Abstract
Phylogenetic analyses inferred from the nuc rDNA ITS1-5.8S-ITS2 (ITS) data set and the combined 2-locus data set [5.8S + nuc 28S rDNA (nLSU)] of taxa of Trechisporales around the world show that Sistotremastrum family forms a monophyletic lineage within Trechisporales. Bayesian evolutionary and divergence time analyses on two data sets of 5.8S and nLSU sequences indicate an ancient divergence of Sistotremastrum family from Hydnodontaceae during the Triassic period (224.25 Mya). Sistotremastrum family is characterized by resupinate and thin basidiomata, smooth, verruculose, or odontoid-semiporoid hymenophore, a monomitic hyphal structure, and generative hyphae bearing clamp connections, the presence of cystidia and hyphidia in some species, thin-walled, smooth, inamyloid, and acyanophilous basidiospores. In addition, four new species, namely, Trechispora dentata, Trechispora dimitiella, Trechispora fragilis, and Trechispora laevispora, are described and illustrated. In addition, three new combinations, namely, Brevicellicium daweishanense, Brevicellicium xanthum, and Sertulicium limonadense, are also proposed.
Keywords: Hydnodontaceae, phylogenetic analysis, Trechispora, taxonomy, wood-rotting fungi
Introduction
Trechisporales K.H. Larss. was established by Hibbett et al. (2007). Most species in this order are corticioid fungi with smooth, grandinioid, odontioid, or hydnoid hymenophores, and others are polypores. All species have a monomitic or dimitic hyphal system with generative hyphae bearing clamp connections, and many species have rhizomorphs (mycelial cords) (Larsson, 2007).
At present, there is only an acknowledged and a named family belonging to Trechisporales, i.e., Hydnodontaceae Jülich. Hydnodontaceae contains 11 genera now, namely, Brevicellicium K.H. Larss. and Hjortstam, Dextrinocystis Gilb. and M. Blackw., Fibrodontia Parmasto, Pteridomyces Jülich, Luellia K.H. Larss. and Hjortstam, Porpomyces Jülich, Scytinopogon Singer, Subulicystidium Parmasto, Suillosporium Pouzar, Trechispora P. Karst., and Tubulicium Oberw (Larsson, 2007; Spirin et al., 2021).
Trechispora is the genus type of Trechisporales and Hydnodontaceae. It is the largest genus in this order, with more than 50 accepted species (Meiras-Ottoni et al., 2021; Zhao and Zhao, 2021). Identification keys for Trechispora species recorded in China and Brazil have been provided by some fungal taxonomists (Chikowski et al., 2020; Meiras-Ottoni et al., 2021; Zong et al., 2021). Trechispora was typified with Trechispora onusta P. Karst. [= Trechispora hymenocystis (Berk. and Broome) K.H. Larss.] (Karsten, 1890). It is characterized by the resupinate basidiomata (a few species have stipitate, flabellate, and effused–reflexed basidiomata) with smooth grandinioid, odontioid, hydnoid, or poroid hymenophores, a monomitic or dimitic hyphal structure with clamped generative hyphae and smooth to verrucose or aculeate basidiospores (Larsson, 1992; Larsson et al., 2004). Most species in Trechispora are soil-dwelling (Larsson et al., 2004). One remarkable character is the presence of ampullate septa on the subicular and especially on some hyphae of the mycelial cords. Above all, ampullate septa are only known from Scytinopogon, Trechispora, and Porpomyces mucidus (Pers.) Jülich within Trechisporales (Furtado et al., 2021; Meiras-Ottoni et al., 2021).
Larsson (2007) used the term “Sistotremastrum family” for the first time to accommodate Sistotremastrum suecicum Litsch. ex J. Erikss. and Sistotremastrum niveocremeum [= Sertulicium niveocremeum (Höhn. and Litsch.) Spirin and K.H. Larss.]. Since then, “Sistotremastrum family” has been adopted by some taxonomists (Telleria et al., 2013; Liu et al., 2019). In this work, the phylogeny of Trechisporales is carried out based on combined 5.8S + nLSU sequences. In addition, Bayesian evolutionary and divergence time analyses are also carried out to indicate the divergence time of Trechisporales, Hydnodontaceae, and Sistotremastrum family. We outline the Sistotremastrum family and discuss the difference between Hydnodontaceae and Sistotremastrum family.
During investigations on the diversity of wood-rotting fungi, seven resupinate specimens were collected from China and Malaysia. Their morphology corresponds to the concept of Trechispora. To confirm their affinity, phylogenetic analyses based on the ITS sequences are carried out. Both morphological characteristics and molecular evidence demonstrate that these seven resupinate specimens represent the four new species of Trechispora.
In addition, we downloaded the type sequences of Trechispora daweishanensis C.L. Zhao, Trechispora xantha C.L. Zhao, and Sistotremastrum limonadense G. Gruhn and P. Alvarado from GenBank. We also studied the type specimens of T. daweishanensis and T. xantha. In conclusion, T. daweishanensis and T. xantha were transferred to Brevicellicium, while S. limonadense was transferred to Sertulicium.
Materials and Methods
Morphological Studies
Macro-morphological descriptions are based on field notes and dry herbarium specimens. Microscopic structures are photographed using a Nikon Digital Sight DS-L3 (Japan) or Leica ICC50 HD (Japan) camera. Microscopic measurements are made from slide preparations of dry tissues stained with 1% Phloxine B (C20H4Br4Cl2K2O5) (Fan et al., 2021). We also use other reagents, such as Cotton Blue and Melzer’s reagent following Dai’s (2010) study. Spore measurements include both with ornamentation and without ornamentation. The following abbreviations are used: KOH = 5% potassium hydroxide; CB = Cotton Blue; CB(+) = weakly cyanophilous; CB− = acyanophilous in Cotton Blue; IKI = Melzer’s reagent; IKI− = neither amyloid nor dextrinoid in Melzer’s reagent; L = mean spore length (arithmetic average of all spores including ornamentation); W = mean spore width (arithmetic average of all spores including ornamentation); Q = a variation in the L/W ratios between the specimens studied; L′ = mean spore length (arithmetic average of all spores excluding ornamentation); W′ = mean spore width (arithmetic average of all spores excluding ornamentation); Q′ = a variation in the L′/W′ ratios between the specimens studied; n (a/b) = the number of spores (a) measured from a given number of specimens (b). When presenting spore size variation, 5% of measurements are excluded from each end of the range and these values are given in parentheses. Special color terms follow Petersen (1996). Herbarium abbreviations follow Thiers (2018). The studied specimens are deposited at the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC), and the herbarium of Southwest Forestry University (SWFC).
DNA Extraction, Polymerase Chain Reaction Amplification, and Sequencing
Total genomic DNA from the dried specimens is extracted by a CTAB rapid plant genome extraction kit (Aidlab Biotechnologies Company Limited, Beijing, China) according to the manufacturer’s instructions with some modifications (Liu and Yuan, 2020; Du et al., 2021). The ITS regions are amplified with the primers ITS4 and ITS5 (White et al., 1990). The nLSU regions are amplified with the primers LR0R and LR7 (Vilgalys and Hester, 1990).
The polymerase chain reaction (PCR) procedure for ITS is as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 58°C for 45 s, and 72°C for 1 min, and a final extension of 72°C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 48°C for 1 min, and 72°C for 1.5 min, and a final extension of 72°C for 10 min (Zhao et al., 2015; Liu and Dai, 2021). The PCR products are purified and sequenced in the Beijing Genomics Institute, China, with the same primers used in the PCR reactions.
Phylogenetic Analyses
Two combined matrices, an ITS1-5.8S-ITS2 (ITS) data set and a two-gene data set (5.8S + nLSU), are used for phylogenetic analyses. Phylogenetic analyses are performed with maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI) methods in the ITS data set. Phylogenetic analyses are performed with ML and BI methods in the combined two-gene data set (5.8S + nLSU). Species and strain sequences are adopted partly from 28S- and ITS-based tree topologies established by Meiras-Ottoni et al. (2021) and Spirin et al. (2021). New sequences generated in this study, along with reference sequences retrieved from GenBank (Table 1), are aligned by MAFFT 7 (Katoh et al., 20191) using the “G-INS-i” strategy and manually adjusted in BioEdit (Hall, 1999). Unreliably aligned sections are removed before analyses and attempts are made to manually inspect and improve alignment. The data matrix is edited in Mesquite v3.70 software (Maddison and Maddison, 2021). The sequence alignment is deposited at TreeBase (submission ID 29141 and 29142). Sequences of Auricularia sp., Exidia recisa (Ditmar) Fr., and Exidiopsis calcea (Pers.) K. Wells are included in phylogenetic analyses. They belong to another order, Auriculariales Bromhead. The order is close to Trechisporales (Sulistyo et al., 2021). We add these three sequences in the combined two-gene data set (5.8S + nLSU) to demonstrate that Trechisporales forms a strongly supported sister clade to Auriculariales. Sequences of Hyphodontia floccosa (Bourdot and Galzin) J. Erikss. and Hyphodontia subalutacea (P. Karst.) J. Erikss. in Hymenochaetales Oberw. obtained from GenBank are used as outgroups to root trees in the 5.8S + nLSU analysis. Two sequences of Brevicellicium atlanticum Melo, Tellería, M. Dueñas and M.P. Martín obtained from GenBank are used as outgroups to root trees in the ITS analysis.
TABLE 1.
