Abstract
CRISPR-Cas9 can be scaled up for large-scale screens in cultured cells, but CRISPR screens in animals have been challenging because generating, validating, and keeping track of large numbers of mutant animals is prohibitive. Here, we report Multiplexed Intermixed CRISPR Droplets (MIC-Drop), a platform combining droplet microfluidics, single-needle en masse CRISPR ribonucleoprotein injections, and DNA barcoding to enable large-scale functional genetic screens in zebrafish. The platform can efficiently identify genes responsible for morphological or behavioral phenotypes. In one application, we show MIC-Drop can identify small molecule targets. Furthermore, in a MIC-Drop screen of 188 poorly characterized genes, we discover several genes important for cardiac development and function. With the potential to scale to thousands of genes, MIC-Drop enables genome-scale reverse-genetic screens in model organisms.
One Sentence Summary:
A rapid, scalable CRISPR screening platform in zebrafish identifies genes responsible for heart development.
Historically, large scale genetic screens in zebrafish have employed forward genetic techniques such as chemical or insertional mutagenesis (Fig. S1A) (1, 2). These screens have proven invaluable in identifying key pathways regulating vertebrate development (3–5) and behavior (6, 7). While impressive in scale, forward genetic techniques are time- and labor-intensive requiring years to link a desired phenotype with the genotype. Reverse genetics approaches such as CRISPR have potential to circumvent some of the issues of forward genetics but are severely limited in throughput (8, 9). Targeting genes-of-interest is typically done one gene at a time—designing individual guide RNAs (gRNA), injecting Cas9-gRNA ribonucleoprotein (RNP) complexes, maintaining, propagating, and genotyping groups of fish—requiring extensive time, labor, and space. The largest such screen to date targeted 128 genes in zebrafish (10, 11). Recent studies using multiplexed gRNAs to generate biallelic F0 mutants that successfully phenocopy germline mutant phenotypes are a welcome step but have not been scaled up for genome-wide CRISPR screens (12–17).
We have developed a platform, Multiplexed Intermixed CRISPR Droplets (MIC-Drop), for performing large-scale reverse-genetic screens in zebrafish (Fig. 1A). The platform uses microfluidics to generate nanoliter-sized droplets, each droplet containing Cas9, multiplexed gRNAs targeting a gene-of-interest, and a unique barcode associated with each target gene. Droplets targeting hundreds to thousands of different genes are intermixed together and injected into zebrafish embryos from a single needle (Movie S1). Embryos are raised en masse, those exhibiting phenotype(s)-of-interest are isolated, and the identities of the perturbed genes are rapidly uncovered by retrieving and sequencing the barcodes. The ability to inject all the intermixed droplets from a single needle circumvents the laborious and wasteful process of filling a separate needle for each gene targeted. The ability to identify perturbed genes by retrieving barcodes circumvents the need to raise each injected animal separately or deconvolute the mutated 25 gene through genome sequencing.
Fig. 1. MIC-Drop enables high-throughput CRISPR screens in zebrafish.
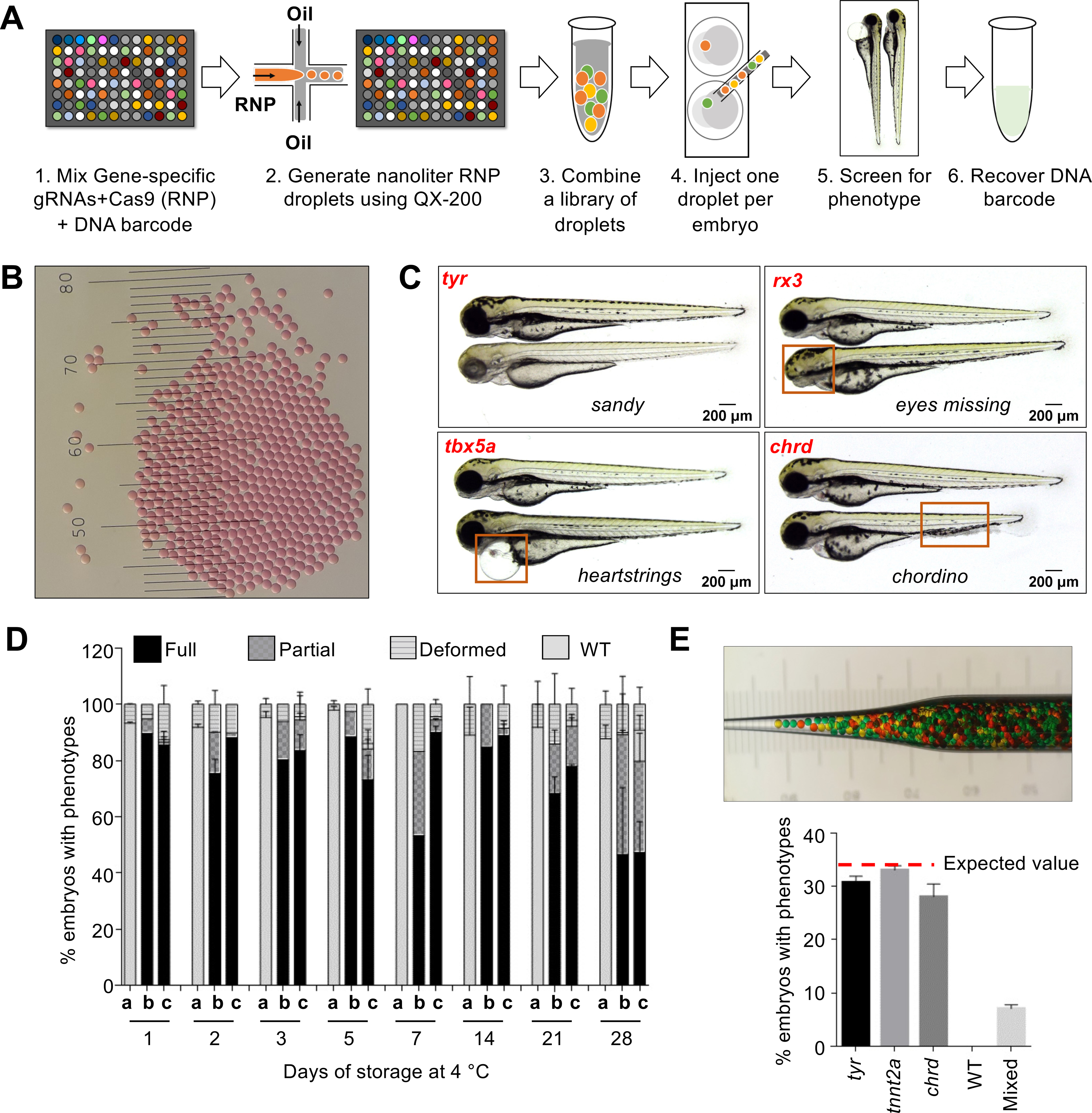
