Abstract
Raman spectroscopy has become an essential tool for chemists, physicists, biologists and materials scientists. In this article, we present the challenges in unravelling the molecule-specific Raman spectral signatures of different biomolecules like proteins, nucleic acids, lipids and carbohydrates based on the review of our work and the current trends in these areas. We also show how Raman spectroscopy can be used to probe the secondary and tertiary structural changes occurring during thermal denaturation of protein and lysozyme as well as more complex biological systems like bacteria. Complex biological systems like tissues, cells, blood serum etc. are also made up of such biomolecules. Using mice liver and blood serum, it is shown that different tissues yield their unique signature Raman spectra, owing to a difference in the relative composition of the biomolecules. Additionally, recent progress in Raman spectroscopy for diagnosing a multitude of diseases ranging from cancer to infection is also presented. The second part of this article focuses on applications of Raman spectroscopy to materials. As a first example, Raman spectroscopy of a melt cast explosives formulation was carried out to monitor the changes in the peaks which indicates the potential of this technique for remote process monitoring. The second example presents various modern methods of Raman spectroscopy such as spatially offset Raman spectroscopy (SORS), reflection, transmission and universal multiple angle Raman spectroscopy (UMARS) to study layered materials. Studies on chemicals/layered materials hidden in non-metallic containers using the above variants are presented. Using suitable examples, it is shown how a specific excitation or collection geometry can yield different information about the location of materials. Additionally, it is shown that UMARS imaging can also be used as an effective tool to obtain layer specific information of materials located at depths beyond a few centimeters.
This paper reviews various facets of Raman spectroscopy. This encompasses biomolecule fingerprinting and conformational analysis, discrimination of healthy vs. diseased states, depth-specific information of materials and 3D Raman imaging.
Introduction
‘VidyaViniyogaVikasah’ (a Sanskrit phrase) means progress comes from proper application of knowledge. This statement is true for the research and development in the field of Raman spectroscopy, which is considered as an evolving technology towards a direct impact to society. Although the discovery happened in 1928, the number of publications remained low until the 1960s; the web of science data indicates that there was an upsurge in the number of publications in the decade following the invention of lasers followed by a steady growth in the number of publications. The 90's saw a revival in Raman research both in theory as well as on instrumentation. Its potential applications were understood immediately as this is a straightforward technique without much need for sample preparation and almost any material, in any state under extreme conditions, can be probed. In addition, due to its non-destructive nature, it can be used as a technique for in situ measurements. Moreover, Raman microscopy has made it possible to probe very small quantity of materials with high spatial resolution. With the advent of techniques like Scanning Near-Field Microscopy (SNOM) and Tip-Enhanced Raman Spectroscopy (TERS), resolution of the order of few nanometres can also be achieved. This advantage makes it an essential and unique technique in the field of biology and materials science to explore the molecular basis of structure and function.1–4
Progress in the field of Raman spectroscopy can be largely attributed to the recent advances made in the areas of instrumentation.5 Historically, analytical applications involving Raman spectroscopy was thought to be difficult, however, developments in the sources such as diode lasers, charge-coupled devices (CCDs), small spectrometers, optics along with interfacing optical fibres with Raman have made it a novel method for analytical applications. Confluence of nanotechnology and Raman spectroscopy which led to the discovery of Surface Enhanced Raman Spectroscopy (SERS) has opened new vistas for detection of analytes at very low concentrations. It is possible to obtain Raman signals of biomolecules, chemicals, explosives, drugs etc. at parts per billion (even at single molecule level) with great sensitivity and specificity6–17
Raman spectroscopy owing to its molecular specificity can be a boon, however, in the case of biological samples, it complicates the analyses. Firstly, all biological systems are composed of biochemicals such as lipids, proteins, nucleic acids, carbohydrates etc. Therefore, vibrations from all the modes comprising the chemicals shall be manifested in the Raman spectra. To an untrained eye, spectra of a cell, tissue and a bacterium may appear very similar. Therefore, analyses of such convoluted complex spectra and assignments can be a challenging endeavour. Despite this, these complex spectra can yield a treasure of information pertaining to the structure and dynamics of the sample under evaluation. Once the assignments are made, the whole world of biochemical processes, such as metabolic pathways or dynamics can be unearthed. The second challenge towards obtaining Raman spectra of biomolecules is the occurrence of peaks from the matrix. For instance, paraffin fixed tissue may show a similar peak to a C–H stretch.18,19 Differentiating the actual spectra from the matrix therefore, becomes an equally important part before analyses. Thirdly, understanding the system under study and making an informed judgement based on the experiments and correlating it with the available data is crucial. Finally, signal processing of a large number of data set can be a contributing factor towards understanding of the system.
The objective of this article is based on the review of our work to demonstrate the challenges and potential of Raman spectroscopy for applications in biology and materials. Emphasis has largely been placed on the challenges of using Raman spectroscopic techniques for studying biomolecules and non-invasive, depth sensitive identification of materials. This laboratory has been working on various facets of Raman spectroscopy for the last three decades. Only a few results pertaining towards research on biological systems and materials have been included in this review which is organized in the following manner:
(1) Discussion on the challenges of probing biomolecules using Raman spectroscopy. Various examples on Raman spectroscopic study of biomolecules such as proteins, lipids etc. have been discussed. Furthermore, examples of Raman spectroscopy for disease diagnosis have been presented. In particular, identifying and assigning Raman spectral bands to specific molecular structures in relation to multitude of complex biomolecules present in a system.
(2) Discussion on depth sensitive Raman measurements such as spatially offset Raman spectroscopy, universal multiple angle Raman spectroscopy towards depth sensitive detection of chemicals (explosives) and transmission Raman spectroscopy. Recent developments and the relative advantages have also been presented.
Results and discussions
(I) Raman spectroscopy towards biological applications
The following sections review how fundamental studies on different types of biomolecules can be carried out and also the complications involved in Raman band assignments using Raman spectroscopy. We also discuss some of the very recent applications of Raman spectroscopy in translational research, citing examples of how this technique can help in understanding and diagnosing different diseases like cancer, malaria, dengue and others.
(a) Raman spectroscopy of biomolecules
Biomolecules are essential building blocks of life. They are central to all life processes and are involved in carrying out important metabolic reactions and maintaining the overall biochemistry of a living organism. Different kinds of biomolecules like nucleic acids, proteins, lipids and carbohydrates are involved in maintaining the functional homeostasis of a cell. Any biological material (cell, tissue etc.) is made up of different composition of various biomolecules. Therefore, it is important to understand the structure and properties of various biomolecules separately. Interestingly, each of these biomolecules has distinct chemical identities and therefore has unique molecular structures. Consequently, these biomolecules generate a signature Raman spectrum.20,21
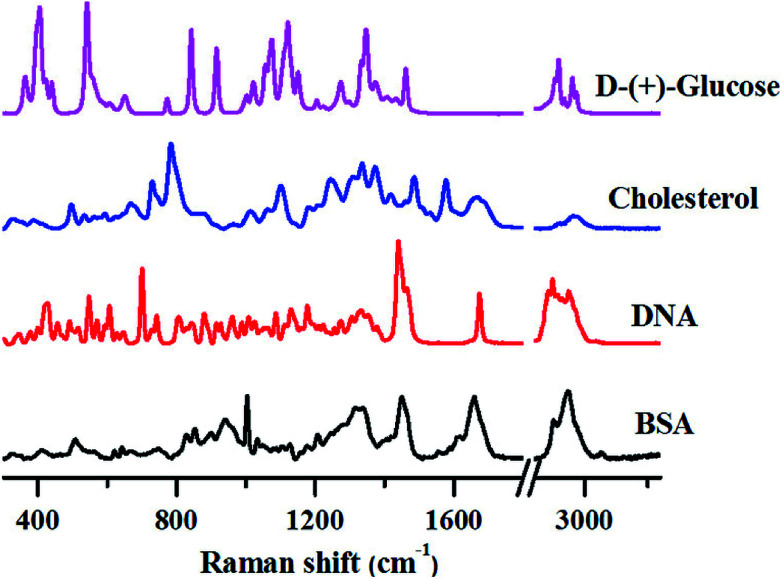
To demonstrate this aspect, we have recorded Raman spectra of pure biomolecules belonging to different classes-protein (Bovine Serum Albumin (BSA)), DNA (calf-thymus DNA), lipids (cholesterol) and carbohydrates (glucose), as shown in Fig. 1.
Fig. 1. Distinctive Raman spectra of various biomolecules. All spectra were collected from powdered samples using 785 nm excitation. Spectra were corrected for cosmic rays and baseline corrected prior to analysis.
Although the Raman spectra shown above are quite complex, several unique marker bands can be identified for each of these biomolecules as explained below.
Proteins
Proteins have numerous functions like catalyzing many reactions, guiding the flow of electrons in reactions, transmitting information between specific cells, controlling the passage of molecules around the cell membrane or providing the filamentous architecture within cells etc. They are characterized by their unique three-dimensional architecture.22 A small change in the conformation can make them biologically inactive and may even be toxic. Various life-threatening diseases have been associated with such denaturation of proteins for e.g., Alzheimer's disease, Parkinson's disease etc.23,24 Raman spectroscopy is an excellent tool to look at the protein structure. Several vibrational modes can be used for the analysis of peptide structure. Most characteristic bands are associated with the CONH group, referred to as amide A (NH stretching, about 3500 cm−1), amide B (NH stretching, about 3100 cm−1) and Amide I to VII.25 Out of all, Amide I & III are frequently used to estimate the secondary structure of proteins. Amide I mode which ranges from 1580 cm−1 to 1700 cm−1 arises from stretching vibration of C O.26 It is very sensitive to the backbone conformation and not affected by the side chains. Amide I band can be deconvoluted with various sub-bands which directly correlate with various secondary structures. Similarly, Amide III band (1200 cm−1–1300 cm−1) which arises from C–N stretching coupled with N–H bending vibrations can also give complementary information about the protein backbone conformations.27,28 Asher's group suggested an empirical relation between Amide III frequency and dihedral angle Φ, which paves the way to analyse conformation of proteins using Amide III band of Raman spectra.29,30
Apart from the secondary structure, characteristic Raman bands have also been identified for various side chains and disulphide regions as well. George J. Thomas resolved the signature for each of the eight cysteines per 666-residue subunit in the trimeric tall spike of an icosahedral bacteriophage P22.31 They demonstrated that a broad range of S–H⋯X interactions is accessible to the protein. The integration of microscope in Raman spectrometer has enabled studying of protein crystals, which led to genesis of a new technique termed as Raman crystallography.32 Raman crystallography can identify and follow the conformations of reaction intermediates in single crystals. Helfand et al. monitored the reactions between three clinically relevant inhibitors, tazobactam, sulbactam, and clavulanic acid, and SHV beta-lactamase (EC 3.5.2.6) in single crystals using a Raman microscope and observed acyl-enzyme formed between Ser70 and the lactam ring's C O group as a key intermediate in the reaction pathway.33 Various characteristic Raman bands for proteins are summarized in Table 1.
Peak assignments for proteinsa.
| Contributing group | Frequency (cm−1) | Assignment |
|---|---|---|
| Backbone | 1670–1680 | β turn, random structure, Amide I26 |
| 1660–1670 | β sheet, Amide I34 | |
| 1650–1655 | α helices, Amide I35 | |
| 1630–1635 | β sheet, Amide I36,37 | |
| 1550 | Amide II25 | |
| 1270–1300 | α helices, Amide III38 | |
| 1240–1250 | Random structure, Amide III39 | |
| 1230–1240 | β sheet, Amide III39 | |
| Disulphide | 505–515 | GGG conformation40,41 |
| 520–530 | GGT, GTG conformation40,41 | |
| 540–545 | TGT conformation40,41 | |
| Side chain | 1615, 830, 643 | Tyrosine42 |
| 850/830 | Hydrogen bonding state of tyrosine;43 | |
| 1550, 1010, 750 | Tryptophan44 | |
| 880/1361 | Ring environment of indole ring of tryptophan45 | |
| 1360/1340 | Hydrophobicity marker for tryptophan44 | |
| 1609, 1205, 1003 | Phenylalanine46 |
α: alpha helices, β: beta sheets, GGG: gauche–gauche–gauche, GGT: gauche–gauche–trans, GTG: gauche–trans–gauche, TGT: trans–gauche–trans.
Nucleic acids
Richard C. Lord and co-workers recorded the Raman spectra of various nucleic acid constituents for the first time in 1960.47 Further improvements in the instrumentation have enabled studying of various problems concerning the structure and interaction in nucleic acids, nucleic acid hydration, interaction with metal ions, DNA melting, DNA damage etc.48–51 Raman spectra of nucleic acids contain a large number of bands originating from various moieties like sugar, phosphate, bases etc. which provide very useful information about backbone conformation, base pairing etc.
Very strong Raman peaks are observed by ring breathing modes of various bases in the region of 600 cm−1 to 800 cm−1.20 Therefore, this region can be used to differentiate various bases. Out of all the bases, thymine has a very distinctive peak at around 1671 cm−1 originating from C O stretching which makes it easily distinguishable from rest of the bases.20 Different conformations of backbone (A-DNA, B-DNA and Z-DNA) can be identified using Raman spectra. J. M. Benevides and co-workers have studied different backbone conformation at various salt conformations. They could qualitatively identify the A, B and Z-DNA using phosphodiester Raman band lying in the region of 750 to 800 cm−1.52 Apart from the above, many important group vibrations have also been assigned for nucleic acids in vibrational spectra, for example, region from 1800–1500 cm−1 has been assigned for nucleobase vibrations appear which are extremely sensitive to base pairing interactions and 1500–1250 cm−1 region have been assigned for vibrational coupling between the base–sugar entities, stacking ligation etc.53 Okotrub et al. (2014) quantified DNA in the cell the nucleus using intensity of phosphate mode at 1096 cm−1 without extraction or dye labelling using Raman spectroscopy.54 Wood's group (2013) probed DNA double-strand breaks (DSBs) with TERS coupled with Atomic Force Microscopy (AFM) and suggested that the O–C bond in the DNA backbone is most sensitive to UVC radiation and its cleavage causes DSBs most of the time.55 Deckert's group recorded the TERS spectra from an artificial DNA single strand with alternating blocks of adenine and cytosine. They could resolve adenine and cytosine bases placed at a distance of 2.1 nm by unique Raman spectral features proving that TERS can offer resolutions upto single nucleobase level.56,57 Various characteristic Raman bands for nucleic acid are summarized in Table 2.
Peak assignments for nucleic acidsa.
| Contributing group | Frequency (cm−1) | Assignment |
|---|---|---|
| Base | 726 | Ring breathing of A58 |
| 668, 746, 785 | Ring breathing of T58 | |
| 668 | Ring breathing of G58 | |
| 781, 785 | Ring breathing of C58 | |
| 785 | Ring breathing of U58 | |
| 1257 | Ring mode (C, T)58 | |
| 1336 | Ring mode (A, G); A form of DNA59 | |
| 1485 | Ring stretching (G, A) A form of DNA59 | |
| 1576 | Ring stretching (A, G); A form of DNA59 | |
| Backbone | 807 | ν as(O–P–O), νs(O–P–O); A-DNA52 |
| 1099 | ν s(PO2−); A-DNA52 | |
| 1415 | df(CH2); A-DNA52 | |
| 784 | ν s(O–P–O); B-DNA52 | |
| 830 | ν as(O–P–O); B-DNA52 | |
| 1090 | ν s(PO2−); B-DNA52 | |
| 1420 | df(CH2); B-DNA52 | |
| 748 | ν s(O–P–O); Z-DNA | |
| 792 | ν as(O–P–O); Z-DNA | |
| 810 | ν as(O–P–O); Z-DNA | |
| 1095 | ν s(PO2−); Z-DNA | |
| 1425 | df(CH2); Z-DNA |
ν: stretching (s: symmetric, as: asymmetric), b: bending, t: twisting, df: deformation, A: adenine, T: thymine, C: cytosine, G: guanine, U: uracil, A-DNA: A-form of DNA, B-DNA: B form of DNA, Z-DNA: Z form of DNA.
