Abstract
Escherichia coli ILT-1, Klebsiella pneumoniae ILT-2, and K. pneumoniae ILT-3 were isolated in May 1999 in Paris, France, from a rectal swab of a hospitalized 5-month-old girl. These isolates had a clavulanic acid-inhibited substrate profile that included expanded-spectrum cephalosporins. The MICs of cefotaxime were higher for E. coli ILT-1 and K. pneumoniae ILT-2 than for K. pneumoniae ILT-3, while the opposite was found for the MICs of ceftazidime. Genetic and biochemical analyses revealed that E. coli ILT-1 and K. pneumoniae ILT-2 produced the CTX-M-18 β-lactamase, while K. pneumoniae ILT-3 produced the CTX-M-19 β-lactamase. The amino acid sequence of the CTX-M-18 β-lactamase differed from that of the CTX-M-9 β-lactamase by an Ala-to-Val change at position 231, while CTX-M-19 possessed an additional Pro-to-Ser change at position 167 in the omega loop of Ambler class A enzymes. The latter amino acid substitution may explain the CTX-M-19-mediated hydrolysis of ceftazidime, which has not been reported for other CTX-M-type enzymes. The blaCTX-M-18 and blaCTX-M-19 genes were located on transferable plasmids that varied in size (ca. 60 and 50 kb, respectively) but that showed similar restriction patterns.
Several clavulanic acid-inhibited Ambler class A expanded-spectrum β-lactamases (ESBLs) have been reported, mostly in members of the family Enterobacteriaceae, in addition to the TEM- and SHV-type ESBLs (21, 22). Among them, the CTX-M-type β-lactamases are typical ESBLs. The designation “CTX” refers to their powerful spectrum of hydrolysis of cefotaxime (32). The CTX-M β-lactamases hydrolyze ceftazidime with a very low catalytic efficiency. Thus, CTX-M β-lactamases confer to both wild-type and laboratory-obtained enterobacterial hosts high levels resistance to cefotaxime, ceftriaxone, and aztreonam but have only marginal effects on the MIC of ceftazidime.
The first CTX-M β-lactamase (CTX-M-1/MEN-1) was characterized in Escherichia coli strains isolated from German and Italian patients (4–6). This family of enzymes now comprises 14 members: CTX-M-1 to CTX-M-12, Toho-1, and Toho-2 (7, 23, 28, 32). These enzymes share 71 to 98% amino acid sequence identities. Escherichia coli and Salmonella enterica serotype Typhimurium are the enterobacterial species most frequently reported to produce the CTX-M β-lactamases (28, 32). The CTX-M-type β-lactamase-producing strains are endemic in Latin America, Japan, and some areas in Eastern Europe (6, 9, 12, 14, 24, 32). Rare reports signal the presence of CTX-M enzymes in enterobacterial isolates from other countries such as France, Kenya, Spain, and Greece (18, 28, 31).
The CTX-M β-lactamase genes are mostly located on plasmids, and Toho-1-like β-lactamase genes have also been found to be located on chromosomes (35). The progenitors of some of these plasmid-mediated enzymes could be the chromosomally encoded β-lactamases of the enterobacterial species Kluyvera ascorbata (GenBank accession number no. AJ251722).
This report identifies two isogenic CTX-M-type β-lactamases from enterobacterial strains isolated from the same patient. One of them was peculiar since it hydrolyzed ceftazidime much more than cefotaxime, a property not reported previously for a CTX-M-type enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present work are listed in Table 1. E. coli ILT-1, Klebsiella pneumoniae ILT-2, and K. pneumoniae ILT-3 clinical isolates were identified by the API 20E system (bioMérieux, Marcy l'Etoile, France).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | endA1 gyrA96 hsdR17 Δ(lac proA) relA recA1 supE44 thi F (lacIqlacZΔM15 proAB+traD36) | Gibco BRL-Life Technologies |
| Rifampin-resistant E. coli JM109 obtained in vitro | Rifampinr | This study |
| E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 | Extended-spectrum cephalosporin-resistant clinical isolates | This study |
| Plasmids | ||
| pPCRScript-Cam (SK+) | Chloramphenicolr | Stratagene Inc. |
| pILT-1, pILT-2, and pILT-3 | Natural plasmids from E. coli ILT-1, K. pneumoniae ILT-2 producing CTX-M-18, and K. pneumoniae ILT-3 producing CTX-M-19 | This study |
| pMA-1 and pMA-2 | Recombinant plasmid with a 900-bp PCR fragment from whole-cell DNA of E. coli ILT-1 and K. pneumoniae ILT-2 containing blaCTX-M-18 into pPCRScript-Cam (SK+) | This study |
| pMA-3 | Recombinant plasmid with a 900-bp PCR fragment from whole-cell DNA of K. pneumoniae ILT-3 containing blaCTX-M-19 into pPCRScript-Cam (SK+) | This study |
A superscript r indicates resistance.
PFGE.
Plug preparation was done according to the instructions of Bio-Rad (Ivry/Seine, France). Whole-cell DNAs from K. pneumoniae ILT-2 and ILT-3 were digested with XbaI at 37°C overnight. Electrophoresis in a 1% agarose gel in 0.5× Tris-borate-EDTA buffer was performed with a CHEF DRII apparatus (Bio-Rad). Migration conditions, staining, and chromosomal fingerprints compared and assigned to pulsed-field gel electrophoresis (PFGE) types and subtypes were as described previously (13).
Susceptibility testing.