(A) Workflow of the MIC-Drop platform. A microfluidics device generates nanoliter-sized droplets, each containing ribonucleoproteins (RNP) targeting a gene-of-interest and a unique DNA barcode associated with the gene. Droplets targeting multiple genes are intermixed, loaded into a single injection needle and injected serially into one-cell zebrafish embryos. Embryos showing phenotypes-of-interest are isolated and the causative genotype is identified by retrieving and sequencing the barcode. (B) Droplets are uniform in size. Distance between bars is 0.1 mm. (C) Injection of droplets containing RNPs targeting tyr, rx3, tbx5a, and chrd genes recapitulates known mutant phenotypes in F0, highlighted by red boxes. (D) RNP-containing droplets are non-toxic and stable for prolonged storage – retaining activity at least 28 days of storage at 4°C. a: Uninjected; b: Traditional RNP injection; c: MIC-Drop injection. N = (Day1: a-118, b-79, c-134; Day2: a-116, b-68, c-59; Day3: a-120, b-88, c-87; Day5: a-129, b-78, c-95; Day7: a-105, b-102, c-107; Day14: a-94, b-53, c-77; Day21: a-123, b-100, c-132; Day28: a-114, b-100, c-94. (E) Single-needle injection of intermixed droplets targeting three different genes (tyr, tnnt2a, chrd) and subsequent phenotyping shows even representation of each droplet with all of the embryos exhibiting only one of the three expected phenotypes, and none (0%) were wildtype. Total N sequenced = 230 from 3 separate injections. Inset: Hundreds of pseudo-colored droplets (used as proxies for droplets targeting different genes) do not fuse when transferred to an injection needle.
After testing different surfactant-oil combinations, we identified a particular combination of fluorinated oil and a fluorosurfactant as optimal for droplet generation using a repurposed BioRad QX-200 droplet generator (see detailed SI methods and Supplementary Text). The droplets generated were uniform, ~100 um in diameter (Fig. 1B). Each droplet contained four gRNAs targeting a gene-of-interest. We found that using four gRNAs per gene recapitulated the phenotypes of homozygous mutants in F0 embryos with high penetrance (Fig. S1B–D and Table S1). Injection of four gRNAs targeting tyr, tnnt2a, tbx5a, rx3, npas4l, chrd, tbx16, and fgf24 resulted in highly efficient biallelic mutagenesis (Fig. S2A–B) and the expected sandy (lack of pigmentation), silent heart, heartstrings, eyes missing, cloche, chordino (tissue ventralization), spadetail, and ikarus (lack of pectoral fins) mutant phenotypes respectively in 70–100% of the F0 embryos. Importantly, no significant toxicity was observed in embryos injected with MIC-Drop except for the low level of non-specific morphological defects typical of traditional RNP injection (Fig. 1C–D, S1E and S3A–B). Droplets were stable during prolonged storage and showed high phenotypic penetrance even after a month of storage at 4 °C (Fig. 1D). Additionally, after generating and injecting a mini-library of MIC-Drops, with each droplet targeting any one of 3–8 different genes, we found that most embryos exhibited a unique phenotype, demonstrating successful injection of a single droplet per embryo (Fig. 1E and S3C–D). Importantly, the frequency of each phenotype was close to the expected value, indicating proportionate representation of each droplet within a mixed pool. Finally, the injected DNA barcodes could be recovered at least up to 7 days post fertilization (dpf) (Fig. S3E). Retrieval and sequencing of the barcode from the injected embryos revealed a high genotype-phenotype correlation. Although we used sgRNA DNA templates as the basis of the barcodes here, other barcoding strategies may be feasible (Supplementary Text).