Information of taxa used in phylogenetic analyses.
| Species | Collector ID (herbarium ID) | GenBank accession no. |
|
| ITS | nLSU | ||
| Auricularia sp. | PBM 2295 | DQ200918 | AY634277 |
| Brevicellicium atlanticum | LISU 178566 (holotype) | NR_119820 | HE963774 |
| Brevicellicium atlanticum | LISU 178590 | HE963775 | HE963776 |
| Brevicellicium daweishanense | CLZhao 18255 (SWFC) | MW302338 | MW293867 |
| Brevicellicium daweishanense | CLZhao 17860 (SWFC, holotype) | MW302337 | MW293866 |
| Brevicellicium exile | MA-Fungi 26554 (holotype) | HE963777 | HE963778 |
| Brevicellicium olivascens | KHL 8571 (GB) | HE963792 | HE963793 |
| Brevicellicium olivascens | MA-Fungi 23496 | HE963787 | HE963788 |
| Brevicellicium xanthum | CLZhao 17781 (SWFC) | MW302340 | MW293869 |
| Brevicellicium xanthum | CLZhao 2632 (SWFC, holotype) | MW302339 | MW293868 |
| Dextrinocystis calamicola | He 5700 (BJFC) | MK204534 | MK204547 |
| Dextrinocystis calamicola | He 5693 (BJFC) | MK204533 | MK204546 |
| Exidia recisa | EL 15-98 (GB) | AF347112 | AF347112 |
| Exidiopsis calcea | MW 331 | AF291280 | AF291326 |
| Fibrodontia alba | TNM F24944 (holotype) | KC928274 | KC928275 |
| Fibrodontia gossypina | AFTOL-ID 599 | DQ249274 | AY646100 |
| Hyphodontia floccosa | Berglund 150-02 (GB) | DQ873618 | DQ873617 |
| Hyphodontia subalutacea | GEL2196 (KAS) | DQ340341 | DQ340362 |
| Porpomyces mucidus | Dai 12692 (BJFC) | KT157833 | KT157838 |
| Porpomyces submucidus | Cui 5183 (BJFC) | KU509521 | KT152145 |
| Pteridomyces galzinii | GB0150230 | LR694188 | LR694210 |
| Pteridomyces galzinii | Bernicchia 8122 (GB) | MN937559 | MN937559 |
| Scytinopogon angulisporus | TFB13611 | – | JQ684661 |
| Scytinopogon chartaceum | FLOR56185 | MK458775 | – |
| Scytinopogon pallescens | He 5192 (BJFC) | – | MK204553 |
| Sertulicium chilense | MA-Fungi 86368 (holotype) | HG315521 | – |
| Sertulicium granuliferum | He 3338 | MK204552 | MK204540 |
| Sertulicium jacksonii | Spirin 10425 (H) | MN987943 | MN987943 |
| Sertulicium lateclavigerum | LY 13467 | MG913225 | – |
| Sertulicium limonadense | LIP 0001683 (holotype) | MT180981 | MT180978 |
| Sertulicium limonadense | He 6276 (BJFC) | OK298489 * | OK298947 * |
| Sertulicium niveocremeum | KHL13727 (GB) | MN937563 | MN937563 |
| Sertulicium vernale | Soderholm 3886 (H, holotype) | MT002311 | MT664174 |
| Sistotremastrum aculeatum | Miettinen 10380.1 (H) | MN991176 | MW045423 |
| Sistotremastrum aculeatum | Cui 8401 (BJFC) | KX081133 | KX081184 |
| Sistotremastrum aculeocrepitans | KHL 16097 (URM) | MN937564 | MN937564 |
| Sistotremastrum confusum | KHL 16004 (URM) | MN937567 | MN937567 |
| Sistotremastrum denticulatum | Motato-Vásquez 894 (SP, holotype) | MN954694 | MW045424 |
| Sistotremastrum fibrillosum | LIP 0001413 (holotype) | NR_161047 | NG_075239 |
| Sistotremastrum fibrillosum s. l. | GUY13-119 (GG) | MG913224 | MG913210 |
| Sistotremastrum fibrillosum s. l. | KHL 16988 (MG) | MN937568 | MN937568 |
| Sistotremastrum geminum | Miettinen 14333 (MAN, holotype) | MN937568 | MN937568 |
| Sistotremastrum induratum | Spirin 8598 (H, holotype) | MT002324 | MT664173 |
| Sistotremastrum mendax | KHL 12022 (O, holotype) | MN937570 | MN937570 |
| Sistotremastrum rigidum | Motato-Vásquez 833 (SP, holotype) | MN954693 | MW045435 |
| Sistotremastrum suecicum | Kunttu 5959 (H) | MT075859 | MT002335 |
| Sistotremastrum suecicum | Miettinen 14550.1 (H) | MT075860 | MT002336 |
| Sistotremastrum suecicum | KHL 11849 (GB) | MN937571 | MN937571 |
| Sistotremastrum vigilans | Fonneland 2011-78 (O, holotype) | MN937572 | MN937572 |
| Sistotremastrum vigilans | Spirin 8778 (H) | MN991182 | MN991182 |
| Subulicystidium tropicum | He 3968 (BJFC) | MK204531 | MK204544 |
| Suillosporium cystidiatum | Spirin 3830 (H) | MN937573 | MN937573 |
| Trechispora alnicola | AFTOL-ID 665 | DQ411529 | AY635768 |
| Trechispora araneosa | KHL8570 (GB) | AF347084 | AF347084 |
| Trechispora bambusicola | CLZhao 3302 (SWFC) | MW544021 | MW520171 |
| Trechispora bispora | CBS 142.63 (holotype) | MH858241 | MH869842 |
| Trechispora cohaerens | TU 110332 | UDB008249 | – |
| Trechispora cohaerens | TU 115568 | UDB016421 | – |
| Trechispora confinis | KHL11064 (GB) | AF347081 | AF347081 |
| Trechispora copiosa | AMO456 | MN701019 | MN687976 |
| Trechispora copiosa | AMO422 (holotype) | MN701013 | MN687971 |
| Trechispora cyatheae | FR-0219442 | UDB024014 | UDB024014 |
| Trechispora cyatheae | FR-0219443 (holotype) | UDB024015 | UDB024015 |
| Trechispora dentata | Dai 22565 (BJFC) | OK298491 * | OM049408 * |
| Trechispora dimitiella | Dai 21931 (BJFC) | OK298492 * | OK298948 * |
| Trechispora dimitiella | Dai 21181 (BJFC) | OK298493 * | OK298949 * |
| Trechispora echinocristallina | FR-0219445 (holotype) | UDB024018 | UDB024019 |
| Trechispora echinocristallina | FR-0219448 | UDB024022 | – |
| Trechispora echinospora | MA-Fungi 82485 (holotype) | JX392845 | JX392846 |
| Trechispora farinacea | KHL 8793 (GB) | AF347089 | AF347089 |
| Trechispora farinacea | KHL 8451 (GB) | AF347082 | AF347082 |
| Trechispora fimbriata | CLZhao 7969 (SWFC) | MW544024 | MW520174 |
| Trechispora fimbriata | CLZhao 4154 (SWFC, holotype) | MW544023 | MW520173 |
| Trechispora fissurata | CLZhao 4571 (SWFC, holotype) | MW544027 | MW520177 |
| Trechispora fissurata | CLZhao 995 (SWFC) | MW544026 | MW520176 |
| Trechispora fragilis | Dai 20535 (BJFC) | OK298494 * | OK298950 * |
| Trechispora gelatinosa | AMO1139 (holotype) | MN701021 | MN687978 |
| Trechispora gelatinosa | AMO824 | MN701020 | MN687977 |
| Trechispora havencampii | SFSU DED8300 (holotype) | NR_154418 | NG_059993 |
| Trechispora hymenocystis | TL 11112 (holotype) | UDB000778 | UDB000778 |
| Trechispora hymenocystis | KHL 8795 (GB) | AF347090 | AF347090 |
| Trechispora incisa | GB0090648 | KU747095 | KU747087 |
| Trechispora incisa | GB0090521 | KU747093 | – |
| Trechispora kavinioides | KGN 981002 (GB) | AF347086 | AF347086 |
| Trechispora laevispora | Dai 21655 (BJFC) | OK298495 * | OM108710 |
| Trechispora minispora | MEXU 28300 (holotype) | MK328886 | MK328894 |
| Trechispora minispora | MEXU 28301 | MK328886 | MK328895 |
| Trechispora mollis | URM 85884 (holotype) | MK514945 | MH280003 |
| Trechispora mollusca | DLL2011-186 (CFMR) | KJ140681 | – |
| Trechispora mollusca | DLL2010-077 (CFMR) | JQ673209 | – |
| Trechispora nivea | GB0102694 | KU747096 | AY586720 |
| Trechispora nivea | MA-Fungi 74044 | JX392832 | JX392833 |
| Trechispora papillosa | AMO713 | MN701022 | MN687979 |
| Trechispora papillosa | AMO795 (holotype) | MN701023 | MN687981 |
| Trechispora regularis | KHL10881 (GB) | AF347087 | AF347087 |
| Trechispora rigida | URM 85754 | MT406381 | MH279999 |
| Trechispora sp. | AMO799 | MN701008 | MN687969 |
| Trechispora sp. | AMO440 | MN701006 | MN687967 |
| Trechispora sp. | KHL16968 (O) | MH290763 | MH290763 |
| Trechispora sp. | Dai 22173 (BJFC) | OK298496 * | OK298951 * |
| Trechispora sp. | Dai 22174 (BJFC) | OK298497 * | OK298952 * |
| Trechispora stevensonii | TU 115499 | UDB016467 | UDB016467 |
| Trechispora stevensonii | MA-Fungi 70669 | JX392841 | JX392842 |
| Trechispora subsphaerospora | KHL 8511 (GB) | AF347080 | AF347080 |
| Trechispora termitophila | AMO396 (holotype) | MN701025 | MN687983 |
| Trechispora termitophila | AMO390 | MN701024 | MN687982 |
| Trechispora torrendii | URM 85886 (holotype) | MK515148 | MH280004 |
| Tubulicium raphidisporum | He 3191 (BJFC) | MK204537 | MK204545 |
*Newly generated sequences for this study. New species and new combinations or putatively new species are in bold.
The MP analysis is applied to the ITS data set sequences. Approaches to phylogenetic analysis follow Liu and Dai (2021), and the tree construction procedure is performed in PAUP* version 4.0 beta 10 software (Swofford, 2002). All characters are equally weighted, and gaps are treated as missing data. Trees are inferred using the heuristic search option with tree bisection and reconnection (TBR) branch swapping, and 1,000 random sequence additions maxtrees are set to 5,000, branches of zero length are collapsed, and all parsimonious trees are saved. Clade robustness is assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) are calculated for each maximum parsimonious tree (MPT) generated.
Maximum likelihood research is conducted with RAxML-HPC v. 8.2.3 (Stamatakis, 2014) and RAxML-HPC through the CIPRES Science Gateway (Miller et al., 20092). Statistical support values (BS) are obtained using nonparametric bootstrapping with 1,000 replicates. The BI analysis is performed with MrBayes 3.2.7a (Ronquist and Huelsenbeck, 2003). Four Markov chains are run for two runs from random starting trees for 4 million generations (ITS) and 8 million generations (5.8S + nLSU) until the split deviation frequency value reaches <0.01, and trees are sampled every 1,000 generations. The first 25% of the sampled trees are discarded as burn-in, and the remaining ones are used to reconstruct a majority rule consensus tree and to calculate Bayesian posterior probabilities (BPP) of the clades.
