Abstract
Cytoplasmic stress granules (SGs) are dynamic foci containing translationally arrested mRNA and RNA binding proteins (RBP) that form in response to a variety of cellular stressors. It has been debated that SGs may evolve into the cytoplasmic inclusions observed in many neurodegenerative diseases. Recent studies have examined the SG proteome by interrogating the interactome of G3BP1. However, it is widely accepted that multiple baits are required to capture the full SG proteome. To gain further insight into the SG proteome, we employed an immunoprecipitation coupled with mass spectrometry approach of endogenous Caprin-1, a RBP implicated in mRNP granules. Overall, we identified 1543 proteins that interact with Caprin-1. Interactors under stressed conditions were primarily annotated to the ribosome, spliceosome, and RNA transport pathways. We validated four Caprin-1 interactors that localized to arsenite-induced SGs: ANKHD1, TALIN-1, GEMIN5, and SNRNP200. We also validated these stress-induced interactions in SH-SY5Y cells and further determined that SNRNP200 also associated with osmotic and thermal induced SGs. Finally, we identified SNRNP200 in cytoplasmic aggregates in ALS spinal cord and motor cortex. Collectively, our findings provide the first description of the Caprin-1 protein interactome, identify novel cytoplasmic SG components, and reveal a SG protein in cytoplasmic aggregates in ALS patient neurons. Proteomic data collected in this study are available via ProteomeXchange with identifier PXD023271.
Keywords: Caprin-1, Stress granules, cytoplasmic inclusions, proteomics, mass spectrometry, RNA binding proteins, amyotrophic lateral sclerosis, protein-protein interactions, cytoplasmic granules
Graphical Abstract
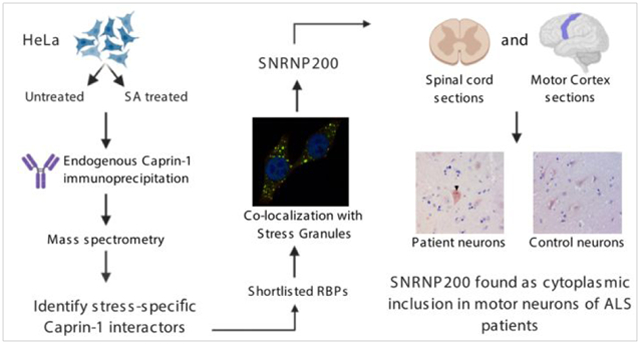
Introduction
In response to environmental stress, eukaryotic cells form non-membranous condensates, termed stress granules (SGs). SGs form upon environmental insults that inhibit global translation such as thermal, oxidative and osmotic stress, and contain untranslated mRNAs as well as a multitude of RNA binding proteins (RBPs)1, 2. SG formation is a transient cellular response and SG dissociation upon stress dissipation is equally important as SG assembly. Upon removal of stress, the release of stalled transcripts, ribosomal components, and other translation initiation machinery that are sequestered in SGs, is vital for cell survival3.
Several RBPs that are associated with SG formation bear low-complexity domains that facilitate liquid-liquid phase separation (LLPS) and aggregate formation4. These aggregates can be reversible as recent studies demonstrate that disaggregation of RBP fibrils can be facilitated by nuclear import receptors5 and such neurotoxic aggregation can be abrogated by RNA binding6. However, it has also been hypothesized that upon prolonged stress, SGs can transform into more solid aggregates which are irreversible7. Multiple neurodegenerative diseases are characterized by the presence of pathological aggregates8 and defects in SG dynamics are thought to seed such pathological aggregates9. A variety of RBPs associated with SGs contain disease-causing mutations that are linked to amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD), and frontotemporal dementia (FTD)10–14.
There has been considerable effort invested to identify and understand the proteomic15, 16 and transcriptomic17, 18 composition of SGs16, 19–22. Most of these studies have used Ras GTPase-activating protein-binding protein 1 (G3BP1), a core SG protein, to define the SG proteome21. It is plausible that some proteins localize to SGs independent of G3BP1 and thus, would not be detected as a SG component by these prior studies. Furthermore, SG composition is likely influenced by type and duration of stress, as well as cellular context. Many of these prior studies used over-expression of SG proteins to identify protein interactors. This may perturb normal protein stoichiometry within SGs potentially leading to erroneous interactions and/or mis-recruitment of proteins to RNP granules. Unravelling the SG proteome is essential to further understanding of disease mechanisms involving SG dynamics. Moreover, probing SGs with multiple baits may be required to fully capture the complete SG proteome.
Caprin-1 is a ubiquitously expressed RBP that exhibits high expression levels in dividing cells and was first isolated in lymphocytes and hematopoietic progenitor cells23. Caprin-1 has been shown to be crucial in regulating cell survival and proliferation in cancer cells24, 25. Caprin-1 is also highly expressed in the brain and associates with messenger RNP complexes26. In neuronal cells, Caprin-1 regulates localized translation at the synaptic terminals of dendrites27. While many prior studies have suggested that G3BP1 and Caprin-1 interact in SGs16, 21, 28, 29, a previous study in our laboratory demonstrated that knockdown of G3BP1 in HeLa cells did not perturb the formation of Caprin-1 containing SGs indicating that formation of these SGs can occur independently of Caprin-1-G3BP1 interaction30.
In this study, we define the Caprin-1 protein interactome in unstressed and sodium arsenite (SA) induced oxidative stress conditions. We utilized immunoprecipitation coupled to mass spectrometry (IP-MS) to identify 1,543 Caprin-1 protein-protein interactions (PPIs) in either unstressed conditions or following SA stress. Our results provide insight into the multitude of Caprin-1 mediated biological functions, including RNA transport and translation. Validation of select interactors resulted in annotating novel SG-localized proteins in HeLa cells. Among them, SNRNP200 was found in SGs induced by oxidative, thermal, as well as osmotic stress in both non-neuronal and neuronal-like cells. Furthermore, we found SNRNP200 in cytoplasmic aggregates within motor neurons of both the lumbar spinal cord and motor cortex of ALS patients. Our work identifies Caprin-1 PPIs and characterizes one that is also a component of SGs assembled in neuronal-like cells.
Experimental section
Cell culture and stress
HeLa cells were maintained in complete media containing DMEM (ThermoFisher) and 10% fetal bovine serum (FBS) (Gibco, Life Technologies) and 1% Penicillin-Streptomycin (ThermoFisher) at 37°C. SH-SY5Y cells were grown in DMEM/F12 media (ThermoFisher) containing 10% FBS (Gibco, Life Technologies). SH-SY5Y cells were differentiated using sequential treatments of retinoic acid (RA) (Cedarlane) and brain-derived neurotrophic factor (BDNF) (Sigma)31. Briefly, cells seeded on collagen coated plates were maintained in 10 μM RA containing media for 5 days with daily media changes. Thereafter, cells were grown in 50 ng/mL BDNF containing media for further 7 days. Stress was performed post day 12 and cells were fixed immediately with 4% paraformaldehyde (PFA, FD NeuroTechnologies) for staining.
Cells were seeded, maintained, and stressed on glass coverslips. To induce oxidative stress, cells were treated with 0.5 mM of SA (Sigma) for 30 mins at 37°C. The media was replaced with fresh media and incubated at 37°C for 60 min to initiate recovery after SA stress30, 32. Heat shock was performed by incubating cells at 43°C for 30 mins. Osmotic stress was performed by incubating the cells in 0.4 M sorbitol (Sigma) containing media at 37°C for 2 hrs. Following stress regimen, all cells were immediately fixed with 4% PFA.
Immunofluorescent staining
Fixed cells were permeabilized with 0.1% triton X-100 solution in 1X PBS. Cells were then blocked using Super Block (Scytek) and subsequently incubated overnight at 4°C with primary antibodies diluted in Super Block. Cells were washed and incubated with Alexa-Fluor secondary antibodies for 1 hour at room temperature. All antibody details are included in Table S1. Nuclei were labelled with either 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher) or Hoechst 333432 (ThermoFisher). Cover slips were mounted on slides using ProLong antifade reagent (Invitrogen). Images were acquired using a 63X oil lens on Zeiss LSM 710 (Zeiss) or a Leica SP5 (Leica Microsystems) confocal microscope. Line scan analyses were performed to assess co-localization using Image J V1.52a33.
Immunostaining of human post-mortem ALS tissues
Paraffin-embedded spinal cord and motor cortex tissue sections from 5 sporadic ALS cases, 2 disease controls (DCs) and 2 non-neurologic diseased controls (NNDCs) were obtained from the Barrow Neurological Institute and Target ALS post-mortem tissue bank cores (subject demographics listed in Table 1). Participants of the post-mortem tissue bank cores provided IRB approved informed consent for the collection of post-mortem tissues. Immunohistochemistry (IHC) was performed as previously described13, 34. Tissue sections were de-paraffinized and rehydrated, and antigen retrieval was performed using the Antigen Retrieval Citra Solution (pH 6) (BioGenex). Tissues were blocked using Super Block supplemented with Avidin (Vector Labs) and incubated with rabbit anti-SNRNP200 antibody (Sigma) diluted in Super Block supplemented with biotin (Vector Labs) overnight. An anti-rabbit biotinylated IgG secondary antibody (Vector Labs) was subsequently added, and immunostaining was visualized using the Vectastain Elite ABC reagent (Vector Labs) and Vector ImmPACT NovaRED peroxidase substrate kit (Vector Labs). Slides were counterstained with hematoxylin (Sigma). Images were acquired using an Olympus BX40 microscope and a minimum of 11 motor neurons were quantified for each case using Image J34. Details on antibodies used for IHC are listed in Table S1.