Antibiotic-containing disks were used for antibiotic susceptibility testing by the disk diffusion assay (Sanofi-Diagnostics Pasteur, Marnes-la-Coquette, France), as described previously (26). The double-disk synergy test was performed with ceftazidime-, cefotaxime-, aztreonam-, and amoxicillin-clavulanic acid-containing disks on Mueller-Hinton (MH) agar plates; and the results were interpreted as described previously (16). MICs were determined by an agar dilution technique on MH agar (Sanofi-Diagnostics Pasteur) with an inoculum of 104 CFU per spot, as described previously (27). All plates were incubated at 37°C for 18 h. The MICs of the β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml) or tazobactam (4 μg/ml). MIC results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards after incubation at 37°C for 18 h (21).
PCR, cloning experiments, recombinant plasmid analysis, and DNA sequencing.
Whole-cell DNAs from E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 were extracted as described previously(27). They were used as templates in standard PCR experiments (29) with sets of laboratory-designed primers for detection of class A β-lactamase genes and their extended-spectrum derivatives, blaTEM, blaSHV, blaPER-1, blaPER-2, blaVEB-1, blaToho-1, blaToho-2, blaSFO-1, blaGES-1, blaCTX-M-2 (26), and blaCTX-M-3 (primer CTXM-A, 5′-CGCTTTGCGATGTGCAG-3′; primer CTXM-B, 5′-ACCGCGATATCGTTGGT-3′).
By using external primers for blaToho-1 (primer PreCTX-M-A, 5′-TGATGTAACACGGATTGACC-3′; primer PreCTX-M-B, 5′-TTACAGCCCTTCGGCGATGA-3′), the fragments obtained by PCR were cloned into the SrfI site of plasmid pPCRScript-Cam SK(+) (Stratagene, Amsterdam, The Netherlands) and expressed in E. coli JM109. Recombinant plasmids were selected on amoxicillin (100 μg/ml)- and chloramphenicol (30 μg/ml)-containing Trypticase soy (TS) agar plates. Recombinant plasmids pMA-1, pMA-2, and pMA-3 were obtained using E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 as templates, respectively. Both strands of the cloned DNA fragments, inserted into recombinant plasmids, were sequenced with an Applied Biosystems sequencer (model ABI 377). The nucleotide and deduced amino acid sequences were analyzed and compared to sequences available over the Internet at the National Center for Biotechnology Information website (http: //www.ncbi.nlm.nih.gov).
Plasmid content, conjugation, and hybridization.
Plasmid DNAs from E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and recombinant clones were extracted with Maxi columns (Qiagen, Courtaboeuf, France). Plasmid DNAs were analyzed by electrophoresis on a 0.8% agarose gel (Gibco BRL-Life Technologies, Cergy-Pontoise, France) containing 0.15 μg of ethidium bromide per ml for 18 h at 90 V. A 1-kb DNA ladder (Gibco BRL-Life Technologies) was used as a reference DNA size standard. Conjugation experiments were performed between E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and nalidixic acid- and rifampin-resistant E. coli JM109 in solid and liquid media at 37°C, as described previously (27). Transconjugants were selected on TS agar plates containing 150 μg of rifampin per ml or 100 μg of nalidixic acid and 100 μg of amoxicillin per ml. In order to locate the β-lactamase genes, plasmid DNAs of E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and their E. coli transconjugants were extracted with a Qiagen Maxi column and restricted with HindIII and BamHI. The DNA fragments were run on a 0.8% agarose gel, transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Orsay, France) by the Southern technique (29), and hybridized with a PCR-generated probe of 550 bp whose sequence was specific for a region internal to the blaCTX-M-3 sequence (primers CTX-M-A and CTX-M-B) and a PCR-generated probe of 850 bp whose sequence was specific for a region internal to the blaTEM sequence (27). The nonradioactive enhanced chemiluminescence random prime system (Amersham Pharmacia Biotech) was used to label the DNA probes and for detection.
β-Lactamase extracts and purification.
Cultures of E. coli JM109 harboring recombinant plasmids pMA-1 and pMA-3 were grown overnight at 37°C in 4 liters of TS broth containing amoxicillin (100 μg/ml) and chloramphenicol (30 μg/ml). The β-lactamase extracts were obtained as described previously (26) and were resuspended in 30 ml of 20 mM Tris-HCl (pH 9.2). Similarly, β-lactamase extracts were obtained from 10-ml cultures of E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and their E. coli transconjugants and were subsequently resuspended in 0.1 ml of sodium phosphate buffer (pH 7).
The β-lactamase extracts of cultures of E. coli JM109(pMA-1) or E. coli JM109(pMA-3) were dialyzed overnight against 20 mM Tris-HCl (pH 9.2) at 4°C in order to eliminate spermine and to adjust the pH to 9.2. The enzyme extracts were loaded onto a preequilibrated Q-Sepharose column (1.6 by 5 cm; Amersham Pharmacia Biotech) with the same buffer. The resulting enzyme extract was recovered in the flowthrough and was dialyzed against 50 mM phosphate buffer (pH 6) overnight at 4°C. This extract was then loaded onto a preequilibrated S-Sepharose column, and the proteins were eluted with a linear NaCl gradient (0 to 0.5 M). The β-lactamase activity was eluted with NaCl at a concentration of 50 mM in phosphate buffer. The fractions with the highest β-lactamase activities were pooled and dialyzed against 50 mM phosphate buffer (pH 7), prior to concentration 10-fold with Centrisart-C30 microcentrifuge filters (Sartorius, Göttingen, Germany). The purified β-lactamase extracts were used for determination of enzymatic activity. Their purity was estimated by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (29).
IEF analysis.
β-Lactamase extracts from cultures of E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and their E. coli transconjugants and purified enzymes from cultures of E. coli JM109 harboring recombinant plasmid pMA-1 or pMA-3 were subjected to analytical isoelectric focusing (IEF) analysis on a polyacrylamide gel with ampholine (pH 3.5 to 9.5), as described previously (27). The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid, Dardilly, France) in 100 mM phosphate buffer (pH 7.0). The pI values were determined and compared to those of known β-lactamases.