The optimal substitution models for the combined data set are determined using the Akaike information criterion (AIC) implemented in MrModeltest 2.3 (Posada and Crandall, 1998; Nylander, 2004) after scoring 24 models of evolution by PAUP* version 4.0 beta 10 software (Swofford, 2002). The selected model applied in the BI analyses and ML analyses is the model GTR + I + G.
Branches that received BT support for ML (BS), MP (BP), and BPP greater than 65% (BS), 70% (BP), and 0.9 (BPP) are considered as significantly supported, respectively. Additionally, the ML analysis results in the best tree, and only the ML tree is presented along with the support values from the MP and BI analyses. FigTree v1.4.4 (Rambaut, 2018) is used to visualize the resulting tree.
Divergence Time Estimation
Divergence time is estimated with the BEAST v2.6.5 software package (Bouckaert et al., 2019) with 5.8S and nLSU sequences representing all main lineages in Basidiomycota (Table 2). Sequences of the species are adopted partly from the topology established by Wang et al. (2021). Neurospora crassa Shear and B.O. Dodge from Ascomycota are designated as outgroup taxon (Wang et al., 2021). A BEAST XML input file is generated with BEATUti v2. The estimation of rates of evolutionary changes at nuclear acids is using ModelTest 3.7 with the GTR substitution model (Posada and Crandall, 1998). A log-normal distribution is employed for molecular clock analysis (Drummond and Rambaut, 2007). A Yule speciation model is selected as prior assuming a constant speciation rate per lineage. Three fossil fungi, viz. Paleopyrenomycites devonicus (Taylor et al., 1999, 2005), Archaeomarasmius leggetti (Hibbett et al., 1995, 1997), and Quatsinoporites cranhamii (Smith et al., 2004; Berbee and Taylor, 2010) are taken from Wang et al.’s (2021) study. An XML file is conducted for 10 billion generations, producing log files and trees files. The log file is analyzed in Tracer 1,3 and a maximum clade credibility (MCC) tree is interpreted in TreeAnnotator by trees file, removing the first 10% of the sampled trees as burn-in, and viewed in FigTree v1.4.2.
TABLE 2.
Information of taxa used in molecular clock analysis.
| Species | Specimen no. | ITS | nLSU |
| Amylocorticium cebennense | HHB-2808 | GU187505 | GU187561 |
| Anomoloma myceliosum | MJL-4413 | GU187500 | GU187559 |
| Athelia arachnoidea | CBS 418.72 | GU187504 | GU187557 |
| Auricularia heimuer | Xiaoheimao | LT716074 | KY418890 |
| Auricularia sp. | PBM 2295 | DQ200918 | AY634277 |
| Australovuilleminia coccinea | MG75 | HM046875 | HM046931 |
| Boletopsis leucomelaena | AFTOL-ID 1527 | DQ484064 | DQ154112 |
| Bondarzewia montana | AFTOL-ID 452 | DQ200923 | DQ234539 |
| Brevicellicium atlanticum | LISU 178566 | NR_119820 | HE963774 |
| Brevicellicium atlanticum | LISU 178590 | HE963775 | HE963776 |
| Brevicellicium daweishanense | CLZhao 17860 | MW302337 | MW293866 |
| Brevicellicium daweishanense | CLZhao 18255 | MW302338 | MW293867 |
| Brevicellicium exile | MA-Fungi 26554 | HE963777 | HE963778 |
| Brevicellicium olivascens | KHL8571 | HE963792 | HE963793 |
| Brevicellicium olivascens | MA-Fungi 23496 | HE963787 | HE963788 |
| Brevicellicium xanthum | CLZhao 17781 | MW302340 | MW293869 |
| Brevicellicium xanthum | CLZhao 2632 | MW302339 | MW293868 |
| Bridgeoporus sinensis | Cui 10013 | KY131832 | KY131891 |
| Calocera cornea | AFTOL-ID 438 | AY789083 | AY701526 |
| Coltricia perennis | Cui 10319 | KU360687 | KU360653 |
| Coltriciella dependens | Dai 10944 | KY693737 | KY693757 |
| Corticium roseum | MG43 | GU590877 | AY463401 |
| Craterocolla cerasi | TUB 020203 | KF061265 | KF061265 |
| Cryptococcus humicola | AFTOL-ID 1552 | DQ645516 | DQ645514 |
| Dacryopinax spathularia | AFTOL-ID 454 | AY854070 | AY701525 |
| Dextrinocystis calamicola | He 5700 | MK204534 | MK204547 |
| Dextrinocystis calamicola | He5693 | MK204533 | MK204546 |
| Exidia recisa | EL 15-98 | AF347112 | AF347112 |
| Exidiopsis calcea | MW 331 | AF291280 | AF291326 |
| Fasciodontia brasiliensis | MSK-F 7245a | MK575201 | MK598734 |
| Fasciodontia bugellensis | MSK-F 5548 | MK575204 | MK598736 |
| Fibrodontia alba | TNMF 24944 | KC928274 | KC928275 |
| Fibrodontia gossypina | AFTOL-ID 599 | DQ249274 | AY646100 |
| Fomitiporia hartigii | MUCL 53551 | JX093789 | JX093833 |
| Fomitiporia mediterranea | AFTOL 688 | AY854080 | AY684157 |
| Gloeophyllum sepiarium | Wilcox-3BB | HM536091 | HM536061 |
| Gloeophyllum striatum | ARIZAN 027866 | HM536092 | HM536063 |
| Grifola frondosa | AFTOL-ID 701 | AY854084 | AY629318 |
| Gymnopilus picreus | ZRL2015011 | LT716066 | KY418882 |
| Hymenochaete rubiginosa | He1049 | JQ716407 | JQ279667 |
| Hyphodontia densispora | LWZ 20170908-5 | MT319426 | MT319160 |
| Hyphodontia zhixiangii | LWZ 20170818-13 | MT319420 | MT319151 |
| Jaapia argillacea | CBS 252.74 | GU187524 | GU187581 |
| Gomphidius roseus | MB 95-038 | DQ534570 | DQ534669 |
| Kneiffiella barba-jovis | KHL 11730 | DQ873609 | DQ873610 |
| Kneiffiella subalutacea | LWZ 20170816-9 | MT319407 | MT319139 |
| Lepiota cristata | ZRL20151133 | LT716026 | KY418841 |
| Leptosporomyces raunkiaeri | HHB-7628 | GU187528 | GU187588 |
| Leucophellinus hobsonii | Cui 6468 | KT203288 | KT203309 |
| Lyomyces macrosporus | LWZ20170817-2 | MT319459 | MT319194 |
| Multiclavula mucida | AFTOL-ID 1130 | DQ521417 | AY885163 |
| Neoantrodiella gypsea | Cui 10372 | KT203290 | MT319396 |
| Neoantrodiella thujae | Dai 5065 | KT203293 | MT319397 |
| Neurospora crassa | OR74A | HQ271348 | AF286411 |
| Nigrofomes melanoporus | JV 1704/39 | MF629835 | MF629831 |
| Nigrofomes sinomelanoporus | Cui 5277 | MF629836 | MT319398 |
| Porodaedalea chinensis | Cui 10252 | KX673606 | MH152358 |
| Porpomyces mucidus | Dai 12692 | KT157833 | KT157838 |
| Porpomyces submucidus | Cui 5183 | KU509521 | KT152145 |
| Pteridomyces galzinii | GB0150230 | LR694188 | LR694210 |
| Pteridomyces galzinii | Bernicchia8122 | MN937559 | MN937559 |
| Ramaria rubella | AFTOL-ID 724 | AY854078 | AY645057 |
| Rigidoporus corticola | ZRL20151459 | LT716075 | KY418899 |
| Rigidoporus ginkgonis | Cui 5555 | KT203295 | KT203316 |
| Scytinopogon angulisporus | TFB13611 | – | JQ684661 |
| Scytinopogon pallescens | He 5192 | – | MK204553 |
| Sertulicium chilense | MA-Fungi 86368 | HG315521 | – |
| Sertulicium granuliferum | He 3338 | MK204552 | MK204540 |
| Sertulicium jacksonii | Spirin 10425 | MN987943 | MN987943 |
| Sertulicium lateclavigerum | LY 13467 | MG913225 | – |
| Sertulicium limonadense | LIP 0001683 | MT180981 | MT180978 |
| Sertulicium niveocremeum | KHL13727 | MN937563 | MN937563 |
| Sertulicium vernale | Soderholm 3886 | MT002311 | MT664174 |
| Sistotremastrum aculeatum | Cui 8401 | KX081133 | KX081184 |
| Sistotremastrum aculeatum | Miettinen 10380.1 | MN991176 | MW045423 |
| Sistotremastrum aculeocrepitans | KHL 16097 | MN937564 | MN937564 |
| Sistotremastrum confusum | KHL 16004 | MN937567 | MN937567 |
| Sistotremastrum denticulatum | MV894 | MN954694 | MW045424 |
| Sistotremastrum fibrillosum | LIP 0001413 | NR_161047 | NG_075239 |
| Sistotremastrum fibrillosum s. l. | GUY13-119 | MG913224 | MG913210 |
| Sistotremastrum fibrillosum s. l. | KHL 16988 | MN937568 | MN937568 |
| Sistotremastrum geminum | Miettinen 14333 | MN937568 | MN937568 |
| Sistotremastrum induratum | Spirin 8598 | MT002324 | MT664173 |
| Sistotremastrum mendax | KHL12022 | MN937570 | MN937570 |
| Sistotremastrum rigidum | MV833 | MN954693 | MW045435 |
| Sistotremastrum suecicum | Kunttu 5959 | MT075859 | MT002335 |
| Sistotremastrum suecicum | Miettinen 14550.1 | MT075860 | MT002336 |
| Sistotremastrum suecicum | KHL 11849 (GB) | MN937571 | MN937571 |
| Sistotremastrum vigilans | Fonneland 2011-78 | MN937572 | MN937572 |
| Sistotremastrum vigilans | Spirin 8778 | MN991182 | MN991182 |
| Subulicystidium tropicum | He3968 | MK204531 | MK204544 |
| Suillosporium cystidiatum | VS3830 | MN937573 | MN937573 |
| Suillus pictus | AFTOL 717 | AY854069 | AY684154 |
| Thelephora ganbajun | ZRL20151295 | LT716082 | KY418908 |
| Trametes versicolor | ZRL20151477 | LT716079 | KY418903 |
| Trechispora hymenocystis | KHL8795 | AF347090 | AF347090 |
| Tremellodendron sp. | PBM2324 | DQ411526 | – |
| Tubulicium raphidisporum | He 3191 | MK204537 | MK204545 |
| Ustilago maydis | AFTOL 505 | AY854090 | AF453938 |
| Xylodon heterocystidiatus | LWZ 20171015-33 | MT319518 | MT319264 |
Results
Phylogenetic Analyses
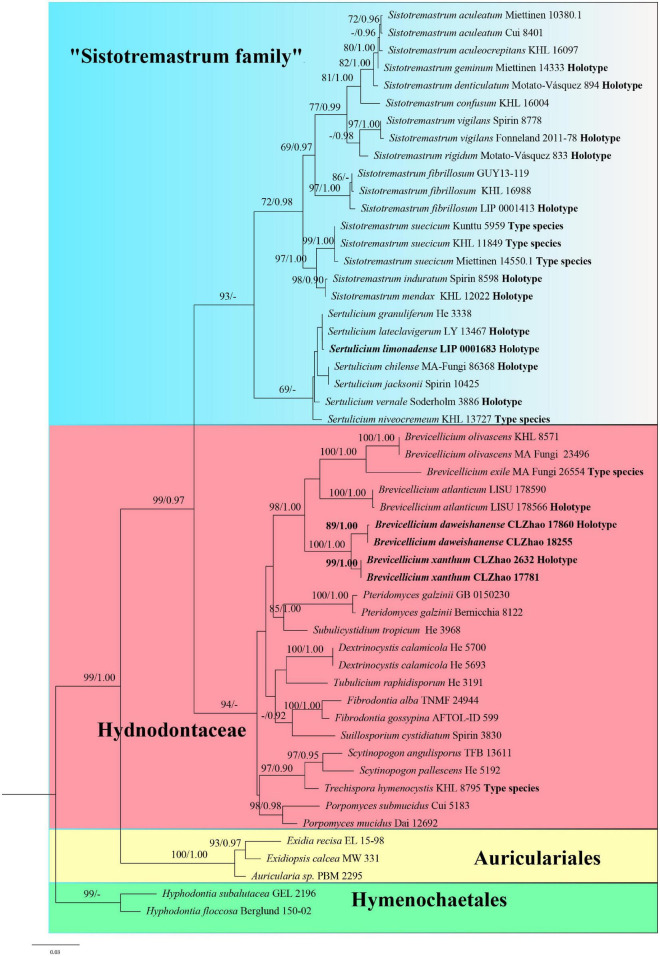
The concatenated 5.8S + nLSU data set contains 50 5.8S and 50 nLSU sequences from 52 fungal specimens representing 35 taxa in Trechisporales. The data set has an aligned length of 1,528 characters, of which 1,126 are constant, 89 are variable but parsimony-uninformative, and 313 are parsimony-informative. The average standard deviation (SD) of split frequencies is 0.005271 (BI). Three new combinations, namely, Brevicellicium daweishanense, Brevicellicium xanthum, and Sertulicium limonadense, are proposed based on the examination of type materials and phylogenetic analyses of type sequences (Figure 1).
