Fig. 1. MIC-Drop enables high-throughput CRISPR screens in zebrafish.
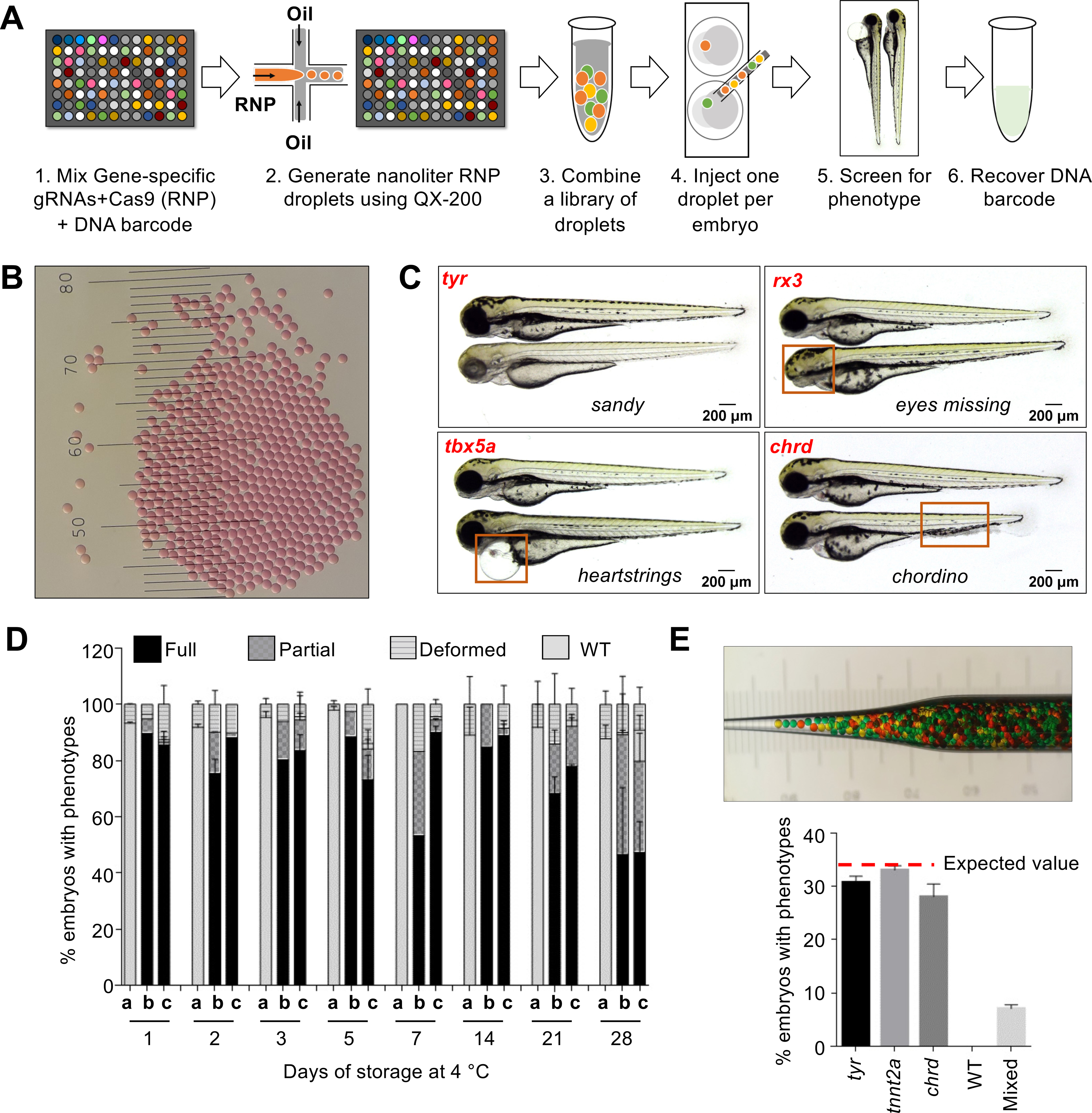
(A) Workflow of the MIC-Drop platform. A microfluidics device generates nanoliter-sized droplets, each containing ribonucleoproteins (RNP) targeting a gene-of-interest and a unique DNA barcode associated with the gene. Droplets targeting multiple genes are intermixed, loaded into a single injection needle and injected serially into one-cell zebrafish embryos. Embryos showing phenotypes-of-interest are isolated and the causative genotype is identified by retrieving and sequencing the barcode. (B) Droplets are uniform in size. Distance between bars is 0.1 mm. (C) Injection of droplets containing RNPs targeting tyr, rx3, tbx5a, and chrd genes recapitulates known mutant phenotypes in F0, highlighted by red boxes. (D) RNP-containing droplets are non-toxic and stable for prolonged storage – retaining activity at least 28 days of storage at 4°C. a: Uninjected; b: Traditional RNP injection; c: MIC-Drop injection. N = (Day1: a-118, b-79, c-134; Day2: a-116, b-68, c-59; Day3: a-120, b-88, c-87; Day5: a-129, b-78, c-95; Day7: a-105, b-102, c-107; Day14: a-94, b-53, c-77; Day21: a-123, b-100, c-132; Day28: a-114, b-100, c-94. (E) Single-needle injection of intermixed droplets targeting three different genes (tyr, tnnt2a, chrd) and subsequent phenotyping shows even representation of each droplet with all of the embryos exhibiting only one of the three expected phenotypes, and none (0%) were wildtype. Total N sequenced = 230 from 3 separate injections. Inset: Hundreds of pseudo-colored droplets (used as proxies for droplets targeting different genes) do not fuse when transferred to an injection needle.