Lipids
Lipids, also called fats, are one of the important classes of biomolecules. They have important role in energy storage, cellular signalling, digestion and absorption of food, building of cellular membranes and various tissues, maintaining body temperature etc. Raman spectroscopy provides an invaluable tool to estimate various structural properties of lipids like the degree of unsaturation, level of E, Z unsaturation etc.60 The stretching vibrations from C O in lipids are observed at around 1749 cm−1 which are quite distinct and far from that of proteins. Similarly, many other markers bands for lipids like Raman bands at 1430 cm−1 (CH2 scissoring), 1443 cm−1 (CH2 and CH3 deformation of lipids and triglycerides), 1453 cm−1 (C–H bending of lipids) have been used to quantify lipids in oral cancer tissues.61 Nieva et al. (2012) characterized the lipid phenotype of breast cancer cells using Raman spectroscopy.62 Raman spectra analysed in the range of 2820–3030 cm−1, where total fatty acid content (TFA) and total unsaturated fatty acids (TUFA) (3015 cm−1) bands were quantified using bands at 2845 cm−1 and 3015 cm−1 respectively. Popp's group (2015) analysed fungal lipids inside living hyphae using Raman spectroscopy.63 Raman spectra from single vesicles were collected and analysed for peak intensity ratio I (1270 cm−1)/I (1445 cm−1) from the signals of CH and –CH2/–CH3 groups, respectively. This ratio is linked to the iodine values of lipids. Raman spectra from lipids can be characterized in seven regions as explained in Table 3 (all assignments have been taken from Czamara et al.60).
Peak assignments for lipidsa.
| Frequency (cm−1) | Assignment | |
|---|---|---|
| Region I | 2800–3100 | ν(CH), νas(CH3), νs(CH3), νas(CH2), νs(CH2) |
| Region II | 1600–1800 | ν(C C), ν(C O) |
| Region III | 1400–1500 | b(CH2/CH3), sc(CH2/CH3), b(CH2) |
| Region IV | 1200–1300 | t(CH2), df(CH) |
| Region V | 1050–1200 | ν(C–C), ν(P–O) |
| Region VI | 800–1050 | b(CH), skeletal(C–O–O), νasN+(CH3)3 |
| Region VII | 500–700 | ν sN+(CH3)3, cholesterol ring df, df(C O–O), b(CH2) ring |
ν: stretching (s: symmetric, as: asymmetric), b: bending, sc: scissoring, t: twisting, df: deformation, skel: skeletal.
Carbohydrates
Carbohydrates are naturally occurring compounds and are essential constituents for living organisms. They have significant functions, like, main source of energy, major constituent of cell wall of plants and microorganisms, molecular agents in cellular recognition etc. Carbohydrates can be classified as monosaccharides, oligosaccharides and polysaccharides. Nonetheless, structural investigations on different carbohydrates are difficult as the basic monomeric unit in the polymeric chains is very similar. Despite this, different nature of H-bonding or the ring configuration gives rise to several marker peaks for carbohydrates. Wiercigroch et al. (2017) have compiled Raman and IR features of 14 carbohydrates in the solid state.64 Arboleda et al. recorded Raman spectra of nine anomerically stable monosaccharides in solutions with concentrations as low as 10 mM and volumes as small as 15 μL. It is observed that Raman spectra are sensitive to the configuration of the carbon centres and unique spectra are obtained of all nine monosaccharides.65 Stuart et al. (2006) demonstrated in vivo quantification of glucose in animal model for the first time using Surface-Enhanced Raman Spectroscopy (SERS).66 Raman spectra of carbohydrates can be categorised in five main regions as given in Table 4. All assignments have been taken from Wiercigroch et al.64
Peak assignments for carbohydratesa.
| Frequency (cm−1) | Assignment | |
|---|---|---|
| Region I | 3100–3600 | ν(OH) |
| Region II | 2800–3100 | ν(CH), νas(CH2), νs(CH2) |
| Region III | 1200–1500 | df(CH/CH2) |
| Region IV | 800–1200 | ν(C–O), ν(C–C) |
| Region V | 100–800 | df(CCO) |
ν: stretching (s: symmetric, as: asymmetric), b: bending, sc: scissoring, t: twisting, df: deformation.
The above work provides extensive evidence that different biomolecules yield distinct Raman spectra. While understanding the absolute structure of any biomolecule can be a confounding task, monitoring changes in the chemical structure in response to some perturbation can be efficiently analysed using Raman spectra. Consequently, Raman spectroscopy has been used to study changes in the structure of various biomolecules with external perturbation, like protein folding, protein–protein interaction, DNA-drug interaction, DNA melting etc.67
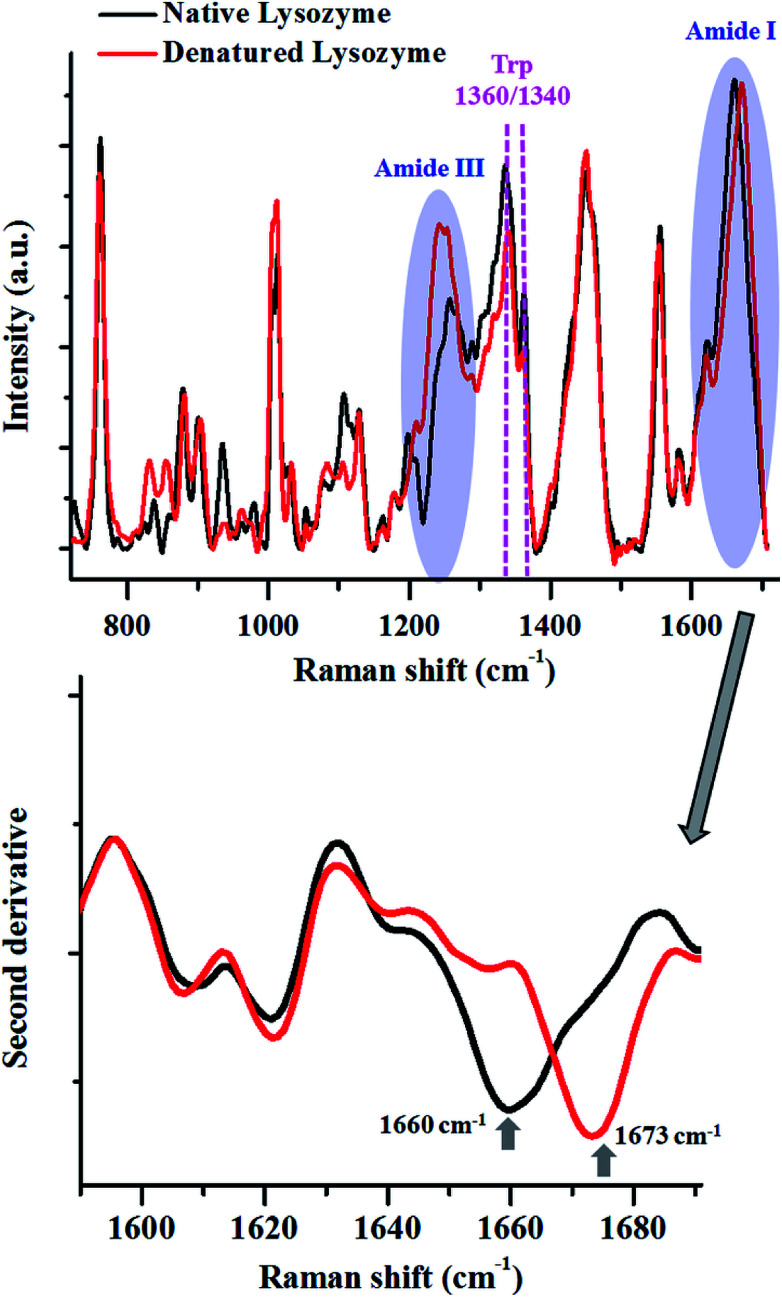
To elucidate further, we performed thermal denaturation studies on the protein, lysozyme and compared the heat-induced changes in the Raman spectra with the native form (Fig. 2).
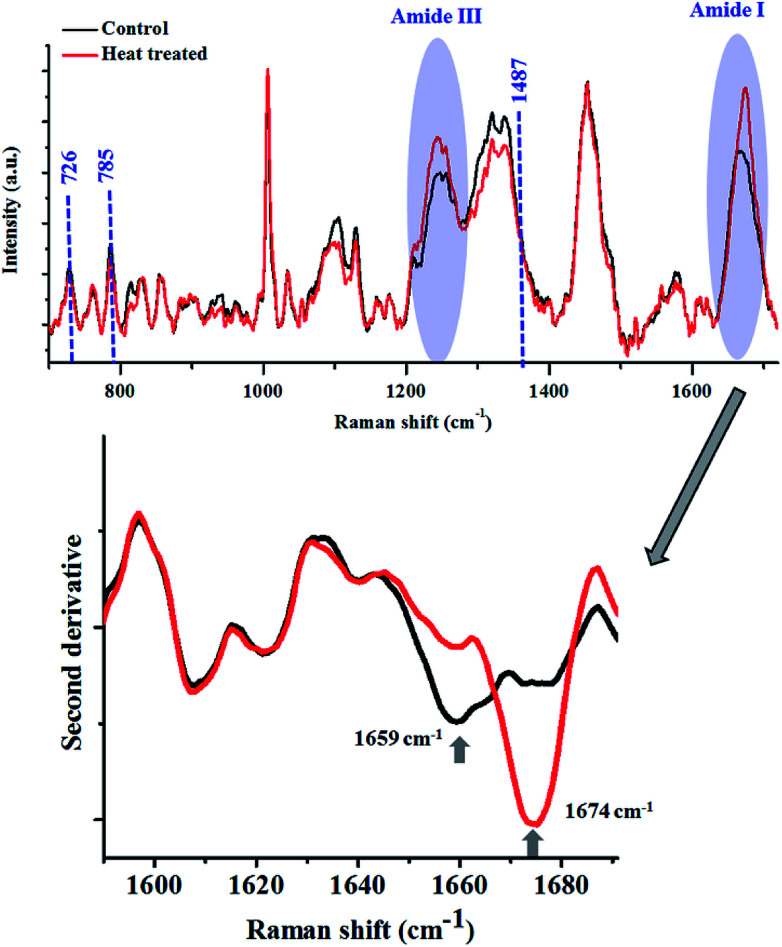
Fig. 2. Raman spectra of lysozyme in its native (black) and thermally denatured (red) state. The major changes in the spectra are highlighted. The shift in the peak maxima of Amide I is shown by the second derivative spectra. Spectra shown in top panel are baseline corrected and vector normalised prior to analysis.
The Amide I peak in a Raman spectrum is a broad band which ranges from 1580–1720 cm−1. It can be deconvoluted into various sub-bands corresponding to different secondary structures like helices, β-sheets, random structures etc. The band at around 1655 cm−1 is usually assigned for helices, 1665 cm−1 for β-sheets and 1675 cm−1 for random structures. Lysozyme in its native state, has 41% helices and 10% β-sheets,68 therefore, Amide I for the native state of this protein had a peak centre around 1660 cm−1. After heat denaturation, the content of β-sheets and random structure increased at the expense of helices. Hence, the intensity of the band corresponding to the helices (∼1655 cm−1) decreased and the intensity of the bands for β-sheets (∼1665 cm−1) and random structures (∼1675 cm−1) increased. This resulted in a change in the band shape of Amide I and a consequent shift in the peak maxima. This shift in the peak maxima was better understood by taking the second derivative of the spectra.
The changes in the Amide III band (∼1200–1300 cm−1) also corroborated this fact. There was a decrease in the intensity of the Amide III band around 1300 cm−1, assigned for alpha-helices. This was coupled with an increase in the intensity of the band between 1200–1260 cm−1, which has contributions from β-sheets and random coils.
The modulations in the Amide I and III bands demonstrated how one can probe the changes in the secondary structure of proteins.
The tertiary structure of the protein can also be probed using various Raman marker bands for side-chains. For instance, the ratio of 1360 cm−1 and 1340 cm−1 serves as a hydrophobicity marker for tryptophan. As a consequence of thermal denaturation of lysozyme, there was a decrease in this ratio, indicating a change in the environment of buried tryptophan residues from hydrophobic to hydrophilic. Similarly, there are several other bands assigned for side chains of proteins which can be analysed using Raman spectroscopy (see Table 1).
This highlights how Raman spectroscopy has proved to be a valuable technique in analysing conformational changes in the proteins as result of different perturbations.
However, it can become a very daunting task when one wants to analyse complex biological systems like cells, tissues, body fluids etc. While biomolecules are ubiquitously present in all biological systems, their relative concentrations can be vastly different. This becomes very evident when the Raman spectra of different tissues and body fluids are compared, since a differential concentration of the biomolecules results in a characteristic Raman spectrum.
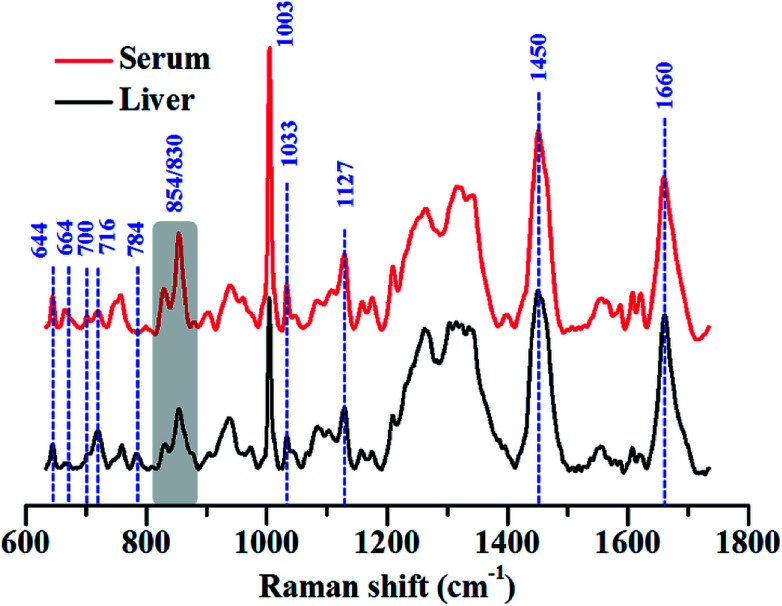
We have highlighted this fact by comparing the Raman spectra of liver tissue and blood serum isolated from healthy mice. Raman spectra were recorded from 50 μm thin sections of liver tissue and ∼2 μL of blood serum isolated from BALB/c mice using 785 nm laser excitation (Fig. 3). The region, 600–1725 cm−1 was analysed since this constitutes the fingerprint region for most biological samples. Intense generic bands with contributions from proteins and lipids, like 1003, 1451 and 1660 cm−1, were observed in the Raman spectra of liver and serum.
Fig. 3. Average Raman spectra of liver and blood serum isolated from BALB/c mice. The major peaks that were different across the samples have been highlighted using arrows and the different peak ratios that were considered are highlighted using grey boxes. Each spectrum shown is an average of 200 spectra collected from 4 mice. All spectra were baseline corrected, vector normalised and smoothed prior to analysis.
The serum is predominantly composed of different kinds of proteins. It contains up to 80 mg mL−1 of protein along with other constituents like amino acids, salts, lipids and sugars like glucose.69 The bulk of the protein content of serum comes from albumins, globulins and lipoproteins. This could also be seen in the Raman spectra of serum which was heavily dominated by Raman bands characteristic of proteins, for example, 644, 830, 854, 1003 and 1033, 1127, 1200–1300 cm−1 (Amide III) and 1660 cm−1 (Amide I) respectively. All assignments have been listed in the preceding tables.
While these bands were also present in the Raman spectra of liver, their relative intensities and the ratios varied across both the samples, indicating differences in the relative distribution of biomolecules (Fig. 4(A) and (B)).
