and are predominantly members of the pentatricopeptide repeat (PPR) family of genes.[46,57,60–63] PPR genes are numerous in plant nuclear genomes (about ~450 Arabidopsis proteins contain PPR motifs[64]), are usually targeted to the mitochondria or other organelles, and often function in RNA editing or other posttranscriptional processing. As expected, putative Rf genes are under strong positive selection in populations with CMS genes,[61,65,66] consistent with an evolutionary arms race between mitochondrial CMS genes and nuclear Rf genes. The result is that many plant populations show no outward signs of CMS due to suppression by locally adapted Rf genes. However, during crosses or migration between populations or species, CMS genes are left unchecked when they are placed against a naïve nuclear background and male sterile individuals (i.e., females) are produced.[67]
While the genetic controls of CMS have received much attention, less is known regarding the physiological mechanisms of male sterility and fertility restoration. The “ATP hypothesis” posits that ATP production is compromised by CMS genes (e.g., because CMS proteins might disturb the formation or the conformation of OXPHOS complexes[68]). Although CMS genes are expressed in all tissues, only developing pollen grains are affected because pollen development is especially energetically expensive.[69,70] This hypothesis would explain how many diverse CMS genes all cause similar aberrant pollen formation phenotypes. However, this assumption has been questioned, as Arabidopsis with knocked down ATP production still produce functional pollen.[71] Another set of hypotheses, collectively termed the “pollen hypothesis”, suggests there is a pollen-specific substance that interacts with mitochondria in anthers that ensure pollen development, and this interaction is altered in CMS lines.[70,72] This hypothesis has received some empirical support, as CMS proteins appear to preferentially accumulate in anthers,[73] but are degraded by a protease in vegetative tissue.[74] Transcriptomic and proteomic CMS studies have identified putative nuclear interactors in pollen grains,[75,76] but these remain to be confirmed. CMS lines also show increased ROS production and altered patterns of programmed cell death in tapetal cells, which are critical to pollen development.[73,77–80] Presumably, Rf genes mainly counteract CMS by post-transcriptional silencing of CMS genes, although CMS restoration at the genomic level and through translational and post-translational mechanisms has also been reported.[54,63,81,82] The exact mechanisms by which CMS genes cause male sterility and how much variation in these processes exist across plants remain important questions.
DUI: SEX-SPECIFIC MITOCHONDRIAL LINEAGES AND SEX-SPECIFIC MITOCHONDRIAL GENES
As in plants, mtDNA is primarily maternally inherited in animals.[83,84] The DUI system in bivalves is one exception, where both parents transmit their mitochondria and mtDNA to their offspring: the female-type (referred to as F-mtDNA) is transmitted by the mother to both daughters and sons, and the male-type (referred to as M-mtDNA) is transmitted by the father to only sons.[25,26] Females are mostly homoplasmic and only possess the F-mtDNA, whereas males are globally heteroplasmic and possess the maternally inherited F-mtDNA in their somatic tissues (with M-mtDNA sometimes present in variable amounts) and exclusively paternally inherited M-mtDNA in their sperm.[29,30,85–88]
The F- and M-mtDNA can be highly divergent, reaching more than 50% divergence in amino acid sequences in some freshwater mussels (e.g., Quadrula, Echyridella menziesi; order Unionoida) and marine clams (e.g., Scrobicularia plana; Cardioida),[32,89,90] and 10%–35% in marine mussels (e.g., Mytilus spp., Geukensia demissa; Modiolus; Mytiloida) and marine clams (e.g., Ruditapes philippinarum; Veneroida).[29,30,91] There is still an ongoing debate regarding DUI origin, that is, a single origin of DUI in an ancestral bivalve lineage, with multiple losses during bivalve phylogenesis, or multiple independent origins of DUI.[29,30] Nevertheless, the existence of a radically different mitochondrial inheritance system raises the questions of how and why DUI has been maintained within bivalves.
The molecular basis of DUI has been investigated mainly through transcriptomic studies,[92–94] and there seems to be a consensus for a role of ubiquitination, a process that is also involved in the strict maternal inheritance (SMI) of mitochondria in other animal species.[84] The question of why DUI has appeared and been maintained in bivalves has also received much attention. One hypothesis is that in the absence of heteromorphic sex chromosomes, sex-specific mtDNAs could be involved in sex determination.[21,22,27,34] This hypothesis has gained strong support with the discovery of mitochondrial sex-specific genes in DUI bivalves with still unknown functions, that is, the F-orf gene (F-specific functional Open Reading Frame) in the F-mtDNA and the M-orf gene (M-specific functional Open Reading Frame) in the M-mtDNA.[21,32,37,39,41,42] Breton and colleagues[21] hypothesized that DUI would be responsible for the maintenance of dioecy in unionoids based on their demonstration that sex-specific mt genomes follow a traditional XY system of sex determination, and closely-related hermaphroditic species do not possess M-type mtDNA. In addition, the remaining maternally-transmitted genome in hermaphroditic species experiences highly divergent evolution in its F-orf gene. Specifically, hermaphrodites possess a highly modified F-orf sequence, called H-orf, relative to their dioecious relatives, and this situation has evolved independently at least four times in freshwater mussels.[21,40,42] However, the link among DUI, sex-specific mt genes, and the maintenance of dioecy remains uncertain.
Finally, DUI species possess other uncommon features regarding mitochondrial genes, particularly the cytochrome c oxidase subunit II gene in M-mtDNA (Mcox2). In most DUI species, including all DUI unionoids and cardioids, and some veneroids and mytiloids, Mcox2 shows a unique 3′-extension or a large in-frame insertion, extending the COX2 protein, which is usually ~250 amino acids long, by ~100 up to 1892 additional amino acids.[32,95–99] In freshwater mussels, this 3′-Mcox2-extention is transcribed, translated and localized in both inner and outer membranes of sperm mitochondria, suggesting that the exposed C-terminus tail of MCOX2 at the mitochondrial surface could act as a specific tag determining the fate of sperm mitochondria in embryos.[95,96] As noted by Stewart and colleagues,[30] gaining novel and/or chimeric genes in the mitogenomes of DUI species might generate new gene products with functions beyond metabolic roles. As explored below, among these putative roles is the possible involvement of the DUI system in sex determination in bivalves, as is the case for the CMS system in plants.
SIMILARITIES BETWEEN CMS AND DUI
Cyto-nuclear sex determination and gynodioecy
The CMS-Rf system is the prevalent sex determination system found in natural gynodioecious plant species,[11] and such nuclear-cytoplasmic sex determination is theoretically expected to occur in the majority of natural gynodioecious species.[100] Interestingly, Bivalvia is one of the rare animal groups in which gynodioecy and trioecy, a system often associated with evolutionary transitioning from gynodioecy to dioecy (or reverse), have been functionally identified, and to our knowledge, mainly involves species with DUI or closely-related species.[16–19,101–103]
In gynodioecious plants, theory predicts that female frequency might be highly variable among populations when sex is determined by interactions between several nuclear and cytoplasmic genetic factors, some of which may not be present in all populations.[104] For example, if a female parent gives rise to all-female progeny when crossed with different pollen-parents, one might suspect that the female carried a type of CMS factor for which nuclear Rf alleles were relatively rare or absent in the population.[104] In other words, within and among CMS plant populations, hermaphrodite individuals may have a male-fertile cytoplasm, or a male-sterile cytoplasm interacting with a nuclear restorer allele, and when the correct complement of nuclear Rf allele(s) is absent, some parental crosses may produce 100% female progeny.[100,104] Interestingly, the sex ratio resulting from parental crosses in several marine and freshwater DUI species can vary from one extreme (100% female progeny) to the other (100% male progeny) depending on the impact of the mother only, suggesting the presence of a key maternal factor in eggs involved in sex determination.[105–109] In Mytilus spp., the proportion of female parents that give rise to all-female progeny can be high [e.g., 25% (6/24) in wild-caught M. galloprovincialis,[105] 29% (14/49) in a pedigreed experiment in M. edulis[106] and 32% (10/31) in wild-caught M. edulis,[35,107]] whereas female parents that give rise to all-male progeny are relatively rare (i.e., < 1%).[35,105,106] In other words, a pattern of producing purely female offspring, as predicted with mitonuclear mismatches and CMS, is common in the DUI-possessing genus Mytilus.