Table 1:
Demographics of patients used for SNRNP200 immunohistochemistry.
| Case | Sex | Age at Death | Age at onset | Site of Onset | Diagnosis | C9 +or− |
|---|---|---|---|---|---|---|
| ALS 1 | Male | 85 | 69 | Limb | Sporadic ALS | - |
| ALS 2 | Male | 74 | N/A | N/A | Sporadic ALS | - |
| ALS 3 | Female | 60 | 57 | Limb | Sporadic ALS | - |
| ALS 4 | Female | 63 | 60 | N/A | Sporadic ALS | - |
| ALS 5 | Female | 69 | 67 | N/A | Sporadic ALS | N/A |
| DC 1 | Male | 81 | N/A | N/A | Alzheimer’s Disease | - |
| DC 2 | Male | 71 | N/A | N/A | Alzheimer’s Disease | - |
| NNDC 1 | Female | 74 | N/A | N/A | Non-Neurologic Disease Control | |
| NNDC 2 | Male | 22 | N/A | N/A | Non-Neurologic Disease Control | - |
N/A – Not assessed
Cell lysis and Immunoprecipitation for Caprin-1
HeLa cells were grown in 100 mm dishes until 95% - 100% confluent. Following stress, cells were crosslinked with 0.1 % formaldehyde and quenched with 1.25 M glycine. Cells were subsequently scraped into IP lysis buffer (50 mM HEPES pH 7.2, 150 mM NaCl, 0.1 % NP-40, 10% Glycerol, 2 mM EDTA, 2 mM EGTA, 1 mM DTT, Protease inhibitor (Sigma Aldrich) and Ribolock (ThermoFisher)), and incubated on a rotator at 4°C for 20 mins. Lysates were centrifuged at 10,000 × g for 15 mins at 4°C and the supernatant was collected. Protein content of the supernatants was determined using the Bradford protein assay (BioRad). Each immunoprecipitation was carried out in quadruplicate. 1 μg of rabbit anti-Caprin-1 (Proteintech) or rabbit anti-IgG (Jackson Immuno Research Laboratories) was added to magnetic protein A Dynabeads (ThermoFisher). Immunoprecipitation was performed with an input of 400 μg of total protein for 1 hr at room temperature. After IP, the beads were washed 6 times with IP lysis buffer (without DTT, protease inhibitors, and Ribolock) and stored at −20°C.
Protein digestion and peptide sample preparation
The immunoprecipitated protein was eluted from the beads with 1X Laemmli buffer (BioRad) by heating at 95°C for 10 mins. Samples were resolved via gel electrophoresis and stained with Bio safe Coomassie stain (BioRad) for 1 hr and destained in double deionized water overnight. Each lane was excised into three individual fractions with IgG heavy (50 kDa) and light (25 kDa) chains excluded. The fractions were cut into 1–2 mm3 cubes and further de-stained, washed and dried. Proteins were reduced by incubating with 10 mM DTT (ThermoFisher) for 30 mins at 60°C and alkylated with 55 mM iodoacetamide (ThermoFisher) for 30 mins at room temperature using a series of hydration and dehydration steps. Digestion was performed using Trypsin gold (Promega) at 1:20 enzyme to protein ratio and incubated overnight at 37°C. After digestion, trypsin was inactivated by adding trifluoroacetic acid (ThermoFisher) to a final concentration of 0.5% (v/v). Peptides were extracted from the gel pieces by hydration and dehydration and further clean-up was performed using C18 StageTips as previously described35, 36.
LC-MS/MS analysis
Data acquisition was performed on a Thermo Orbitrap Fusion Lumos interfaced with a U3000 RSLCnano UHPLC operating at a flow rate of 300 nL/min, with Solvent A (water, 0.1% FA) and Solvent B (acetonitrile, 0.1% FA). Peptides in 5 μL of loading solvent (98% water, 2% acetonitrile, 0.1% FA) spiked with 25 femtomoles of synthetic peptides (Pierce Retention Time Calibrator Mix, ThermoFisher) were directly loaded on a 15 cm C18 EasySpray column (ES800, ThermoFisher) maintained at 45°C. Peptides were separated over 60 mins using the following gradient: 2–19% B in 42 mins, 19–45% B in 6 mins and then to 90% B in 0.5 min, isocratic at 90% B for 1 min followed by return to initial conditions in 0.5 min and column equilibration for 10 mins. Data-Dependent Acquisition (DDA) of eluted peptides was performed using top-speed mode with a cycle time of 3 secs. MS1 scans were acquired in the Orbitrap at a resolution of 120K, with a mass range of 400–1500 m/z, keeping the AGC target at 4E5 and with a max ion injection time to 50 millisecs. Most abundant precursor ions with a charge state between 2 and 7 were selected with an isolation window of 1.6 Da and fragmented using High Energy Collision Dissociation (HCD), followed by detection in the ion trap. A dynamic exclusion of 60 secs was used to prevent resampling of the same precursors during the elution of chromatographic peaks.
Data processing
Mass spectra were aligned across replicate bands for each IP and condition, within Progenesis QI for Proteomics v4.1.6 (Nonlinear Dynamics, A Waters Company) using default parameters for automated processing. Aligned and filtered spectra were searched against a human database (Swissprot/UniprotKB, 2017) in Mascot v2.6 (Matrix Science) with trypsin cleavage rules, and a maximum of 2 missed cleavages. Allowed dynamic modifications were oxidation (methionine) and acetylation (N-term, lysine). Cysteine carbamidomethylation was set as a fixed modification. Matched spectra were imported into Progenesis QI and only peptides with a Mascot homology score >21 were retained. Gel bands were recombined for protein identification and quantitation.
Proteins were first filtered for their presence in >75% of samples and then normalized using the cBioconductor DEP package in R37. Average probabilities of interaction (AvgP) were calculated using the SAINTexpress algorithm38 against IgG controls. SAINTexpress uses a Bayesian estimation model to calculate interaction probability distributions between bait-prey pairs. These distributions are then compared between the test samples and negative controls to elucidate true interactions. High confidence interactors (AvgP >0.7 and presence in 3 out of 4 replicates) were compared between the stressed and unstressed cell lines to determine unique Caprin-1 interactors for each condition. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository39 with the dataset identifier PXD023271.
StringDB, gene ontology, pathway analysis
High confidence proteins unique to either the stressed or unstressed condition were imported into StringDB (1.6.0) through Cytoscape (v3.8.0) to reveal protein clusters and associations previously annotated in literature. Networks were created from all interaction evidence in the String database, and filtered for a String score >0.7. Gene ontology and KEGG pathway analysis was performed using DAVID V6.8. with default settings40.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 8. For the motor neuron quantification, a Mann-Whitney test was performed to compare differences between ALS and controls with p < 0.05 considered significant.
Results
Caprin-1 immunoprecipitation and mass spectrometry
In order to interrogate the interactome of endogenous Caprin-1 (Figure 1A), we employed immunoprecipitation (IP) of endogenous Caprin-1 from HeLa cells exposed to SA stress and compared against unstressed controls. We followed a paradigm of 30 mins of acute SA stress followed by 60 mins of recovery which captures the secondary assembly of SGs, as previously described32. After stress, cells were cross-linked with formaldehyde to preserve all interactions. Immunoblot using anti-Caprin-1 antibody confirmed enrichment of Caprin-1 in the Caprin-1 IP lanes but not in the IgG control IP lanes (Figure 1B and Figure S1 for the full blot). The IP fractions were separated by SDS page, and subjected to reduction, alkylation and trypsin digestion and subsequently analyzed by data-dependent LC-MS/MS. Overall, we identified 1,543 proteins for Caprin-1 IPs and 525 proteins from the IgG control pull-downs (Figure 1C, 1D, Table S2) with known SG components also being identified in both the stressed and unstressed Caprin-1 IPs (e.g. Ataxin-2, USP10, and TIAR).
Figure 1: Caprin-1 IP-MS methods and analysis of the interactome.
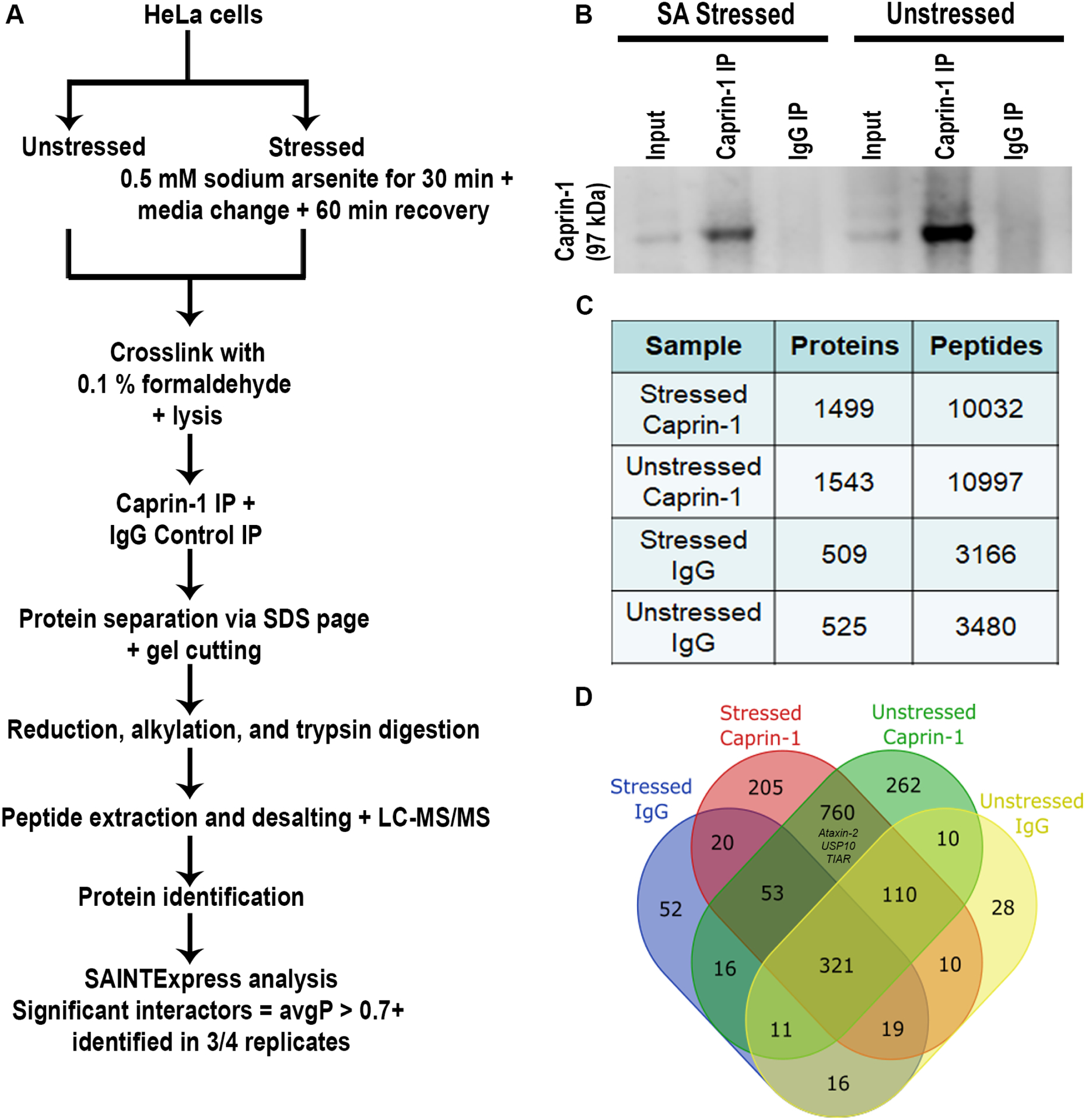
A) Schematic of the methodology employed in this study. B) Western blot of Caprin-1 and IgG IP fractions from lysates derived from both stressed and unstressed conditions demonstrating the presence of Caprin-1 only in the Caprin-1 IP and not in the IgG control. C) Summary of the proteins and peptides identified by LC-MS/MS from quadruplicate IP experiments per condition. D) Venn diagram demonstrating the overlap of all of the proteins identified across all comparisons.