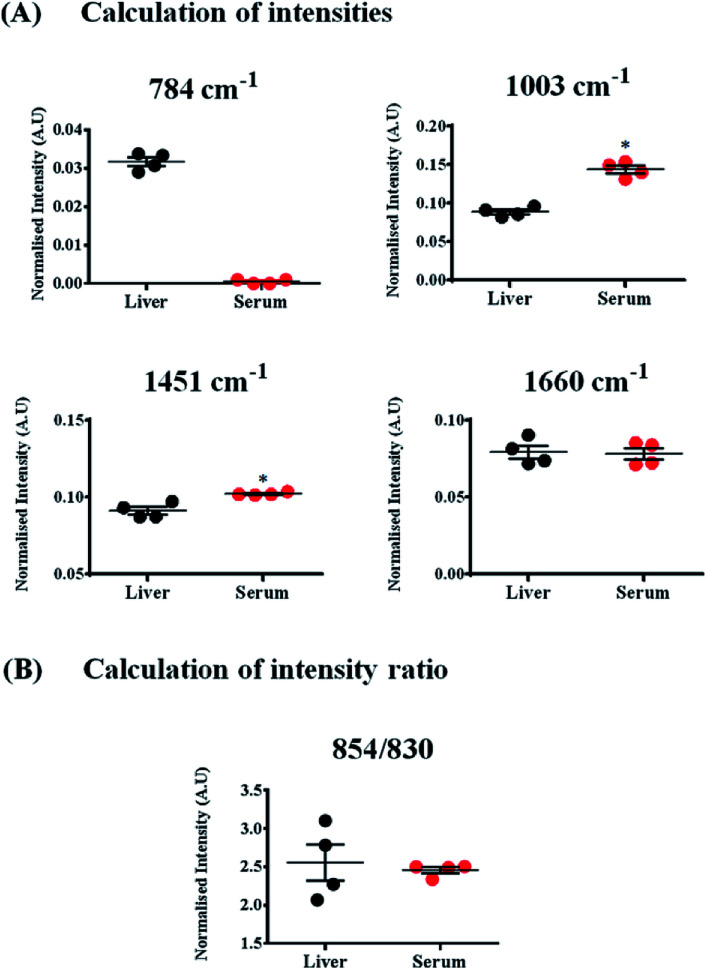
Fig. 4. Calculation of Raman band intensities and intensity ratios pertaining to different biomolecules in liver and serum. Marker bands were chosen for each of the biomolecules; (A) DNA (784 cm−1), proteins and lipids (1003, 1451 and 1660 cm−1). (B) Ratiometric analysis of two Raman bands – 830 and 854 cm−1. Each circle in the figure represents an individual mouse. Spectra obtained from a total of 4 mice were used for enumeration. Statistical analysis was performed using Mann–Whitney test where * indicates p < 0.05.
The Raman band for DNA at 784 cm−1 was observed only in the spectra of liver and was completely absent in the serum indicating negligible amounts of DNA present in the serum (Fig. 4(A)). Liver, being a tissue is made up of a large number of cells containing nucleic acids like DNA and RNA. On the contrary, blood serum is largely cell-free and thus devoid of any quantifiable amounts of DNA under normal conditions.
Other peaks corresponding to biomolecules such as cholesterol (700 cm−1) and phosphatidylcholine (716 cm−1) could also be observed in the Raman spectra of both the samples. The Raman bands for glycogen could be seen at 854 and 937 cm−1.70,71 These bands were more intense in the spectra of liver, which is quite expected, since liver is the main organ that stores glycogen.
Quantifying ratios of Raman bands can be very advantageous as they are least affected by background fluctuations and pre-processing methods.72 The ratio of two Raman bands at 830 and 854 cm−1 has been shown in Fig. 4(B). These bands have been famously called as the tyrosine Fermi doublet, originating from proteins. The ratio has extensive significance and can be a very good marker for the H-bonding status of the phenolic OH of tyrosine.69 In serum, these two bands can be assigned to tyrosine since serum is a rich source of proteins. However, in case of liver, the assignment of the band at 854 cm−1 can be ambiguous. In liver, the main contribution to this band comes from glycogen, since it is this organ where glycogen is primarily stored. This point brings to one's notice that most Raman bands can have overlapping contributions from several biomolecules and therefore, understanding the system under study is essential before band assignments are made.
As already mentioned that although the Raman spectra of such biological entities are quite complex, certain marker Raman bands can still be assigned for different classes of biomolecules in such systems.51,73,74 This can help us monitor the changes induced as a result of any stress or change in the native environment. This point can be proven through an example by monitoring the biomolecular changes induced in bacteria as a consequence of heat treatment.
Heat treatment of the bacterial cells induced several changes in the Raman spectra (Fig. 5), as highlighted in the figure above. The major changes observed were in the Amide I and III bands, similar to what was noted in the lysozyme-denaturation experiment shown in Fig. 2. This emphasises that these Raman bands can be unambiguously assigned to proteins, even in complex systems like bacteria.
Fig. 5. Raman spectra of bacterial cells before (black) and after (red) heat treatment. E. coli WT cells were grown for 8 h in shaking condition at 37 °C and 160 rpm. Bacteria were heat treated at 85 °C for 15 minutes. The shift in the peak maxima of Amide I is shown by the second derivative spectra. All spectra in the top panel were corrected for cosmic rays, background subtracted and vector normalised prior to analysis.
After heat treatment, a prominent increase in the intensity of the Raman band at 1674 cm−1 was observed, which indicated the increase in the content of β-sheets/random coils. This was also highlighted by the second derivative spectra. Complementary information was obtained from the analysis of the Amide III band. Accumulation of β-sheet/random coils is highlighted by the increase in intensity of Raman bands in the range of 1200–1260 cm−1 at the expense of helical structure (1300 cm−1). Another biomolecule which showed changes as a result of heat treatment was DNA. All the major Raman bands for DNA (726, 785, 1487 cm−1) showed a marginal decrease in the heat treated bacterial cells.
These changes showcase how complex systems can also be studied using Raman spectroscopy by analysing the unique features assigned for different biomolecules. In the field of microbiology such information is deemed useful for classification of different bacterial species or understanding biochemical changes in bacteria due to external perturbations in the environment.73,75 This paves the way for monitoring other perturbations like diseases in more complex living systems like human beings.76–78 Any disease can also be considered as a perturbation to the native composition of these biomolecules in different tissues and body fluids.
Every disease is associated with a change in biochemistry. This principle is the central doctrine for modern medicine. The biochemical changes can either be a cause for disease manifestation or may be a consequence of the disease itself. Since Raman spectroscopy probes bond vibrations, the spectra generated are very molecule-specific. Any change in the concentration or the conformation of these biomolecules as a result of a disease, however subtle, is reflected in a Raman spectrum. This principle has formed the basis for a Raman-based diagnostic tool. Though Raman scattering is a weak phenomenon, technological advancements over the past few decades have led to shorter acquisition times and improved signal-to noise ratio, making the use of Raman spectrometers in a clinical setting easier. The clinical practicality has also been enhanced by the advent of fibre-optic probes for delivery of light as well as collection of Raman scattered photons. The use of advanced chemometric methods for handling Raman data sets has also made the diagnosis and classification process user-friendly.79
Different methods are available that help the user in deriving meaningful information from a complex spectral dataset. Principal component analysis (PCA) is one such technique that reduces the spectra into a defined number of principal components (PCs) that account for spectral variance. A detailed analysis of cluster analysis and PCA in spectral data analysis is demonstrated by Bonnier and Byrne.80 PCA is an unsupervised method and hence cannot be used for classification and prediction purposes. Hence, supervised methods like Support Vector Machines, Artificial Neural Network, Linear Discriminant Analysis etc. are employed. A concise description of different chemometric methods used for Raman spectral data analysis has been given in the review by Butler et al.81
In the following sections, we discuss examples of how Raman spectroscopy has been used to understand some pathological conditions of the human body like cancer, infection and inflammation.
Cancer
Of the several diseases and pathological conditions that have been studied using Raman spectroscopy, cancer tops the list. In 2015, around 8.8 million deaths were due to cancer. As is true for any disease, early and rapid diagnosis of cancer is of utmost need. Traditionally, cancer diagnosis occurs through histopathology which requires a biopsy to be taken from the suspected area. This method of detection is highly subjective and requires the availability of a trained pathologist. Other methods of diagnosis include imaging based modalities like Positron Emission Tomography (PET) scan, Magnetic Resonance Imaging (MRI), Computed Tomography (CT) etc. However, these methods are difficult to be included in intra-operative procedures as most of these techniques require extensive labelling. Cancer, like most other diseases, is accompanied by distinct modulations in biochemistry which are expected to be reflected in the Raman spectrum. Consequently, Raman spectroscopy has been used for the detection of various types of cancers. Kopec et al. used a combination of Raman and AFM imaging to determine the biochemical composition as well as the mechanical properties of blood vessels in a tumour mass of human breast tissue. They found an increase in glycogen and lactic acid in breast cancer patients coupled with an increase in the collagen–fibroblast–glycocalyx network.82 An insight into how Raman spectroscopy can function as a tool for personalised radiation therapy was given by Van Nest et al. They used human breast adenocarcinoma xenografts after radiation therapy and found spectral changes linked to different biomolecules like proteins, lipids and nucleic acids on a temporal scale, highlighting the potential of Raman spectroscopy for assessing radiation-responses of tumours.83 The use of cancerous cell lines as a platform to differentiate invasive vs. non-invasive breast cancer on the basis of Raman spectral markers was done by Chaturvedi et al. The Raman bands at 1447 cm−1 (CH2 bending from lipids), 1003 cm−1 (phenylalanine) and 876 cm−1 (tryptophan) were found to be higher in invasive cell lines vis-à-vis the non-invasive ones. Their work is in agreement with other reports that have correlated increased lipid levels to more aggressive and metastatic cancers and, therefore, a poor prognosis.84 The work of Kim S. et al., shows how cancer detection can be improved by coupling Raman spectroscopy to other methods like pH sensing. It is known that pH levels in cancerous tissues are much lower than that of normal tissues. They exploited the chemical information revealed by Raman spectroscopy as well as the metabolic information obtained by the pH sensor for improving the specificity of cancer diagnosis. Their study was done on human breast tissue samples and both the Raman and pH sensors were fibre-optic probes.85 A recent study undertaken by Winnard et al., has beautifully captured the potential of Raman spectroscopy to characterise organ-specific metastatic lesions at a molecular level, to give insights into metastatic progression. A combinatorial approach of Raman spectroscopy and metabolomics was employed in their study.86 In another study, stromal adaptations occurring in pre-metastatic lungs that were primed by breast cancer were analysed using Raman spectroscopy. This work was done in mouse models where the mice were xenografted with human breast cancer cells of varying metastatic potentials. Changes in the extra-cellular matrix of pre-metastatic lungs, such as increase in collagen (859 cm−1) and proteoglycan (1061 cm−1) were identified and this was a direct correlation of the metastatic potential of the breast cancer cells used.74
Desroches et al., utilized the higher wavenumber Raman signatures for designing, developing and testing of an in situ intraoperative system for brain cancer detection. They engineered this Raman-based system into a commercially available biopsy system for analysis before the tissue was extracted. Using this approach, they could achieve sensitivity and specificity of brain cancer detection in a swine model system, upto 80 and 90% respectively.87 The reprogramming of lipid biosynthesis and proteins in high-grade medulloblastoma was demonstrated by Abramczyk and Imiela very recently. Medulloblastoma is a term given to high grade cancers of the central nervous system. Such samples showed a higher content of β-sheets and lower levels of α-helix when compared to normal tissues. The ratio of the Raman intensities at 2930 and 2845 cm−1 was also indicative of cancer progression. For all the cancerous samples, this ratio had a value of 1.99 in comparison to the value of 1.45 for healthy tissue.88
Murali Krishna's group demonstrated the use of Raman spectroscopy for identification of sites in the oral cavity that have a higher propensity to become cancerous even before the manifestation of clinical symptoms of oral cancer recurrence. This study was performed on 99 patients of oral cancer and spectra were recorded from the cancerous sites as well as contralateral normal mucosa.89 Nasopharyngeal cancer has also been diagnosed using Raman spectroscopy by Ming et al. in 2017. They demonstrated the use of a fibre-optic Raman probe for in vivo surveillance of patients suffering from nasopharyngeal cancer. This form of cancer is often very difficult to diagnose owing to the deep anatomical location. This study had grouped patients under three categories – normal, cancerous and post-radiation. All experiments were performed using a 1.8 mm probe, which is currently the smallest probe used for Raman-based diagnosis. A specificity of ∼96% was observed in distinguishing normal, cancerous and post-radiation patients.90
Almond et al. utilised Raman spectroscopy and translated it into an endoscopic tool for the early detection of dysplasia in patients suffering from Barret's oesophagus. A high-grade dysplasia (HGD) often results into oesophageal malignancy. Tissues from a cohort of 62 patients were taken for the analysis and sensitivity and specificity of 86 and 88% respectively were obtained for detecting HGD and adenocarcinoma.91
Another useful application of Raman spectroscopy in cancer research was highlighted by the work of Moradi et al., where they tested the potential of this method to discriminate between human ovarian cancer cells that were sensitive or resistant to cisplatin. An increase in the relative concentration of proteins and glutathione was observed in the resistant cells compared to the sensitive ones.92 The ability of Raman spectroscopy-based tool to serve an intra-operative device for detecting distant invasive grade 2–4 gliomas was shown by Jermyn et al. in 2016. They found that Raman spectroscopy could detect as few as 6 cancer cells per 0.0625 mm2, which was much better than traditionally used methods like MRI. All the Raman measurements were performed during brain tumour resections.93 A detailed review on intra-operative Raman spectroscopy has been given by Brusatori et al.94
The use of body fluids like blood plasma/serum, urine, tears etc. can pave way for a diagnostic method that is minimally invasive. Body fluids can be obtained easily and therefore repeated samplings can be done. Unlike tissues and cells, biofluids can be used for routine monitoring as well as under intra-operative conditions. They contain several biomolecules and therefore can be very good indicators of the health status of a person or the underlying disease. The major limitation in using bio fluids for Raman spectroscopy is the relatively low concentrations of biomolecules when compared to tissues and cells. This problem can be easily circumvented by the use of Drop Coated Deposition Raman Spectroscopy (DCDRS) where the biomolecules are concentrated.95–97 Harvey et al. in 2008 demonstrated the use of urine for the detection of prostate cancer where increased proteins and nucleic acids were associated with the cancerous cells.98 The work of Corsetti et al. demonstrated the efficacy of Raman spectroscopy in distinguishing hormone (androgen)-resistant prostate cancer cells. Bands specific to amino acids like arginine were found to be diminished in the cancerous cells whereas those with contributions from phenylalanine and tyrosine were found to be increased. The androgen-resistant cancer cells also had increased DNA and Amide III bands when compared to the sensitive cells.99 O'Malley et al., in their study, have highlighted Raman-based lipid quantification to be of diagnostic value for prostate cancer. Increased lipid amounts are directly correlated to the tumour stage.100
The use for serum-based cancer diagnosis has also been explored for different types of cancers. Ovarian cancer was studied using a combined approach of ATR-FTIR and Raman spectroscopy on human ovarian cancer patients by Owens et al. in 2014.101 The basis for differentiation using Raman spectroscopy was the spectral shifts in the Raman bands at 711 cm−1 (DNA) and 913 cm−1 (sphingolipids).
Infectious, inflammatory and other diseases
While a plethora of information can be found for Raman-based cancer diagnosis, infectious and inflammatory diseases also require significant attention since they constitute a large percentage of the global burden on the health care system.