Plant-like mtDNAs in bivalves
CMS has been linked to the recombinogenic and repetitive nature of plant mitogenomes, two characteristics that may cause genomic rearrangements that are a source of novel ORFs (ORFans that do not match a known coding DNA sequence in other species) or chimeric genes, which are key drivers in the evolution of CMS.[10,110–112] Genomic rearrangements are thus frequently observed within a species (also among closely-related species) for CMS-inducing genomes versus “fertile” ones. These characteristics, that is, recombination, repeats and rearrangements, have also been reported in bivalve mtDNAs,[113,114] including in DUI species.[15] Furthermore, genomic rearrangements with novel ORFs or chimeric genes have been observed among closely-related DUI and non-DUI species,[21,40,115] as well as within DUI species (e.g., Mytilus).[116–118] For example, the F-orf and M-orf genes both occur in DNA regions corresponding to gene order rearrangements observed among F or M mtDNAs or between F and M mtDNAs in freshwater mussels.[21,42] In particular, the atp8-nad4L region, which contains the M-orf gene in the M mtDNA of freshwater mussels, is known as a hotspot for significant rearrangements, gene duplications and “gene chimerization”[40,90] (see also below). Another observation congruent with plants is recombination events between the F and M mtDNAs in marine mussels Mytilus.[29] These events appear to primarily occur in the control regions (CRs), in which sex specific F- and M-orfs are housed,[38,39] and often result in the introduction of the M-orf gene (i.e., M-type CR portions containing M-orf) in an otherwise F-type mtDNA. To our knowledge, the reverse (i.e., the introduction of F-type CR portions and F-orf in an M-type mtDNA) has yet to be reported. Interestingly, the acquisition of the M-orf gene by an F mtDNA also often results in the “masculinization” of the F mtDNA, which is subsequently transmitted through sperm[29,117,119,120] (see Box 1). It thus seems that the acquisition of an M-orf is a prerequisite for a mtDNA that functioned as an F-type genome to become a functional M-type genome, although this has not been fully demonstrated.[29,121]
Box 1: Role reversal of the F mitogenome.
Given that F and M mtDNAs regularly coexist in male embryos of DUI species, intermolecular mitochondrial recombination between both genomes has been frequently reported or inferred, but only fully described and characterized in Mytilus mussels.[29] Intermolecular recombination in Mytilus spp. often results in a “role-reversal” event where a F mtDNA invades the male route of inheritance and becomes transmitted through sperm, effectively reversing its sex-specific role.[29,119–121] This phenomenon has been highlighted following the discovery of genomes recovered from male gonads but with coding sequences almost identical to the F mtDNA.[29] Specifically, role-reversed mtDNAs are mostly F-type genomes but with mosaic control regions (CRs) consisting of M- and F-type segments.[29,119–121] It thus seems that the switch from F to M function is mediated through intermolecular recombination introducing M-like CRs (and M-orfs) in an otherwise F-type mtDNA, although this hypothesis still need to be clearly demonstrated.[29,121] Because they reset M-mtDNAs to be nearly identical to F-mtDNAs (except for the control region), role-reversal events have further complicated DUI origination hypotheses.[27,29,30,146,147]
ORFans and chimeric ORFs in CMS and DUI systems
As seen above, CMS is associated with ORFans or chimeric genes that contain parts of typical mitochondrial genes. In other words, CMS genes in plant mitochondria are often composed of pieces of duplicated OXPHOS genes. Evidence suggests a similar situation in some DUI bivalves. Sex-specific F-orfs and M-orfs in DUI bivalves are speculated to have originated from endogenization of viral genes[39,42] or mitochondrial gene duplication.[42] In freshwater mussels in particular, in silico analyses suggested that the F-orf might have originated from a nad2 mitochondrial gene duplication, and the M-orf from an atp8 mitochondrial gene duplication, with the M-orf clearly containing segments of atp8 in some species.[40,42,90]
CMS genes are usually known to encode small transmembrane proteins (10-35 kDa) that sometimes show specific spatiotemporal accumulation in male parts of the flower such as anthers and microspores to interfere with pollen development.[82] Moreover, mitochondrially-encoded CMS proteins may sometimes localize outside of mitochondria and exert their function via interactions with cellular components other than the mitochondrion.[122] In DUI species, F-orf and M-orf genes (and H-orf genes in hermaphroditic unionoids) are also known to encode transmembrane proteins that localize inside but also outside mitochondria.[21,39,41,42,123,124] While spatiotemporal expression has not been studied for H-ORF proteins in hermaphroditic freshwater mussels, the available data for F-ORF proteins in dioecious freshwater mussels indicate a presence in mitochondria and nuclei of eggs,[21] as well as sperm mitochondria.[123] In the marine mussel M. edulis, the F-ORF protein seems to accumulate preferentially in male gonads and sperm mitochondria.[41] Because gametes are usually homoplasmic for their sex-specific mtDNA in DUI bivalves,[86] this suggests that the mtDNA-encoded F-ORF protein is exported from F-type mitochondria and imported into sperm mitochondria.[41] The data available for M-ORF proteins indicate a presence in male gonads in freshwater mussels,[37] and a progressive accumulation in mitochondria and nuclei of spermatogenic cells (and an absence in eggs) in the marine clam Ruditapes philippinarum.[124]
To date, the functions of F-ORF, M-ORF, and H-ORF proteins are unknown. However, it has been hypothesized that the M-ORF protein could be a masculinizing factor whereas the F-ORF protein could be a feminizing factor.[39,42] In freshwater DUI mussels in particular, hermaphroditism has evolved independently multiple times and in each of the hermaphroditic lineages, the M genome is apparently lost, and the F-orf of the F genome experiences highly divergent evolution and becomes an H-orf.[21,40,42,90] Therefore, it has been suggested that the F-ORF protein could inhibit testicular development in embryos that will become females in dioecious species, while the extreme modifications seen in the H-ORF protein (or its absence) could explain why the development of some testicular tissue is not completely inhibited in closely-related hermaphroditic species.[30,42] This hypothesis is also supported by the observation in marine mussels Mytilus that recombinant mtDNAs that have transitioned from F- to M-types (see point 3 above and Box 1), always contain truncated F-orfs or no F-orf in addition to having acquired an M-orf gene.[38,121] This suggests that either a complete, intact F-ORF protein might be necessary to avoid producing males and/or a complete, intact M-ORF protein might be necessary to produce sperm.