FIGURE 1.
Phylogeny of Trechisporales generated by maximum likelihood (ML) analyses based on combined 5.8S + nLSU sequences. Branches are labelled with ML bootstrap (BT) >65%, and Bayesian posterior probabilities (BPP) >0.90, respectively. New combinations, the sequence origin from holotype and the type status of the species in the genus are indicated in bold.
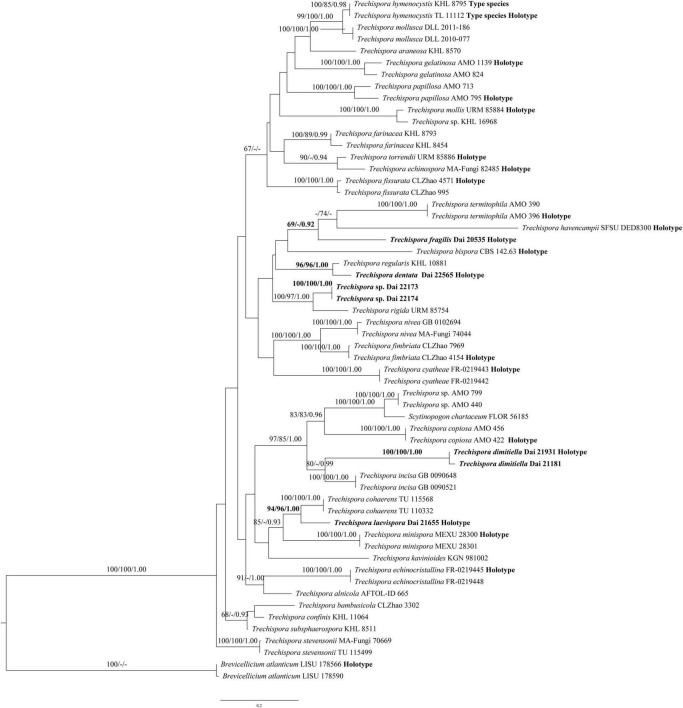
The ITS data set contains sequences from 58 fungal specimens representing 36 Trechispora taxa (4 new species and another 32 Trechispora taxa). The data set has an aligned length of 753 characters, of which 284 are constant, 72 are variable but parsimony-uninformative, and 397 are parsimony-informative. MP analysis yields 13 equally parsimonious trees (TL = 2,318, CI = 0.398, RI = 0.638, RC = 0.254, and HI = 0.602). The average SD of split frequencies in BI analyses is 0.006959 (BI). The phylogenetic tree (Figure 2) reveals four new and independent lineages represented by our specimens, indicating that they are phylogenetically distinct from the species currently known in the genus. In addition, another taxon (Dai 22173 and Dai 22174) is treated as Trechispora sp.
FIGURE 2.
Phylogeny of Trechispora generated by ML analyses based on combined ITS sequences. Branches are labelled with ML BT >65%, Parsimony Bootstrap Proportions >70%, and BPP >0.90, respectively. Putatively new species, the sequence origin from the holotype, and the type status of the species in the genus are indicated in bold.
The combined data set for the molecular clock analysis includes 100 collections, of which 47 belonged to Trechisporales. This data set results in a concatenated alignment of 1,588 characters with GTR as the best-fit evolutionary model. The MCC tree is used to study divergence time. The tree shows that Trechisporales occurs in a mean stem age of 270.85 Mya with a 95% highest posterior density (HPD) of 234.1–307.93 Mya (Figure 3). The tree also shows that the Sistotremastrum family and Hydnodontaceae occur in a mean stem age of 224.25 Mya [posterior probabilities (PP) = 0.8] with a 95% HPD of 182.47–266.75 Mya.
FIGURE 3.
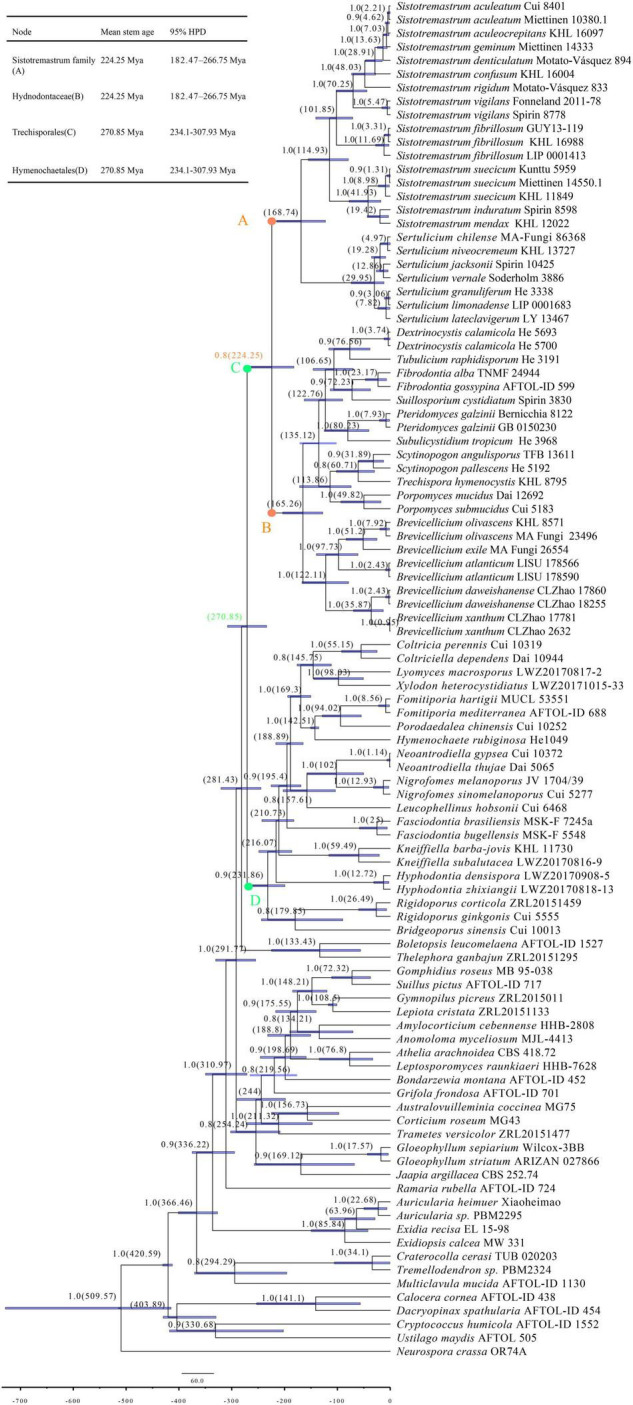
Maximum clade credibility (MCC) chronogram and estimated divergence times of all main lineages in Basidiomycota inferred from the combined data set of 5.8S and LSU regions. The estimated divergence times of 95% highest posterior density (HPD) for all clades are indicated as node bars. The colored dots refer to the positions of the mean stem age of Sistotremastrum family, Hydnodontaceae, Trechisporales, and Hymenochaetales. The BPP above 0.8 and the mean divergence times of clades are labelled above and below the branches, respectively, at the nodes.
Taxonomy
Sistotremastrum family
“Type genus”: Sistotremastrum J. Erikss.
Habitat: It grows on rotten angiosperm and gymnosperm wood.