Infection with the dengue virus claims several lives especially in the developing countries. Current methods for diagnosis include measuring serum IgG and IgM levels and the Ns1 antigen. Mahmood et al., demonstrated the use of Raman spectroscopy for profiling dengue-associated biochemical changes occurring in the blood plasma of 17 infected patients. Changes were seen mostly in the Raman bands associated with lipids (608 cm−1, 875 cm−1, 1297 cm−1, 1719 cm−1 and 1736 cm−1) which increased with infection. The peak intensity of the Amide III band at 1239 cm−1 was also higher in the infected samples. An increase in the Raman band at 1409 cm−1 was also observed which can be assigned to IgG. The method showed specificity and sensitivity of 95 and 98% respectively.102 Dengue-associated spectral changes were also studied by Bilal et al. and they found the peaks for lactate at 750, 830 and 1450 cm−1 to be higher in intensity in all 50 dengue patients when compared to 20 healthy controls. They also spiked the serum of healthy controls with lactate in a controlled manner and found that indeed the abovementioned peaks were due to lactate.103 Ahmed's group from Pakistan has tried to develop a Raman-based diagnostic method for dengue using several multivariate approaches like Support Vector Machines (SVM), random forest method, etc.104
The same group has also conducted Raman studies on human malarial samples and found 10 distinct spectral signatures that could differentiate malarial samples from healthy controls as well as dengue samples and patients suffering from non-malarial fever. A multivariate model based on partial least square (PLS) regression was used and the accuracy of detection was 86%.105
Malaria-induced changes in the spleen of mice were evaluated using Raman spectroscopy by Frame et al. and they found an increase in the heme-associated Raman bands. This directly correlated to the hemozoin detection with peaks at ∼1370, 1529, 1588, and 1628 cm−1 which matched closely with the reference spectra.106 A comprehensive review on the potential of vibrational spectroscopy to diagnose and study malaria and the malarial parasite was written very recently by Perez-Guaita et al.107
One of the major causes of death in developing countries is tuberculosis (TB). In India alone, around 400 000 people die annually because of TB infections. While tuberculosis is mainly pulmonary in origin, extra-pulmonary TB also exists and is more difficult to diagnose. Tuberculous meningitis is one of the forms of extra-pulmonary TB and requires culturing of Mycobacterium for a precise diagnosis. Satyavathi et al., used Raman spectroscopy on cerebrospinal fluid of patients suffering from tuberculous meningitis and found that the silicate Raman signature from M. tb positive cases was a very useful indicator of the underlying infection.108
Another leading cause of mortality in the Intensive Care Units of hospitals is sepsis. Sepsis can be triggered by infection of any kind and results in an uncontrolled immune response known as the Systemic Inflammatory Response Syndrome (SIRS). SIRS, however, can be a manifestation of other conditions like burns, trauma etc. Presently, no reliable marker exists that can efficiently distinguish SIRS from sepsis. In order to achieve this, Raman spectroscopy combined with multivariate approaches was used by Popp's group and they found that SIRS could be distinguished from sepsis with a predictive accuracy of 80%.109
Different inflammatory conditions like ulcerative colitis have also been studied using Raman spectroscopy. Ding et al. investigated the colon mucosal composition in vivo using an endoscope-coupled Raman probe in patients suffering from ulcerative colitis and age and BMI-matched healthy controls. A decrease in the phosphatidylcholine (720 cm−1) and total lipids (1303 cm−1) was observed in the inflamed colon tissue. The degree of lipid unsaturation, determined by calculating the ratio between Raman bands at 1657 and 1447 cm−1, was found to be high in the patient group. These differences were also seen when active colitis patients were compared with the quiescent ones.110 Their results were corroborated by the work of Addis et al., which also showed reduction in phospholipids to be of diagnostic value for ulcerative colitis.111
Inflammatory bowel disease (IBD) was also diagnosed by Mahadevan-Jansen's group where a colonoscopy-coupled fibre optic Raman probe was used. IBD includes ulcerative colitis (UC) and Crohn's disease (CD) and the motive in this study was to determine the spectral signatures of both UC and CD.112
Inflammation in the oral cavity as a result of periodontal disease was also assessed using Raman spectroscopy by Camerlingo et al. They used gingival crevicular fluid from patients with periodontal disease and found an increase in the carotene content to be diagnostic of the condition (1537 cm−1). They also studied the chemical and structural changes occurring in the periodontal ligament after orthodontic force was applied and found changes in the secondary structures of proteins (Amide I band) to change with application of the force.113
In addition to the above mentioned inflammatory diseases, Raman spectroscopy has been extensively used to study and diagnose neurodegenerative diseases like Alzheimer's disease (AD). Till date, there is no cure for such diseases primarily due to their delayed diagnosis which occurs when the extent of brain damage is already quite pronounced. In fact, a definitive diagnosis can only be established post-mortem. It is now known that changes in the brain of such individuals begin much earlier than the appearance of the first symptoms. This can pave way for an early diagnosis which may make therapeutic interventions more successful. Protein misfolding and aggregation are central to most neurodegenerative diseases. Raman spectroscopy is an excellent tool to study protein conformation and therefore, has significant potential in diagnosing such diseases. This has been efficiently shown in the work of Paraskevaidi et al., where blood plasma was used to successfully discriminate Alzheimer's disease from healthy controls as well as from patients suffering from Dementia with Lewy Bodies (DLB). It is important to highlight that patients suffering from DLB are often misdiagnosed as AD and hence the choice of the patient cohorts is well justified. The same study also showed that patients suffering from early and late stages of AD could be differentiated from healthy controls with a sensitivity of 84%.114
In another study, Ryzhikova et al., demonstrated the use of Raman microspectroscopy on blood serum for diagnosis of AD using a cohort of 20 AD patients, 18 from other neurodegenerative diseases and 10 healthy controls with specificity and sensitivity of ∼95%.115
(II) Raman spectroscopic studies on materials
Preceding discussions revolved around the use of Raman spectroscopy in the field of biology. It should be emphasized that biomolecules have a low Raman cross-section and as a consequence show weak Raman signals. Probing such weak scatterers has been made possible due to the introduction of Raman microscope in the early 70s.7–9 The other advantages which Raman spectroscopy offers over other analytical tools are its non-invasive and non-destructive nature. This has resulted in its prolific use in the field of materials such as studies on carbon allotropes, explosives, semiconductors, pharmaceuticals, art, painting and archaeology. Raman spectroscopy has been an indispensable tool for characterization of different allotropes of carbon. Ranging from 3D diamond or graphite to 2D materials such as graphene or 1D materials like carbon nanotubes (CNT), Raman spectroscopy provides information pertaining to lattice vibrations, defects, orientations etc.116,117 For instance, single walled and multi walled CNTs can be distinguished using this technique. The radial breathing modes which are unique to single walled CNT are absent in multi walled CNT.118 Additionally, various sp2 carbon can also be differentiated.119 One of the more recent interests on carbon allotropes has been in graphene research. Here, Raman spectroscopy has proved to be an integral tool to determine number of layers of graphene along with orientational information, edges, strains, defects etc.120,121 Enhancement of Raman signals of analytes using carbon materials has also been explored.122–125 In the field of explosives detection, surface enhanced Raman spectroscopy has been employed for trace level detection.126,127 Furthermore, studies on feasibility of Raman spectroscopy for stand-off detection have also been explored.128,129 The subsequent sections provide a different aspect of this technique to demonstrate the usefulness of Raman spectroscopy towards studies on explosives. The first example shows how Raman spectroscopy can be used to monitor melt cast explosives formulations. In the second case, detection of concealed materials in non-metallic containers using depth sensitive Raman spectroscopy such as spatially offset Raman spectroscopy (SORS), transmission Raman and universal multiple angle Raman spectroscopy (UMARS) has been demonstrated.
(a) Raman spectroscopy as an effective tool in process monitoring of explosives formulation
The first use of explosive dates back as early as 220 B.C. when some Chinese alchemists were trying to separate gold from silver and met with accident. The material was ‘black powder’, which is a mixture of charcoal, sulphur and potassium nitrate.130,131 An explosive in the literature is defined as ‘a substance which when suitably triggered, releases a large amount of heat and pressure by way of a very rapid self-sustaining exothermic decomposition reaction.130 The temperature of such a reaction is approximately equal to the temperature at the Sun's surface ∼5000 °C. Secondary explosives are a class of explosives which are comparatively insensitive to heat, friction and impact. They require more energy to initiate the detonation process. Typical secondary explosives are 2,4,6-trinitro toluene (TNT), pentaerythritol tetranitrate (PETN), cyclotrimethylenetrinitramine (RDX), tetrahexaminetetranitramine (HMX) and more recently 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane (HNIW or CL-20), and mixtures of these explosives in different proportions. Explosives in a nutshell, have been described as a ‘devil ’ during war and considered as an ‘evil’ during processing, handling, transportation and storage, have proved to be an ‘angel’ due to their tremendous impact on economy and industries and their innumerable applications in all walks of life.130 Most of the explosives formulations are composed of binders, oxidizers and explosives. For instance, stabilizers are added to ester based rocket-propellants for the formation of a stable product.132 Raman spectroscopy technique can be used as an effective tool for non-destructive and rapid evaluation and monitoring of the individual components of the formulation. Additionally, the depletion of the components over time can be monitored during ageing studies of stabilizers. Similarly, different polymorphic forms of explosives can also be determined using Raman spectroscopy.133,134 Recent trends in the development of high explosives molecules have shifted towards engineering co-crystals of explosives molecules to distinct properties such as greater detonation velocities without compromising with impact or friction sensitivities.135–137 Raman spectroscopic technique can be used to monitor the structures of such co-crystals.137
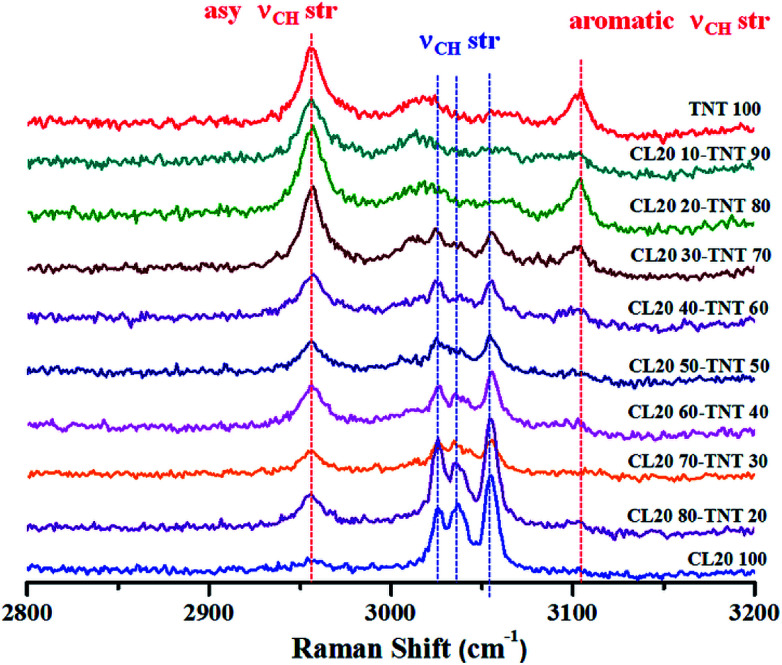
High explosives are used in military applications to achieve the terminal effect i.e. destruction of tactical and strategic targets. Typical examples of industrial explosives are blasting gelatine and ammonium nitrate slurries. However, high explosives are useful in civilian applications as advanced demolition devices (ADDs) for demolition of structures with precision and least collateral damage. Most of the ADDs comprise of composition-B (RDX/TNT 60-40) or similar compositions. These are usually manufactured using melt cast technology.138 TNT forms the work horse for such formulation owing to its low melting point (∼80 °C) which allows the ease of mixing and homogenization of other explosive ingredients. Therefore, the entire process requires constant monitoring to ensure thorough mixing and formation of a uniform melt. Explosives are aliphatic, aromatic or nitramine based organic molecules which are rich in Raman signatures. Since melt cast compositions are physical mixtures, the structures of the molecules are unaltered (in most of the cases). Therefore, Raman spectroscopy can be a potential tool for in situ monitoring of such processes. Fig. 6 depicts an example of an explosives formulation comprising of CL-20 (nitramine class) and TNT (nitroaromatic class) at different weight ratios. Raman spectra were acquired for different compositions at higher wavenumber regions as these molecules have unique spectral features which can be attributed to C–H stretches.139–141 Fibre optics based Raman spectroscopy can be employed for online monitoring of such processes which least human intervention. Although a less explored area, Raman spectroscopy can be a useful tool in the area of explosives research.
Fig. 6. Raman spectroscopic signatures at higher wavenumber range for CL-20 and TNT melt cast composition at different weight percentages. As the % of one of the component increases, intensity of the corresponding Raman peaks also increases. Data was obtained using 514.5 nm Ar+ ion laser with 1.25 mW power and 10 s acquisition time.
(b) Deep Raman spectroscopy for detection of sub-surfaces
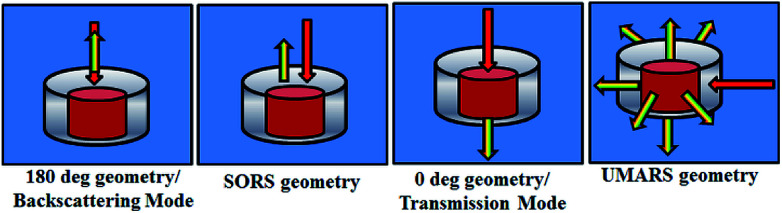
Historically, Raman spectroscopic studies were carried out using backscattering (180°), 90°, 135° and transmission (0°) geometries respectively. The choice of illumination to collection geometry is decided by the application and the type of the sample to be studied. For better spatial resolution, confocal backscattering geometry is a preferred choice which forms the basis of Raman microspectroscopy. On the contrary, for transparent and liquid samples, 90° and 135° are the preferred geometries. These geometries work fine for pure materials. Furthermore, Raman microspectroscopy provides spatial information of 2D heterogeneous materials up to a diffraction limited spot size (λ/2). However, when the material of interest is layered or when it is concealed by other layers, backscattering geometry may not yield complete structural information due to the low scattering volume of backscattering geometry (depending upon the depth of field). Such situations demand for exploration of new methods for studying layered materials. In 2005, a new method was introduced for obtaining sub-surface information of layered materials and was termed as spatially offset Raman spectroscopy (SORS).142 The principal of this method is to introduce an offset with respect to the illumination followed by collection of the Raman signals. The authors noted that at no offset, there is a strong propensity of Raman signal of the outer material. As the collection offset is increased, Raman signal from the inner layer becomes prominent. The intensity ratio of the inner vs. the outer layer improves significantly with offset. This technique offers discrimination of Raman signals of layered materials.143–145 Since this technique depends upon the point of illumination, the depth of penetration may be restricted upto a few centimetres. However, one can also use transmission geometry to obtain bulk signature of materials.146 This method has been successfully employed in pharmaceutical sector for quality control.147–151 The next challenge is in the case of materials which are hidden beyond a few tens of millimetres. For instance, in the case of strongly scattering media, such as biological tissues, retrieval of Raman signal from a tissue buried under overlying layers non-invasively can be a daunting experimental task. If our target material is hidden deeper inside a scattering sample, the Raman signal will be dominated by that of the surface layer. Therefore, new Raman-based methods need to be developed for depth-specific measurements of such materials. Our laboratory has focused its attention towards exploration of novel methods to address the challenge. Understanding of photon migration studies shows that that the excitation and Raman photons inside turbid media undergo multiple scattering events.152–156 This leads to randomization of photons. As a consequence, Raman photons from any angle or geometry can be collected to identify substances lying deep within such media. This technique has been termed as Universal Multiple Angle Raman Spectroscopy (UMARS). Therefore, the collection geometry is independent of the illumination geometry. Additionally, Raman signals are collected from all sides and angles around the material under study. Fig. 7 shows the schematic of different geometries used in Raman spectroscopy. The subsequent sections will describe examples of SORS, backscattering, transmission and UMARS methods.
Fig. 7. Schematic of different experimental geometries employed for obtaining depth specific Raman spectroscopic signatures. Left panel to right depict backscattering, spatially offset Raman spectroscopy (SORS), transmission and Universal Multiple Angle Raman Spectroscopy (UMARS) geometries respectively.
