Abstract
Almost a century after the devastating pandemic of the Spanish flu, humankind is facing the relatively comparable global outbreak of COVID‐19. COVID‐19 is an infectious disease caused by SARS‐CoV‐2 with an unprecedented transmission pattern. In the face of the recent repercussions of COVID‐19, many have argued that the clinical experience with influenza through the last century may have tremendous implications in the containment of this newly emerged viral disease. During the last 2 years, from the emergence of COVID‐19, tremendous advances have been made in diagnosing and treating coinfections. Several approved vaccines are available now for the primary prevention of COVID‐19 and specific treatments exist to alleviate symptoms. The present review article aims to discuss the pathophysiology, diagnosis, and treatment of SARS‐CoV‐2 and influenza A virus coinfection while delivering a bioinformatics‐based insight into this subject matter.
Keywords: bioinformatics perspective, coinfection, COVID‐19, influenza, SARS‐CoV‐2
1. INTRODUCTION
A newly emerged menace to public health, coronavirus disease 2019, or COVID‐19, was first reported in the Huanan South China Seafood Market located in Wuhan, China. Ever since the first report in the country of origin, COVID‐19 has continued to pose a severe health issue in almost all parts of the world (Heidari Nia et al., 2022; Sheervalilou, Shirvaliloo, Sargazi, Bahari, et al., 2021; Sheervalilou, Shirvaliloo, Sargazi, Shirvalilou, et al., 2021; Sivasankarapillai et al., 2020). A seasonal disease of the respiratory tract, influenza was the eighth leading cause of mortality among Americans before the massive outbreak of COVID‐19 (Jain et al., 2015). The recent 2019–2020 influenza outbreak had resulted in tens of millions of patients in the United States before the unwelcome reign of COVID‐19 started (Singer, 2020). As of February 18, 2022, there have been 418,650,474 confirmed cases of COVID‐19, including 5,856,224 deaths, in the world. As of February 14, 2022, a total of 10,279,668,555 doses of vaccines have been administered (https://covid19.who.int/).
The newly identified viral strain called severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) causes significant morbidity and mortality as an impending consequence of acute respiratory distress syndrome (Sheervalilou et al., 2020; Sheervalilou, Shirvaliloo, Sargazi, Shirvalilou, et al., 2021). This is why we now live in a distinctly different world with a redefined lifestyle (Debby & David, 2020). In the absence of relevant information regarding the pathogenesis of SARS‐CoV‐2, the rational alternative was to turn to our previous experience with outbreaks such as SARS, MERS, and influenza. There were conclusive libraries of information particularly concerned with the pathogenesis of these causative agents (Debby & David, 2020; Y. Wang, Wang, et al., 2020). However, the overwhelmingly high fatality rate of COVID‐19 urged the scientific society to learn about this new calamity as quickly as possible (Sheervalilou, Shirvaliloo, Sargazi, Bahari, et al., 2021; Y. Wang, Wang, et al., 2020).
To date, there has been officially approved specific medication for the treatment of COVID‐19 as well as vaccines; hence, the need to comply with the tried‐and‐true methods such as self‐isolation and prevention of infection. This is because developing novel therapeutic agents, or even repurposing the old ones, is a rather challenging task for clinicians. They first need to make an evident lab‐based diagnosis of the disease due to the many similarities it shares with other viral infections (Zhu et al., 2020). It is well established that the primary intervention in the case of any contagious disease is immunization of individuals against the causative agents through vaccination (Azizi & Azizi, 2020). Several vaccines have been approved for emergency use against COVID‐19 (Aruru et al., 2021), including vaccines developed by Pfizer and BioNTech. Nevertheless, researchers are still trying to design a more effective vaccine versus the SARS‐CoV‐2 36. Table 1 displays various vaccine platforms utilized versus CoVs (Anderson et al., 2020; Buchholz et al., 2004; H.‐W. Chen et al., 2016; D. D., 2020; Graham et al., 2018; Hashemi et al., 2021; Jimenez‐Guardeño et al., 2015; Kalita et al., 2020; T. W. Kim et al., 2004; D. Kim et al., 2020; J. Lu et al., 2020; Menachery et al., 2018; Spruth et al., 2006; Takashima et al., 2011; S.‐F. Wang et al., 2014; Zakhartchouk et al., 2007; Zhang et al., 2020).
Table 1.
Approved vaccines against coronaviruses
| Vaccine type | Virus | Vaccine target | Reference |
|---|---|---|---|
| Live‐attenuated virus | Coronaviruses | All virus proteins | Graham et al. (2018); Menachery et al. (2018) |
| Inactivated virus | Coronaviruses | Whole structural protein of the virus | Jimenez‐Guardeño et al. (2015); Spruth et al. (2006) |
| RNA‐based vaccines | SARS‐CoV‐2 | Spike protein | Anderson et al. (2020) |
| Virus‐like vaccines | Coronaviruses | N/A | H.‐W. Chen et al. (2016); J. Lu et al. (2020) |
| DNA‐based vaccines | Coronaviruses containing SARS‐CoV‐2 | S glycoprotein | T. W. Kim et al. (2004); S.‐F. Wang et al. (2014); Zhang et al. (2020) |
| Protein‐based vaccines | NVX‐(COV2373)/respiratory syncytial virus | S glycoprotein and peptides | Kalita et al. (2020); Zakhartchouk et al. (2007) |
| Recombinant protein‐based vaccines | Coronaviruses containing SARS‐CoV‐2 | All virus proteins: Spike protein, membrane protein nucleocapsid protein | Buchholz et al. (2004); D. D. (2020); E. Kim et al. (2020); Takashima et al. (2011) |
It is a pressing concern to find that many investigations focus on isolated infection with SARS‐CoV‐2 while overlooking instances of simultaneous infection with another pathogen or even pathogens. This is an important issue as coinfection with several pathogens may interfere with an accurate diagnosis of COVID‐19 and result in discrepancies in the therapeutic window related to either one of the infections (Zhu et al., 2020). Before the SARS‐CoV‐2 outbreak, respiratory viral coinfections were reported in about 40% of patients with flu‐like symptoms (Pinky & Dobrovolny, 2020). More recently, a meta‐analysis concluded that nearly 3% of patients hospitalized with the diagnosis of COVID‐19 were simultaneously infected with other respiratory pathogens, with influenza A being the secondary infection in most of the cases (Lansbury et al., 2020). As the more recent studies suggest, there has been a descending trend in the detection of coinfection in adults diagnosed with COVID‐19 (Blasco et al., 2020; D. Kim et al., 2020; Nowak et al., 2020; Siordia, 2020; Xing et al., 2020). Still, the same statement cannot be made for pediatric patients with COVID‐19, as they showed a 40% coinfection rate with another pathogen along SARS‐CoV‐2 in several investigations (Xia et al., 2020). It is unclear whether coinfection with another pathogen may result in unfavorable therapeutic outcomes in patients infected with SARS‐CoV‐2. According to two separate investigations, SARS‐CoV‐2/influenza coinfection did not result in worse clinical outcomes (Ding et al., 2020). Moreover, this condition decreased the mortality rate among patients. Interestingly, coinfection with influenza virus in COVID‐19 patients might render them less vulnerable to morbidities associated with COVID‐19, and hence, a better prognosis overall (G. Wang, Xie, et al., 2020). On average, more than 90% of patients with COVID‐19 are simultaneously infected with other bacteria, viruses, and/or fungi, according to an investigation. While the number of cases with SARS‐CoV‐2/influenza coinfection might not appear as staggering, the relatively high prevalence of seasonal influenza can increase the risk of coinfection in patients with COVID‐19 (Zhu et al., 2020). Though, the present theory needs to be further evaluated by prospective investigations, as in some instances, coinfection might very well worsen patients' clinical condition (Khorramdelazad et al., 2021).