Next, we tested whether MIC-Drop could identify genes responsible for a particular phenotype from a list of candidate genes (Fig. 2A). We spiked droplets targeting the tyr or npas4l genes into a larger pool of droplets containing scrambled gRNAs such that the tyr or npas4l MIC-drops each represented 2% of the total. Hundreds of embryos were injected with the intermixed droplets and the frequency of albino and cloche phenotypes among the injected embryos was assessed. We observed frequencies of (1.7 ± 0.8) % and (2.2 ± 0.8) % for the albino and cloche phenotypes respectively (Fig. 2A inset), comparable to the theoretical expected frequency of 2%, thereby indicating MIC-Drop screens are sensitive and may be a useful platform for a variety of applications requiring identification of genotype-phenotype relationships in vertebrates on a large scale.
Fig. 2. MIC-Drop enables large-scale phenotypic screens and small molecule target identification.
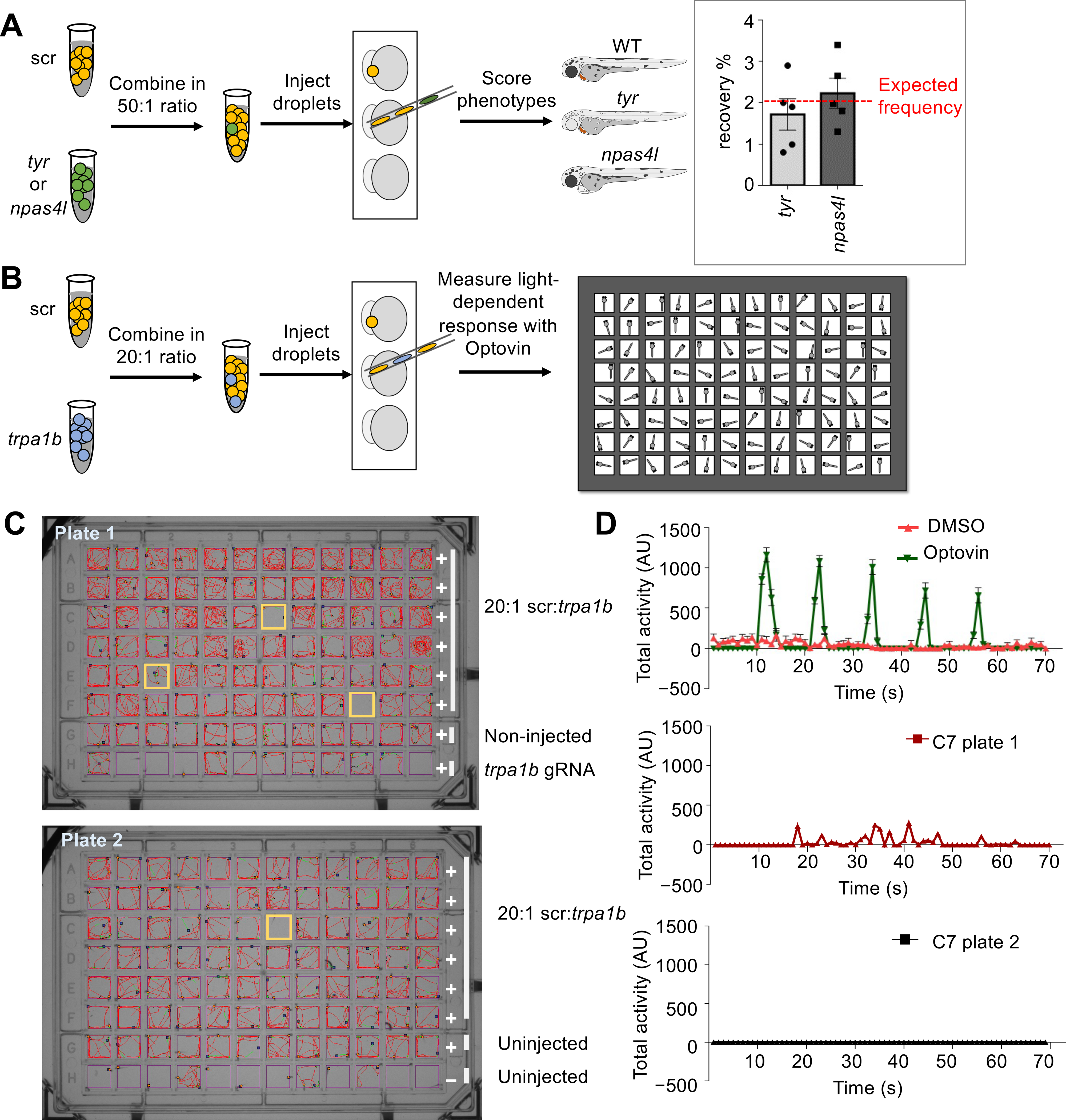
Schematic of a spike-in (A) phenotypic and (B) behavioral screen to test robustness of the MIC-Drop platform. (A) For the phenotypic screen, droplets targeting either tyr or npas4l were intermixed with droplets containing non-targeting scrambled gRNAs (scr) in a 1:50 ratio. After single-needle droplet injection, the percentage of embryos showing albino or cloche phenotypes was scored. Inset: The albino and cloche phenotypes are recovered at a frequency of ~2%, which is the expected frequency from a 1:50 ratio mix. (B) Similar to A, except droplets targeting trpa1b were intermixed with scr droplets in a 1:20 ratio. Following injection, embryos were arrayed in a multi-well plate, treated with optovin, and assayed for light-dependent motor response. (C) Tracking and (D) quantitation of zebrafish movement showed that embryos injected with droplets targeting trpa1b are refractory to optovin- and light-induced motion response.
Identifying the protein targets of small-molecules remains one of the major challenges in chemical biology and pharmacology (18, 19). We hypothesized that MIC-Drop could be used to identify the targets of small molecules that result in complex behavioral phenotypes in the zebrafish. As proof-of-principle, we utilized optovin, a small molecule agonist of the trpa1b channel that allows photo-activatable behavioral modifications in zebrafish (20). We spiked droplets targeting the trpa1b channel into a collection of droplets containing scrambled gRNAs in a 1:20 ratio (Fig. 2B). Droplet-injected embryos were arrayed into 96-well plates, treated with optovin and exposed to violet light