Basidioma are resupinate, thin, pruinose, or waxy. Hymenophores are smooth, verruculose, or odontioid-semiporoid. The hyphal structure is monomitic; generative hyphae bear clamp connections, CB(+). Cystidia and hyphidia are present in some species. Basidia are clavate or cylindrical, often with a median constriction, mostly with 2–4 or 4–6 sterigmata, and rarely with 6–8 sterigmata. Basidiospores are narrowly ellipsoid, ovoid, or cylindrical, thin-walled (but the wall is distinct), smooth, inamyloid, and acyanophilous.
Notes: Sistotremastrum family accommodates the genera Sistotremastrum and Sertulicium in the order Trechisporales based on its distinct lineage in the phylogenetic analysis. The combined phylogeny of two-gene data (Figure 1) demonstrates that Sistotremastrum family forms a supported sister clade to Hydnodontaceae. Basidia of most species in the Sistotremastrum family have more than four sterigmata, and basidiospores are smooth, while basidia of species in Hydnodontaceae have four sterigmata and their basidiospores are smooth to verrucose or aculeate. In addition, ampullate septa are only present in Scytinopogon, Trechispora, and P. mucidus in Hydnodontaceae.
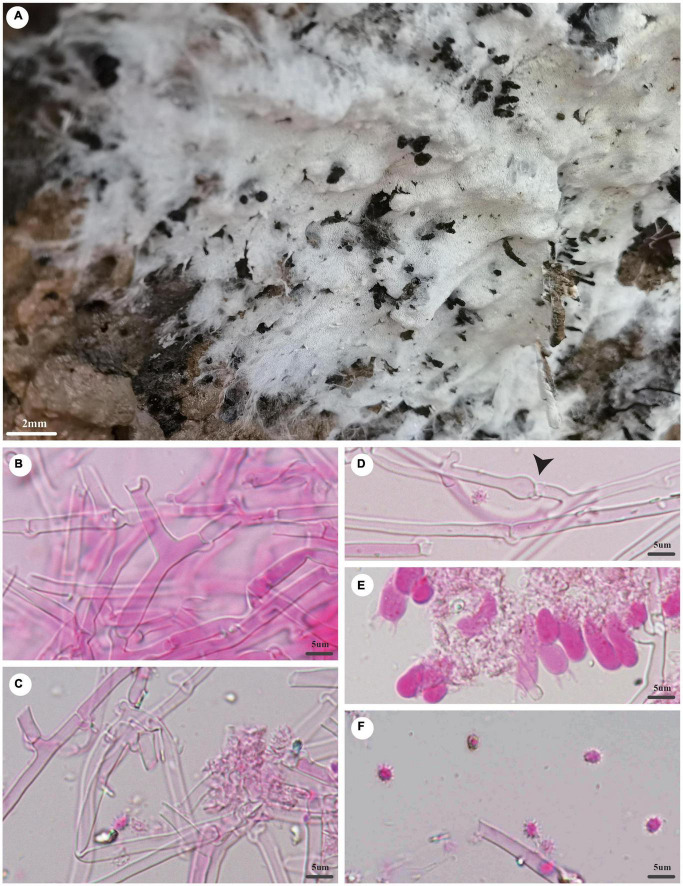
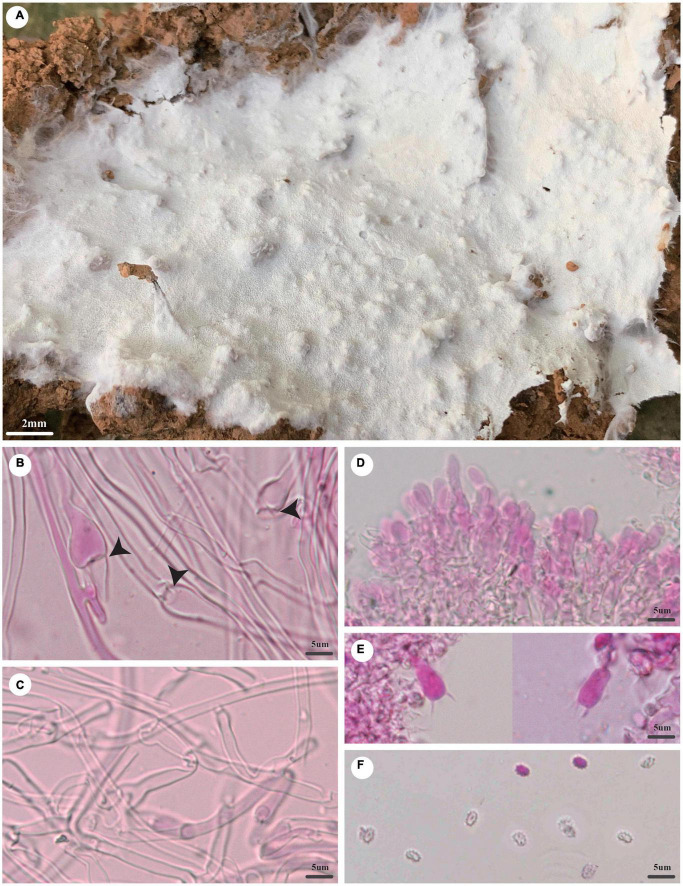
Trechispora dentata Z.B. Liu and Yuan Yuan, sp. November Figure 4
FIGURE 4.
Trechispora dentata (holotype, Dai 22565). (A) A basidioma, (B) hyphae from subiculum, (C) hyphae from trama, (D) hyphae with ampullate septa (black arrow), (E) basidia and basidioles, and (F) basidiospores. Photo by Ya-Ping Lian and Zhan-Bo Liu.
MycoBank number: MB 842865.
Type: China, Yunnan province, Sipsongpanna, Mengla County, XiShuangBanNa Tropical Botanical Garden, on soil, in southwestern China, ca. E 101° 25′, N 21° 41′, alt. 570 m. The vegetation is a natural tropical forest. 4 July 2021, Y.C. Dai 22565 (holotype BJFC 037139).
Etymology: Dentata (Lat.): It refers to the species having a dentate hymenophore.
Basidioma: They are annual, resupinate, soft when fresh, fragile when dry, easily separable from the substratum, up to 2.5-cm long, 2-cm wide, and less than 1-mm thick at the center; hymenial surface irpicoid, white when fresh, becoming cream (4A2/3) when dry; margin indistinct and fimbriate, mycelial cords absent; pores or aculei 3–4/mm; hymenophore lacerate to dentate; subiculum very thin to almost absent; tubes or aculei concolorous with a hymenial surface, less than 1 mm long.
Hyphal structure: Hyphal system is monomitic; generative hyphae bear clamp connections; ampullate septa occasionally present in subiculum and trama, up to 5-μm wide; all hyphae IKI−, CB− are unchanged in KOH; rhomboidal calcium oxalate crystals are scattered.
Subiculum: Generative hyphae hyaline, thin- to thick-walled, frequently branched, loosely interwoven, 2–4 μm in diameter.
Tubes or aculei: Generative hyphae in trama hyaline, thin- to thick-walled, frequently branched, loosely interwoven, 2–3 μm in diameter; cystidia and cystidioles are absent; basidia are clavate or barrel-shaped, hyaline, bearing four sterigmata and a basal clamp connection, 10–15 × 4–5 μm; basidioles are similar to basidia in shape but slightly shorter.
Basidiospores: They are ellipsoid, hyaline, thick-walled, aculeate, occasionally with one guttule, IKI−, CB−, (4−)4.1–5 × (3−)3.2–4(−4.1) μm (including ornamentation), L = 4.46 μm, W = 3.66 μm, Q = 1.22 (n = 60/1); (2.2−)2.6–3.7(−3.8) × 2–2.5 μm (excluding ornamentation), L′ = 3.17 μm, W′ = 2.23 μm, and Q′ = 1.42 (n = 60/1).
Notes: T. dentata was discovered in the Yunnan Province of China. Phylogenetically, T. dentata is close to Trechispora regularis (Murrill) Liberta with strong support (96% BS, 96% BP, 1.00 BPP; Figure 2). However, T. regularis is strictly poroid (Liberta, 1973), and basidiospores of T. dentata are smaller than that of T. regularis [4.1–5 × 3.2–4 μm vs. 4–5.5 × 3.5–5 μm in T. regularis (including ornamentation); Liberta, 1973].
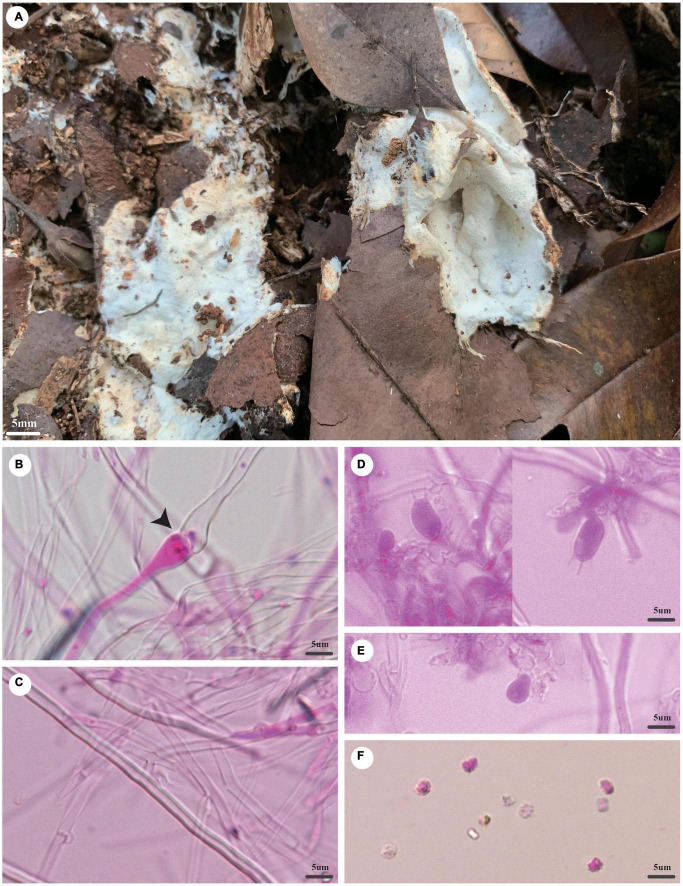
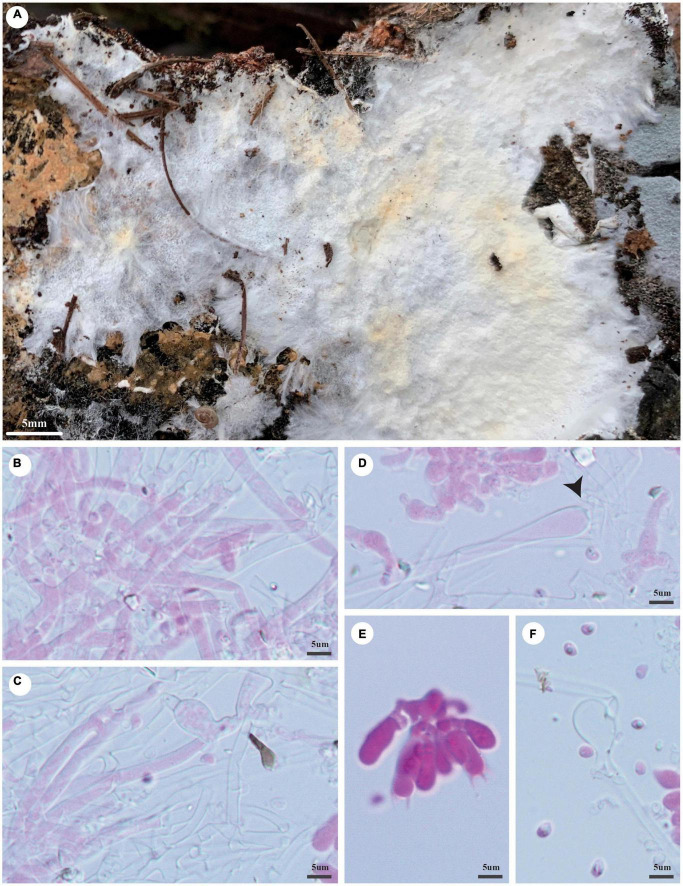
Trechispora dimitiella Z.B. Liu and Yuan, sp. November Figure 5
FIGURE 5.