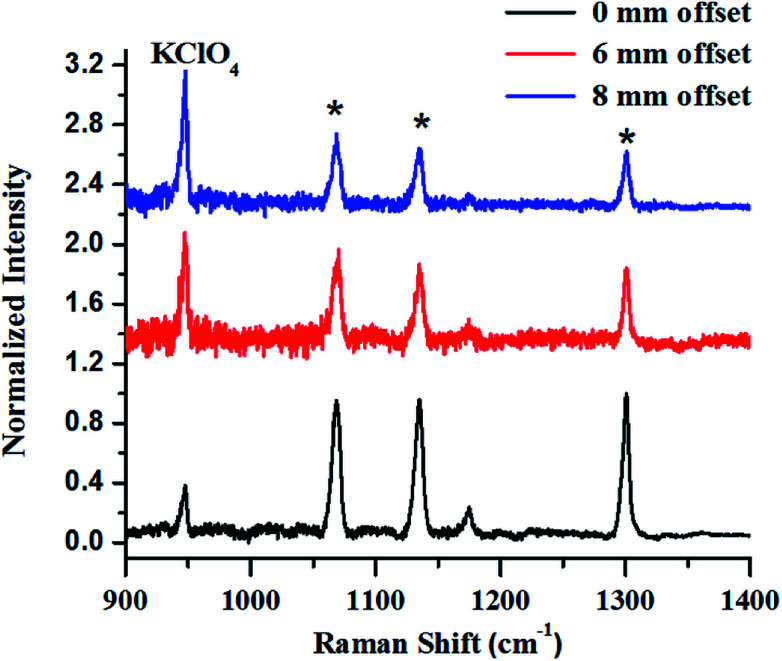
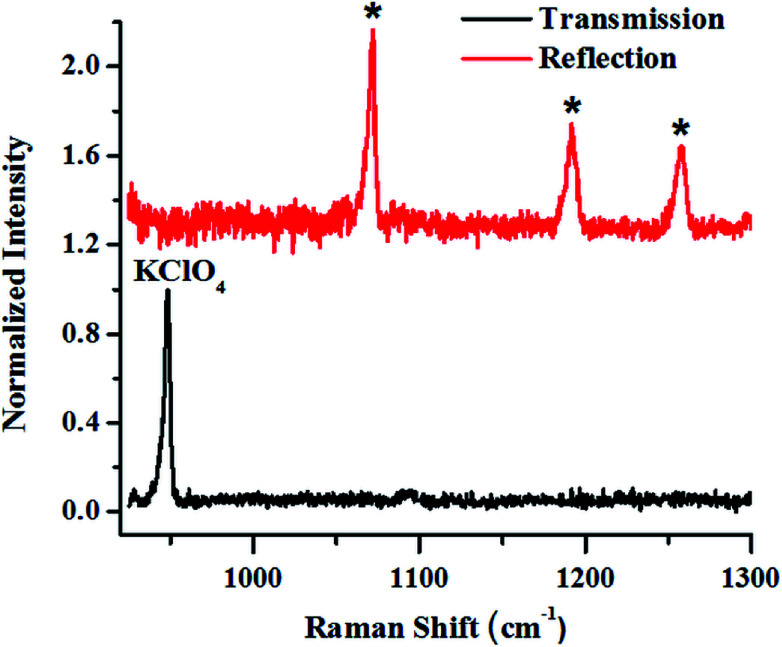
The first example describes SORS for detection of KClO4, an oxidizer used towards making explosive formulations (Fig. 8). In this study, KClO4 was taken inside a 3 mm thick high-density polyethylene (HDPE) container. Experiments revealed that a certain offset from the excitation point was necessary to discriminate the Raman signals of the content from that of the container. It may be noted that at no offset, the signal from the container was pronounced (peaks with asterisk). As the offset was increased, the Raman signal intensity of KClO4 improved with respect to the container, while that of the container did not show any appreciable increase.
Fig. 8. (a) Raw SORS signals from KClO4 powder filled in 25 mm square HDPE container with a wall thickness of 3 mm with 0 mm (a), 6 mm (b) and 8 mm (c) offsets respectively. Laser beam was focused on to the container. As the offset is increased to 8 mm, the signal of the sub-surface material improves.
In the next set of studies, experiments were performed using transmission and backscattering geometry on the same material by using a focused beam of incident light (Fig. 9). We noted, however, that the transmission geometry yielded better content signal quality for the same container wall thickness (3 mm), as shown in the figure, and the signal from the container was remarkably suppressed. In this case the signal was collected at the back of the 25 mm thick square container, opposite to the point of excitation. This is possible in transmission geometry because of the long migration paths of Raman photons in non-absorbing powder media.147
Fig. 9. Raman signals of layers of KClO4 explosive in a 3 mm thick HDPE container obtained in reflection (backscattering) and right transmission mode.
From the above-mentioned geometry-specific Raman experiments it is clear that backscattering mode yielded appreciable Raman signals of the top layer. This was overcome by using spatial offsets. However, transmission geometry provided Raman signals with even less interference from the top layer signals. It should be noted that transmission geometry is limited by the path length of the system due to the fact that after a certain depth, the photons would attenuate and low Raman signals can be acquired beyond that. In the cases where the material thickness exceeds a few centimetres, retrieving Raman signatures from material located at a depth will be a challenge by utilizing the above-mentioned geometries. It is worth mentioning that direct evidence of the extra pathlength for Raman photons from deeper layer can be obtained via time resolved Raman spectroscopic techniques.142 For instance, Petterson et al., have reported detection of explosives concealed behind opaque, diffusely scattering materials using picoseconds time resolved Raman spectroscopy.157
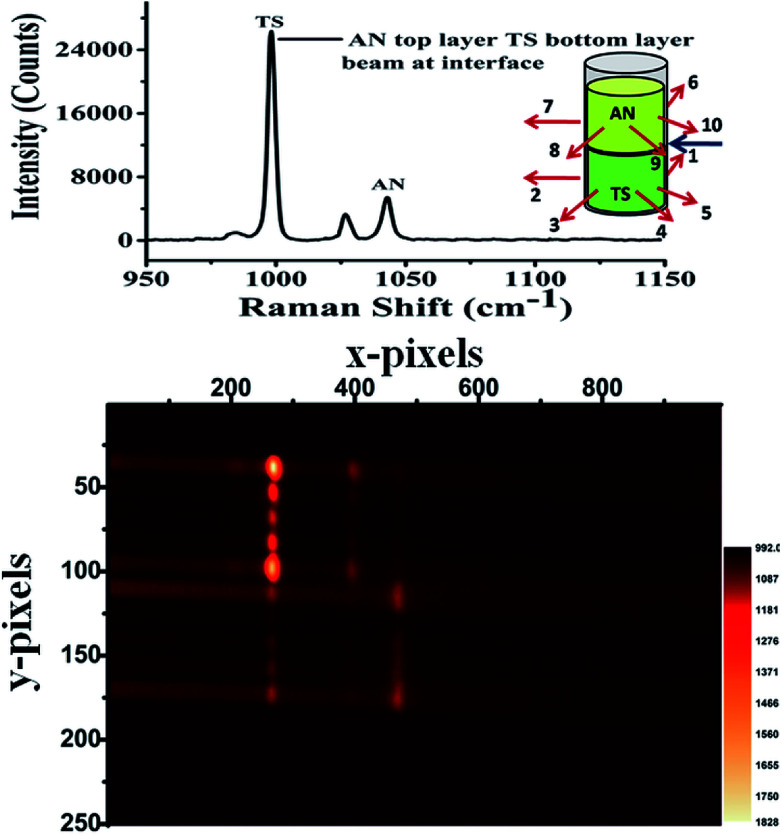
Our laboratory has demonstrated UMARS for the first time where we have obtained Raman signals in strongly scattering samples beyond a few centimetres.125,158–162 As a demonstration, imaging experiments using the UMARS technique were conducted for bilayered solid compounds; in this case, ammonium nitrate (AN) was taken as a top layer and t-stilbene (TS) as a bottom layer. The excitation fibre was placed at the interface of the two layers, collection fibres 1–5 were placed around the bottom layer, TS, while fibres 6–10 were placed around the top layer, i.e., AN. The experimental arrangement is shown schematically in the inset of Fig. 10. Before recording the image, Raman spectra were recorded for the separate components. Peaks centred at 992 cm−1 and 1044 cm−1 correspond to TS & AN, respectively. TS, being a very strong Raman scatterer, showed a higher peak intensity as compared to AN. When the CCD was used in imaging mode, different collection fibres produced separate Raman spectra at different heights of the CCD. Y axis of the image shows the projection of 10 separate collection fibres on different pixel rows. Spectrum at the top of the figure corresponds to the sum of all y pixels, i.e., a full vertical binning of the signal. Since fibres 1–5 were placed around TS, those spots appeared on the top section of the CCD, while the bottom section contained mostly spots of AN. It was observed that in addition to the AN signal, fibres 6–10 also collected some signals from TS. This indicates that TS photons migrated to the top layer and contributed to the Raman signal. These results show that the spatial locations of the species present in the layers could be identified based on the position (image) of the fibres. In this setup, spectra of the individual fibres are collected simultaneously and can be evaluated separately. This is an important attribute of UMARS, as spatial information will be very important in the case of medical diagnostics. As expected, Rayleigh scattering was strongest in fibres 1 and 5 and fibres 6 and 9; closest to the point of excitation. The Raman signal of the bilayer clearly shows the TS and AN peaks.
Fig. 10. UMARS imaging of bilayer solid powders ammonium nitrate (AN) & trans-stilbene (TS) using 10 optical fibres (830 nm diode laser, laser power: 300 mW, acquisition time: 100 s). The imaging experiments have been carried out by keeping the excitation beam at the interface of the two layers while fibre numbers 1 through 5 were placed around the bottom layer while fibres 6 through were placed around the top layer. TS being a strong Raman scatterer showed intense spots (fibres 1–5, top) on the CCD as compared to AN. Identification of the spatial position is possible with UMARS imaging. Y axis: projection of 10 separate collection fibres on different pixel rows. Spectrum at the top of the figure corresponds to the sum of all y pixels (full vertical binning).
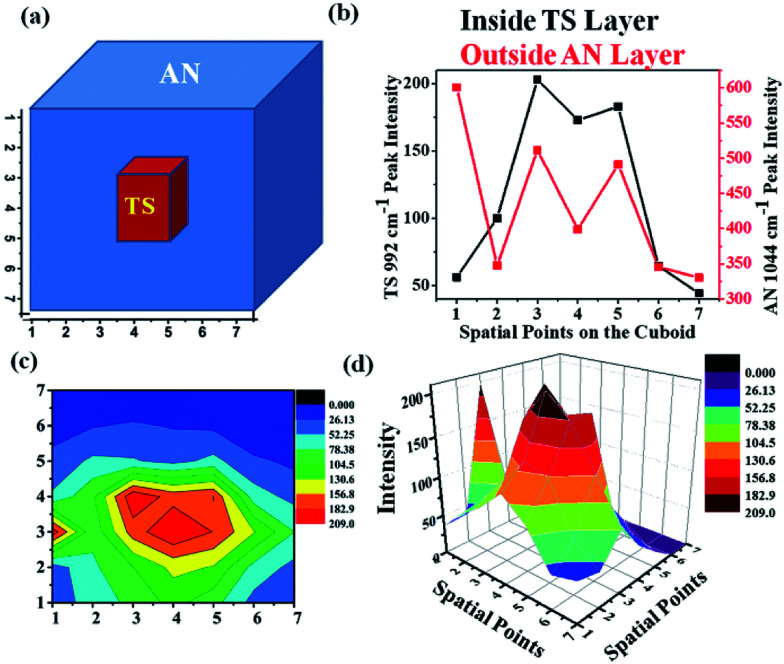
In order to ascertain the location of the inner layer, a special experiment was designed. An HDPE cuboid container was taken and it was filled with AN as an outer layer and TS as an inner layer as shown in Fig. 11(a). The cuboid was divided into a 7 × 7 grid. The excitation fibre was placed at each point in the grid, while the collection fibres were placed around the periphery maintaining the same plane as the input fibre. UMARS signals were collected from all the 49 points and the obtained spectra were analysed to retrieve depth specific molecular information. At first, the Raman spectral intensity variation of TS & AN were recorded by keeping the y-axis constant (y = 4) i.e., at the centre of the cuboid and by varying the x at the y level, i.e., from x = 1 to x = 7 (Fig. 11b). It was observed that the Raman intensity of AN was strongest at x = 1 while in the case of TS, the signal was weak at x = 1, followed by an increase in Raman intensity to reach a plateau and finally decreased. It was also noted from the spatial points 3 to 5, the intensity of TS remained more or less same. This implied that TS was present in higher amounts close to those spatial points as compared to other points. To get a clear picture, a contour and a 3D surface map was plotted for the TS Raman intensity (992 cm−1 peak) for the 7 × 7 matrix (49 spectral points) (Fig. 11c and d). This confirmed the presence of TS at the center of the cuboid. This example provides sufficient evidence of the capability of UMARS as a non-invasive, depth sensitive technique. Further experiments on phantoms and biological samples are being conducted in our laboratory to understand the potential of UMARS towards 3D imaging. We anticipate that UMARS would pave way towards Raman tomography for disease diagnosis in the future.
Fig. 11. Depth sensitive detection of bilayer solid powders ammonium nitrate (outer layer) and t-stilbene (inner layer) using UMARS where, (a) depicts the bilayer cuboid sample schematically, (b) is the Raman intensity variation of AN & TS as the signals were collected by keeping the y-axis constant (y = 4) i.e., at the center of the cuboid and by varying the x at that y, i.e., from x = 1 to x = 7; (c) shows the contour of the bilayer mapped with respect to TS Raman intensity where red colours corresponds to maximum TS intensity, blue corresponds to a minimum value; and (d) shows the 3D surface plot of the variation of TS Raman intensity.
Experimental
Biomolecules
Bovine Serum Albumin (BSA), calf thymus DNA, D-(+)-glucose and cholesterol were purchased from Sigma Aldrich and used without further purification. Spectra were recorded from all the samples in powdered form using 785 nm laser and 50× L objective (N. A = 0.5) using Renishaw InVia Raman Microscope.
Thermal denaturation of lysozyme
Hen egg white lysozyme (lysozyme) was purchased as lyophilized powder from Sigma and was dissolved in sterile MilliQ water to obtain a working concentration of 30 mg mL−1. For thermal denaturation studies, the protein solution was heated to a temperature of 85 °C for 8 hours. Spectra were recorded from the native and denatured protein solution using 785 nm laser and 50× L objective using Renishaw InVia Raman Microscope at room temperature.
Liver and serum studies
6–8 weeks old healthy BALB/c mice were sacrificed and liver and blood were harvested. For collection of serum, blood was allowed to clot at 37 °C for 4 hours, followed by centrifugation at 4000 rpm for 5 min. For recording Raman spectra of liver, ∼50 μm thin cryo-sections were used. All Raman experiments were conducted using Renishaw in Via system using a 785 laser and 50× objective (N. A = 0.75).
Bacterial heat treatment
E. coli MG1655 K-12 strain was cultured in a shaking condition at 37 °C and 160 rpm for 8 hours. 1 mL of the culture was subjected to heat shock at a temperature of 85 °C for 15 minutes. The control and the heated samples were then washed with sterile water three times to remove any residual media components; ∼3 μL of the bacterial sample was drop cast onto a magnesium fluoride coverslip before collecting Raman spectra. All Raman experiments were performed at room temperature using Renishaw in Via system using a 785 laser and 100× objective (N. A = 0.85).
Data analysis
All spectra were processed using Wire 4.2, Origin 8.5 and MATLAB softwares. Statistical analysis (Mann–Whitney test) was performed using GraphPad Prism 6.
Raman spectroscopic studies on materials
CL-20 and TNT were provided by DRDO laboratory for conducting the Raman experiments. The CL-20-TNT melt cast formulations were prepared in milligram quantities in the laboratory. Required quantity of TNT was first melted followed by addition of appropriate amount of CL-20 and homogenized using a wooden pestle. All chemicals used in SORS, reflection, transmission and UMARS experiments except ammonium nitrate (Nice Chemicals Pvt. Ltd.), were procured from Sigma Aldrich. The solid powder samples used in the experiments were either filled in commercial high-density polyethylene (HDPE) containers, quartz cuvettes or glass vials of assorted sizes.
The Raman experiments on explosives formulation was conducted using Renishaw in Via system using a 514.5 nm Ar+ laser. The power at the sample was 2.5 mW and the spectra were acquired for 10 seconds.
The SORS, reflection and transmission Raman experiments were carried out using the 830 nm laser diode. The spectrometer was a high resolution imaging spectrometer (Horiba Jobin Yvon, iHR320) fitted with a LN2-cooled CCD camera. Grating with a groove density of 1200 lines per mm was used for the experiments. In the case of UMARS experiments, the laser was delivered to the sample through an optical fibre of core diameter 400 μm and a numerical aperture of 0.22 and the Raman signals were collected using 10 fibres of the same core diameter and NA placed on the sample container sides at various angles. The ten collection fibres terminated in a vertical linear array, placed in front of a telescopic arrangement fixed to the entrance slit of the spectrometer. The power of the laser was maintained between 250–450 mW. The acquisition time for the experiments varied from 10 s to a few minutes.
The acquired data were processed using Wire 4.2 and Labspec softwares and were plotted using Origin 8.5 and Origin 9.1.