It would be sensible to elucidate how SARS‐CoV‐2 might interact with other pathogens, especially respiratory agents, in severe cases of coinfection (Pinky & Dobrovolny, 2020). A pathogen of interest, the influenza virus, commonly results in signs and symptoms that may not be readily distinguished from that of COVID‐19 in all instances. Hence, our current lack of knowledge could beget a wave of coinfection, rendering us incapable of containing the situation (Azekawa et al., 2020). In terms of clinical reasoning, SARS‐CoV‐2 is a beguiling pathogen that most deceptively mimics the influenza virus in numerous regards, including clinical manifestations and transmission mode as well. Ergo, coinfection by both viruses might not be as far‐fetched as expected (Cuadrado‐Payán et al., 2020) (Table 2).
Table 2.
Differences between influenza and COVID‐19
| Difference | Flu | COVID‐19 | References |
|---|---|---|---|
| Virus characteristics | Segmented genome with negative‐sense ss‐RNA chain | Unsegmented genome with positive‐sense ss‐RNA chain | B. Singh et al. (2020); Konala et al. (2020); Balla et al. (2020) |
| Transmission | Contact/Respiratory droplets | Contact/Respiratory droplets | |
| Asymptomatic or symptomatic | Asymptomatic Patients due to herd immunity | Patients with symptom developments within 2 days of infection | |
| Signs and symptoms | Fever, headache, myalgia, malaise, cough, sore throat, nasal discharge, gastrointestinal illness (vomiting and diarrhea in 10%–20% infected children) | Fever, cough, dyspnea, nasal discharge, myalgias, common diarrhea, and smell or taste disorders, conjunctivitis/dermatologic manifestations, maculopapular, urticarial, vesicular eruptions, transient livedo reticularis | |
| Incubation period | 1–4 days (average 2 days) | Within 14 days after exposure (most cases 4–5 days after exposure) | |
| Viral shedding | 5–10 days | Up to 14 days or longer | |
| Severity of illness | Mild to moderate | Mild to severe | |
| Mortality | <1% | Approximately 3%–4% | |
| Diagnostics | Antigen detection assays, RT‐PCR, multiplex PCR, rapid molecular assays | NAAT most commonly RT‐PCR assay | |
| Laboratory findings | Leukocyte counts: normal/low early in the illness elevated later in the illness | Lymphopenia, elevated AT levels, elevated LDH, elevated inflammatory markers (ferritin, CRP, ESR), abnormal coagulation tests | |
| Chest X‐ray findings | Bilateral reticular or reticulonodular opacities with or without superimposed consolidation | Consolidation and GGO | |
| Vaccines | FDA‐licensed influenza vaccines available; with variable efficacy from season to season | No available vaccine, clinical trials in progress Convalescent blood therapy proposed | |
| Treatments | FDA‐approved antiviral drugs: oseltamivir, zanamivir, peramivir, baloxavir | No treatment available, clinical trials in progress. Proposed antiviral agent remdesivir and dexamethasone | |
| Complications | ARDS (less common), rhabdomyolysis, acute myocardial infarction, myocarditis and pericarditis, toxic shock syndrome, Guillain–Barre syndrome, transverse myelitis, encephalopathy | ARDS (more common), myocarditis, heart failure, acute coronary syndrome, arrhythmias, cardiogenic shock, thromboembolic complications (pulmonary embolism, acute limb ischemia, mesenteric thrombosis, acute stroke), multisystem inflammatory syndrome, and Guillain–Barre syndrome. |
Abbreviations: ARDS, acute respiratory distress syndrome; AT, aminotransaminase; COVID‐19, coronavirus diseases 2019; CRP, C‐reactive protein; ESR, erythrocyte sedimentation rate; Flu, influenza; GGO, ground‐glass opacities; LDH, lactate dehydrogenase levels; NAAT, nucleic acid amplification testing; RT‐PCR, reverse‐transcription polymerase chain reaction; ss, single strand.
Coinfections contribute to the morbidity and mortality accompanied by influenza infection (Dunning et al., 2020). Early data from China showed that nearly 50% of the patients who deceased due to COVID‐19 disease were also infected with the influenza virus at the time (Huang et al., 2020). Viral infections of the respiratory tract predispose individuals to more severe forms of COVID‐19. Cases of coinfection with SARS‐CoV‐2 and influenza A virus have been frequently reported, bringing about certain challenges in the diagnosis and treatment of concordant infections disease (Khorramdelazad et al., 2021). There are also other reports with more lenient results, suggesting a 10% rate of coinfection at the highest (N. Chen et al., 2020; Table 3). Additionally, as of December 30, 2021, Israel has coined the term “Florona” to describe coinfection with SARS‐CoV‐2 and influenza A virus. In Israel, a pregnant woman was reported to have “Florona” without a vaccination history (https://www.ndtv.com/world-news/israel-detects-first-case-of-florona-disease-report-2681965). Deductively, Florona is not caused by a new variant of SARS‐CoV‐2. It is a term referring to simultaneous infection with COVID‐19 and influenza (Meena, 2019).
Table 3.