Kinetic parameters.
Purified β-lactamase extracts were used for kinetic measurements performed at 30°C in 100 mM sodium phosphate (pH 7.0). The initial rates of hydrolysis were determined with an ULTROSPEC 2000 UV spectrophotometer (Amersham Pharmacia Biotech) and were analyzed by computer with Swift II software (Amersham Pharmacia Biotech). The kcat and Km values were determined by analyzing β-lactam hydrolysis under initial rate conditions by using the Eadie-Hoffstee linearization of the Michaelis-Menten equation, as described previously (27).
The 50% inhibitory concentrations (IC50s) were determined for clavulanic acid, tazobactam, and sulbactam. Various concentrations of these inhibitors were preincubated with the purified enzyme for 3 min at 30°C to determine the concentrations that reduced the rate of hydrolysis of 1 μM cephaloridine by 50%. Results are expressed in units of micromolar. The specific activities of the purified β-lactamase from E. coli JM109 harboring pMA-1 and E. coli JM109 harboring pMA-3 were obtained as described previously (27). One unit of enzyme activity was defined as the activity which hydrolyzed 1 μM benzylpenicillin per min per mg of protein. Specific activities were determined with 100 μM benzylpenicillin as the substrate. Additionally, the specific activity of nonpurified β-lactamase extract from cultures of E. coli JM109 harboring pMA-1 or pMA-3 was determined with 100 μM piperacillin, a substrate that was hydrolyzed similarly by β-lactamases from cultures of E. coli JM109(pMA-1) or E. coli JM109(pMA-3). The protein content was measured by the Bio-Rad DC Protein assay.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the CTX-M-18 and CTX-M-19 β-lactamases reported in this paper have been assigned GenBank nucleotide sequence database accession nos. AF325133 and AF325134, respectively.
RESULTS
Characterization of clinical isolates and preliminary antibiotic susceptibility testing.
E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 were isolated from a 5-month-old girl in May 1999 at the Robert Debré Hospital (Paris, France) as a result of systematic rectal screening of patients admitted to the pediatric intensive care unit (ICU) for multidrug-resistant bacteria. The child was born in Vietnam and was transferred to the cardiovascular unit of Insitute Jacques Cartier (Paris, France), where she was hospitalized for 15 days for surgical cure of intraventricular communication. During the follow-up after surgery, she developed cardiovascular failure, for which she received treatment with cefotaxime, vancomycin, and netilmicin for 1 week. Rectal screening for multidrug-resistant bacteria performed 2 weeks after her admission to the ICU was positive for ESBL-producing isolates, but the rectal screening performed on the day of her admission was negative. No other enterobacterial isolate with a similar ESBL resistance profile was identified in the same ICU concomitantly or during the following 4-month period (data not shown).
Antibiotic susceptibility testing by disk diffusion suggested that the extended-spectrum cephalosporin resistance profile was due to the presence of an ESBL. Synergies were observed between clavulanate and ceftazidime, cefotaxime, and aztreonam (data not shown). E. coli ILT-1 and K. pneumoniae ILT-2 were more resistant to cefotaxime than to ceftazidime (Table 2). On the other hand, K. pneumoniae ILT-3 was more resistant to ceftazidime than to cefotaxime (Table 2). E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 were also resistant to amikacin, gentamicin, kanamycin, netilmicin, tobramycin, and chloramphenicol and were susceptible to sulfonamides and fluoroquinolones. E. coli ILT-1 and K. pneumoniae ILT-2 were susceptible to rifampin, while K. pneumoniae ILT-3 was resistant to rifampin (data not shown).
TABLE 2.
MICs of β-lactams for clinical isolates, their E. coli JM109 transconjugants, E. coli JM109 harboring recombinants plasmids, and reference strain E. coli JM109
| β-Lactam(s)a | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| E. coli ILT-1 (CTX-M-18 and TEM-1) | K. pneumoniae ILT-2 (CTX-M-18 and TEM-1 and SHV-1 like) | K. pneumoniae ILT-3 (CTX-M-19 and TEM-1 and SHV-1 like) | E. coli JM109 (pILT-1 and pILT-2) (CTX-M-18 and TEM-1) | E. coli JM109 (pILT-3) (CTX-M-19 and TEM-1) | E. coli JM109 (pMA-1 or pMA-2) (CTX-M-18) | E. coli JM109 (pMA-3) (CTX-M-19) | E. coli JM109 | |
| Amoxicillin | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 2 |
| Amoxicillin + CLA | 64 | 32 | 32 | 32 | 8 | 128 | 128 | 1 |
| Ticarcillin | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 2 |
| Ticarcillin + CLA | 256 | 64 | 64 | 64 | 32 | 256 | 256 | 2 |
| Ticarcillin + TZB | 32 | 16 | 64 | 16 | 64 | 64 | 64 | 2 |
| Piperacillin | 512 | 512 | 512 | 512 | 128 | >512 | >512 | 1 |
| Piperacillin + CLA | 8 | 8 | 4 | 4 | 2 | 16 | 8 | 1 |
| Piperacillin + TZB | 4 | 4 | 8 | 2 | 2 | 4 | 4 | 1 |
| Cephalothin | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 4 |
| Cefuroxime | 512 | 256 | 128 | >512 | 256 | >512 | >512 | 1 |
| Cefoxitin | 16 | 4 | 8 | 16 | 16 | 4 | 4 | 4 |
| Ceftriaxone | 128 | 32 | 32 | 64 | 4 | 64 | 4 | 0.06 |
| Cefotaxime | 256 | 32 | 8 | 8 | 1 | 64 | 4 | 0.06 |
| Cefotaxime + CLA | 4 | 2 | <0.06 | 0.5 | 0.12 | 0.5 | 0.5 | 0.06 |
| Cefotaxime + TZB | 1 | 0.5 | 0.25 | 0.12 | 0.12 | 0.25 | 0.25 | 0.06 |
| Ceftazidime | 4 | 1 | 512 | 0.5 | 64 | 2 | 128 | 0.25 |
| Ceftazidime + CLA | 1 | 0.5 | 16 | 0.25 | 2 | 0.5 | 16 | 0.25 |
| Ceftazidime + TZB | 0.5 | 0.06 | 32 | 0.25 | 4 | 0.5 | 32 | 0.25 |
| Cefepime | 32 | 4 | 4 | 4 | 0.5 | 16 | 4 | 0.03 |
| Aztreonam | 32 | 8 | 8 | 32 | 4 | 64 | 4 | 0.12 |
| Aztreonam + CLA | 2 | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.12 | 0.12 |
| Moxalactam | 0.5 | 0.5 | 2 | 0.5 | 2 | 0.5 | 2 | 0.25 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 | 0.25 | 0.25 | 0.25 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
PFGE analysis of XbaI-restricted DNAs of K. pneumoniae ILT-2 and ILT-3 showed that they have distinguishable patterns and thus were not epidemiologically related (data not shown).