To further characterize the interactome of Caprin-1 under stressed and unstressed conditions, we employed SAINTexpress, an algorithm premised on label free IP-MS experiments to detect PPIs41 that we have successfully used to define the protein interactome for other disease related proteins42, 43. Proteins identified in at least 3 of the 4 replicates and demonstrating an average probability of interaction (AvgP) greater than 0.7 were considered high confidence interactors of Caprin-1 (Figure 1A). Based on this criterion, we identified 281 and 326 proteins that interact with Caprin-1 under stressed and unstressed conditions respectively, with 38% (166/441) overlap between the two conditions (Figure 2A, Table S3). Of the 166 commonly identified proteins in both stressed and unstressed conditions, 10 proteins have been previously reported to play a functional role in SGs. This includes core SG proteins like G3BP1 and G3BP2b which reflects that pre-stress interactions between SG proteins exist and perhaps is the reason for rapid dynamics in SG formation at the onset of stress. Other SG components like USP10, FMR1, Ataxin2-like (ATXN2L) and UPF1 are also pre-stress interactors of Caprin-1. Multiple RNA helicases like DDX1, DDX3X, and DDX5 which have known roles in RNP granules (including SGs and P-bodies) are also pre-stress interactors of Caprin-1 (Figure 2A). CNOT7, which is localized to P-bodies and participates in mRNA decay, was also detected as a Caprin-1 interactor in both conditions.
Figure 2: Analysis of the unstressed interactome of high confidence Caprin-1 interactors revealed associations with ribosomes, RNA transport, and spliceosome pathways.
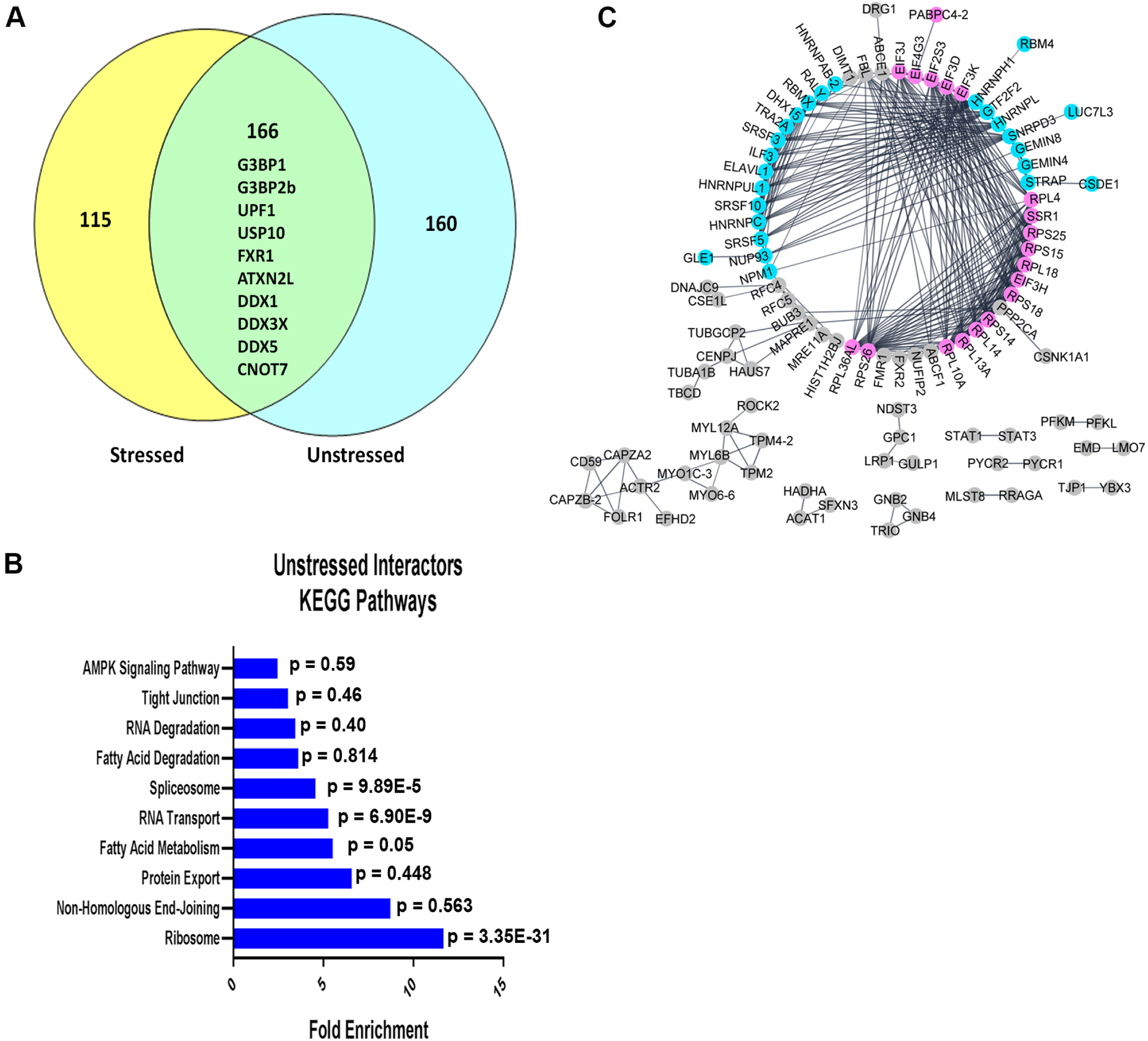
A) Venn Diagram comparing high confidence interactors of Caprin-1 in both stressed and unstressed conditions, as determined by SAINTexpress. A 38% overlap was observed between conditions. Notable proteins common between the two conditions are indicated. B) KEGG pathway analysis of the unstressed Caprin-1 interactors. C) StringDB analysis demonstrating protein-protein interactions between the unique interactors of Caprin-1 in unstressed conditions, with distinct groups annotated as RNA binding proteins (pink) and splicing factors (blue). All other interactors are labeled in gray.
Caprin-1 interactome in unstressed conditions
To identify biological pathways in which Caprin-1 functions in unstressed conditions, we employed KEGG pathway analysis using all 326 high confidence interactors of Caprin-1 and found significant enrichment of pathways related to ribosomes, RNA transport, and the spliceosome suggesting that Caprin-1 participates in these pathways in unstressed conditions (Figure 2B, Table S4). Gene ontology enrichment analyses of the top 15 biological processes and molecular functions revealed similar findings as KEGG pathway analysis, with terms broadly related to ribosomes and splicing (Figure S2). More specifically, this analysis suggested a functional role of Caprin-1 in translation initiation, mRNA splicing, and RNA binding (Figure S2A, S2B). To investigate known PPIs between Caprin-1 and the unstressed interactors, StringDB analysis was employed on 160 uniquely interacting proteins in the unstressed conditions (Figure 2A). Within the largest interaction network, we identified two groups which correspond to splicing factors (highlighted in blue) and ribosome and translation related proteins (highlighted in pink) that interact with Caprin-1 (Figure 2C). This complementary analysis recapitulated the gene ontology analysis, re-affirming that the primary functions of Caprin-1 are associated with translation and RNA splicing.
Caprin-1 interactome following sodium arsenite stress
We next interrogated the Caprin-1 interactome in SA stress conditions to understand how the interactome of Caprin-1 changes when it predominantly localizes to SGs (281 proteins in Figure 2A). KEGG pathway analysis demonstrated similar enrichment of pathways as compared to the unstressed interactors of Caprin-1 where the stress-dependent interactors were annotated to the ribosome, RNA transport, and spliceosome, and additionally to protein export (Figure 3A, Table S5). Interestingly, nearly half (131/281) of the high confidence interactors were annotated to known pathways (Figure 3B, Table S5). As Caprin-1 is primarily localized to SGs upon stress29 and SGs are known to modulate translation3, it was not surprising that several translation-related proteins and ribosomal subunits were among Caprin-1 interactors. StringDB analysis of the unique stress-dependent interactors also demonstrated two interaction networks corresponding to splicing factors and ribosomal proteins as Caprin-1 interactors (Figure S3).
Figure 3: Analysis of the stressed Caprin-1 interactome and shortlisted hits for validation.
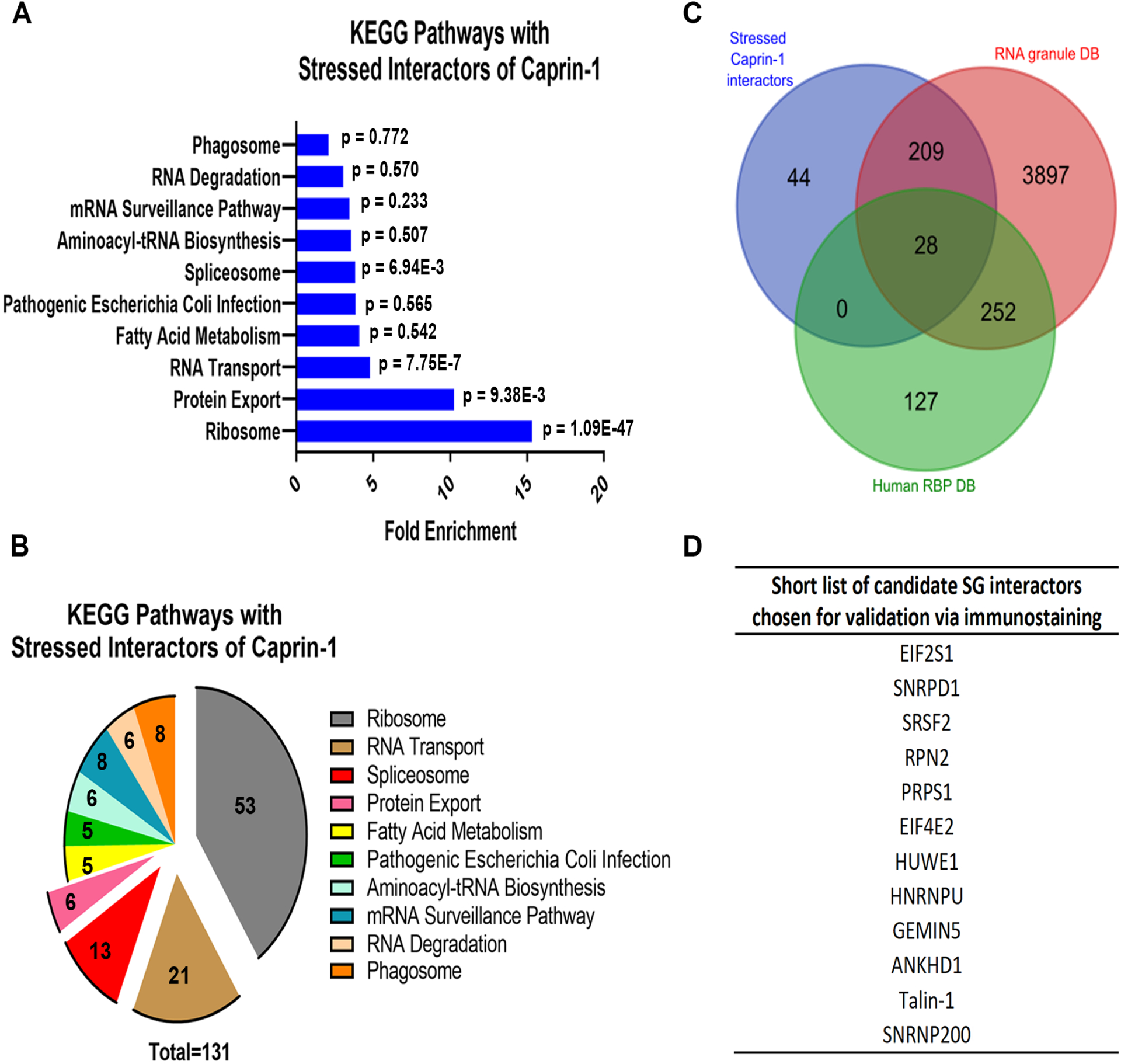
A) Pathway analysis of Caprin-1 interactors in stressed conditions (326 proteins). B) Pie chart demonstrating the number of Caprin-1 stressed interactors annotated to each pathway. C) Venn Diagram demonstrating the overlap between the stress-dependent Caprin-1 interactors from this study (blue) and RNA binding proteins from Human RBPs (green, http://rbpdb.ccbr.utoronto.ca/) and RNA Granule DB (red, http://rnagranuledb.lunenfeld.ca/). D) Short list of 12 candidate Caprin-1 interactors derived from pathway and RBP analyses that were further interrogated by immunostaining to assess co-localization with Caprin-1.