Coinfection studies
| Patient/case | Coinfection | Characteristics | Medical history | Diagnostics | Therapeutics | Outcome | Reference |
|---|---|---|---|---|---|---|---|
| A 66‐year‐old woman, ex‐smoker | Flu A and COVID‐19 | Syncopal episode: fever (38.9°C), nonproductive cough, shortness of breath, decreased appetite, ABG: pH 7.3, PO2: 59 mmHg, PCO2: 45 mmHg, Hb: 13.8 g/dl, Creatinine phosphokinase: 89U/I | Ischemic cardiomyopathy, T2DM, hypertension, CAD, and CKD (baseline Cr: 1.3) |
Flu A test: positive, COVID‐19 nasopharyngeal swab testing: positive CXR: right lower lobe infiltrate |
Tamiflu 30 mg by mouth, twice a day for 5 days, azithromycin, ceftriaxone, hydroxychloroquine, Losartan, IV normal saline, transferred to ICU, high‐flow oxygen, intubated/ventilated | Renal failure, dehydrated, Urine output increased, improved Cr, ventilator‐dependent with minimal settings with an FiO2 of 30%, tracheostomy, and percutaneous gastrostomy tube placement | Konala et al. (2020) |
| 78‐year‐old woman, nonsmoker | Flu A virus and SARS‐CoV‐2 | General malaise, anorexia, SpO2: 98%–95%, cough, max temp: 37.7°C, RR: 20 breaths/min, heart rate: 106 beats/min, BP: 139/63 mmHg; elevated CRP, AST: 106 U/L, ALT: 80 U/L, GGT: 153 U/L, ALP: 372 U/L, LDH: 383 U/L | Dyslipidemia and hypothyroidism |
CXR: bilateral reticular shadow CT: GGO adjacent to the pleura PCR: positive |
Oseltamivir with ceftriaxone 2 g/day, azithromycin 500 mg/day | Afebrile, worsened general malaise. Improvements in GGO more like a consolidation, improvement in clinical symptoms and CT findings, positive PCR, negative PCR, patient discharged, not require oxygen therapy | Azekawa et al. (2020) |
| P1–3: men aged 53, 78, and 56 years, P4: a woman aged 81 years | SARS‐CoV‐2 and Flu A/B | Nonproductive cough, fever, dyspnea |
All: hypertension: P1 and P4: CKD on hemodialysis P2 and P4: T2D |
Physical examination: all patients (except P3): tachypnea and bronchospasm, low oxygen saturation CXR: P2: bilateral infiltrates, P4: right bilobar pneumonia Rapid NA amplification test: P1 and P2: positive for Flu A, P3: positive for both Flu A and B, P4: positive for Flu B RT‐PCR: All positive for SARS‐COV‐2 |
P1, P2, P4: acute respiratory deterioration, orotracheal intubation, mechanical ventilation Lopinavir–Ritonavir 400/100 mg twice a day hydroxychloroquine 200 mg twice a day oseltamivir 150 mg twice a day, subcutaneous interferonβ−1b 8MU every 48 h |
P3: discharged after 48 h without treatment or any complication P1: clinical improvement with minimal oxygen requirements P1 and P4 remained under mechanical ventilation |
Cuadrado‐Payán et al. (2020) |
| 1 male and 2 females with mean age of 59.6 years | Flu and COVID‐19 | Cough, fever, shortness of breath, myalgia, positive blood culture for Enterococcus faecium, elevated Inflammatory markers (ESR, CRP, IL‐6) | Hypertension and DM |
Positive nasopharyngeal swab RT‐PCR for COVID‐19. Rapid antigen assay: P1, P2: Flu B; P3: Flu A. CXR: P1: bilateral patchy infiltrates P2: multilobar infiltrates, P3: bilateral patchy infiltrates CT: P1: diffuse scattered areas of GGO and mixed attenuating opacities P2: diffuse bilateral GGO infiltrates |
All patients: hydroxychloroquine, azithromycin, ceftriaxone for COVID‐19 and oseltamivir for influenza | All discharged in stable condition | B. Singh et al. (2020) |
| A 4‐month‐old infant | Flu A and SARS‐COV‐2 | Fever (38.2°C), cough, nasal congestion, clear rhinorrhea | ‐ | Flu rapid immunochromatographic assay testing, RT‐PCR |
Satisfying oral fluid intake and excretions. Oseltamivir for 5 days |
‐ | Wehl et al. (2020) |
| 4 patients: a 74‐year‐old woman; 40‐year‐old healthy man; a 64‐year‐old man; 50‐year‐old healthy man | SARS‐CoV‐2 and Flu A |
Pneumonia symptoms P1: dry cough, malaise, body pain, subjective fever, headache, anorexia, dyspnea, orthopnea, BP: 70/50 mmHg, body temp: 38.7°C, PR: 89 beat/min, RR: 26 breath/min, O2 saturation: 90% P2: headache, fever, developed sweating, chills, cough, severe compressive chest pain, dyspnea, orthopnea, body pain, diarrhea, chest pain worsened, respiratory distress, sweating, a low‐grade fever, severe compressive chest pain, orthopnea, body pain, BP: 110/70 mmHg, body temp: 35.4°C, PR: 77 beat/min, RR: 20 breath/min, O2 saturation: 97% P3: dry cough, malaise, headache, subjective fever, dyspnea, BP:130/80 mmHg, body temp: 37.7°C, PR: 110 beat/min, RR: 19 breath/min, O2 saturation: 87% P4: fever, dry cough, and dyspnea, BP: 120/65 mm, body temp: 38.0°C, PR: 85 beat/min, RR: 18 breath/min, O2 saturation: 93% |
P1: ischemic cerebrovascular accident and hypertension |
RT‐PCR test: P1, P2, P3, P4 Flu viruses test: Flu A: P1, P2, P3, P4 CXR: P1: diffuse infiltrates in both lungs, P2: diffuse and bilateral infiltration in the lungs, P3: diffuse and bilateral infiltration in the lungs, P4: diffuse infiltrates in both lungs |
‐ | ‐ | Khodamoradi et al. (2020) |
| 69‐year‐old man, China | Covid‐19 and influenza |
Cough and fever, a ground‐glass consolidation lesion in the right inferior lobe of lungs, No Leukopenia 1 week later: persistent fever and worsening dyspnea , leukopenia |
No underlying diseases |
RT‐PCR: negative SARS‐CoV‐2, Xpert Flu/RSV Xpress assay of the nasopharyngeal swab: positive influenza A and negative for SARS‐CoV‐2 1 week later: RT‐PCR: positive SARS |
‐ | ‐ | Wu et al. (2020) |
| 4 patients, Spain | Covid‐19 and influenza | Persistent nonproductive cough, fever, dyspnea | Diabetes and severe kidney disorder |
Rapid NA amplification assay: positive influenza type A and B RT‐PCR: positive for SARS‐CoV‐2 |
‐ | ‐ | Cuadrado‐Payán et al. (2020) |
| A 66‐year‐old woman, African American | Covid‐19 and influenza | Fever (38.9°C), nonproductive cough, anorexia, shortness of breath | Chronic kidney disease, diabetes, coronary artery disease, and hypertension | Laboratory tests: positive COVID‐19, positive influenza | ‐ | ‐ | Konala et al. (2020) |
| 115 patients, China | Covid‐19 and influenza |
Pneumonia, during the admission; Lymphopenia: during the remission Total lymphocyte count was gradually raised 5 coinfected patients with fever, cough, fatigue, and headache; unusual symptoms such as a nasal tampon, pharyngalgia, diarrhea, and mild hemoptysis |
No underlying diseases | 115 patients with positive SARS‐CoV‐2 infection, 5 patients with positive influenza virus | ‐ | ‐ | Ding et al. (2020); N. Chen et al. (2020); D. Wang, Hu et al. (2020) |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BP, blood pressure; CAD, coronary artery disease; CKD, chronic kidney disease; Cr, creatinine; CRP, C‐reactive protein; CT, computed tomography; DM, diabetes mellitus; Flu, influenza; GGO, ground‐glass opacities; GGT, g‐glutamyltransferase; Hb, Hemoglobin; ICU, intensive care unit; IV, intravenous; LDH, lactate dehydrogenase; max temp, maximum temperature; NA, nucleic acid; P, patient; PCR, polymerase chain reaction; PR, pulse rate; RR, respiratory rate; T2D, type 2 diabetes; T2DM, type 2 diabetes mellitus.
An important issue with the molecular tests used currently to diagnose COVID‐19 is the uncertainty regarding the adequacy of the samples obtained from patients. Another challenge is the false‐negative test results for COVID‐19 in cases of simultaneous infection with another respiratory virus (Khorramdelazad et al., 2021). It seems that distinguishing COVID‐19 from seasonal influenza based on clinical manifestation is another challenge, as both diseases present with fever, nonproductive cough, and dyspnea. To avoid this, it is recommended that RT‐PCR and other tests based on nucleic acids be taken in all cases with an indeterminate respiratory infection. This could also help prevent further disease propagation by misdiagnosed patients (Wu et al., 2020).
