Abstract
The molecular characterization of patients with Lynch syndrome (LS) involves germline testing to detect a deleterious mutation in one of the genes of the mismatch repair (MMR) pathway. To date, however, a large proportion of patients with a clinical suspicion of LS who undergo genetic testing do not show a germline pathogenetic variant in these genes. Germline DNA from 73 patients with a clinical suspicion of LS was examined with next-generation sequencing methods, using a multigene custom panel designed and standardized by our research group, that targets a set of 15 genes. Deleterious variants were identified in 5.6% of index cases, while unclassified variants were identified in 80.3% of probands. To evaluate the pathogenicity of these uncertain variants, the American College of Medical Genetics and Genomics criteria was used, also considering wherever possible the microsatellite instability (MSI) status detected on tumor tissues as pathogenic criterion. In this manner, 8 of these uncertain significance variants were classified as likely pathogenic variants. Notably, some of these likely pathogenetic variants were also identified in the MLH3 gene that is a gene not routinely analyzed for cases with a clinical suspicion of LS. The present study highlighted the importance of verifying the pathogenicity of the numerous variants of unknown significance identified in patients for whom heredity is already clinically confirmed suggesting the importance of considering the MSI-H status on the tumor of patients carrying an uncertain variant to evaluate its pathogenicity. Moreover, the present study also suggested analyzing other MMR genes, such as MLH3, in panels used for the molecular screening of LS.
Keywords: Lynch syndrome, mismatch repair genes, MLH3 gene, uncertain variants, rare variants, in silico analysis, microsatellite instability-high status, next-generation sequencing, custom panel, colorectal cancer genes
Introduction
Lynch syndrome (LS) is an autosomal hereditary form of colorectal cancer (CRC) with high penetrance and an incidence of 3-5% of all CRC cases (1-3). This hereditary form of CRC is clinically distinguishable from familial adenomatous polyposis (FAP), another hereditary form of CRC that develops from numerous polyps, as FAP is associated with APC mutations (4). Individuals with LS are characterized by a high lifetime risk of development of CRC, as well as endometrial, gastric and ovarian cancers, and other extracolonic tumors (such as small intestine, brain, skin, hepatobiliary, and urinary tract) (5,6). This hereditary syndrome is due to germline mutations in DNA mismatch repair (MMR) genes, mainly MLH1, MSH2, MSH6 and PMS2 (7-11).
The molecular characterization of these patients involves germline testing to detect a deleterious mutation in MLH1, MSH2, MSH6 or PMS25 (11), by direct sequencing and multiplex ligation-dependent probe amplification (MLPA), commonly used for the detection of large rearrangements (12). Numerous pathogenic mutations have been detected in MLH1 and MSH2 (13,14), while germline mutations in MSH6 and PMS2 are responsible for the disease only in a minority of cases (15-18). DNA MMR deficiency determines the presence of microsatellite instability (MSI) or loss of MMR protein expression at the somatic level (19). To identify patients with LS suitable for genetic testing, MSI testing and/or the lack of MMR protein expression (dMMR) according to immunohistochemical staining of colorectal or endometrial tumoral tissues is recommended for universal tumor screening, and these should be conducted as the first step (20,21). However, the Amsterdam criteria (AC) still remain a valid tool to allow clinicians to raise a clinical suspicion of LS in young patients with CRC and a significant family history of cancer (22).
To date, however, a large proportion of patients that exhibit the MSI/dMMR phenotype and undergo genetic testing do not show germline pathogenic variants in any of the MMR genes (MLH1, MSH2, MSH6, or PMS2). Unfortunately, the molecular mechanisms underlying MSI/dMMR CRC remain poorly understood (23). With the introduction of a next-generation sequencing (NGS) approach (24,25) that allows the concurrent investigation of multiple genes (25), the evaluation of inherited CRC is changing. For example, previous high-throughput sequencing studies suggested that tumor-suppressed genes such as TP53 and cyclin-dependent kinase inhibitor 2A (CDKN2A) could be responsible for genomic instability in numerous sporadic cancers (26,27). In the last few decades, NGS applications have led to the discovery of mutations in several predisposing CRC genes and to LS-related cancers. Germline mutations in other DNA-repair genes, such as ATM, PALB2, MRE11, and CHEK2, expressed throughout the cell cycle in response to double-stranded DNA breaks, have been associated with susceptibility to CRC (28,29). Furthermore, variants in CDH1, a gene normally associated with gastric cancer, could be a risk factor for CRC (30); a reduction of its expression is associated with tumoral dedifferentiation, lymphatic vessel invasion, and metastatic processes in CRC (31). In several previous studies, common MSH3 polymorphisms and rare variants were found to be significantly associated with CRC and prostate cancer as low-penetrance risk alleles (32-35). Moreover, biallelic MSH3 germline mutations appear to cause an additional rare recessively-inherited subtype of colorectal adenomatous polyposis (36). Evidence for an involvement of MLH3 includes a recent publication by Olkinuora et al (37) showing that a biallelic MLH3-truncating variant causes classical or attenuated adenomatous polyposis and possibly extracolonic tumors, while rare heterozygous variants of the MLH3 gene are associated with the LS phenotype (38). Finally, individuals with constitutional MMR deficiency (CMMRD) often have a high risk of developing a broad spectrum of malignancies and frequently display features reminiscent of neurofibromatosis type 1 (NF1) (39).
However, despite the NGS applications involving the multiple and simultaneous investigation of the various genes hitherto associated with MSI/dMMR CRC, the number of pathogenic variants identified following these analyses remains almost unchanged. Instead the variants of uncertain significance (VUS) that are detected in these patients who exhibit the MSI/dMMR phenotype are increasingly numerous.
Therefore, the aim of the present study was to elucidate the molecular basis of predisposition to the development of hereditary LS-related cancers in a cohort of 73 patients with a clinical suspicion of LS, using an NGS multigene panel designed and standardized by our research group and evaluating the pathogenicity of the numerous VUS identified, by applying criteria well known in the literature in order to obtain conclusive interpretation.
Materials and methods
Patient selection
A total of 73 patients with suspected LS meeting AC and/or Bethesda guidelines (BG) were recruited from Federico II University Hospital, National Cancer Institute IRCCS G. Pascale Foundation, and Luigi Vanvitelli University Hospital, all in Napoli, in southern Italy between January 2006 and December 2019, after study of their clinical characteristics and MSI and/or MMR protein expression in tumors.
Personal and family histories were obtained from each proband and written informed consent was provided by all patients. The present study was approved (protocol no. 120/10) by the local ethics committee 'Comitato Etico per le Attività Biomediche 'Carlo Romano' ('Carlo Romano' Ethics Committee for Biomedical Activities) at the University of Naples Federico II (Napoli, Italy).
Genomic DNA extraction
Genomic DNA was isolated from 3 ml of peripheral blood lymphocytes using the Nucleon BACC2 kit (Cytiva), according to the manufacturer's protocol. DNA quantity was assessed using a NanoDrop OneC spectrophotometer (reading at 260 nm and ratio 260/280 and 260/230 nm) and an Invitrogen Qubit 4 fluorometer (both from Thermo Fisher Scientific, Inc.). DNA quality was evaluated by 1% agarose gel electrophoresis and visualized with ethidium bromide.