Prior studies have shown that Caprin-1 interacts with G3BP1 in SGs16, 21, 29 suggesting that there may be some degree of overlap among their respective interactomes. Therefore, we compared our stress dependent Caprin-1 interactors (281 proteins) with previously published SG proteomes (Figure S4). BioID based profiling of Caprin-1 interactors demonstrated only 14 interactors that overlapped with our Caprin-1 interactors. Interestingly, of these 14, USP10, FXR1, UPF1 are known SG components (Figure S4A). There was also a minor overlap with the BioID G3BP1-interactome and notable SG components such as PABPC1, PABPC4, ATXN2L which were common interactors of both Caprin-1 and G3BP1 (Figure S4B). While there were only 14 proteins that overlapped between our Caprin-1 interactome and either BioID-Caprin-1 or BioID-G3BP1 interactomes, four proteins were common to all the three datasets (PABPC4, FXR1, UPF1 and USP10) (Table S6). Comparison of our Caprin-1 interactors with recent publications probing the SG proteome with GFP-G3BP1 also showed minimal overlap, including multiple RBPs such as SYNCRIP, IGF2BP1, and HNRNPA3 (Figure S4C, Table S6). Lastly, only 28 proteins overlapped between our Caprin-1 dataset and that obtained using Apex-mediated proximity labelling of G3BP1 as bait, with notable translation-related SG components such as EIF3L, EIF3A, TAF15 being common (Figure S4D, Table S6).
As Caprin-1 is an integral SG protein that may interact with multiple RBPs, we also compared our list of high confidence interactors with known RBPs listed in two independent databases: Human RBP DB v1.3.1 (http://rbpdb.ccbr.utoronto.ca/) and RNA Granule DB v1 (http://rnagranuledb.lunenfeld.ca/) (Figure 3C, Table S7). Our results demonstrate 84% overlap (237/281) between proteins within our stressed candidate list (281 proteins from Figure 2A) and known RBPs. Combining protein lists from both the significantly enriched KEGG pathways as well as those that overlap between our interactors in stressed conditions and known RBPs, we shortlisted 12 Caprin-1 interactors that had not been previously associated with SGs to assess their localization in SGs (Figure 3D). We chose to focus on RBPs with no prior evidence of localization to SGs as well as those that annotated to the spliceosome and RNA transport pathways since RNG105 (Xenopus ortholog of human Caprin-1) has been previously shown to concentrate in ribosomal-associated granules27 and the presence of various translation-related proteins to SGs is widely reported44.
Validation of Caprin-1 interactors in HeLa cells
To assess if our Caprin-1 interacting candidate proteins co-localize with SGs, we performed double label immunofluorescence microscopy in HeLa cells following SA stress. Among the 12 candidates we prioritized for validation, ANKHD1, Talin-1, GEMIN5, and SNRNP200 demonstrated co-localization with Caprin-1 labeled SGs following exposure to SA (Figure 4). While one prior study has identified ANKHD1 as an interactor of Caprin-120, this is the first report of ANKHD1 being visualized in SGs by immunostaining. The remaining 8 candidates failed to co-localize with SA-induced SGs (Figure S5). As SG composition can vary with different stressors45, we assessed the potential of ANKHD1, Talin-1, GEMIN5, and SNRNP200 to co-localize with SGs induced by thermal (heat shock) and osmotic (sorbitol) stressors. Under these conditions, only SNRNP200 (Figure 5B) co-localized with SGs while ANKHD1, Talin-1 and GEMIN5 did not (Figures 5A, 5C, 5D). Note, although GEMIN5 did not colocalize with SGs, we did observe GEMIN5 puncta that were frequently located adjacent to SGs.
Figure 4: Arsenite stress induced Caprin-1 interactors; ANKHD1, Talin-1, SNRNP200, and GEMIN5 co-localize with SGs in HeLa cells.
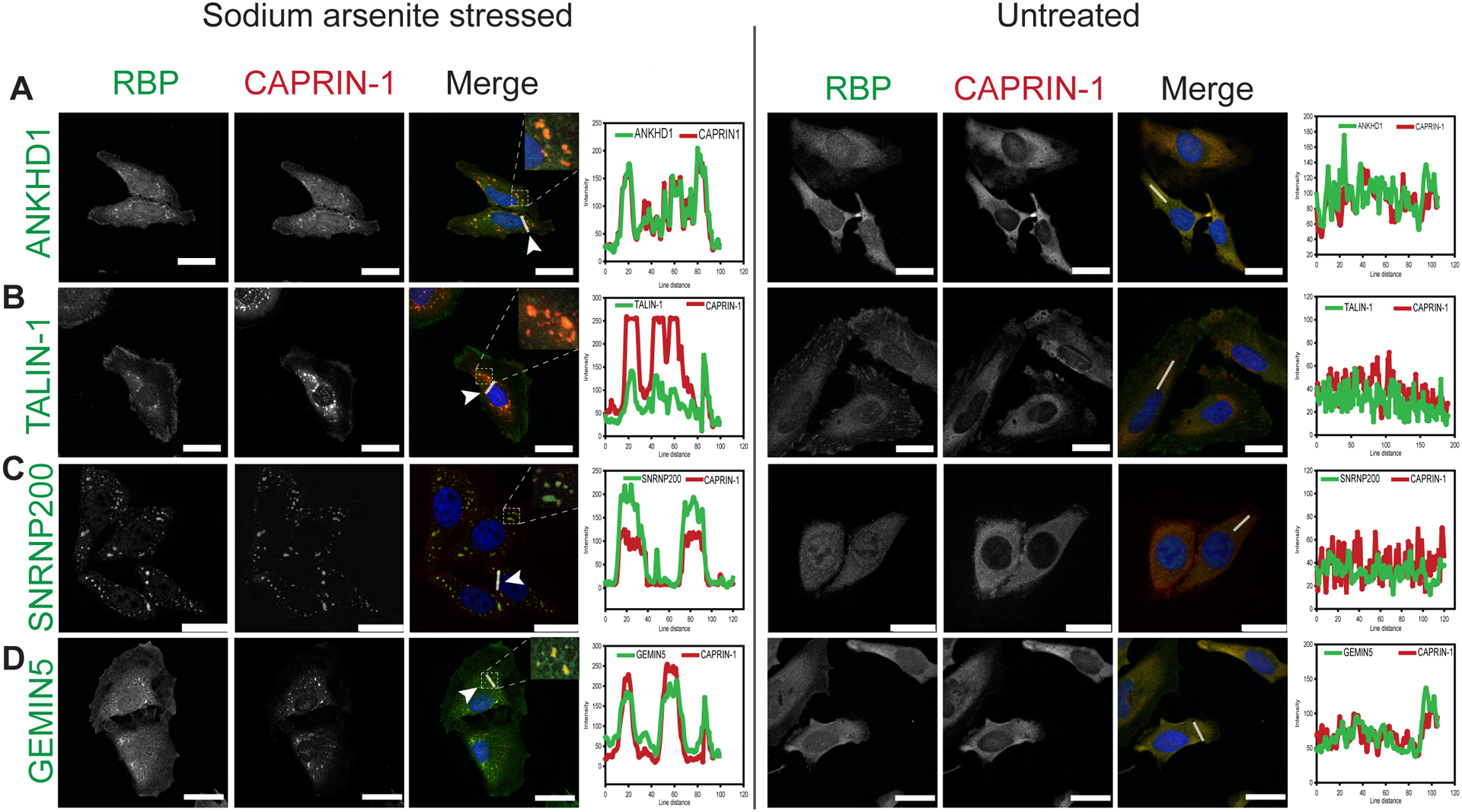
HeLa cells were stressed with either 0.5mM SA (left panel) or, left untreated (right panel). Representative images of immunofluorescent labelling for Caprin-1 and A) ANKHD1, B) TALIN-1, C) SNRNP200, and D) GEMIN5. Nuclei were visualized with DAPI. Scale bar, 25 μm. Line scan-based co-localization analysis is plotted as histograms.
Figure 5: Caprin-1 interactor SNRNP200 co-localize with SGs in HeLa cells subjected to thermal and osmotic stressors.