Conclusion
Raman spectroscopy has proved to be a versatile tool in studying different diseases occurring because of the altered biochemistry in the body. Various advancements have been made in the field of Raman spectroscopy with respect to instrumentation and data handling methods. In this review, we have attempted to provide a glimpse of some of the research activities which are being pursued in our laboratory in the area of biophotonics, explosives detection and development of novel Raman techniques. We started by showing Raman spectra of pure molecules which comprise of the building blocks of cells and tissues. Examples of Raman spectroscopy of tissues are presented to understand the unique spectral features along with the complexity of the Raman signals. The second part of the review deals with Raman spectroscopic applications towards materials by taking explosives as a representative example. The last part of the review discusses the variants of Raman spectroscopy like SORS, transmission and UMARS for depth sensitive detection of layered materials. Finally, it is shown that UMARS imaging is possible and that chemical as well as spatial information of the materials lying underneath other materials can be clearly obtained using UMARS when the fibres are separately imaged on the CCD. The day is not far when Raman spectroscopy-based diagnostic methods will be routinely prescribed in clinics and public health care systems. One of the most anticipated uses of Raman spectroscopy is in clinical diagnostics. Most of the research on Raman is focused on biological research and imaging. It remains to be seen whether Raman spectroscopy would take off as portable equipment for point of care application. In his concluding remarks of his review paper, Prof Kiefer wrote, “Sometimes I am asking myself what cannot be done with Raman spectroscopy – a wonderful analytical tool for science and technology which was discovered by Sir Chandrashekhar Venkata Raman and Krishnan already eight decades ago at the Indian Association for the Cultivation of Science in Calcutta, India. I am looking forward for many more years of significant advancements in this research field.163”
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Department of Science & Technology, Defence Research & Development Organization, Council of Scientific & Industrial Research, Indian Institute of Science, Bangalore for their financial support. S. U. acknowledges the J. C. Bose fellowship from DST.
Biographies
Biography
Nikki Kuhar.

Nikki Kuhar is a Senior Research Fellow, pursuing her PhD from Department of Inorganic and Physical Chemistry, IISc Bangalore. She received her master's degree from IIT Bombay and bachelor's degree from St Stephen's College, Delhi University. Her research interest includes Raman and Raman optical activity studies of protein, FTIR-imaging of tissues and Molecular Dynamic Simulations.
Biography
Sanchita Sil.

Dr Sanchita Sil is a Scientist at DEBEL-DRDO. She received her PhD in Chemistry in the area of Raman spectroscopy from the Indian Institute of Science, India. Prior to joining Defence Bioengineering & Electromedical Laboratory (DEBEL), she was a Scientist at High Energy Materials Research Laboratory (HEMRL), India. She has more than 13 years of research experience. Her research interests include Raman spectroscopy, SERS, development of novel Raman based instrumentation towards detection of materials, air quality management, catalysis, carbon based materials, adsorption and nanoparticles.
Biography
Taru Verma.

Taru Verma received her Master of Science in Biotechnology from Nagpur University, India. Currently, she is a PhD student in the Centre for BioSystems Science and Engineering (BSSE) at the Indian Institute of Science, Bangalore, India. Her research work mainly involves studying bacteria and infectious diseases using vibrational microspectroscopy.
Biography
Siva Umapathy.

Siva Umapathy is a professor in the Department of Inorganic & Physical Chemistry and Instrumentation & Applied Physics, Indian Institute of Science, Bangalore, India and an Honorary Professor at the Department of Chemistry, University of Nottingham, United Kingdom. He secured his PhD from the University of Otago, New Zealand. His major Research interests are Ultrafast Raman Spectroscopy, Resonance Raman Spectroscopy, understanding Molecular Dynamics, and Raman & Infrared microscopy application to biology, medicine and materials research & the development of novel instrumentation for detection and diagnostics.
References
- McCreery R. L., Raman Spectroscopy for Chemical Analysis, Wiley, 2005 [Google Scholar]
- Das R. S. Agrawal Y. K. Raman spectroscopy: Recent advancements, techniques and applications. Vib. Spectrosc. 2011;57:163–176. doi: 10.1016/j.vibspec.2011.08.003. [DOI] [Google Scholar]
- Long D. A., Raman Spectroscopy, McGraw-Hill, New York, 1977 [Google Scholar]
- Lyon L. A. Keating C. D. Fox A. P. Baker B. E. He L. Nicewarner S. R. et al., Raman spectroscopy. Anal. Chem. 1998;70:341R–61R. doi: 10.1021/a1980021p. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P. Edwards H. G. M. Moens L. A decade of Raman spectroscopy in art and archaeology. Chem. Rev. 2007;107:675–686. doi: 10.1021/cr068036i. [DOI] [PubMed] [Google Scholar]
- Butler H. J. Fogarty S. W. Kerns J. G. Martin-Hirsch P. L. Fullwood N. J. Martin F. L. Gold nanoparticles as a substrate in bio-analytical near-infrared surface-enhanced Raman spectroscopy. Analyst. 2015;140:3090–3097. doi: 10.1039/C4AN01899K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbillat J. Dhamelincourt P. Delhaye M. Dasilva E. Raman Confocal Microprobing, Imaging and Fiberoptic Remote-Sensing – a Further Step in Molecular Analysis. J. Raman Spectrosc. 1994;25:3–11. doi: 10.1002/jrs.1250250103. [DOI] [Google Scholar]
- Delhaye M. Dhamelincourt P. Raman Microprobe and Microscope with Laser Excitation. J. Raman Spectrosc. 1975;3:33–43. doi: 10.1002/jrs.1250030105. [DOI] [Google Scholar]
- Mccreery R. L. Fleischmann M. Hendra P. Fiber Optic Probe for Remote Raman Spectrometry. Anal. Chem. 1983;55:146–148. doi: 10.1021/ac00252a039. [DOI] [Google Scholar]
- Albrecht M. G. Creighton J. A. Anomalously Intense Raman-Spectra of Pyridine at a Silver Electrode. J. Am. Chem. Soc. 1977;99:5215–5217. doi: 10.1021/ja00457a071. [DOI] [Google Scholar]
- Campion A. Kambhampati P. Surface-enhanced Raman scattering. Chem. Soc. Rev. 1998;27:241–250. doi: 10.1039/A827241Z. [DOI] [Google Scholar]
- Fleischmann M. Hendra P. J. Mcquillan A. J. Raman-Spectra of Pyridine Adsorbed at a Silver Electrode. Chem. Phys. Lett. 1974;26:163–166. doi: 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- Jeanmaire D. L. Vanduyne R. P. Surface Raman Spectroelectrochemistry 1 Heterocyclic, Aromatic, and Aliphatic-Amines Adsorbed on Anodized Silver Electrode. J. Electroanal. Chem. 1977;84:1–20. doi: 10.1016/S0022-0728(77)80224-6. [DOI] [Google Scholar]
- Kneipp K. Wang Y. Kneipp H. Perelman L. T. Itzkan I. Dasari R. et al., Single molecule detection using surface-enhanced Raman scattering (SERS) Phys. Rev. Lett. 1997;78:1667–1670. doi: 10.1103/PhysRevLett.78.1667. [DOI] [Google Scholar]
- Le Ru E. C. Etchegoin P. G. Single-Molecule Surface-Enhanced Raman Spectroscopy. Annu. Rev. Phys. Chem. 2012;63:65–87. doi: 10.1146/annurev-physchem-032511-143757. [DOI] [PubMed] [Google Scholar]
- Nie S. M. Emery S. R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- Xie W. Schlucker S. Medical applications of surface-enhanced Raman scattering. Phys. Chem. Chem. Phys. 2013;15:5329–5344. doi: 10.1039/C3CP43858A. [DOI] [PubMed] [Google Scholar]
- Ibrahim O. Maguire A. Meade A. D. Flint S. Toner M. Byrne H. J. et al., Improved protocols for pre-processing Raman spectra of formalin fixed paraffin preserved tissue sections. Anal. Methods. 2017;9:4709–4717. doi: 10.1039/C6AY03308C. [DOI] [Google Scholar]
- Downey F., Cade N., Cook R., Springall R., Gillet C. and Richards D., et al., Use of Raman Spectroscopy in Characterizing Formalin-Fixed, Paraffin-Embedded Breast Tumor Samples (abstract), AIP Conf. Proc., 2009, vol. 1119, p. 211 [Google Scholar]
- De Gelder J. De Gussem K. Vandenabeele P. Moens L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007;38:1133–1147. doi: 10.1002/jrs.1734. [DOI] [Google Scholar]
- Carey P. R., Biochemical applications of Raman and resonance Raman spectroscopies, Academic Press, New York, 1982 [Google Scholar]
- Schirmer R. H. and Sa G. E., Principles of Protein Structure, Springer-Verlag, Berlin-Heidelberg-New York, 1979 [Google Scholar]
- Chaudhuri T. K. Paul S. Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J. 2006;273:1331–1349. doi: 10.1111/j.1742-4658.2006.05181.x. [DOI] [PubMed] [Google Scholar]
- Hartl F. U. Protein Misfolding Diseases. Annu. Rev. Biochem. 2017;86:21–26. doi: 10.1146/annurev-biochem-061516-044518. [DOI] [PubMed] [Google Scholar]
- Rygula A. Majzner K. Marzec K. M. Kaczor A. Pilarczyk M. Baranska M. Raman spectroscopy of proteins: a review. J. Raman Spectrosc. 2013;44:1061–1076. doi: 10.1002/jrs.4335. [DOI] [Google Scholar]
- Maiti N. C. Apetri M. M. Zagorski M. G. Carey P. R. Anderson V. E. Raman spectroscopic characterization of secondary structure in natively unfolded proteins: alpha-synuclein. J. Am. Chem. Soc. 2004;126:2399–2408. doi: 10.1021/ja0356176. [DOI] [PubMed] [Google Scholar]
- Lippert J. L. Tyminski D. Desmeules P. J. Determination of the secondary structure of proteins by laser Raman spectroscopy. J. Am. Chem. Soc. 1976;98:7075–7080. doi: 10.1021/ja00438a057. [DOI] [PubMed] [Google Scholar]
- Williams R. W. Protein Secondary Structure-Analysis Using Raman Amide-I and Amide-III Spectra. Methods Enzymol. 1986;130:311–331. doi: 10.1016/0076-6879(86)30016-8. [DOI] [PubMed] [Google Scholar]
- Ianoul A. Boyden M. N. Asher S. A. Dependence of the peptide Amide III vibration on the phi dihedral angle. J. Am. Chem. Soc. 2001;123:7433–7434. doi: 10.1021/ja0023128. [DOI] [PubMed] [Google Scholar]
- Mikhonin A. V. Bykov S. V. Myshakina N. S. Asher S. A. Peptide secondary structure folding reaction coordinate: correlation between uv raman Amide III frequency, Psi Ramachandran angle, and hydrogen bonding. J. Phys. Chem. B. 2006;110:1928–1943. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- Thomas G. J. New structural insights from Raman Spectroscopy of proteins and their assemblies. Biopolymers. 2002;67:214–225. doi: 10.1002/bip.10105. [DOI] [PubMed] [Google Scholar]
- Carey P. R. Raman crystallography and other biochemical applications of Raman microscopy. Annu. Rev. Phys. Chem. 2006;57:527–554. doi: 10.1146/annurev.physchem.57.032905.104521. [DOI] [PubMed] [Google Scholar]
- Helfand M. S. Totir M. A. Carey M. P. Hujer A. M. Bonomo R. A. Carey P. R. Following the reactions of mechanism-based inhibitors with beta-lactamase by Raman crystallography. Biochemistry. 2003;42:13386–13392. doi: 10.1021/bi035716w. [DOI] [PubMed] [Google Scholar]
- Hamodrakas S. J. Asher S. A. Mazur G. D. Regier J. C. Kafatos F. C. Laser Raman studies of protein conformation in the silkmoth chorion. Biochim. Biophys. Acta. 1982;703:216–222. doi: 10.1016/0167-4838(82)90051-6. [DOI] [PubMed] [Google Scholar]
- Mangialardo S. Piccirilli F. Perucchi A. Dore P. Postorino P. Raman analysis of insulin denaturation induced by high-pressure and thermal treatments. J. Raman Spectrosc. 2012;43:692–700. doi: 10.1002/jrs.3097. [DOI] [Google Scholar]
- Williams R. W. Dunker A. K. Determination of the secondary structure of proteins from the Amide I band of the laser Raman spectrum. J. Mol. Biol. 1981;152:783–813. doi: 10.1016/0022-2836(81)90127-3. [DOI] [PubMed] [Google Scholar]
- Tuma R. Raman spectroscopy of proteins: from peptides to large assemblies. J. Raman Spectrosc. 2005;36:307–319. doi: 10.1002/jrs.1323. [DOI] [Google Scholar]
- Bandekar J. Amide Modes and Protein Conformation. Biochim. Biophys. Acta. 1992;1120:123–143. doi: 10.1016/0167-4838(92)90261-B. [DOI] [PubMed] [Google Scholar]
- Cai S. Singh B. R. Identification of beta-turn and random coil Amide III infrared bands for secondary structure estimation of proteins. Biophys. Chem. 1999;80:7–20. doi: 10.1016/S0301-4622(99)00060-5. [DOI] [PubMed] [Google Scholar]
- Nakamura K. Era S. Ozaki Y. Sogami M. Hayashi T. Murakami M. Conformational changes in seventeen cystine disulfide bridges of bovine serum albumin proved by Raman spectroscopy. FEBS Lett. 1997;417:375–378. doi: 10.1016/S0014-5793(97)01326-4. [DOI] [PubMed] [Google Scholar]
- Dong A. Huang P. Caughey W. S. Protein secondary structures in water from second-derivative Amide I infrared spectra. Biochemistry. 1990;29:3303–3308. doi: 10.1021/bi00465a022. [DOI] [PubMed] [Google Scholar]
- Hernandez B. Coic Y. M. Pfluger F. Kruglik S. G. Ghomi M. All characteristic Raman markers of tyrosine and tyrosinate originate from phenol ring fundamental vibrations. J. Raman Spectrosc. 2016;47:210–220. doi: 10.1002/jrs.4776. [DOI] [Google Scholar]
- Siamwiza M. N. Lord R. C. Chen M. C. Takamatsu T. Harada I. Matsuura H. et al., Interpretation of the doublet at 850 and 830 cm−1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry. 1975;14:4870–4876. doi: 10.1021/bi00693a014. [DOI] [PubMed] [Google Scholar]
- Takeuchi H. Raman structural markers of tryptophan and histidine side chains in proteins. Biopolymers. 2003;72:305–317. doi: 10.1002/bip.10440. [DOI] [PubMed] [Google Scholar]
- Miura T. Takeuchi H. Harada I. Characterization of Individual Tryptophan Side-Chains in Proteins Using Raman-Spectroscopy and Hydrogen-Deuterium Exchange Kinetics. Biochemistry. 1988;27:88–94. doi: 10.1021/bi00401a015. [DOI] [PubMed] [Google Scholar]
- Hernandez B. Pfluger F. Kruglik S. G. Ghomi M. Characteristic Raman lines of phenylalanine analyzed by a multiconformational approach. J. Raman Spectrosc. 2013;44:827–833. doi: 10.1002/jrs.4290. [DOI] [Google Scholar]
- Lord R. C. Thomas Jr G. J. Raman studies of nucleic acids II Aqueous purine and pyrimidine mixtures. Biochim. Biophys. Acta. 1967;142:1–11. doi: 10.1016/0005-2787(67)90510-2. [DOI] [PubMed] [Google Scholar]