Mutational analysis of coding regions of MMR genes
Genomic rearrangements in the MMR genes were analyzed by MLPA using SALSA-MSH6 P072 and SALSA-PMS2 P008 C1 kits (MRC-Holland BV), according to the manufacturer's protocol.
Targeted NGS
Library construction
Patient DNA samples were examined using an AmpliSeq Custom Panel (Illumina, Inc.), targeting 15 genes (Table I) involved in the MMR pathway or associated with CRC and other well-characterized cancer syndromes. This panel was developed based on the literature (26-39) to include genes associated with an increased risk of developing colon cancer. The kit (cat. no. 20020495; Illumina, Inc.) includes 470 amplicon regions that cover 87,353 bp, all the exonic and flanking intronic regions of these genes.
Table I.
Lynch syndrome full-exome panel.
| Gene name | RefSeq | Band Chr | Genomic size | RNA size | Exons |
|---|---|---|---|---|---|
| MLH1 | NM_000249.3 | 3p22.2 | 57497 | 2662 | 19 |
| MSH2 | NM_000251.2 | 2p21 | 80162 | 3226 | 16 |
| MSH6 | NM_000179.2 | 2p16.3 | 23872 | 4435 | 10 |
| PMS2 | NM_000535.5 | 7p22.1 | 35868 | 2851 | 15 |
| MLH3 | NM_001040108.1 | 14q24.3 | 37769 | 7911 | 13 |
| MSH3 | NM_002439.4 | 5q14.1 | 222168 | 4472 | 24 |
| EPCAM | NM_002354.2 | 2p21 | 17881 | 1731 | 9 |
| CDH1 | NM_004360.3 | 16q22.1 | 98250 | 4815 | 16 |
| TP53 | NM_000546.5 | 17p13.1 | 19149 | 2591 | 11 |
| ATM | NM_000051.3 | 1p34.1 | 11229 | 1945 | 16 |
| CHEK2 | NM_001005735.1 | 22q12.1 | 54092 | 1991 | 16 |
| PALB2 | NM_024675 | 16p12.2 | 38196 | 4069 | 13 |
| MRE11A | NM_005591 | 11q21 | 76572 | 5141 | 20 |
| NF1 | NM_001042492 | 17q11.2 | 282751 | 12444 | 58 |
| CDKN2A | NM_000077 | 9p21.3 | 7382 | 1267 | 3 |
DNA was diluted to a final concentration of 4 ng/µl using Low Tris-EDTA buffer (included in the kit) and re-quantitated with the fluorometric quantification method (Invitrogen Qubit 4). The standard input was 20 ng of DNA per sample. Briefly, the workflow involved multiplex PCR to amplify target regions of each DNA sample according to the procedure for two primer pools from an AmpliSeq Illumina Custom DNA Panel. FuPa reagent (included in the kit) was used to partially digest amplicons and each library was mixed and ligated with a unique index-specific combination. Subsequently, a second amplification step ensured sufficient quantity for the final sequencing analysis. The quality and quantity of the libraries obtained were assessed using a TapeStation 4200 System (Agilent Technologies, Inc.) with an Agilent DNA 1000 kit. The sequencing was performed on a MiSeqSystem (Illumina, Inc.) with a Nano V.2 flow cell (300 cycles) reagent kit according to the manufacturer's protocol. The raw data generated by this analysis are available on site Mendeley (Elsevier; https://data.mendeley.com/datasets/mxp536twnw/draft?a=a642b2f6-f2374e78-802c-c5cab21ef866.
Bioinformatics analysis
The sequencing data was analyzed using a BaseSpace Sequence Hub (Illumina, Inc.). Primary data analysis involved the detection and analysis of raw data (signal analysis), targeting sequencing reads (base calling) and scoring base quality. FASTQ files (generated by MiSeq Reporter Software 1.3.17; Integrative Genomics Viewer) were the outputs from this primary analysis. A demultiplexing process was subsequently required to produce separated sequencing read files, according to the single index used for each sample. In a secondary data analysis, FASTQ files for each sample were aligned against an entire reference genome specified in the manifest file with a DNA Amplicon Analysis App on BaseSpace Sequence Hub (version 2.1.1).
In silico analysis of unclassified variants
The following variant calling step had the main objective of identifying variants using a post-processed BAM file (https://basespace.illumina.com/analyses; BAM metrics version 0.0.22). A default value of 10 was used to define the Variant Caller Depth Filter level. Lower filter values may cause further false positive variants to pass the filter.
Output VCard File (VCF) was finally used for downstream analysis on a Variant Interpreter App (https://variantinterpreter.informatics.illumina.com; version 2.16.0.235), integrated with a BaseSpace Sequence Hub, that provided variant classification and reporting.
The BaseSpace Variant Interpreter is a cloud-based platform that uses the following annotation sources: Single Nucleotide Polymorphism Database (dbSNP) (https://www.ncbi.nlm.nih.gov/snp/?cmd=search), Catalogue of Somatic Mutations in Cancer (COSMIC) (https://cancer.sanger.ac.uk/cosmic), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), 1000 Genomes (https://www.internationalgenome.org), Exome Variant Server (EVS) (https://evs.gs.washington.edu/EVS/), (ExAC) (https://gnomad.broadinstitute.org), Polymorphism Phenotyping v.2 (PolyPhen-2), and Sorting Intolerant from Tolerant (SIFT). Such software classify germline variants as pathogenic, likely pathogenic, VUS, likely benign and benign. These categories follow The American College of Medical Genetics and Genomics (ACMG) guidelines (40). A total of 7 different complementary in silico programs were subsequently used for functional impact predictions of the identified variants.
Human Splicing Finder (HSF) (www.umd.be/HSF/) for silent and intron variants is a tool designed to predict the effects of mutations on splicing signals or to identify splicing motifs in human sequences. It contains all available matrices for auxiliary sequence prediction and presents a novel position weight matrix to assess the strength of 5′ and 3′ splice sites and branch points (41).
SIFT (http://blocks.fhcrc.org/sift/SIFT.html), PolyPhen (http://genetics.bwh.harvard.edu/pph/) (42), and PredictProtein server (http://www.predictprotein.org) (43) are prediction tools based on a combination of phylogenetic, structural and sequence annotation information characterizing a substitution with its position in the protein.
Mutation Taster (http://www.mutationtaster.org/) (44) and Align-Grantham Variation Grantham deviation (A-GVGD) (http://agvgd.hci.utah.edu/) (45) were employed in the study of missense variants. Briefly, Mutation Taster analyses comprise evolutionary conservation, splice-site changes, loss of protein features, and changes that may affect the amount of mRNA; moreover, the A-GVGD method can be used to identify sets of missense substitutions that are either enriched for deleterious variants or enriched for neutral variants.
Finally, Protein Variation Effect Analyzer (PROVEAN) is useful software for predicting whether nonsynonymous or indel variants are functionally important (http://provean.jcvi.org/index.php); its performance is comparable to that of SIFT or PolyPhen-2.
All variants identified were annotated according to the nomenclature recommendations from the Human Genome Variation Society (www.hgvs.org/mutnomen).
Variant analysis by Sanger sequencing
The coding regions corresponding to 22 variants (pathogenic variants and deleterious variants by in silico analysis) were amplified using customized primer sets (Table II). The PCR products were separated on a 1-2% agarose gel to check for unspecific amplicons. Subsequently, the PCR products were sequenced in both the forward and reverse directions using an ABI 3100 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Table II.