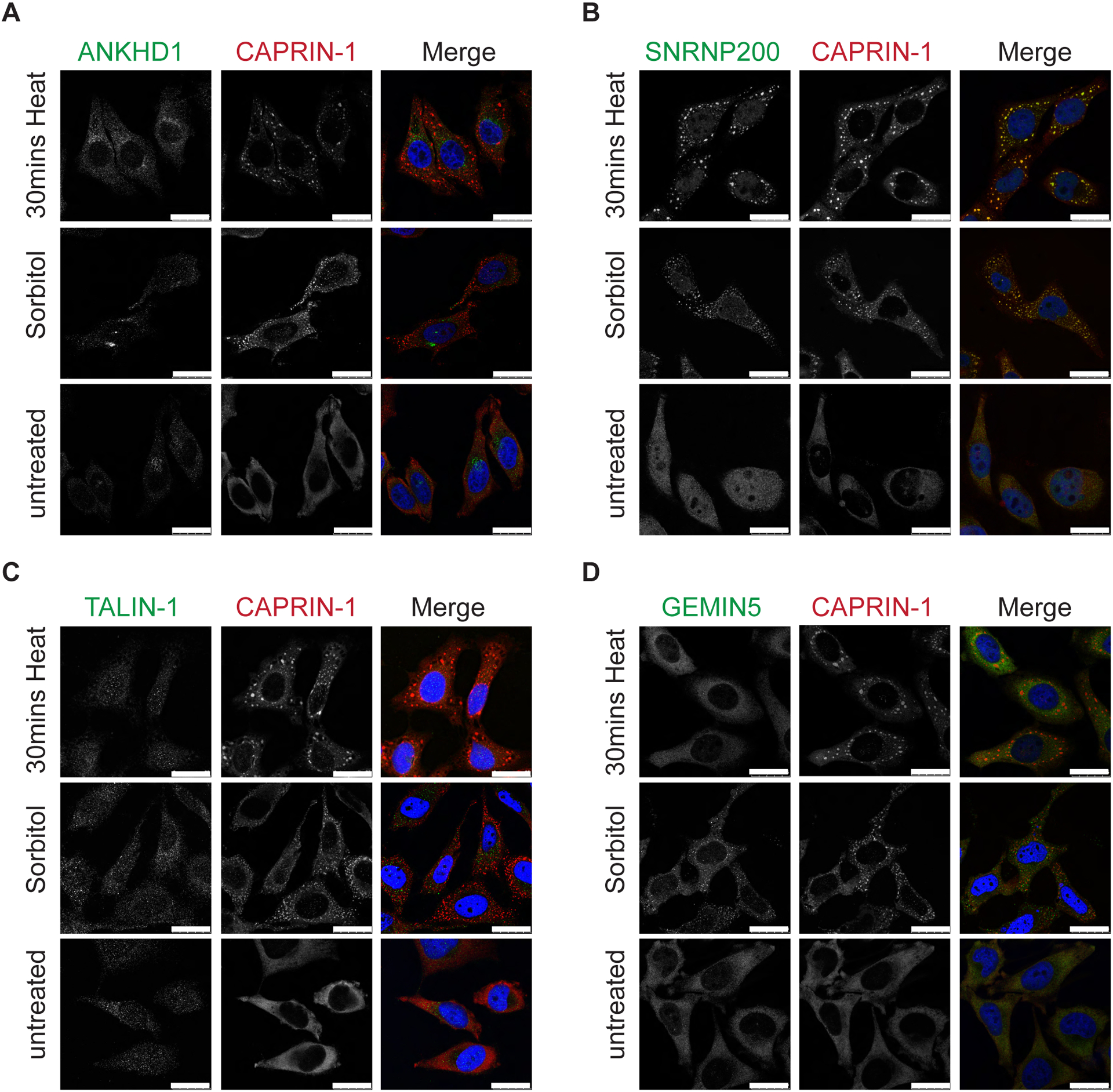
HeLa cells were stressed with either 30 mins heat stress at 43°C or 0.4M sorbitol for 2 hrs or, left untreated. Representative images shown after co-labelling for Caprin-1 and A) ANKHD1, B) SNRNP200, C) TALIN-1, and D) GEMIN5. Nuclei were visualized with Hoeschst. Scale bar, 25 μm.
SNRNP200 co-localizes with Caprin-1 in SGs in neuronal-like cells
It is reported that SG components can differ among various cell types, with neuronal cells having a more diverse SG composition21. To determine the propensity of the four novel Caprin-1 interactors (ANKHD1, TALIN-1, GEMIN5, and SNRNP200) to co-localize with SGs in a neuronal-like context, we utilized the SH-SY5Y human neuroblastoma cell line. SNRNP200 co-localized with Caprin-1 labelled SGs following SA or heat shock induced stress (Figure 6A). In contrast, ANKHD1, TALIN-1 and GEMIN5 did not form any punctate structures in SH-SY5Y cells during thermal or SA stress (Figure S6A–C). This further demonstrates the variability of SG composition between cell types. As the SG composition may vary in terminally differentiated cells, we also assessed SNRNP200 in SH-SY5Y cells differentiated with BDNF. We validated the differentiation of SH-SY5Y cells using the neuronal marker Tubb3/Tuj1 (Figure S6D). Indeed, we found SNRNP200 co-localizes with Caprin-1 labelled SGs in terminally differentiated SH-SY5Y cells (Figure 6B). Line scans to evaluate co-localization indicate clear overlap of both the Caprin-1 and SNRNP200 signals following acute thermal or SA stress (Figure 6B, right panel; Table 2).
Figure 6: SNRNP200 co-localizes with SGs in neuronal-like cells.
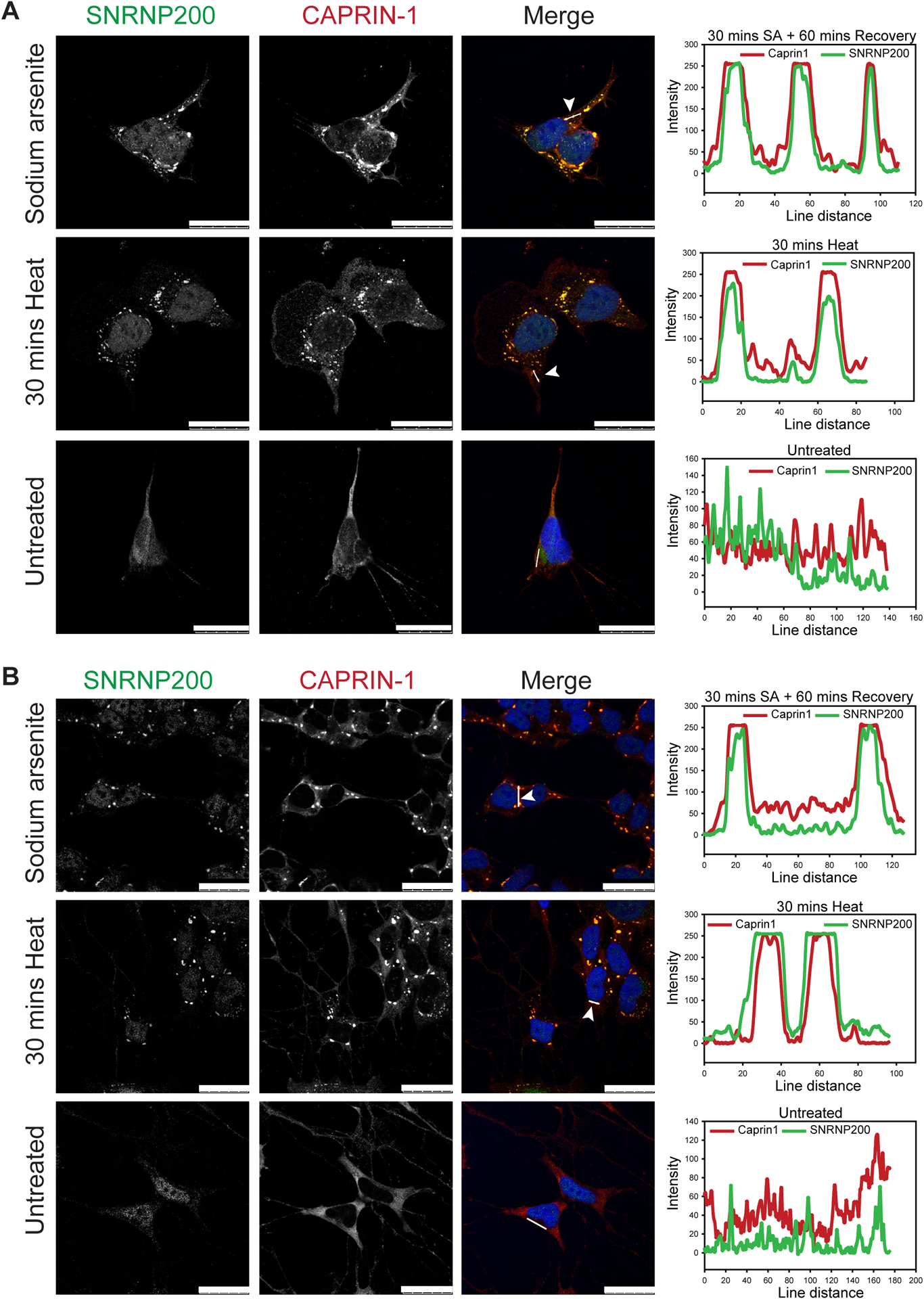
Representative images of SNRNP200 with Caprin-1 co-labelling in A) undifferentiated or B) differentiated SH-SY5Y neuronal-like cells following SA (0.5mM, 30 mins stress with 60mins recovery) or heat stress (43°C, 30 mins). Nuclei are visualized with Hoechst. Line scan analysis indicates co-localization. Scale bar, 25 μm.