- Piredda P. Berning M. Boukamp P. Volkmer A. Subcellular Raman Microspectroscopy Imaging of Nucleic Acids and Tryptophan for Distinction of Normal Human Skin Cells and Tumorigenic Keratinocytes. Anal. Chem. 2015;87:6778–6785. doi: 10.1021/acs.analchem.5b01009. [DOI] [PubMed] [Google Scholar]
- Talari A. C. S. Evans C. A. Holen I. Coleman R. E. Rehman I. U. Raman spectroscopic analysis differentiates between breast cancer cell lines. J. Raman Spectrosc. 2015;46:421–427. doi: 10.1002/jrs.4676. [DOI] [Google Scholar]
- Denbigh J. L. Perez-Guaita D. Vernooij R. R. Tobin M. J. Bambery K. R. Xu Y. et al., Probing the action of a novel anti-leukaemic drug therapy at the single cell level using modern vibrational spectroscopy techniques. Sci. Rep. 2017;7:2649. doi: 10.1038/s41598-017-02069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. Gautam R. Chatterjee S. Ariese F. Sikdar S. K. Umapathy S. Ascorbate protects neurons against oxidative stress: a Raman microspectroscopic study. ACS Chem. Neurosci. 2015;6:1794–1801. doi: 10.1021/acschemneuro.5b00106. [DOI] [PubMed] [Google Scholar]
- Benevides J. M. Thomas G. J. Characterization of DNA Structures by Raman-Spectroscopy – High-Salt and Low-Salt Forms of Double Helical Poly(Dg-Dc) in H2O and D2O Solutions and Application to B-DNA, Z-DNA and A-DNA. Nucleic Acids Res. 1983;11:5747–5761. doi: 10.1093/nar/11.16.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid J. Bloomfield V. A. Benevides J. Thomas G. J. Raman-Spectroscopy of DNA-Metal Complexes. 1. Interactions and Conformational Effects of the Divalent-Cations – Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993;65:1916–1928. doi: 10.1016/S0006-3495(93)81263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okotrub K. A. Surovtsev N. V. Semeshin V. F. Omelyanchuk L. V. Raman Spectroscopy for DNA Quantification in Cell Nucleus. Cytometry, Part A. 2015;87A:68–73. doi: 10.1002/cyto.a.22585. [DOI] [PubMed] [Google Scholar]
- Lipiec E. Sekine R. Bielecki J. Kwiatek W. M. Wood B. R. Molecular Characterization of DNA Double Strand Breaks with Tip-Enhanced Raman Scattering. Angew. Chem., Int. Ed. 2014;53:169–172. doi: 10.1002/anie.201307271. [DOI] [PubMed] [Google Scholar]
- Bailo E. Deckert V. Tip-enhanced Raman spectroscopy of single RNA strands: Towards a novel direct-sequencing method. Angew. Chem., Int. Ed. 2008;47:1658–1661. doi: 10.1002/anie.200704054. [DOI] [PubMed] [Google Scholar]
- Treffer R. Bohme R. Deckert-Gaudig T. Lau K. Tiede S. Lin X. et al., Advances in TERS (tip-enhanced Raman scattering) for biochemical applications. Biochem. Soc. Trans. 2012;40:609–614. doi: 10.1042/BST20120033. [DOI] [PubMed] [Google Scholar]
- Hlaing M. M. Wood B. McNaughton D. Ying D. Y. Augustin M. A. Raman spectroscopic analysis of Lactobacillus rhamnosus GG in response to dehydration reveals DNA conformation changes. J. Biophotonics. 2017;10:589–597. doi: 10.1002/jbio.201600046. [DOI] [PubMed] [Google Scholar]
- Benevides J. M. Overman S. A. Thomas G. J. Raman, polarized Raman and ultraviolet resonance Raman spectroscopy of nucleic acids and their complexes. J. Raman Spectrosc. 2005;36:279–299. doi: 10.1002/jrs.1324. [DOI] [Google Scholar]
- Czamara K. Majzner K. Pacia M. Z. Kochan K. Kaczor A. Baranska M. Raman spectroscopy of lipids: a review. J. Raman Spectrosc. 2015;46:4–20. doi: 10.1002/jrs.4607. [DOI] [Google Scholar]
- Singh S. P. Krishna C. M. Raman spectroscopic studies of oral cancers: correlation of spectral and biochemical markers. Anal. Methods. 2014;6:8613–8620. doi: 10.1039/C4AY01615G. [DOI] [Google Scholar]
- Nieva C. Marro M. Santana-Codina N. Rao S. Petrov D. Sierra A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: towards a stratification of malignancy. PLoS One. 2012;7:e46456. doi: 10.1371/journal.pone.0046456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchberg U. Wagner L. Rohrer C. Voigt K. Rosch P. Jahreis G. et al., Quantitative assessment of the degree of lipid unsaturation in intact Mortierella by Raman microspectroscopy. Anal. Bioanal. Chem. 2015;407:3303–3311. doi: 10.1007/s00216-015-8544-2. [DOI] [PubMed] [Google Scholar]
- Wiercigroch E. Szafraniec E. Czamara K. Pacia M. Z. Majzner K. Kochan K. et al., Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta, Part A. 2017;185:317–335. doi: 10.1016/j.saa.2017.05.045. [DOI] [PubMed] [Google Scholar]
- Arboleda P. H. Loppnow G. R. Raman spectroscopy as a discovery tool in carbohydrate chemistry. Anal. Chem. 2000;72:2093–2098. doi: 10.1021/ac991389f. [DOI] [PubMed] [Google Scholar]
- Stuart D. A. Yuen J. M. Shah N. Lyandres O. Yonzon C. R. Glucksberg M. R. et al., In vivo glucose measurement by surface-enhanced Raman spectroscopy. Anal. Chem. 2006;78:7211–7215. doi: 10.1021/ac061238u. [DOI] [PubMed] [Google Scholar]
- Jakubek R. S. Handen J. White S. E. Asher S. A. Lednev I. K. TrAC, Trends Anal. Chem. 2018;103:223–229. doi: 10.1016/j.trac.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk P. J. Blake C. C. F. Rice D. W. Wilson K. S. The structures of the monoclinic and orthorhombic forms of hen egg-white lysozyme at 6 A resolution. Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1982;38:778–783. doi: 10.1107/S0567740882004075. [DOI] [Google Scholar]
- Jaros J. A. Guest P. C. Bahn S. Martins-de-Souza D. Affinity depletion of plasma and serum for mass spectrometry-based proteome analysis. Methods Mol. Biol. 2013;1002:1–11. doi: 10.1007/978-1-62703-360-2_1. [DOI] [PubMed] [Google Scholar]
- Gautam R. Vanga S. Madan A. Gayathri N. Nongthomba U. Umapathy S. Raman spectroscopic studies on screening of myopathies. Anal. Chem. 2015;87:2187–2194. doi: 10.1021/ac503647x. [DOI] [PubMed] [Google Scholar]
- Movasaghi Z. Rehman S. Rehman I. U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007;42:493–541. doi: 10.1080/05704920701551530. [DOI] [Google Scholar]
- Kumar S. Verma T. Mukherjee R. Ariese F. Somasundaram K. Umapathy S. Raman and infra-red microspectroscopy: towards quantitative evaluation for clinical research by ratiometric analysis. Chem. Soc. Rev. 2016;45:1879–1900. doi: 10.1039/C5CS00540J. [DOI] [PubMed] [Google Scholar]
- Kumar S. Matange N. Umapathy S. Visweswariah S. S. Linking carbon metabolism to carotenoid production in mycobacteria using Raman spectroscopy. FEMS Microbiol. Lett. 2015;362:1–6. doi: 10.1093/femsle/fnu048. [DOI] [PubMed] [Google Scholar]
- Paidi S. K. Rizwan A. Zheng C. Cheng M. Glunde K. Barman I. Label-Free Raman Spectroscopy Detects Stromal Adaptations in Premetastatic Lungs Primed by Breast Cancer. Cancer Res. 2017;77:247–256. doi: 10.1158/0008-5472.CAN-16-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil S. Mukherjee R. Kumar N. S. Aravind S. Kingston. J. K. Singh U. Detection and classification of Bacteria using Raman Spectroscopy Combined with Multivariate Analysis. Def. Life Sci. J. 2017;2:435–441. doi: 10.14429/dlsj.2.12275. [DOI] [Google Scholar]
- Sil S., Gautam R. and Umapathy S., Applications of Raman and Infrared Microscopy to Materials and Biology, Molecular and Laser Spectroscopy, Gupta V. P., Elsevier, 2018, ch. 6, pp. 117–146 [Google Scholar]
- Singh B. Gautam R. Kumar S. Kumar B. N. V. Nongthomba U. Nandi D. et al., Application of vibrational microspectroscopy to biology and medicine. Curr. Sci. 2012;102:232–244. [Google Scholar]
- Gautam R. Samuel A. Sil S. Chaturvedi D. Dutta A. Ariese F. et al., Raman and mid-infrared spectroscopic imaging: applications and advancements. Curr. Sci. 2015;108:341–356. [Google Scholar]
- Gautam R. Vanga S. Ariese F. Umapathy S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Techniques and Instrumentation. 2015;2:8. doi: 10.1140/epjti/s40485-015-0018-6. [DOI] [Google Scholar]
- Bonnier F. Byrne H. J. Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems. Analyst. 2012;137:322–332. doi: 10.1039/C1AN15821J. [DOI] [PubMed] [Google Scholar]
- Butler H. J. Ashton L. Bird B. Cinque G. Curtis K. Dorney J. et al., Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016;11:664–687. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- Kopec M. Abramczyk H. Angiogenesis – a crucial step in breast cancer growth, progression and dissemination by Raman imaging. Spectrochim. Acta, Part A. 2018;198:338–345. doi: 10.1016/j.saa.2018.02.058. [DOI] [PubMed] [Google Scholar]
- Van Nest S. J. Nicholson L. M. DeVorkin L. Brolo A. G. Lum J. J. Jirasek A. Raman Spectroscopic Signatures Reveal Distinct Biochemical and Temporal Changes in Irradiated Human Breast Adenocarcinoma Xenografts. Radiat. Res. 2018;189:497–504. doi: 10.1667/RR15003.1. [DOI] [PubMed] [Google Scholar]
- Chaturvedi D. Balaji S. A. Bn V. K. Ariese F. Umapathy S. Rangarajan A. Different Phases of Breast Cancer Cells: Raman Study of Immortalized, Transformed, and Invasive Cells. Biosensors. 2016;6:57. doi: 10.3390/bios6040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Lee S. H. Min S. Y. Byun K. M. Lee S. Y. Dual-modal cancer detection based on optical pH sensing and Raman spectroscopy. J. Biomed. Opt. 2017;22:1–6. doi: 10.1117/1.JBO.22.10.105002. [DOI] [PubMed] [Google Scholar]
- Winnard Jr P. T. Zhang C. Vesuna F. Kang J. W. Garry J. Dasari R. R. et al., Organ-specific isogenic metastatic breast cancer cell lines exhibit distinct Raman spectral signatures and metabolomes. Oncotarget. 2017;8:20266–20287. doi: 10.18632/oncotarget.14865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desroches J. Jermyn M. Pinto M. Picot F. Tremblay M. A. Obaid S. et al., A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy. Sci. Rep. 2018;8:1792. doi: 10.1038/s41598-018-20233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramczyk H. Imiela A. The biochemical, nanomechanical and chemometric signatures of brain cancer. Spectrochim. Acta, Part A. 2018;188:8–19. doi: 10.1016/j.saa.2017.06.037. [DOI] [PubMed] [Google Scholar]
- Malik A. Sahu A. Singh S. P. Deshmukh A. Chaturvedi P. Nair D. et al., In vivo Raman spectroscopy-assisted early identification of potential second primary/recurrences in oral cancers: An exploratory study. Head Neck. 2017;39:2216–2223. doi: 10.1002/hed.24884. [DOI] [PubMed] [Google Scholar]
- Ming L. C. Gangodu N. R. Loh T. Zheng W. Wang J. Lin K. et al., Real time near-infrared Raman spectroscopy for the diagnosis of nasopharyngeal cancer. Oncotarget. 2017;8:49443–49450. doi: 10.18632/oncotarget.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almond L. M. Hutchings J. Lloyd G. Barr H. Shepherd N. Day J. et al., Endoscopic Raman spectroscopy enables objective diagnosis of dysplasia in Barrett's esophagus. Gastrointest. Endosc. 2014;79:37–45. doi: 10.1016/j.gie.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Moradi H. Ahmad A. Shepherdson D. Vuong N. H. Niedbala G. Eapen L. et al., Raman micro-spectroscopy applied to treatment resistant and sensitive human ovarian cancer cells. J. Biophotonics. 2017;10:1327–1334. doi: 10.1002/jbio.201600211. [DOI] [PubMed] [Google Scholar]
- Jermyn M. Desroches J. Mercier J. St-Arnaud K. Guiot M. C. Leblond F. et al., Raman spectroscopy detects distant invasive brain cancer cells centimeters beyond MRI capability in humans. Biomed. Opt. Express. 2016;7:5129–5137. doi: 10.1364/BOE.7.005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusatori M. Auner G. Noh T. Scarpace L. Broadbent B. Kalkanis S. N. Intraoperative Raman Spectroscopy. Neurosurg. Clin. N. Am. 2017;28:633–652. doi: 10.1016/j.nec.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Klener J. Hofbauerova K. Bartos A. Ricny J. Ripova D. Kopecky V. Instability of cerebrospinal fluid after delayed storage and repeated freezing: a holistic study by drop coating deposition Raman spectroscopy. Clin. Chem. Lab. Med. 2014;52:657–664. doi: 10.1515/cclm-2013-0800. [DOI] [PubMed] [Google Scholar]
- Kocisova E. Antalik A. Prochazka M. Drop coating deposition Raman spectroscopy of liposomes: role of cholesterol. Chem. Phys. Lipids. 2013;172–173:1–5. doi: 10.1016/j.chemphyslip.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Kocisova E. Prochazka M. Vaculciakova L. Drop-Coating Deposition Raman (DCDR) Spectroscopy as a Tool for Membrane Interaction Studies: Liposome-Porphyrin Complex. Appl. Spectrosc. 2015;69:939–945. doi: 10.1366/14-07836. [DOI] [PubMed] [Google Scholar]
- Harvey T. J. Faria E. C. Henderson A. Gazi E. Ward A. D. Clarke N. W. et al., Spectral discrimination of live prostate and bladder cancer cell lines using Raman optical tweezers. J. Biomed. Opt. 2008;13:064004. doi: 10.1117/1.2999609. [DOI] [PubMed] [Google Scholar]
- Corsetti S. Rabl T. McGloin D. Nabi G. Raman spectroscopy for accurately characterizing biomolecular changes in androgen-independent prostate cancer cells. J. Biophotonics. 2018;11:e201700166. doi: 10.1002/jbio.201700166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley J. Kumar R. Kuzmin A. N. Pliss A. Yadav N. Balachandar S. et al., Lipid quantification by Raman microspectroscopy as a potential biomarker in prostate cancer. Cancer Lett. 2017;397:52–60. doi: 10.1016/j.canlet.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G. L. Gajjar K. Trevisan J. Fogarty S. W. Taylor S. E. Da Gama-Rose B. et al., Vibrational biospectroscopy coupled with multivariate analysis extracts potentially diagnostic features in blood plasma/serum of ovarian cancer patients. J. Biophotonics. 2014;7:200–209. doi: 10.1002/jbio.201300157. [DOI] [PubMed] [Google Scholar]
- Mahmood T. Nawaz H. Ditta A. Majeed M. I. Hanif M. A. Rashid N. et al., Raman spectral analysis for rapid screening of dengue infection. Spectrochim. Acta, Part A. 2018;200:136–142. doi: 10.1016/j.saa.2018.04.018. [DOI] [PubMed] [Google Scholar]
- Bilal M. Ullah R. Khan S. Ali H. Saleem M. Ahmed M. Lactate based optical screening of dengue virus infection in human sera using Raman spectroscopy. Biomed. Opt. Express. 2017;8:1250–1256. doi: 10.1364/BOE.8.001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. Ullah R. Khan A. Sohail A. Wahab N. Bilal M. et al., Random Forest-Based Evaluation of Raman Spectroscopy for Dengue Fever Analysis. Appl. Spectrosc. 2017;71:2111–2117. doi: 10.1177/0003702817695571. [DOI] [PubMed] [Google Scholar]