flashes while simultaneously recording embryo movement. Treatment of wild-type zebrafish embryos with optovin results in a light-dependent motor response (Fig. S4A–C and Movie S2). Embryos that showed reduced or no movement in the assay were isolated, and their barcodes sequenced for genotype verification. We found that 2–3% of embryos showed a complete loss of photo-induced motion (Fig. 2B, S4D). Barcode sequencing revealed 100% of the unresponsive embryos were of trpa1b genotype. An additional ~2% of the embryos showed photo-induced motor response despite being of the trp1ab genotype, likely due to incomplete loss of trpa1b function (Fig. S4D). Thus, we were able to use the MIC-Drop platform to identify the target of optovin from among a library of non-target candidates.
We envisioned that MIC-Drop can be used to rapidly perform large-scale, reverse-genetic screens to uncover genes responsible for important phenotypes such as developmental defects in the cardiovascular system. Congenital heart disease (CHD) is the most common form of birth defect in humans, affecting nearly 1% of all live births (21). Genetic factors play a strong causal role in the development of CHD, however, a comprehensive understanding of all the genes responsible for CHD is still lacking. We used publicly available RNAseq datasets to curate a list of 188 poorly characterized genes that are enriched in the zebrafish embryonic heart tissue relative to skeletal muscle (Fig. 3A–B, S5A–B, and Tables S2–S4) (22, 23) and postulated these genes might be important in vertebrate heart development. We generated a library containing MIC-Drops for all 188 genes, plus several control genes (Fig. 3C and Table S5). When two ohnologs of a gene were identified, two gRNAs for each ohnolog were combined in a single droplet. Morphological phenotyping of zebrafish embryos at 48–72 hpf after MIC-Drop injection identified 13 novel genes, the loss of which result in a variety of specific cardiac or blood phenotypes (Fig. 3D–E and Table S5). Secondary validation of these “hits” corroborated the findings of the initial screen, with 10/13 genes showing phenotypic penetrance in >20% of F0 embryos (Fig. 3E). Interestingly, the screen identified gene mutations responsible for a range of phenotypes including 1 mutation (alad) causing porphyria, 2 mutations (gstm.3 and atp6v1c1) causing arrhythmia, and 7 mutations (actb2, clec19a, gse1, ppan, sf3b4, cox8a, and ddah2) causing abnormal cardiac development, including defects in ventricular morphogenesis, cardiac looping, and formation of the atrioventricular valve and cardiac jelly.