Table 2:
Summary of co-immunostaining to assess co-localization of the candidate RBP with SGs.
| Candidate RNA binding protein |
In HeLa cells | In SH-SY5Y cells | In Differentiated SH-SH5Y cells | ||||
|---|---|---|---|---|---|---|---|
| Sodium arsenite stress | 43°C heat stress | 0.4M Sorbitol stress |
Sodium arsenite stress | 43°C heat stress | Sodium arsenite stress | 43°C heat stress | |
| HUWE1 | No | N/A | N/A | N/A | N/A | N/A | N/A |
| HNRNPU | No | N/A | N/A | N/A | N/A | N/A | N/A |
| ANKHD1 | Yes | No | No | No | No | No | No |
| TALIN-1 | Yes | No | No | No | No | No | No |
| GEMIN5 | Yes | No | No | No | No | No | No |
| SNRPD1 | No | N/A | N/A | N/A | N/A | N/A | N/A |
| EIF2S1 | No | No | N/A | N/A | N/A | N/A | N/A |
| PRPS1 | No | N/A | N/A | N/A | N/A | N/A | N/A |
| RPN2 | No | N/A | N/A | N/A | N/A | N/A | N/A |
| SRSF2 | No | N/A | N/A | N/A | N/A | N/A | N/A |
| SNRNP200 | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
N/A: not assessed
SNRNP200 is found in aggregates in post-mortem ALS neurons
Our in vitro studies demonstrated that SNRNP200 co-localized with Caprin-1 containing SGs in both HeLa as well as differentiated and undifferentiated SH-SY5Y cells. Interestingly, SNRNP200 has been previously identified as an interactor of TDP-43 in HEK293 cells46 and Caprin-1 has been reported as localized within cytoplasmic inclusions in sporadic ALS patient spinal motor neurons13. Therefore, we speculated that SNRNP200 may also be re-distributed into cytoplasmic aggregates in motor neurons of ALS patients. To address this, we performed immunostaining on post-mortem lumbar spinal cord and motor cortex tissues from 5 sporadic ALS cases, 2 neurologic disease controls (DCs), and 2 non-neurologic disease controls (NNDCs). SNRNP200 labeling was diffuse in most motor neurons of both DC and NNDC controls (Figure 7). In contrast, SNRNP200 labeling was found as cytoplasmic inclusions within some motor neurons of both the lumbar spinal (Figure 7AI, black arrowhead) and motor cortex (Figure 7AIV, black arrowhead) of ALS cases. Quantification revealed a trend for the fraction of spinal cord motor neurons displaying SNRNP200 inclusions to be higher in ALS cases relative to controls, but did not reach statistical significance (Figure 7B, 18% in ALS vs. 1% in controls, p = 0.0635). In addition to the SNRNP200 inclusions, we also observed a punctate staining pattern in the gray matter parenchyma (Figure 7I-III, red arrowheads) outside of the motor neurons across all ALS cases and controls which we hypothesize to be SNRNP200 localized at synaptic terminals.
Figure 7: SNRNP200 was identified in cytoplasmic inclusions in ALS post mortem tissues.
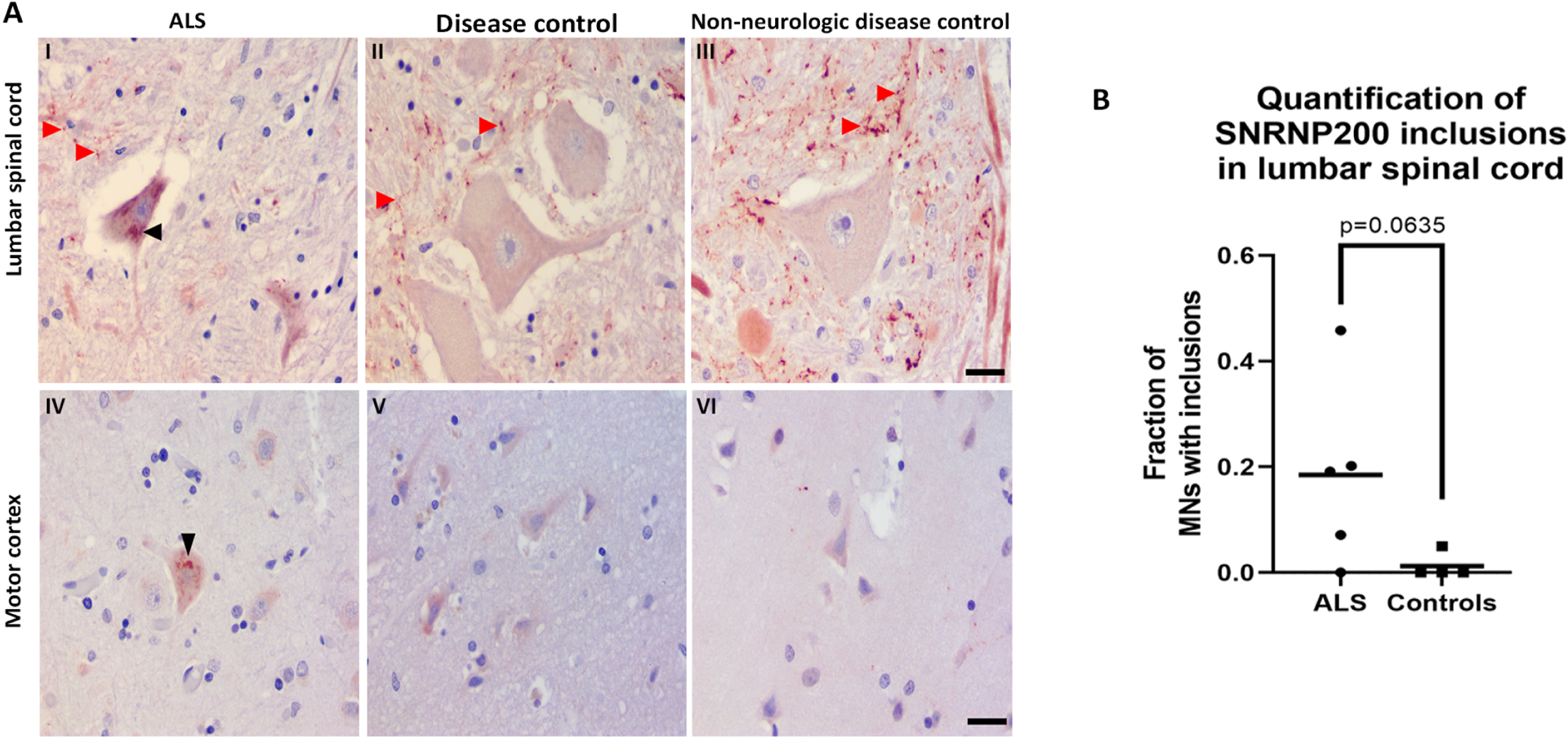
A) Immunostaining for SNRNP200 on post mortem lumbar spinal cord tissues from I) sporadic ALS, II) disease controls, and III) non-neurologic disease controls. Immunostaining for SNRNP200 on post mortem motor cortex tissues from IV) sporadic ALS, V) disease controls, and VI) non-neurologic disease controls. Scale bar, 25 μm for both spinal cord and motor cortex. Black arrowhead denotes SNRNP200 cytoplasmic inclusions. Red arrowheads denote the punctate staining pattern in the gray matter parenchyma. B) Quantification of SNRNP200 cytoplasmic inclusions in ALS and controls lumbar spinal cord sections. Mann Whitney U test was used to assess statistical differences between the two groups. At least 2 fields of view were quantified per case with quantification of a minimum of 11 motor neurons.
Discussion
In this study, we investigated the Caprin-1 interactome to provide new insight into the proteome of SGs. Prior SG proteome studies were performed primarily using G3BP1 as the bait protein. These studies usually employ either overexpression16 or the integration of tagged sequences20, 21, each of which may perturb SG protein stoichiometry or structure. While these studies have provided tremendous insight, characterization of SGs at physiological levels is lacking. Our study addresses this gap through the IP of endogenous Caprin-1, providing insight into the native proteome of SGs. In addition to the stressed interactome, we also examined the unstressed interactome of Caprin-1. Our pathway analyses demonstrated that the interactors in both the stressed and unstressed conditions annotated to similar pathways. There was a considerable overlap (166 proteins; Table S3) between the unstressed and stressed interactors suggesting pre-stress interactions exist between Caprin-1 and ribosomal, RNA binding, and spliceosome proteins. Similar pre-stress interactions have also been observed with the G3BP1 interactome20, 21. These pre-stress complexes may serve as seeds to facilitate rapid nucleation of SGs under stressed conditions, as previously proposed20, 21. Interestingly, Caprin-1 interacts with G3BP1 in both stressed and unstressed conditions, therefore, it is reasonable to posit similarities in their interactomes. However, investigation of the overlap between our stress dependent Caprin-1 interactor list and three published G3BP1 proteomics studies indicate minimal overlap (Figure S4; Table S6). These discrepancies may be due to the different cell lines and stress regimes used in the studies. Moreover, overexpression or proximity labeling approaches capture the most abundant or nearby interactors, respectively, which may also account for the minimal overlap found between our study and previously published G3BP1 interactomes. The interactions within RNP granules are dynamic and not all proteins interact within the same time scale during SG assembly and disassembly21. Interactions captured during acute SA stress (30 mins) as used in the BioID labeling approaches20 may be altered during the recovery phase, as performed in our study. In summary, probing the SG interactome at various time points will be required to capture the dynamism of SG-related PPIs. Moreover, the use of multiple bait proteins will be necessary to catalogue the complete SG proteome, as one study has recently reported22.
From our IP-MS results, we selected 12 candidate RBPs to further validate as SG proteins, and four proteins (SNRNP200, ANKHD1, TALIN-1, GEMIN5) co-localized with SA induced SGs in HeLa cells (Figures 4 and S5). Our IP-MS methodology used whole cell lysates, thus these data suggest that the other eight candidate RBPs may interact with Caprin-1 in diffuse cytoplasmic complexes. Future studies may modify our IP-MS protocol to fractionate the cell lysate similar to the protocol used to isolate SG cores16. Only SNRNP200 co-localized with SGs in neuronal-like cells (Figure 6), highlighting that the SG proteome may differ between cell types or in vivo. Although a prior study has demonstrated GEMIN5 to be associated with EIF4G labelled SGs in both SA and thermal stress conditions in HeLa cells47, we did not observe GEMIN5 co-localization with Caprin-1 following thermal stress nor in SH-SY5Y cells in response to any stress. Curiously, in sorbitol-stressed HeLa cells, we did observe GEMIN5 puncta upon stress which were independent of SGs. Given their size and proximity to SGs, we speculate that these puncta represent processing bodies. In summary, SNRNP200 was the only candidate that exhibited co-localization to SGs irrespective of the stressor and cell type used.