Heteroplasmy and substochiometric shifting
Mitochondria in plants with CMS are frequently heteroplasmic, often containing substoichiometric mtDNA molecules at low copy number in addition to an abundant primary mitogenome.[125] Sometimes, these coexisting substoichiometric mtDNAs are amplified and take on the role of the primary molecule, thus being responsible for “substoichiometric shifting” or rapid and dramatic changes in relative copy number of substoichiometric molecules within a generation.[125,126] Genomic shifting usually involves a single substoichiometric molecule, often containing recombination-derived chimeric sequences, and can alter plant phenotype by activating or silencing mtDNA sequences located on the shifted molecule. Thus, if the mitochondrial population consists of both CMS and “male-fertile” mtDNAs, substoichiometric shifting might be responsible for transition between the CMS and hermaphroditic condition and back again.[125,126] This heteroplasmy may result from recombination and/or mutations but also from paternal leakage of mtDNA.[125,127] Also, intra-individual segregation of different mitotypes may result in plants with mosaic phenotypes. For example, the segregation of one mitotype conditioning development of the male flower function and another inhibiting it was proposed to explain the spatial distribution of female and hermaphrodite flowers found on the same Silene vulgaris plants.[128] It was also proposed that the proportion of the mitotypes could change progressively during plant growth, leading to different sexual phenotypes in different parts of the plant.[125] These phenomena could result from an interaction between particular nuclear genes and mitotypes that are under selection to guard specific mtDNA configurations and eliminate or reduce the copy number of others.[125]
Interestingly, recent investigations on the DUI bivalve R. philippinarum revealed the presence of heteroplasmy (i.e., presence of both F- and M-mtDNAs) at the organelle level in undifferentiated germ cells of both sexes, whereas homoplasmy was observed for the F mtDNA in eggs and for the M mtDNA in sperm.[86] It is tempting to speculate that a mechanism exists in DUI species that is similar to what has been proposed in plants, where changes in the proportion of the mitotypes in undifferentiated germ cells change during development leads to different sexual phenotypes. The recent report of a hermaphroditic species with DUI (Semimytilus algosus), which possesses a sperm producing gonad (that contains M-mtDNA) located on one half of the body and an egg producing gonad (that contains F-mtDNA) located on the opposite side of the body,[103] supports this. In this species, cell-specific segregation and/or preferential replication of mitotypes could lead to different sexual phenotypes in different parts of the animal, similar to the spatial distribution of hermaphrodite and female flowers in some plants.
A HYPOTHESIS FOR DUI ORIGINATING AS A NUCLEAR-CYTOPLASMIC SEX DETERMINING SYSTEM
It has been hypothesized that DUI first emerged in an ancestral hermaphroditic species, in which both eggs and sperm were produced in an ovotestis.[21,27,124,129] Our hypothetical scenario for the origin of DUI as a nuclear-cytoplasmic sex determining system would start with the origin of a CMS-causing gene in this ancestral hermaphroditic lineage, that is, with the origin of the“feminizing” F-orf gene[42] (Figure 2). The presence of this CMS factor causing gynodioecy and female-biased sex ratios then selected for paternal transmission (leakage) of the male-fertile mitochondrial genome (i.e., without the F-orf). This idea of selection for paternal mitochondrial leakage has been proposed by Burt and Trivers[130] and McCauley[53] for the following reason: in order to transmit genes via sperm, it is evolutionary advantageous for a nuclear genome to be paired with a male-fertile mitotype. In a gynodioecious system, paternal mitochondrial leakage enhances the probability of producing a male-fertile phenotype since a sperm donor must be a hermaphrodite that is more likely to carry a fertile phenotype, while a sperm recipient could be either hermaphroditic or female. As suggested by McCauley,[53] leakage of male-fertile mtDNAs through sperm produced by hermaphrodites can eventually offset the added productivity often associated with females carrying CMS. In early bivalves, this situation might have set the stage for the evolution of pure paternal inheritance or DUI (with further genes, likely nuclear, involved in the process), together with the invasion of gynodioecious populations by female-sterility mutation(s) or factors (i.e., masculinizing factors). If these mutations/factors increase male function sufficiently to compensate for the loss of female function, then “pure males” will successfully establish and lead to subdioecy or dioecy (Figure 2). We propose that such female-sterile or “masculinizing” factors could be the chimeric M-orf gene, and/or the elongated cox2 gene in the paternally-transmitted M mtDNAs of DUI bivalves. This is in line with the recent demonstration that sperm and their M-type mitochondria express a “M-specific phenotype” in DUI species, which is characterized by low OXPHOS rates and an almost null spare capacity of the cytochrome c oxidase complex compared with F-type mitochondria, a phenotype that could potentially be detrimental in somatic tissues or eggs.[131,132]
FIGURE 2.
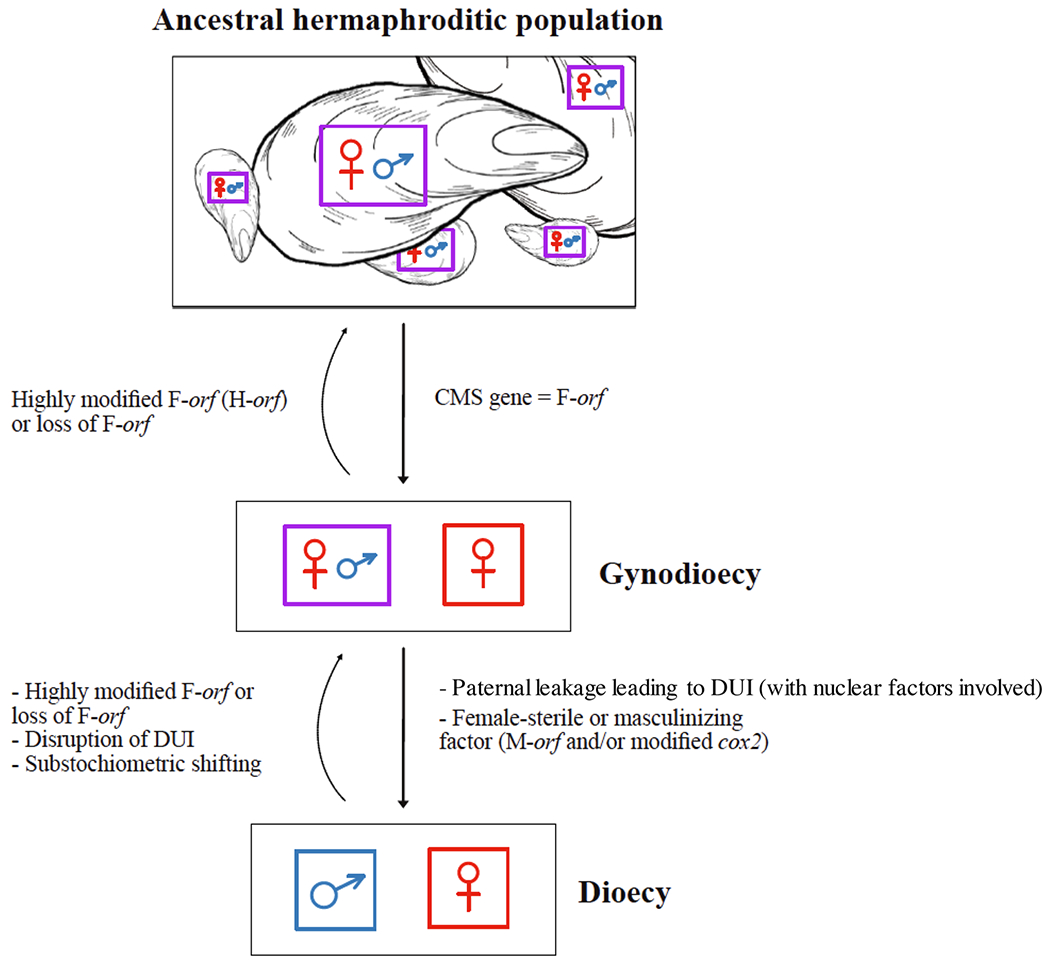
Hypothetical scenario for the origin of DUI as a nuclear-cytoplasmic sex determining system
Our hypothetical scenario is somewhat different than the one proposed by Milani and colleagues[124] to describe how DUI and dioecy might have evolved together in Ruditapes philippinarum. Based on in silico evidence that the M-orf might be of viral origin in some bivalve species,[39] Milani and colleagues[124] proposed that a viral infection of some mitochondria would have conferred upon them the ability to avoid degradation in embryos, and to be preferentially transmitted through generations (i.e., segregation distortion). Specifically, in a population of hermaphroditic R. philippinarum, infected mitochondria would have spread more efficiently through sperm, allowing paternal inheritance of mitochondria, selection for mtDNA mutations that increase male fitness and emergence of males in the population. This transition from hermaphroditism to androdioecy would have set up a condition for selective pressure favoring egg production in hermaphrodites, eventually leading to the evolution of dioecy (with DUI) from androdioecy.[124] While gynodioecy, not androdioecy, precedes dioecy in our hypothesis, viral infections could have led to the establishment of M-orf genes in our hypothesis, and the two hypotheses might have complementary components. At this moment, the two hypothetical scenarios need further evaluation.