Primers, annealing PCR and amplicon sizes of PCR products corresponding to the pathogenic variants and variants resulted deleterious by in silico analysis.
| Gene variants | Primer sequences (5′→3′) | TM (°C) | Amplicon (bp) | Exon |
|---|---|---|---|---|
| MLH1 c.350C>T | F: GTGACCCAGCAGTGAGTTTT | 58 | 245 | 4 |
| R: AGCCTCACTTTTACCCTCTCT | ||||
| MSH6 c.3311_3312del | F: AGCCTCACTTTTACCCTCTCT | 58 | 398 | 5 |
| R: TGGCTGACTTTTATGTAACTGTG | ||||
| MSH6 c.892C>T | F: TGGTGGCTCTGATGTGGAAT | 59 | 206 | 4 |
| R: TTGCTTGTTTGGTGGCTGAG | ||||
| ATM c.3802delG | F: TGCTACTGAACAAGGTCCCA | 59 | 448 | 26 |
| R: CTCTCTTTGCTGTGCCATCC | ||||
| MLH1 c.376T>A | F: GTGACCCAGCAGTGAGTTTT | 58 | 245 | 4 |
| R: ACGTACTCAAGATCTCTGCCA | ||||
| EPCAM c.332A>G | F: TGATGAAGGCAGAAATGAATGG | 58 | 250 | 3 |
| R: ACAAGTAGTATAGGCAGCCCC | ||||
| MSH3 c.1778G>A | F: CCTGGGCATTAGAGTGGGAA | 58 | 264 | 13 |
| R: TGTCCTCAAGCTGAAGAACAC | ||||
| MLH3 c.470T>C | F: TATGGTTTCCGAGGAGAGGC | 59 | 232 | 2 |
| R: CCAGTCTAGGGTCCATGCAT | ||||
| MLH3 c.3440A>T | F: TCAGTTTGTGCAGAAAGAGGT | 58 | 250 | 3 |
| R: GAGTTAGGTGGTACGATGTGT | ||||
| ATM c.7475T>G | F: AGGAAGGTGTGTGAATTGCA | 58 | 465 | 50 |
| R: CCTGACATCAAGGGGCTTATG | ||||
| ATM c.1178G>T | F: GGCAACAACAGCGAAACTCT | 58 | 396 | 9 |
| R: TGTCATGGCAATCACATATCCC | ||||
| ATM c.8734A>G | F: ATTAGCTGTCAAACCTCCTAACT | 58 | 221 | 60 |
| R: TGCCCAGCCCATGTAATTTT | ||||
| CHEK2 c.911T>C | F: TGTCTTCTGTCCAAGTGCGT | 59 | 245 | 9 |
| R: GGTCCCTCGATTTCTGCCTA | ||||
| PMS2 c.1004A>G | F: AAAGTGAATTTGGCTGGGCG | 59 | 498 | 10 |
| R: TGGCTGCTGACTGACATTTAGCTTG | ||||
| PMS2 c.2249G>A | F: TCTCAGGAAGTTTTGTGACACT | 59 | 295 | 13 |
| R: CACCCAGCCGCTATAGTTCT | ||||
| PMS2 c.1253C>T | F: GACCCTCTTCTCCGTCCAC | 58 | 491 | 11 |
| R: GAGAGTCCACATGTTCCTGC | ||||
| MLH1 c.589-9_589-6delGTTT | F: GTTTGCTGGTGGAGATAAGGT | 58 | 392 | 8 |
| R: ACGCCACAGAATCTAGGAGA | ||||
| MRE11 c.1783+7A>G | F: ACTTTCTCCTTCTTCTCCCTCT | 59 | 410 | 15 |
| R: TGTCAGAACTGCCTTAAAGACTG | ||||
| CDH1 c.585A>C | F: TTCTCTGGGAGGGATTTGGC | 59 | 293 | 5 |
| R: CCCGGTGTCAACAAGCTTC | ||||
| CDH1 c.344C>T | F: GAAGATTGCACCGGTCGAC | 58 | 250 | 3 |
| R: CAACAGCGAACTTCTCAGAAAA | ||||
| NF1 c.4445T>C | F: CTGGGTGTATCTGGTGTTGAAAA | 58 | 486 | 34 |
| R: GGATCTATAACAATCTGCAAGCC | ||||
| CHEK2 c.688G>T | F: CTTGAAGTGGACCCAGGAGT | 58 | 242 | 6 |
| R: TGGGAAGTTATGAAGACGTGTT |
F, forward; R, reverse.
Results
A total of 73 patients with a clinical suspicion of LS were analyzed in the present study. The patients were selected as follows: 42 patients based on the AC (5), and 31 patients according to MMR deficiency detected in tumoral tissue with respect to the Bethesda guidelines (BG) (20). Individuals undergoing analysis were all affected by CRC, and two had MMR-deficient endometrial cancer. These patients were also affected by ovarian, bladder, breast, prostate, melanoma and renal cancers. The mean age at diagnosis, MSI-high (MSI-H) status analysis and sex of patients are outlined in Table III.
Table III.
Clinical and molecular characteristics of the 73-patient cohort.
| Characteristics | Amsterdam criteria | Bethesda guidelines | Total |
|---|---|---|---|
| Sex | |||
| Female | 21 | 14 | 35 |
| Male | 21 | 17 | 38 |
| Age at diagnosis, years (mean ± SD) | 33.47±11.96 | 47.59±16.93 | 44.89±14.26 |
| Tumor type | |||
| CRC | 40 | 31 | 71 |
| Breast | 3 | 3 | 6 |
| Endometrium | 3 | 4 | 9 |
| Other tumors | 2 | 3 | |
| MSI status | |||
| MSI | 9 | 31 | 40 |
| Unknown | 31 | 31 |
MSI, microsatellite instability.
All patients were previously shown to be negative for pathogenic variants in the MLH1 and MSH2 genes detected by single-gene analyses [denaturing high-pressure liquid chromatography (dHPLC) followed by Sanger sequencing] and large rearrangements (by MLPA).
Germline DNA samples from the patients were tested with NGS methods using a multigene custom panel developed by our research group that targets a set of 15 genes (Table I), MLH1, MSH2, MSH6, PMS2, MSH3, MLH3, CHEK2, MRE11, EPCAM, ATM, TP53, CDKN2A, PALB2, CDH1 and NF1. These genes are involved in the MMR pathway or associated with CRC and/or other well-characterized cancer syndromes (26-39).
Paired-end NGS generated an average of 705,000 reads, 93.93% of which mapped against the human genome (version GRCh38). Variants with 99.42% of exons covered were only selected, labelled as 'PASS' by the filter applied, with an estimated average amplicon depth of coverage of 1,350 reads. In this manner, an overall number of 724 variants were identified, of which only four (0.55%) were pathogenic variants already known, reported in the international databases Insight-Group and ClinVar (http://insight-database.org; https://www.ncbi.nlm.nih.gov/clinvar/) as pathogenetic variants. Most of the remaining variants (87.8%) were already known in the literature as being benign or polymorphic variants; 86 variants (11.88%) had been classified as variants that are likely benign or of uncertain pathogenic significance.