More recently, bioinformatics analyses have been considered in various fields. Molecular docking studies are conducted for unraveling the binding of protein–ligand complexes (R. Singh et al., 2022). These investigations have been of great value in the case of repurposing old drugs and identifying new therapeutic targets for COVID‐19, such as the SARS‐CoV‐2 main protease (Rangsinth et al., 2021; Zaki et al., 2022), papain‐like protease (R. Singh et al., 2022), spike protein, and RNA‐dependent RNA polymerase (Rangsinth et al., 2021; Zaki et al., 2022). In 2021, Singh and colleagues suggested Tea (Camellia sinensis), theaflavin, BCH derivates (BCH10, BCH15, BCH16, and BCH17), dicaffeoylquinic acid, diacetylcurcumin, curcumin, and diacetylcurcumin candidate therapeutic agents for inhibiting nonstructural protein 16 (NSP16), nonstructural protein 1 (Nsp1), the Spike receptor‐binding domain, and RNA‐dependent RNA polymerase (RdRp) of SARS‐CoV‐2, respectively (R. Singh et al., 2022; R. Singh, Bhardwaj, & Purohit, 2021; R. Singh, Bhardwaj, Das, et al., 2021; R. Singh, Bhardwaj, Sharma, et al., 2021). The same year, Sharma et al. (2021) confirmed the inhibitory potential of three tea derivates (barrigenol, kaempferol, and myricetin) against nonstructural protein 15 (Nsp15) of SARS‐CoV2. Later, Bhardwaj and colleagues indicated that the DSPD derivate (DSPD‐2, DSPD‐6, DSPD‐5) bound effectively to the S1 subunit of SARS‐CoV‐2 S protein, as well as Oolonghomobisflavan‐A that could bind and neutralize the SARS‐CoV‐2 main protease (Bhardwaj, Singh, Das, et al., 2021; Bhardwaj, Singh, Sharma, et al., 2021a). Further investigations identified three bioactive molecules from tea (epicatechin‐3,5‐di‐O‐gallate, epigallocatechin‐3,5‐di‐O‐gallate, and epigallocatechin‐3,4‐di‐O‐gallate) that displayed more effective binding with the enzyme RdRp than antiviral drugs remdesivir and favipiravir (Bhardwaj, Singh, Sharma, et al., 2021b). Figure 1 represents the schematic procedure of the present study.
Figure 1.
The schematic overview of present article
2. THERE ARE MANY CONCERNS
2.1. What can we expect as the COVID‐19 outbreak continues?
The H1N1pdm09 virus caused the last influenza outbreak in 2009, which led to approximately 60 million cases of infection, over 274,000 hospitalizations, and 12,500 deaths in the United States throughout the following year (Shrestha et al., 2011). Almost a decade later, the virus can still be contracted even though it has been included in the influenza vaccine ever since the original outbreak. In fact, H1N1pdm09 was the dominant viral strain during the 2019–2020 annual outbreak of influenza. The same, however, cannot be applied to coronaviruses, as the causative strain behind the last human coronavirus epidemic, known as SARS, was successfully contained as a result of rigorous containment procedures before the complete development of a vaccine. Concerning the rebellious course of the COVID‐19 outbreak and the timeline of vaccine development programs, it is safe to say that SARS‐CoV‐2 will not follow the abrupt termination of its predecessor. Instead, likely, SARS‐CoV‐2 will still be contracted in communities as we face the subsequent influenza epidemic (Singer, 2020).
2.2. What are the odds of coinfection with COVID‐19 and seasonal influenza?
According to D. Kim et al. (2020), nearly 20% of patients infected with SARS‐CoV‐2 were concurrently infected with at least one other respiratory pathogen, most notably the influenza virus. There are critical diagnostic implications to this issue. In patients with respiratory symptoms, detecting any respiratory pathogen other than SARS‐CoV‐2 might not be a valid alternative to rule out COVID‐19 infection, especially in areas with limited laboratory resources for SARS‐CoV‐2 detection in the first place. These findings imply the urgent need for on‐demand access to diagnostic tools to detect SARS‐CoV‐2 and other respiratory pathogens (Singer, 2020).
2.3. How can the COVID‐19 epidemic and seasonal influenza aid us in developing strategies for proper preparation?
Both the influenza virus and SARS‐CoV‐2 spread primarily via droplets expelled from the respiratory tract. Thus, self‐isolation and social distancing methods suggested for the containment of COVID‐19 transmission can also be useful against influenza (Fong et al., 2020). Accordingly, should the transmission rate of COVID‐19 spike in the fall of this year, the corresponding preventive measures are most likely to taper the spread of the influenza virus (Singer, 2020). In terms of higher‐level prevention, influenza is thought to be adequately amenable to antiviral therapy in the majority of patients (Dunning et al., 2020). SARS‐CoV‐2, on the contrary, is still being investigated for target pathways in its pathogenesis that might render the virus vulnerable to antiviral agents (S. Lu, 2020). In the case of influenza, vaccine efficacy is a determining factor that depends on the inclusion of the most recently extracted antigens in the vaccine. The effectiveness of the 2019–2020 influenza vaccine was reported to be 45% in the United States, which is not ideal considering that the vaccine contained the circulating influenza antigen in that year (Dawood et al., 2020). Despite recommendations for global vaccination, the Influenza vaccine merely covered 45% of adults during the last season in the United States (Centers for Disease Control and Prevention, 2017). Then again, it is encouraged to pursue research and development programs on COVID‐19 vaccines. Efforts must be focused on devising strategies to contain potentially infectious agents that might superimpose on SARS‐CoV‐2, such as the influenza A virus (Singer, 2020).
2.4. Bioinformatics provide us with such mathematical models
Mathematical models have long been of particular value for understanding the mechanisms and dynamics of infections. One of the first iterations of such models was developed by Perelson et al. in 1996 to explore the dynamics of human immunodeficiency virus (HIV) infection (Perelson et al., 1996), which was later adapted by Nowak et al. and Neumann et al. to investigate hepatitis B (HBV) (Nowak et al., 1996) and hepatitis C viruses (HCV) (Neumann et al., 1998). Later, HCV was further investigated by Wodarz et al. (in 2003) in terms of adaptive immunity (Wodarz, 2003). In 2015, Hattaf and colleagues adapted these models to study HIV, Ebola, and Zika viruses (Hattaf & Yousfi, 2016; Hattaf et al., 2015). It is recommended that several conditions, known as delays, be considered in the study of infectious diseases. Infection of the host cell and virus replication are two such delays that occur instantaneously. Equations developed to address these delays are termed delay differential equations (Hattaf & Yousfi, 2020). Such equations were used by Zhuo (2012) for studying HBV infection with noncytolytic loss of infected cells and by Hattaf and Yousfi (2016), Raid (2016), and Mahrouf et al. (2017) as well. The equation introduced by Hattaf and Yousfi (2016) was a partial differential equation that allowed them to explain the evolution in time and space of infectious diseases. This model assumed that normal and infected cells, along with viral particles and antibodies are mixed together, and ignored their mobility. However, it is thought that the motion of these entities follows the Fickian diffusion. In this connection, the fluxes of these entities are correlated with their concentration gradient, flowing from the regions of high concentration to the regions of low concentration (Hattaf & Yousfi, 2020).