EVALUATING THE CMS/DUI HYPOTHESIS
As stated above, the unique mode of inheritance and sex-biased mt architecture in DUI bivalves has led to the hypothesis that DUI is a critical component of sex determination.[21,22,29] Herein, we outline some methods to test our hypothesis that the DUI system in bivalves might have originated as a CMS system, in which mitochondrial factors interact with nuclear factors to determine the production of hermaphrodites, females, or males. While we acknowledge that this original system might have been replaced during bivalve evolution, we argue that mitonuclear sex determining mechanisms might still operate in multiple bivalve lineages or might be able to be resurrected in contemporary bivalves.
One way to determine if there are mitochondrial CMS genes in a species of interest is by comparing offspring sex ratios from reciprocal crosses, where pairs of parents are used to produce two sets of offspring, one set with one parent being the “egg parent” and the other the sperm donor, and the second set where their roles are reversed.[100] The two sets of offspring will have the same distribution of nuclear genes but will differ for cytoplasmic genes that are primarily maternally inherited, which will have a major effect on sex ratios in the presence (or absence) of CMS.[100] For DUI bivalves however, this strategy is somewhat problematic because the paternal mtDNA is also transmitted. Moreover, DUI bivalve species that are currently gynodioecious or dioecious might have evolved from populations with mitochondrial sex determining genes but for which sex determination has now come under complete nuclear control. For example, recent studies suggest extremely low within-species variability for the F-orf gene in bivalves.[133,134] That said, freshwater mussels might offer an opportunity to investigate the genetic basis of sex determination through reciprocal crosses because of the presence of gynodioecious species with SMI of mtDNA that are closely related to dioecious DUI species.[16,17,135] Specifically, a cross using eggs from a dioecious (DUI) species and sperm from a closely related hermaphrodite species could be performed,[135,136] which in theory, should lead to all female progeny if the F-orf is acting as a CMS gene. Reciprocal crosses could also be done using pairs of SMI hermaphrodites, and if the offspring sex ratios differ, sex determination in the species would be under nuclear-cytoplasmic control.[100] Crossing distant DUI populations or even closely-related DUI species with divergent F-orf genes could also be considered. For example, we could expect that a female would produce male embryos when crossed with males from the same population but only (or predominantly) female embryos when crossed with males from distant populations (i.e., if the F-orf acts as a CMS gene and restorers are absent in the distant population).
Another approach would be to silence the F-orf gene (e.g., RNA interference or RNAi) and see if this results in hermaphroditic or male individuals, or if it causes downstream dysregulation of specific genes (e.g., genes in sex determination/differentiation pathways). An RNAi-like mechanism has been proven to operate in the mitochondria in an Ago2-dependent manner.[137] However, the technique is relatively untested with respect to the study of mitochondrial biology and has limitations of its own, for example, transfected small interference RNAs (siRNAs) are able to enter the matrix of mitochondria and affect mRNA levels, but their translational effect seems to be only recordable on relatively unstable proteins.[137] That said, RNAi has been widely used in bivalves,[138–142] although not at the mitochondrial level. An alternative approach would be to overexpress the F-orf or M-orf gene. In this case, we would expect sperm sterility with an overexpression of F-orf in male individuals (and female sterility with overexpression of M-orf), and potentially also cytotoxicity following heterologous gene expression, an effect that has been reported several times for CMS genes introduced in Escherichia coli or yeast cells.[110,111,143] Although the above-mentioned approaches might never convincingly confirm the hypothesis that DUI originally arose via CMS, they could provide a framework for testing the hypothesis that mitonuclear sex determining mechanisms still operate or might be reactivated in DUI bivalves. It is worth mentioning that the origin of DUI via CMS and mitochondrial sex determination in extant bivalve species could be two different, non-mutually exclusive hypotheses. In either case, this would indicate, for the first time, that mitochondrial genetic elements might be involved in sex determination in animals. Many of the above-mentioned studies are currently underway.
CONCLUSION
There are many intriguing parallels between bivalves with DUI and CMS in plants. Based on these similarities, we present a novel and testable hypothesis, namely that the DUI system in bivalves might have originated as a CMS system, in which mitochondrial factors interact with nuclear factors to determine sex, and that mitonuclear sex determining mechanisms could still perform their roles or might be reactivated in extant bivalve species. It is conceivable that nuclear-cytoplasmic sex determination might be more common than previously thought in animal species.[144,145] In order to further uncover and fully appreciate the implications of mitochondria in sex determination (and sex-ratio distortion), we must expand taxonomic sampling in a comprehensive manner, for example starting with animal species with atypical sexual systems. Such studies will significantly contribute to a better understanding of fundamental evolutionary processes, such as the role of intergenomic conflict in sex determination.
ACKNOWLEDGMENTS
The authors thank Fabrizio Ghiselli, Liliana Milani, Mélanie Tassé, Thierry Niaison, Stefano Bettinazzi, Laura Kienzle, Thierry Choquette and Chiara Babinski for helpful discussions. The authors also gratefully thank Kerstin Brachhold and two anonymous reviewers for their useful comments. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) [grant numbers 217175 and 435656]; and the National Institutes of Health (NIH) [1R35GM142836]. S Breton holds the Canadian Research Chair in Evolutionary Mitochondrial Biology.