ALS and FTD are pathologically characterized by phosphorylated TDP-43 (pTDP-43) containing cytoplasmic inclusions in motor neurons and glia48. Previous studies have suggested that TDP-43 cytoplasmic aggregates co-localize with SG markers in ALS and FTD postmortem tissues49. However, the protein composition of cytoplasmic inclusions in patient derived tissues remains unclear. Our study identified SNRNP200 as a novel SG protein in our in vitro data, and also a component of some cytoplasmic inclusions within the motor neurons of ALS patient lumbar spinal cord and motor cortex. SNRNP200 is a member of the DEXH-box family of RNA helicases, is ubiquitously expressed in cells, and is a core component that plays a critical role in the formation of the spliceosome50. It is responsible for unwinding U4/U6 U5 snRNA and the pre-mRNA required for activation of the spliceosome51. To the best of our knowledge, there is no prior evidence of SNRNP200 localization to SGs or cytoplasmic inclusions in ALS. Abnormal accumulation of TDP-43 and FUS in cytoplasmic inclusions has been linked to RNA processing deficits in ALS52–54. Interestingly, SNRNP200 has also been identified as a significant interactor of proteins associated with ALS including members of the FET family (FUS, TAF15, and EWSR1) and Matrin 355. While the functional consequences of these interactions remained to be fully validated, prior evidence indicates that defects in nucleocytoplasmic transport leads to increased cytoplasmic accumulation of U2 SNRNPs and its RNA components in response to oxidative stress56. Furthermore, the cytoplasmic accumulation correlated with altered RNA splicing of downstream target genes56. Additionally, over-expression of FUS mutations in ALS patient fibroblasts is associated with cytoplasmic accumulation of U1 SNRNPs57. Collectively, these results suggest defective nucleocytoplasmic transport as well as ALS causing mutations can lead to cytoplasmic accumulation of spliceosomal components, possibly leading to RNA splicing deficits. Therefore, the SNRNP200 cytoplasmic inclusions that we observed may also occur as a result of these transport deficits. Whether these inclusions cause perturbations in alternative splicing and motor neuron deficits warrant further investigation. Interestingly, prior evidence indicates that knockdown of U1 SNRNPs in zebrafish results in truncated motor axons, suggesting that loss of SNRNP functions may cause motor neuron defects57. Additionally, it remains unclear if these inclusions are part of pTDP-43 or p62 inclusions that are characteristic of ALS, and thus future studies are required to determine co-localization with SNRNP200. Our data also demonstrated a lack of nuclear immunostaining for SNRNP200 in motor neurons in the lumbar spinal cord or motor cortex of all cases examined. While novel, this requires further validation in subsequent studies. Little is known about the expression and subcellular distribution of SNRNP200 in the adult human central nervous system, although a recent report indicates SNRNP200 gene expression is downregulated in ALS spinal cord relative to controls58. This study utilized two publicly available transcriptomics datasets in which RNA-seq was performed from whole tissue extracts of spinal cord, so it remains unclear which cells in the spinal cord exhibited reduced SNRNP200 expression. Nevertheless, our results are suggestive of a functional depletion of SNRNP200 protein, possibly from the nuclei of affected motor neurons in ALS patients, which may indicate splicing defects in ALS. However, it is also possible that low levels of SNRNP200 protein are present in adult motor neurons and therefore undetectable by IHC staining. This would explain the differences in nuclear SNRNP200 immunostaining between our cell culture experiments using proliferating cells versus non-dividing adult motor neurons in the spinal cord and motor cortex. Alternatively, the lack of nuclear staining could also be attributed to the use of two different SNRNP200 antibodies for IHC staining of human tissues and IF staining for HeLa cells where the antibody used for IHC failed to detect the epitope within the nuclei of human motor neurons.
In addition to some SNRNP200 labelling of cytoplasmic inclusions, we also observed a punctate staining pattern within the parenchyma of the lumbar spinal cord and along the periphery of motor neurons in both the ALS and control cases used in this study. Prior work in rats has demonstrated RNG105 (ortholog of human Caprin-1) granule structures that accumulate near post-synaptic terminals in the dendrites of hippocampal neurons27. Additionally, another study profiled the postsynaptic density protein 95 (PSD95) interactome in the anterior cingulate cortex and determined that SNRNP200 was among the list of identified interactors upregulated in bipolar patients relative to controls59. We hypothesize that the SNRNP200 puncta we observed may be localized to pre/post synaptic terminals. Our initial attempt to validate was unsuccessful due to difficulties in finding suitable antibodies compatible with post mortem human tissues. Further studies are required to address this hypothesis.
Taken together, we characterized the Caprin-1 protein interactome in both unstressed and stressed conditions. Our results highlighted that Caprin-1, a component not regulated by TDP-43 or G3BP130, interacts with components of the ribosome, RNA transport proteins, spliceosome components in unstressed conditions. Additionally, these associations persist and coalesce into SGs when subjected to various stressors. One Caprin-1 interactor, SNRNP200, is a novel SG component in vitro in neuronal and non-neuronal cells, and also exhibited cytoplasmic aggregation within the motor neurons of post-mortem ALS patient lumbar spinal cord and motor cortex. While the cause/consequence of these inclusions remains to be elucidated, these results highlight Caprin-1 SGs and its binding partners as potential contributors to altered RNA metabolism in neurodegenerative disease.
Supplementary Material
Table S1- List of antibodies used.
Table S2 - Table of all proteins identified from each IP experiment and their overlap from Figure 1D.
Table S3 - Protein lists corresponding to the Venn diagram in Figure 2A.
Table S4 - Protein lists corresponding to pathway analysis in Figure 2B.
Table S5 - Protein lists for pathway analysis, corresponding to in Figure 3A.
Table S6 - Protein lists corresponding to Venn diagrams in Figure S4A – S4D.
Table S7 - Protein lists corresponding to the Venn diagram comparing RNA binding proteins and significant high confidence Caprin-1 interactors from Figure 3C.
Figure S1 - Western blot membrane of the Caprin-1 and IgG immuno-precipitated (IP) fractions.
Figure S2 - Gene ontology analysis using DAVID on the unstressed interactors of Caprin-1.
Figure S3 - Known protein-protein interactions for SA-stress dependent Caprin-1 interactors.
Figure S4 - Comparison of stress-dependent Caprin-1 interactome with previously published SG interactomes.
Figure S5 - Shortlisted Caprin-1 interactors which did not co-localize with SGs in HeLa cells subjected to oxidative stress.
Figure S6 - Imaging of ANKHD1, TALIN-1 and GEMIN5 in SHSY-5Y cells upon thermal and osmotic stress.
Acknowledgements
We thank Patrick Dion and Jay Ross for important discussions and the CRCHUM Cellular Imaging platform. The authors thank the Target ALS Human Postmortem Tissue Core for access to human tissue samples. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding Information
We gratefully acknowledge funding from the ALS Canada/Brain Canada Arthur J. Hudson Translational Team grant (CVV, RB) and the Fein Family Foundation (RB, LV). We thank Ray and Amy Thurston for their generous funding contribution (PP), and research reported in this publication includes work performed in the mass spectrometry core supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. HS is supported by an FRQS Doctoral Studentship. CVV is an FRQS Senior Research Scholar.
References
- 1.Anderson P; Kedersha N, Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008, 33 (3), 141–50. [DOI] [PubMed] [Google Scholar]
- 2.Aulas A; Vande Velde C, Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front Cell Neurosci 2015, 9, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan JR; Parker R, Eukaryotic stress granules: the ins and outs of translation. Molecular cell 2009, 36 (6), 932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuechler ER; Budzyńska PM; Bernardini JP; Gsponer J; Mayor T, Distinct Features of Stress Granule Proteins Predict Localization in Membraneless Organelles. Journal of Molecular Biology 2020, 432 (7), 2349–2368. [DOI] [PubMed] [Google Scholar]
- 5.Guo L; Kim HJ; Wang H; Monaghan J; Freyermuth F; Sung JC; O’Donovan K; Fare CM; Diaz Z; Singh N; Zhang ZC; Coughlin M; Sweeny EA; DeSantis ME; Jackrel ME; Rodell CB; Burdick JA; King OD; Gitler AD; Lagier-Tourenne C; Pandey UB; Chook YM; Taylor JP; Shorter J, Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 2018, 173 (3), 677–692 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann JR; Gleixner AM; Mauna JC; Gomes E; DeChellis-Marks MR; Needham PG; Copley KE; Hurtle B; Portz B; Pyles NJ; Guo L; Calder CB; Wills ZP; Pandey UB; Kofler JK; Brodsky JL; Thathiah A; Shorter J; Donnelly CJ, RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 2019, 102 (2), 321–338 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molliex A; Temirov J; Lee J; Coughlin M; Kanagaraj AP; Kim HJ; Mittag T; Taylor JP, Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163 (1), 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YR; King OD; Shorter J; Gitler AD, Stress granules as crucibles of ALS pathogenesis. The Journal of cell biology 2013, 201 (3), 361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidibe H; Vande Velde C, RNA Granules and Their Role in Neurodegenerative Diseases. Adv Exp Med Biol 2019, 1203, 195–245. [DOI] [PubMed] [Google Scholar]
- 10.Baradaran-Heravi Y; Van Broeckhoven C; van der Zee J, Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiology of Disease 2020, 134, 104639. [DOI] [PubMed] [Google Scholar]
- 11.Mackenzie IR; Nicholson AM; Sarkar M; Messing J; Purice MD; Pottier C; Annu K; Baker M; Perkerson RB; Kurti A; Matchett BJ; Mittag T; Temirov J; Hsiung GR; Krieger C; Murray ME; Kato M; Fryer JD; Petrucelli L; Zinman L; Weintraub S; Mesulam M; Keith J; Zivkovic SA; Hirsch-Reinshagen V; Roos RP; Zuchner S; Graff-Radford NR; Petersen RC; Caselli RJ; Wszolek ZK; Finger E; Lippa C; Lacomis D; Stewart H; Dickson DW; Kim HJ; Rogaeva E; Bigio E; Boylan KB; Taylor JP; Rademakers R, TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 2017, 95 (4), 808–816 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami T; Qamar S; Lin JQ; Schierle GS; Rees E; Miyashita A; Costa AR; Dodd RB; Chan FT; Michel CH; Kronenberg-Versteeg D; Li Y; Yang SP; Wakutani Y; Meadows W; Ferry RR; Dong L; Tartaglia GG; Favrin G; Lin WL; Dickson DW; Zhen M; Ron D; Schmitt-Ulms G; Fraser PE; Shneider NA; Holt C; Vendruscolo M; Kaminski CF; St George-Hyslop P, ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 2015, 88 (4), 678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakkar N; Kovalik T; Lorenzini I; Spangler S; Lacoste A; Sponaugle K; Ferrante P; Argentinis E; Sattler R; Bowser R, Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis. Acta Neuropathol 2018, 135 (2), 227–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolozin B; Ivanov P, Stress granules and neurodegeneration. Nature reviews. Neuroscience 2019, 20 (11), 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isabelle M; Gagné J-P; Gallouzi I-E; Poirier GG, Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. Journal of Cell Science 2012, 125 (19), 4555–4566. [DOI] [PubMed] [Google Scholar]