2.5. Can mathematical models be of help in estimating viral coinfection amid the COVID‐19 pandemic?
Mathematical models have had an essential role in extending our understanding of the dynamics of viral respiratory infections (Baccam et al., 2006). They have also been of particular interest to scientists studying coinfections (Pinky & Dobrovolny, 2020). According to one such mathematic model developed recently, viral strains with a faster growth rate can dominate strains with a slower growth rate. This theory might yield innovative insights into the issue of viral coinfection amid the COVID‐19 pandemic (Pinky & Dobrovolny, 2016). The model mentioned earlier was designed in a recent study to investigate SARS‐CoV‐2 coinfection with several other viruses, such as influenza A virus, parainfluenza virus, respiratory syncytial virus, human rhinovirus, and human metapneumovirus. The study found that other strains readily suppress the replication of SARS‐CoV‐2. This would happen because SARS‐CoV‐2 had a significantly lower growth rate (1.8/day) than other studied strains. These findings may have implications for the severity and timing of a potential prospective second wave of infection, should there be any. It is hypothesized that once the primary infection with SARS‐CoV‐2 is established, secondary infection with another strain may not result in the suppression of SARS‐CoV‐2 to undetectable levels (Pinky & Dobrovolny, 2020). Importantly, it has been suggested that the angiotensin‐converting enzyme 2 (ACE2) plays a fundamental role in SARS‐CoV‐2 replication (Huang et al., 2020). It is possible that several other strains may target this molecule, and thus, result in suppression of SARS‐CoV‐2. The influenza virus, for instance, has been reported to downregulate ACE2 (Liu et al., 2014) (Figure 1). Nonetheless, more investigations are warranted for the exact determination of SARS‐CoV‐2 growth rate and its interaction with other respiratory viruses to see if coinfection with influenza might actually result in the mitigation of a potential second wave of COVID‐19 outbreak (Pinky & Dobrovolny, 2020) (Figure 2).
Figure 2.
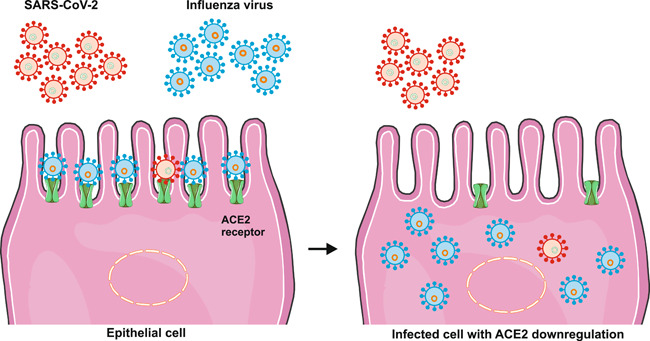
SARS‐CoV‐2 and influenza coinfection of lung epithelial cells based on bioinformatics modeling. SARS‐CoV‐2 showed a significantly slower growth rate than that of other strains. It was reported that other virus strains readily suppress the SARS‐CoV‐2 replication. It is possible that several strains, as well as the influenza virus, may target ACE2 molecules, and thus, result in suppression of SARS‐CoV‐2 by downregulating ACE2. Because it has been suggested that the ACE2 plays a fundamental role in SARS‐CoV‐2 replication.
3. CONCLUSION
Humankind has struggled with influenza for a long time, and the new reality of COVID‐19 will only complicate matters in the ongoing pandemic situation. Of paramount importance are the preventive measures at every level that will help dwindle the burden of viral respiratory infection in the coming days. With lessons learned from the several influenza outbreaks through the last century, we now have sufficient clinical experience to attain similar success in the containment of the recent outburst of COVID‐19. Cases of coinfection with the two strains in question can be easily diagnosed and treated thanks to the development of different vaccines as a means of primary prevention of COVID‐19. In addition, repurposing several drugs as well as employing bioinformatics‐based models for the influenza A virus have significantly improved clinical management of SARS‐CoV‐2 infection. Thus, we strongly recommend that all individuals at a greater level of risk get vaccinated against influenza in due time to reduce the likelihood of respiratory coinfections in the coming fall.
AUTHOR CONTRIBUTIONS
All authors participated in the investigation, design, interpretation of the studies and analysis of the data, and review of the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENT
This study was supported by the Zahedan University of Medical Sciences (Ethical code: IR.ZAUMS.REC.1399.197) (Webpage of ethical approval code is: https://ethics.research.ac.ir/EthicsProposalViewEn.php?id=149801).
Ghaznavi, H. , Shirvaliloo, M. , Sargazi, S. , Mohammadghasemipour, Z. , Shams, Z. , Hesari, Z. , Shahraki, O. , Nazarlou, Z. , Sheervalilou, R. , & Shirvalilou, S. (2022). SARS‐CoV‐2 and influenza viruses: Strategies to cope with coinfection and bioinformatics perspective. Cell Biology International, 46, 1009–1020. 10.1002/cbin.11800
Contributor Information
Roghayeh Sheervalilou, Email: sheervalilour@tbzmed.ac.ir.
Sakine Shirvalilou, Email: shirvaliloo.s@tak.iums.ac.ir.
DATA AVAILABILITY STATEMENT
All data are available.
REFERENCES
- Anderson, E. J. , Rouphael, N. G. , Widge, A. T. , Jackson, L. A. , Roberts, P. C. , Makhene, M. , Chappell, J. D. , Denison, M. R. , Stevens, L. J. , Pruijssers, A. J. , McDermott, A. B. , Flach, B. , Lin, B. C. , Doria‐Rose, N. A. , O'Dell, S. , Schmidt, S. D. , Corbett, K. S. , Swanson, P. A. , Padilla, M. , … mRNA Study Group. (2020). Safety and immunogenicity of SARS‐CoV‐2 mRNA‐1273 vaccine in older adults. New England Journal of Medicine, 383(25), 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruru, M. , Truong, H.‐A. , & Clark, S. (2021). Pharmacy Emergency Preparedness and Response (PEPR): A proposed framework for expanding pharmacy professionals' roles and contributions to emergency preparedness and response during the COVID‐19 pandemic and beyond. Research in social and administrative pharmacy. Journal of Pharmacy Education and Practice, 17(1), 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azekawa, S. , Namkoong, H. , Mitamura, K. , Kawaoka, Y. , & Saito, F. (2020). Co‐infection with SARS‐CoV‐2 and influenza A virus. IDCases, 20, e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi, S. A. , & Azizi, S.‐A. (2020). Neurological injuries in COVID‐19 patients: Direct viral invasion or a bystander injury after infection of epithelial/endothelial cells. Journal of Neurovirology, 26(5), 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam, P. , Beauchemin, C. , Macken, C. A. , Hayden, F. G. , & Perelson, A. S. (2006). Kinetics of influenza A virus infection in humans. Journal of Virology, 80(15), 7590–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla, M. , Merugu, G. P. , Patel, M. , Koduri, N. M. , Gayam, V. , Adapa, S. , Naramala, S. , & Konala, V. M. (2020). COVID‐19, modern pandemic: A systematic review from front‐line health care providers' perspective. Journal of Clinical Medicine Research, 12(4), 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K. , Singh, R. , Das, P. , & Purohit, R. (2021). Evaluation of acridinedione analogs as potential SARS‐CoV‐2 main protease inhibitors and their comparison with repurposed anti‐viral drugs. Computers in Biology and Medicine, 128, 104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K. , Singh, R. , Sharma, J. , Rajendran, V. , Purohit, R. , & Kumar, S. (2021a). Identification of bioactive molecules from tea plant as SARS‐CoV‐2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 39(10), 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K. , Singh, R. , Sharma, J. , Rajendran, V. , Purohit, R. , & Kumar, S. (2021b). Bioactive molecules of Tea as potential inhibitors for RNA‐dependent RNA polymerase of SARS‐CoV‐2. Frontiers in Medicine, 8, 684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco, M. L. , Buesa, J. , Colomina, J. , Forner, M. J. , Galindo, M. J. , Navarro, J. , Noceda, J. , Redón, J. , Signes‐Costa, J. , & Navarro, D. (2020). Co‐detection of respiratory pathogens in patients hospitalized with Coronavirus viral disease‐2019 pneumonia. Journal of Medical Virology, 92, 1799–1801. [DOI] [PubMed] [Google Scholar]