Abbreviations:
- CMS
cytoplasmic male sterility
- CR
control region
- DUI
doubly uniparental inheritance
- ORF
open reading frame
- OXPHOS
oxidative phosphorylation
- PPR
pentatricopeptide repeat family of genes
- Rf
restorer-of-fertility genes
- RNAi
RNA interference
- SMI
strict maternal inheritance
- siRNA
small interference RNA
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1.Leonard JL (Ed.). (2018). Transitions between sexual systems: Understanding the mechanisms of, and pathways between, dioecy, hermaphroditism and other sexual systems (pp. 1–58), Springer International Publishing. [Google Scholar]
- 2.Casimiro-Soriguer I, Buide ML, & Narbona E (2015). Diversity of sexual systems within different lineages of the genus Silene. AoB PLANTS, 7, plv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth B, & Charlesworth D (1978). A model for the evolution of dioecy and gynodioecy. American Naturalist, 112, 975–997. [Google Scholar]
- 4.Bawa KS, & Beach JH (1981). Evolution of sexual systems in flowering plants. Annals of the Missouri Botanical Garden, 68, 254–274. [Google Scholar]
- 5.Weeks SC (2012). The role of androdioecy and gynodioecy in mediating evolutionary transitions between dioecy and hermaphroditism in the animalia. Evolution; International Journal of Organic Evolution, 66, 3670–3686. [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth D (1999). In Geber MA, Dawson TE, & Delph LF (Eds.). Gender and sexual dimorphism in flowering plants (pp. 33–60), Springer. [Google Scholar]
- 7.Fruchard C, & Marais GAB (2017). In Nuno de la Rosa L & Müller G (Eds.). Evolutionary developmental biology (pp. 1–14), Springer International Publishing. [Google Scholar]
- 8.Delph LF (2009). Sex allocation: Evolution to and from dioecy. Current Biology, 19, R249–R251. [DOI] [PubMed] [Google Scholar]
- 9.Jarne P, & Charlesworth D (1993). The evolution of the selfing rate in functionally hermaphrodite plants and animals. Annual Review of Ecology and Systematics, 24, 441–466. [Google Scholar]
- 10.Chase CD (2007). Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends in Genetics, 23, 81–90. [DOI] [PubMed] [Google Scholar]
- 11.Delph LF, Touzet P, & Bailey MF (2007). Merging theory and mechanism in studies of gynodioecy. Trends in Ecology & Evolution, 22, 17–24. [DOI] [PubMed] [Google Scholar]
- 12.Hanson MR, & Bentolila S (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell, 16(Suppl): S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morley SA, & Nielsen BL (2017). Plant mitochondrial DNA. Frontiers in Bioscience, 22, 1023–1032. [DOI] [PubMed] [Google Scholar]
- 14.Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, & Passamonti M (2014). A resourceful genome: Updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends in Genetics, 30, 555–564. [DOI] [PubMed] [Google Scholar]
- 15.Ghiselli F, Gomes-dos-Santos A, Adema CM, Lopes-Lima M, Sharbrough J, & Boore JL (2021). Molluscan mitochondrial genomes break the rules. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 376, 20200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heard WH (1975). Sexuality and other aspects of reproduction in Anodonta (Pelecypoda: Unionidae). Malacologia, 15, 81–103. [Google Scholar]
- 17.Kat PW. (1983). Sexual selection and simultaneous hermaphroditism among the Unionidae (Bivalvia: Mollusca). Journal of Zoology, 201, 395–416. [Google Scholar]
- 18.Byrne M (1998). Reproduction of river and lake populations of Hyridella depressa (Unionacea: Hyriidae) in New South Wales: Implications for their conservation. Hydrobiologia, 389, 29–43. [Google Scholar]
- 19.Oyarzún PA, Nuñez JJ, Toro JE, & Gardner JPA (2020). Trioecy in the marine mussel semimytilus algosus (Mollusca, Bivalvia): Stable sex ratios across 22 degrees of a latitudinal gradient. Frontiers in Marine Science, 348. [Google Scholar]
- 20.Collin R (2013). Phylogenetic patterns and phenotypic plasticity of molluscan sexual systems. Integrative and Comparative Biology, 53, 723–735. [DOI] [PubMed] [Google Scholar]
- 21.Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, & Hoeh WR (2011). Novel protein genes in animal mtDNA: A new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Molecular Biology and Evolution, 28, 1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breton S, Capt C, Guerra D, & Stewart DT (2018). In Leonard JL (Ed.). Transitions between sexual systems: Understanding the mechanisms of, and pathways between, dioecy, hermaphroditism and other sexual systems (pp. 165–192), Springer International Publishing. [Google Scholar]
- 23.Fisher C, & Skibinski DO (1990). Sex-biased mitochondrial DNA heteroplasmy in the marine mussel Mytilus. Proceedings of the Royal Society B: Biological Sciences, 242, 149–156. [Google Scholar]
- 24.Hoeh WR, Blakley KH, & Brown WM (1991). Heteroplasmy suggests limited biparental inheritance of Mytilus mitochondrial DNA. Science, 251, 1488–1490. [DOI] [PubMed] [Google Scholar]
- 25.Skibinski DO, Gallagher C, & Beynon CM (1994). Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics, 138, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zouros E, Oberhauser Ball A, Saavedra C, & Freeman KR (1994). An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proceedings of the National Academy of Sciences of the United States of America, 91, 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breton S, Doucet-Beaupré H, Stewart DT, Hoeh WR, & Blier PU (2007). The unusual system of doubly uniparental inheritance of mtDNA: Isn’t one enough? Trends in Genetics, 23, 465–474. [DOI] [PubMed] [Google Scholar]
- 28.Passamonti M, & Ghiselli F (2009). Doubly uniparental inheritance: Two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA and Cell Biology, 28, 79–89. [DOI] [PubMed] [Google Scholar]
- 29.Zouros E (2013). Biparental inheritance through uniparental transmission: The Doubly Uniparental Inheritance (DUI) of mitochondrial DNA. Evolutionary Biology, 40, 1–31. [Google Scholar]
- 30.Stewart DT, Breton S, Chase EE, Robicheau BM, Bettinazzi S, Pante E, Youssef N, & Garrido-Ramos MA (2020). An unusual evolutionary strategy: The origins, genetic repertoire, and implications of doubly uniparental inheritance of mitochondrial DNA in bivalves. In Pontarotti P (Ed.). Evolutionary Biology–A Transdisciplinary Approach (pp. 301–323). Springer International Publishing. [Google Scholar]
- 31.Gusman A, Lecomte S, Stewart DT, Passamonti M, & Breton S (2016). Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ, 4, e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capt C, Bouvet K, Guerra D, Robicheau BM, Stewart DT, Pante E, & Breton S (2020). Unorthodox features in two venerid bivalves with doubly uniparental inheritance of mitochondria. Science Reports, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soroka M (2020). Doubly uniparental inheritance of mitochondrial DNA in freshwater mussels: History and status of the European species. Journal of Zoological Systematics and Evolutionary Research, 58, 598–614. [Google Scholar]
- 34.Zouros E (2000). The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes & Genetic Systems, 75, 313–318. [DOI] [PubMed] [Google Scholar]
- 35.Yusa Y, Breton S, & Hoeh WR (2013). Population genetics of sex determination in Mytilus mussels: Reanalyses and a model. Journal of Heredity, 104, 380–385. [DOI] [PubMed] [Google Scholar]