Fig. 3. A genetic screen to identify novel regulators of cardiovascular development.
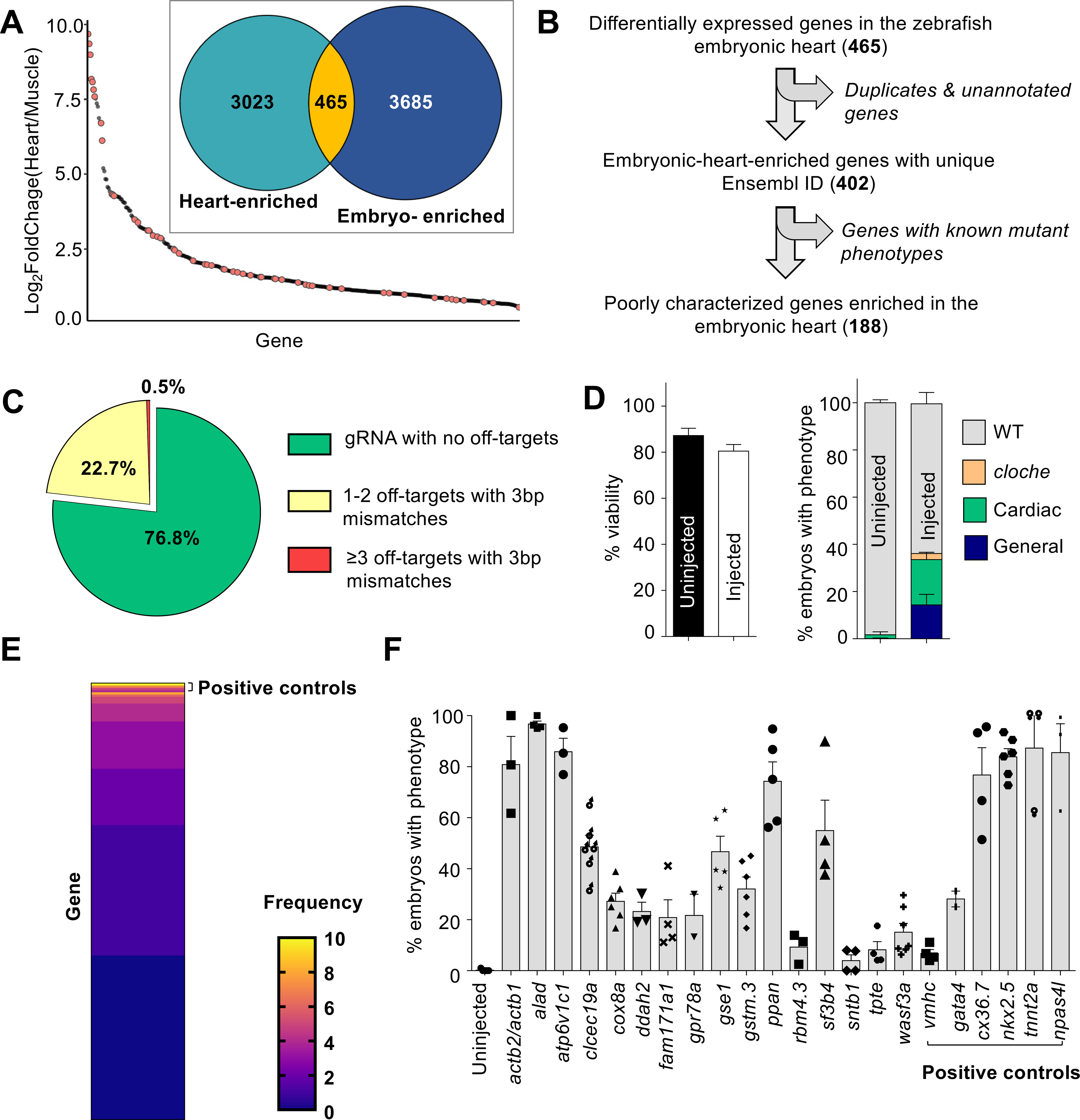
(A) A publicly available dataset was used to populate a list of candidate genes enriched in the embryonic zebrafish heart. ~14% of the genes (orange dots) have reported cardiac phenotypes in ZFIN suggesting enrichment of genes important in heart development. (B) Filtering to remove genes with known mutant phenotypes yields 188 poorly-characterized genes potentially important for cardiovascular development in zebrafish. (C) gRNA sequences with fewer off-targets were prioritized. (D) MIC-Drop screen of the 188 candidate genes and subsequent phenotyping shows no significant differences in viability between uninjected and droplet-injected embryos by 3 dpf N = (Left panel: Uninjected-1801, Injected-2502; Right panel: Uninjected-1571, Injected-2013). Embryos with gross morphological defects at 3 dpf (~15%) were removed and the barcodes of those with cardiac defects were sequenced. Droplets targeting npas4l were spiked-in at 2% proportion as positive control. (E) Barcode sequencing of embryos displaying any cardiac phenotype (e.g. looping defect, chamber dysmorphogenesis, valve defect, arrhythmia, etc.) yields “hit” candidates. Heat map shows the observed frequency of each barcode. As positive controls, barcodes for tnnt2a, nkx2.5, and npas4l are enriched in the set of embryos exhibiting any visible cardiac phenotype. Genes with barcode frequency of ≥ 4 (Binomial probability ≤ 0.05) or with consistent cardiac phenotypes were considered for secondary validation. (F) Secondary validation by direct RNP injection corroborates screening results and identifies a dozen novel genes, the loss of which results in cardiac phenotypes in at least 20% of F0 embryos. N = (WT-238; actb2/actb1-124; alad-137; atp6v1c1-135; clec19a-182; cox8a-130; ddah2-126; fam171a1-259; gpr78a-186; gse1-266; gstm.3-199; ppan-304; rbm4.3-153; sf3b4-307; sntb1-107; tpte-209; wasf3a/b-130; vmhc-107; gata4-106; cx36.7-147; nkx2.5-186; tnnt2a-124; npas4l-103).