- Buchholz, U. J. , Bukreyev, A. , Yang, L. , Lamirande, E. W. , Murphy, B. R. , Subbarao, K. , & Collins, P. L. (2004). Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences, 101(26), 9804–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Flu vaccination coverage, United States, 2018–19 influenza season. https://cdc.gov/flu/fluvaxview/coverage-1819estimates.htm
- Chen, H.‐W. , Huang, C. Y. , Lin, S. Y. , Fang, Z. S. , Hsu, C. H. , Lin, J. C. , Chen, Y. I. , Yao, B. Y. , & Hu, C. M. J. (2016). Synthetic virus‐like particles prepared via protein corona formation enable effective vaccination in an avian model of coronavirus infection. Biomaterials, 106, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , Han, Y. , Qiu, Y. , Wang, J. , Liu, Y. , Wei, Y. , Xia, J. , Yu, T. , Zhang, X. , & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado‐Payán, E. , Montagud‐Marrahi, E. , Torres‐Elorza, M. , Bodro, M. , Blasco, M. , Poch, E. , Soriano, A. , & Piñeiro, G. J. (2020). SARS‐CoV‐2 and influenza virus co‐infection. Lancet (London, England), 395(10236), e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado‐Payán, E. , Montagud‐Marrahi, E. , Torres‐Elorza, M. , Bodro, M. , Blasco, M. , Poch, E. , Soriano, A. , & Piñeiro, G. J. (2020). SARS‐CoV‐2 and influenza virus co‐infection. Lancet, 395(10236), e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D. D . (2020). Dynavax and Clover Biopharmaceuticals announce research collaboration to evaluate coronavirus (COVID‐19) vaccine candidate with CpG 1018 adjuvant, DT Corporation. https://investors.dynavax.com/news-releases/news-release-details/dynavax-and-clover-biopharmaceuticals-announce-research
- Dawood, F. S. , Chung, J. R. , Kim, S. S. , Zimmerman, R. K. , Nowalk, M. P. , Jackson, M. L. , Jackson, L. A. , Monto, A. S. , Martin, E. T. , Belongia, E. A. , McLean, H. Q. , Gaglani, M. , Dunnigan, K. , Foust, A. , Sessions, W. , DaSilva, J. , Le, S. , Stark, T. , Kondor, R. J. , … Flannery, B. (2020). Interim estimates of 2019–20 seasonal influenza vaccine effectiveness—United States, February 2020. MMWR. Morbidity and Mortality Weekly Report, 69, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debby, B. , & David, H. D. (2020). 100 years of influenza research seen through the lens of Covid‐19. Mucosal Immunology, 13, 561–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q. , Lu, P. , Fan, Y. , Xia, Y. , & Liu, M. (2020). The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. Journal of Medical Virology, 92, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, J. , Thwaites, R. S. , & Openshaw, P. J. (2020). Seasonal and pandemic influenza: 100 years of progress, still much to learn. Mucosal Immunology, 13, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, M. W. , Gao, H. , Wong, J. Y. , Xiao, J. , Shiu, E. , Ryu, S. , & Cowling, B. J. (2020). Nonpharmaceutical measures for pandemic influenza in nonhealthcare settings‐social distancing measures. Emerging Infectious Diseases, 26, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, R. L. , Deming, D. J. , Deming, M. E. , Yount, B. L. , & Baric, R. S. (2018). Evaluation of a recombination‐resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Communications Biology, 1(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi, B. , Akram, F. A. , Amirazad, H. , Dadashpour, M. , Sheervalilou, M. , Nasrabadi, D. , Ahmadi, M. , Sheervalilou, R. , Ameri Shah Reza, M. , Ghazi, F. , & Roshangar, L. (2021). Emerging importance of nanotechnology‐based approaches to control the COVID‐19 pandemic; focus on nanomedicine iterance in diagnosis and treatment of COVID‐19 patients. Journal of Drug Delivery Science and Technology, 67, 102967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattaf, K. , Khabouze, M. , & Yousfi, N. (2015). Dynamics of a generalized viral infection model with adaptive immune response. International Journal of Dynamics and Control. 3(3), 253–261. [Google Scholar]
- Hattaf, K. , & Yousfi, N. (2016). A class of delayed viral infection models with general incidence rate and adaptive immune response. International Journal of Dynamics and Control, 4(3), 254–265. [Google Scholar]
- Hattaf, K. , & Yousfi, N. (2020). Mathematical modeling in virology, in Emerging and reemerging viral pathogens. (Vol. 2, pp. 325–339). Elsevier. [Google Scholar]
- Heidari Nia, M. , Rokni, M. , Mirinejad, S. , Kargar, M. , Rahdar, S. , Sargazi, S. , Sarhadi, M. , & Saravani, R. (2022). Association of polymorphisms in tumor necrosis factors with SARS‐CoV‐2 infection and mortality rate: A case‐control study and in silico analyses. Journal of Medical Virology, 94(4), 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , Zhang, L. , Fan, G. , Xu, J. , Gu, X. , Cheng, Z. , Yu, T. , Xia, J. , Wei, Y. , Wu, W. , Xie, X. , Yin, W. , Li, H. , Liu, M. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S. , Self, W. H. , Wunderink, R. G. , Fakhran, S. , Balk, R. , Bramley, A. M. , Reed, C. , Grijalva, C. G. , Anderson, E. J. , Courtney, D. M. , Chappell, J. D. , Qi, C. , Hart, E. M. , Carroll, F. , Trabue, C. , Donnelly, H. K. , Williams, D. J. , Zhu, Y. , Arnold, S. R. , … CDC EPIC Study Team. (2015). Community‐acquired pneumonia requiring hospitalization among U.S. adults. New England Journal of Medicine, 373, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez‐Guardeño, J. M. , Regla‐Nava, J. A. , Nieto‐Torres, J. L. , DeDiego, M. L. , Castaño‐Rodriguez, C. , Fernandez‐Delgado, R. , Perlman, S. , & Enjuanes, L. (2015). Identification of the mechanisms causing reversion to virulence in an attenuated SARS‐CoV for the design of a genetically stable vaccine. PLOS Pathogens, 11(10), e1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita, P. , Padhi, A. K. , Zhang, K. Y. J. , & Tripathi, T. (2020). Design of a peptide‐based subunit vaccine against novel coronavirus SARS‐CoV‐2. Microbial Pathogenesis, 145, 104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodamoradi, Z. , Moghadami, M. , & Lotfi, M. (2020). Co‐infection of coronavirus disease 2019 and influenza: A report from Iran. Archives of Iranian Medicine, 23(4), 239–243. [DOI] [PubMed] [Google Scholar]