PCR experiments and cloning of β-lactamase genes.
Preliminary PCR amplification experiments with primers designed to amplify several internal fragments of ESBL genes gave positive results for blaTEM and blaCTX-M-3 for each clinical strain and positive results for blaSHV for the two K. pneumoniae clinical isolates. External primers for blaCTX-M-3 were used to PCR amplify a 900-bp fragment of a blaCTX-M-3-like gene by using whole-cell DNAs of E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 as templates. The corresponding PCR amplimers were cloned into the SrfI site of pPCRScript-Cam (SK+), giving rise to recombinant plasmids pMA-1, pMA-2, and pMA-3, respectively.
DNA sequencing.
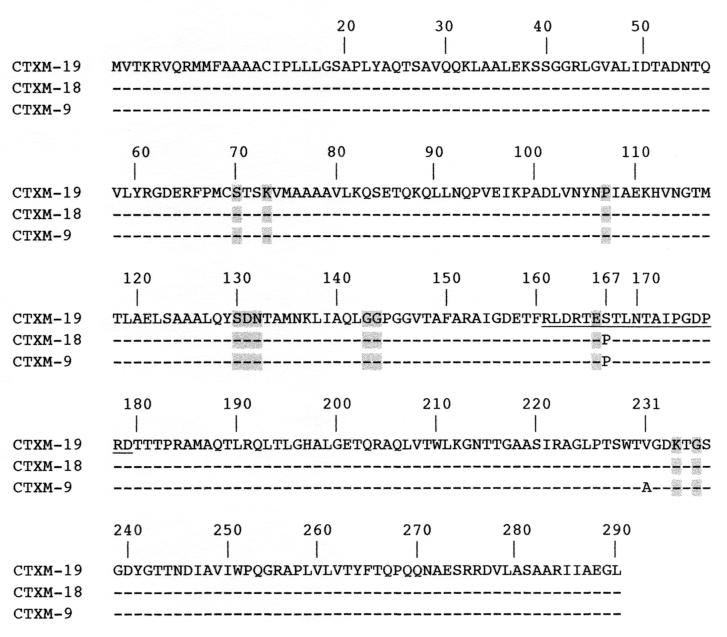
Analysis of the inserted nucleotide sequences from recombinant plasmids pMA-1, pMA-2, and pMA-3 showed in each case an 876-bp open reading frame encoding a 291-amino-acid protein. On the basis of the protein alignments, two CTX-M-type enzymes were identified: CTX-M-18 encoded by pMA-1 and pMA-2 and CTX-M-19 encoded by pMA-3. CTX-M-18 differed from the previously characterized CTX-M-9 enzyme by an alanine-to-valine substitution at position 231, according to the designation of class A enzymes of Ambler et al. (1), and CTX-M-19 had an additional proline-to-serine substitution at position 167 (Fig. 1). The latter substitution lay close to conserved residue Glu166 in the omega loop of class A β-lactamases. CTX-M-18 and CTX-M-19, like CTX-M-9, share 88% amino acid identity with the most closely related plasmid-mediated CTX-M-type enzyme, Toho-2, and 80% amino acid identity with the chromosome-encoded penicillinase of K. ascorbata (GenBank accession number no. AJ251722).
FIG. 1.
Comparison of the amino acid sequences of CTX-M-18 and CTX-M-19 to that of CTX-M-9. Dashes indicate identical amino acids. The numbering is according to the designation of Ambler et al. (1). Highlighted amino acids are those strictly conserved in class A β-lactamases (17), and the amino acids of the omega loop are underlined.
Since PCR experiments also gave positive results for blaTEM with clinical strains, the PCR amplicons were sequenced. In all cases, the same blaTEM-1 gene was identified (data not shown).
Kinetic parameters.
The specific activities of the purified CTX-M-18 and CTX-M-19 β-lactamases from cultures of E. coli JM109(pMA-1) and E. coli JM109(pMA-3) were 30 and 10 μmol · min−1 · mg of protein−1, respectively, when 100 μM benzylpenicillin was used as the substrate. Their overall rate of recovery was 70% with 60-fold purification. Their purities were estimated to be 90%. The kinetic parameters for the purified CTX-M-18 β-lactamase showed that it had strong activity against most β-lactams including cefotaxime, ceftriaxone, cefepime, and cefpirome (Table 3). The catalytic activity of CTX-M-19 was lower than that of CTX-M-18 for all substrates except piperacillin and ceftazidime. The activity of CTX-M-18 against ceftazidime was not detectable (Table 3). In contrast, kinetic parameters could be calculated for ceftazidime hydrolysis for CTX-M-19. However, the specific activity of CTX-M-19 for ceftazidime remained low (0.5 mU/mg). The catalytic activity of CTX-M-19 against aztreonam, cefepime, and cefpirome was not detectable.