- 16.Jain S; Wheeler JR; Walters RW; Agrawal A; Barsic A; Parker R, ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164 (3), 487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padrón A; Iwasaki S; Ingolia NT, Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Molecular cell 2019, 75 (4), 875–887.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khong A; Matheny T; Jain S; Mitchell SF; Wheeler JR; Parker R, The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Molecular cell 2017, 68 (4), 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boncella AE; Shattuck JE; Cascarina SM; Paul KR; Baer MH; Fomicheva A; Lamb AK; Ross ED, Composition-based prediction and rational manipulation of prion-like domain recruitment to stress granules. Proceedings of the National Academy of Sciences 2020, 117 (11), 5826–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youn JY; Dunham WH; Hong SJ; Knight JDR; Bashkurov M; Chen GI; Bagci H; Rathod B; MacLeod G; Eng SWM; Angers S; Morris Q; Fabian M; Cote JF; Gingras AC, High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Molecular cell 2018, 69 (3), 517–532 e11. [DOI] [PubMed] [Google Scholar]
- 21.Markmiller S; Soltanieh S; Server KL; Mak R; Jin W; Fang MY; Luo EC; Krach F; Yang D; Sen A; Fulzele A; Wozniak JM; Gonzalez DJ; Kankel MW; Gao FB; Bennett EJ; Lecuyer E; Yeo GW, Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172 (3), 590–604 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marmor-Kollet H; Siany A; Kedersha N; Knafo N; Rivkin N; Danino YM; Moens TG; Olender T; Sheban D; Cohen N; Dadosh T; Addadi Y; Ravid R; Eitan C; Toth Cohen B; Hofmann S; Riggs CL; Advani VM; Higginbottom A; Cooper-Knock J; Hanna JH; Merbl Y; Van Den Bosch L; Anderson P; Ivanov P; Geiger T; Hornstein E, Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Molecular cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grill B; Wilson GM; Zhang K-X; Wang B; Doyonnas R; Quadroni M; Schrader JW, Activation/Division of Lymphocytes Results in Increased Levels of Cytoplasmic Activation/Proliferation-Associated Protein-1: Prototype of a New Family of Proteins. The Journal of Immunology 2004, 172 (4), 2389–2400. [DOI] [PubMed] [Google Scholar]
- 24.Qiu YQ; Yang CW; Lee YZ; Yang RB; Lee CH; Hsu HY; Chang CC; Lee SJ, Targeting a ribonucleoprotein complex containing the caprin-1 protein and the c-Myc mRNA suppresses tumor growth in mice: an identification of a novel oncotarget. Oncotarget 2015, 6 (4), 2148–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong B; Hu H; Chen J; Cao S; Yu J; Xue J; Chen F; Cai Y; He H; Zhang L, Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother 2013, 67 (7), 629–36. [DOI] [PubMed] [Google Scholar]
- 26.Angenstein F; Evans AM; Ling SC; Settlage RE; Ficarro S; Carrero-Martinez FA; Shabanowitz J; Hunt DF; Greenough WT, Proteomic characterization of messenger ribonucleoprotein complexes bound to nontranslated or translated poly(A) mRNAs in the rat cerebral cortex. The Journal of biological chemistry 2005, 280 (8), 6496–503. [DOI] [PubMed] [Google Scholar]
- 27.Shiina N; Shinkura K; Tokunaga M, A Novel RNA-Binding Protein in Neuronal RNA Granules: Regulatory Machinery for Local Translation. The Journal of Neuroscience 2005, 25 (17), 4420–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon S; Xu Y; Wang B; David MD; Schubert P; Kennedy D; Schrader JW, Distinct Structural Features ofCaprin-1 Mediate Its Interaction with G3BP-1 and Its Induction of Phosphorylation of Eukaryotic Translation InitiationFactor 2α, Entry to Cytoplasmic Stress Granules, and Selective Interaction with a Subset of mRNAs. Molecular and Cellular Biology 2007, 27 (6), 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedersha N; Panas MD; Achorn CA; Lyons S; Tisdale S; Hickman T; Thomas M; Lieberman J; McInerney GM; Ivanov P; Anderson P, G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. The Journal of cell biology 2016, 212 (7), 845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aulas A; Caron G; Gkogkas CG; Mohamed NV; Destroismaisons L; Sonenberg N; Leclerc N; Parker JA; Vande Velde C, G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. The Journal of cell biology 2015, 209 (1), 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Encinas M; Iglesias M; Liu Y; Wang H; Muhaisen A; Cena V; Gallego C; Comella JX, Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 2000, 75 (3), 991–1003. [DOI] [PubMed] [Google Scholar]
- 32.Aulas A; Stabile S; Vande Velde C, Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Molecular Neurodegeneration 2012, 7 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aulas A; Fay MM; Szaflarski W; Kedersha N; Anderson P; Ivanov P, Methods to Classify Cytoplasmic Foci as Mammalian Stress Granules. J Vis Exp 2017, (123). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vu L; An J; Kovalik T; Gendron T; Petrucelli L; Bowser R, Cross-sectional and longitudinal measures of chitinase proteins in amyotrophic lateral sclerosis and expression of CHI3L1 in activated astrocytes. Journal of Neurology, Neurosurgery & Psychiatry 2020, 91 (4), 350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappsilber J; Mann M; Ishihama Y, Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007, 2 (8), 1896–906. [DOI] [PubMed] [Google Scholar]
- 36.Rappsilber J; Ishihama Y; Mann M, Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003, 75 (3), 663–70. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X; Smits AH; van Tilburg GB; Ovaa H; Huber W; Vermeulen M, Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nature protocols 2018, 13 (3), 530–550. [DOI] [PubMed] [Google Scholar]
- 38.Teo G; Liu G; Zhang J; Nesvizhskii AI; Gingras AC; Choi H, SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics 2014, 100, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang da W; Sherman BT; Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009, 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- 41.Choi H; Glatter T; Gstaiger M; Nesvizhskii AI, SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J Proteome Res 2012, 11 (4), 2619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y; Collins M; An J; Geiser R; Tegeler T; Tsantilas K; Garcia K; Pirrotte P; Bowser R, Immunoprecipitation and mass spectrometry defines an extensive RBM45 protein-protein interaction network. Brain research 2016, 1647, 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehringer A; Garcia-Mansfield K; Singh G; Bakkar N; Pirrotte P; Bowser R, ALS Associated Mutations in Matrin 3 Alter Protein-Protein Interactions and Impede mRNA Nuclear Export. Sci Rep 2017, 7 (1), 14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimball SR; Horetsky RL; Ron D; Jefferson LS; Harding HP, Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. American journal of physiology. Cell physiology 2003, 284 (2), C273–84. [DOI] [PubMed] [Google Scholar]
- 45.Aulas A; Fay MM; Lyons SM; Achorn CA; Kedersha N; Anderson P; Ivanov P, Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 2017, 130 (5), 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freibaum BD; Chitta RK; High AA; Taylor JP, Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. Journal of proteome research 2010, 9 (2), 1104–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Battle DJ; Kasim M; Wang J; Dreyfuss G, SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. The Journal of biological chemistry 2007, 282 (38), 27953–9. [DOI] [PubMed] [Google Scholar]
- 48.Neumann M; Sampathu DM; Kwong LK; Truax AC; Micsenyi MC; Chou TT; Bruce J; Schuck T; Grossman M; Clark CM; McCluskey LF; Miller BL; Masliah E; Mackenzie IR; Feldman H; Feiden W; Kretzschmar HA; Trojanowski JQ; Lee VM-Y, Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314 (5796), 130–133. [DOI] [PubMed] [Google Scholar]
- 49.Liu-Yesucevitz L; Bilgutay A; Zhang YJ; Vanderweyde T; Citro A; Mehta T; Zaarur N; McKee A; Bowser R; Sherman M; Petrucelli L; Wolozin B, Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PloS one 2010, 5 (10), e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruzickova S; Stanek D, Mutations in spliceosomal proteins and retina degeneration. RNA Biol 2017, 14 (5), 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agafonov DE; Kastner B; Dybkov O; Hofele RV; Liu WT; Urlaub H; Luhrmann R; Stark H, Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016, 351 (6280), 1416–20. [DOI] [PubMed] [Google Scholar]
- 52.Butti Z; Patten SA, RNA Dysregulation in Amyotrophic Lateral Sclerosis. Front Genet 2018, 9, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polymenidou M; Lagier-Tourenne C; Hutt KR; Bennett CF; Cleveland DW; Yeo GW, Misregulated RNA processing in amyotrophic lateral sclerosis. Brain research 2012, 1462, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polymenidou M; Lagier-Tourenne C; Hutt KR; Huelga SC; Moran J; Liang TY; Ling SC; Sun E; Wancewicz E; Mazur C; Kordasiewicz H; Sedaghat Y; Donohue JP; Shiue L; Bennett CF; Yeo GW; Cleveland DW, Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 2011, 14 (4), 459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi B; O’Connell JD; Yamazaki T; Gangopadhyay J; Gygi SP; Reed R, Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II machinery and contains multiple ALS/SMA-causative proteins. Scientific Reports 2018, 8 (1), 8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossi S; Rompietti V; Antonucci Y; Giovannini D; Scopa C; Scaricamazza S; Scardigli R; Cestra G; Serafino A; Carrì MT; D’Ambrosi N; Cozzolino M, UsnRNP trafficking is regulated by stress granules and compromised by mutant ALS proteins. Neurobiology of Disease 2020, 138, 104792. [DOI] [PubMed] [Google Scholar]
- 57.Yu Y; Chi B; Xia W; Gangopadhyay J; Yamazaki T; Winkelbauer-Hurt ME; Yin S; Eliasse Y; Adams E; Shaw CE; Reed R, U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res 2015, 43 (6), 3208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Cognata V; Gentile G; Aronica E; Cavallaro S, Splicing Players Are Differently Expressed in Sporadic Amyotrophic Lateral Sclerosis Molecular Clusters and Brain Regions. Cells 2020, 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Focking M; Dicker P; Lopez LM; Hryniewiecka M; Wynne K; English JA; Cagney G; Cotter DR, Proteomic analysis of the postsynaptic density implicates synaptic function and energy pathways in bipolar disorder. Transl Psychiatry 2016, 6 (11), e959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1- List of antibodies used.
Table S2 - Table of all proteins identified from each IP experiment and their overlap from Figure 1D.
Table S3 - Protein lists corresponding to the Venn diagram in Figure 2A.
Table S4 - Protein lists corresponding to pathway analysis in Figure 2B.
Table S5 - Protein lists for pathway analysis, corresponding to in Figure 3A.
Table S6 - Protein lists corresponding to Venn diagrams in Figure S4A – S4D.
Table S7 - Protein lists corresponding to the Venn diagram comparing RNA binding proteins and significant high confidence Caprin-1 interactors from Figure 3C.
Figure S1 - Western blot membrane of the Caprin-1 and IgG immuno-precipitated (IP) fractions.
Figure S2 - Gene ontology analysis using DAVID on the unstressed interactors of Caprin-1.
Figure S3 - Known protein-protein interactions for SA-stress dependent Caprin-1 interactors.
Figure S4 - Comparison of stress-dependent Caprin-1 interactome with previously published SG interactomes.
Figure S5 - Shortlisted Caprin-1 interactors which did not co-localize with SGs in HeLa cells subjected to oxidative stress.
Figure S6 - Imaging of ANKHD1, TALIN-1 and GEMIN5 in SHSY-5Y cells upon thermal and osmotic stress.