- 36.Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, & Passamonti M (2014). Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biology and Evolution, 6, 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breton S, Doucet-Beaupré H, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, & Hoeh WR (2009). Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: Gender-specific open reading frames and putative origins of replication. Genetics, 183, 1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, & Hoeh WR (2011). Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine mussels (Bivalvia: Mytilidae). Plos One, 6, e19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milani L, Ghiselli F, Guerra D, Breton S, & Passamonti M (2013). A comparative analysis of mitochondrial ORFans: New clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biology and Evolution, 5, 1408–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerra D, Lopes-Lima M, Froufe E, Gan HM, Ondina P, Amaro R, Klunzinger MW, Callil C, Prié V, Bogan AE, Stewart DT, & Breton S (2019). Variability of mitochondrial ORFans hints at possible differences in the system of doubly uniparental inheritance of mitochondria among families of freshwater mussels (Bivalvia: Unionida). BMC Evolutionary Biology, 19, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouimet P, Kienzle L, Lubosny M, Burzyński A, Angers A, & Breton S (2020). The ORF in the control region of the female-transmitted Mytilus mtDNA codes for a protein. Gene, 725, 144161. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell A, Guerra D, Stewart DT, & Breton S (2016). In silico analyses of mitochondrial ORFans in freshwater mussels (Bivalvia: Unionoida) provide a framework for future studies of their origin and function. BMC Genomics, 17, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda GP, lannello M, McConie HJ, Ghiselli F, & Havird JC (2021). Relaxed selection on male mitochondrial genes in DUI bivalves eases the need for mitonuclear coevolution. Journal of Evolutionary Biology, 00, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnable PS, & Wise RP (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends in Plant Science, 3, 175–180. [Google Scholar]
- 45.Budar F, Touzet P, & De Paepe R (2003).The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica, 117, 3–16. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y-J, & Zhang D (2018). Molecular control of male fertility for crop hybrid breeding. Trends in Plant Science, 23, 53–65. [DOI] [PubMed] [Google Scholar]
- 47.Cosmides LM, & Tooby J (1981). Cytoplasmic inheritance and intragenomic conflict. Journal of Theoretical Biology, 89, 83–129. [DOI] [PubMed] [Google Scholar]
- 48.Frank SA, & Hurst LD (1996). Mitochondria and male disease. Nature, 383, 224. [DOI] [PubMed] [Google Scholar]
- 49.Gemmell NJ, Metcalf VJ, & Allendorf FW (2004). Mother’s curse: The effect of mtDNA on individual fitness and population viability. Trends in Ecology & Evolution, 19, 238–244. [DOI] [PubMed] [Google Scholar]
- 50.Havird JC, Forsythe ES, Williams AM, Werren JH, Dowling DK, & Sloan DB (2019). Selfish mitonuclear conflict. Current Biology, 29, R496–R511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis D (1941). Male sterility in natural populations of hermaphrodite plants: The equilibrium between females and hermaphrodites to be expected with different types of inheritance. The New phytologist, 40, 56–63. [Google Scholar]
- 52.McCauley DE, Olson MS, Emery SN, & Taylor DR (2000). Population structure influences sex ratio evolution in a gynodioecious plant. American Naturalist, 155, 814–819. [DOI] [PubMed] [Google Scholar]
- 53.McCauley DE (2013). Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. The New phytologist, 200, 966–977. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, & Liu Y-G (2014). Male sterility and fertility restoration in crops. Annual Review of Plant Biology, 65, 579–606. [DOI] [PubMed] [Google Scholar]
- 55.Adhikari B, Caruso CM, & Case AL (2019). Beyond balancing selection: Frequent mitochondrial recombination contributes to high-female frequencies in gynodioecious Lobelia siphilitica (Campanulaceae). The New Phytologist, 224, 1381–1393. [DOI] [PubMed] [Google Scholar]
- 56.Chevigny N, Schatz-Daas D, Lotfi F, & Gualberto JM (2020). DNA repair and the stability of the plant mitochondrial genome. International Journal of Molecular Sciences, 21, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang H, Zheng X, Li C, Xie X, Chen Y, Chen L, Zhao X, Zheng H, Zhou J, Ye S, Guo J, & Liu YG (2017). Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Research, 27, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, & Song K (2000). Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proceedings of the National Academy of Sciences of the United States of America, 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christensen AC (2014). Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biology and Evolution, 6, 1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bentolila S, Alfonso AA, & Hanson MR (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proceedings of the National Academy of Sciences of the United States of America, 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujii S, Bond CS, & Small ID (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proceedings of the National Academy of Sciences of the United States of America, 108, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaborieau L, Brown GG, & Mireau H (2016). The Propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Frontiers in Plant Science, 7, 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubo T, Arakawa T, Honma Y, & Kitazaki K (2020). What does the molecular genetics of different types of restorer-of-fertility genes imply? Plants, 9, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Toole N, Hattori M, Andres C, lida K, Lurin C, Schmitz-Linneweber C, Sugita M, & Small I (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Molecular Biology and Evolution, 25, 1120–1128. [DOI] [PubMed] [Google Scholar]
- 65.Touzet P, & Budar F (2004). Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends in Plant Science, 9, 568–570. [DOI] [PubMed] [Google Scholar]
- 66.Case AL, Finseth FR, Barr CM, & Fishman L (2016). Selfish evolution of cytonuclear hybrid incompatibility in Mimulus. Proceedings of the Royal Society B: Biological Sciences, 283, 20161493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roux F, Mary-Huard T, Barillot E, Wenes E, Botran L, Durand S, Villoutreix R, Martin-Magniette ML, Camilleri C, & Budar F (2016). Cytonuclear interactions affect adaptive traits of the annual plant Arabidopsis thaliana in the field. Proceedings of the National Academy of Sciences of the United States of America, 113, 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Pandeya D, Jo YD, Liu W, & Kang B-Y (2013). Reduced activity of ATP synthase in mitochondria causes cytoplasmic male sterility in chili pepper. Planta, 237, 1097–1109. [DOI] [PubMed] [Google Scholar]
- 69.Warmke HE, & Lee SL (1978). Pollen abortion in T cytoplasmic male-sterile corn (Zea mays): A suggested mechanism. Science, 200, 561–563. [DOI] [PubMed] [Google Scholar]
- 70.Touzet P, & Meyer EH (2014). Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion, 19, 166–171. [DOI] [PubMed] [Google Scholar]
- 71.Robison MM, Ling X, Smid MPL, Zarei A, & Wolyn DJ (2009). Antisense expression of mitochondrial ATP synthase subunits OSCP (ATP5) and gamma (ATP3) alters leaf morphology, metabolism and gene expression in Arabidopsis. Plant & Cell Physiology, 50, 1840–1850. [DOI] [PubMed] [Google Scholar]
- 72.Flavell R (1974). A model for the mechanism of cytoplasmic male sterility in plants, with special reference to maize. Plant Science Letters, 3, 259–263. [Google Scholar]
- 73.Luo D, Xu H, Liu Z, Guo J, Li H, Chen L, Fang C, Zhang Q, Bai M, Yao N, Wu H, Wu H, Ji C, Zheng H, Chen Y, Ye S, Li X, Zhao X, Li R, & Liu YG (2013). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nature Genetics, 45, 573–577. [DOI] [PubMed] [Google Scholar]
- 74.Sarria R, Lyznik A, Vallejos CE, & Mackenzie SA (1998). A cytoplasmic male sterility-associated mitochondrial peptide in common bean is post-translationally regulated. Plant Cell, 10, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xing M, Sun C, Li H, Hu S, Lei L, & Kang J (2018). Integrated analysis of transcriptome and proteome changes related to the Ogura cytoplasmic male sterility in cabbage. Plos One, 13, e0193462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan Q, Song C, Gao L, Zhang H, Yang C, Sheng J, Ren J, Chen D, & Wang Y (2018). Transcriptome de novo assembly and analysis of differentially expressed genes related to cytoplasmic male sterility in onion. Plant Physiology and Biochemistry, 125, 35–44. [DOI] [PubMed] [Google Scholar]
- 77.Balk J, & Leaver CJ (2001).The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell, 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li S, Wan C, Kong J, Zhang Z, Li Y, & Zhu Y (2004). Programmed cell death during microgenesis in a Honglian CMS line of rice is correlated with oxidative stress in mitochondria. Functional Plant Biology, 31, 369–376. [DOI] [PubMed] [Google Scholar]
- 79.Jiang P, Zhang X, Zhu Y, Zhu W, Xie H, & Wang X (2007). Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Reports, 26, 1627–1634. [DOI] [PubMed] [Google Scholar]