TABLE 3.
Steady-state kinetic parameters of CTX-M-18 and CTX-M-19 β-lactamases
| Substrate | CTX-M-18
|
CTX-M-19
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) | |
| Benzylpenicillin | 30 | 29 | 1,000 | 5 | 15 | 300 |
| Amoxicillin | 10 | 105 | 100 | 1 | 100 | 10 |
| Ticarcillin | 3 | 17 | 160 | 1 | 30 | 40 |
| Piperacillin | 15 | 23 | 600 | 8 | 10 | 750 |
| Cephaloridine | 7 | 216 | 30 | 30 | 123 | 250 |
| Cefuroxime | 40 | 70 | 600 | 8 | 40 | 170 |
| Cefoxitin | ND | ND | ND | ND | ND | ND |
| Ceftazidime | ND | ND | ND | 0.02 | 25 | 0.1 |
| Cetriaxone | 20 | 20 | 850 | 0.1 | 80 | 1 |
| Cefotaxime | 20 | 54 | 370 | 3 | 60 | 55 |
| Cefepime | 20 | 525 | 40 | ND | ND | ND |
| Cefpirome | 65 | 650 | 100 | ND | ND | ND |
| Imipenem | ND | ND | ND | ND | ND | ND |
| Aztreonam | 2 | 286 | 10 | ND | ND | ND |
Values are means of three independent measures (the standard deviations of the values were within 15%).
ND, not determinable (the initial rate of hydrolysis was lower than 0.001 μM−1 · s−1).
Inhibition studies as measured by determination of the IC50s with cephaloridine as the substrate showed that CTX-M-18 and CTX-M-19 were inhibited by clavulanic acid (60 and 90 nM, respectively) and sulbactam (500 and 150 nM, respectively). However, tazobactam inhibited more CTX-M-18 activity (IC50, 5 nM) than CTX-M-19 activity (IC50, 40 nM).
Transfer of antibiotic resistance, plasmid analysis, and hybridizations.
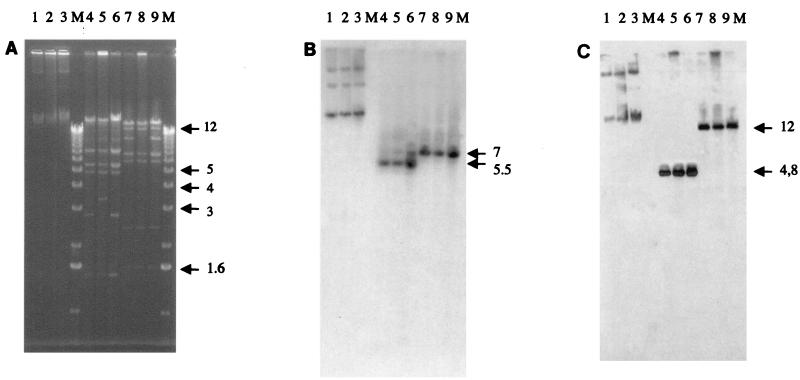
E. coli transconjugants with E. coli ILT-1 and K. pneumoniae ILT-2 and ILT-3 as donors were obtained at high frequencies. Each transconjugant showed an ESBL phenotype. Transconjugants of E. coli ILT-1 and K. pneumoniae ILT-2 harbored plasmids pILT-1 and pILT-2 of ca. 60 kb, respectively, with indistinguishable restriction patterns (Fig. 2A). Transconjugants of K. pneumoniae ILT-3 had plasmid pILT-3 of ca. 50 kb. Plasmid pILT-3 was related to plasmids pILT-1 and pILT-2 but differed by at least deletions of 1.6- and 3-kb HindIII-restricted fragments, deletion of an additional 3-kb HindIII-restricted fragment, and deletion of ca.10 kb of a BamHI-restricted fragment (Fig. 2A). Plasmids pILT-1, pILT-2, and pILT-3 conferred the same pattern of resistance to non-β-lactam antibiotics, including resistance to gentamicin, kanamycin, netilmicin, tobramycin, and chloramphenicol. HindIII- and BamHI-restricted plasmids pILT-1, pILT-2, and pILT-3 hybridized with an internal probe for blaCTX-M on DNA fragments of different sizes. However, for a given restriction digest, the hybridizing DNA fragments were identical for plasmids pILT-1 to pILT-3 (Fig. 2B). Similarly, hybridization of HindIII- and BamHI-restricted plasmids pILT-1 to pILT-3 with an internal probe for blaTEM-1 gave different hybridizing DNA fragments that were identical for all plasmids for a given restriction digest (Fig. 2C). These results showed that restricted DNA fragments of pILT-1, pILT-2, and pILT-3 that contained either the blaTEM or the blaCTX-M gene were different and that these natural plasmids were structurally related. Additionally, probes for blaTEM or blaCTX-M hybridized at the migration position of chromosomal DNA (Fig. 2B and C).
FIG. 2.