Clinically significant variants: Pathogenic variants
At least one clinically significant variant was identified in four patients (5.6%) of the cohort: One nonsense variant in ATM, two nonsense variants in MSH6, and one missense variant in MLH1 (Table IV). These had not been previously found by traditional methods (dHPLC and Sanger sequencing). These variants were validated by Sanger sequencing (Fig. 1).
Table IV.
Clinically significant variants identified.
| ID of patient | Gene | Variant | Variant classification | InSiGHT | ClinVar | PHENOTYPE (age onset) |
|---|---|---|---|---|---|---|
| 07.19 | MLH1 | c.350C>T p. (Thr117Met) | Pathogenetic | Pathogenetic | Pathogenetic | Index case k-co (34), MSI-H; father k-co (37) and his father's brother succumbed to k-co (65) |
| 07.13 | MSH6 | c.3311_3312del p. (Phe1104TrpfsTer3) | Pathogenetic | Pathogenetic | Pathogenetic | Index case k-co (49), MSI-H; father k-st (50), died k-co (58), his father's brother k-co (70), his cousin k-st (61) and colon polyps |
| 14.07 | MSH6 | c.892C>T p. (Arg298Ter) | Pathogenetic | Pathogenetic | Pathogenetic | Index case k-end (32), MSI-H; father leukemia (72), and her father's mother succumbed to leukemia (45) |
| 13.68 | ATM | c.3802delG p. (Val1268Ter) | Pathogenetic | - | Pathogenetic | Index case k-co (36) and several colon polyps; Sister k-co (75), brother k-pro (60) and father died k-pan (70) |
k-, cancer; -co, colon; -st, stomach; end, endometrium; -pan, pancreas; -pro, prostate.
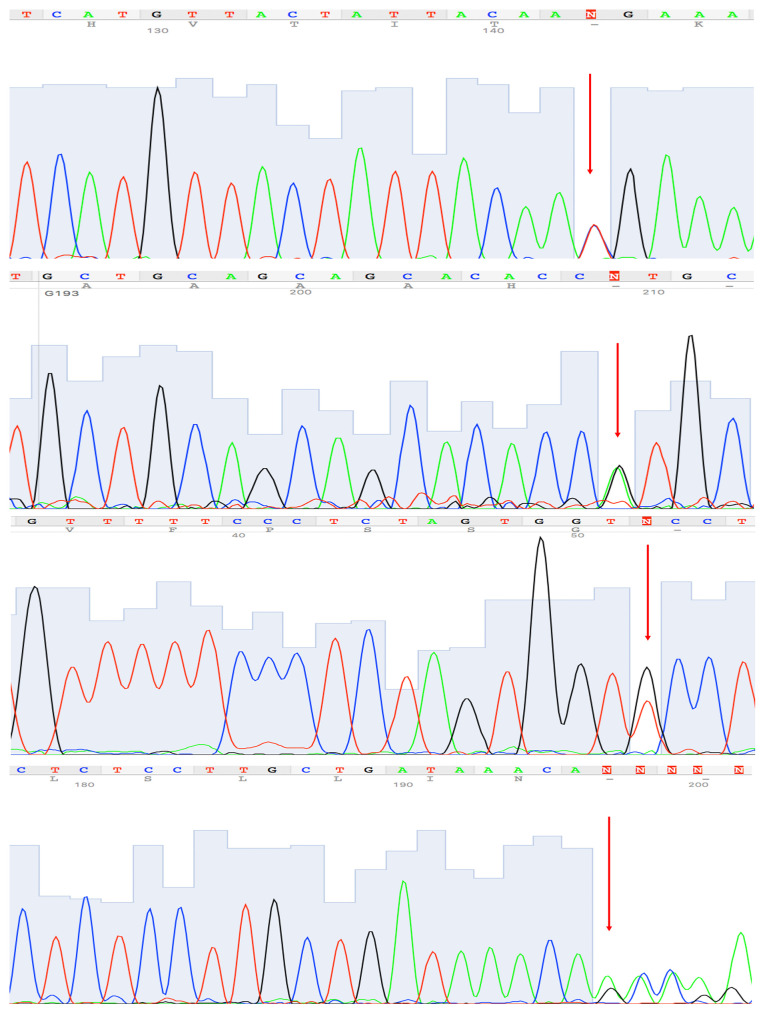
Figure 1.
Certain representative electropherograms by Sanger-sequencing of variants identified in the present study. (1) c.350C>T, p. (Thr117Met), MLH1 gene. (2) c.2149G>A, p. (Val717Met), PMS2 gene. (3) c.688G>T, p. (Ala230Ser), CHEK2 gene. (4) c.589-9_589-6delGTTT, MLH1 gene.
Unclassified and likely benign variants: Pathogenicity assessment
For the present study, 86 variants were considered as VUS. These were the variants that showed a minor allele frequency (MAF) that was very low (<0.1%) or not reported, some known in the international database (as ClinVar and Insight) and others novel. At least one of these VUS was identified in 57 patients (80.3%) in the cohort. These variants were distributed among the Lynch full-exome panel genes as follows: 4 in MLH1, 2 in MSH2, 4 in MSH6, 12 in PMS2, 12 in MSH3, 4 in MLH3, 3 in CHEK2, 5 in MRE11, 3 in EPCAM, 18 in ATM, 3 in TP53, 5 in PALB2, 5 in CDH1 and 6 in NF1. For each VUS identified in the present study, multiple bioinformatics analyses were performed using several software programs described in Table V and in the Materials and methods section. For silent and intronic variants, an ad hoc in silico analysis was used (as described in Table V and in the Materials and methods). Most of these variants presented discordant results from the computational algorithms applied. All lines of computational evidence supported a deleterious effect for 13 of these 86 variants. In addition, five others showed a deleterious effect in all but one or two of the computational algorithms used (Table V). These variants were validated by Sanger sequencing (Fig. 1).
Table V.
Variants identified in the panel genes and predicted in silico to have a deleterious effect.