- Bilal M. Saleem M. Amanat S. T. Shakoor H. A. Rashid R. Mahmood A. et al., Optical diagnosis of malaria infection in human plasma using Raman spectroscopy. J. Biomed. Opt. 2015;20:017002. doi: 10.1117/1.JBO.20.1.017002. [DOI] [PubMed] [Google Scholar]
- Frame L. Brewer J. Lee R. Faulds K. Graham D. Development of a label-free Raman imaging technique for differentiation of malaria parasite infected from non-infected tissue. Analyst. 2017;143:157–163. doi: 10.1039/C7AN01760J. [DOI] [PubMed] [Google Scholar]
- Perez-Guaita D. Marzec K. M. Hudson A. Evans C. Chernenko T. Matthaus C. et al., Parasites under the Spotlight: Applications of Vibrational Spectroscopy to Malaria Research. Chem. Rev. 2018;118:5330–5358. doi: 10.1021/acs.chemrev.7b00661. [DOI] [PubMed] [Google Scholar]
- Sathyavathi R. Dingari N. C. Barman I. Prasad P. S. Prabhakar S. Narayana Rao D. et al., Raman spectroscopy provides a powerful, rapid diagnostic tool for the detection of tuberculous meningitis in ex vivo cerebrospinal fluid samples. J. Biophotonics. 2013;6:567–572. doi: 10.1002/jbio.201200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer U. Trenkmann S. Bocklitz T. Schmerler D. Kiehntopf M. Popp J. Fast differentiation of SIRS and sepsis from blood plasma of ICU patients using Raman spectroscopy. J. Biophotonics. 2014;7:232–240. doi: 10.1002/jbio.201400010. [DOI] [PubMed] [Google Scholar]
- Ding H. Dupont A. W. Singhal S. Scott L. D. Guha S. Younes M. et al., In vivo analysis of mucosal lipids reveals histological disease activity in ulcerative colitis using endoscope-coupled Raman spectroscopy. Biomed. Opt. Express. 2017;8:3426–3439. doi: 10.1364/BOE.8.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis J. Mohammed N. Rotimi O. Magee D. Jha A. Subramanian V. Raman spectroscopy of endoscopic colonic biopsies from patients with ulcerative colitis to identify mucosal inflammation and healing. Biomed. Opt. Express. 2016;7:2022–2035. doi: 10.1364/BOE.7.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence I. J. Beaulieu D. B. Horst S. N. Bi X. Herline A. J. Schwartz D. A. et al., Clinical characterization of in vivo inflammatory bowel disease with Raman spectroscopy. Biomed. Opt. Express. 2017;8:524–535. doi: 10.1364/BOE.8.000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerlingo C. d'Apuzzo F. Grassia V. Perillo L. Lepore M. Micro-Raman spectroscopy for monitoring changes in periodontal ligaments and gingival crevicular fluid. Sensors. 2014;14:22552–22563. doi: 10.3390/s141222552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevaidi M. Morais C. L. M. Halliwell D. E. Mann D. M. A. Allsop D. Martin-Hirsch P. L. et al., Raman Spectroscopy to Diagnose Alzheimer's Disease and Dementia with Lewy Bodies in Blood. ACS Chem. Neurosci. 2018 doi: 10.1021/acschemneuro.8b00198. [DOI] [PubMed] [Google Scholar]
- Ryzhikova E. Kazakov O. Halamkova L. Celmins D. Malone P. Molho E. et al., Raman spectroscopy of blood serum for Alzheimer's disease diagnostics: specificity relative to other types of dementia. J. Biophotonics. 2015;8:584–596. doi: 10.1002/jbio.201400060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokobza L. Bruneel J.-L. Couzi M. Raman spectroscopy as a tool for the analysis of carbon-based materials (highly oriented pyrolitic graphite, multilayer graphene and multiwall carbon nanotubes) and of some of their elastomeric composites. Vib. Spectrosc. 2014;74:57–63. doi: 10.1016/j.vibspec.2014.07.009. [DOI] [Google Scholar]
- Wang Y. Alsmeyer D. C. McCreery R. L. Raman spectroscopy of carbon materials: structural basis of observed spectra. Chem. Mater. 1990;2:557–563. doi: 10.1021/cm00011a018. [DOI] [Google Scholar]
- Dresselhaus M. S. Dresselhaus G. Saito R. Jorio A. Raman spectroscopy of carbon nanotubes. Phys. Rep. 2005;409:47–99. doi: 10.1016/j.physrep.2004.10.006. [DOI] [Google Scholar]
- Dresselhaus M. S. Jorio A. Hofmann M. Dresselhaus G. Saito R. Perspectives on Carbon Nanotubes and Graphene Raman Spectroscopy. Nano Lett. 2010;10:751–758. doi: 10.1021/nl904286r. [DOI] [PubMed] [Google Scholar]
- Ferrari A. C. Meyer J. C. Scardaci V. Casiraghi C. Lazzeri M. Mauri F. et al., Raman Spectrum of Graphene and Graphene Layers. Phys. Rev. Lett. 2006;97:187401. doi: 10.1103/PhysRevLett.97.187401. [DOI] [PubMed] [Google Scholar]
- Ferrari A. C. Basko D. M. Raman spectroscopy as a versatile tool for studying the properties of graphene. Nat. Nanotechnol. 2013;8:235. doi: 10.1038/nnano.2013.46. [DOI] [PubMed] [Google Scholar]
- Kagan M. R. Mccreery R. L. Reduction of Fluorescence Interference in Raman-Spectroscopy Via Analyte Adsorption on Graphitic Carbon. Anal. Chem. 1994;66:4159–4165. doi: 10.1021/ac00095a008. [DOI] [Google Scholar]
- Ling X. Xie L. M. Fang Y. Xu H. Zhang H. L. Kong J. et al., Can Graphene be used as a Substrate for Raman Enhancement? Nano Lett. 2010;10:553–561. doi: 10.1021/nl903414x. [DOI] [PubMed] [Google Scholar]
- Xie L. Ling X. Fang Y. Zhang J. Liu Z. Graphene as a Substrate To Suppress Fluorescence in Resonance Raman Spectroscopy. J. Am. Chem. Soc. 2009;131:9890–9891. doi: 10.1021/ja9037593. [DOI] [PubMed] [Google Scholar]
- Sil S. Kuhar N. Acharya S. Umapathy S. Is Chemically Synthesized Graphene 'Really' a Unique Substrate for SERS and Fluorescence Quenching? Sci. Rep. 2013;3:3336. doi: 10.1038/srep03336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lopez M. Merk V. Garcia-Ruiz C. Kneipp J. Surface-enhanced Raman spectroscopy for the analysis of smokeless gunpowders and macroscopic gunshot residues. Anal. Bioanal. Chem. 2016;408:4965–4973. doi: 10.1007/s00216-016-9591-z. [DOI] [PubMed] [Google Scholar]
- Sil S. Chaturvedi D. Krishnappa K. B. Kumar S. Asthana S. N. Umapathy S. Density functional theoretical modeling, electrostatic surface potential and surface enhanced Raman spectroscopic studies on biosynthesized silver nanoparticles: observation of 400 PM sensitivity to explosives. J. Phys. Chem. A. 2014;118:2904–2914. doi: 10.1021/jp4090266. [DOI] [PubMed] [Google Scholar]
- National Research Council, Committee on the Review of Existing and Potential Standoff Explosives Detection Techniques, The National Academies Press, Washington, DC, 2004 [Google Scholar]
- Hirschfeld T. Schildkraut E. R. Tannenbaum H. Tannenbaum D. Remote Spectroscopic Analysis of Ppm-Level Air Pollutants by Raman Spectroscopy. Appl. Phys. Lett. 1973;22:38–40. doi: 10.1063/1.1654465. [DOI] [Google Scholar]
- Agrawal J. P., High Energy Materials: Propellants, Explosives and Pyrotechnics, 2010 [Google Scholar]
- Akhavan J., The Chemistry of Explosives, 3rd edn, 2011
- Mills J. Farley C. Kassu A. Curley M. Sharma A. Ruffin P. et al., Raman monitoring and evaluation of the aging effects of rocket propellant stabilizers. Proc. SPIE. 2017:10382. [Google Scholar]
- Ghosh M. Venkatesan V. Sikder N. Sikder A. K. Quantitative Analysis of alpha-CL-20 Polymorphic Impurity in epsilon-CL-20 using Dispersive Raman Spectroscopy. Cent. Eur. J. Energ. Mater. 2013;10:419–438. [Google Scholar]
- Dreger Z. A. Gupta Y. M. Raman Spectroscopy of High-Pressure-High-Temperature Polymorph of Hexahydro-1,3,5-trinitro-1,3,5-triazine (epsilon-RDX) J. Phys. Chem. A. 2010;114:7038–7047. doi: 10.1021/jp102668d. [DOI] [PubMed] [Google Scholar]
- Thottempudi V. Shreeve J. M. Synthesis and promising properties of a new family of high-density energetic salts of 5-nitro-3-trinitromethyl-1H-1,2,4-triazole and 5,5′-bis(trinitromethyl)-3,3′-azo-1H-1,2,4-triazole. J. Am. Chem. Soc. 2011;133:19982–19992. doi: 10.1021/ja208990z. [DOI] [PubMed] [Google Scholar]
- Landenberger K. B. Matzger A. J. Cocrystal Engineering of a Prototype Energetic Material Supramolecular Chemistry of 2,4,6-Trinitrotoluene. Cryst. Growth Des. 2010;10:5341–5347. doi: 10.1021/cg101300n. [DOI] [Google Scholar]
- Kent R. V. Wiscons R. A. Sharon P. Grinstein D. Frimer A. A. Matzger A. J. Cocrystal Engineering of a High Nitrogen Energetic Material. Cryst. Growth Des. 2018;18:219–224. doi: 10.1021/acs.cgd.7b01126. [DOI] [Google Scholar]
- Ravi P. Badgujar D. M. Gore G. M. Tewari S. P. Sikder A. K. Review on Melt Cast Explosives. Propellants, Explos., Pyrotech. 2011;36:393–403. doi: 10.1002/prep.201100047. [DOI] [Google Scholar]
- Kholod Y. Okovytyy S. Kuramshina G. Qasim M. Gorb L. Leszczynski J. An analysis of stable forms of CL-20: A DFT study of conformational transitions, infrared and Raman spectra. J. Mol. Struct. 2007;843:14–25. doi: 10.1016/j.molstruc.2006.12.031. [DOI] [Google Scholar]
- Goede P. Latypov N. V. Ostmark H. Fourier transform Raman Spectroscopy of the four crystallographic phases of alpha, beta, gamma and epsilon 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazatetracyclo[5.5.0.0(5,9).0(3,11)]dodecane (HNIW, CL-20) Propellants, Explos., Pyrotech. 2004;29:205–208. doi: 10.1002/prep.200400047. [DOI] [Google Scholar]
- Farhadian A. H. Tehrani M. K. Keshavarz M. H. Darbani S. M. R. Raman spectroscopy combined with principle component analysis to investigate the aging of high energy materials. Laser Phys. 2017;27:075701. doi: 10.1088/1555-6611/aa7485. [DOI] [Google Scholar]
- Matousek P. Clark I. P. Draper E. R. C. Morris M. D. Goodship A. E. Everall N. et al., Subsurface probing in diffusely scattering media using spatially offset Raman spectroscopy. Appl. Spectrosc. 2005;59:393–400. doi: 10.1366/0003702053641450. [DOI] [PubMed] [Google Scholar]
- Stevens O. Petterson I. E. I. Day J. C. C. Stone N. Developing fibre optic Raman probes for applications in clinical spectroscopy. Chem. Soc. Rev. 2016;45:1919–1934. doi: 10.1039/C5CS00850F. [DOI] [PubMed] [Google Scholar]
- Stokes R. J., Bailey M., Bonthron S., Stone T., Maskall G. and Presly O., et al., New capability for hazardous materials ID within sealed containers using a portable spatially offset Raman spectroscopy (SORS) device, Optics and Photonics for Counterterrorism, Crime Fighting, and Defence Xii, 2016, vol. 9995 [Google Scholar]
- Matousek P. Conti C. Realini M. Colombo C. Micro-scale spatially offset Raman spectroscopy for non-invasive subsurface analysis of turbid materials. Analyst. 2016;141:731–739. doi: 10.1039/C5AN02129D. [DOI] [PubMed] [Google Scholar]
- Schrader B. Bergmann G. Die Intensität des Ramanspektrums polykristalliner Substanzen. Fresenius' J. Anal. Chem. 1967;225:230–247. doi: 10.1007/BF00983673. [DOI] [Google Scholar]
- Matousek P. Parker A. W. Bulk Raman analysis of pharmaceutical tablets. Appl. Spectrosc. 2006;60:1353–1357. doi: 10.1366/000370206779321463. [DOI] [PubMed] [Google Scholar]
- Eliasson C. Macleod N. A. Jayes L. C. Clarke F. C. Hammond S. V. Smith M. R. et al., Non-invasive quantitative assessment of the content of pharmaceutical capsules using transmission Raman spectroscopy. J. Pharm. Biomed. Anal. 2008;47:221–229. doi: 10.1016/j.jpba.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Johansson J. Sparen A. Svensson O. Folestad S. Claybourn M. Quantitative transmission Raman spectroscopy of pharmaceutical tablets and capsules. Appl. Spectrosc. 2007;61:1211–1218. doi: 10.1366/000370207782597085. [DOI] [PubMed] [Google Scholar]
- Edinger M. Knopp M. M. Kerdoncuff H. Rantanen J. Rades T. Lobmann K. Quantification of microwave-induced amorphization of celecoxib in PVP tablets using transmission Raman spectroscopy. Eur. J. Pharm. Sci. 2018;117:62–67. doi: 10.1016/j.ejps.2018.02.012. [DOI] [PubMed] [Google Scholar]
- Andrews D. Geentjens K. Igne B. McGeorge G. Owen A. Pedge N. et al., Analytical Method Development Using Transmission Raman Spectroscopy for Pharmaceutical Assays and Compliance with Regulatory Guidelines—Part I: Transmission Raman Spectroscopy and Method Development. J. Pharm. Innov. 2018;13:121–132. doi: 10.1007/s12247-018-9311-7. [DOI] [Google Scholar]
- Hartmann M. Light scattering by small particles. By H. C. van de Hulst. New York (John Wiley and Sons), London (Chapman and Hall), 1957. Pp. xiii, 470; 103 Figs.; 46 Tables. 96s. Q. J. R. Metereol. Soc. 1958;84:198–199. [Google Scholar]
- Bohren C. F. and Huffman D. R., Absorption and Scattering of Light by Small Particles, Wiley, 1998 [Google Scholar]
- Ishimaru A., Wave Propagation and Scattering in Random Media-Multiple Scattering, Turbulence, Rough Surfaces, and Remote Sensing, Academic Press, 1978 [Google Scholar]
- Sasic S. and Ozaki Y., Raman, Infrared, and Near-Infrared Chemical Imaging, 2010 [Google Scholar]
- Pine D. J. Weitz D. A. Chaikin P. M. Herbolzheimer E. Diffusing-Wave Spectroscopy. Phys. Rev. Lett. 1988;60:1134–1137. doi: 10.1103/PhysRevLett.60.1134. [DOI] [PubMed] [Google Scholar]
- Petterson I. E. Lopez-Lopez M. Garcia-Ruiz C. Gooijer C. Buijs J. B. Ariese F. Noninvasive detection of concealed explosives: depth profiling through opaque plastics by time-resolved Raman spectroscopy. Anal. Chem. 2011;83:8517–8523. doi: 10.1021/ac2018102. [DOI] [PubMed] [Google Scholar]
- Umapathy S., Sil S., Kiran J., Method and a system for detection of hazardous chemicals in a non-metallic container, Ind. Pat., IN2311CH2013, 2013, US Pat. 9,606,062B2, 2017
- Umapathy S., Sil S., Dhal G., Ariese F., Chemical signature resolved detection of concealed objects, Ind. Pat., IN6626/CHE/2014, 2014, US Pat. App. 15/122,170, 2016
- Umapathy S., Sil S., Kiran J., Method and an apparatus for obtaining sample specific signatures, Ind. Pat., IN2312CH2013, 2013, US Pat., US9983136B2, 2018
- Periyasamy V. Sil S. Dhal G. Ariese F. Umapathy S. Pramanik M. Experimentally validated Raman Monte Carlo simulation for a cuboid object to obtain Raman spectroscopic signatures for hidden material. J. Raman Spectrosc. 2015;46:669–676. doi: 10.1002/jrs.4709. [DOI] [Google Scholar]
- Sil S. Umapathy S. Raman spectroscopy explores molecular structural signatures of hidden materials in depth: Universal Multiple Angle Raman Spectroscopy. Sci. Rep. 2014;4:5308. doi: 10.1038/srep05308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer W. Recent advances in linear and nonlinear Raman spectroscopy II. J. Raman Spectrosc. 2008;39:1710–1725. doi: 10.1002/jrs.2171. [DOI] [Google Scholar]