Several reports have demonstrated that F0 “crispant” phenotypes reliably reflect loss of target gene function, but non-specific or off-target phenotypes are theoretically possible (Supplementary Text). To ensure the phenotypes we observed are due to on-target gene knockout, we performed phenotype rescue with mRNA injection. alad crispants showed a complete loss of hemoglobin synthesis which was rescued by injection of alad mRNA (Fig. 4A and S6A), indicating a role for alad in erythropoiesis.
Fig. 4. CRISPR screen using MIC-Drop identifies novel genes responsible for cardiovascular development.
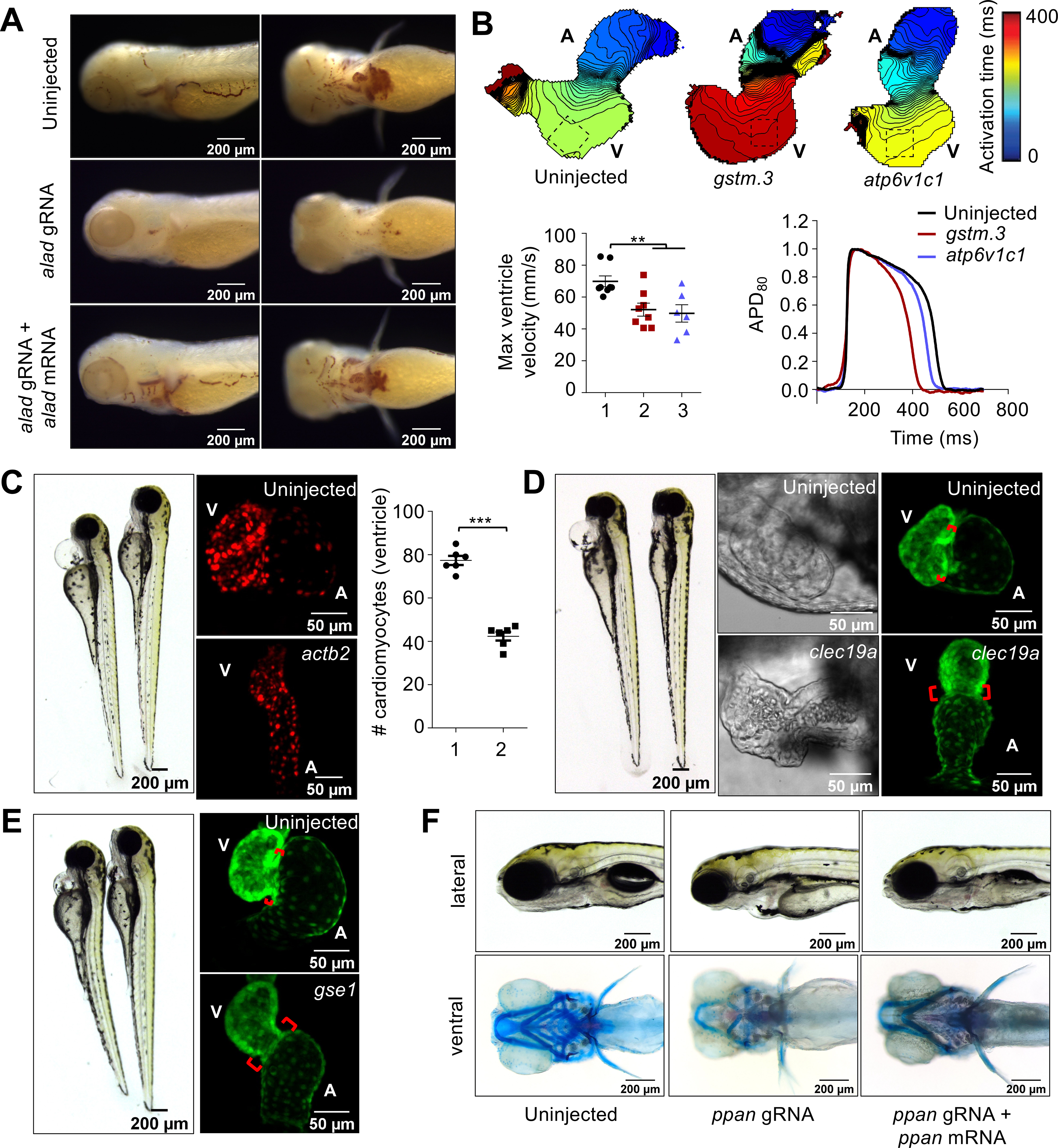
(A) o-dianisidine staining shows loss of alad results in porphyria, which can be rescued by co-injection of alad mRNA. (B) Loss of gstm.3 or atp6v1c1 results in abnormal cardiac electrophysiology. Isochronal maps and action potential measurements reveal reduced conduction velocities, and shorter ventricular action potential duration in the gstm.3 and atp6v1c1 crispants relative to uninjected controls 1: Uninjected; 2: gstm.3 crispants; 3. atp6v1c1 crispants. Data are presented as mean ± sem (** = p≤0.01; *** = p≤0.001). Loss of (C) actb2 (D) clec19a (E) gse1, and (F) ppan result in distinct cardiac malformations. actb2 crispants have a small ventricle with reduced number of ventricular cardiomyocytes 1: Control; 2: actb2-targeting gRNAs. (C). Loss of clec19a and gse1 result in abnormal morphogenesis and an extended atrioventricular canal relative to wildtype embryos (D-E). Alcian blue staining of ppan crispants shows abnormal jaw and skull development, which is rescued by ppan mRNA injection. The embryos also display cardiac edema, and a silent ventricle (F).