Plasmid analysis of E. coli JM109 transconjugants harboring natural plasmids pILT-1, pILT-3, and pILT-2 from E. coli ILT-1, K. pneumoniae ILT-3, and K. pneumoniae ILT-2, respectively. (A) Agarose gel electrophoresis of crude DNA lysates. Lane 1, pILT-1; lane 2, pILT-3; lane 3, pILT-2; lane 4, HindIII-restricted pILT-1; lane 5, HindIII-restricted pILT-3; lane 6, HindIII-restricted pILT-2; lane 7, BamHI-restricted pILT-1; lane 8, BamHI-restricted pILT-3; lane 9, BamHI-restricted pILT-2; lane M, molecular size marker. (B) Corresponding autoradiograph after Southern transfer and hybridization with the blaCTX-M probe. (C) Corresponding autoradiograph after Southern transfer and hybridization with the blaTEM probe. Hybridizing DNA fragments and their sizes are indicated by horizontal arrows. Numbers on the right of each gel are in kilobases.
IEF analysis.
IEF analysis showed that E. coli ILT-1, K. pneumoniae ILT-2 and ILT-3, and their transconjugants expressed two β-lactamases with pIs of 5.4 and 8. The β-lactamase with a pI of 5.4 corresponded to TEM-1. The β-lactamase with a pI of 8 corresponded to CTX-M-18 for E. coli ILT-1, K. pneumoniae ILT-2, their transconjugants, E. coli JM109 harboring recombinant plasmid pMA-1, and E. coli JM109 harboring recombinant plasmid pMA-2 and to CTX-M-19 for K. pneumoniae ILT-3, its transconjugant, and E. coli JM109(pMA-3). An additional β-lactamase with a pI of 7.6 likely corresponded to the chromosomally encoded SHV-type β-lactamase of K. pneumoniae for isolates ILT-2 and ILT-3.
Detailed analysis of β-lactam susceptibility.
The MICs of β-lactams for the CTX-M-18- or CTX-M-19-producing strains are shown in Table 2. These results indicate that CTX-M-18- and CTX-M-19-producing strains were resistant to penicillins and restricted- and expanded-spectrum cephalosporins. For the CTX-M-18-producing strains (E. coli ILT-1, K. pneumoniae ILT-2, their E. coli transconjugants, E. coli JM109 harboring pMA-1, and E. coli JM109 harboring pMA-2), the MICs of cefotaxime and aztreonam were higher than those of ceftazidime. On the contrary, the MICs of ceftazidime were higher than the MICs of cefotaxime and aztreonam for the CTX-M-19 producers [K. pneumoniae ILT-3, its transconjugant, and E. coli JM109(pMA-3)]. Moreover, the MICs of cefepime and cefpirome for the CTX-M-18 producers were higher than those for the CTX-M-19 producers. The addition of clavulanic acid and tazobactam strongly reduced the MICs of penicillins and cephalosporins for the CTX-M-18 and CTX-M-19 producers. The differences in the MICs of β-lactams for the CTX-M-18 and CTX-M-19 producers could be related not only to the catalytic activities of the enzymes but also to differences in folding or the stabilities of the proteins. Indeed, although the kinetic parameters for CTX-M-18 and CTX-M-19 were very similar for piperacillin (Table 3), the specific activities of nonpurified extracts of cultures of E. coli JM109(pMA-1/CTX-M-18) and of E. coli JM109(pMA-3/CTX-M-19) were 840 and 433 mU · mg of protein−1, respectively, with 100 μM piperacillin as the substrate.
DISCUSSION
The work described here further characterized biochemically two CTX-M-type enzymes, CTX-M-18 in E. coli and K. pneumoniae and CTX-M-19 in K. pneumoniae. CTX-M enzymes have been identified mostly from E. coli and Salmonella sp. strains and rarely from K. pneunomiae, whereas the TEM- and SHV-type extended-spectrum derivatives have been identified mostly from K. pneumoniae (22, 32). Identification of blaCTX-M-18 in E. coli and K. pneumoniae signals the interspecies transfer of a plasmid-mediated CTX-M gene between isolates in the fecal flora of the same patient.
The CTX-M β-lactamases have been reported in Europe, mostly in Eastern Europe. CTX-M-9 was recently identified in an E. coli isolate in Spain and was also likely present in 22 additional E. coli strains and in 1 S. enterica serotype Virchow strain in the same hospital in Barcelona, Spain (28). Although CTX-M-1 (MEN-1) was identified from a clinical sample of an Italian patient hospitalized in France (4), CTX-M-3-producing Enterobacter cloacae was the first CTX-M-producing isolate recovered in France from a French patient (11). In the present study, the enterobacterial isolates were from a Vietnamese patient hospitalized in France several times and consecutively in different French hospitals. Thus, it is difficult to define reliably the geographical origins of the isolated strains.
On the basis of their amino acid sequences, CTX-M enzymes may be divided into three clusters (28, 32) that differ by 20 to 25%. The first cluster groups CTX-M-1, CTX-M-3, CTX-M-10, and CTX-M-12; the second cluster groups CTX-M-2, CTX-M-4 to CTX-M-7, and Toho-1; and the third cluster groups Toho-2, CTX-M-9, and now CTX-M-18 and CTX-M-19. The plasmid-mediated CTX-M-18 and CTX-M-19 enzymes are related to the chromosomal penicillinase of K. ascorbata, underlining their possible origin. All characteristic substitutions assumed to be implicated in expanded-spectrum cephalosporin hydrolysis of the CTX-M enzymes are also present in CTX-M-18 and CTX-M-19, for example, Ser237, Thr244, and Arg276 (32). Our work has characterized the biochemical properties of CTX-M-18, which were found to be similar to those of the other cefotaxime-hydrolyzing CTX-M enzymes and which may correspond to those partially reported for CTX-M-9 (28). Tazobactam exhibited a stronger inhibitory activity than clavulanic acid against CTX-M-18 and CTX-M-19, as previously reported for the CTX-M enzymes (32). The catalytic efficiencies of CTX-M-18 and CTX-M-19 for several β-lactams remained lower than those reported for CTX-M-1 and CTX-M-5, the only other two CTX-M enzymes for which detailed kinetic data have been reported (4, 9).