- 80.Wang K, Gao F, Ji Y, Liu Y, Dan Z, Yang P, Zhu Y, & Li S (2013). ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. The New Phytologist, 198, 408–418. [DOI] [PubMed] [Google Scholar]
- 81.Kitazaki K, Arakawa T, Matsunaga M, Yui-Kurino R, Matsuhira H, Mikami T, & Kubo T (2015). Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). The Plant Journal, 83, 290–299. [DOI] [PubMed] [Google Scholar]
- 82.Melonek J, Duarte J, Martin J, Beuf L, Murigneux A, Varenne P, Comadran J, Specel S, Levadoux S, Bernath-Levin K, Torney F, Pichon JP, Perez P, & Small I (2021). The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nature Communication, 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Breton S, & Stewart DT (2015). Atypical mitochondrial inheritance patterns in eukaryotes. Genome, 58, 423–431. [DOI] [PubMed] [Google Scholar]
- 84.Sato K, & Sato M (2017). Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. Journal of Biochemistry, 162, 247–253. [DOI] [PubMed] [Google Scholar]
- 85.Breton S, Bouvet K, Auclair G, Ghazal S, Sietman BE, Johnson N, Bettinazzi S, Stewart DT, & Guerra D (2017). The extremely divergent maternally- and paternally-transmitted mitochondrial genomes are co-expressed in somatic tissues of two freshwater mussel species with doubly uniparental inheritance of mtDNA. Plos One, 12, e0183529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghiselli F, Maurizii MG, Reunov A, Ariño-Bassols H, Cifaldi C, Pecci A, Alexandrova Y, Bettini S, Passamonti M, Franceschini V, & Milani L (2019). Natural heteroplasmy and mitochondrial inheritance in bivalve molluscs. Integrative and Comparative Biology, 59, 1016–1032. [DOI] [PubMed] [Google Scholar]
- 87.Iannello M, Bettinazzi S, Breton S, Ghiselli F, & Milani L (2021). A naturally heteroplasmic clam provides clues about the effects of genetic bottleneck on paternal mtDNA. Genome Biology and Evolution, 13, evab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mioduchowska M, Kaczmarczyk A, Zając K, Zając T, & Sell J (2016). Gender-associated mitochondrial DNA heteroplasmy in somatic tissues of the endangered freshwater mussel unio crassus (Bivalvia: Unionidae): Implications for sex identification and phylogeographical studies. Journal of Experimental Zoology. Part A, Ecological Genetics and Physiology, 325, 610–625. [DOI] [PubMed] [Google Scholar]
- 89.Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, & Hoeh WR (2010). Mitochondrial phylogenomics of the Bivalvia (Mollusca): Searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evolutionary Biology, 10, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guerra D, Plazzi F, Stewart DT, Bogan AE, Hoeh WR, & Breton S (2017). Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida). Science Reports, 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robicheau BM, Powell AE, Del Bel L, Breton S, & Stewart DT (2017). Evidence for extreme sequence divergence between the male- and female-transmitted mitochondrial genomes in the bivalve mollusk, Modiolus (Mytilidae). Journal of Zoological Systematics and Evolutionary Research, 55, 89–97. [Google Scholar]
- 92.Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, Nuzhdin SV, & Passamonti M (2012). De novo assembly of the manila clam Ruditapes philippinarum transcriptome provides new insights into expression bias, mitochondrial doubly uniparental inheritance and sex determination. Molecular Biology and Evolution, 29, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Capt C, Renaut S, Ghiselli F, Milani L, Johnson NA, Sietman BE, Stewart DT, & Breton S (2018). Deciphering the link between doubly uniparental inheritance of mtDNA and sex determination in bivalves: Clues from comparative transcriptomics. Genome Biology and Evolution, 10, 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Capt C, Renaut S, Stewart DT, Johnson NA, & Breton S (2019). Putative mitochondrial sex determination in the Bivalvia: Insights from a hybrid transcriptome assembly in freshwater mussels. Frontiers in Genetics, 10, 840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chakrabarti R, Walker JM, Chapman EG, Shepardson SP, Trdan RJ, Curole JP, Watters GT, Stewart DT, Vijayaraghavan S, & Hoeh WR (2007). Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Letters, 581, 5213–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chapman EG, Piontkivska H, Walker JM, Stewart DT, Curole JP, & Hoeh WR (2008). Extreme primary and secondary protein structure variability in the chimeric male-transmitted cytochrome c oxidase subunit II protein in freshwater mussels: Evidence for an elevated amino acid substitution rate in the face of domain-specific purifying selection. BMC Evolutionary Biology, 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Passamonti M, Ricci A, Milani L, & Ghiselli F (2011). Mitochondrial genomes and Doubly Uniparental Inheritance: New insights from Musculista senhousia sex-linked mitochondrial DNAs (Bivalvia Mytilidae). BMC Genomics, 12, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bettinazzi S, Plazzi F, & Passamonti M (2016). The complete female- and male-transmitted mitochondrial genome of Meretrix lamarckii. Plos One, 11, e0153631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lubosny M, Smietanka B, Przylucka A, & Burzynski A (2020). Highly divergent mitogenomes of Geukensiademissa (Bivalvia, Mytilidae) with extreme AT content. Journal of Zoological Systematics and Evolutionary Research„ 58, 571–580. [Google Scholar]
- 100.Bailey MF, & Delph LF (2007). A field guide to models of sex-ratio evolution in gynodioecious species. Oikos, 116, 1609–1617. [Google Scholar]
- 101.Heller J (1993). Hermaphroditism in molluscs. Biological Journal of the Linnean Society, 48, 19–42. [Google Scholar]
- 102.Dublinowska M, Smolarz K, Zabrzańska S, Larsson J, & Czerniawska N (2016). Intersexuality in the blue mussel mytilus edulis complex (Mytilidae) from the baltic sea and the danish strait. American Malacological Bulletin, 34, 28–39. [Google Scholar]
- 103.Lubosny M, Przylucka A, Smietanka B, & Burzynski A (2020). Semimytilus algosus: First known hermaphroditic mussel with doubly uniparental inheritance of mitochondrial DNA. Science Reports, 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dudle DA, Mutikainen P, & Delph LF (2001). Genetics of sex determination in the gynodioecious species Lobelia siphilitica: Evidence from two populations. Heredity, 86, 265–276. [DOI] [PubMed] [Google Scholar]
- 105.Saavedra C, Reyero M-I, & Zouros E (1997). Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex-ratio in the mussel mytilus galloprovincialis. Genetics, 145, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kenchington E, MacDonald B, Cao L, Tsagkarakis D, & Zouros E (2002). Genetics of mother-dependent sex ratio in blue mussels (Mytilus spp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics, 161, 1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kenchington EL, Hamilton L, Cogswell A, & Zouros E (2009). Paternal mtDNA and maleness are co-inherited but not causally linked in mytilid mussels. Plos One, 4, e6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghiselli F, Milani L, & Passamonti M (2011). Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Molecular Biology and Evolution, 28, 949–961. [DOI] [PubMed] [Google Scholar]
- 109.Machordom A, Araujo R, Toledo C, Zouros E, & Ladoukakis ED (2015). Female-dependent transmission of paternal mtDNA is a shared feature of bivalve species with doubly uniparental inheritance (DUI) of mitochondrial DNA. Journal of Zoological Systematics and Evolutionary Research„ 53, 200–204. [Google Scholar]
- 110.Hu J, Huang W, Huang Q, Qin X, Yu C, Wang L, Li S, Zhu R, & Zhu Y (2014). Mitochondria and cytoplasmic male sterility in plants. Mitochondrion, 19, 282–288. [DOI] [PubMed] [Google Scholar]
- 111.Horn R, Gupta KJ, & Colombo N (2014). Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion, 19, 198–205. [DOI] [PubMed] [Google Scholar]
- 112.Chen Z, Zhao N, Li S, Grover CE, Nie H, Wendel JF, Hua J (2017). Plant mitochondrial genome evolution and cytoplasmic male sterility. Critical Reviews in Plant Sciences, 36, 55–69. [Google Scholar]
- 113.Calcino A, Baranyi C, & Wanninger A (2020). Heteroplasmy and repeat expansion in the plant-like mitochondrial genome of a bivalve mollusc. bioRxiv, 2020.09.23.310516. [Google Scholar]