| Variants | ID Cancer (age onset) selected criteria | MAF% | Mutation Taster | Poly Phen 2 [0-1] | Align GVGD | Predict protein | Provean (c. off 2.5) | Sift (c.off 0.05) | HSF | Source |
|---|---|---|---|---|---|---|---|---|---|---|
| c.376T>A p. (Tyrl26Asn) MLH1 | 19.39 endometrial (26) BG | 0.004% | Disease-causing | 0.911 (POSS DAM) | CLASS C65 | 95 | DEL (-7.69) | DAM (0) | Insight-group ClmVar | |
| c.332A>G p. (AsnlllSer) EPCAM | 18.33 colon (40) AC | Disease-causing | 1.00 (DAM) | CLASS C45 | 71 | DEL (-4.13) | DAM (0.004) | - | ClinVar | |
| c,1778G>A p. (Arg593Gln) MSH3 | 14.42 breast (38) colon (38) AC | - | Disease-causing | 1.00 (DAM) | CLASS C35 | 59 | DEL (-3.90) | DAM (0.001) | - | None |
| c.470T>C p. (Vall57Ala) MLH3 | 18.17 breast (37) colon (38), melanoma AC | Disease-causing | 1.00 (DAM) | CLASS C55 | 49 | DEL (-3.66) | DAM (0) | - | None | |
| c.3440A>T p. (Asnll47 Ile) MLH3 | 17.14 colon (52) AC | 0.03% | Disease-causing | 1.00 (DAM) | CLASS C65 | DEL (-6.77) | DAM (0) | - | ClinVar | |
| c.7475T>G p. (Leu2492 Arg) ATM | 18.17 (as above) 06.10 colon (53) AC | 0.02% | Disease-causing | 1.00 (DAM) ' | CLASS C6f | DEL (-4.53) | DAM (0.003) | - | None | |
| c,1178G>T p. (Trp393Leu) ATM | 19.24 colon (29) AC | 0.002% | Disease-causing | 1.00 (DAM) | CLASS C55 | DEL (-4.86) | DAM (0) | None | ||
| c.8734A>G p. (Arg2912 Gly)AW | 18.08 rectum 16 BG | 0.02% | Disease-causing | 1.00 (DAM) | CLASS C65 | DEL (-5.83) | DAM (0) | ClinVar | ||
| c.911T>C p. (Met304Thr) CHEK2 | 11.33 sigma (51) AC | 0.0004% | Disease-causing | 0.999 (DAM) | CLASS C65 | 86 | DEL (-3.76) | DAM (0.014) | - | ClinVar |
| c.1004a>g p. (Asn335Ser) PMS2 | 18.21 breast (49) colon (50) BG | 0.1% | Disease-causing | 1.00 (DAM) | CLASS C45 | 48 | DEL (-4.47) | DAM (0.006) | Insight-group ClinVar | |
| c.2149G>A p. (Val717Met) PMS2 | 18.36 colon (38) AC | 0.08% | Disease-causing | 0.982 (DAM) | CLASS C15 | 14 | NEU (-1-79) | DAM (0.010) | - | ClinVar |
| c.1253C>T p. (Ser418Phe) PMS2 | 10.17 colon (49) AC | 0.0008% | Disease-causing | 0.977 (DAM) | CLASS C65 | -6 | DEL (-3.04) | DAM (0.004) | Insight-group ClmVar | |
| c.589-9_589-6delGTTT MLH1 | 18.08 (as above) | 0.01% | Activation of an intronic cryptic acceptor site. Potential alteration of spicing. | Insight-group ClinVar | ||||||
| c. 1783+7A> G MRE11 | 09.10 colon (34) BG | 0.004% | - | - | - | - | - | - | Activation of an intronic cryptic donor site. Potential alteratior of spicing. | None 1 |
| c.585A>C p. (Gln195His) CDH1 | 11.25 colon prostate (69) BG | - | Disease-causing | 0.873 (POSS DAM) | CLASS C 15 | -65 | DEL (-2.59) | TOL (0.059) | - | ClinVar |
| c.344C>T p. (Thrll5Met) CDH1 | 19.59 colon, kidney (36) AC | 0.0024% | Disease-causing | 0.992 (DAM) | CLASS C 15 | -81 | NEUT (-1.93) | TOL (0.080) | - | ClinVar |
| c.4445t>c p. (Ilel482Thr) NF1 | 06.08 colon (28) AC | - | Disease-causing | 0.393 (BEN) | CLASS C65 | DEL (-4.15) | DAM (0.002) | - | ClinVar | |
| c.688G>T p. (Ala230Ser) CHEK2 | 18.30 sigma 24 BG | 0.0008% | Disease-causing | 0.076 (BEN) | CLASS C 65 | 64 | DEL (-2.76) | DAM (0.006) | - | ClinVar |
Poss Dam, possibly damaging; Dam, damaging; Del, deleterious; Neut, neutral; Tol, tolerated; Ben, benign.
Some of these rare variants are not present in healthy controls (as they are not reported in The Genome Aggregation Database, Exome Aggregation Consortium, or 1,000 Genome Projects database) and they are present in genes for which an association with a predisposition to developing colorectal tumors (or LS-related cancers) is well known. Therefore, the criteria reported in ACMG and the Association for Molecular Pathology guidelines were applied for the interpretation of sequence variants, (Table VI) (40). The MSI/dMMR status on tumor tissue was evaluated as a strong evidence of pathogenicity comparable with the results of well-established in vivo functional studies supportive of a damaging effect of variant on the gene or gene product only for the rare variants identified in the MMR genes. In this manner, eight of these 18 variants could be considered to be 'likely pathogenic' and for some of these variants the analysis of segregation in family environment was also performed (Table VI). Of these, two variants, (in the MLH3 and ATM genes) are novel and not previously reported in the literature. For the remaining 78 variants, the criteria for being benign and pathogenic were contradictory; therefore, these variants remain classified as having an uncertain significance.
Table VI.
Variants defined as 'likely pathogenic'.
| Variants | Gene | ID Cancer (age onset) Familial history (Segregation analysis) | Somatic dMMR | The Genome Aggregation Database (GnomAD) | Exome Aggregation Consortium (ExAC) | 1000 Genomes Project | Additional information | ACMG Criteria (a) | New classification variant | INSIGHT/CLINVAR |
|---|---|---|---|---|---|---|---|---|---|---|
| c.376T>A p.(Tyr126Asn) | MLH1 | 19.39 endometrial (26) Not affected- (N.D.) BG |
MS1-HNO
MLH1 NO PMS2 |
0.00004 | 0.00007 | 0.00020 | This variant is located in the N-terminal ATPase domain | PS3.PM1, PP2, PP3, PP4 | Likely pathogenic | Reported as VUS, as pathogenic and six times as benign/VUS |
| c.470T>C p.(Val157Ala) | MLH3 | 18.17 breast and colon (37, 38) other affected-breast and colon (N.D.) AC | MS1-H | No frequency | No frequency | No frequency | The valine residue is highly conserved. | PS3, PM2, PP2, PP3, PP4 | Likely pathogenic | Not reported/VUS |
| c.3440A>T p.(Asn1147Ile) | MLH3 | 17.14 colon (52) other affected- endometrial and colon (N.D.) AC | MS1-H | 0.00040 | 0.00044 | 0.00060 | No relevant information | PS3, PP2, PP3, PP4 | Likely pathogenic | Not reported/conflicting interpretation |
| c.1178G>T p.(Trp393Leu) | ATM | 19.24 colon (29) other affected-endometrial and gastric (N.D.) AC | ND | No frequency | No frequency | No frequency | No information | PM2, PM5, PP2, PP3, PP4 | Likely pathogenic | Not reported |
| c.911T>C p.(Met304Thr) | CHEK2 | 11.33 sigma (51) other affected- colon (also in affected brother) AC | MS1-H | No frequency | No frequency | No frequency | The methionine residue is highly conserved. This variant has been reported to affect CHEK2 protein function (PMID: 30851065). | PM2.PP1, PP2, PP3, PP4 | Likely pathogenic | Not reported/VUS |
| c.1253C>T p.(Ser418Phe) | PMS2 | 10.17 colon (49) other affected-colon (also in affected brother) AC |
MSI-H NO
MLH1 NO PMS2 |
No frequency | No frequency | No frequency | The seine residue is moderately conserved. | PS3, PM2, PP2, PP3, PP4 | Likely pathogenic | Not reported/VUS |
| c.589-9_589-6delGTTT | MLH1 | 18.08 Colon (18) other affected-colon and pancreas (also in affected father and two uncle) BG | MSI-H | No frequency | No frequency | No frequency | This variant could affect mRNA splicing (HSF software) | PS3, PM2, PP3, PP4 | Likely pathogenic | Reported seven times as VUS/conflicting interpretation |
| c.2149G>A p.Val717Met | PMS2 | 19.46 Colon (38) not affected-(N.D.) BG |
MSI-H NO
MLH1 NO PMS2 |
0.00076 | 0.00091 | 0.00020 | The variant is located within the MutL C-terminal, and dimerisation functional domain. | PS3, PM2, PP3, PP4 | Likely pathogenic | Not reported/conflicting interpretation |
PS, strong evidence of pathogenicity (class 1-4); PM, moderate evidence of pathogenicity (class 1-6); PP, Supporting evidence of pathogenicity (class 1-5); adapted from Ref (45). dMMR, deficient mismatch repair; N.D., not determined; BG, Bethesda guidelines; AC, Amsterdam criteria; VUS, variants of uncertain significance.