- Khorramdelazad, H. , Kazemi, M. H. , Najafi, A. , Keykhaee, M. , Zolfaghari Emameh, R. , & Falak, R. (2021). Immunopathological similarities between COVID‐19 and influenza: Investigating the consequences of co‐infection. Microbial Pathogenesis, 152, 104554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Quinn, J. , Pinsky, B. , Shah, N. H. , & Brown, I. (2020). Rates of co‐infection between SARS‐CoV‐2 and other respiratory pathogens. Journal of the American Medical Association, 323, 2085–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. , Erdos, G. , Huang, S. , Kenniston, T. W. , Balmert, S. C. , Carey, C. D. , Raj, V. S. , Epperly, M. W. , Klimstra, W. B. , Haagmans, B. L. , Korkmaz, E. , Falo LD, J. r , & Gambotto, A. (2020). Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine, 55, 102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. W. , Lee, J. H. , Hung, C. F. , Peng, S. , Roden, R. , Wang, M. C. , Viscidi, R. , Tsai, Y. C. , He, L. , Chen, P. J. , Boyd, D. A. K. , & Wu, T. C. (2004). Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. Journal of Virology, 78(9), 4638–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konala, V. M. , Adapa, S. , Gayam, V. , Naramala, S. , Daggubati, S. R. , Kammari, C. B. , & Chenna, A. (2020). Co‐infection with Influenza A and COVID‐19. European Journal of Case Reports in Internal Medicine, 7(5), 001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury, L. , Lim, B. , Baskaran, V. , & Lim, W. S. (2020). Co‐infections in people with COVID‐19: A systematic review and meta‐analysis. Journal of Infection, 81, 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Yang, N. , Tang, J. , Liu, S. , Luo, D. , Duan, Q. , & Wang, X. (2014). Downregulation of angiotensin‐converting enzyme 2 by the neuraminidase protein of influenza A (H1N1) virus. Virus Research, 185, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Lu, G. , Tan, S. , Xia, J. , Xiong, H. , Yu, X. , Qi, Q. , Yu, X. , Li, L. , Yu, H. , Xia, N. , Zhang, T. , Xu, Y. , & Lin, J. (2020). A COVID‐19 mRNA vaccine encoding SARS‐CoV‐2 virus‐like particles induces a strong antiviral‐like immune response in mice. Cell Research, 30(10), 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. (2020). Timely development of vaccines against SARS‐CoV‐2. Emerging microbes infections, 9(1), 542–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrouf, M. , Hattaf, K. , & Yousfi, N. (2017). Dynamics of a stochastic viral infection model with immune response. Mathematical Modelling of Natural Phenomena, 12(5), 15–32. [Google Scholar]
- Meena, M. V. A. (2019). COVID‐19 to omicron—An overview. COVID 19 impact response, Vol. VII, p. 61. https://www.bhumipublishing.com/wp-content/uploads/2022/01/COVID-19-Impact-and-Response-Volume-VII.pdf#page=67
- Menachery, V. D. , Gralinski, L. E. , Mitchell, H. D. , Dinnon, K. H. , Leist, S. R. , Yount BL, J. r , McAnarney, E. T. , Graham, R. L. , Waters, K. M. , & Baric, R. S. (2018). Combination attenuation offers strategy for live attenuated coronavirus vaccines. Journal of Virology, 92(17), e00710–e00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, A. U. , Lam, N. P. , Dahari, H. , Gretch, D. R. , Wiley, T. E. , Layden, T. J. , & Perelson, A. S. (1998). Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon‐α therapy. Science, 282(5386), 103–107. [DOI] [PubMed] [Google Scholar]
- Nowak, M. A. , Bonhoeffer, S. , Hill, A. M. , Boehme, R. , Thomas, H. C. , & McDade, H. (1996). Viral dynamics in hepatitis B virus infection. Proceedings of the National Academy of Sciences, 93(9), 4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, M. D. , Sordillo, E. M. , Gitman, M. R. , & Paniz Mondolfi, A. E. (2020). Co‐infection in SARS‐CoV‐2 infected patients: Where are influenza virus and rhinovirus/enterovirus? Journal of Medical Virology, 92, 1699–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson, A. S. , Neumann, A. U. , Markowitz, M. , Leonard, J. M. , & Ho, D. D. (1996). HIV‐1 dynamics in vivo: Virion clearance rate, infected cell life‐span, and viral generation time. Science, 271(5255), 1582–1586. [DOI] [PubMed] [Google Scholar]
- Pinky, L. , & Dobrovolny, H. M. (2016). Coinfections of the respiratory tract: Viral competition for resources. PLOS One, 11(5), e0155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinky, L. , & Dobrovolny, H. M. (2020). SARS‐CoV‐2 coinfections: Could influenza and the common cold be beneficial? Journal of Medical Virology, 92, 2623–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangsinth, P. , Sillapachaiyaporn, C. , Nilkhet, S. , Tencomnao, T. , Ung, A. T. , & Chuchawankul, S. (2021). Mushroom‐derived bioactive compounds potentially serve as the inhibitors of SARS‐CoV‐2 main protease: An in silico approach. Journal of Traditional and Complementary Medicine, 11(2), 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riad, D. (2016). Dynamics of capital‐labour model with Hattaf‐Yousfi functional response. British Journal of Mathematics & Computer Science, 18(5), 1–7. [Google Scholar]
- Sharma, J. , Bhardwaj, V. K. , Singh, R. , Rajendran, V. , Purohit, R. , & Kumar, S. (2021). An in‐silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein‐15 of SARS‐CoV‐2. Food Chemistry, 346, 128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheervalilou, R. , Shirvaliloo, M. , Dadashzadeh, N. , Shirvalilou, S. , Shahraki, O. , Pilehvar‐Soltanahmadi, Y. , Ghaznavi, H. , Khoei, S. , & Nazarlou, Z. (2020). COVID‐19 under spotlight: A close look at the origin, transmission, diagnosis, and treatment of the 2019‐nCoV disease. Journal of Cellular Physiology, 235(12), 8873–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheervalilou, R. , Shirvaliloo, M. , Sargazi, S. , Bahari, S. , Saravani, R. , Shahraki, J. , Shirvalilou, S. , Shahraki, O. , Nazarlou, Z. , Shams, Z. , & Ghaznavi, H. (2021). Convalescent blood: Current perspective on the efficacy of a legacy approach in COVID‐19 treatment. Blood Purification, 51, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheervalilou, R. , Shirvaliloo, M. , Sargazi, S. , Shirvalilou, S. , Shahraki, O. , Pilehvar‐Soltanahmadi, Y. , Sarhadi, A. , Nazarlou, Z. , Ghaznavi, H. , & Khoei, S. (2021). Application of nanobiotechnology for early diagnosis of SARS‐CoV‐2 infection in the COVID‐19 pandemic. Applied Microbiology and Biotechnology, 105(7), 2615–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, S. S. , Swerdlow, D. L. , Borse, R. H. , Prabhu, V. S. , Finelli, L. , Atkins, C. Y. , Owusu‐Edusei, K. , Bell, B. , Mead, P. S. , Biggerstaff, M. , Brammer, L. , Davidson, H. , Jernigan, D. , Jhung, M. A. , Kamimoto, L. A. , Merlin, T. L. , Nowell, M. , Redd, S. C. , Reed, C. , … Meltzer, M. I. (2011). Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009–April 2010). Clinical Infectious Diseases, 52(Suppl 1), S75–S82. [DOI] [PubMed] [Google Scholar]