Analysis of the crystal structure of Toho-1 shows that the amino acids of the omega loop play the main role in its substrate profile for cephalosporins (15). The omega loop amino acids (residues 161 to 179) of CTX-M-9 and CTX-M-18 are identical to those of Toho-1. Thus, results of analysis of the crystal structure of the Glu166Ala mutant of Toho-1 (15) may be applied to analysis of CTX-M-9 and CTX-M-18. The Toho-1 β-lactamase has fewer hydrogen bond interactions between the omega loop and the α-β domain in the vicinity of Asn170 and Asp240 compared to the numbers of interactions for the restricted-spectrum β-lactamase of Bacillus licheniformis (15). Moreover, no hydrogen bond connects both the N and the C termini of the omega loop between the amino acid at position 160 and the threonine at position 181. The CTX-M-9, CTX-M-18, and CTX-M-19 β-lactamases, like the Toho-1 β-lactamase, have a Phe residue at position 160 that cannot form a hydrogen bond with the threonine at position 181. The same Phe residue cannot interact with Asp157. The CTX-M-9, CTX-M-18, and CTX-M-19 β-lactamases, like the Toho-1 β-lactamase, retain a hydrogen bond between Asp179 and Arg164, as in some narrow-spectrum class A β-lactamases; in addition, they retain the interactions between Lys73 and Glu166 and between Asn136 and Glu166 that help to maintain the structural integrity of the omega loop. The expanded-spectrum hydrolytic activity of the CTX-M-9, CTX-M-18, and CTX-M-19 β-lactamases, like that of the Toho-1 β-lactamase, may be related to an increased flexibility of the omega loop.
The most interesting aspect of our work is the CTX-M-19-mediated resistance to ceftazidime. The proline-to-serine change in CTX-M-19 occurs in the omega loop structure of class A β-lactamases (Fig. 1). No natural class A β-lactamases have a serine residue at position 167 like CTX-M-19 does. Amino acid substitutions in the omega loop of extended-spectrum TEM- and SHV-type enzymes selected in vivo were identified only at position 164 or 179, breaking the hydrogen bond between Arg164 and Asp179 (2, 8, 19, 20, 33). Site-specific mutagenesis experiments showed that a Pro-to-Gly substitution at position 167 in TEM-1 and PSE-4 increased the hydrolytic activities of the enzymes against ceftazidime (25, 30). It is possible that Ser167 in CTX-M-19 may enlarge the binding site for ceftazidime in the catalytic site of the enzyme. Interestingly, Ser167 in CTX-M-19 is located next to Glu166, an amino acid residue that is known to promote activation of the hydrolytic water molecule for hydrolysis of the acyl enzyme intermediate in class A β-lactamases (3).
The substrate profile of CTX-M-19 was extended to ceftazidime but its catalytic efficiency for the other extended-spectrum cephalosporins was lower than that obtained for the other CTX-M-type enzymes. The variabilities of the substrate profiles of expanded-spectrum cephalosporins (except ceftazidime) have been reported for CTX-M enzymes such as CTX-M-2 and its derivative with a point mutation, CTX-M-4 (12). The kinetic parameters of CTX-M-19 against ceftazidime revealed a surprisingly low catalytic efficiency. A similar discrepancy between MIC results and kinetic data has been reported in other cases, such as for ceftazidime and OXA-10 derivatives of Ambler class D (10). Interestingly, misfolding was found for a TEM-1 β-lactamase with a Pro167Thr mutation (34). Similarly, the Pro167Ser change in CTX-M-19 may the explain instability of this protein and the rapid loss of its activity (data not shown)
The blaCTX-M-18 and blaCTX-M-19 genes were located on self-transferable and structurally related plasmids of enterobacterial isolates from the same patient, thus suggesting the in vivo selection of a CTX-M-19 producer that conferred resistance to ceftazidime. The blaCTX-M-18 and blaCTX-M-19 genes were associated with blaTEM-1 on structurally related plasmids. Although clinical strains that produce two β-lactamase genes have been reported, plasmids that carry two β-lactamases genes have not been extensively described. Interestingly, the probes for blaCTX-M and blaTEM also hybridized at the chromosomal position (Fig. 2B and C). This result may indicate a transposon location of the blaCTX-M-18 and blaCTX-M-19 genes, as suggested for Toho-1-like genes (35), which is being studied in our laboratory.
Finally, this work showed once again that novel ESBLs can be identified from clinical specimens of patients hospitalized in ICUs and that the substrate profiles of the CTX-M enzymes may be extended to ceftazidime. Thus, detection of CTX-M producers should not be based solely on the fact that the MICs of cefotaxime are higher than those of ceftazidime.
ACKNOWLEDGMENTS
This work was financed by a grant from the Ministères de l'Education Nationale et de la Recherche (grant UPRES, JE-2227), Université Paris XI, Paris, and the French network on β-lactamase research, “Les β-Lactamases; de l'Observation Clinique à la Structure.”