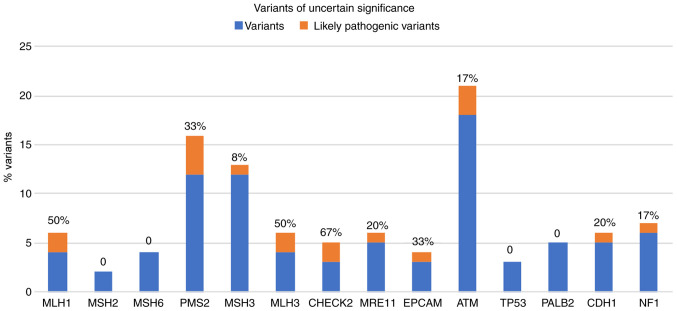
In Fig. 2, the percentage of these 'likely pathogenic variants' of the total uncertain variants identified for each gene analyzed, are presented. Notably, 50% of the gene variants identified in MLH1 and MLH3 are classified as likely pathogenic.
Figure 2.
Graphical representation of the number of rare variants (minor allele frequency <0.1%) identified in the genes involved in the Lynch-Full-exome custom panel. Percent value (%) refers to the variants found in each gene analyzed that were predicted to be likely pathogenic by The American College of Medical Genetics and Genomics criteria.
Discussion
The study of hereditary forms of CRC has increased the importance of genetic testing. However, the limited capacity of old genetic screening methods, due to their low sensitivity and small number of genes studied, has left numerous gaps in identifying variants conveying a predisposition to cancer. Indeed, more than half of CRC cases with a clinical suspicion of LS that are referred for genetic testing remain without a clear molecular diagnosis. In the present study, an NGS multigene custom panel was designed and standardized; it included, beyond the four MMR genes classically associated with LS, other MMR genes such as MLH3 and MSH3, and other genes that predispose to hereditary CRC or LS-related cancer according to the literature, (26-39) such as CHEK2, MRE11, EPCAM, ATM, TP53, PALB2, CDH1, NF1 and CDKN2A. This NGS multigene panel was used to analyze 73 patients with a clinical suspicion of LS selected according to the BG and AC. The patients had already been shown to be negative for pathogenetic variants in the MLH1 and MSH2 genes. This is probably the reason why the number of pathogenic variants clearly identified in the present study was so low. Only four pathogenic variants reported in the literature were identified in the cohort, of which one related to a gene not included among the MMR genes. Indeed, one patient (ID 13.68) was found to carry a variant, c.3802delG p. (Val1268Ter), in the ATM gene. This patient developed a right colon adenocarcinoma at an early age of onset (36 years) and was selected by AC due to a strong family history of early onset of colon tumors. Unfortunately for this patient, MSI could not be performed on the tumor tissue; however, it was certainly a case that could be confused with a clinical suspicion of LS. Variants in the ATM gene are associated in the recessive form with ataxia-telangiectasia and in the dominant form with breast cancer susceptibility and more generally with hereditary cancers (46). In the present study, this pathogenetic variant, c.3802delG p. (Val1268Ter), appeared to be associated with CRC.
The remaining pathogenic variants were identified in MMR genes (MLH1 and MSH6) in patients who met the AC and showed a typical LS phenotype with MSI-H status in cancer tissues (Table III). No other variant that was already classified in the literature as pathogenic was identified in the cohort, although the patients were selected according to very specific criteria. However, numerous genetic variants beyond these four clearly pathogenic variants were identified in the present study. The majority of these were benign or polymorphic variants, but numerous variants of unclear pathogenic significance were also found. Unfortunately, these variants were difficult to clinically interpret, which poses a significant barrier to the broad utility of genetic testing and carrier screening. In LS, nearly 90% of the identified genetic variants that are not included as nonsense or indel variants are deemed 'variants of uncertain significance' (47,48). To clarify the pathogenetic significance of such variants, it would have been useful to perform functional assays on proteins; recently, a massive parallel screen in human cells has been proposed to identify loss of function missense VUS in the MSH2 gene (49).
In the present study, 86 variants were identified that were already classified in international databases (Insight-group and/or ClinVar) as likely benign or of uncertain pathogenic significance, and four novel variants that showed a MAF that was very low or not reported. According to suggestions reported in the ACMG guidelines for the classification of variants (40), an interpretation of these variants was performed using multiple bioinformatics analyses, investigating seven different types of software for the pathogenic prediction of each variant. When the results of the in silico analyses were all in agreement as to the pathogenicity of the variants, it was examined whether these variants also respected the other ACMG criteria (population, computational, functional and segregation data) for establishment of the pathogenicity. Furthermore, the MSI/dMMR status (where this had been determined), found on the tumor tissues of the patients carrying these variants, was considered as a fundamental part for the interpretation of the pathogenicity. To the best of our knowledge, the MSI status in the majority of hereditary CRCs is associated to pathogenic variants in MMR genes. Thus, for the rare uncertain significance variants identified in the MMR genes that by bioinformatics analyses were resulted as pathogenic variants, it was hypothesized that the likely lack of function of corresponding protein at the somatic level, could be confirmed by the MSI/dMMR status showed on tumoral tissue of patients carrying these variants (Table VI). Therefore, our classification has arisen by the combination of the molecular and clinical data of each patient, in particular data from segregation and MSI analyses, applying the criteria provided from the guidelines for the classification of variants established by the ACMG (40). In this manner, it was possible to classify eight of these 86 variants as 'likely pathogenic' variants (Table VI). Thus, applying the ACMG criteria (40) our variant interpretation differs from classifications reported in public databases, such as ClinVar which reports these variants in majority as uncertain significance. Surely, further studies are needed to establish the real pathogenic role of these variants; however, at present it can be hypothesized that these variants could be the cause of disease in eight patients of our cohort. Thus, in light of the results obtained in the present study, the importance of establishing for the variants identified in MMR genes, a correlation with a deficient MMR system at the tumor level, is suggested, thus strengthening the evaluation of pathogenicity (Table VI). The interpretation of the VUS represents a crucial step in clinical decision-making, improving risk assessment, and promoting appropriate medical management, including variant-specific cascade testing for relatives. Therefore, an accurate assessment of the predictions of the clinical significance of the VUS is needed. The rule-based classification of the ACMG as it was performed in the present study, can represent a valid alternative to functional studies of VUS, which remain the most reliable tool to support the pathogenicity or benignity of the variant studied.