- Singer, B. D. (2020). COVID‐19 and the next influenza season. Science Advances, 6(31), eabd0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, B. , Kaur, P. , Reid, R. J. , Shamoon, F. , & Bikkina, M. (2020). COVID‐19 and influenza co‐infection: Report of three cases. Cureus, 12(8), 9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Bhardwaj, V. K. , Sharma, J. , Purohit, R. , & Kumar, S. (2022). In‐silico evaluation of bioactive compounds from tea as potential SARS‐CoV‐2 nonstructural protein 16 inhibitors. Journal of Traditional and Complementary Medicine, 12(1), 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Bhardwaj, V. K. , Das, P. , & Purohit, R. (2021). A computational approach for rational discovery of inhibitors for non‐structural protein 1 of SARS‐CoV‐2. Computers in Biology and Medicine, 135, 104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Bhardwaj, V. K. , & Purohit, R. (2021). Potential of turmeric‐derived compounds against RNA‐dependent RNA polymerase of SARS‐CoV‐2: An in‐silico approach. Computers in Biology and Medicine, 139, 104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Bhardwaj, V. K. , Sharma, J. , Kumar, D. , & Purohit, R. (2021). Identification of potential plant bioactive as SARS‐CoV‐2 spike protein and human ACE2 fusion inhibitors. Computers in Biology and Medicine, 136, 104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siordia J. A. Jr. (2020). Epidemiology and clinical features of COVID‐19: A review of current literature. Journal of Clinical Virology, 127, 104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankarapillai, V. S. , Pillai, A. M. , Rahdar, A. , Sobha, A. P. , Das, S. S. , Mitropoulos, A. C. , Mokarrar, M. H. , & Kyzas, G. Z. (2020). On facing the SARS‐CoV‐2 (COVID‐19) with combination of nanomaterials and medicine: Possible strategies and first challenges. Nanomaterials, 10(5), 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruth, M. , Kistner, O. , Savidis‐Dacho, H. , Hitter, E. , Crowe, B. , Gerencer, M. , Brühl, P. , Grillberger, L. , Reiter, M. , Tauer, C. , Mundt, W. , & Barrett, P. N. (2006). A double‐inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine, 24(5), 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima, Y. , Osaki, M. , Ishimaru, Y. , Yamaguchi, H. , & Harada, A. (2011). Artificial molecular clamp: A novel device for synthetic polymerases. Angewandte Chemie International Edition, 50(33), 7524–7528 [DOI] [PubMed] [Google Scholar]
- Wang, D. , Hu, B. , Hu, C. , Zhu, F. , Liu, X. , Zhang, J. , Wang, B. , Xiang, H. , Cheng, Z. , Xiong, Y. , Zhao, Y. , Li, Y. , Wang, X. , & Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Journal of the American Medical Association, 323(11), 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Xie, M. , Ma, J. , Guan, J. , Song, Y. , Wen, Y. , Fang, D. , Wang, M. , Tian, D.‐A. , & Li, P. (2020). Is co‐infection with influenza virus a protective factor of COVID‐19? Available at SSRN. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3576904
- Wang, S.‐F. , Tseng, S. P. , Yen, C. H. , Yang, J. Y. , Tsao, C. H. , Shen, C. W. , Chen, K. H. , Liu, F. T. , Liu, W. T. , Chen, Y. M. , & Huang, J. C. (2014). Antibody‐dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochemical and Biophysical Research Communications, 451(2), 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Wang, Y. , Chen, Y. , & Qin, Q. (2020). Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. Journal of Medical Virology, 92(6), 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehl, G. , Laible, M. , & Rauchenzauner, M. (2020). Co‐infection of SARS CoV‐2 and influenza A in a pediatric patient in Germany. Klinische Pädiatrie, 232(04), 217–218. [DOI] [PubMed] [Google Scholar]
- Wodarz, D. (2003). Hepatitis C virus dynamics and pathology: The role of CTL and antibody responses. Journal of General Virology, 84(7), 1743–1750. [DOI] [PubMed] [Google Scholar]
- Wu, D. , Lu, J. , Liu, Y. , Zhang, Z. , & Luo, L. (2020). Positive effects of COVID‐19 control measures on influenza prevention. International Journal of Infectious Diseases, 95, 345–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Cai, Y. , Huang, X. , Yu, X. , Zhao, L. , Wang, F. , Li, Q. , Gu, S. , Xu, T. , Li, Y. , Lu, B. , & Zhan, Q. (2020). Co‐infection with SARS‐CoV‐2 and influenza A virus in patient with pneumonia, China. Emerging Infectious Diseases, 26(6), 1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, W. , Shao, J. , Guo, Y. , Peng, X. , Li, Z. , & Hu, D. (2020). Clinical and CT features in pediatric patients with COVID‐19 infection: Different points from adults. Pediatric Pulmonology, 55(5), 1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Q. , Li, G.‐J. , Xing, Y.‐H , Chen, T. , Li, W.‐J. , Ni, W. , Deng, K. , Gao, R.‐Q. , Chen, C.‐Z. , Gao, Y. , Li, Q. , Yu, G.‐L. , Tong, J.‐N. , Li, Wei , Hao, G.‐L. , Sun, Y. , Zhang, A. , Wu, Q. , Li, Z.‐P. , & Pan, S.‐L (2020). Precautions are needed for COVID‐19 patients with coinfection of common respiratory pathogens. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3550013
- Zakhartchouk, A. N. , Sharon, C. , Satkunarajah, M. , Auperin, T. , Viswanathan, S. , Mutwiri, G. , Petric, M. , See, R. H. , Brunham, R. C. , Finlay, B. B. , Cameron, C. , Kelvin, D. J. , Cochrane, A. , Rini, J. M. , & Babiuk, L. A. (2007). Immunogenicity of a receptor‐binding domain of SARS coronavirus spike protein in mice: Implications for a subunit vaccine. Vaccine, 25(1), 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. A. , Ashour, A. , Elhady, S. S. , Darwish, K. M. , & Al‐Karmalawy, A. A. (2022). Calendulaglycoside A showing potential activity against SARS‐CoV‐2 main protease: Molecular docking, molecular dynamics, and SAR studies. Journal of Traditional and Complementary Medicine, 12(1), 16–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Zeng, H. , Gu, J. , Li, H. , Zheng, L. , & Zou, Q. (2020). Progress and prospects on vaccine development against SARS‐CoV‐2. Vaccines, 8(2), 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Ge, Y. , Wu, T. , Zhao, K. , Chen, Y. , Wu, B. , Zhu, F. , Zhu, B. , & Cui, L. (2020). Co‐infection with respiratory pathogens among COVID‐2019 cases. Virus Research, 285, 198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, X. (2012). IEEE. Analysis of a HBV infection model with non‐cytolytic cure process. in 2012. IEEE 6th International Conference on Systems Biology (ISB) .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available.