Trechispora dimitiella (holotype, Dai 21931). (A) A basidioma, (B) hyphae with ampullate septa from subiculum (black arrow), (C) hyphae from tubes, (D) basidia, (E) basidioles, and (F) basidiospores. Photo by Ya-Ping Lian and Zhan-Bo Liu.
MycoBank number: MB 842866.
Type: China, Hainan Province, Haikou, Jinniuling Park, on a rotten leaf, in southwestern China, ca. E 110° 19′, N 20° 1′, alt. 17 m. The vegetation is a plantation in tropical China. 7 November 2020, Y.C. Dai 21931 (holotype BJFC 035830).
Etymology: Dimitiella (Lat.): It refers to the species having a dimitic hyphal system.
Basidioma: They are annual, resupinate, soft when fresh, fragile when dry, easily separable from the substratum, up to 6-cm long, 4-cm wide, and approximately 3-mm thick at the center; the hymenial surface is poroid, pore surface white to cream (4A2/3) when fresh, becoming white to buff-yellow (4A4) when dry; margin indistinct, often with emerging mycelial cords; pores angular, 5–6/mm; dissepiments thin, lacerate; subiculum up to 1 mm thick; tubes concolorous with a poroid surface, up to 2 mm long.
Hyphal structure: Hyphal system is dimitic; generative hyphae bear clamp connections; ampullate septa occasionally present in subiculum and trama, up to 4.5 μm wide; all hyphae IKI−, CB− are unchanged in KOH; rhomboidal calcium oxalate crystals are scattered.
Subiculum: Generative hyphae hyaline, thin-walled, rarely branched, 2–3 μm in diameter; skeletal hyphae thick-walled with a wide lumen, unbranched, loosely interwoven, 2–4 μm diameter.
Tubes: Generative hyphae hyaline, thin-walled, rarely branched, 1.5–2.5 μm in diameter; skeletal hyphae thick-walled with a wide lumen, unbranched, loosely interwoven, 2–3 μm in diameter; cystidia and cystidioles are absent; basidia are barrel-shaped, hyaline, bearing four sterigmata and a basal clamp connection, 9.5–12 × 4–5 μm; basidioles are similar to basidia in shape but slightly shorter.
Basidiospores: They are ellipsoid, hyaline, thick-walled, aculeate, IKI−, CB−, (3.5−)3.6–4(−4.2) × (2.5–)2.7–3.1(−3.2) μm (including ornamentation), L = 3.84 μm, W = 2.92 μm, Q = 1.31–1.33 (n = 60/2); (2.6−)2.7–3.4(−3.7) × 2–2.6(−2.9) μm (excluding ornamentation), L′ = 3.04 μm, W′ = 2.18 μm, and Q′ = 1.38–1.4 (n = 60/2).
Additional specimen examined (paratypes): China, Yunnan Province, Jinghong, Primeval Forest Park, on soil, 7 July 2021, Y.C. Dai 22601 (BJFC), Dai 22602 (BJFC). Malaysia, Selangor, Kota Damansara, Community Forest Reserve, on rotten angiosperm wood, 7 December 2019, Y.C. Dai 21181 (BJFC 032835).
Notes: T. dimitiella was discovered in China and Malaysia. Most species in Trechispora are corticioid fungi with a monomitic hyphal structure, but T. dimitiella is different. Morphologically, T. dimitiella and Trechispora brasiliensis (Corner) K.H. Larss. share the poroid hymenophore with a dimitic hyphal system and aculeate basidiospores. However, the basidiospores of T. dimitiella are smaller than that of T. brasiliensis [3.6–4 × 2.7–3.1 μm vs. 4–4.5 × 3–4 μm in T. brasiliensis (including ornamentation), Larsson, 1992]. Phylogenetically, T. dimitiella is close to Trechispora incisa K.H Larss. (80% BS, 0.99 BPP; Figure 2), but T. dimitiella can be easily distinguished from T. incisa due to its poroid hymenophore with a dimitic hyphal system because T. incisa has arachnoid to farinose or minutely granulose hymenophore with a monomitic hyphal system (Larsson, 1996).
Trechispora fragilis Z.B. Liu and Yuan Yuan, sp. November Figure 6
FIGURE 6.
Trechispora fragilis (holotype, Dai 20535). (A) A basidioma, (B) hyphae with ampullate septa from subiculum (black arrows), (C) hyphae from aculei, (D) hymenium with basidioles, (E) basidia, and (F) basidiospores. Photo by Ya-Ping Lian and Zhan-Bo Liu.
MycoBank number: MB 842867.
Type: China, Yunnan Province, Sipsongpanna, Mengla County, XiShuangBanNa Tropical Botanical Garden, on the ground of the forest, in southwestern China, ca. E 101° 25′, N 21° 41′, alt. 570 m. The vegetation is a natural tropical forest. 18 August 2019, Y.C. Dai 20535 (holotype BJFC 032203).
Etymology: Fragilis (Lat.): It refers to the species having fragile basidiocarps.
Basidioma: They are annual, resupinate, soft when fresh, fragile when dry, easily separable from the substratum, up to 3 cm long, 2 cm wide, and less than 1 mm thick at the center; the hymenial surface is odontoid, white when fresh, becoming cream (4A2/3) to buff-yellow (4A4) when dry; margin is indistinct and fimbriate, often with emerging mycelial cords; aculei sparse, 4–6/mm; subiculum very thin to almost absent; aculei concolorous with a hymenial surface, less than 1 mm long.
Hyphal structure: Hyphal system monomitic; generative hyphae bear clamp connections; ampullate septa occasionally present in subiculum and aculei, up to 7 μm wide; all hyphae IKI−, CB− are unchanged in KOH; rhomboidal calcium oxalate crystals are scattered.
Subiculum: Generative hyphae hyaline, thin- to thick-walled, frequently branched, loosely interwoven, 1.5–4 μm in diameter.
Aculei: Generative hyphae in trama hyaline, thin- to thick-walled, frequently branched, loosely interwoven, 1.5–3 μm in diameter; cystidia and cystidioles are absent; basidia are clavate shaped, hyaline, bearing four sterigmata, and a basal clamp connection, 12–14 × 3.5–4 μm; basidioles are similar to basidia in shape but slightly shorter.
Basidiospores: Ellipsoid, hyaline, thick-walled, aculeate, IKI−, CB−, (3.2−)3.8–4(−4.2) × (2.4−)2.5–3 μm (including ornamentation), L = 3.53 μm, W = 2.79 μm, Q = 1.27 (n = 60/1); (2.6−)2.8–3.7(−4) × (1.9−)2–2.7(−3.1) μm (excluding ornamentation), L′ = 3.16 μm, W′ = 2.26 μm, and Q′ = 1.40 (n = 60/1).
Notes: T. fragilis was discovered in the Yunnan Province of China. Phylogenetically, T. fragilis groups with Trechispora termitophila Meiras-Ottoni and Gibertoni and Trechispora havencampii (Desjardin and B.A. Perry) Meiras-Ottoni and Gibertoni (69% BS, 0.92 BPP; Figure 2). T. termitophila can be easily distinguished from T. fragilis due to its coralloid basidioma. In addition, the basidiospores of T. fragilis are smaller than that of T. termitophila [6.5–7.5 μm vs. 4.5–5 μm in T. termitophila (including ornamentation), Meiras-Ottoni et al., 2021]. T. havencampii can also be easily distinguished from T. fragilis due to its coralloid basidioma. In addition, basidiospores of T. fragilis are smaller than that of T. havencampii [3.8–4 × 2.5–3 μm vs. 5.2–6.5 × 3.5–4.2 μm in T. havencampii (including ornamentation), Desjardin and Perry, 2015].
Trechispora laevispora Z.B. Liu, Y.D. Wu and Yuan Yuan, sp. November Figure 7
FIGURE 7.
Trechispora laevispora (holotype, Dai 21655). (A) A basidioma, (B,C) hyphae from subiculum, (D) subicular hyphae with ampullate septa (black arrow) and a piece of hymenium, (E) basidia and basidioles, and (F) basidiospores. Photo by Ya-Ping Lian and Zhan-Bo Liu.
MycoBank number: MB 842868.
Type: China, Inner Mongolia Autonomous Region, Arxan, Bailang Feng Scenic Spot, on the charred trunk of Larix, in southwestern China, ca. E 119° 56′, N 47° 10′, alt. 1,511 m. The vegetation is a natural boreal forest. 25 August 2020, Y.C. Dai 21655 (holotype BJFC 035556).
Etymology: Laevispora (Lat.): It refers to the species having smooth basidiospores.
Basidioma: They are annual, resupinate, soft when fresh and dry, up to 8 cm long, 3 cm wide, and less than 1 mm thick at the center; the hymenial surface is smooth, white when fresh and dry; margin is indistinct and fimbriate, often with emerging mycelial cords; subiculum very thin to almost absent.