Furthermore, it is interesting to note that two of the eight likely pathogenic variants were found in the MLH3 gene (Table VI), the c.470T>C p. (Val157Ala) and c.3440A> T p. (Asn1147Ile) variants. The first is a novel variant that was identified in a woman (ID 18.17) with adenocarcinoma in situ of the colon with an MSI-H phenotype and with breast cancer. This patient was also negative for pathogenetic variants in BRCA1 and 2, MutYH and APC. The second variant was identified in a woman (ID 17.14) who developed peritoneal adenocarcinoma with MSI-H at age 52. Both women were selected for the present study since they met the AC. Both these patients did not exhibit MLH1 hypermethylation, and were also carriers of other two sequence variants in the MLH3 gene, the c.2476A>G and c.2531C>T, already described as benign in the ClinVar database. Finally, in these patients no other significant variants were identified in the genes included in the panel. The MLH3 gene is not routinely analyzed in patients with a clinical suspicion of LS. However, a previous study on MLH3-knockout mice highlighted the early onset of tumors in the abdominal sphere (50). Moreover, previous studies revealed a possible involvement of the MLH3 gene in LS (17,51). Loukola et al (51) and Wu et al (17) reported data on missense mutations and intronic substitutions in families meeting the AC, but without germline mutations in the MLH1 and MSH2 genes. Nonetheless, this gene is currently considered to be of low risk for a predisposition to the development of tumors on the LS spectrum. Unfortunately, functional assays were unable to be performed for either variant to clarify the pathogenetic effects, and no family segregation studies could be carried out due to a lack of interest from these patients. However, it is important to point out that both patients showed an MSI-H phenotype at the tumor level.
The results herein revealed that the use of the custom panel allowed the identification of variants in genes not routinely analyzed for cases with a clinical suspicion of LS, mainly variants in the MLH3 gene, but also rare variants identified in genes such as CHEK2, ATM, MSH3 and NF1. Although these results do not offer any evidence for a disease-causing role, they indicated the importance of deepening the study of all rare variants, to define their pathogenicity and to clarify the involvement of non-canonical genes in the pathogenesis of LS. The assessment of rare uncertain variants in genetic counseling could improve the risk estimate in those families that remain without a clear molecular diagnosis to provide precision medicine for this pathogenic condition (52).
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by 'Funding ATENEODuraturo 2018', Federico II Naples, Italy.
Availability of data and materials
The data generated and/or analyzed during the current study are available on site Mendeley (Elsevier; https://data.mendeley.com/datasets/z9gkb9vd9j/draft?a=5466cd6c-f760-4e6d-ba6d-3033b599b6bd.
Authors' contributions
FD conceptualized the study. FD, RL, ML and AN were involved in the literature search, in the data interpretation and critical reviewing of the manuscript. FD, AN and ML were involved in the preparation of the draft of the manuscript. FD, MDR and PI were involved in critically revising and editing the manuscript for important intellectual content. FD, RL, ML and AN confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. 120/10) by the local ethics committee, 'Carlo Romano' Ethics Committee for Biomedical Activities of the University of Naples, Federico II (Napoli, Italy). Written informed consent was obtained by all patients who participated in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohlmann W. Lynch syndrome and breast cancer risk: Weighing the data. JCO Precis Oncol. 2020 Feb 26; doi: 10.1200/PO.19.00376. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerretelli G, Ager A, Arends MJ, Frayling IM. Molecular pathology of lynch syndrome. J Pathol. 2020;250:518–531. doi: 10.1002/path.5422. [DOI] [PubMed] [Google Scholar]
- 4.Dodaro C, Grifasi C, Florio J, Santangelo ML, Duraturo F, De Rosa M, Izzo P, Renda A. The role of mutation analysis of the APC gene in the management of FAP patients. A controversial issue. Ann Ital Chir. 2016;87:321–325. [PubMed] [Google Scholar]
- 5.Duraturo F, Liccardo R, De Rosa M, Izzo P. Genetics, diagnosis and treatment of lynch syndrome: Old lessons and current challenges. Oncol Lett. 2019;17:3048–3054. doi: 10.3892/ol.2019.9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duraturo F, Liccardo R, Cavallo A, De Rosa M, Rossi GB, Izzo P. Multivariate analysis as a method for evaluating the pathogenicity of novel genetic MLH1 variants in patients with colorectal cancer and microsatellite instability. Int J Mol Med. 2015;36:511–517. doi: 10.3892/ijmm.2015.2255. [DOI] [PubMed] [Google Scholar]
- 7.Liccardo R, De Rosa M, Rossi GB, Carlomagno N, Izzo P, Duraturo F. Incomplete segregation of MSH6 frameshift variants with phenotype of lynch syndrome. Int J Mol Sci. 2017;18:999. doi: 10.3390/ijms18050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liccardo R, De Rosa M, Izzo P, Duraturo F. Novel MSH2 splice-site mutation in a young patient with lynch syndrome. Mol Med Rep. 2018;17:6942–6946. doi: 10.3892/mmr.2018.8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liccardo R, Della Ragione C, Mitilini N, De Rosa M, Izzo P, Duraturo F. Novel variants of unknown significance in the PMS2 gene identified in patients with hereditary colon cancer. Cancer. Manag Res. 2019;18:6719–6725. doi: 10.2147/CMAR.S167348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liccardo R, De Rosa M, Izzo P, Duraturo F. Novel implications in molecular diagnosis of lynch syndrome. Gastroenterol Res Pract. 2017;2017:2595098. doi: 10.1155/2017/2595098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, Church JM, Dominitz JA, Johnson DA, Kaltenbach T, et al. Guidelines on genetic evaluation and management of lynch syndrome: A consensus statement by the US multi-society task force on colorectal cancer. Gastroenterology. 2014;147:502–526. doi: 10.1053/j.gastro.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Duraturo F, Cavallo A, Liccardo R, Cudia B, De Rosa M, Diana G, Izzo P. Contribution of large genomic rearrangements in italian lynch syndrome patients: Characterization of a novel Alu-mediated deletion. Biomed Res Int. 2013;2013:219897. doi: 10.1155/2013/219897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tognetto A, Pastorino R, Castorina S, Condorelli DF, DeCensi A, De Vito C, Magnano A, Scaldaferri F, Villari P, Genuardi M, Boccia S. The current practice of lynch syndrome diagnosis and management in Italy: A qualitative assessment. Public Health Genomics. 2019;22:189–207. doi: 10.1159/000504305. [DOI] [PubMed] [Google Scholar]
- 14.Lynch PM. The hMSH2 and hMLH1 genes in hereditary nonpolyposis colorectal cancer. Surg Oncol Clin N Am. 2009;18:611–624. doi: 10.1016/j.soc.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, Sandkuijl L, Møller P, Genuardi M, Van Houwelingen H, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: Impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 16.Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, Lindblom A, Lagerstedt K, Thibodeau SN, Lindor NM, et al. The clinical phenotype of lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Berends MJ, Sijmons RH, Mensink RG, Verlind E, Kooi KA, van der Sluis T, Kempinga C, van dDer Zee AG, Hollema H, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29:137–138. doi: 10.1038/ng1001-137. [DOI] [PubMed] [Google Scholar]