Voltage mapping of the gstm.3 and atp6v1c1 crispants showed slowed atrial and ventricular conductions and altered action potential duration (Fig. 4B and S6B). We identified atp6v1c1b as the ohnolog responsible for the ventricular arrhythmia phenotype (Fig. S6C). GSTM3 was recently identified as a risk factor in Brugada syndrome with increased susceptibility to sudden cardiac death (24). Germline gstm.3 zebrafish mutants exhibited ventricular arrhythmia corroborating our results observed in MIC-Drop crispants. Loss of function of several genes resulted in cardiac development defects. β-actin (actb1 and actb2) crispants showed cardiac edema, a small, silent ventricle with reduced cardiomyocytes, leaky blood vessels, as well as gross craniofacial defects (Fig. 4C and Movie S3). Interestingly, loss of actb2 alone was sufficient to recapitulate the cardiac phenotypes without the gross morphological defects, suggesting actb2 and actb1 have non-overlapping roles (Fig. 4C and S6D–E). clec19a, a c-type lectin protein with unknown functions was identified as important for the normal development of cardiac jelly and the atrioventricular valve in 3 dpf zebrafish embryos (Fig. 4D and Movie S4). Additionally, cox8a, a component of the mitochondrial electron transport chain and ddah2, an arginine metabolizing enzyme, were shown to be important for normal cardiac looping and ventricular morphogenesis (Fig. S7A). Finally, three other genes with limited annotation of their functions were identified as being important in heart development. Loss of ppan, gse1, and sf3b4 resulted in cardiac abnormalities along with other development defects such as malformed bones/cartilage in the jaw and pharyngeal arches (ppan), bent trunk (gse1 and sf3b4), and craniofacial defects (sf3b4) causing embryonic lethality (Fig. 4E–F, S7B- and Movie S5–6). Phenotypes associated with actb2, ppan, and sf3b4 MIC-Drops were also rescued by co-injection of actb2, ppan, and sf3b4 mRNAs (Fig. 4F and S6–S7), suggesting the phenotypes are caused by loss of function of the targeted genes. Therefore, MIC-Drop enabled a highly efficient reverse-genetic CRISPR screen in an intact vertebrate, leading to the discovery of several genes that contribute to diverse aspects of cardiac development or function.