Hyphal structure: Hyphal system monomitic; generative hyphae bear clamp connections; ampullate septa frequently present in subiculum and hymenium, up to 7 μm wide; all hyphae IKI−, CB− are unchanged in KOH; rhomboidal calcium oxalate crystals are abundant.
Subiculum: Generative hyphae hyaline, thin-walled, frequently branched, loosely interwoven, 1.5–3 μm in diameter.
Hymenium: Generative hyphae in subhymenium hyaline, thin-walled, frequently branched, 1.5–3 μm in diameter; cystidia and cystidioles are absent; basidia are clavate shaped, hyaline, bearing four sterigmata and a basal clamp connection, 11.5–15 × 4–5 μm; basidioles are similar to basidia in shape but slightly shorter.
Basidiospores: Ellipsoid, hyaline, thin-walled, smooth, IKI−, CB−, (2.5−) 2.6–3.2(−3.3) × (1.8−)1.9–2.2(−2.5) μm, L = 2.92 μm, W = 2.04 μm, and Q = 1.43 (n = 60/1).
Notes: T. laevispora was discovered in the Inner Mongolia Autonomous Region of China. Phylogenetically, T. laevispora groups with Trechispora cohaerens (Schwein.) Jülich and Stalpers with strong support (94% BS, 96% BP, 1.00 BPP; Figure 2). Both species share a smooth hymenophore, a monomitic hyphal system with smooth basidiospores. However, basidiospores of T. cohaerens are thick-walled and larger than that of T. laevispora (3.5–4 × 2.2–2.5 μm in T. cohaerens; Larsson, 1992).
B. daweishanense (C.L. Zhao) Z.B. Liu and Yuan Yuan, comb. November
MycoBank number: MB 842869.
Basionym: T. daweishanensis C.L. Zhao, Phytotaxa 479(2): 153 (2021).
Type: China. Yunnan Province, Honghe, Pingbian County, Daweishan National Nature Reserve, on the fallen branch of angiosperms, 1 August 2019, CLZhao 17860 (holotype SWFC).
Description: See Zong et al. (2021, as T. daweishanensis).
B. xanthum (C.L. Zhao) Z.B. Liu and Yuan Yuan, comb. November
MycoBank number: MB 842870.
Basionym: T. xantha C.L. Zhao, Phytotaxa 479(2): 155 (2021).
Type: China. Yunnan Province, Yuxi, Xinping County, Mopanshan National Forestry Park, on the trunk of Albizia julibrissin, 20 August 2017, CLZhao 2632 (holotype SWFC).
Description: See Zong et al. (2021, as T. xantha).
Notes: Zong et al. (2021) described T. daweishanensis and T. xantha as new species. However, in our phylogeny, they belong to the genus Brevicellicium (98% BS, 1.00 BPP; Figure 1). The type specimens of abovementioned species are studied [CLZhao 17860 (SWFC); CLZhao 2632 (SWFC)]. We do not observe ampullate hyphae from type materials as mentioned by Zong et al. (2021). We suppose that Zong et al. (2021) confused basidioles with ampullate hyphae (ampullate septa on some generative hyphae), which are remarkable characters of Trechispora. In fact, T. daweishanensis and T. xantha have a smooth hymenophore, a monomitic hyphal structure with clamped generative hyphae, and the absence of ampullate septa. They fit Brevicellicium well. Herein, we combine these two species in Brevicellicium based on morphological and phylogenetic evidence (Figure 1).
S. limonadense (G. Gruhn and P. Alvarado) Z.B. Liu and Yuan Yuan, comb. November
MycoBank number: MB 842871.
Basionym: S. limonadense G. Gruhn and P. Alvarado, Phytotaxa 498(1): 36 (2021).
Type: French Guiana. On the bark of an unidentified dead trunk lying on the ground, October 22, 2013, LIP 0001683 (holotype).
Description: See Gruhn and Alvarado (2021, as S. limonadense).
Notes: Gruhn and Alvarado (2021) described S. limonadense as a new species. However, at the same time, Spirin et al. (2021) segregated the species around S. niveocremeum (Höhn. and Litsch.) J. Erikss. into the new genus Sertulicium. In our phylogeny, S. limonadense groups with Sertulicium granuliferum (Hallenb.) Spirin and Volobuev Sertulicium lateclavigerum (Boidin and Gilles) Spirin and Viner (Figure 1). We did not study specimens, but S. limonadense is characterized by smooth to tuberculate hymenophore and basidia have 6–8 sterigmata (Gruhn and Alvarado, 2021) and fits Sertulicium better. Hence, we transfer S. limonadense to Sertulicium.
Discussion
Larsson (2007) showed that S. suecicum and S. niveocremeum (= S. niveocremeum) formed a strongly supported sister clade (94% BS, 1.00 BPP) to Hydnodontaceae within Trechisporales. However, in his phylogenetic analysis of 5.8S + nLSU, there were a few species in Hydnodontaceae and Sistotremastrum to establish a new family for S. suecicum and S. niveocremeum. Hence, Larsson (2007) named this clade Sistotremastrum family. The same strongly supported topology was recovered by Telleria et al. (2013); Gruhn et al. (2018), and Meiras-Ottoni et al. (2021) by the nLSU phylogenetic analysis. Spirin et al. (2021) presented a comprehensive study of Sistotremastrum and Sertulicium with 17 species. They used the nLSU region to perform phylogenetic analyses of 16 species in the two genera (Figure 1 in Spirin et al., 2021), except for Sertulicium chilense (Telleria, M. Dueñas and M.P. Martín) Spirin and Volobuev because the nLSU sequences of S. chilense were absent. However, they were not able to generate high support values for the node connecting Sistotremastrum and Sertulicium (87% BS, 0.87 BPP, Figure 1 in Spirin et al., 2021). As a result, they gave up establishing a new family too.
ITS1-5.8S-ITS2 is an important marker used for the barcoding of fungal species (Liu et al., 2021; Wangsawat et al., 2021). However, the difficulty in aligning ITS sequences for fungi in Trechisporales is evident because it is a data set covering taxa in distinct taxonomic levels (Larsson, 2007). Therefore, it is not a good idea to run combined analyses of ITS + nLSU, so we use the most stable and conservative portion of ITS (5.8S) and nLSU to our phylogenetic analyses of Sistotremastrum and Sertulicium (5.8S + nLSU) (Figure 1). We add S. chilense and S. limonadense to phylogenetic analyses. Our results of the Sistotremastrum are the same as phylogenetic analyses by Spirin et al. (2021, Figure 1). However, our phylogenetic analyses of Sertulicium are a bit different from that by Spirin et al. (2021, Figure 1) because the data sets used in both studies are different. Above all, we generate high support values for the node connecting Sistotremastrum and Sertulicium from ML analysis (93% BS) based on 5.8S and nLSU sequences; however, BI fails to provide support for the node (0.76 BPP).
Divergence time is estimated with 5.8S and nLSU sequences representing all main lineages in Basidiomycota (Figure 3). The MCC tree shows that Basidiomycota occurs in a mean stem age of 509.57 Mya. Trechisporales occurs in a mean stem age of 270.85 Mya. The tree also shows that the Sistotremastrum family and Hydnodontaceae occur in a mean stem age of 224.25 Mya (PP = 0.8). Zhao et al. (2017) indicate that the divergence times of Basidiomycota are 530 Mya (the mean stem age). He et al. (2019) indicate that the divergence times of Trechisporales and Hydnodontaceae are 259 Mya (the mean stem age). Our experimental results agree with them. In this paper, we update the divergence times of Trechisporales and Hydnodontaceae and define the divergence time of the Sistotremastrum family.
Bayesian phylogenetic inference fails to provide support for the node of Sistotremastrum and Sertulicium, so we use the term “Sistotremastrum family” for the two genera without a formal description of the new family. In the future, we will sequence additional DNA regions or whole genomes, for a more robust phylogenetic analysis.
At present, there are only two species in the Sistotremastrum family ever been recorded from China, i.e., Sistotremastrum aculeatum Miettinen and Viner (Cui 8401) and S. granuliferum (He 3338; CLZhao 5531, 9771). Recently, we collected a specimen from the Yunnan Province of China (He 6276), and its morphological and DNA data demonstrated the specimen is S. limonadense. The species is a new record in China, and we have uploaded ITS and nLSU sequences of the specimen (He 6276) to GenBank. Above all, we study all the Chinese specimens of species in the Sistotremastrum family seriously, and their morphology fits the descriptions of Gruhn and Alvarado (2021) and Spirin et al. (2021). We also collected a specimen from the Hainan Province of China (Dai 17696). The ITS (OK298490) region is different from Sistotremastrum fibrillosum G. Gruhn and P. Alvarado by 6%, and morphologically it is similar to S. fibrillosum. However, we only have a single specimen, so for the time being we regard Dai 17696 as Sistotremastrum sp.
In this article, we use the whole ITS region in analyses of Trechispora to visualize the genetic distances among new taxa and those already described. T. dentata, T. dimitiella, T. fragilis, and T. laevispora are described as new to science based on morphological characteristics and molecular evidence (Figure 2). Most of these new species are found in subtropical or tropical Asia and conform to the phenomenon that subtropical or tropical Asia harbors high taxonomic diversity for all wood-decaying fungi (Dai, 2012; Cui et al., 2019). We also collected two resupinate specimens (Dai 22173 and Dai 22174) from the Hainan Province of China. The morphology of the two specimens corresponds to the concept of Trechispora and forms a distinct lineage within the Trechispora clade (100% BS, 1.00 BPP; Figure 2). However, these specimens are sterile, so we regard Dai 22173 and Dai 22174 as Trechispora spp. temporarily here.
Molecular phylogenetic analyses in the present study show that Brevicellicium forms a monophyletic clade in which all Brevicellicium species are included (98% BS, 1.00 BPP; Figure 1). However, when we add sequences of T. xantha and T. daweishanensis, we find sequences of a two-species cluster with Brevicellicium with high support (100% BS, 1.00 BPP; Figure 1). We request and examine type specimens from Zhao and find T. xantha and T. daweishanensis corresponding to the concept of Brevicellicium and they should be transferred to the genus Brevicellicium (see the notes of B. daweishanense).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
Z-BL: design of the research, performance of the research, and writing and revising this manuscript. Z-BL, HZ, Y-PL, Y-RW, C-GW, and W-LM: data analysis and interpretation. Z-BL, YY, and Y-DW: a collection of the materials. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Dr. Chang-Lin Zhao (SWFC, China) and Prof. Yu-Cheng Dai for allowing us to study their specimens.