- 18.Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, Hopper JL, Le Marchand L, Gallinger S, Newcomb PA, et al. Identification of lynch syndrome among patients with colorectal cancer. JAMA. 2012;17:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S, Provenzale D, Llor X, Halverson AL, Grady W, Chung DC, Haraldsdottir S, Markowitz AJ, Slavin TP, Jr, Hampel H, et al. NCCN guidelines insights: Genetic/familial high-risk assessment: Colorectal, version 2. J Natl Compr Canc Netw. 2019;2019;17:1032–1041. doi: 10.6004/jnccn.2019.0044. [DOI] [PubMed] [Google Scholar]
- 20.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 21.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) Proposed By the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/S0016-5085(99)70510-X. [DOI] [PubMed] [Google Scholar]
- 22.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turano M, Costabile V, Cerasuolo A, Duraturo F, Liccardo R, Delrio P, Pace U, Rega D, Dodaro CA, Milone M, et al. Characterisation of mesenchymal colon tumour-derived cells in tumourspheres as a model for colorectal cancer progression. Int J Oncol. 2018;53:2379–2396. doi: 10.3892/ijo.2018.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fecteau H, Vogel KJ, Hanson K, Morrill-Cornelius S. The evolution of cancer risk assessment in the era of next generation sequencing. J Genet Couns. 2014;23:633–639. doi: 10.1007/s10897-014-9714-7. [DOI] [PubMed] [Google Scholar]
- 25.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 26.Lin EI, Tseng LH, Gocke CD, Reil S, Le DT, Azad NS, Eshleman JR. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget. 2015;6:42334–42344. doi: 10.18632/oncotarget.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JC, Choi JS, Roh SA, Cho DH, Kim TW, Kim YS. Promoter methylation of specific genes is associated with the phenotype and progression of colorectal adenocarcinomas. Ann Surg Oncol. 2010;17:1767–1776. doi: 10.1245/s10434-009-0901-y. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Wang QS, Wang YJ. The CHEK2 I157T variant and colorectal cancer susceptibility: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13:2051–2055. doi: 10.7314/APJCP.2012.13.5.2051. [DOI] [PubMed] [Google Scholar]
- 29.Han P, Liu G, Lu X, Cao M, Yan Y, Zou J, Li X, Wang G. CDH1 rs9929218 Variant at 16q22.1 contributes to colorectal cancer susceptibility. Oncotarget. 2016;26:47278–47286. doi: 10.18632/oncotarget.9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shenoy S. CDH1 (E-Cadherin) mutation and gastric cancer: Genetics, molecular mechanisms and guidelines for management. Cancer Manag Res. 2019;13:10477–10486. doi: 10.2147/CMAR.S208818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elzagheid A, Buhmeida A, Laato M, El-Faitori O, Syrjänen K, Collan Y, Pyrhönen S. Loss of E-cadherin expression predicts disease recurrence and shorter survival in colorectal carcinoma. APMIS. 2012;120:539–548. doi: 10.1111/j.1600-0463.2011.02863.x. [DOI] [PubMed] [Google Scholar]
- 32.Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ. Mismatch repair polymorphisms and the risk of colorectal cancer. Int J Cancer. 2007;120:1548–1554. doi: 10.1002/ijc.22510. [DOI] [PubMed] [Google Scholar]
- 33.Orimo H, Nakajima E, Yamamoto M, Ikejima M, Emi M, Shimada T. Association between single nucleotide polymorphisms in the hMSH3 gene and sporadic colon cancer with microsatellite instability. J Hum Genet. 2000;45:228–230. doi: 10.1007/s100380070031. [DOI] [PubMed] [Google Scholar]
- 34.Jafary F, Salehi M, Sedghi M, Nouri N, Jafary F, Sadeghi F, Motamedi S, Talebi M. Association between mismatch repair gene MSH3 Codons 1036 and 222 polymorphisms and sporadic prostate cancer in the Iranian population. Asian Pac J Cancer Prev. 2012;13:6055–6057. doi: 10.7314/APJCP.2012.13.12.6055. [DOI] [PubMed] [Google Scholar]
- 35.Duraturo F, Liccardo R, Cavallo A, De Rosa M, Grosso M, Izzo P. Association of low-risk MSH3 and MSH2 variant alleles with lynch syndrome: Probability of synergistic effects. Int J Cancer. 2011;129:1643–1650. doi: 10.1002/ijc.25824. [DOI] [PubMed] [Google Scholar]
- 36.Adam R, Spier I, Zhao B, Kloth M, Marquez J, Hinrichsen I, Kirfel J, Tafazzoli A, Horpaopan S, Uhlhaas S, et al. Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet. 2016;99:337–351. doi: 10.1016/j.ajhg.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olkinuora A, Nieminen TT, Mårtensson E, Rohlin A, Ristimäki A, Koskenvuo L, Lepistö A, Swedish Extended Genetic Analysis of Colorectal Neoplasia (SWEN) Study Group. Gebre-Medhin S, Nordling M, Peltomäki P. Biallelic germline nonsense variant of MLH3 underlies polyposis predisposition. Genet Med. 2019;21:1868–1873. doi: 10.1038/s41436-018-0405-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duraturo F, Liccardo R, Izzo P. Coexistence of MLH3 germline variants in colon cancer patients belonging to families with lynch syndrome-associated brain tumors. J Neurooncol. 2016;129:577–578. doi: 10.1007/s11060-016-2203-0. [DOI] [PubMed] [Google Scholar]
- 39.Titze S, Peters H, Währisch S, Harder T, Guse K, Buske A, Tinschert S, Harder A. Differential MSH2 promoter methylation in blood cells of Neurofibromatosis type 1 (NF1) patients. Eur J Hum Genet. 2010;18:81–87. doi: 10.1038/ejhg.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: An online bio-informatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost B, Yachdav G, Liu J. The predictprotein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. Mutationtaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 45.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhunussova G, Afonin G, Abdikerim S, Jumanov A, Perfilyeva A, Kaidarova D, Djansugurova L. Mutation spectrum of cancer-associated genes in patients with early onset of colorectal cancer. Front Oncol. 2019;9:673. doi: 10.3389/fonc.2019.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liccardo R, Nolano A, Lambiase M, Della Ragione C, De Rosa M, Izzo P, Duraturo F. MSH2 overexpression due to an unclassified variant in 3-Untranslated region in a patient with colon cancer. Biomedicines. 2020;8:167. doi: 10.3390/biomedicines8060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arora S, Huwe PJ, Sikder R, Shah M, Browne AJ, Lesh R, Nicolas E, Deshpande S, Hall MJ, Dunbrack RL, Jr, Golemis EA. Functional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methods. Cancer Biol Ther. 2017;18:519–533. doi: 10.1080/15384047.2017.1326439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia X, Burugula BB, Chen V, Lemons RM, Jayakody S, Maksutova M, Kitzman JO. Massively parallel functional testing of MSH2 missense variants conferring lynch syndrome risk. Am J Hum Genet. 2021;108:163–175. doi: 10.1016/j.ajhg.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–5408. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol. 2000;157:347–352. doi: 10.1016/S0002-9440(10)64546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turano M, Delrio P, Rega D, Cammarota F, Polverino A, Duraturo F, Izzo P, De Rosa M. Promising colorectal cancer biomarkers for precision prevention and therapy. Cancers (Basel) 2019;11:1932. doi: 10.3390/cancers11121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated and/or analyzed during the current study are available on site Mendeley (Elsevier; https://data.mendeley.com/datasets/z9gkb9vd9j/draft?a=5466cd6c-f760-4e6d-ba6d-3033b599b6bd.