In conclusion, we have developed a microfluidics-based platform for large-scale CRISPR screens in a vertebrate. CRISPR screens have previously been performed in cultured cells (25, 26), but genome editing in vertebrates has primarily been done one gene at a time. The few small-scale CRISPR screens reported in vertebrates were enabled by brute force scaling of single-gene methods for generating, tracking, and analyzing individual genes, with little economy of scale (10, 11, 27). By intermixing droplets targeting many genes and by incorporating a barcode for retrospective target identification, the MIC-drop platform enables zebrafish to be injected, housed, and analyzed en masse, with rapid identification of the target genes in individuals exhibiting phenotypes of interest. The pilot screen reported here quickly discovered several genes important for cardiovascular development and function. This screen of 188 genes was completed efficiently by a single investigator and could readily be scaled to thousands of genes or even to full genome scale (Supplementary Text). Moreover, MIC-Drop is versatile and conceptually can be used not just for gene knockout but for other screens such as CRISPR activation/inactivation screens, screens that mutate promoter or regulatory elements to create ‘transcript-less’ mutants (28), and functional screens of non-coding genetic elements (Supplementary Text). Finally, the platform can be adapted for use in other model organisms including Xenopus and mouse embryos where F0 crispants are shown to recapitulate known germline mutant phenotypes (29, 30) and likely in other model organisms amenable to microinjection, such as sea urchins, fruit flies, and nematodes. Thus, the MIC-Drop platform enables in vivo vertebrate CRISPR experiments to be performed with the speed, efficiency, and scale previously available only to in vitro systems.
Supplementary Material
Acknowledgments:
We appreciate Angie Serrano for assistance in imaging, Brent Bisgrove for advice on phenotyping and providing a list of candidate cardiac development genes, Bradley Demarest for advice on RNAseq data analysis. We acknowledge the Centralized Zebrafish Animal Resource (CZAR) for providing zebrafish husbandry and microinjection equipment. We thank members of the Peterson Lab for their insights and critical feedback at all stages of the project.
Funding:
We acknowledge support from NIH (5R01GM134069-02 to R.T.P. and J.J.Y., and UM1 HL098160 to H.J.Y.) and an American Heart Association postdoctoral fellowship to SP.
Footnotes
Competing interests: S.B., J.J.Y., and R.T.P. have applied for a patent covering technology described herein. R.T.P. has received honoraria (NIH) and expert testimony fees (Sterne Kessler/University of California) for work related to CRISPR technology.
Data and materials availability: All data are available in the main text or the supplementary materials. RNAseq data analysis files are available at https://github.com/chelseaherdman/MIC-drop. Reagents will be made available upon request to randall.peterson@pharm.utah.edu.
Supplementary Materials:
References (31)
References and Notes:
- 1.Patton EE, Zon LI, The art and design of genetic screens: zebrafish. Nature Reviews Genetics 2, 956–966 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam A et al. , Identification of 315 genes essential for early zebrafish development. Proceedings of the National Academy of Sciences of the United States of America 101, 12792–12797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driever W et al. , A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Haffter P et al. , The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Weinstein BM et al. , Hematopoietic mutations in the zebrafish. Development 123, 303–309 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Sison M, Cawker J, Buske C, Gerlai R, Fishing for genes influencing vertebrate behavior: zebrafish making headway. Lab Animal 35, 33–39 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Jain RA et al. , A Forward Genetic Screen in Zebrafish Identifies the G-Protein-Coupled Receptor CaSR as a Modulator of Sensorimotor Decision Making. Current biology: CB 28, 1357–1369.e1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang WY et al. , Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei W et al. , Guided genetic screen to identify genes essential in the regeneration of hair cells and other tissues. npj Regenerative Medicine 3, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varshney GK et al. , High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome research 25, 1030–1042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu RS et al. , A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Developmental Cell 46, 112–125.e114 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Kroll F et al. , A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. eLife 10, e59683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB, Rapid reverse genetic screening using CRISPR in zebrafish. Nature methods 12, 535–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshijima K et al. , Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev Cell 51, 645–657.e644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagnon JA et al. , Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLOS ONE 9, e98186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burger A et al. , Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025–2037 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Jost M, Weissman JS, CRISPR Approaches to Small Molecule Target Identification. ACS Chemical Biology 13, 366–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE, Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov 16, 89–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokel D et al. , Photochemical activation of TRPA1 channels in neurons and animals. Nature chemical biology 9, 257–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi S, Brueckner M, Genetics and Genomics of Congenital Heart Disease. Circulation research 120, 923–940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Ma X, Xu X, Zhang Y, Systematic identification and characterization of cardiac long intergenic noncoding RNAs in zebrafish. Scientific reports 7, 1250–1250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih YH et al. , Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ Cardiovasc Genet 8, 261–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juang J-MJ et al. , GSTM3 variant is a novel genetic modifier in Brugada syndrome, a disease with risk of sudden cardiac death. EBioMedicine 57, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Wei JJ, Sabatini DM, Lander ES, Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W et al. , Genetic Control of Collective Behavior in Zebrafish. iScience 23, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Brolosy MA et al. , Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blitz IL, Biesinger J, Xie X, Cho KWY, Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system. genesis 51, 827–834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panáková D, Werdich AA, MacRae CA, Wnt11 patterns a myocardial electrical gradient via regulation of the L-type Ca2+ channel. Nature 466 (7308) 874–878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