Footnotes
Funding
The research is supported by the National Natural Science Foundation of China (Project Nos. 31870007 and 32011540380).
References
- Berbee M. L., Taylor J. W. (2010). Dating the molecular clock in fungi – how close are we? Fungal Biol. Rev. 24 1–16. 10.1016/j.fbr.2010.03.001 [DOI] [Google Scholar]
- Bouckaert R., Vaughan T. G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. 10.1371/journal.pcbi.1006650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikowski R., Larsson K. H., Gibertoni T. B. (2020). Taxonomic novelties in Trechispora (Trechisporales, Basidiomycota) from Brazil. Mycol. Prog. 19 1403–1414. 10.1007/s11557-020-01635-y [DOI] [Google Scholar]
- Cui B. K., Li H. J., Ji X., Zhou J. L., Song J., Si J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97 137–392. 10.1007/s13225-019-00427-4 [DOI] [Google Scholar]
- Dai Y. C. (2010). Hymenochaetaceae (Basidiomycota) in China. Fungal Divers. 45 131–343. 10.1007/s13225-010-0066-9 [DOI] [Google Scholar]
- Dai Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53 49–80. 10.1007/s10267-011-0134-3 [DOI] [Google Scholar]
- Desjardin D. E., Perry B. A. (2015). A new species of Scytinopogon from the island of príncipe, republic of são tomé and príncipe, West Africa. Mycosphere 6 434–441. 10.5943/mycosphere/6/4/5 [DOI] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214–221. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Cao T. X., Wu Y. D., Zhou M., Liu Z. B. (2021). Two new species of Hymenochaetaceae on Dracaena cambodiana from tropical China. MycoKeys 80 1–17. 10.3897/mycokeys.80.63997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. F., Alvarenga R. L. M., Gibertoni T. B., Wu F., Dai Y. C. (2021). Four new species in the Tremella fibulifera complex (Tremellales, Basidiomycota). MycoKeys 82 33–56. 10.3897/mycokeys.82.63241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Furtado A. N. M., Danils P. P., Reck M. A., Neves M. A. (2021). Scytinopogon caulocystidiatus and S. foetidus spp. nov. and five other species recorded from Brazil. Mycotaxon 136 107–130. 10.5248/136.107 30528588 [DOI] [Google Scholar]
- Gruhn G., Alvarado P., Hallenberg N., Roy M., Courtecuisse R. (2018). Contribution to the taxonomy of Sistotremastrum (Trechisporales, Basidiomycota) and the description of two new species, S. fibrillosum and S. aculeocrepitans. Phytotaxa 379:1. 10.11646/phytotaxa.379.1.2 [DOI] [Google Scholar]
- Gruhn G., Alvarado P. (2021). Sistotremastrum limonadense sp. nov. from French Guiana. Phytotaxa 498 35–43. 10.11646/phytotaxa.498.1.4 [DOI] [Google Scholar]
- Hall T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- He M. Q., Zhao R. L., Hyde K. D., Dominik B., Martin K., Andrey Y., et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99 105–367. 10.1007/s13225-019-00435-4 [DOI] [Google Scholar]
- Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., et al. (2007). A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Hibbett D. S., Grimaldi D., Donoghue M. J. (1995). Cretaceous mushrooms in amber. Nature 377:487. 10.1038/377487a0 [DOI] [Google Scholar]
- Hibbett D. S., Grimaldi D., Donoghue M. J. (1997). Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. Am. J. Bot 1997 981–991. 10.2307/2446289 [DOI] [PubMed] [Google Scholar]
- Karsten P. A. (1890). Fragmenta mycologica XXIX. Hedwigia 29 147–149. [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics 20 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson K. H. (1992). The Genus Trechispora. (Corticiaceae, Basidiomycetes). Ph.D. thesis. Gothenburg: University of Göteborg. [Google Scholar]
- Larsson K. H. (1996). New species and combination in Trechispora (Corticiaceae, Basidiomycotina). Nordic J. Bot. 16 83–98. 10.1111/j.1756-1051.1996.tb00218.x [DOI] [Google Scholar]
- Larsson K. H. (2007). Re-thinking the classification of corticioid fungi. Mycol. Res. 111 1040–1063. 10.1016/j.mycres.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Larsson K. H., Larsson E., Kõljalg U. (2004). High phylogenetic diversity among corticioid Homobasidiomycetes. Mycol. Res. 108 983–1002. [DOI] [PubMed] [Google Scholar]
- Liberta A. E. (1973). The genus Trechispora (Basidiomycetes, Corticiaceae). Botany 51 1871–1892. 10.1139/b73-240 [DOI] [Google Scholar]
- Liu S. L., Ma H. X., He S. H., Dai Y. C. (2019). Four new corticioid species in Trechisporales (Basidiomycota) from East Asia and notes on phylogeny of the order. MycoKeys 48 97–113. 10.3897/mycokeys.48.31956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. B., Dai Y. C. (2021). Steccherinum fragile sp. nov. and S. subcollabens comb. nov. (Steccherinaceae, Polyporales), evidenced by morphological characters and phylogenetic analysis. Phytotaxa 483 106–116. 10.11646/phytotaxa.483.2.3 [DOI] [Google Scholar]
- Liu Z. B., Yuan Y. (2020). Luteoporia citriniporia sp. nov. (Polyporales, Basidiomycota), evidenced by morphological characters and phylogenetic analysis. Phytotaxa 461 31–39. 10.11646/phytotaxa.461.1.4 [DOI] [Google Scholar]
- Liu Z. B., Zhou M., Yuan Y., Dai Y. C. (2021). Global diversity and taxonomy of Sidera (Hymenochaetales, Basidiomycota): four new species and keys to species of the genus. J. Fungi 7:251. 10.3390/jof7040251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R. (2021). Mesquite: A Modular System For Evolutionary Analysis. Version 3.70. Available online at: https://www.mesquiteproject.org/ (Accessed March, 2022). [Google Scholar]
- Meiras-Ottoni A. D., Larsson K. H., Gibertoni T. B. (2021). Additions to Trechispora and the status of Scytinopogon (Trechisporales, Basidiomycota). Mycol. Prog. 20 203–222. 10.1007/s11557-021-01667-y [DOI] [Google Scholar]
- Miller M. A., Holder M. T., Vos R., Midford P. E., Liebowitz T., Chan L., et al. (2009). The CIPRES Portals. CIPRES. Available online at: http://www.phylo.org/sub_sections/portal. (Accessed Aug 4, 2009). (Archived by WebCite® at: http://www.webcitation.org/5imQlJeQa) [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest v2: Program Distributed By The Author. Uppsala: Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Petersen J. H. (1996). The Danish Mycological Society’s colour-chart. Greven: Foreningen til Svampekundskabens Fremme, 1–6. [Google Scholar]
- Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2018). Molecular Evolution, Phylogenetics and Epidemiology. FigTree ver. 1.4.4 Software. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed March, 2022). [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). Mrbayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Smith S. Y., Currah R. S., Stockey R. A. (2004). Cretaceous and Eocene poroid hymenophores from Vancouver Island, British Columbia. Mycologia 96 180–186. 10.2307/3762001 [DOI] [PubMed] [Google Scholar]
- Spirin V., Volobuev S., Viner I., Miettinen O., Vlasák J., Schoutteten N., et al. (2021). On Sistotremastrum and similar-looking taxa (Trechisporales, Basidiomycota). Mycol. Prog. 20 453–476. 10.1007/s11557-021-01682-z [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML Version 8: a tool for phylogenetic analyses and post analyses of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistyo B. P., Larsson K. H., Haelewaters D., Ryberg M. (2021). Multigene phylogeny and taxonomic revision of Atheliales s.l.: reinstatement of three families and one new family, Lobuliciaceae fam. nov. Fungal Biol. 125 239–255. 10.1016/j.funbio.2020.11.007 [DOI] [PubMed] [Google Scholar]
- Swofford D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (*And Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Taylor T. N., Hass H., Kerp H. (1999). The oldest fossil ascomycetes. Nature 399:648. 10.1038/21349 [DOI] [PubMed] [Google Scholar]
- Taylor T. N., Hass H., Kerp H., Krings M., Hanlin R. T. (2005). Perithecial ascomycetes from the 400 million year old Rhynie chert: an example of ancestral polymorphism. Mycologia 97 269–285. 10.1080/15572536.2006.11832862 [DOI] [PubMed] [Google Scholar]
- Telleria M. T., Melo I., Dueñas M., Larsson K. H., Martín P. (2013). Molecular analyses confirm Brevicellicium in Trechisporales. IMA Fungus 4 21–28. 10.5598/imafungus.2013.04.01.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers B. (2018). Index Herbariorum: A Global Directory Of Public Herbaria And Associated Staff. New York, NY: New York Botanical Garden’s Virtual Herbarium. [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W., Tom W. M., Liu X. L., Zhou L. W. (2021). Towards a natural classification of hyphodontia sensu lato and the trait evolution of basidiocarps within hymenochaetales (Basidiomycota). J. Fungi 7:478. 10.3390/jof7060478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangsawat N., Ju Y. M., Phosri C., Whalley A. J. S., Suwannasai N. (2021). Twelve new taxa of xylaria associated with termite nests and soil from northeast thailand. Biology 10:575. 10.3390/biology10070575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, A Guide To Methods And Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (New York, NY: Academic Press; ), 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Zhao C. L., Cui B. K., Song J., Dai Y. C. (2015). Fragiliporiaceae, a new family of Polyporales (Basidiomycota). Fungal Divers. 70 115–126. 10.1007/s13225-014-0299-0 [DOI] [Google Scholar]
- Zhao R. L., Li G. J., Sanchez-Ramirez S., Stata M., Yang Z. L., Wu G., et al. (2017). A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84 43–74. 10.1007/s13225-017-0381-5 [DOI] [Google Scholar]
- Zhao W., Zhao C. L. (2021). The phylogenetic relationship revealed three new wood-inhabiting fungal species from genus Trechispora. Front. Microbiol. 12:650195. 10.3389/fmicb.2021.650195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong T. K., Liu C. M., Wu J. R., Zhao C. L. (2021). Trechispora daweishanensis and T. xantha spp. nov. (Hydnodontaceae, Trechisporales) found in Yunnan Province of China. Phytotaxa 479 147–159. 10.11646/phytotaxa.479.2.1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.