- 114.Kong L, Li Y, Kocot KM, Yang Y, Qi L, Li Q, & Halanych KM (2020). Mitogenomics reveals phylogenetic relationships of Arcoida (Mollusca, Bivalvia) and multiple independent expansions and contractions in mitochondrial genome size. Molecular Phylogenetics and Evolution, 150, 106857. [DOI] [PubMed] [Google Scholar]
- 115.Ghiselli F, Milani L, lannello M, Procopio E, Chang PL, Nuzhdin SV, & Passamonti M (2017). The complete mitochondrial genome of the grooved carpet shell, Ruditapes decussatus (Bivalvia, Veneridae). PeerJ, 5, e3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breton S, Burger G, Stewart DT, & Blier PU (2006). Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.). Genetics, 172, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Burzyński A, Zbawicka M, Skibinski DOF, & Wenne R (2006). Doubly uniparental inheritance ss associated with high polymorphism for rearranged and recombinant control region haplotypes in Baltic Mytilus trossulus. Genetics, 174, 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mizi A, Zouros E, & Rodakis GC (2006). Multiple events are responsible for an insertion in a paternally inherited mitochondrial genome of the mussel Mytilus galloprovincialis. Genetics, 172, 2695–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Burzynski A (2003). Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Molecular Biology and Evolution, 20, 388–392. [DOI] [PubMed] [Google Scholar]
- 120.Stewart DT, Breton S, Blier PU, & Hoeh WR (2009). Masculinization events and Doubly Uniparental Inheritance of mitochondrial DNA: A model for understanding the evolutionary dynamics of gender-associated mtDNA in mussels. In: Pontarotti P (Ed.). Evolutionary biology. (pp. 163–173), Springer. ISBN 978-3-642-00951-8. [Google Scholar]
- 121.Zbawicka M, Wenne R, & Burzynski A (2014). Mitogenomics of recombinant mitochondrial genomes of Baltic Mytilus mussels. Molecular Genetics and Genomics, 289, 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.He S, Abad AR, Gelvin SB, & Mackenzie SA (1996). A cytoplasmic male sterility-associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proceedings of the National Academy of Sciences of the United States of America, 93, 11763–11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shepardson SP, Heard WH, Breton S, & Hoeh WR (2012). Light and transmission electron microscopy of two spermatogenic pathways and Unimorphic Spermatozoa in Venustaconcha ellipsiformis (Conrad, 1836) (Bivalvia: Unionoida). Malacologia, 55, 263–284. [Google Scholar]
- 124.Milani L, Ghiselli F, & Passamonti M (2016). Mitochondrial selfish elements and the evolution of biological novelties. Current Zoology, 62, 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Woloszynska M (2010). Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes–though this be madness, yet there’s method in’t. Journal of Experimental Botany, 61, 657–671. [DOI] [PubMed] [Google Scholar]
- 126.Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, & Mackenzie SA (2006). Mitochondrial genome dynamics in plants and animals: Convergent gene fusions of a MutS homologue. Journal of Molecular Evolution, 63, 165–173. [DOI] [PubMed] [Google Scholar]
- 127.McCauley DE, Bailey MF, Sherman NA, & Darnell MZ (2005). Evidence for paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity, 95, 50–58. [DOI] [PubMed] [Google Scholar]
- 128.Andersson H (1999). Female and hermaphrodite flowers on a chimeric gynomonoecious Silene vulgaris plant produce offspring with different genders: A case of heteroplasmic sex determination? Journal of Heredity, 90, 563–565. [Google Scholar]
- 129.Davison A (2006). The ovotestis: An underdeveloped organ of evolution. Bioessays, 28, 642–650. [DOI] [PubMed] [Google Scholar]
- 130.Burt A, & Trivers R (2006). Genes in conflict: The biology of selfish genetic elements. CambridgeUniversity Press. [Google Scholar]
- 131.Bettinazzi S, Rodríguez E, Milani L, Blier PU, & Breton S (2019). Metabolic remodelling associated with mtDNA: Insights into the adaptive value of doubly uniparental inheritance of mitochondria. Proceedings of the Royal Society B, 286, 20182708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bettinazzi S, Milani L, Blier PU, & Breton S (2021). Bioenergetic consequences of sex-specific mitochondrial DNA evolution. Proceedings of the Royal Society B, 288, 20211585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Robicheau BM, Chase EE, Hoeh WR, Harris JL, Stewart DT, & Breton S (2018). Evaluating the utility of the female-specific mitochondrial f-orf gene for population genetic, phylogeographic and systematic studies in freshwater mussels (Bivalvia: Unionida). PeerJ, 6, e5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stewart DT, Stephenson CM, Stanton LM, Chase EE, Robicheau BM, Hoeh WR, & Breton S (2021). A proposed method for analyzing molecular signatures to detect hermaphroditism in freshwater mussels: A case study using the eastern floater (Pyganodon cataracta). Canadian Journal of Zoology, 99, 450–458. [Google Scholar]
- 135.Hoeh WR (1991). The evolution and consequences of simultaneous hermaphroditism in the freshwater mussel genus Utterbackia (Bivalvia: Unionidae). PhD dissertation thesis. [Google Scholar]
- 136.Riccardi N, Froufe E, Bogan AE, Zieritz A, Teixeira A, Vanetti I, Varandas S, Zaccara S, Nagel K-O, & Lopes-Lima M (2019). Phylogeny of European Anodontini (Bivalvia: Unionidae) with a redescription of Anodonta exulcerata. Zoological journal of the Linnean Society, 189, 745–761. [Google Scholar]
- 137.Gao K, Cheng M, Zuo X, Lin J, Hoogewijs K, Murphy MP, Fu XD, & Zhang X (2021). Active RNA interference in mitochondria. Cell Research, 31, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fabioux C, Corporeau C, Quillien V, Favrel P, & Huvet A (2009). In vivo RNA interference in oyster-vasa silencing inhibits germ cell development. FEBS Journal, 276, 2566–2573. [DOI] [PubMed] [Google Scholar]
- 139.Choi SH, Jee BY, Lee SJ, Cho MY, Lee SJ, Kim JW, Jeong HD, & Kim KH (2013). Effects of RNA interference-mediated knock-down of hypoxia-inducible factor-α on respiratory burst activity of the Pacific oyster Crassostrea gigas hemocytes. Fish & Shellfish Immunology, 35, 476–479. [DOI] [PubMed] [Google Scholar]
- 140.Wang X, Song X, Wang T, Zhu Q, Miao G, Chen Y, Fang X, Que H, Li L, & Zhang G (2013). Evolution and functional analysis of the Pif97 gene of the Pacific oyster Crassostrea gigas. Current Zoology, 59, 109–115. [Google Scholar]
- 141.Wu C, Jiang Q, Wei L, Cai Z, Chen J, Yu W, He C, Wang J, Guo W, & Wang X (2018). A rhodopsin-like gene may be associated with the light-sensitivity of adult Pacific oyster Crassostrea gigas. Frontiers in Physiology, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qi P, Huang H, Guo B, Liao Z, Liu H, Tang Z, & He Y (2019). A novel interleukin-1 receptor-associated kinase-4 from thick shell mussel Mytilus coruscus is involved in inflammatory response. Fish & Shellfish Immunology, 84, 213–222. [DOI] [PubMed] [Google Scholar]
- 143.Peng X, Li F, Li S, & Zhu Y (2009). Expression of a mitochondrial gene orfH79 from the CMS-HongLian rice inhibits Saccharomyces cerevisiae growth and causes excessive ROS accumulation and decrease in ATP. Biotechnology Letters, 31, 409–414. [DOI] [PubMed] [Google Scholar]
- 144.Clancy DJ, Hime GR, & Shirras AD (2011). Cytoplasmic male sterility in Drosophila melanogaster associated with a mitochondrial CYTB variant. Heredity, 107, 374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Macey JR, Pabinger S, Barbieri CG, Buring ES, Gonzalez VL, Mulcahy DG, DeMeo DP, Urban L, Hime PM, Prost S, Elliott AN, & Gemmell NJ (2021). Evidence of two deeply divergent coexisting mitochondrial genomes in the Tuatara reveals an extremely complex genomic organization. Communications Biololgy, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hoeh WR, Stewart DT, Saavedra C, Sutherland BW, & Zouros E (1997). Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia: Mytilidae). Molecular Biology and Evolution, 14, 959–967. [DOI] [PubMed] [Google Scholar]
- 147.Hoeh WR, Stewart DT, & Guttman SI (2002). High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evolution; Internation Journal of Organic Evolution, 56, 2252–2261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