REFERENCES
- 1.Ambler R P, Coulson A F W, Frère J-M, Ghuysen J M, Joris B, Forsman M, Lévesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol Lett. 1997;152:163–167. doi: 10.1016/s0378-1097(97)00196-1. [DOI] [PubMed] [Google Scholar]
- 3.Barnerjee S, Pieper U, Kapadia G, Pannell L K, Herzberg O. Role of omega-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry. 1998;37:3286–3296. doi: 10.1021/bi972127f. [DOI] [PubMed] [Google Scholar]
- 4.Barthélémy M, Peduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 6.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet R, Sampaio J L M, Labia R, De Champs C, Sirot D, Chanal C, Sirot J. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob Agents Chemother. 2000;44:1936–1942. doi: 10.1128/aac.44.7.1936-1942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonomo R A, Rudin S D, Shlaes D M. OHIO-1 β-lactamase mutants: Asp179Gly mutation confers resistance to ceftazidime. FEMS Microbiol Lett. 1997;152:275–278. doi: 10.1111/j.1574-6968.1997.tb10439.x. [DOI] [PubMed] [Google Scholar]
- 9.Bradford P A, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5: a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danel F, Hall L M C, Duke B, Gür D, Livermore D M. OXA-17, a further extended-spectrum variant of OXA-10 β-lactamase isolated from P. aeruginosa. Antimicrob Agents Chemother. 1999;43:1362–1366. doi: 10.1128/aac.43.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucet-Populaire F, Ghnassia J C, Bonnet R, Sirot J. First isolation of a CTX-M-3-producing Enterobacter cloacae in France. Antimicrob Agents Chemother. 2000;44:3239–3240. doi: 10.1128/aac.44.11.3239-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girlich D, Karim A, Poirel L, Cavin M-H, Verny C, Nordmann P. Molecular epidemiology of an outbreak due to IRT-2 β-lactamase-producing strains of Klebsiella pneumoniae in a geriatric department. J Antimicrob Chemother. 2000;45:467–473. doi: 10.1093/jac/45.4.467. [DOI] [PubMed] [Google Scholar]
- 14.Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibuka A, Taguchi A, Ishiguro M, Fushinobu S, Ishii Y, Kamitori S, Okuyama K, Yamaguchi K, Konno M, Matsuzawa H. Crystal structure of the E166A mutant of extended-spectrum β-lactamase Toho-1 at 1.8 Å resolution. J Mol Biol. 1999;285:2079–2087. doi: 10.1006/jmbi.1998.2432. [DOI] [PubMed] [Google Scholar]
- 16.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 17.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J-M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariuki S, Corkill J E, Revathi G, Musoke R, Hart C A. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob Agents Chemother. 2001;45:2141–2143. doi: 10.1128/AAC.45.7.2141-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurokawa H, Yagi T, Shibata N, Shibayama K, Kamichi K, Arakawa Y. A new SHV-derived extended-spectrum β-lactamase (SHV-24) that hydrolyzes ceftazidime through a single-amino-acid substitution (D179G) in the omega loop. Antimicrob Agents Chemother. 2000;44:1725–1727. doi: 10.1128/aac.44.6.1725-1727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matagne A, Lamotte-Brasseur J, Frère J-M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. National Committee for Clinical Laboratory Standards, Wayne, Pa. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7–A4. [Google Scholar]
- 22.Nordmann P. Trends in β-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27:S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 23.Oliver A, Pérez-Diaz J C, Coque T M, Baquero F, Canton R. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob Agents Chemother. 2001;45:616–620. doi: 10.1128/AAC.45.2.616-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palucha A, Mikiewicz B, Hryniewicz W, Gniadkowski M. Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J Antimicrob Chemother. 1999;44:489–499. doi: 10.1093/jac/44.4.489. [DOI] [PubMed] [Google Scholar]
- 25.Palzkill T, Le Q Q, Venkatachalam K V, LaRocco M, Ocera H. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 26.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabaté M, Tarrago R, Navarro F, Miro E, Vergés C, Barbé J, Prats G. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1970–1973. doi: 10.1128/aac.44.7.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Therrien C, Sanschagrin F, Palzkill T, Lévesque R C. Roles of amino acids 161 to 179 in the PSE-4 omega loop in substrate specificity and in resistance to ceftazidime. Antimicrob Agents Chemother. 1998;42:2576–2583. doi: 10.1128/aac.42.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzouvelekis L S, Gazouli M, Markigiannakis A, Paraskaki E, Legakis N J, Tzelepi E. Emergence of resistance to third-generation cephalosporins amongst Salmonella typhimurium isolates in Greece: report of the first three cases. J Antimicrob Chemother. 1998;42:273–275. doi: 10.1093/jac/42.2.273. [DOI] [PubMed] [Google Scholar]
- 32.Tzouvelekis L S, Tzelepi E, Tassios P T, Legakis N J. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int J Antimicrob Agents. 2000;14:137–142. doi: 10.1016/s0924-8579(99)00165-x. [DOI] [PubMed] [Google Scholar]
- 33.Vakulenko S B, Taibi-Tronche P, Toth M, Massova I, Lerner S A, Mobashery S. Effects on substrate profile by mutational substitutions at positions 164 and 179 of the class A TEMpUC19 β-lactamase from Escherichia coli. J Biol Chem. 1999;274:23052–23060. doi: 10.1074/jbc.274.33.23052. [DOI] [PubMed] [Google Scholar]
- 34.Vanhove M, Raquet X, Palzkill T, Pain R H, Frère J M. The rate limiting step in the folding of the cis-Pro167Thr mutant of TEM-1 β-lactamase is the trans to cis isomerization of a non-proline peptide bond. Proteins. 1996;25:104–111. doi: 10.1002/(SICI)1097-0134(199605)25:1<104::AID-PROT8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 35.Yagi T, Kurokawa H, Senda K, Ichiyama S, Ito H, Ohsuka S, Shibayama K, Shimokata K, Kato N, Ohta M, Arakawa Y. Nosocomial spread of cephem-resistant Escherichia coli strains carrying multiple Toho-1-like β-lactamase genes. Antimicrob Agents Chemother. 1997;41:2606–2611. doi: 10.1128/aac.41.12.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]